Significance
Large-scale neural recordings in freely moving animals are important for understanding how patterns of activity across a population of neurons generates animal behavior. Previously, recordings have been limited to either small brain regions or to immobilized or anesthetized animals exhibiting limited behavior. This work records from neurons with cellular resolution throughout the entire brain of the nematode Caenorhabditis elegans during free locomotion. Neurons are found whose activity correlates with behaviors including forward and backward locomotion and turning. A growing body of evidence suggests that animal behavior is sometimes generated by the collective activity of many neurons. It is hoped that methods like this will provide quantitative datasets that yield insights into how brain-wide neural dynamics encode animal action and perception.
Keywords: calcium imaging, large-scale recording, behavior, C. elegans, microscopy
Abstract
The ability to acquire large-scale recordings of neuronal activity in awake and unrestrained animals is needed to provide new insights into how populations of neurons generate animal behavior. We present an instrument capable of recording intracellular calcium transients from the majority of neurons in the head of a freely behaving Caenorhabditis elegans with cellular resolution while simultaneously recording the animal’s position, posture, and locomotion. This instrument provides whole-brain imaging with cellular resolution in an unrestrained and behaving animal. We use spinning-disk confocal microscopy to capture 3D volumetric fluorescent images of neurons expressing the calcium indicator GCaMP6s at 6 head-volumes/s. A suite of three cameras monitor neuronal fluorescence and the animal’s position and orientation. Custom software tracks the 3D position of the animal’s head in real time and two feedback loops adjust a motorized stage and objective to keep the animal’s head within the field of view as the animal roams freely. We observe calcium transients from up to 77 neurons for over 4 min and correlate this activity with the animal’s behavior. We characterize noise in the system due to animal motion and show that, across worms, multiple neurons show significant correlations with modes of behavior corresponding to forward, backward, and turning locomotion.
How do patterns of neural activity generate an animal’s behavior? To answer this question, it is important to develop new methods for recording from large populations of neurons in animals as they move and behave freely. The collective activity of many individual neurons appears to be critical for generating behaviors including arm reach in primates (1), song production in zebrafinch (2), the choice between swimming or crawling in leech (3), and decision-making in mice during navigation (4). New methods for recording from larger populations of neurons in unrestrained animals are needed to better understand neural coding of these behaviors and neural control of behavior more generally.
Calcium imaging has emerged as a promising technique for recording dynamics from populations of neurons. Calcium-sensitive proteins are used to visualize changes in intracellular calcium levels in neurons in vivo which serve as a proxy for neural activity (5). To resolve the often weak fluorescent signal of an individual neuron in a dense forest of other labeled cells requires a high magnification objective with a large numerical aperture that, consequently, can image only a small field of view. Calcium imaging has traditionally been performed on animals that are stationary from anesthetization or immobilization to avoid imaging artifacts induced by animal motion. As a result, calcium imaging studies have historically focused on small brain regions in immobile animals that exhibit little or no behavior (6).
No previous neurophysiological study has attained whole-brain imaging with cellular resolution in a freely behaving unrestrained animal. Previous whole-brain cellular resolution calcium imaging studies of populations of neurons that involve behavior investigate either fictive locomotion (3, 7), or behaviors that can be performed in restrained animals, such as eye movements (8) or navigation of a virtual environment (9). One exception has been the development of fluorescence endoscopy, which allows recording from rodents during unrestrained behavior, although imaging is restricted to even smaller subbrain regions (10).
Investigating neural activity in small transparent organisms allows one to move beyond studying subbrain regions to record dynamics from entire brains with cellular resolution. Whole-brain imaging was performed first in larval zebrafish using two-photon microscopy (7). More recently, whole-brain imaging was performed in Caenorhabditis elegans using two-photon (11) and light-field microscopy (12). Animals in these studies were immobilized, anesthetized, or both and thus exhibited no gross behavior.
C. elegans’ compact nervous system of only 302 neurons and small size of only 1 mm make it ideally suited for the development of new whole-brain imaging techniques for studying behavior. There is a long and rich history of studying and quantifying the behavior of freely moving C. elegans dating back to the mid-1970s (13, 14). Many of these works involved quantifying animal body posture as the worm moved, for example as in ref. 15. To facilitate higher-throughput recordings of behavior, a number of tracking microscopes (16–18) or multiworm imagers were developed (19, 20) along with sophisticated behavioral analysis software and analytical tools (21, 22). Motivated in part to understand these behaviors, calcium imaging systems were also developed that could probe neural activity in at first partially immobilized (23) and then freely moving animals, beginning with ref. 24 and and then developing rapidly (17, 18, 25–29). One limitation of these freely moving calcium imaging systems is that they are limited to imaging only a very small subset of neurons and lack the ability to distinguish neurons that lie atop one another in the axial direction of the microscope. Despite this limitation, these studies, combined with laser-ablation experiments, have identified a number of neurons that correlate or affect changes in particular behaviors including the AVB neuron pair and VB-type motor neurons for forward locomotion; the AVA, AIB, and AVE neuron pairs and VA-type motor neurons for backward locomotion; and the RIV, RIB, and SMD neurons and the DD-type motor neurons for turning behaviors (17, 18, 25, 26, 28, 30, 31). To move beyond these largely single-cell studies, we sought to record simultaneously from the entire brain of C. elegans with cellular resolution and record its behavior as it moved about unrestrained.
Results
Volumetric Imaging in a Freely Moving Worm.
We generated transgenic animals that express both a genetically encoded calcium indicator, GCaMP6s, and a calcium-insensitive fluorescent protein RFP, in the nucleus of each neuron (strain AML14; Materials and Methods). The transgenic worms exhibited normal behavior compared with WT as measured by the animal’s velocity, turn rate, pause rate, and run length (SI Appendix, Fig. S1 and Table S1).
To record fluorescent images of all neurons in the worm’s head while simultaneously recording the animal’s behavior, we developed a custom dual-objective 3D tracking system. A spinning disk confocal microscope records images of the head of a worm through a 40× objective using both 488- and 561-nm excitation laser light while a second imaging path records the worm’s position and behavior through a 10× objective (Fig. 1). In contrast to previous dual-objective worm trackers (18), this system operates in three dimensions. Custom real-time software uses fluorescence imaging to identify the location of the worm’s brain in three dimensions and adjusts a motorized stage and piezoelectric objective mount to follow the brain as it moves and to scan through its brain volume as it crawls.
Fig. 1.
Simultaneous imaging of whole-brain calcium activity and behavior, simplified schematic. A worm crawls freely on a motorized stage under near-infrared (NIR) dark-field illumination. A spinning disk confocal microscope acquires volumetric fluorescent images of the worm’s brain by scanning a 40× objective along the imaging axis (z) to acquire 6 brain volumes/s. A low-magnification 10× objective images the animal’s posture and behavior. Custom 3D real-time tracking software feeds back on the fluorescence images and adjusts the motorized stage and z-piezo stage accordingly to keep the worm’s head centered in the high magnification objective’s field of view.
To acquire 3D image stacks of the worm’s head, a piezoelectric stage translates the 40× objective up and down along the imaging axis such that the focal plane passes through the animal’s head six times per second. The confocal spinning disk rejects out-of plane light and provides optical sectioning. A high-speed, high-sensitivity Scientific CMOS (sCMOS) camera records fluorescent images at 200 fps, thereby capturing 6 head volumes/s, each consisting of ∼33 z-slices, 1.5–1.6 µm apart, depending on the size of the individual animal’s head. Each z-stack spans a volume that is sized to the head of the individual worm but is usually µm in length and width and 48–51 µm in depth. This field is sufficiently large to image all neuronal cell bodies in the head of the animal, including those located from the tip of the nose, through the nerve ring, to the ventral nerve cord immediately posterior of the pharyngeal terminal bulb. To capture both the calcium dynamics and information about the location of cell bodies, red- and green-channel fluorescent images are recorded side-by-side on the sCMOS sensor simultaneously (Fig. 2 C–F).
Fig. 2.
Simultaneous recording of four video streams showing behavior and neural activity. (A) The worm’s posture and behavior are recorded via infrared (IR) darkfield imaging through the 10× objective at 42 fps. (B) Fluorescence from neurons in the worm’s head is tracked in real time to keep the worm centered in the field of view, via the 10× objective at 67 fps. (Scale bar, 100 μm.) Orange line indicates the worm’s centerline. (C–F) Fluorescence images of neuronal nuclei are simultaneously recorded through the 40× objective at 200 fps as the objective scans through the worm’s head along the axial imaging axis, z. A 3D volume is reconstructed from a z-stack of acquired images. (Scale bars, 10 μm.) (C and E) Individual , , and slices are shown for (C) RFP and (E) GCaMP6s. White dashed line indicates approximate outline of the worm’s head. (D and F) Maximum intensity projection of the same volume is shown for (D) RFP and (F) GCaMP6s.
An animal’s head moves dramatically during crawling, which poses challenges for volumetric imaging. Worms crawl with a center of mass velocity of up to 0.25 mm/s and in 1 s the head can swing side-to-side by as much as 70 µm, corresponding to nearly two head diameters. To keep the worm’s head centered in the field of view of the high magnification objective, we used a second, low-magnification microscope located on top of the worm to track its movements in real time and to adjust a motorized stage to compensate for the animal’s motion. A high-speed CMOS camera records large depth-of-field fluorescence images of the neurons in the worm’s head through a 10× objective at 67 fps (Fig. 2B). Custom software based on the MindControl codebase (32), written in C, finds the x-y position of the animal’s head from the low-magnification fluorescent images and adjusts the velocity of the motorized stage so as to keep the head of the worm centered. This x-y feedback loop updates at 67 Hz and keeps the worm within the field of view for the duration of our recordings, even when the worm touches itself or crosses over itself. During recordings, the tracker regularly accommodated worm velocities in excess of 0.15 mm/s and turn rates of more than 3 turns/min without losing the animal (SI Appendix, Fig. S1 and Table S1).
The worm also moves its head along the imaging axis (z) as it crawls. This movement occurs when the worm raises its head off of the agarose pad or when the worm crawls over agarose of varying thickness. To keep the worm’s head in the center of the z-scanning range, we analyze fluorescent images from the high magnification objective in real time to find the center of the worm’s head in each z-stack. We then adjust the piezo scan range to compensate for any head motion. This z feedback loop is part of the same LabView code that records high magnification fluorescence. The z feedback updates at 3 Hz and keeps the worm within the scan range. The feedback loop compensated for changes in the worm’s head position of as large as 8 µm in z, or 20% of its head diameter, in the course of 333 ms.
To study neural coding of behavior, it is critically important to record the animal’s full body posture. A third camera in our system records the worm’s posture and behavior at 42 fps through the 10× objective using near-infrared (NIR) dark-field illumination. Images from this camera are analyzed off-line to define the body shape over time. Images from the four simultaneously recorded video feeds are visible in Fig. 2 and Movies S1 and S2.
Whole-Brain Neural Dynamics with Cellular Resolution During Locomotion.
As a proof of principle, we recorded the neural activity and behavior of four individual worms (strain AML14) crawling freely on agarose pads for up to 5 min. We quantified their behavior by calculating the worm’s center-of-mass velocity and body centerline at each time point. The center-of-mass velocity was used to define the worm velocity relative to the body orientation such that positive velocity corresponds to forward locomotion and negative velocity to backward locomotion (Fig. 3). Body centerline information was used to automatically classify animal behavior via an eigenworm analysis (21) (SI Appendix, Methods) into four behavioral modes: forward crawling, backward crawling, pausing, and turning. Note that on average worms exhibited slightly slower locomotion and had higher turning rates when in the imaging arena than on an open agar plate (SI Appendix, Fig. S1 and Tables S1 and S2).
Fig. 3.
The worm’s behavior is recorded during imaging. Data from worm 1 is shown. The worm’s (A) center-of-mass trajectory, body shape, and (B) velocity in the anterior direction are shown. (C) An ethogram describing the behavior is generated automatically from the worm’s posture and behavior (SI Appendix, Methods).
As the worm crawled we recorded the location of each neuron in the worm’s head. Only fluorescence from the calcium insensitive RFP was used to identify the location of each cell. Nuclear regions were identified through a human-supervised semiautomated process. 77, 68, 57, and 56 neuronal nuclei were observed in the four moving worms, respectively, accounting for slightly less than half of the 181 neurons we would expect to be in our field of view based on electron microscopy data (33) (caltech.wormbase.org/virtualworm/). The number of neurons we observe in our moving worms is comparable to the (mean ± SD) neurons identified in a recent whole-brain imaging study of immobilized and anesthetized worms (11). A number of factors likely account for the discrepancy between the mean of 64.5 neurons per worm that we observe and the 181 neurons expected from electron microscopy. Constraints associated with imaging freely moving animals accounts for some of the discrepancy. When worms of the same strain are immobilized and imaged under more favorable conditions, such as longer exposure times, shorter recordings, and thus less photobleaching, up to 90 neurons are observed (SI Appendix, Fig. S2 and Table S3). Most of the discrepancy however, is likely due to the expression pattern of the pan-neuronal promotor and the mosaic inheritance of the extrachromsomal array through development.
To extract calcium dynamics, we measured the fluorescence intensity of GCaMP6s, which was coexpressed with RFP in the nuclei of each neuron. The GCaMP6s fluorescence was measured in a segmented volume defined by the fluorescence from the RFP channel. The segmented region was identified by searching within a sphere with radius 2 μm centered around the centroid of the neuron’s nucleus. The ratio of the fluorescent intensities of GCaMP6s to RFP provided a robust signal of the relative intracellular calcium in the neuron.
This nuclear-localized ratiometric approach allowed us to record calcium dynamics even as the worm turned, reversed, touched itself, or reoriented (Fig. 4, SI Appendix, Fig. S3, and Movies S3 and S4). The most active neurons exhibited calcium transients with a of nearly 200% (SI Appendix, Fig. S4), where is the ratio of the intensity of the GCaMP6s fluorescence, , to the intensity of the RFP fluorescence, ; and is the neuron-specific ratiometric fluorescence baseline (SI Appendix, Methods).
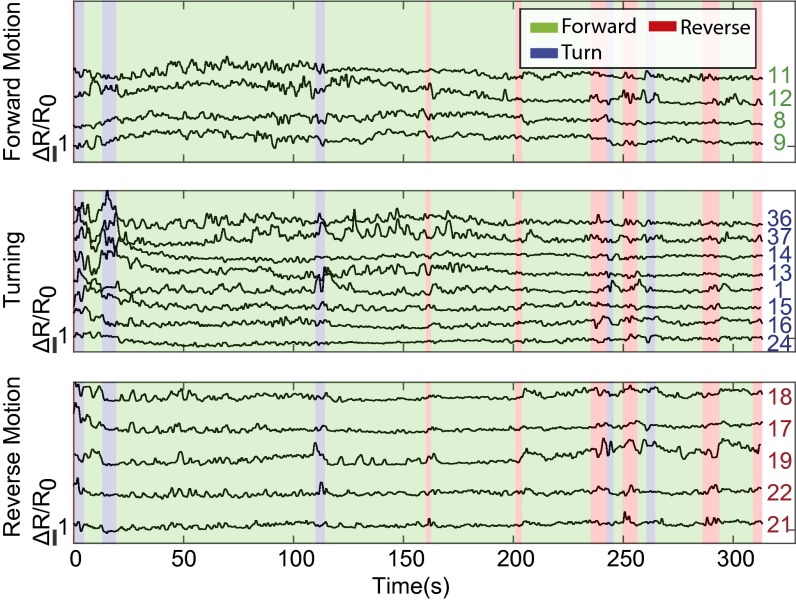
Fig. 4.
Recordings from two of four individuals are shown containing neural activity from 56 and 77 neurons, respectively. Additional recordings are shown in SI Appendix, Fig. S3. The ratio of GCaMP6s fluorescence to RFP fluorescence for each neuron is plotted. The ratio of fluorescent intensities, R, is represented as the fractional change in , where the baseline is defined for each neuron as the lower 20th percentile value. Neurons are sorted via a hierarchical clustering algorithm. Corresponding correlations are shown in Fig. 5. Behavioral ethograms are shown. Neural data from worm 1 corresponds to the same recording as in Fig. 3. Note, neural ID numbers do not indicate correspondence across worms. Occasional white gaps in the data represent instances when the neuron is obscured or transiently leaves the field of view (SI Appendix, Methods).
The fluorescence intensity of each neuron was sampled six times in a second (SI Appendix, Methods), similar to the 4–6 Hz used in previous whole-brain imaging experiments on immobilized and anesthetized worms (11). This rate is sufficient to capture a vast majority of the calcium dynamics probed with GCaMP6s (SI Appendix, Fig. S5).
The correlations of calcium transients between neurons were also calculated. The resulting block structure is suggestive of functionally connected neural units (Fig. 5 and SI Appendix, Fig. S6). In the future, these functional units may serve as a starting point for investigating interactions between microcircuits and whole-brain dynamics.
Fig. 5.
Correlations between calcium activity of neurons from two worms are shown. Data correspond to neural activity shown in Fig. 4. Correlation values are hierarchically clustered using a Euclidean distance metric so that neurons with similar activity are organized together. Additional worms are shown in SI Appendix, Fig. S6.
Observed GCaMP Fluorescence Cannot Be Explained by Motion Alone.
Artifacts created by the animal’s motion pose particular concern because they could be misinterpreted as calcium signals. For example, as the worm moves, individual neurons might compress or expand thereby changing the density of fluorescent protein, which could potentially create the appearance of a calcium transient. Additional artifacts could arise from motion blur, or from spatial inhomogeneities in illumination.
We used a GFP control worm to directly measure changes in our apparent calcium signal due to motion and other artifacts. A transgenic worm was made to express calcium insensitive GFP in place of the calcium sensitive GCaMP (strain AML18; Materials and Methods). RFP expression remained the same and both fluorophores were targeted to neuronal nuclei. We recorded from this GFP control worm as it crawled freely and subjected the recordings to the same analysis pipeline used for GCaMP worms. Any apparent change in fluorescence ratio R in this GFP control worm should be due to artifacts from motion or from other measurement noise in the system because neither the GFP nor RFP has calcium-dependent fluorescence. Extracted fluorescence signals from the GFP control worm showed that noise remains present in our system and motion artifact is likely a source (Fig. 6). Nonetheless, the GFP control worm exhibited a lower mean amplitude and a lower variance compared with the calcium-sensitive GCaMP (Fig. 7A). Note that this population of neurons likely includes many silent as well as active neurons. The mean and SD for GCaMP was 0.2, , whereas for GFP, it was 0.14, . This discrepancy suggests that the GCaMP recordings contain neural activity that is unlikely to be attributed purely to motion or noise.
Fig. 6.
Transients observed from 63 neurons in a freely moving GFP control worm. Fractional change from baseline, , of the ratio between green- and red-channel fluorescence of 63 neurons in a control worm expressing GFP and RFP in the nuclei is shown. Neural activity was extracted using the same analysis pipeline as for GCaMP worms. Color map is identical to that in Fig. 4 and SI Appendix, Fig. S3.
Fig. 7.
Comparison between GCaM6s individuals and GFP control worm. (A) The distribution of fractional change of ratios of fluorescent intensity values, , of all neurons recorded during free behavior is shown for the four GCaMP6s worms (red) and a control GFP worm (blue). Note, this population of neurons likely includes silent as well as active neurons. GCaMP6s had larger mean and SD in its time-varying fluctuations of ratios of fluorescent intensity 0.2, than that of the GFP control worm 0.14, . This is consistent with GCaMP6s’ role as a calcium indicator and suggests that the time varying fluorescence we observe is not merely due to motion artifact. (B) Fraction of observed neurons whose activity correlated with forward (green), turning (blue), or reverse behavior (red) above a significance threshold is shown for the four GCaMP6s individuals and the GFP control worm. Numerals refer to the number of neurons that correlated with each behavior. Number of neurons that correlate with any of the three behaviors out of total neurons observed is shown above each bar.
The nuclear-localized ratiometric imaging approach used here was selected to minimize artifact from motion and to maximize signal. Initially, we also explored alternate imaging approaches, including recording from worms expressing GCaMP in the cytoplasm and RFP in the nucleus, using either ratiometric or nonratiometric imaging. To evaluate the susceptibility of these various approaches to motion artifact, we compared the power spectra of two GFP control worms: a “nuclear” strain, AML18, which expressed both GFP and RFP in the nucleus, and a “cytoplasmic” strain AML10, which expressed GFP in the cytoplasm and RFP in the nucleus. These strains were compared for both ratiometric and nonratiometric imaging. Any apparent neural activity observed in either control worm should be due to artifact from motion or from other noise in the system. We expect that artifact due to animal motion would be most prominent at the frequency of the worm’s sinusoidal body oscillations. Therefore, we compared the power spectra of the observed fluorescent transients of the two control worms to the power spectra of the animal’s postural oscillations and sought the imaging approach that gave fluorescent transients with the lowest power at the frequencies of animal motion. A clear peak corresponding to animal motion was visible in the control worms for each imaging approach except for the nuclear-localized ratiometric approach (SI Appendix, Fig. S7). The absence of this peak suggests that a nuclear-localized ratiometric approach is less susceptible to artifact from animal motion.
We furthered explored the choice of ratiometric vs. nonratiometric imaging for our nuclear GCaMP recordings by calculating the percentage of our GCaMP recordings that can confidently be attributed to signal generally and not noise for both ratiometric and nonratiometric approaches (SI Appendix, Fig. S8 and Table S4). The all-nuclear-localized ratiometric imaging approach was found to have a higher portion of apparent neural activity that could be attributed to signal than a nonratiometric approach.
We speculate that a nuclear localized ratiometric approach performs most favorably because when GCaMP and RFP are both expressed in the same spatial patterns, such as in the nuclei, they are both affected similarly by many sources of noise, including motion. In these cases the calcium-insensitive RFP acts as a built-in neuron-specific control and taking the ratio of the GCaMP and RFP fluorescence intensities mitigates any noise common to both signals.
Motion artifact should affect neighboring neurons similarly. We were therefore reassured to observe in our GCaMP recordings neighboring neurons that show qualitatively different temporal dynamics (see, for example, neurons 32″ and 7″ in SI Appendix, Fig. S4). This observation further suggests that GCaMP recordings are capturing calcium dynamics and that observed changes in fluorescent intensity cannot merely be explained by motion artifact.
Specific Neurons Correlate with Each of the Three Observed Behaviors.
We next calculated the correlation between the dynamics of individual neurons and changes in locomotory behaviors. The roughly neurons we observed in each worm displayed a varying degree of correlation with the behaviors we measured. To reveal neurons with significant behavioral correlations that are also unlikely to arise from motion artifact, we applied two statistical thresholds, one related to the strength of the correlation and another related to the change in fluorescent intensity. The thresholds were selected based on intrinsic properties of noise observed in the neural signals and were calibrated to the motion artifacts observed in our GFP control recordings. Briefly, we calculated the Pearson’s correlation coefficient r between each neural trace and three individual behaviors: forward, backward, and turning. To be considered significant, we required that each neuron’s correlation r be statistically larger than 0 as determined by bootstrapping (SI Appendix, Methods) and further that r always be greater than . To be considered significant, we also required that the change of the signal’s intensity during the behavior exceed a threshold based on the intensities observed in the GFP control worm, which arise solely due to noise, including motion artifact (SI Appendix, Methods). Always at least 8 neurons and as many as 18 neurons in a worm passed our criteria for significance and correlated with either of three behaviors, forward, backward, and turning, with an average coefficient of (Figs. 7B and 8 and SI Appendix, Fig. S9).
Fig. 8.
All neurons whose activity correlated with either forward, turning, or reverse behavior above a significance threshold are shown for worm 1. The significance threshold was calibrated to the noise observed in the GFP control recordings. Vertical scale bar indicates of 1 (100%). Each neuron’s ID number is indicated. Note neuron numbers do not indicate correspondence across worms. Additional individuals are shown in SI Appendix, Fig. S9.
We repeated the same analysis on the GFP control worm recordings and were reassured to find that of 63 neurons not a single one had fluorescence intensity dynamics that met the threshold criteria for significance in its correlation with behavior (Fig. 7B).
Comparison of Neural Position.
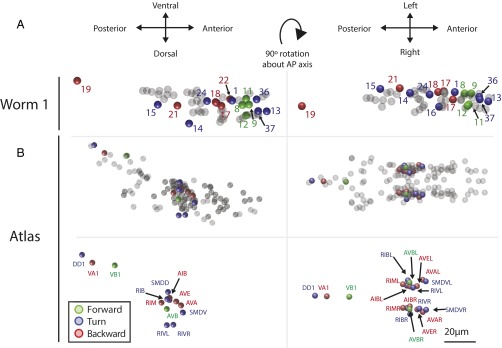
We sought to investigate the spatial location of those neurons that had calcium transients that correlated with behavior (Fig. 9). Plotting the 3D neuronal positions of those neurons that passed our significance threshold, we occasionally see a correspondence in location between the behavior-specific neurons across worms. For example reversal neurons 17 and 18 in worm 1 are in strikingly similar position to reversal neuron 7″ and 8″ in worm 3 (SI Appendix, Fig. S10), suggesting that these may be the same neuron across worms.
Fig. 9.
The position of recorded neurons for worm 1 is plotted in 3D and compared with an atlas. (Left) Dorsal-ventral plane view of the animal. (Right) Left-right plane view of the animal. (A) Neurons whose activity correspond to forward, turning, or reverse are colored and labeled with a neuron ID corresponding to Fig. 4. Remaining neurons are shaded gray. Additional worms are shown in SI Appendix, Fig. S10. (B) The position of all known neurons are plotted using reference data culled from the WormBase Virtual Worm Project. Neurons that have previously been reported in the literature as implicated in a behavior are colored accordingly (SI Appendix, Table S5) and labeled with their neuron name. Colored neurons are shown both in and out of context of all other neurons for clarity.
To compare recordings across worms or to compare with the literature, it would be informative to know the identity of each neuron in our recordings, e.g., which neuron is AVA, AVB, etc. We wondered whether neuron position information alone would be sufficient to systematically match neurons across worms or to assign an identity to each neuron with respect to the known neuroanotomical atlas from electron microscopy data (33). We designed an experiment that measured the accuracy with which neurons in one worm could be matched to neurons in another based on position information alone. An integrated transgenic line was generated that expressed RFP in the nuclei of all neurons and expressed GFP in a genetically defined subset of neurons of which 11 could be unambiguously determined by eye (strain AML5; Materials and Methods). Two adults of similar age were immobilized and imaged. We tested various 3D alignment, registration, or nonlinear warping algorithms that relied only on neural position information to match neurons between the two animals. Candidate algorithms were given position information of all neurons observed in the head (127 neurons for worm A and 128 neurons for worm B). Matchings were validated by inspecting whether the subset of 11 unambiguous GFP labeled-neurons, whose identities are known, were matched correctly. Simply aligning the two worms and matching nearest neighbors resulted in correctly matching 3 of the 11 known GFP neurons (SI Appendix, Fig. S11). Of seven algorithms from the literature that were tested (SI Appendix, Methods), a nonrigid point set registration algorithm (34) was found to perform best but still only correctly matched 6 of the 11 known GFP-labeled neurons (SI Appendix, Fig. S11).
Our neural identity validation experiments suggest that position information alone is insufficient to correctly assign neural identities across worms. Therefore, we do not know the true identities of the neurons we observe (Discussion). As in previous whole-brain imaging studies (11, 12), we instead turn to evidence from the literature to reveal likely candidate neural identities to assign to our neurons based on a combination of observed neural activity, previously reported neural function and neuroanotomical position. We found reports in the literature of nonsensory neurons implicated in the three locomotory behaviors of interest (SI Appendix, Table S5). We plotted these candidate neurons’ position in three dimensions (Fig. 9 and SI Appendix, Fig. S10) using data culled from the WormBase Virtual Worm Project (caltech.wormbase.org/virtualworm/), which in turn is based on electron microscopy data from ref. 33. Comparing this atlas to data from our three worms provides plausible candidate neural identities for those neurons we observed who had activity correlated with forward, turning, and reversing. For example, the position and activity of neurons 17 and 18 in worm 1 is consistent with these neurons being AIBL or AVEL.
Discussion
The imaging system presented here allows for whole-brain recording of calcium activity with cellular resolution in a freely moving C. elegans. We used this system to investigate dynamics of neurons in the worm whose activity correlates with forward, backward, and turning modes of locomotion. We observed a number of neurons in each worm whose activity correlated with behavior. Some of these neurons compare favorably with results from the literature, whereas others appear to have previously unidentified roles in locomotory behavior. This study is the first example, to our knowledge, of recorded calcium dynamics from nonsensory neurons in the head that correlate with turning. We also characterize noise in the system due to motion or other artifacts and show that the calcium activity of the neurons who are most correlated with behavior are unlikely to be explained by motion artifact. We believe this work represents a significant advance toward studying how population dynamics of a brain-sized neural network generates behavior.
Of nearly 80 recorded neurons, at least 8 and as many as 18 neurons in each worm were observed to have neural signals that correlate with behavior and exceed our noise threshold based on an analysis of the GFP control worm. Aggregate, 23% of neurons were found to correlate significantly with behavior in our GCaMP worms, whereas none were found in our GFP control. We suspect that there may be other additional neurons that have activity that also correlate with behavior but that their signals are too weak to unambiguously rule out contributions from noise, even using a ratiometric approach. Revealing the weakest of neural signals is a challenge that will need to be addressed in future work.
The system currently records intracellular calcium levels from within neuronal nuclei. Recording from the nuclei is advantageous because the spatially distinct nuclei allow each cell to be distinguished from its neighbors. Moreover, compared with a cytoplasmic approach, the all-nuclear approach is less susceptible to artifacts from animal motion. Nuclear recordings were successfully used previously in two whole-brain imaging studies of anesthetized and immobilized C. elegans, and those works argue that nuclear calcium levels capture the calcium dynamics of the rest of the neuron (11, 12). Recordings from the cell body are the most prevalent in the literature and have previously been used to study activity of a number of neurons relating to locomotory behavior (17, 18, 25–28). Because C. elegans nuclei are thought to be porous to calcium and because the nuclei take up a large portion of the cell body, nuclear recordings used here likely capture calcium transients observed in the cell body. There are, however, known neurons where calcium signals in the processes are different from those in the cell body and, by extension, the nuclei (24, 35–37). It remains to be seen whether independent calcium dynamics in the processes are unique to these few specific neurons or are a more general phenomenon. For example, a recent investigation of three C. elegans interneurons found that calcium activity peaked more quickly in the process than the cell body but was otherwise similar (29).
Although it is tempting to try to explore neural dynamics in the processes by expressing a calcium indicator in the cytoplasm of all neurons, pan-neuronal cytoplasmic expression of a fluorophore is impractical for segmenting individual processes. Currently single-cell recordings remain the most viable method for probing neural dynamics in the processes. Future work is needed to solve the problem of segmenting densely labeled neuronal processes in freely moving animals.
This system successfully identifies and tracks neurons through time within a given worm. One of the advantages of working in C. elegans is that its neural anatomy and connectivity has been mapped (33). Unfortunately, when all neurons are uniformly labeled there is currently no systematic and reliable way of relating identified neurons across worms or to the known atlas. This challenge has plagued all previous pan-neuronal imaging studies in adult C. elegans (11, 12) and remains unresolved here. Conventional wisdom in the field suggests that the worm’s neuroanatomy is highly stereotyped across adults, and this conventional wisdom is supported by a comparison of neurons in the tail of three worms (38). The neural position validation experiments presented here, however, have led us to conclude that worm-to-worm variability in neuronal position in the head is large enough to pose a formidable challenge for neuron identification. Future work is needed to solve this neural identity challenge. One potential approach would be to develop worms that have multicolored fiducial references that can be used for 3D warping and registration.
The approach here records images of the entire worm’s body and posture as the animal crawls. We use the body posture to define four behaviors via an eigenworm analysis (21). Future work could use the video recordings of the animal’s entire body to correlate neural activity with a variety of other behaviors that are also captured by our instrument. For example, full-body recordings have been used to enumerate many more C. elegans behaviors (22) and to study such physiologically relevant behaviors as deep ventral bends (30), foraging head oscillations (39), coiling (13), pumping (40), defecating (41), and egg-laying (42), all of which exist in our datasets. Recent work in flies has shown the value of capturing images of full body posture for analyzing behavior (43). An analysis similar to that work, but which also combines population neural activity, could be performed in C. elegans using the methods presented here in the future.
The current system uses a spinning disk confocal imaging modality. Our system is fully modular, and the 3D tracking microscopy and off-line image analysis are well suited to take advantage of other imaging modalities such as light-field or two-photon microscopy, as well as advances in genetically encoded calcium indicators (44) and voltage indicators (45–47).
Materials and Methods
Strains.
We cultivated transgenic worms on nematode growth medium (NGM) plates with OP50 bacteria. OP50 plates were made by seeding 6-cm NGM plates with 250 µL of a suspension of OP50 bacteria in lysogeny broth. All worms used in this study were young adult hermaphrodites. Worms were cultivated and handled in the dark or under red-light illumination to minimize potential photobleaching from ambient light.
Strain AML14 (wtfEx4[Prab-3::NLS::GCaMP6s, Prab-3::NLS::tagRFP]) was generated by coinjecting into an N2 background a Prab-3::NLS::tagRFP plasmid (gift of O. Hobert, Columbia University, New York) (48) with a derivative of that plasmid where NLS::tagRFP was replaced with a GCaMP6s sequence flanked by nuclear localization sequences described in (11). Strain AML18 (wtfIs3[Prab-3::NLS::GFP, Prab-3::NLS::tagRFP]) was generated by coinjecting into an N2 background a Prab-3::NLS::RFP plasmid (gift of O. Hobert) (48) with a derivative of that plasmid where RFP was replaced with a GFP sequence and then integrated via UV irradiation. Strain AML10 (otIs45[Punc-119::GFP]; otIs355[Prab-3::NLS::tagRFP]) was generated by crossing a male of strain QW1155 (otIs355[Prab-3::NLS::tagRFP]) (gift of M. Alkema, University of Massachusetts Worcester, Worcester, MA) with a hermaphrodite of strain OH441 (otIs45[Punc-119::GFP]) (gift of C. Murphy, Princeton University, Princeton). Strain AML5 (otIs355[Prab-3::NLS::tagRFP]; KyIs51[Podr-2b::GFP]) was generated by crossing a male from strain QW1155 (otIs355[Prab-3::NLS::tagRFP]) (gift of M. Alkema), with a hermaphrodite of strain CX3300 (KyIs51[Podr-2b::GFP]) (gift of C. Bargmann, Rockefeller University, New York). Strain N2 bristol was used for WT.
Strain AML14 was used for all calcium imaging experiments. Individual animals of extrachromasomal strain AML14 were selected for bright and uniform expression on a fluorescent dissection scope before use.
Additional Methods.
For additional methods, please see SI Appendix, Methods.
Supplementary Material
Acknowledgments
We thank M. Zimmer, G. Berman, and B. Bratton for productive discussions. We thank the following for gifts of transgenic nematodes used here or used in preliminarily versions of this work: O. Hobert, C. Bargmann, M. Zimmer, C. Clark, J. Pirri, M. Alkema, A. Sylvain, and C. Murphy. We thank the following for assistance with data analysis: K. Mizes, D. Mazumder, J. Chinchilla, and L. Novak. Some strains were provided by the CGC, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by Simons Foundation Grant SCGB 324285 (to A.M.L.) and Princeton University’s Inaugural Dean’s Innovation Fund for New Ideas in the Natural Sciences (to J.W.S. and A.M.L.). A.N.L. is supported by a National Institutes of Health institutional training grant through the Princeton Neuroscience Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Datasets S1–S16 provide the extracted neural activity and behavior data. Raw imaging data (>200 GB) will be provided to any researcher on request.
See Commentary on page 1965.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1507110112/-/DCSupplemental.
References
- 1.Maynard EM, et al. Neuronal interactions improve cortical population coding of movement direction. J Neurosci. 1999;19(18):8083–8093. doi: 10.1523/JNEUROSCI.19-18-08083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long MA, Jin DZ, Fee MS. Support for a synaptic chain model of neuronal sequence generation. Nature. 2010;468(7322):394–399. doi: 10.1038/nature09514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Briggman KL, Abarbanel HDI, Kristan WBJ., Jr Optical imaging of neuronal populations during decision-making. Science. 2005;307(5711):896–901. doi: 10.1126/science.1103736. [DOI] [PubMed] [Google Scholar]
- 4.Harvey CD, Coen P, Tank DW. Choice-specific sequences in parietal cortex during a virtual-navigation decision task. Nature. 2012;484(7392):62–68. doi: 10.1038/nature10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499(7458):295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwan AC. What can population calcium imaging tell us about neural circuits? J Neurophysiol. 2008;100(6):2977–2980. doi: 10.1152/jn.91037.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahrens MB, et al. Brain-wide neuronal dynamics during motor adaptation in zebrafish. Nature. 2012;485(7399):471–477. doi: 10.1038/nature11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubo F, et al. Functional architecture of an optic flow-responsive area that drives horizontal eye movements in zebrafish. Neuron. 2014;81(6):1344–1359. doi: 10.1016/j.neuron.2014.02.043. [DOI] [PubMed] [Google Scholar]
- 9.Dombeck DA, Khabbaz AN, Collman F, Adelman TL, Tank DW. Imaging large-scale neural activity with cellular resolution in awake, mobile mice. Neuron. 2007;56(1):43–57. doi: 10.1016/j.neuron.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flusberg BA, et al. High-speed, miniaturized fluorescence microscopy in freely moving mice. Nat Methods. 2008;5(11):935–938. doi: 10.1038/nmeth.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schrödel T, Prevedel R, Aumayr K, Zimmer M, Vaziri A. Brain-wide 3D imaging of neuronal activity in Caenorhabditis elegans with sculpted light. Nat Methods. 2013;10(10):1013–1020. doi: 10.1038/nmeth.2637. [DOI] [PubMed] [Google Scholar]
- 12.Prevedel R, et al. Simultaneous whole-animal 3D imaging of neuronal activity using light-field microscopy. Nat Methods. 2014;11(7):727–730. doi: 10.1038/nmeth.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croll NA. Behavioural analysis of nematode movement. Adv Parasitol. 1975;13:71–122. doi: 10.1016/s0065-308x(08)60319-x. [DOI] [PubMed] [Google Scholar]
- 14.Croll N. Components and patterns in the behavior of the nematode Caenorhabditis elegans. J Zool. 1975;176:159–176. [Google Scholar]
- 15.Pierce-Shimomura JT, Morse TM, Lockery SR. The fundamental role of pirouettes in Caenorhabditis elegans chemotaxis. J Neurosci. 1999;19(21):9557–9569. doi: 10.1523/JNEUROSCI.19-21-09557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cronin CJ, Feng Z, Schafer WR. Automated imaging of C. elegans behavior. Methods Mol Biol. 2006;351:241–251. doi: 10.1385/1-59745-151-7:241. [DOI] [PubMed] [Google Scholar]
- 17.Ben Arous J, Tanizawa Y, Rabinowitch I, Chatenay D, Schafer WR. Automated imaging of neuronal activity in freely behaving Caenorhabditis elegans. J Neurosci Methods. 2010;187(2):229–234. doi: 10.1016/j.jneumeth.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 18.Faumont S, et al. An image-free opto-mechanical system for creating virtual environments and imaging neuronal activity in freely moving Caenorhabditis elegans. PLoS One. 2011;6(9):e24666. doi: 10.1371/journal.pone.0024666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramot D, Johnson BE, Berry TL, Jr, Carnell L, Goodman MB. The Parallel Worm Tracker: A platform for measuring average speed and drug-induced paralysis in nematodes. PLoS One. 2008;3(5):e2208. doi: 10.1371/journal.pone.0002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swierczek NA, Giles AC, Rankin CH, Kerr RA. High-throughput behavioral analysis in C. elegans. Nat Methods. 2011;8(7):592–598. doi: 10.1038/nmeth.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stephens GJ, Johnson-Kerner B, Bialek W, Ryu WS. Dimensionality and dynamics in the behavior of C. elegans. PLOS Comput Biol. 2008;4(4):e1000028. doi: 10.1371/journal.pcbi.1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown AEX, Yemini EI, Grundy LJ, Jucikas T, Schafer WR. A dictionary of behavioral motifs reveals clusters of genes affecting Caenorhabditis elegans locomotion. Proc Natl Acad Sci USA. 2013;110(2):791–796. doi: 10.1073/pnas.1211447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faumont S, Lockery SR. The awake behaving worm: Simultaneous imaging of neuronal activity and behavior in intact animals at millimeter scale. J Neurophysiol. 2006;95(3):1976–1981. doi: 10.1152/jn.01050.2005. [DOI] [PubMed] [Google Scholar]
- 24.Clark DA, Biron D, Sengupta P, Samuel ADT. The AFD sensory neurons encode multiple functions underlying thermotactic behavior in Caenorhabditis elegans. J Neurosci. 2006;26(28):7444–7451. doi: 10.1523/JNEUROSCI.1137-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piggott BJ, Liu J, Feng Z, Wescott SA, Xu XZS. The neural circuits and synaptic mechanisms underlying motor initiation in C. elegans. Cell. 2011;147(4):922–933. doi: 10.1016/j.cell.2011.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawano T, et al. An imbalancing act: Gap junctions reduce the backward motor circuit activity to bias C. elegans for forward locomotion. Neuron. 2011;72(4):572–586. doi: 10.1016/j.neuron.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Larsch J, Ventimiglia D, Bargmann CI, Albrecht DR. High-throughput imaging of neuronal activity in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2013;110(45):E4266–E4273. doi: 10.1073/pnas.1318325110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shipley FB, Clark CM, Alkema MJ, Leifer AM. Simultaneous optogenetic manipulation and calcium imaging in freely moving C. elegans. Front Neural Circuits. 2014;8:28. doi: 10.3389/fncir.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordus A, Pokala N, Levy S, Flavell SW, Bargmann CI. Feedback from network states generates variability in a probabilistic olfactory circuit. Cell. 2015;161(2):215–227. doi: 10.1016/j.cell.2015.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Donnelly JL, et al. Monoaminergic orchestration of motor programs in a complex C. elegans behavior. PLoS Biol. 2013;11(4):e1001529. doi: 10.1371/journal.pbio.1001529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray JM, Hill JJ, Bargmann CI. A circuit for navigation in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102(9):3184–3191. doi: 10.1073/pnas.0409009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leifer AM, Fang-Yen C, Gershow M, Alkema MJ, Samuel ADT. Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans. Nat Methods. 2011;8(2):147–152. doi: 10.1038/nmeth.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314(1165):1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- 34. Bing Jian, Vemuri BC (2011) Robust Point Set Registration Using Gaussian Mixture Models. IEEE Trans Pattern Anal Mach Intell 33(8):1633–1645. [DOI] [PubMed]
- 35.Chalasani SH, et al. Dissecting a circuit for olfactory behaviour in Caenorhabditis elegans. Nature. 2007;450(7166):63–70. doi: 10.1038/nature06292. [DOI] [PubMed] [Google Scholar]
- 36.Li Z, Liu J, Zheng M, Xu XZS. Encoding of both analog- and digital-like behavioral outputs by one C. elegans interneuron. Cell. 2014;159(4):751–765. doi: 10.1016/j.cell.2014.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hendricks M, Ha H, Maffey N, Zhang Y. Compartmentalized calcium dynamics in a C. elegans interneuron encode head movement. Nature. 2012;487(7405):99–103. doi: 10.1038/nature11081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall DH, Russell RL. The posterior nervous system of the nematode Caenorhabditis elegans: Serial reconstruction of identified neurons and complete pattern of synaptic interactions. J Neurosci. 1991;11(1):1–22. doi: 10.1523/JNEUROSCI.11-01-00001.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pirri JK, McPherson AD, Donnelly JL, Francis MM, Alkema MJ. A tyramine-gated chloride channel coordinates distinct motor programs of a Caenorhabditis elegans escape response. Neuron. 2009;62(4):526–538. doi: 10.1016/j.neuron.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song BM, Avery L. Serotonin activates overall feeding by activating two separate neural pathways in Caenorhabditis elegans. J Neurosci. 2012;32(6):1920–1931. doi: 10.1523/JNEUROSCI.2064-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas JH. Genetic analysis of defecation in Caenorhabditis elegans. Genetics. 1990;124(4):855–872. doi: 10.1093/genetics/124.4.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ringstad N, Horvitz HR. FMRFamide neuropeptides and acetylcholine synergistically inhibit egg-laying by C. elegans. Nat Neurosci. 2008;11(10):1168–1176. doi: 10.1038/nn.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berman GJ, Choi DM, Bialek W, Shaevitz JW. Mapping the stereotyped behaviour of freely moving fruit flies. J R Soc Interface. 2014;11(99):20140672. doi: 10.1098/rsif.2014.0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thestrup T, et al. Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat Methods. 2014;11(2):175–182. doi: 10.1038/nmeth.2773. [DOI] [PubMed] [Google Scholar]
- 45.Hochbaum DR, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nat Methods. 2014;11(8):825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jin L, et al. Single action potentials and subthreshold electrical events imaged in neurons with a fluorescent protein voltage probe. Neuron. 2012;75(5):779–785. doi: 10.1016/j.neuron.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gong Y, Li JZ, Schnitzer MJ. Enhanced archaerhodopsin fluorescent protein voltage indicators. PLoS One. 2013;8(6):e66959. doi: 10.1371/journal.pone.0066959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stefanakis N, Carrera I, Hobert O. Regulatory Logic of Pan-Neuronal Gene Expression in C. elegans. Neuron. 2015;87(4):733–750. doi: 10.1016/j.neuron.2015.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.