Significance
Recent studies showing that both high-accuracy protein synthesis and low-accuracy protein synthesis are critical for cellular growth and survival under different growth conditions suggest that the translation quality-control machinery may function in cellular processes other than protein synthesis. We explored this possibility and found that inhibition of misacylated-tRNA quality control suppressed the synthesis of the starvation-inducible second messenger guanosine tetraphosphate and limited the induction of stringent response-dependent gene expression. These findings show that translation quality control is a single checkpoint for both accurate protein synthesis and transcriptional responses to stress.
Keywords: translation, quality control, stress, stringent response
Abstract
Gene expression relies on quality control for accurate transmission of genetic information. One mechanism that prevents amino acid misincorporation errors during translation is editing of misacylated tRNAs by aminoacyl-tRNA synthetases. In the absence of editing, growth is limited upon exposure to excess noncognate amino acid substrates and other stresses, but whether these physiological effects result solely from mistranslation remains unclear. To explore if translation quality control influences cellular processes other than protein synthesis, an Escherichia coli strain defective in Tyr-tRNAPhe editing was used. In the absence of editing, cellular levels of aminoacylated tRNAPhe were elevated during amino acid stress, whereas in the wild-type strain these levels declined under the same growth conditions. In the editing-defective strain, increased levels of aminoacylated tRNAPhe led to continued synthesis of the PheL leader peptide and attenuation of pheA transcription under amino acid stress. Consequently, in the absence of editing, activation of the phenylalanine biosynthetic operon becomes less responsive to phenylalanine limitation. In addition to raising aminoacylated tRNA levels, the absence of editing lowered the amount of deacylated tRNAPhe in the cell. This reduction in deacylated tRNA was accompanied by decreased synthesis of the second messenger guanosine tetraphosphate and limited induction of stringent response-dependent gene expression in editing-defective cells during amino acid stress. These data show that a single quality-control mechanism, the editing of misacylated aminoacyl-tRNAs, provides a critical checkpoint both for maintaining the accuracy of translation and for determining the sensitivity of transcriptional responses to amino acid stress.
Accurate translation of mRNA into the corresponding amino acid sequence is an essential step during gene expression. Translational fidelity depends on correct tRNA–codon pairing by the ribosome as well as attachment of the proper amino acids to their respective tRNAs by aminoacyl-tRNA synthetases (aaRSs). The ability of aaRS enzymes to distinguish between cognate and noncognate amino acids is a major determinant of the fidelity with which the genetic code is translated (1). Discrimination against noncognate amino acid substrates is particularly challenging for aaRSs, because a near-cognate amino acid may differ from the cognate substrate by as little as a single methyl or hydroxyl group. Generally, highly specific aaRS enzymes and the widespread existence of editing mechanisms that proofread noncognate amino acids prevent errors in amino acid recognition from compromising the overall accuracy of translation (2). For example, in the class I isoleucyl-tRNA synthetase a posttransfer editing site for deacylation of misacylated Val-tRNAIle exists 35 Å from the active site. Similarly, the class II phenylalanine-tRNA synthetase (PheRS) edits misacylated Tyr-tRNAPhe at a hydrolytic editing site ∼30 Å from the synthetic active site. Bacterial PheRS editing prevents meta-tyrosine (m-Tyr), a readily activated nonprotein amino acid and product of Phe oxidation, from being misincorporated into the proteome (3). These and other aaRS proofreading activities provide quality-control checkpoints against noncognate amino acid incorporation during translation and thereby help prevent the formation of an aberrant proteome.
Despite their role in accurately translating the genetic code, aaRS editing pathways are not conserved, and their activities have varying effects on cell viability. Mycoplasma mobile, for example, tolerates relatively high error rates during translation and apparently has lost both PheRS and leucyl-tRNA synthetase proofreading activities (4). Similarly, the editing activity of Streptococcus pneumoniae IleRS is not robust enough to compensate for its weak substrate specificity, leading to the formation of misacylated Leu-tRNAIle and Val-tRNAIle species (5). Also, in contrast to its cytoplasmic and bacterial counterparts, Saccharomyces cerevisiae mitochondrial PheRS completely lacks an editing domain and instead appears to rely solely on stringent Phe/Tyr discrimination to maintain specificity (6). The divergent range of mechanisms used to discriminate against noncognate amino acids illustrates how the requirements for translation quality control vary with cellular physiology and among environmental niches (7). Although translation quality control by aaRS editing has not been shown to be essential in any organisms, both its presence and absence can be critical for optimal growth and cellular fitness under various stress conditions in different organisms (2). These highly varied effects on cellular physiology suggest that quality control of misacylated tRNA synthesis may play other important roles in the cell in addition to maintaining proteome integrity.
In addition to acting as substrates for protein synthesis, aminoacyl-tRNAs have numerous other functions in the cell, e.g., as amino acid donors in processes including lipid and protein modification, antibiotic biosynthesis, and heme metabolism (8). Another key role of aminoacyl-tRNA is to serve as signaling molecules for a number of starvation sensors (9). The levels of aminoacyl-tRNA and deacylated tRNA are, respectively, signals for stimulating and limiting translation in response to amino acid stress. Most bacteria contain transcriptional attenuation mechanisms or translational controls that monitor the aminoacylation state of tRNA as the signal for regulating the expression of genes required for amino acid biosynthesis and import. These mechanisms include ribosomal attenuation, T-box riboswitches, and occlusion of the ribosome binding site, such as the feedback regulation of the E. coli threonyl-tRNA synthetase gene (thrS) leader sequence that resembles tRNAThr (10–12). An example of ribosome attenuation occurs during translation of the E. coli leader peptide encoded by pheL when ribosome stalling stimulates the transcription of pheA, which encodes the chorismate mutase required for phenylalanine biosynthesis.
Deacylated tRNA is an important signaling molecule in bacteria, most notably during activation of the highly conserved stringent response pathway (13). The stringent response in E. coli is activated under a number of stresses, including nutrient deprivation, and helps promote cell survival under unfavorable growth conditions. The paralogous enzymes RelA and SpoT both mediate the stringent response through the synthesis of the alarmones guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) collectively known as “(p)ppGpp.” SpoT has been proposed to be responsible for synthesis of (p)ppGpp as part of the general cellular stress response to conditions including fatty acid starvation, carbon starvation, and osmotic shock. Alternatively amino acid starvation also may lead to ribosome stalling and binding of deacylated tRNA in the ribosomal A site, resulting in the release of RelA and production of (p)ppGpp. Therefore, levels of deacylated tRNA are critical for regulating the activation of the stringent response under conditions of amino acid limitation in E. coli. The rise in alarmone levels in the cell is rapid and inhibits transcription initiation at the promoters of genes needed for the transcription and translation machineries, including rRNAs, aaRSs, and tRNAs, through direct binding to the β′–Ω subunit interface of E. coli RNA polymerase (14–16). The initiation of the transcription of some ribosomal protein operons is regulated via ppGpp, although regulation of these genes largely occurs through translational feedback (17, 18). At the same time the transcription of genes required for stress survival, virulence, and antibiotic resistance and those encoding metabolic enzymes, particularly glycolytic and amino acid biosynthesis enzymes, is up-regulated upon activation of the stringent response (19). Given that tRNA aminoacylation levels directly regulate both transcriptional attenuation of pheA and the stringent response, we investigated the possible role of misacylated tRNA editing in determining the responsiveness of cellular responses to amino acid stress. The sensitivities to amino acid stress of both transcription attenuation and the stringent response were significantly reduced in the absence of editing, revealing an important physiological role for this translation quality-control pathway.
Results
Ablation of PheRS Editing Alters the Cellular Ratio of Aminoacylated to Deacylated tRNAPhe.
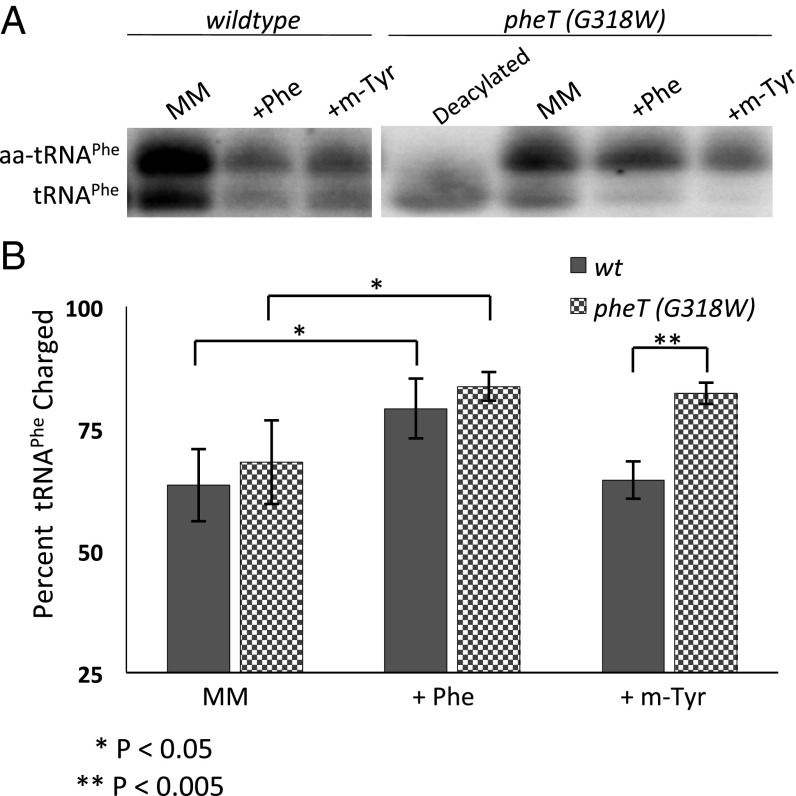
To investigate possible roles of editing outside translation, we first assessed the degree to which misaminoacylation by PheRS alters the overall intracellular levels of deacylated and aminoacylated tRNAPhe (aa-tRNAPhe). Our previously constructed PheRS editing-deficient strain in a MG1655 background was used to misacylate tRNAPhe in vivo (3). This strain contains a βG318W substitution in the editing site of PheRS, which blocks aa-tRNA access and abolishes posttransfer editing. Using acid-urea polyacrylamide gel separation and Northern blotting, we assessed the in vivo levels of aminoacylated tRNAPhe relative to deacylated tRNAPhe in E. coli in the presence and absence of PheRS editing. In M9 minimal medium the percentage of aminoacylated tRNAPhe is ∼65% for the wild-type and editing-deficient strains, and the addition of 0.1 mM Phe to the culture increased this percentage to ∼80% in both cases (Fig. 1). These tRNA aminoacylation levels are comparable to those previously reported in other bacterial systems using similar methods, as is the observed increase in deacylated tRNA upon starvation (20–22). When the strains were grown under amino acid stress conditions in the presence of physiologically relevant levels of the natural noncognate PheRS substrate m-Tyr (3, 23, 24), levels of aa-tRNAPhe were 23% higher in the editing-deficient strain than in the wild type. These results also revealed that the cellular ratio of aminoacylated to deacylated tRNA increases significantly in the absence of PheRS editing, particularly under amino acid stress conditions (Fig. 1). This result, in turn, suggests that regulation of the stress response pathways for which aminoacylated and deacylated-tRNAs serve as signaling molecules also may be perturbed in the absence of editing.
Fig. 1.
Aminoacylated and deacylated tRNA levels are altered in the absence of quality control by PheRS editing. (A) Representative Northern blot probed with a 32P-5′-end–labeled tRNAPhe probe in which aminoacylated and nonaminoacylated species are separated by an acid urea gel before transfer. (B) Percent tRNA charging levels in vivo in wild-type cells (solid bars) and in editing-deficient pheT(G318W) PheRS cells (checkered bars) grown to late log in M9 minimal medium containing no Phe, 100 µM Phe, or 10 µM m-Tyr. Error bars represent the SD from three independent experiments.
PheRS Editing Is Required for Proper Regulation of Phenylalanine Biosynthesis in E. coli.
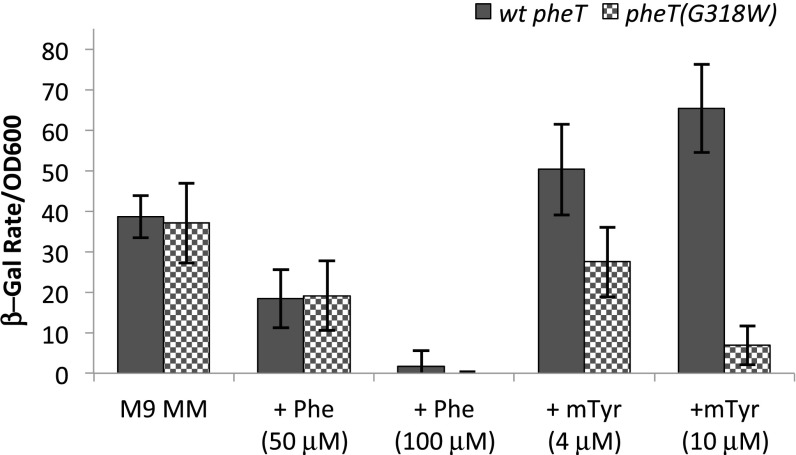
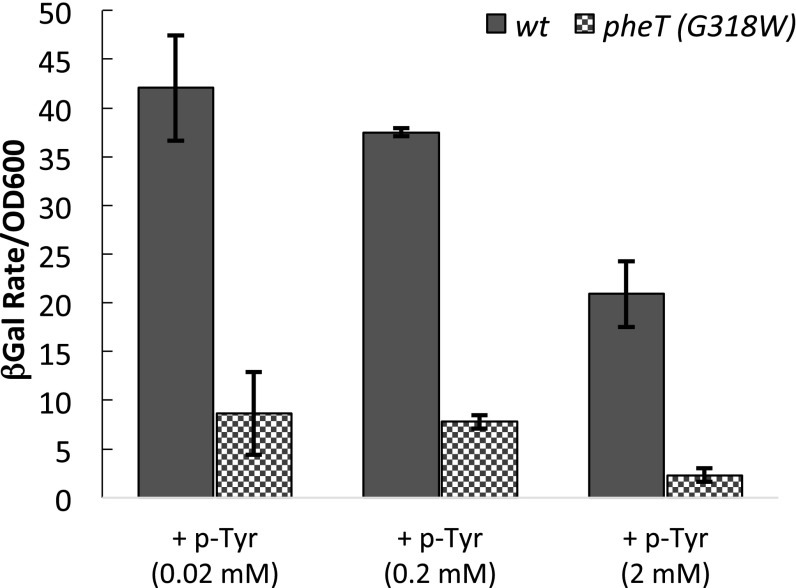
To decipher the transcription regulatory effects of noncognate aa-tRNAPhe accumulation under amino acid stress conditions, regulation of the pheA leader region was examined. Expression of the pheA phenylalanine biosynthesis operon in E. coli is regulated by transcription attenuation via synthesis of the leader peptide PheL (25). Here, pheA′-lacZ reporter strains were constructed using a λ vector (Materials and Methods). A markerless deletion of the endogenous lacIZYA operon was made in the wild-type pheT and pheT(G318W) PheRS editing-deficient E. coli strains, and the lacZ fusion reporter was integrated into the attB site of these lac− strains. Both strains showed similar levels of lacZ expression upon Phe starvation and comparable down-regulation of pheA′ upon the addition of 0.2 mM cognate Phe to the growth medium, indicating that regulation by the cognate amino acid Phe is identical in the two strains (Fig. 2). Growth in the presence of limiting Phe and increasing concentrations of the noncognate amino acid m-Tyr, the natural substrate for PheRS editing, had substantially different effects on pheA′-lacZ expression in the two strains. The wild-type pheT strain showed increased expression of pheA′-lacZ in the presence of m-Tyr (Fig. 2), suggesting that aa-tRNAPhe levels are reduced upon m-Tyr addition, although the changes were not sufficient to be detected by conventional analyses (Fig. 1). m-Tyr is readily activated by PheRS (3); thus its addition leads to competition with cognate Phe for binding to the active site, which in turn reduces the rate of cognate Phe-tRNAPhe synthesis. For the pheT(G318W) strain the addition of noncognate m-Tyr shut off expression of lacZ from the pheA′ reporter, indicating that in the absence of PheRS editing misacylated aa-tRNAPhe accumulates to a level that leads to transcription attenuation despite Phe limitation (Fig. 2). Given that attenuation of pheA′ transcription via formation of a terminator hairpin first requires synthesis of the PheL leader peptide, our data indicate that EF-Tu and the ribosome can use misacylated m-Tyr-tRNAPhe efficiently for translation of PheL. This finding is consistent both with previous in vitro translation data and with detection of m-Tyr in the proteome of E. coli deficient in PheRS editing (3). The concentration of m-Tyr that leads to detectable attenuation of the pheA transcriptional switch is about 10-fold lower than seen for cognate Phe. Consistent with these findings, higher concentrations of p-Tyr, a poorer PheRS substrate than m-Tyr, were required to attenuate pheA′ transcription (Fig. S1).
Fig. 2.
Transcription of the pheA leader under amino acid stress is reduced in the absence of quality control by PheRS editing. β-Galactosidase activity from an E. coli pheA leader-lacZ expression transcriptional reporter was monitored in wild-type PheT and editing-defective PheT(G318W) strains. β-Gal activity is reported as rate (Δ absorbance at 420 nm time) as a function of OD600 and was determined for both wild-type PheT (solid bars) and PheT(G318W) (checkered bars) strains at late log phase. Activity from cultures grown in M9 minimal medium containing no Phe, 50 µM and 100 µM Phe, and 4 µM and 10 µM m-Tyr are shown. Error bars represent the SD from three independent experiments.
Fig. S1.
Transcription of the pheA leader under amino acid stress is reduced in the absence of quality control by PheRS editing. β-Galactosidase activity was monitored as described in the Materials and Methods and was determined for both wild-type pheT (solid bars) and pheT (G318W) (checkered bars) strains at late log phase. Activity from cultures grown in M9 minimal medium containing 20 µM, 200 µM Phe, and 2 mM p-Tyr are shown. Error bars represent the SD from three independent experiments.
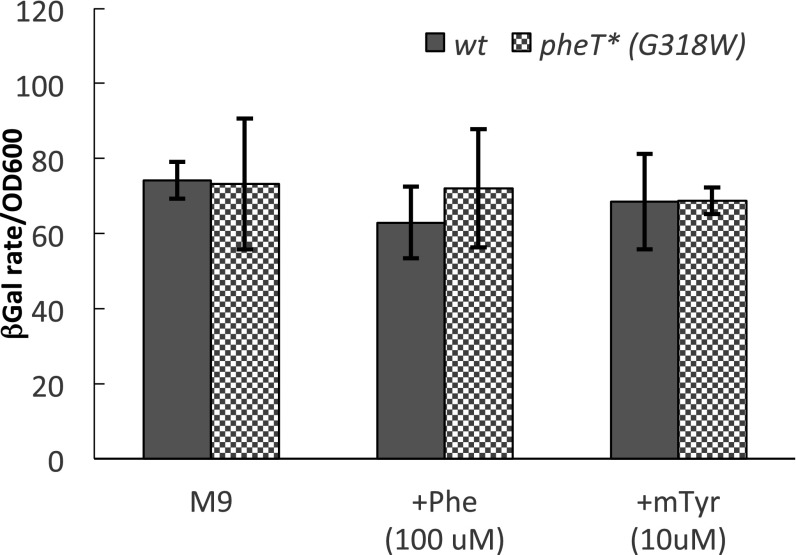
To ensure that the reduced pheA′-lacZ expression in the absence of PheRS editing was not caused by nonspecific perturbation of transcription arising from protein mistranslation or indirect regulation of another tRNA sensor, a pheA reporter was constructed in which a stop codon was inserted at the sixth residue of PheL (F6/stop). The insertion of a stop codon in pheL leads to constitutive pheA expression regardless of growth conditions by preventing completion of PheL synthesis and the subsequent formation of a transcription terminator (25). β-Galactosidase activity for the F6/stop reporter was found to independent of amino acid addition or pheT mutation, indicating that the repression of pheA in the editing-deficient strain is specifically caused by mistranslation of the pheL leader peptide. (Fig. S2).
Fig. S2.
PheRS editing-dependent reduction of pheA expression is caused by the translation of the PheL gene past residue 6. β-Galactosidase activity from a F6/stop mutant pheA′-lacZ reporter was monitored as described in the Materials and Methods and was determined for both the wild-type pheT (solid bars) and pheT(G318W) (checkered bars) strains at late log phase. Activity from cultures grown in M9 minimal medium containing 100 µM Phe or 10 µM m-Tyr is shown. Error bars represent the SD from three independent experiments.
PheRS Editing Is Required for Activation of the Stringent Response in E. coli.
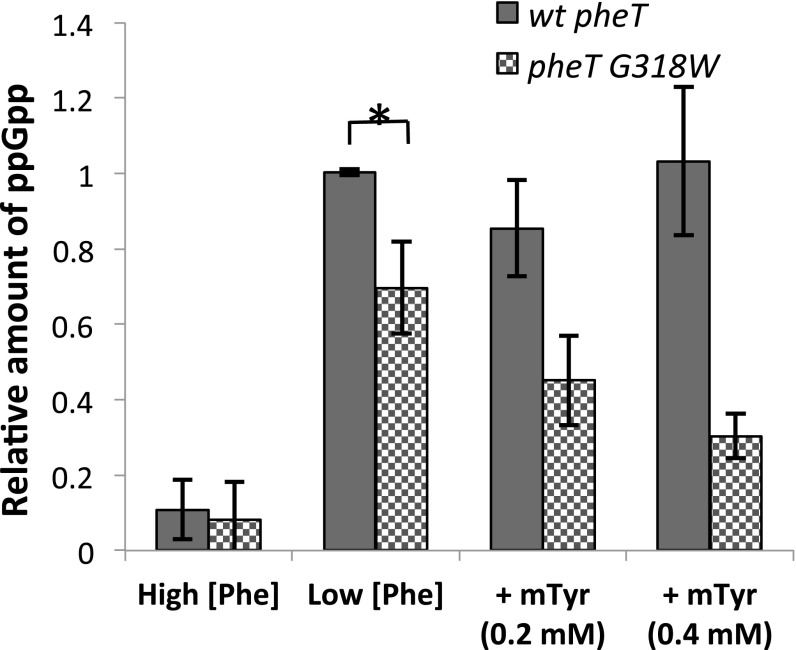
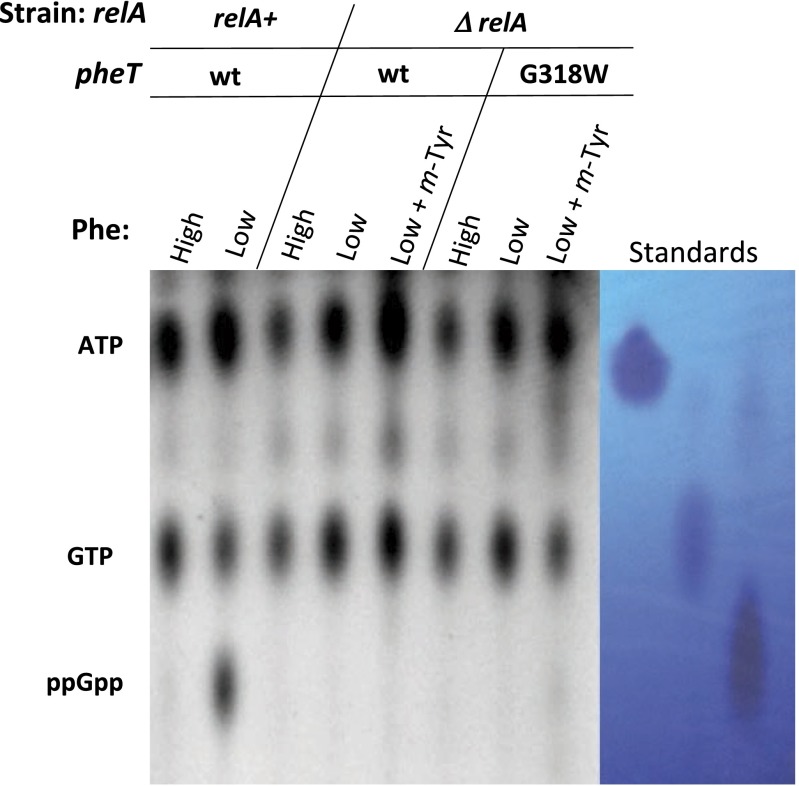
To explore how misacylated tRNA levels affect other cellular stress responses, we monitored the regulation of the stringent response, which depends on the accumulation of deacylated tRNA in the cell for proper activation during nutrient deprivation. Activation of the stringent response during amino acid starvation in E. coli strains lacking PheRS editing was investigated by measuring in vivo levels of 32P-labeled ppGpp. In the wild-type strain background only low levels of ppGpp could be detected using 32P labeling, presumably as a result of endogenous Phe biosynthesis. To address this issue, auxotrophic strains (ΔpheA::kan) were constructed using the wild-type pheT and pheT(G318W) editing-deficient E. coli strains. In the auxotrophic strains, the level of ppGpp increased significantly in conditions of low Phe (10 µM) relative to conditions with a high Phe concentration (0.2 mM). For wild-type strain a 10-fold increase in ppGpp synthesis was observed under Phe limitation, whereas only a sevenfold increase was observed for the PheRS editing-defective strain (Fig. 3). The lower level of ppGpp in the editing-defective strain indicates a reduction in deacylated tRNA accumulation in this strain compared with the wild-type strain; however, this reduction was not detected in our Northern analysis (Fig. 1). This inconsistency may be caused by the absence of endogenous Phe biosynthesis or may indicate that the Northern analyses are not sensitive enough to detect small differences that nevertheless lead to changes in RelA activity. With increasing concentrations of m-Tyr the levels of ppGpp in the PheRS mutant strain are reduced even further to less than half that seen in the wild-type strain under the same growth conditions (Fig. 3). To determine if the observed changes in ppGpp synthesis are dependent on RelA activity, relA also was deleted from our strains (Materials and Methods). The production of ppGpp could not be detected in either relA− strain under any of the conditions tested, including Phe starvation (Fig. S3). These results indicate that in the experiment shown in Fig. 3 ppGpp synthesis and its suppression by m-Tyr-tRNAPhe misacylation are dependent on the activity of RelA. These data indicate that in the absence of editing, the accumulation of misacylated m-Tyr-tRNAPhe, as seen in Fig. 1, is accompanied by significantly reduced levels of deacylated tRNA, leading to suppression of the stringent response under conditions in which the cell is starving for phenylalanine.
Fig. 3.
ppGpp synthesis is reduced in the absence of quality control by PheRS editing. ppGpp levels were normalized to OD600 in pheA auxotrophic strains with either wild-type PheT (solid bars) or editing-deficient PheT(G318W) (checkered bars) strains grown to early log phase in dropout medium containing high Phe (0.5 mM), low Phe (10 µM), or low Phe and m-Tyr (0.2 or 0.4 mM). Error bars represent the SD from three independent experiments.
Fig. S3.
Reduction in ppGpp synthesis in the absence of quality control by PheRS editing is dependent on RelA. (Left) Representative TLC showing undetectable in vivo 32P-ppGpp levels in ΔrelA, pheA auxotrophic strains with either wild-type pheT or editing-deficient pheT G318W) strains grown to early log phase in dropout medium containing high Phe (0.5 mM), low Phe (10 µM), or low Phe and m-Tyr (0.4 mM). (Right) Cold standards were run in parallel and visualized with UV light.
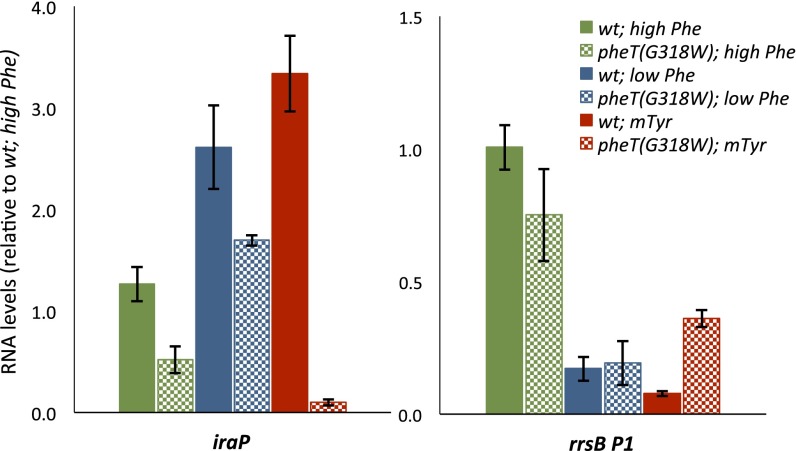
To investigate further the effects of translation quality control on the bacterial stringent response, the regulation of downstream transcriptional targets of ppGpp was investigated using quantitative RT-PCR (qRT-PCR). The relative abundance of rrsB P1 and iraP transcripts was determined and compared in our mutants and under varying concentrations of Phe and m-Tyr. The qPCR primers used were complementary to the rrsB P1 leader and were within the iraP mRNA (Table S1). These stress-response genes are direct targets of ppGpp-regulated transcription and are responsible for the expression of rRNAs and tRNAs and RpoS accumulation, respectively (26, 27). Expression of iraP has been shown to increase in response to increased ppGpp levels, whereas expression from the rrsB P1 promoter decreases during the stringent response (27–29). When normalized to levels of ompA, a transcript unaffected by the stringent response (30), iraP expression increased in both the wild-type and editing-deficient PheRS strains grown at low concentrations of Phe relative to the levels in strains grown at high Phe concentrations (Fig. 4). In contrast, in the presence of the noncognate substrate m-Tyr, the editing-deficient PheRS strain showed 30-fold lower expression of iraP relative to wild type. As expected, levels of rrsB P1 transcript are lower in both wild-type and editing-deficient PheRS strains grown in low-Phe medium than in these strains grown in high-Phe medium. However, in the presence of m-Tyr, levels of the rrsB P1 transcript are almost fourfold higher in the editing-deficient PheRS strain than in wild type. These findings, together with the ppGpp data (Fig. 3), further support the conclusion that the stringent response is suppressed in the presence of substantial tRNA misacylation and the accompanying reduction in deacylated tRNA levels.
Table S1.
Primer sequences used in this study
| Gene | qPCR primer sequences | |
| Forward | Reverse | |
| ompA | 5′-GACACTGGTTTCATCAACAACAATGGCC | 5′-CGGCATACGACCTAACCAGTCGTAAC |
| iraP | 5′-CATTGCTGAGTTGTTATTTAAGCTTGCCC | 5′-GCATTGCAGTGACGATAATCTCCAAAGC |
| rrsB P1 | 5′-CGGAACAACGGCAAACACGCC | 5′-CCTTCCCGCTACAGAGTCAAGCAT |
| rpoH | 5′-GATTCCTACATCCGGGCAGCTAACG | 5′-CTGAATCAAATCCGCCTGTGGCAG |
| dnak | 5′-CAACTCTTGTGTAGCGATTATGGATGGC | 5′-TACTTCTTCGTCCTGGAAGCGGC |
| mopA | 5′-GGCGTAAACGTACTGGCAGATGC | 5′-CTTCTTTCACCATCTGCGCACCC |
| rpoE | 5′-GCGAGCAGTTAACGGACCAGG | 5′-GGAACGAATCCAGCGCACGATAG |
| ptr | 5′-GGCAGAAACGGGATGGCAGC | 5′-GCGCCGAGAGCGATTTAACTGC |
| htrA | 5′-AACGGCGGCTGAGACTTCTTCAG | 5′-TCCTGGCAGAACGGAGAATCATCACC |
Fig. 4.
Downstream transcriptional effects of the stringent response are repressed by tRNA mischarging. qRT-PCR analyses of iraP and rrnB P1 transcripts are normalized to ompA levels and are shown relative to levels in the wild-type pheT strain in high-Phe conditions. Cultures of pheA auxotrophic strains with either wild-type PheT (solid bars) or editing-deficient PheT(G318W) (checkered bars) strains grown to early log phase in Mops minimal medium containing high Phe (0.5 mM) (green bars), low Phe (10 µM) (blue bars), or low Phe and m-Tyr (0.4 mM) (red bars). Errors bars represent the SD of three independent biological replicates.
The Integrity of Protein Synthesis Is Reduced in the Absence of PheRS Editing.
To assess further how increased cellular levels of m-Tyr-tRNAPhe affect the global integrity of the proteome, transcript levels of several components of the sigma 32-(rpoH)– and sigma E-(rpoE)–dependent protein stress regulons were measured (Table 1). Consistent with increased levels of misfolded proteins, genes encoding components of the general heat shock response (rpoH, dnaK, and mopA) were up-regulated in the presence of m-Tyr in the PheRS editing-deficient strain (31). In contrast, transcript levels of two components of the sigma E envelope stress response (rpoE and ptr) were unchanged under the conditions studied, but a third Sigma E-dependent transcript, htrA, was 32-fold higher in the editing-deficient strain in the presence of m-Tyr (32). Taken together these data indicate that in the absence of PheRS editing of misacylated tRNAPhe the structure and function of the cellular proteome is significantly compromised under amino acid stress conditions.
Table 1.
Transcriptional responses to protein stress are altered in the absence of PheRS quality control
| Gene | Wild-type pheT | pheT(G318W) | ||||
| HF | LF | +m-Tyr | HF | LF | +m-Tyr | |
| rpoH | 1.3 ± 0.2 | 1.5 ± 0.4 | 1.3 ± 0.4 | 1.5 ± 0.4 | 2.6 ± 0.4 | 7.3 ± 0.9 |
| dnaK | 1.3 ± 0.1 | 0.5 ± 0.2 | 0.6 ± 0.3 | 1.0 ± 0.2 | 0.6 ± 0.2 | 8.9 ± 2.3 |
| mopA | 1.3 ± 0.1 | 0.6 ± 0.1 | 0.7 ± 0.3 | 1.1 ± 0.3 | 0.6 ± 0.1 | 3.8 ± 0.5 |
| rpoE | 1.3 ± 0.2 | 1.3 ± 0.5 | 1.9 ± 0.6 | 0.9 ± 0.1 | 1.6 ± 0.5 | 0.7 ± 0.2 |
| ptr | 1.3 ± 0.4 | 1.9 ± 0.3 | 3.0 ± 0.7 | 0.6 ± 0.1 | 1.7 ± 0.5 | 2.7 ± 1.1 |
| htrA | 1.3 ± 0.4 | 1.6 ± 0.5 | 1.8 ± 0.9 | 1.2 ± 0.3 | 1.4 ± 0.4 | 32 ± 7.6 |
Listed are relative transcript levels for representative genes of the sigma 32 and sigma E regulons measured by qPCR and normalized to ompA levels for either wild-type pheT or editing-deficient pheT (G318W) grown to early log phase in Mops minimal medium containing high Phe (HF, 0.5 mM), low Phe (LF, 10 µM), or low Phe and m-Tyr (+m-Tyr, 0.4 mM). Results are shown as mean ± SD of three independent biological replicates.
Discussion
Misacylated tRNA Editing Increases the Sensitivity and Specificity of tRNA-Dependent Stress Responses.
The editing of misacylated tRNAs prevents the accumulation of noncognate substrates that otherwise could lead to miscoding errors during mRNA translation. In the absence of misacylated tRNA editing, the frequency of errors in protein synthesis increases significantly and can lead to substantial losses in cellular viability (4, 33, 34). Additionally, the production of abnormal proteins in general leads to induction of the heat shock responses in E. coli (reviewed in ref. 35). The data reported above support the assertion that in the absence of aaRS-dependent editing misfolded proteins accumulate readily under amino acid stress, leading to the increased expression of several genes involved in the E. coli protein stress response. Although the detrimental effects of aaRS-induced decoding errors on proteome integrity are well documented, far less is known about how the failure to eliminate misacylated tRNAs might affect other cellular processes. In particular, the accumulation of misacylated tRNAs has the potential to disrupt the regulation of stress responses that monitor changes in either acylated or deacylated tRNA levels as measures of cellular fitness (9). Our results demonstrate that under amino acid stress, and in the absence of editing, the accumulation of misacylated species such as m-Tyr-tRNAPhe can significantly impact the total levels of both acylated and deacylated tRNAPhe. This impact, in turn, shuts down the expression of genes encoding the biosynthetic pathway for Phe, further reducing levels of cognate Phe-tRNAPhe species in the cell and increasing mistranslation of the proteome under amino acid stress. These data show that editing helps minimize mistranslation in two ways: directly, by eliminating noncognate aminoacyl-tRNAs before they reach the ribosome, and indirectly, by ensuring that amino acid biosynthesis responds to the correct physiological cues and thereby maintains substrate pools in the cell. In addition to limiting errors in protein synthesis, the editing of misacylated tRNA is integral to the down-regulation of global translation in response to amino acid stress. Reduction in total deacylated tRNA levels in the editing-deficient mutant prevented proper activation of the stringent response, resulting in reduced ppGpp synthesis and widespread changes in gene expression. It is unclear if reduced ppGpp synthesis also led to downstream translational effects that helped activate protein stress responses or if this activation resulted solely from misincorporation of m-Tyr at Phe codons. Although in most instances the observed changes in transcript levels were consistent with previous studies, htrA behaved atypically, suggesting that editing contributes to the control of a specific, and not yet fully defined, stress-response regulon. More detailed investigations are now underway into the global effects on gene expression of tRNA misacylation and mistranslation and the broader roles editing plays in the corresponding stress responses.
Overall our findings show that efficient editing of misacylated tRNAs is critical to ensure that tRNA-dependent cellular responses to amino acid stress are both sensitive, as exemplified by the stringent response, and substrate specific, as demonstrated for Phe synthesis. In addition to uncovering the role of editing in bacterial stress responses, this study provides some insights into the potential advantages of detecting amino acid stress via tRNAs compared with direct receptor binding of amino acids. One possible advantage is the ability to increase substrate specificity by exploiting an existing translational proofreading mechanism, as seen here where editing prevents misregulation of pheA by near-cognate amino acids. In addition, controlling Phe biosynthesis via changes in Phe-tRNAPhe rather than by detecting the free amino acid ensures that regulation is adjusted to maintain an adequate supply of Phe specifically for protein synthesis. Perhaps the greatest benefit of tRNA-dependent signaling is the ability to detect amino acid starvation positively via the accumulation of deacylated tRNAs, a strategy also used in eukaryotes (36). Monitoring deacylated tRNA levels provides both a highly sensitive mechanism to detect amino acid depletion and also an indication that the supply of amino acids is not sufficient to meet the demands of protein synthesis. Overall the use of tRNA-dependent rather than free amino acid sensing provides the cell with a means to accurately match substrate supply and demand during protein synthesis under different growth conditions.
Increased tRNA Misacylation Provides a Possible Mechanism to Promote Survival Independent of Mistranslation.
In addition to the numerous examples of lost or reduced cellular viability that often accompany mistranslation, several studies have ascribed beneficial effects to protein synthesis errors. In the pathogenic yeast Candida albicans an ambiguity in the genetic code results in the systematic miscoding of ∼3% of all Leu codons as Ser. This programmed miscoding event generates an array of proteins originating from a single gene that can be beneficial in some cases, for example by generating increased cell surface variation (37, 38). Similar potential mistranslation benefits have been proposed in mammalian and bacterial cells in which errors in protein synthesis have been linked with increased diversity of antigen presentation (4, 39, 40). Recently, the broader notion that adaptive mistranslation can provide a potentially beneficial stress response has been proposed (41) and is based in part on observations of stress-induced loss of editing and increased tRNA misacylation (42–44). For example, enhanced misacylation of tRNA with Met has been suggested to provide cells with transient protection against oxidative and other stresses by increasing the structural and functional diversity of the proteome (41). In the case of oxidative stress, increased Met incorporation may be beneficial by protecting proteins against damage from excess reactive oxygen species, but how nongenetic diversification of the proteome could provide other benefits is not yet known. Although increases in misacylated tRNA levels have been correlated with stress under various conditions, in most cases the subsequent responses that promote growth and survival have been linked only indirectly to adaptive mistranslation. Our findings now suggest that, in addition to mistranslation, other mechanisms exist by which decreased accuracy of tRNA aminoacylation under stress conditions can help to promote survival.
Materials and Methods
Strains and Media.
Wild-type E. coli pheT (BL4074) and a pheT(G318W) PheRS editing-deficient strain [BL4073 (3)] were used to construct pheA′-lacZ reporter derivatives by P1 transduction. First, a ΔproC::kan P1 lysate (provided by N. Ruiz, The Ohio State University, Columbus, OH) was used to make proC− auxotrophic strains. Then the XTB28 P1 lysate (proC+ ΔlacIZYA::FRT) was used to delete the endogenous lac operon, and proline protrophy (growth on M9 minimal medium) was used for selection. Strain constructions were confirmed by screening for loss of growth on X-Gal–containing plates and via PCR and sequencing of the modified allele. The pheA′-lacZ reporter then was constructed via PCR amplification of the pheA′ leader region from MG1655 genomic DNA and subcloning into the EcoRI and BamHI sites of pRS552 (Simons). Transduction into the attB site of each pheT background strain was carried out using the lambda vector system λRS45 (45). Single lysogens were confirmed via PCR as described (46).
The wild-type pheT and the pheT(G318W) PheRS editing-deficient E. coli strains were converted to make phenylalanine auxotrophs (ΔpheA::kan) using standard P1 transduction methods and the donor Keio collection strain 41-0-9 (47, 48). The kan cassette was flipped out using pCP20 (49). For the relA deletion controls, relA782(del)::kan strains were constructed in our wild-type pheT, ΔpheA, and pheT(G318W), ΔpheA backgrounds using P1 transduction of the Keio collection strain JW2755. All deletions were confirmed with PCR. M9 minimal medium contains 1× M9 salts, 2 g/L glucose,1 mg/L thiamine,1 mM MgSO4, and 0.1 mM CaCl2 (50). The 3-(N-morpholino)propanesulfonic acid (Mops) Phe and Tyr drop-out medium contains 1× Mops Mixture, 0.8% glucose, 0.2 mM K2HPO4, and 3× amino acids mix (University of Wisconsin Genome Project protocol minus Phe and Tyr).
Quantification of Aminoacylated and Deacylated tRNAPhe.
Total RNA was isolated from E. coli cells under acidic conditions on ice as described (51). Cells were harvested at late log phase (OD600 ∼1.2), the same point used for β-galactosidase assays, by centrifugation (5,000 × g for 10 min at 4 °C). The cell pellet was suspended in 0.5 mL cold lysis buffer [0.3 M sodium acetate (pH 4.5), 10 mM EDTA]. Suspended cells were mixed with an equal volume of phenol:chloroform:isoamyl-alcohol (1:1:1) (pH 4.5) and were vortexed vigorously three times for 30 s each, followed by centrifugation at 18,600 × g for 20 min at 4 °C. The aqueous phase was removed and placed into a new tube, and the extraction step was repeated. The final aqueous phase was transferred to a new tube, and RNA was precipitated by the addition of 2.7 volumes of cold 100% ethanol followed by incubation at −20 °C for 1 h. RNA was pelleted by centrifugation at 18,600 × g for 30 min at 4 °C and then was suspended in 20 L of cold 10 mM NaOAc (pH 4.5). The stability of 14C-Phe-tRNAPhe (native) was assessed following incubation for 4 h at room temperature under the same buffer conditions; after 4 h incubation the aminoacylation level of the tRNA was not affected.
Northern blotting and acid/urea gel electrophoresis were performed essentially as follows. Loading dye [7 M urea, 0.3 M sodium acetate (pH 5.2), 0.5 µg/mL bromophenol blue] was added to isolated aminoacylated-tRNA (10 μg) and to a deacylated control [isolated aminoacylated-tRNA incubated at 42 °C in Tris⋅HCl (pH 9.0) for 1 h]. Samples were loaded on 20-cm gels [11% acrylamide, 8 M urea, 0.1 M sodium acetate (pH 5.2)] and then were run at 12 W for 20 h at 4 °C. Electrophoretic transfer of RNA to Zeta-probe cationized nylon membrane (Bio-Rad) was performed in transfer buffer [10 mM Tris-acetate (pH 7.8), 5 mM sodium acetate, 0.5 mM EDTA] for 2 h at 44 V at 4 °C. RNA then was UV cross-linked to the membrane followed by prehybridization for 2–4 h and hybridization for 16 h at 50 °C with a [32P]-5′-end–labeled probe (5′-AATCGAACCAAGGACACGGGG) and finally was washed according to the manufacturer’s instructions. Visualization and quantification were performed by phosphor imaging.
Measurement of β-Galactosidase Activity.
E. coli cultures were grown to saturation, pelleted, and washed two times in M9 minimal medium and were used to inoculate the assay medium. For assays cultures were inoculated to an OD600 of 0.02 in M9 minimal medium supplemented with varying amounts of amino acids. Liquid cultures were grown at 37 °C, and 625-µL samples were harvested at late log phase (OD600 of 1.2) by centrifugation (8,000 × g for 2 min at 4 °C). Cell pellets were frozen at −20 °C. After all samples for assaying had been collected, frozen cell pellets were suspended in an equal volume of cold Z buffer (60 mM Na2HPO4·7H2O, 40 mM NaH2PO4·H2O, 10 mM KCl, 1 mM MgSO4·7 H2O, 50 mM β-mercaptoethanol). After measurement of OD600, 500-µL aliquots of the suspended cells were permeabilized by the addition of 30 µL chloroform and 10 µL 0.1% SDS and vortexing for 15 s followed by 15-min incubation at room temperature. Assays were carried out in 96-well microtiter plates. To each well, 200 µL of permeabilized cells were added to 50 µL CPRG (chlorophenol red–β-d-galactopyranoside) substrate (5 mg/mL) freshly made in Z buffer. The rate of β-gal activity at 28 °C was measured as absorbance readings at 420 nm at 1-min intervals in the linear range of activity. These rates then were normalized to the OD600 of suspended cells before permeabilization.
Determination of Cellular ppGpp Levels.
Levels of ppGpp were assayed as previously described (52). Saturated E. coli cultures were first pelleted and washed two times in prewarmed Mops dropout medium and then were used to inoculate assay cultures. Liquid cultures (2 mL) were inoculated to an OD600 of 0.05 in prewarmed Mops dropout medium with 50 µCi 32P (carrier free; Perkin-Elmer) and appropriate amounts of Phe and Tyr as indicated. Cultures were grown at 37 °C, and 50-µL samples were taken at early log phase (OD600 ∼0.2), added to 10 µL of formic acid on ice, and frozen immediately at −80 °C. Cells were lysed by freeze-thawing three times, with vigorous vortexing after each thaw cycle. Cell debris was pelleted at 13,000 × g for 5 min at 4 °C, and lysates were appropriately diluted (normalized by comparison with the OD600 of the cultures upon sample collection). Equal volumes of normalized lysate were spotted on PEI cellulose plates (EMD). Labeled nucleotides were separated by TLC developed in 1.5 M NaOAc (pH 3.5). The plates were dried, placed in methanol for 5 min, dried again, and visualized by phosphor imaging. Unlabeled nucleotides (0.5 µL, 100 mM) were used as TLC migration standards and were detected using UV light.
Total RNA Purification, Reverse Transcription, and qPCR.
Purification of total RNA from early log-phase cells was done using Trizole (Ambion) as described in the manufacturer’s protocol. Genomic DNA was removed using the Turbo DNA-free Kit (Life Technologies), and total RNA was assessed and quantified using the Agilent 2100 Bioanalyzer with the RNA 6000 Nano Kit. Reverse transcription of 100 ng of total RNA was carried out using 300 nM of random hexamer primer (Promega) and SuperScript III reverse transcriptase (Invitrogen). The reaction was performed as directed by the manufacturer using reaction parameters of 50 °C for 60 min. qPCR was carried out using gene-specific primers (Table S1) and IQ SYBR Green Supermix (Bio-Rad). Equal volumes of the reverse-transcription reactions were used for each sample (1:400 final dilution) in 20-µL reaction volumes. qPCR reactions were performed on a Bio-Rad CFX real-time PCR detection system with cycling parameters as follows: 95 °C for 2 min, followed by 40 cycles of 95 °C for 20 s, 55 °C for 20 s, (read) 72 °C for 20 s. Negative controls including no reverse transcriptase and no template, as well as standard curves of cDNA dilutions used to determine the PCR efficiency, were included in each set of experiments. Two technical replicates for each of three biological replicates were performed. Data analysis was performed by the Pfaffl method of relative quantification using Bio-Rad CFX Manager 3.0 software (53).
Acknowledgments
We thank Natasha Ruiz and Kurt Frederick for strains, helpful insights about strain construction, and use of the lambda reporter system and Susan Gottesman, Beth Lazazzera, and Kyle Mohler for comments on the manuscript. This work was supported by National Science Foundation Grant MCB 1412611 and Army Research Office Grant W911NF1510105.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1525206113/-/DCSupplemental.
References
- 1.Yadavalli SS, Ibba M. Quality control in aminoacyl-tRNA synthesis its role in translational fidelity. Adv Protein Chem Struct Biol. 2012;86:1–43. doi: 10.1016/B978-0-12-386497-0.00001-3. [DOI] [PubMed] [Google Scholar]
- 2.Reynolds NM, Lazazzera BA, Ibba M. Cellular mechanisms that control mistranslation. Nat Rev Microbiol. 2010;8(12):849–856. doi: 10.1038/nrmicro2472. [DOI] [PubMed] [Google Scholar]
- 3.Bullwinkle TJ, et al. Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code. eLife. 2014;3:02501. doi: 10.7554/eLife.02501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L, et al. Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc Natl Acad Sci USA. 2011;108(23):9378–9383. doi: 10.1073/pnas.1016460108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shepherd J, Ibba M. Relaxed substrate specificity leads to extensive tRNA mischarging by Streptococcus pneumoniae class I and class II aminoacyl-tRNA synthetases. MBio. 2014;5(5):e01656–e14. doi: 10.1128/mBio.01656-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reynolds NM, et al. Cell-specific differences in the requirements for translation quality control. Proc Natl Acad Sci USA. 2010;107(9):4063–4068. doi: 10.1073/pnas.0909640107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yadavalli SS, Ibba M. Selection of tRNA charging quality control mechanisms that increase mistranslation of the genetic code. Nucleic Acids Res. 2013;41(2):1104–1112. doi: 10.1093/nar/gks1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Banerjee R, et al. tRNAs: Cellular barcodes for amino acids. FEBS Lett. 2010;584(2):387–395. doi: 10.1016/j.febslet.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet. 2014;5:171. doi: 10.3389/fgene.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Springer M, Graffe M, Butler JS, Grunberg-Manago M. Genetic definition of the translational operator of the threonine-tRNA ligase gene in Escherichia coli. Proc Natl Acad Sci USA. 1986;83(12):4384–4388. doi: 10.1073/pnas.83.12.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graffe M, et al. The specificity of translational control switched with transfer RNA identity rules. Science. 1992;255(5047):994–996. doi: 10.1126/science.1372129. [DOI] [PubMed] [Google Scholar]
- 12.Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: How RNA provides instructions for transcription termination/antitermination decisions. BioEssays. 2002;24(8):700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 13.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13(5):298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11(2):100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Zuo Y, Wang Y, Steitz TA. The mechanism of E. coli RNA polymerase regulation by ppGpp is suggested by the structure of their complex. Mol Cell. 2013;50(3):430–436. doi: 10.1016/j.molcel.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross W, Vrentas CE, Sanchez-Vazquez P, Gaal T, Gourse RL. The magic spot: A ppGpp binding site on E. coli RNA polymerase responsible for regulation of transcription initiation. Mol Cell. 2013;50(3):420–429. doi: 10.1016/j.molcel.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 18.Lemke JJ, et al. Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc Natl Acad Sci USA. 2011;108(14):5712–5717. doi: 10.1073/pnas.1019383108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou YN, Jin DJ. The rpoB mutants destabilizing initiation complexes at stringently controlled promoters behave like “stringent” RNA polymerases in Escherichia coli. Proc Natl Acad Sci USA. 1998;95(6):2908–2913. doi: 10.1073/pnas.95.6.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Varshney U, Lee CP, RajBhandary UL. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J Biol Chem. 1991;266(36):24712–24718. [PubMed] [Google Scholar]
- 21.Sørensen MA. Charging levels of four tRNA species in Escherichia coli Rel(+) and Rel(-) strains during amino acid starvation: A simple model for the effect of ppGpp on translational accuracy. J Mol Biol. 2001;307(3):785–798. doi: 10.1006/jmbi.2001.4525. [DOI] [PubMed] [Google Scholar]
- 22.Dittmar KA, Sørensen MA, Elf J, Ehrenberg M, Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6(2):151–157. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klipcan L, Moor N, Kessler N, Safro MG. Eukaryotic cytosolic and mitochondrial phenylalanyl-tRNA synthetases catalyze the charging of tRNA with the meta-tyrosine. Proc Natl Acad Sci USA. 2009;106(27):11045–11048. doi: 10.1073/pnas.0905212106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertin C, et al. Grass roots chemistry: Meta-tyrosine, an herbicidal nonprotein amino acid. Proc Natl Acad Sci USA. 2007;104(43):16964–16969. doi: 10.1073/pnas.0707198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gavini N, Pulakat L. Role of translation of the pheA leader peptide coding region in attenuation regulation of the Escherichia coli pheA gene. J Bacteriol. 1991;173(15):4904–4907. doi: 10.1128/jb.173.15.4904-4907.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barker MM, Gaal T, Josaitis CA, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305(4):673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 27.Paul BJ, Berkmen MB, Gourse RL. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci USA. 2005;102(22):7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bougdour A, Gottesman S. ppGpp regulation of RpoS degradation via anti-adaptor protein IraP. Proc Natl Acad Sci USA. 2007;104(31):12896–12901. doi: 10.1073/pnas.0705561104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Battesti A, Majdalani N, Gottesman S. Stress sigma factor RpoS degradation and translation are sensitive to the state of central metabolism. Proc Natl Acad Sci USA. 2015;112(16):5159–5164. doi: 10.1073/pnas.1504639112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190(3):1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nonaka G, Blankschien M, Herman C, Gross CA, Rhodius VA. Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev. 2006;20(13):1776–1789. doi: 10.1101/gad.1428206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006;4(1):e2. doi: 10.1371/journal.pbio.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, et al. Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc Natl Acad Sci USA. 2014;111(49):17570–17575. doi: 10.1073/pnas.1420196111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443(7107):50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 35.Gottesman S. Genetics of proteolysis in Escherichia coli*. Annu Rev Genet. 1989;23:163–198. doi: 10.1146/annurev.ge.23.120189.001115. [DOI] [PubMed] [Google Scholar]
- 36.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu Rev Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 37.Miranda I, et al. Candida albicans CUG mistranslation is a mechanism to create cell surface variation. MBio. 2013;4(4):e00285–e00213. doi: 10.1128/mBio.00285-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bezerra AR, et al. Reversion of a fungal genetic code alteration links proteome instability with genomic and phenotypic diversification. Proc Natl Acad Sci USA. 2013;110(27):11079–11084. doi: 10.1073/pnas.1302094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yewdell JW. DRiPs solidify: Progress in understanding endogenous MHC class I antigen processing. Trends Immunol. 2011;32(11):548–558. doi: 10.1016/j.it.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yewdell JW, Nicchitta CV. The DRiP hypothesis decennial: Support, controversy, refinement and extension. Trends Immunol. 2006;27(8):368–373. doi: 10.1016/j.it.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 41.Pan T. Adaptive translation as a mechanism of stress response and adaptation. Annu Rev Genet. 2013;47:121–137. doi: 10.1146/annurev-genet-111212-133522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones TE, Alexander RW, Pan T. Misacylation of specific nonmethionyl tRNAs by a bacterial methionyl-tRNA synthetase. Proc Natl Acad Sci USA. 2011;108(17):6933–6938. doi: 10.1073/pnas.1019033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ling J, Söll D. Severe oxidative stress induces protein mistranslation through impairment of an aminoacyl-tRNA synthetase editing site. Proc Natl Acad Sci USA. 2010;107(9):4028–4033. doi: 10.1073/pnas.1000315107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Netzer N, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462(7272):522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 46.Powell BS, Rivas MP, Court DL, Nakamura Y, Turnbough CL., Jr Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 1994;22(25):5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1972. [Google Scholar]
- 49.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 51.Zaborske J, Pan T. Genome-wide analysis of aminoacylation (charging) levels of tRNA using microarrays. J Vis Exp. 2010;(40):2007. doi: 10.3791/2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rojiani MV, Jakubowski H, Goldman E. Effect of variation of charged and uncharged tRNA(Trp) levels on ppGpp synthesis in Escherichia coli. J Bacteriol. 1989;171(12):6493–6502. doi: 10.1128/jb.171.12.6493-6502.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]