Significance
These studies show a physiological role for Notch signaling in female reproduction. The fact that both loss and gain of function of Notch signaling result in the impairment of early pregnancy identifies Notch1 signaling as a critical regulator of endometrial function. We also provide the first evidence, to our knowledge, that Notch signaling can regulate methylation of exon 1 of the progesterone receptor (Pgr) gene through its target PU.1, which provides novel insight into the role of Notch in steroid hormone regulation. This mechanism also provides an opportunity for future studies in identifying the cause of progesterone resistance in gynecological pathologies in women, such as endometriosis and adenomyosis, in which the hypermethylation of Pgr has been reported.
Keywords: mouse uterus, Notch1, Pgr, methylation, infertility
Abstract
In mammalian reproduction, implantation is one of the most critical events. Failure of implantation and the subsequent decidualization contribute to more than 75% of pregnancy losses in women. Our laboratory has previously reported that inhibition of Notch signaling results in impaired decidualization in both women and a transgenic mouse model. In this study, we generated a Notch gain-of-function transgenic mouse by conditionally overexpressing the Notch1 intracellular domain (N1ICD) in the reproductive tract driven by a progesterone receptor (Pgr) -Cre. We show that the overexpression of N1ICD in the uterus results in complete infertility as a consequence of multiple developmental and physiological defects, including the absence of uterine glands and dysregulation of progesterone and estrogen signaling by a Recombination Signal Binding Protein Jκ-dependent signaling mechanism. We further show that the inhibition of progesterone signaling is caused by hypermethylation of its receptor Pgr by Notch1 overexpression through the transcription factor PU.1 and DNA methyltransferase 3b (Dnmt3b). We have generated a mouse model to study the consequence of increased Notch signaling in female reproduction and provide the first evidence, to our knowledge, that Notch signaling can regulate epigenetic modification of the Pgr.
Implantation is one of the most critical and highly regulated processes during mammalian reproduction. In reproductive aged women, there is only a 15% chance of pregnancy each cycle (1), and 75% of failed pregnancies are caused by implantation failure (2). The window of uterine receptivity is defined as the optimum time when the uterine endometrium is able to accept a blastocyst to implant. Uterine transition into the receptive phase requires priming with progesterone (P4) superimposed with estrogen [mainly 17β-estradiol (E2)], which has functions that are mediated primarily by nuclear receptors progesterone receptor (Pgr) and estrogen receptor isoform 1 (Esr1), respectively (3). Dysregulation of these two signaling pathways leads to defective uterine receptivity and failed implantation (4).
Notch signaling is a highly conserved pathway across species and present in most multicellular organisms. It plays vital roles in cellular survival, communication, and differentiation throughout development from embryonic to adult life (5). Canonical Notch signaling is initiated after the interaction of Notch transmembrane receptors (Notch1–Notch4) with cell-bound ligands (δ-like 1, 3, or 4 or Jagged 1 or 2), which leads to a cleavage cascade of Notch involving ADAM proteases and γ-secretase (6). Subsequently, the cleaved Notch intracellular domain (NICD) translocates to the nucleus, where it interacts with transcriptional repressor Recombination Signal Binding Protein Jκ (RBP-Jκ; also known as CBF-1) and converts it into a transcriptional activator of downstream target genes, such as hairy enhancer of split and hairy enhancer of split-related transcription factor families (7, 8). However, recent studies have revealed the existence of several other modes of Notch signaling generally referred to as noncanonical Notch signaling (6).
Notch signaling is critical for maternal–fetal communication during implantation and placentation (9). Our laboratory has previously reported that both the conditional deletion of Notch1 in the mouse uterus and NOTCH1 silencing in Human Uterine Fibroblasts (HuFs) inhibit decidualization (10, 11). In the pathological condition of endometriosis, the decrease in NOTCH1 in eutopic endometrium results in impaired decidualization of endometrial stromal cells from patients with the disease (12). NOTCH2 has also been shown as a regulator of decidualization (13). Subsequently, expression levels of Notch1 are decreased in the mouse endometrial stroma and HuF cells on completion of transition to the decidual phenotype (10, 11). It is unclear whether down-regulation of Notch1 expression is required for completion of decidualization. To determine the mechanism(s) of Notch1-mediated effects in endometrial physiology, we generated a reproductive tract-specific, constitutively activated Notch1 mouse model, in which the intracellular domain of Notch1 (N1ICD) is overexpressed specifically in Pgr-positive cells within the reproductive tract.
In this study, we show that constitutively activated Notch signaling in the mouse uterus compromises uterine receptivity through multiple mechanisms, including the loss of uterine glands and the inhibition of P4 signaling. We further show that the suppression of P4 signaling is a result of the hypermethylation of its receptor Pgr. Our findings further indicate the importance of Notch signaling during early pregnancy.
Results
N1ICD Overexpression Mice Are Completely Infertile Because of Impaired Uterine Receptivity.
Generation of N1ICD overexpression (OEx) mice (Pgrcre/+Rosa26N1ICD/+) is shown in Fig. S1A. After confirming the overexpression of N1ICD (Fig. S1 B–F), we tested the fertility of N1ICD OEx mice. For control females, both homozygous and heterozygous animals had normal litters (n = 5 and n = 3, respectively). However, both homozygous and heterozygous OEx female mice are completely infertile as determined with a 6-mo breeding test (n = 4 in both genotypes) (Table S1). Furthermore, N1ICD OEx mice displayed no visible implantation sites at 4.5 d postconception (dpc) in contrast to control mice, which displayed normal implantation (Fig. 1A). Successful implantation requires both a competent embryo and a receptive uterus (3). To bypass potential embyrotrophic causes for infertility, we transferred blastocysts collected from WT donors into the uterine lumen of pseudopregnant OEx and control mice. Two days after transfer, the control recipients displayed evidence of implantation; blastocysts were attached to the uterine luminal epithelium, and the decidual reaction could be observed surrounding the implanted embryo (Fig. 1B, control). In contrast, no implantation sites were observed in the OEx recipients. Blastocysts remained free-floating in the uterine lumen, and the stromal cells showed no evidence of decidualization (Fig. 1B, OEx). Furthermore, OEx females showed no response to an artificial decidualization stimulus, whereas control mice exhibited a clear decidual response (Fig. 1C). Uterine weight and expression of decidualization markers bone morphogenetic protein 2 (Bmp2) and wingless-type MMTV integration site family member 4 (Wnt4) in the stimulated horn of control mice were significantly increased compared with those of the nonstimulated horn, but no difference was evident between the stimulated and nonstimulated horns of OEx mice (Fig. 1 D–F). Collectively, N1ICD OEx mice are infertile because of defective uterine receptivity and impaired ability of uterine stromal cells to undergo decidualization.
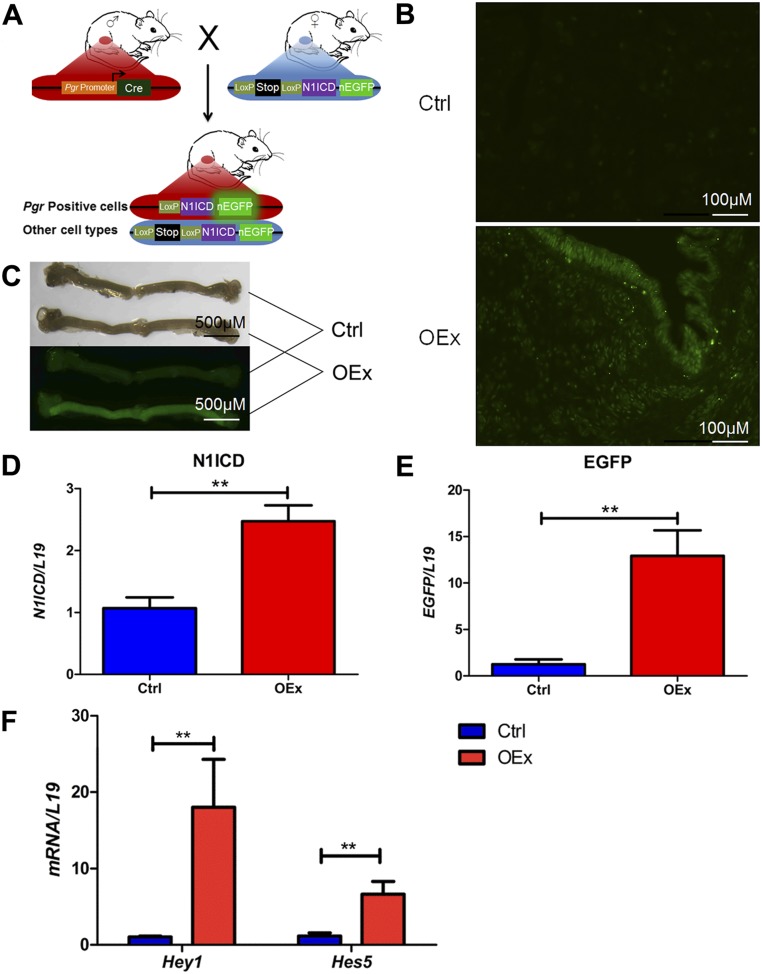
Fig. S1.
Generation of N1ICD OEx mice. (A) The Pgr+/+Rosa26N1ICD/+ (control) and OEx mice were generated by crossing PgrCre/+ mice, in which Cre recombinase is driven by Pgr promoter, with Rosa26N1ICD/N1ICD mice, in which N1ICD together with a nuclear-localized EGFP are inserted to the Rosa26 locus after an loxP-flanked transcription termination fragment (STOP). (B) Overexpressed EGFP is visible in both epithelial and stromal cells of the uterus in OEx mice but not in control mice at 3.5 dpc. (C) Fluorescent uterine horns are visible as early as postnatal day 10 in OEx mice. (D and E) Significantly higher expression of N1ICD and EGFP mRNA confirms success of the overexpression. (F) Up-regulation of Notch signaling targets hairy enhancer of split 5 (Hes5) and hairy enhancer of split-related 1 (Hey1) confirms that overexpression of N1ICD is functional. Ctrl, control. **P < 0.01.
Table S1.
Fertility test
| Genotype | No. of mice | No. of litters | No. of pups | Mean pups per litter | Mean litter per mouse |
| Pgr+/+Rosa26N1ICD/N1ICD | 5 | 27 | 203 | 7.53 ± 0.49 | 5.40 ± 0.54 |
| Pgr+/+Rosa26N1ICD/+ | 3 | 17 | 137 | 8.06 ± 0.96 | 5.67 ± 0.58 |
| PgrCre/+Rosa26N1ICD/N1ICD | 4 | 0 | 0 | 0 | 0 |
| PgrCre/+Rosa26N1ICD/+ | 4 | 0 | 0 | 0 | 0 |
Note the 6-mo breeding protocol.
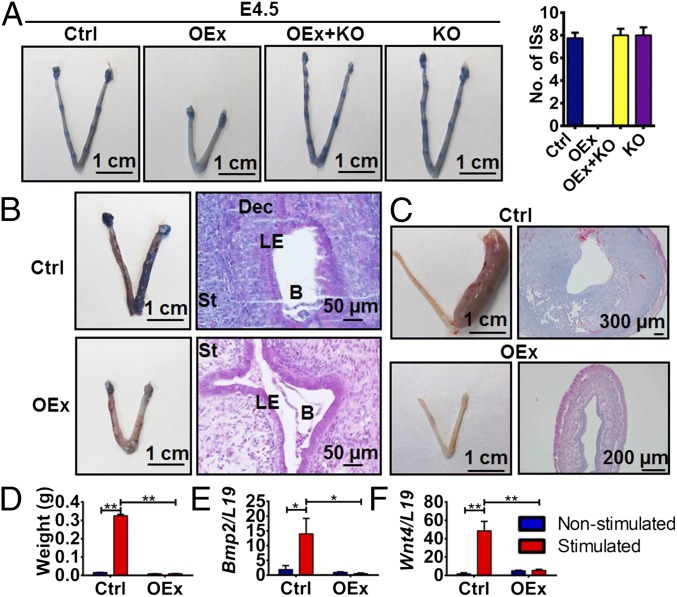
Fig. 1.
Defective uterine receptivity in N1ICD OEx mice. (A) No implantation sites (ISs) are detected in N1ICD OEx mice at 3.5 dpc, whereas control, OEx + KO, and Rbpj KO mice have normal numbers of ISs. The number of ISs (n = 3 of each group) is shown in the histogram. (B) The right uterine horns, which received WT embryos, have ISs in control mice but not in OEx mice, whereas the left horns served as controls without embryo transfer. Histological staining shows an implanted blastocyst with decidualized stromal cells surrounding it in control mice and a free-floating blastocyst in uterine lumen without decidualization in OEx mice. (C) Artificial decidualization stimulus induces a decidual response in control mice vs. no decidualization response in OEx mice. Histological staining shows cross-sections of stimulated horns (right horns) in both two groups. Left horns are nonstimulated controls. (D) Weight of stimulated horns in control mice is >20 times higher than that of nonstimulated horns in control mice, but there is no difference between stimulated and nonstimulated horns of OEx mice (n = 4 of each group). Decidualization markers (E) Bmp2 and (F) Wnt4 are significantly increased in the stimulated horns of control mice, whereas no significant induction is evident in OEx mice (n = 4 of each group). B, blastocyst; Ctrl, control; Dec, decidua; LE, luminal epithelium; St, stroma. *P < 0.05; **P < 0.01.
N1ICD OEx Mice Fail to Develop Uterine Glands.
The uterine size and weight of N1ICD OEx mice were significantly lower than those of control mice at 3.5 dpc (Fig. S2 A and B). Histological analysis revealed that the uteri of the OEx mice were completely devoid of uterine glands, whereas their control littermates had normal glandular structures (Fig. 2A). Forkhead box A2 (Foxa2), a marker of uterine glands (14, 15), was used to confirm the loss of glands in the OEx mice. The glandular epithelial (GE) cells of control mice expressed Foxa2 protein as expected, but surprisingly, luminal epithelial (LE) cells of OEx mice showed strong Foxa2 staining (Fig. 2B). Uterine gland secretion of leukemia inhibitory factor (Lif) is critical for implantation in mice (16, 17). The LE staining of Foxa2 in OEx mice suggested a glandular phenotype of LE cells, which we further confirmed by mRNA expression of Lif by in situ hybridization. Lif mRNA was observed in the luminal epithelium of OEx mice, whereas only GE cells expressed Lif mRNA in control mice (Fig. 2C). At 3.5 dpc, expressions of Lif mRNA (Fig. S2D) and Lif protein (Fig. 2D) in OEx mice were comparable with those of controls as measured by quantitative PCR (qPCR) and Western blot.
Fig. S2.
(A and B) N1ICD mice have smaller uterine sizes, and the weights of uteri are significantly lower in OEx mice compared with other genotypes at 3.5 dpc. (C) Confirmation of overexpression of N1ICD in OEx mice and KO of Rbpj in Rbpj KO mice by qPCR at 3.5 dpc. (D) Expression level of Lif mRNA in N1ICD OEx mice is not significantly different compared with control mice at 3.5 dpc (n = 3–4 in each group). Ctrl, control; Hey1, hairy enhancer of split-related 1. *P < 0.05; **P < 0.01.
Fig. 2.
N1ICD OEx mice have no uterine glands. (A) Unlike control, OEx + KO, and KO mice, there are no glands in the uteri of N1ICD OEx mice at 3.5 dpc as detected by H&E staining. (B) The gland marker Foxa2 is expressed in the glandular epithelium of control, OEx + KO, and KO mice, whereas its expression is only present in the luminal epithelium of OEx mice. (C) Expression of Lif mRNA in GE cells of control mice and LE cells of N1ICD OEx mice at 3.5 dpc. (D) N1ICD OEx mice express comparable levels of Lif protein to control mice as measured by Western blot at 3.5 dpc. Ctrl, control. (Scale bar: 100 µm.)
E2 and P4 Signaling Is Altered in N1ICD OEx Mice.
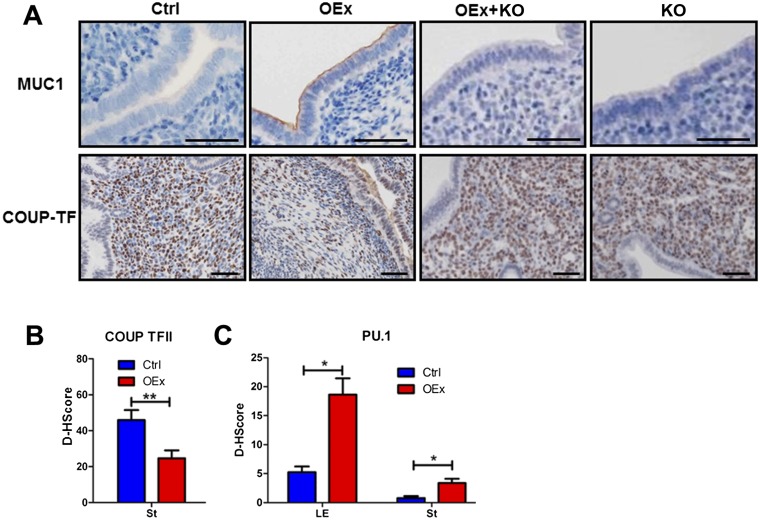
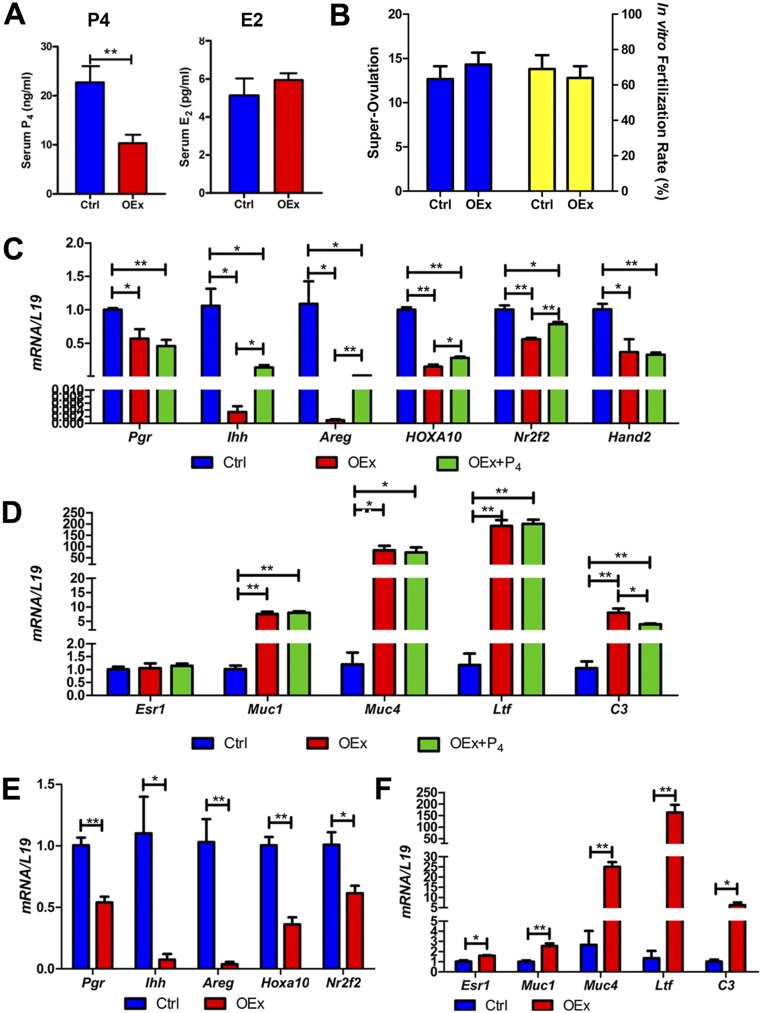
Uterine receptivity in the mouse is mainly regulated by two ovarian steroid hormones, E2 and P4, which act by binding to their cognate receptors, Esr1 and the Pgr, respectively. The expression patterns of both receptors are critical for controlling receptivity in the mouse uterus. In OEx mice, expression of Pgr was markedly decreased in both epithelial and stromal cells compared with control mice, and expression of Esr1 was significantly increased in LE cells, especially its active phosphorylated form (p-Esr1) (Fig. 3 A and B) at 3.5 dpc. Pgr mRNA was significantly reduced in OEx mice, whereas Esr1 mRNA expression was not changed (Fig. 3 C and D). Immunostaining for antigen Ki67 (Ki67) also showed that stromal cell proliferation was significantly decreased in N1ICD OEx mice, whereas epithelial cell proliferation was significantly increased compared with control mice at 3.5 dpc (Fig. 3 A and B). The expressions of Pgr-regulated genes Indian hedgehog (Ihh) (18), amphiregulin (Areg) (19), homobox A10 (Hoxa10) (20), nuclear receptor subfamily 2, group F, member 2 (Nr2f2) (18), and heart and neural crest derivatives expressed transcript 2 (Hand2) (21) were significantly decreased, and E2 target genes mucin 1 (Muc1), Muc4, lactotransferrin (Ltf), and complement component 3 (C3) (21, 22) were significantly increased in OEx mice compared with control mice (Fig. 3 C and D). Protein levels of Muc1 and chicken ovalbumin upstream promoter transcriptional factors II (COUP-TFII) (encoded by the Nr2f2 gene) were detected by immunohistochemistry on 3.5 dpc. Muc1 was observed on the luminal surface of the uterus in OEx mice but not control mice, and COUP-TFII expression was lower in stromal cells of OEx mice (Fig. S3 A and B). The expression pattern of target genes in N1ICD OEx mice uteri indicated that P4 signaling was inhibited and that E2 signaling was enhanced, which correlate with the expressions of their respective receptors, suggesting that overexpression of N1ICD in the mouse uterus effects uterine receptivity.
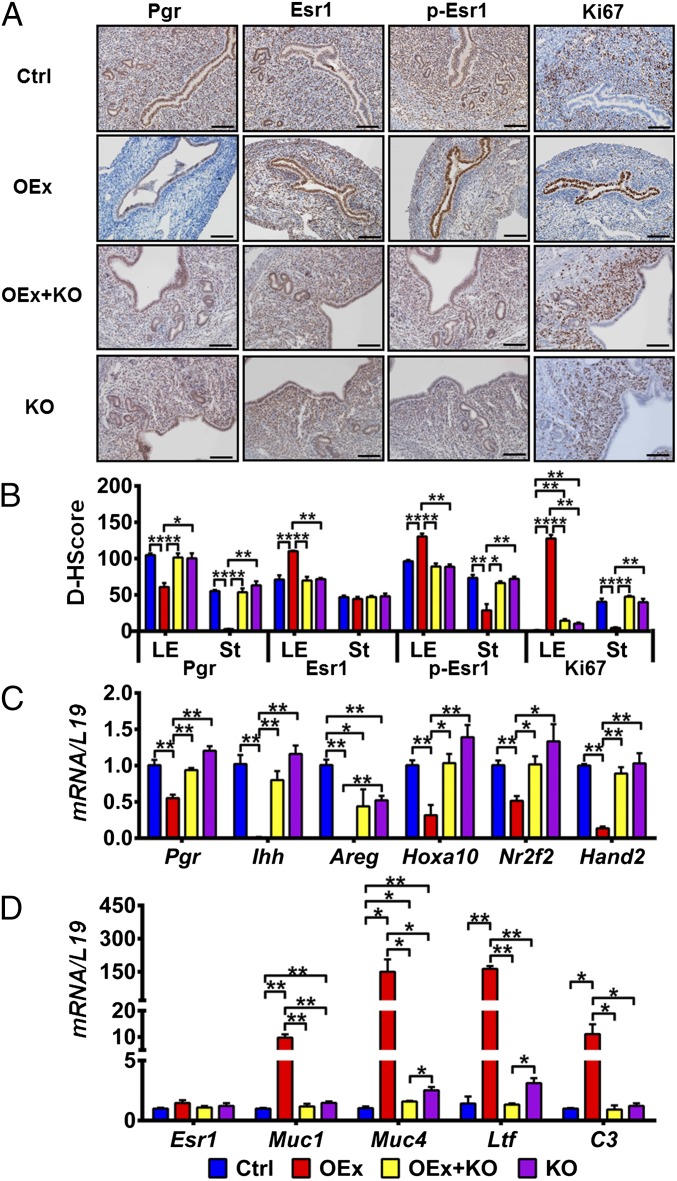
Fig. 3.
Dysregulation of P4 and E2 signaling and proliferation pattern in N1ICD OEx mice at 3.5 dpc. (A) Immunohistochemistry shows that the expression patterns of Pgr, Esr1, p-Esr1, and Ki67 (proliferation marker) are altered in N1ICD OEx mice compare with control, OEx + KO, and KO mice. Quantitative expression levels of these proteins are shown in B. (C) mRNA levels of Pgr together with P4 target genes Ihh, Areg, Hoxa10, Nr2f2, and Hand2 are significantly decreased in OEx mice compared with control mice but rescued, at least partially, in OEx + KO mice. Expression levels in KO mice are similar to those of OEx + KO mice. (D) Quantitative PCR expression of Esr1 has no significant difference among the four different genotypes of mice, but E2 targets Muc1, Muc4, Ltf, and C3 are dramatically up-regulated in OEx mice compared with control mice. The up-regulation of E2 targets is abolished in OEx + KO mice (n = 3–4 in each group). Ctrl, control; D-HScore, digital HScore; St, stroma. *P < 0.05; **P < 0.01. (Scale bar: 100 µm.)
Fig. S3.
Dysregulation of P4 and E2 signaling in N1ICD OEx mice at 3.5 dpc. (A) Immunohistochemistry identifies up-regulation of Muc1 expression and down-regulation of COUP-TFII in OEx mice at 3.5 dpc. Expression of these two proteins was not different in OEx + KO and KO mice compared with control mice. (B) Quantitative expression level of COUP-TFII in the control and OEx mice shown in B. (C) Quantitative expression level of PU.1 immunohistochemistry in control and OEx mice, which are shown in Fig. 4B (n = 3–4 in each group). Ctrl, control. *P < 0.05; **P < 0.01.
However, decreased expression of Pgr is not the only factor that contributes to the inhibition of P4 signaling in N1ICD OEx mice. Serum P4 levels in OEx mice at 3.5 dpc were significantly lower than those of control mice, whereas E2 levels were not altered (Fig. S4A). These data show that both ligand and receptor for P4 signaling were altered in N1ICD OEx mice, although ovulation and the fertilization ability of oocytes are not different between the two groups of mice (Fig. S4B). Supplemental P4 injections from 2.5 to 3.5 dpc significantly up-regulated expression of P4 target genes compared with vehicle-treated OEx mice but were still significantly lower than those of vehicle-treated control mice (Fig. S4C). Supplemental P4 injections had no effect on E2 targets (Fig. S4D). Exogenous hormones (P4 and E2) were used to prime the uterus of ovariectomized mice to mimic the receptivity of the uterus at 3.5 dpc, independent of the effects of ovarian factors (22). This treatment was capable of inducing a decidualization response after an artificial stimulation. All ovarian steroid hormone receptor target genes displayed the same expression patterns after this exogenous hormone treatment as those observed in the OEx and control pregnant mice at 3.5 dpc (Fig. S4 E and F), indicating that the disordered expression pattern of Pgr and Esr1 is sufficient for the dysregulation of their target genes and the failure of uterine receptivity in N1ICD OEx mice.
Fig. S4.
(A) N1ICD OEx mice have lower levels of serum P4 but not E2 compared with control mice at 3.5 dpc. (B) Ovulation and fertilizing ability of oocytes showed no difference between N1ICD OEx mice and control mice. (C) Injection (s.c.) of P4 from 2.5 to 3.5 dpc can partially rescue expression of P4 targets in N1ICD OEx mice, but it is still significantly lower than that in control mice without supplemental P4 injection. (D) Supplemental P4 also does not alter estrogen signaling in OEx mice. (E) In the ovariectomized mouse model of exogenous treatment with P4 and E2 to mimic 3.5 dpc, the expressions of P4 targets are still significantly lower in OEx mice than in control mice. E2 targets are increased in OEx mice in the model described in F (n = 3–4 in each group). Ctrl, control. *P < 0.05; **P < 0.01.
PU.1 Mediates Hypermethylation of the Pgr Promoter in N1ICD OEx Mice.
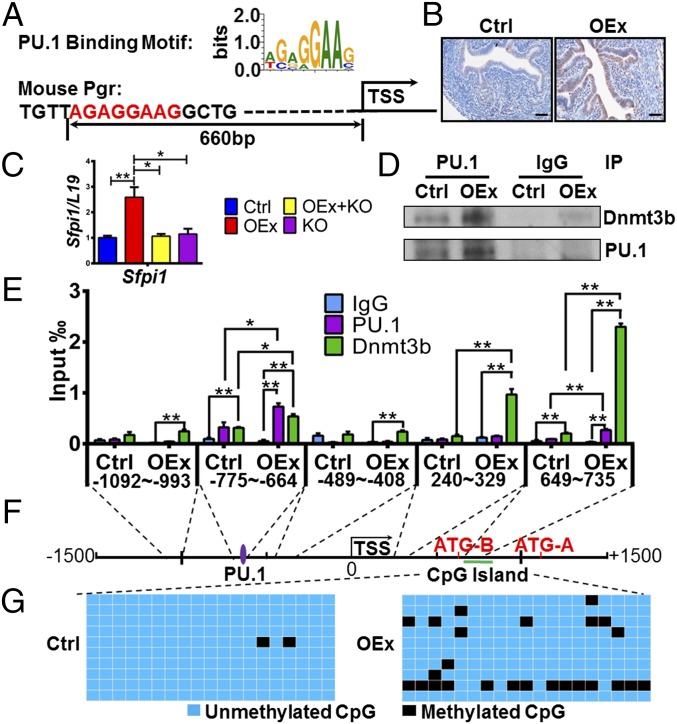
To determine the mechanism by which Pgr is down-regulated in N1ICD OEx mice, transcription factor binding sites on the Pgr promoter were identified using MotifMap (motifmap.ics.uci.edu). Potential binding sites for the transcription factor PU.1 [encoded by the spleen focus forming virus (SFFV) proviral integration oncogene (Sfpi1)] were identified on the promoter region of mouse Pgr ∼660 bp 5′ of the transcription start site (TSS) (Fig. 4A). PU.1 has been reported to be a direct target of Notch signaling (23). In our study, expression of PU.1 was increased in OEx mice as well as Sfpi1 mRNA (Fig. 4 B and C and Fig. S3C). Previous studies suggested that PU.1 can hypermethylate its target genes by recruiting DNA methyltransferase 3b (DNMT3b) to the promoter region of target genes in the human during osteoclastogenesis (24). Therefore, we hypothesized that methylation of the Pgr promoter in N1ICD OEx mice could be altered by a PU.1/Dnmt3b-mediated pathway and further leads to decreased Pgr expression.
Fig. 4.
DNA hypermethylation of the Pgr gene mediated by PU.1/Dnmt3b. (A) Predicted PU.1 binding site on the Pgr promoter. PU.1 is up-regulated in N1ICD OEx mice by (B) immunohistochemistry and (C) quantitative PCR; quantitative expression levels of PU.1 (immunohistochemistry) are shown in Fig. S3C. (D) The PU.1 and Dnmt3b interaction is stronger in OEx mice than that in control mice (n = 3). (E) Binding of PU.1 and Dnmt3b protein on the Pgr promoter was detected by ChIP (n = 3). Binding efficiency of both PU.1 and Dnmt3b is significantly higher in OEx mice than control mice at positions of the predicted PU.1 motif (−755 to −644) and the CpG island (649–735). (F) Relative positions of the PU.1 binding site, the Pgr transcription start site, and the CpG island. (G) The number of methylated CpGs (black cells) is much higher in OEx mice than in control mice. Blue cells are unmethylated CpGs. All data are collected at 3.5 dpc. Ctrl, control; IP, immunoprecipitation. *P < 0.05; **P < 0.01. (Scale bar: 100 µm.)
To confirm our hypothesis, we first showed an interaction of PU.1 and Dnmt3b proteins in our mice by immunoprecipitation. PU.1 and Dnmt3b interacted with each other in uterine tissues of N1ICD OEx mice, and this interaction was much stronger than that observed in control mice (Fig. 4D). Using ChIP, binding of both PU.1 and Dnmt3b on predicted DNA regions on the Pgr promoter was verified. Binding efficiency of both PU.1 and Dnmt3b was significantly higher in OEx mice than in control mice (Fig. 4E, −755 to −644). Next, we identified the presence of cytosine–phosphate–guanine (CpG) islands by using the UCSC(University of California Santa Cruz) Genome Browser (https://genome.ucsc.edu/). Surprisingly, the CpG islands appear in exon 1 of the Pgr gene instead of the promoter (Fig. S5). The relative positions of the PU.1 binding site, Pgr TSS, and CpG islands are diagrammatically illustrated in Fig. 4F. The methylation of CpG islands on exon 1 of Pgr gene was identified by bisulfite sequencing. Our data showed that Pgr is hypermethylated in the uterus of N1ICD OEx mice compared with control mice at 3.5 dpc (Fig. 4G).
Fig. S5.
The CpG island (green arrow) in the UCSC Genome Browser and the fragment that we amplified (black arrow) using the bisulfite sequencing PCR experiment.
However, the hypermethylated CpG islands were localized at ∼1,300 bp downstream from the PU.1/Dnmt3b binding site (Fig. 4F). To further investigate if there are more PU.1/Dnmt3b binding sites close to the hypermethylated CpG islands, additional ChIP primers were designed to scan PU.1 and Dnmt3b binding along the region in proximity to the predicted binding site and the CpG islands (Fig. 4 E and F). Interestingly, the presence of another PU.1/Dnmt3b binding site in the region of the CpG islands was detected, supporting our hypothesis that the hypermethylation of Pgr exon 1 is associated with the DNA binding of PU.1/Dnmt3b complexes (Fig. 4E, 649–735).
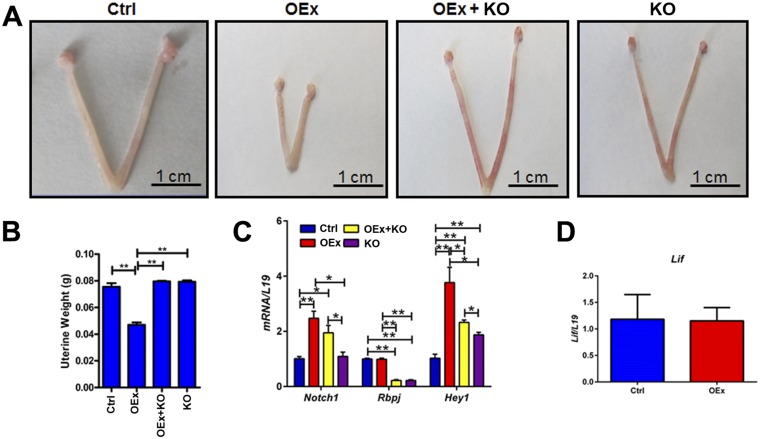
Ablation of RBP-Jκ in N1ICD OEx Mice Rescues Infertility.
To determine whether the overexpression of the N1ICD-induced infertile phenotype was mediated solely by RBP-Jκ–dependent signaling mechanisms, we performed the N1ICD OEx experiments in the absence of RBP-Jκ [PgrCre/+Rosa26N1ICD/+Rbpjflox/flox (OEx + KO; Fig. S2C)]. The implantation failure observed in the N1ICD OEx was rescued in the absence of RBP-Jκ. The number of implantation sites at 4.5 dpc in OEx + KO mice was comparable with that in control mice and markedly higher than that in OEx mice with intact RBP-Jκ (Fig. 1A). Smaller uterine size, as seen in N1ICD OEx mice, was also rescued in OEx + KO mice (Fig. S2 A and B). Glandular development was detected in uteri of OEx + KO mice with Foxa2 staining evident (Fig. 2 A and B). Expression patterns of Pgr and Esr1 showed no difference in OEx + KO mice compared with control mice at 3.5 dpc, both of which were significantly different from the OEx mice (Fig. 3 A and B). Expression levels of all target genes of P4 and E2 signaling were also rescued when RBP-Jκ was deleted (Fig. 3 C and D and Fig. S3A). The altered proliferation pattern of OEx mice was also rescued by the deletion of RBP-Jκ (Fig. 3 A and B). Most importantly, the expression of Sfpi1 (PU.1) also returned to the basal level when Rbpj was deleted in N1ICD OEx mice (Fig. 4C). These data in the OEx + KO mice collectively show that the phenotype associated with N1ICD overexpression occurs in an RBP-Jκ–dependent manner.
Discussion
Implantation is one of the most critical events in the establishment of pregnancy in rodents and primates. Successful implantation requires a competent blastocyst and a receptive uterus during a specific window of time during the cycle to initiate the bilateral communication and establish a successful pregnancy (3). Here, we show that uterine receptivity has been compromised by aberrant activation of Notch signaling in the mouse uterus. The uteri of OEx mice neither respond to artificial decidualization nor accept transferred WT embryos for implantation. The absence of uterine glands contributes significantly to the defective uterine receptivity. Brief exposure of female pups to P4 during neonatal days 2–10 results in the failure to develop uterine glands and further leads to infertility (25–27). Loss of certain genes, such as Foxa2, Ctnnb1 (β-catenin), Wnt4, Wnt5a, Wnt7a, and Lef1, also results in reduction or absence of uterine glands (reviewed in ref. 28). In this study, N1ICD OEx mice also failed to develop their uterine glands, similar to the studies described above. However, there is a marked difference between our OEx mice and data reported in previous studies: the LE cells of OEx mice exhibit a glandular phenotype, including the expression of GE markers Foxa2 and Lif. Uterine gland secretion of Lif is essential for blastocyst implantation in mouse (16, 17). Lif is also the main mediator of uterine receptivity failure in the absence of uterine glands: an intrauterine injection of Lif can partially rescue the lack of a decidual response in the Foxa2 null mouse uterus (14). Our N1ICD OEx mice completely failed to decidualize, although they produce a comparable amount of Lif to that of control mice. Therefore, we deduced that the lack of uterine glands is not the only component that contributes to uterine receptivity in OEx mice.
P4 signaling is a highly regulated cellular pathway that plays a critical role in the initiation and maintenance of pregnancy. Without functional P4 signaling, as seen in Pgr KO mice, pregnancy is unable to occur because of implantation and decidualization failure (29, 30). In our study, the expression of Pgr is significantly decreased in N1ICD OEx mice compared with control mice, which correlates with the dramatic down-regulation of P4 target genes (Ihh, Nr2f2, Areg, Hoxa10, and Hand2) in both epithelial and stromal cells. These data suggest that P4 signaling is significantly reduced when Notch signaling is aberrantly activated. During and after embryo implantation, the surrounding uterine stromal cells undergo decidualization, during which stromal cells proliferate and differentiate into decidual cells to promote the growth of the embryo (3). Stromal proliferation is significantly decreased in OEx mice as indicated by Ki67 staining, suggesting an impaired potential of these cells to decidualize. Furthermore, the inhibitory P4 signaling in N1ICD OEx mice is because of not only the decrease in Pgr but also, the lower serum P4 levels, which suggest impaired ovarian P4 synthesis. However, supplemental P4 can only rescue the expression of P4 target genes by 10–30% compared with the expression levels found in control mice. Dysregulated E2 signaling and E2-induced LE proliferation associated with infertility can result from a lack of P4 receptor and target gene expression, including Ihh, Nr2f2, and Hand2 (21, 29–32). In our study, decreased P4 signaling is associated with abnormally up-regulated E2 signaling in uterine epithelial cells of OEx mice as evidenced by increased expression of Esr1 and its phosphorylated form p-Esr1; up-regulation of target genes Muc1, Muc4, Ltf, and C3; and the increased proliferation of epithelial cells, which is driven by E2 signaling. These data suggest that the dysregulation of epithelial E2 signaling in OEx mice is, at least partially, caused by suppression of P4 signaling. However, loss of inhibition by decreased P4 signaling is not the only factor responsible for increased E2 signaling and its induced proliferation, because induction of target genes in OEx mice is much higher (∼10 times) than induction of these genes in Ihh, Nr2f2, and Hand2 null mice (21, 31, 32). In ESR1-positive breast cancer cells, N1ICD stimulates ESR1-dependent transcription, even in the absence of E2, by recruiting p300 and IKKα to ESR1 binding sites on chromatin (33). This effect requires RBP-Jκ, because canonical Notch1 transcriptional complexes form in proximity to ESR1 binding sites (33). The possibility that overactivated Notch1 signaling may interact with E2 signaling directly in endometrial epithelial cells through a similar mechanism will be the focus of future studies.
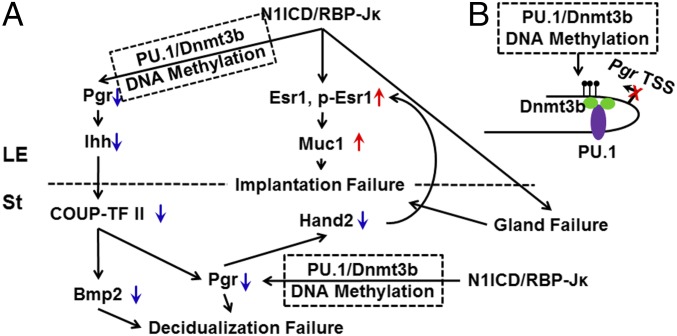
To our knowledge, regulation of DNA methylation by Notch signaling has not been reported. Sfpi1 or transcription factor PU.1, a direct target of Notch1 (23, 34), has been reported as a DNA methylation mediator through interaction with DNA methyltransferase Dnmt3b (24). For the first time, to our knowledge, we show that overactivation of Notch signaling can induce hypermethylation of exon 1 of the Pgr gene and further lead to the inhibition of P4 signaling. This process is associated with increased binding of PU.1 as well as its interacting protein Dnmt3b, which directly mediates DNA methylation. According to a previous study, the PU.1/Dnmt3b complex can methylate DNA regions within 500 bp of their binding site (24). In the case of our uterine-specific N1ICD OEx mouse, the verified PU.1 binding motif predicted by MotifMap exists about 1,300 bp away from the hypermethylated CpG islands, which is farther than previously reported. However, the presence of a second binding site for the PU.1/Dnmt3b complex in the DNA region of CpG islands showed the correlation between PU.1/Dnmt3b binding and CpG island hypermethylation in N1ICD OEx mice. Furthermore, the binding of Dnmt3b to the CpG island region (649–735) is much stronger than its binding to the PU.1 binding motif on the Pgr promoter (−755 to −644). Similarly, the binding of PU.1 to its binding motif (−755 to −644) is significantly higher than that of the CpG island region (649–735). These data suggest that Dnmt3b directly binds to the CpG island region (649–735) and binds to the PU.1 binding motif (−755 to −644) indirectly through its interaction with PU.1. However, the PU.1 binding that we observed from the −755 to −644 region is likely direct binding, whereas the CpG islands binding site occurs indirectly through interaction with Dnmt3b. Therefore, we created a spatial model based on our findings (Fig. 5B). First, PU.1 binds to the Pgr promoter and enriches Dnmt3b through their direct interaction. Second, the enriched Dnmt3b binds to the CpG island 1.3 kb away from the promoter through a looped DNA structure and hypermethylates this region.
Fig. 5.
Working model. (A) Overexpression of N1ICD, working through RBP-Jκ, inhibits P4 signaling and overactivates E2 signaling. Dysregulation of P4 and E2 signaling contributes to implantation and decidualization failure and further leads to defective uterine receptivity as a consequence of the altered expression of their target genes. Overactivation of canonical Notch signaling decreases Pgr through hypermethylation of exon 1 by the PU.1/Dnmt3b complex. (B) The PU.1/Dnmt3b spatial working model. PU.1 first binds on the promoter of Pgr and then recruits Dnmt3b through their direct interaction. Dnmt3b binds to the CpG island from 1.3 kb away from the PU.1 binding site through a looped DNA structure and hypermethylates this region.
Notch signaling occurs through canonical or noncanonical pathways. In canonical Notch signaling, the NICD translocates to the nucleus and binds directly to RBP-Jκ, converting it from a transcriptional suppressor to an activator and inducing the transcription of downstream target genes (7). In contrast, noncanonical Notch signaling does not require activation of RBP-Jκ (6). In this study, the fact that the deletion of Rbpj completely rescues the phenotypes of N1ICD OEx mice indicates that the abnormalities that we observe when N1ICD is overexpressed are occurring through canonical pathway signaling. Deletion of Rbpj would result in derepression of genes normally repressed by RBP-Jκ in the absence of NICD. Our data indicate that genes actively transactivated by N1ICD through RBP-Jκ are responsible for the phenotype of N1ICD OEx mice. Recently, an N1ICD/RBP-Jκ cobinding site was identified ∼2.6 kb upstream from the Sfpi1 gene (supplemental data in ref. 35), suggesting that Notch signaling directly regulates PU.1 expression in an RBP-Jκ–dependent manner, which supported by our data that PU.1-mediated Pgr suppression is rescued by deletion of Rbpj.
Our study shows the first genetic evidence, to our knowledge, that overactivation of canonical Notch signaling leads to hypermethylation, suggesting that Notch signaling plays a role in regulating an epigenetic modification of gene expression. In addition, hypermethylation of Pgr has been reported to contribute to decreased expression of Pgr in pathological conditions, such as endometriotic ectopic lesions (36) and adenomyosis (37). Our finding that Notch signaling hypermethylates the Pgr gene provides a novel direction for understanding gynecological pathologies, which could lead to therapeutic avenues for these diseases.
In previous studies, we reported that inhibition or deletion of Notch signaling results in impaired decidualization in both women and a transgenic mouse model because of failure of cell survival before differentiation (10, 11). In this study, we showed that constitutively active Notch1 signaling also impairs decidualization in mouse uterus as well as HuFs (Fig. S6). These studies suggest that Notch signaling plays two distinct roles during decidualization. Silencing of NOTCH1 during the initiation of decidualization inhibits stromal cell differentiation, indicating that activation of Notch1 signaling is required only at the initiation of the decidualization process, which is also associated with the induction of FOXO1 (12). As decidualization progresses, Notch1 is down-regulated (10, 11), and this down-regulation is necessary to permit differentiation into the decidual phenotype, which was shown in a previous study (11) and is also shown in this study. Decidualization is dependent on cAMP stimulation, sustained PKA activity, and cAMP response element-binding protein (CREB) activation (38, 39), and N1ICD sequesters nuclear CREB and inhibits cAMP/PKA-mediated signaling (40). When N1ICD is overexpressed, cAMP/PKA-mediated signaling is inhibited. This inhibition also prevents the ability of the stromal cells to differentiate and further illustrates the importance of Notch1 down-regulation in the physiological context during decidualization. Furthermore, in addition to preventing stromal cell differentiation and decidualization, N1ICD overexpression contributes to epithelial defects, such as the dysregulation of both P4 and E2 signaling and absence of uterine glands, all of which combined contribute to decidualization failure in the N1ICD OEx mice.
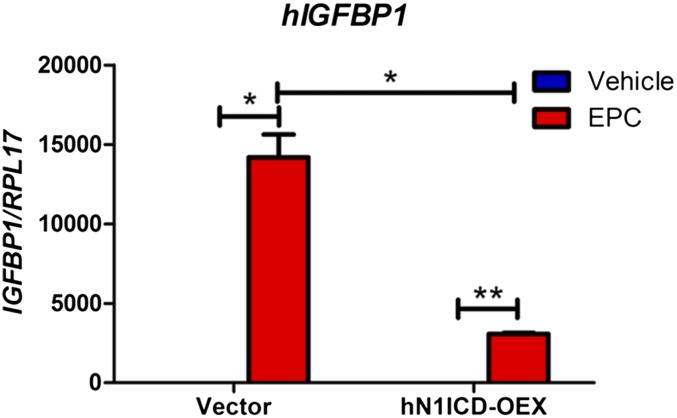
Fig. S6.
Overexpression of N1ICD in HuF cells inhibits in vitro decidualization as determined by the mRNA expression of the decidualization marker, IGF binding protein-1 (IGFBP1). *P < 0.05.
In summary, we have investigated the effects of constitutively activated Notch signaling in female reproduction. We found that increased Notch signaling in the mouse reproductive tract leads to infertility because of the failure of multiple reproductive processes, including dysregulation of P4 and E2 signaling through their nuclear receptors in a canonical RBP-Jκ–dependent manner. We also showed that the inhibition of P4 signaling is a consequence of both hypermethylation of Pgr by N1ICD complexing with PU.1 and Dnmt3b and lower levels of P4 synthesized by ovaries. However, the defect of P4 signaling is not the sole cause for the implantation and decidualization failure. Other physiological defects, such as the hyperactivation of E2 signaling in part caused by the decrease in Pgr and the absence of uterine glands in N1ICD OEx mice, could also contribute to implantation and decidualization failure. The mechanisms associated with this process are summarized in Fig. 5. The mechanisms by which N1ICD OEx causes glandular development failure and hyperactivation of E2 signaling will be investigated in future studies.
Materials and Methods
All antibodies and primers used in this study are listed in Tables S2 and S3, respectively. More descriptions of methods are in SI Materials and Methods.
Table S2.
List of antibodies
| Antibody | Type | Manufacturer | Catalog no. | Application(s) |
| Pgr | Rabbit IgG | Dako | A0098 | IHC |
| Esr1 | Mouse IgG | Vector Laboratories | VP-E613 | IHC |
| p-Esr1 (S118) | Rabbit IgG | Abcam | ab31477 | IHC |
| Ki67 | Mouse IgG | BD Biosciences | 550609 | IHC |
| Muc1 | Rabbit IgG | Abcam | ab15481 | IHC |
| COUP-TFII | Mouse IgG | Perseus Proteomics | PD-H7147-00 | IHC |
| PU.1 | Rabbit IgG | Santa Cruz Biotechnology | sc-352 | IHC, ChIP, immunoprecipitation |
| Dnmt3b | Rabbit IgG | Abcam | ab2851 | ChIP, immunoprecipitation |
| Lif | Goat IgG | Santa Cruz Biotechnology | Sc-1336 | WB |
| β-Actin | Mouse IgG | Sigma-Aldrich | A-5441 | WB |
IHC, immunohistochemistry; WB, Western blot.
Table S3.
List of primers
| Gene | Primer application | Forward sequence | Reverse sequence |
| Notch1 | qPCR intracellular domain (N1ICD) | 5′-GGAGGTGGATGCTGACTG-3′ | 5′-TCACTGTTGCCTGTCTCAAG-3′ |
| EGFP | qPCR | 5′-AAGCTGACCCTGAAGTTCATCTGC-3′ | 5′-CTTGTAGTTGCCGTCGTCCTTGAA-3′ |
| Hey1 | qPCR | 5′-ACTGTCTCCACCGCTGCTCT-3′ | 5′-GCTTTCCCCTCCCTTGTTCT-3′ |
| Hes5 | qPCR | 5′-GAGAAGATGCGTCGGGACC-3′ | 5′-GGCGAAGGCTTTGCTGTG-3′ |
| Bmp2 | qPCR | 5′-CGCAGCTTCCATCACGAA-3′ | 5′-GCTTCCTGTATCTGTTCCCG-3′ |
| Wnt4 | qPCR | 5′-AGTGCCAATACCAGTTCCG-3′ | 5′-CACACTTCTCCAGTTCTCCAC-3′ |
| Lif | qPCR | 5′-TTCCCATCACCCCTGTAAATG-3′ | 5′-GAAACGGCTCCCCTTGAG-3′ |
| Pgr | qPCR | 5′-TGAGCCTGATGGTGTTTGG-3′ | 5′-ACAGCGAGTAGAATGACAGC-3′ |
| Ihh | qPCR | 5′-CCCAACTACAATCCCGACATC-3′ | 5′-TCACCCGCAGTTTCACAC-3′ |
| Areg | qPCR | 5′-AGATACATCGAGAACCTGGAGG-3′ | 5′-AGAGACAAAGATAGTGACAGCTAC-3′ |
| Hoxa10 | qPCR | 5′-GAAAACAGTAAAGCTTCGCCG-3′ | 5′-GAAACTCCTTCTCCAGCTCC-3′ |
| Nr2f2 | qPCR | 5′-GCCATAGTCCTGTTCACCTC-3′ | 5′-GCTCCTAACGTACTCTTCCAAAG-3′ |
| Hand2 | qPCR | 5′-CAGATACATCGCCTACCTCATG-3′ | 5′-GGCCTTTGGTTTTCTTGTCG-3′ |
| Esr1 | qPCR | 5′-AACCGCCCATGATCTATTCTG-3′ | 5′-AGATTCAAGTCCCCAAAGCC-3′ |
| Muc1 | qPCR | 5′-TTCCAACCCAGGACACCTAC-3′ | 5′-ATTACCTGCCGAAACCTCCT-3′ |
| Muc4 | qPCR | 5′-AATGTTCCTGCCTATACTGCC-3′ | 5′-TTGTATGGTTCCTGGGTCAC-3′ |
| Ltf | qPCR | 5′-ATCTCTGTGCCCTGTGTATTG-3′ | 5′-ACATTTCCTGCCTTCTCAGC-3′ |
| C3 | qPCR | 5′-AGACTGCCTGACCTTCAAAG-3′ | 5′-CATCCCATCGTCCTTCTCTG-3′ |
| Sfpi1 | qPCR | 5′-CAACGTCCAATGCATGACTAC-3′ | 5′-ATCTGTTCCAGCTCCATGTG-3′ |
| Rpl19 (L19) | qPCR | 5′-TCATGGAGCACATCCACAAGCTGA-3′ | 5′-CGCTTTCGTGCTTCCTTGGTCTTA-3′ |
| Rpl19 (L19) | qPCR | TaqMan Probe | Catalog no. 4331182 |
| Rbpj | qPCR | TaqMan Probe | Catalog no. 4351372 |
| Lif | Template for cRNA probe | 5′-GGTCCTTAGCAGTTCAGC-3′ | 5′-GGTAGGTGGTTGCGTTG-3′ |
| Pgr | ChIP: −1,092 to −993 from TSS | 5′-GAAACACTCAAAGGCATTTCCCAAAC-3′ | 5′-CCGTCATAAGCTGTCCACTCTCAAGC-3′ |
| Pgr | ChIP: −755 to −644 from TSS | 5′-CCCATACAGGCAGCGATGTGTTAC-3′ | 5′-TGTCCAGCCTTCCTCTAACACGC-3′ |
| Pgr | ChIP: −489 to −408 from TSS | 5′-TTGCTGCTCCTGCAGAGAACCC-3′ | 5′-GTGAGAGCCCTCAGAGGTGAAGTTG-3′ |
| Pgr | ChIP: 240–329 from TSS | 5′-CAAACATAAGAAGTCCGGAGATAGCG-3′ | 5′-TTCGTGTTTCTTCTTGAATGGGAGAC-3′ |
| Pgr | ChIP: 649–735 from TSS | 5′-AGGCAAAGGATCCGCAGGTTC-3′ | 5′-AGTCCAAGCGTGCAAGCAAGG-3′ |
| Pgr | Bisulfite sequencing PCR | 5′-TTTAGGAGATAGGGGAGGAGAAA-3′ | 5′-ATAACTTCTACCCCAAAAAAAACTC-3′ |
Hey1, hairy enhancer of split-related 1; Hes5, hairy enhancer of split 5.
Animals.
PgrCre/+ mice (41), Rosa26N1ICD/N1ICD mice (The Jackson Laboratory), and Rbpjflox/flox mice (42) were maintained in the designated animal care facility according to the Michigan State University institutional guidelines. All animal procedures were approved by the Institutional Animal Care and Use Committee of Michigan State University.
Human Tissue Collection.
Placental tissues were obtained with informed consent using a protocol approved by the Institutional Review Board at Michigan State University and Spectrum Health System.
Embryo Transfer.
All embryos used were collected from WT C57BL/6 females. Pseudopregnant recipients were induced by mating with vasectomized males. Seven blastocysts were transferred into the uterine lumen of one horn in the afternoon at 2.5 dpc, and implantation status was detected at 4.5 dpc by tail vein injection of Chicago Sky Blue (Sigma-Aldrich).
Artificial Decidualization Model.
As described in ref. 10, mice were ovariectomized and treated with E2 and P4 (Sigma-Aldrich). A scratch stimulus was then performed on the uterine luminal epithelium of the antimesometrium side of one uterine horn to induce decidualization. The unscratched horn served as a hormonal control. Animals were killed 5 d after the scratch, and both horns were collected for analysis.
Statistical Analysis.
Data are expressed as means ± SEMs. Data were analyzed using the Student’s t test and one-way ANOVA followed by Tukey’s posthoc multiple-range test. Values were considered significant if P was <0.05. All statistical analyses were performed by GraphPad Prism 5.0 (GraphPad Software).
SI Materials and Methods
Animals.
PgrCre/+ mice (41), Rosa26N1ICD/N1ICD mice (The Jackson Laboratory), and Rbpjflox/flox mice (42) were maintained in the designated animal care facility according to the Michigan State University institutional guidelines. All animal procedures were approved by the Institutional Animal Care and Use Committee of Michigan State University. PgrCre/+ mice were crossed with Rosa26N1ICD/N1ICD mice, Rbpjflox/flox mice, or both strains to generate PgrCre/+Rosa26N1ICD/+, PgrCre/+Rbpjflox/flox, or OEx + KO mice for this study. Pgr+/+Rosa26N1ICD/+ mice were used as controls. Females were caged with fertile males to induce pregnancy, and the morning of a vaginal plug was designated as 0.5 dpc. For supplemental P4 treatment, P4 (1 mg per mouse per day; Sigma-Aldrich) was injected s.c. from 2.5 to 3.5 dpc, and mice were killed at 3.5 dpc. For the fertility test, females were housed together with fertile males for 6 months, and the date and pup number of each litter were recorded.
Embryo Transfer.
All embryos used were collected from WT C57BL/6 females. Pseudopregnant recipients were induced by mating with vasectomized males. Seven blastocysts were transferred into the uterine lumen of one horn in the afternoon of 2.5 dpc, and implantation status was detected at 4.5 dpc by tail vein injection of Chicago Sky Blue (Sigma-Aldrich).
Artificial Decidualization Model.
As described in ref. 10, mice were ovariectomized and treated with E2 (100 ng; Sigma-Aldrich) for 3 d and P4 (1 mg) plus E2 (6.7 ng) for another 3 d, with 2 d of rest in between to mimic the hormonal environment of normal early pregnancy. Six hours after the last injection, a scratch stimulus was performed on the uterine luminal epithelium of the antimesometrium side of one uterine horn to induce decidualization. The unscratched horn served as a hormonal control. Daily injections of P4 plus E2 were continued for 5 d to maximize the decidual response. Mice that were used to mimic receptivity at 3.5 dpc were killed at 30 h after the first E2 + P4 injection.
Measurement of Serum E2 and P4 Levels.
Serum levels of E2 and P4 at 3.5 dpc of pregnancy were measured by enzyme immunoassays, which was performed by the University of Virginia Ligand Assay and Analysis Core.
Histological Analysis and Immunohistochemistry.
Four percent paraformaldehyde-fixed, paraffin-embedded uterine tissues were sectioned (6 µm). After deparaffinization and hydration, sections were stained with H&E for histological analysis. For immunohistochemistry, sections were subjected to antigen retrieval in citrate buffer and hydrogen peroxide treatment, and sections were incubated with primary and secondary antibodies. HRP and diaminobenzidine (Vector Laboratories) were used to visualize antigens. Sections were counterstained with hematoxylin. All primary antibodies used in this study were listed in Table S2.
In Situ Hybridization.
In situ hybridization was performed according to the work in ref. 44. Briefly, frozen sections (10 µm) were fixed in 4% (wt/vol) paraformaldehyde, permeabilized, and hybridized with a digoxigenin (DIG) -labeled Lif cRNA probe at 55 °C overnight. Sections were then washed and incubated with anti-DIG antibody. The signal was visualized with 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium, and the positive signal was visualized as dark brown. All of the sections were counterstained with nuclear fast red. The DIG-labeled cRNA probe was transcribed in vitro using the DIG RNA Labeling Kit (Roche) from Lif cDNA (primers are listed in Table S3).
Western Blot.
Total protein was isolated from frozen tissues using lysis buffer, and the concentrations were calculated using the bicinchoninic acid (BCA) method (Thermo Fischer Scientific). Ten micrograms protein was separated on SDS/PAGE gels (Invitrogen) and transferred onto a PVDF membrane (Millipore). The membranes were then blocked and incubated overnight with primary antibodies at 4 °C. After incubation with respective secondary antibodies labeled with HRP, immunocomplexes were visualized by enhanced chemiluminescence (GE Amersham). Primary antibodies used in this study are listed in Table S2.
Quantitative PCR.
Total RNA was isolated from frozen tissue or cultured cells using TRIzol Reagent (Invitrogen) and then, reverse-transcribed to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Quantitative PCR was then performed to measure gene expression levels with SYBR Green PCR Master Mix (Applied Biosystems) using the ViiA 7 qPCR System (Applied Biosystems). Rpl19 (L19) was used for normalization. Primer sequences used for qPCR are listed in Table S3.
Immunoprecipitation.
Whole uteri were cut into small pieces, and cell lysates were extracted and precleaned by normal rabbit IgG (Bethyl Laboratories) with protein A-agarose beads (Thermo Fisher Scientific). After bead removal, the supernatant protein solution (1 mg/mL) was immunoprecipitated with an antibody to PU.1 or normal rabbit IgG overnight and pulled down by protein A-agarose beads the next day. Immunocomplexes were then subjected to SDS/PAGE, transferred onto PVDF membranes, and subsequently, incubated with PU.1 and Dnmt3b antibodies as described above. Primary antibodies used in this study are listed in Table S2.
ChIP.
ChIP was performed by using the ChIP-It High Sensitivity Kit (Active Motif) according to the manufacturer’s specifications. Briefly, fresh uterine tissues of each group were cut into small pieces, fixed in formaldehyde solution, and homogenized, and chromatin was prepared. Chromatin samples were then processed by sonication to be fragmented into 200- to 500-bp size fragments. Next, immunoprecipitation was performed using normal rabbit IgG, PU.1, or Dnmt3b antibodies. Pulled down DNA was then reverse cross-linked, purified, and quantitated by qPCR using the primers listed in Table S3.
Bisulfite Sequencing PCR.
Primers for bisulfite sequencing PCR of CpG islands on the Pgr promoter/gene were designed by Methprimer (www.urogene.org/) (listed in Table S3) (43). Whole-uterine tissue of each group was cut into small pieces, and genomic DNA was extracted; 500 ng genomic DNA was then converted by using the EZ DNA Methylation-Gold Kit (Zymo Research) followed by PCR amplification of CpG island regions. PCR products were purified, inserted into PCR2.1-TOPO cloning vector (Invitrogen), and sequenced. CpG methylation levels were identified by using Bisulfite Sequencing DNA Methylation Analysis (services.ibc.uni-stuttgart.de/BDPC/BISMA).
Isolation of HuF Cells.
Stromal fibroblasts from the decidua parietalis maintain a proliferating population of nondifferentiated fibroblastic cells, which closely resemble endometrial stromal cells (45). HuF cells were isolated from the decidua parietalis dissected from the placental membrane after normal-term vaginal delivery. Isolation and culture of HuF cells were completed as previously described (45). Cells isolated from each patient were grown individually and maintained for study between passages 3 and 5. Placental tissues were obtained with informed consent using a protocol approved by the Institutional Review Board at Michigan State University and Spectrum Health System.
Lentivirus Packaging and Transfection.
Lentivirus plasmids encoding N1ICD and the control empty plasmid were provided by Meenhard Herlyn (The Wistar Institute, Philadelphia) and packaged using the Trans-Lentiviral Packaging Kit (TLP5912; Thermo) in the 293FT cell line (Invitrogen) according to the manufacturer’s instructions. Briefly, each plasmid was transfected into 293FT cells together with the packaging mix for the plasmids. Cells were then cultured for 58 h, and viral particle-containing supernatants were harvested and stored at −80 °C until further use. HuF cells were cultured in 60-mm dishes with 3 mL growth media until ∼60% confluence; 1 mL viral particle-containing supernatant was then added for transfection, and media were changed back to growth media after 24 h.
In Vitro Decidualization.
To induce in vitro decidualization, cells were treated with an EPC mixture (36 nM E2, 1 μM medroxyprogesterone acetate, and 0.1 mM dibutyryl-cAMP; Sigma-Aldrich) or vehicle for 6–8 d. Media were replaced every 3 d. All experiments were repeated a minimum of three times, and each experiment was done in duplicate. Cells were then collected for additional RNA or protein isolation.
Acknowledgments
The authors thank Dr. John Lydon (Baylor College of Medicine) and Dr. Francesco DeMayo (National Institute of Environmental Health Sciences) for providing the PgrCre/+ mice and Dr. Nadia Carlesso (Indiana University) for providing the Rbpjflox/flox mice. The authors also thank Ms. Samantha Bond for excellent technical assistance and Mr. Mark Olson for his help in manuscript preparation. This study was funded by NIH Grants F30 HD082951 (to M.R.S.) and R01 HD042280 (to A.T.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1520441113/-/DCSupplemental.
References
- 1.Hjollund NH, et al. Spontaneous abortion and physical strain around implantation: A follow-up study of first-pregnancy planners. Epidemiology. 2000;11(1):18–23. doi: 10.1097/00001648-200001000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Edwards RG. Human implantation: The last barrier in assisted reproduction technologies? Reprod Biomed Online. 2006;13(6):887–904. doi: 10.1016/s1472-6483(10)61039-5. [DOI] [PubMed] [Google Scholar]
- 3.Cha J, Sun X, Dey SK. Mechanisms of implantation: Strategies for successful pregnancy. Nat Med. 2012;18(12):1754–1767. doi: 10.1038/nm.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasquez YM, DeMayo FJ. Role of nuclear receptors in blastocyst implantation. Semin Cell Dev Biol. 2013;24(10-12):724–735. doi: 10.1016/j.semcdb.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rizzo P, et al. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008;68(13):5226–5235. doi: 10.1158/0008-5472.CAN-07-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Souza B, Meloty-Kapella L, Weinmaster G. Canonical and non-canonical Notch ligands. Curr Top Dev Biol. 2010;92:73–129. doi: 10.1016/S0070-2153(10)92003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kopan R, Ilagan MX. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iso T, Kedes L, Hamamori Y. HES and HERP families: Multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194(3):237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 9.Cuman C, et al. Fetal-maternal communication: The role of Notch signalling in embryo implantation. Reproduction. 2014;147(3):R75–R86. doi: 10.1530/REP-13-0474. [DOI] [PubMed] [Google Scholar]
- 10.Afshar Y, et al. Notch1 mediates uterine stromal differentiation and is critical for complete decidualization in the mouse. FASEB J. 2012;26(1):282–294. doi: 10.1096/fj.11-184663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Afshar Y, Miele L, Fazleabas AT. Notch1 is regulated by chorionic gonadotropin and progesterone in endometrial stromal cells and modulates decidualization in primates. Endocrinology. 2012;153(6):2884–2896. doi: 10.1210/en.2011-2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su RW, et al. Decreased Notch pathway signaling in the endometrium of women with endometriosis impairs decidualization. J Clin Endocrinol Metab. 2015;100(3):E433–E442. doi: 10.1210/jc.2014-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Otti GR, et al. Notch2 controls prolactin and insulin-like growth factor binding protein-1 expression in decidualizing human stromal cells of early pregnancy. PLoS One. 2014;9(11):e112723. doi: 10.1371/journal.pone.0112723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeong JW, et al. Foxa2 is essential for mouse endometrial gland development and fertility. Biol Reprod. 2010;83(3):396–403. doi: 10.1095/biolreprod.109.083154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Filant J, Lydon JP, Spencer TE. Integrated chromatin immunoprecipitation sequencing and microarray analysis identifies FOXA2 target genes in the glands of the mouse uterus. FASEB J. 2014;28(1):230–243. doi: 10.1096/fj.13-237446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhatt H, Brunet LJ, Stewart CL. Uterine expression of leukemia inhibitory factor coincides with the onset of blastocyst implantation. Proc Natl Acad Sci USA. 1991;88(24):11408–11412. doi: 10.1073/pnas.88.24.11408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart CL, et al. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature. 1992;359(6390):76–79. doi: 10.1038/359076a0. [DOI] [PubMed] [Google Scholar]
- 18.Lee K, et al. Indian hedgehog is a major mediator of progesterone signaling in the mouse uterus. Nat Genet. 2006;38(10):1204–1209. doi: 10.1038/ng1874. [DOI] [PubMed] [Google Scholar]
- 19.Das SK, et al. Amphiregulin is an implantation-specific and progesterone-regulated gene in the mouse uterus. Mol Endocrinol. 1995;9(6):691–705. doi: 10.1210/mend.9.6.8592515. [DOI] [PubMed] [Google Scholar]
- 20.Benson GV, et al. Mechanisms of reduced fertility in Hoxa-10 mutant mice: Uterine homeosis and loss of maternal Hoxa-10 expression. Development. 1996;122(9):2687–2696. doi: 10.1242/dev.122.9.2687. [DOI] [PubMed] [Google Scholar]
- 21.Li Q, et al. The antiproliferative action of progesterone in uterine epithelium is mediated by Hand2. Science. 2011;331(6019):912–916. doi: 10.1126/science.1197454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurihara I, et al. COUP-TFII mediates progesterone regulation of uterine implantation by controlling ER activity. PLoS Genet. 2007;3(6):e102. doi: 10.1371/journal.pgen.0030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen PM, et al. Down-regulation of Notch-1 expression decreases PU.1-mediated myeloid differentiation signaling in acute myeloid leukemia. Int J Oncol. 2008;32(6):1335–1341. doi: 10.3892/ijo_32_6_1335. [DOI] [PubMed] [Google Scholar]
- 24.de la Rica L, et al. PU.1 target genes undergo Tet2-coupled demethylation and DNMT3b-mediated methylation in monocyte-to-osteoclast differentiation. Genome Biol. 2013;14(9):R99. doi: 10.1186/gb-2013-14-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cooke PS, et al. Brief exposure to progesterone during a critical neonatal window prevents uterine gland formation in mice. Biol Reprod. 2012;86(3):63. doi: 10.1095/biolreprod.111.097188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filant J, Zhou H, Spencer TE. Progesterone inhibits uterine gland development in the neonatal mouse uterus. Biol Reprod. 2012;86(5):146. doi: 10.1095/biolreprod.111.097089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Filant J, Spencer TE. Endometrial glands are essential for blastocyst implantation and decidualization in the mouse uterus. Biol Reprod. 2013;88(4):93. doi: 10.1095/biolreprod.113.107631. [DOI] [PubMed] [Google Scholar]
- 28.Cooke PS, Spencer TE, Bartol FF, Hayashi K. Uterine glands: Development, function and experimental model systems. Mol Hum Reprod. 2013;19(9):547–558. doi: 10.1093/molehr/gat031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lydon JP, et al. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 1995;9(18):2266–2278. doi: 10.1101/gad.9.18.2266. [DOI] [PubMed] [Google Scholar]
- 30.Franco HL, et al. Epithelial progesterone receptor exhibits pleiotropic roles in uterine development and function. FASEB J. 2012;26(3):1218–1227. doi: 10.1096/fj.11-193334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee DK, et al. Suppression of ERalpha activity by COUP-TFII is essential for successful implantation and decidualization. Mol Endocrinol. 2010;24(5):930–940. doi: 10.1210/me.2009-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franco HL, et al. Ablation of Indian hedgehog in the murine uterus results in decreased cell cycle progression, aberrant epidermal growth factor signaling, and increased estrogen signaling. Biol Reprod. 2010;82(4):783–790. doi: 10.1095/biolreprod.109.080259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao L, et al. Notch-1 activates estrogen receptor-alpha-dependent transcription via IKKalpha in breast cancer cells. Oncogene. 2010;29(2):201–213. doi: 10.1038/onc.2009.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroeder T, Kohlhof H, Rieber N, Just U. Notch signaling induces multilineage myeloid differentiation and up-regulates PU.1 expression. J Immunol. 2003;170(11):5538–5548. doi: 10.4049/jimmunol.170.11.5538. [DOI] [PubMed] [Google Scholar]
- 35.Geimer Le Lay AS, et al. The tumor suppressor Ikaros shapes the repertoire of notch target genes in T cells. Sci Signal. 2014;7(317):ra28. doi: 10.1126/scisignal.2004545. [DOI] [PubMed] [Google Scholar]
- 36.Wu Y, Strawn E, Basir Z, Halverson G, Guo SW. Promoter hypermethylation of progesterone receptor isoform B (PR-B) in endometriosis. Epigenetics. 2006;1(2):106–111. doi: 10.4161/epi.1.2.2766. [DOI] [PubMed] [Google Scholar]
- 37.Jichan Nie, Xishi Liu, Guo SW. Promoter hypermethylation of progesterone receptor isoform B (PR-B) in adenomyosis and its rectification by a histone deacetylase inhibitor and a demethylation agent. Reprod Sci. 2010;17(11):995–1005. doi: 10.1177/1933719110377118. [DOI] [PubMed] [Google Scholar]
- 38.Gellersen B, Brosens JJ. Cyclic decidualization of the human endometrium in reproductive health and failure. Endocr Rev. 2014;35(6):851–905. doi: 10.1210/er.2014-1045. [DOI] [PubMed] [Google Scholar]
- 39.Kusama K, et al. The role of exchange protein directly activated by cyclic AMP 2-mediated calreticulin expression in the decidualization of human endometrial stromal cells. Endocrinology. 2014;155(1):240–248. doi: 10.1210/en.2013-1478. [DOI] [PubMed] [Google Scholar]
- 40.Hallaq R, et al. The Notch intracellular domain represses CRE-dependent transcription. Cell Signal. 2015;27(3):621–629. doi: 10.1016/j.cellsig.2014.11.034. [DOI] [PubMed] [Google Scholar]
- 41.Soyal SM, et al. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis. 2005;41(2):58–66. doi: 10.1002/gene.20098. [DOI] [PubMed] [Google Scholar]
- 42.Han H, et al. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14(6):637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- 43.Li LC, Dahiya R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics. 2002;18(11):1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 44.Ni H, Sun T, Ding NZ, Ma XH, Yang ZM. Differential expression of microsomal prostaglandin e synthase at implantation sites and in decidual cells of mouse uterus. Biol Reprod. 2002;67(1):351–358. doi: 10.1095/biolreprod67.1.351. [DOI] [PubMed] [Google Scholar]
- 45.Jasinska A, Strakova Z, Szmidt M, Fazleabas AT. Human chorionic gonadotropin and decidualization in vitro inhibits cytochalasin-D-induced apoptosis in cultured endometrial stromal fibroblasts. Endocrinology. 2006;147(9):4112–4121. doi: 10.1210/en.2005-1577. [DOI] [PubMed] [Google Scholar]