Abstract
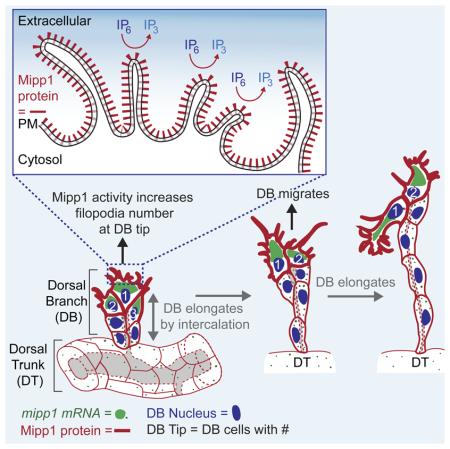
Multiple Inositol Polyphosphate Phosphatase – Mipp, a highly conserved but poorly understood histidine phosphatase, dephosphorylates higher order IPs (IP6 to IP4) to IP3. To gain insight into the biological roles of these enzymes, we have characterized Drosophila mipp1. mipp1 is dynamically expressed in the embryonic trachea, specifically in the leading cells of migrating branches at late stages, where Mipp1 localizes to the plasma membrane and filopodia. FGF signaling activates mipp1 expression in these cells, where extensive filopodia form to drive migration and elongation by cell intercalation. We show that Mipp1 faciliates formation and/or stabilization of filopodia in leading cells through its extracellular activity. mipp1 loss decreases filopodia number, whereas mipp1 overexpression increases filopodia number in a phosphatase-activity dependent manner. Importantly, expression of Mipp1 gives cells a migratory advantage for the lead position in elongating tracheal branches. Altogether, these findings suggest that extracellular pools of inositol polyphosphates affect cell behavior during development.
Keywords: epithelial migration, filopodia, inositol polyphosphates, mipp, trachea
INTRODUCTION
Inositol polyphosphates (IPs) are important second messengers involved in diverse cellular processes. Cytosolic IPs control ion channel physiology, membrane dynamics and cell migration (Irvine and Schell, 2001), whereas nuclear IPs have functions in mRNA export and gene regulation (York, 2006). The roles of extracellular IPs have been less thoroughly explored. Intracellular IPs are synthesized in the cell and extracellular IPs are obtained from the diet (Grases et al., 2005). For both intracellular and extracellular IPs, inositol hexakisphosphate (IP6) or phytate, is the most abundant, reaching intracellular concentrations of 10 to 100 μM (Sasakawa et al., 1995). IP6 can form tight insoluble complexes with a number of cations, including calcium, magnesium, iron and zinc, and thus serves as a storage molecule for inositol, phosphorus, and minerals (Kumar et al., 2010; Vucenik and Shamsuddin, 2006). IP6 is a well-known anti-tumor agent found in high-fiber foods and sold in many health food stores (Vucenik and Shamsuddin, 2003). High concentrations of extracellular IP6 inhibit cell migration and proliferation in human breast cancer cells (Shamsuddin et al., 1997) and induce G1 arrest and apoptotic death of pancreatic cancer cells (Singh et al., 2003). Maintenance of IP6 at critical threshold levels is important for mouse development (Verbsky et al., 2005).
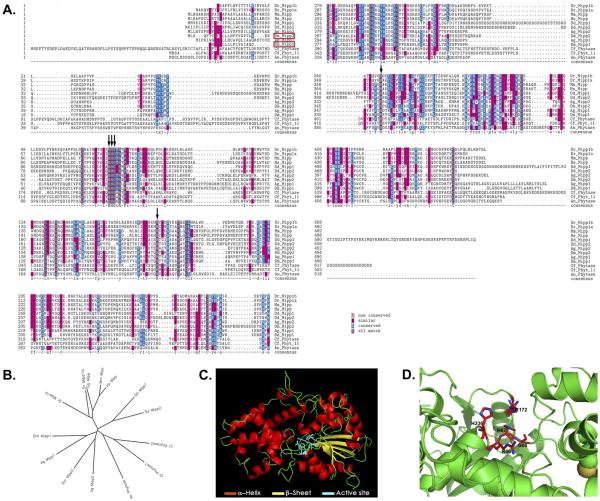
Mipp (Multiple Inositol Polyphosphate Phosphatase) is the major phosphatase that dephosphorylates IP6 to lower order IPs (IP5, IP4, and IP3). Mipp is a highly conserved histidine phosphatase with clear orthologues in all animals (Figures 1A-1B). The key catalytic residues of the Mipp histidine phosphatase are RHG-R-H, which are brought together into a catalytic pocket (Figures 1C-1D). Mipp homologues in plants and microorganisms are known as phytases, secreted histidine phosphatases that dephosphorylate extracellular IP6. Mammalian Mipp localizes to the ER lumen (Ali et al., 1993; Chi et al., 2000) as well as to the erythrocyte plasma membrane (Estrada-Garcia et al., 1991). Since manipulation of Mipp levels in mouse embryonic fibroblasts changes the cytosolic IP profile, Mipp may have access to cytosolic pools (Chi et al., 2000). Recent studies reveal that mammalian Mipp, like phytase, is secreted in certain cells (Windhorst et al., 2013), suggesting that Mipp may have intracellular and extracellular functions.
Figure 1. Mipp1 encodes a highly conserved histidine phosphatase with active site residues dispersed throughout the coding region.
(A-B) Textshade alignment (A) and Unrooted Phylip Tree (B) of Mipps and Phytases from representative model organisms.
(C) Phyre2 predicted structure of Drosophila Mipp1.
(D) Phyre2 predicted configuration of active site residues in the active site pocket of Drosophila Mipp1. Acronyms: Dr = Danio Rerio; Mm = Mus musculus; Hs = Homo sapiens; Dd = Dictyostelium discoideum; Ag = Anopheles gambiae; Dm = Drosophila melanogaster; Cf = Cryptococcus neoformans; An = Aspergillis nigers
Mipp is ubiquitously expressed in mammalian tissues, with elevated levels in growth plate chondrocytes, suggesting a potential role in chondrocyte differentiation (Craxton et al., 1995; Craxton et al., 1997). Although Mipp is the major phosphatase known to convert IP6 to IP3, mipp knockout mice are viable and fertile, without obvious defects (Chi et al., 2000). The characterization of erythrocytes from these mutants, however, suggests that Mipp activity is compensated by another unknown enzyme (Chi et al., 2000).
The observation that all animals encode at least one Mipp gene argues for a fundamental, undiscovered biological function for this enzyme. Drosophila encodes two mipp genes: mipp1 and mipp2. Whereas mipp2 is ubiquitously expressed (data not shown), mipp1 shows tissue-specific expression, including high and dynamic Trachealess-dependent expression in the developing embryonic trachea (embryonic stages (st) 10-15) (Figure S1A-S1O) (Chung et al., 2011). Tracheal development initiates with ten epithelial plates (placodes) of ~40 cells each on both sides of the embryo (Maruyama and Andrew, 2012). Each placode invaginates as tracheal cells undergo their final mitotic division to generate an internalized tracheal sac of ~80 cells. Five primary branches subsequently form in each tracheal segment (metamere), including the dorsal branch (DB), dorsal trunk (DT), visceral branch (VB), lateral trunk (LT), and ganglionic branch (GB). FGF signaling at the ends of branches drives filopodia formation to facilitate branch formation, elongation and migration (Figure S2) (Klambt et al., 1992; Ohshiro et al., 2002; Okenve-Ramos and Llimargas, 2014; Ribeiro et al., 2002; Sutherland et al., 1996). During st14, cells at the ends of the DT in each segment meet up and fuse with their partner cells in anterior and posterior segments to form an interconnected branch that runs along the length of the embryo. Other branches continue to migrate and elongate until they reach their final destinations; these branches elongate by transforming from multicellular tubes into unicellular tubes though a cell rearrangement process known as stalk cell intercalation or SCI (Figure S2B) (Ribeiro et al., 2004). By the end of development, the trachea has formed an elaborate interconnected branched network to provide gas exchange for every tissue of the animal.
To gain insight into the biological function of Mipps, we characterized Drosophila Mipp1. We showed that Mipp1 is dynamically expressed in migrating tracheal branches with FGF-dependent enrichment at the distal tip, where filopodia form. We learned that Mipp1 localizes to the filopodia and to the plasma membrane, where it converts extracellular IP6 to IP3. To determine the extracellular functions of Mipp1 in vivo, we created a null allele of mipp1 and constructs for overexpression. Our analysis revealed that Mipp1 facilitates filopodia formation and that expression of Mipp1 gives migrating tracheal cells a competitive advantage for the lead position in an extracellular phosphatase activity dependent manner.
RESULTS
mipp1 mRNA is dynamically expressed and enriched at the distal tip of migrating tracheal branches
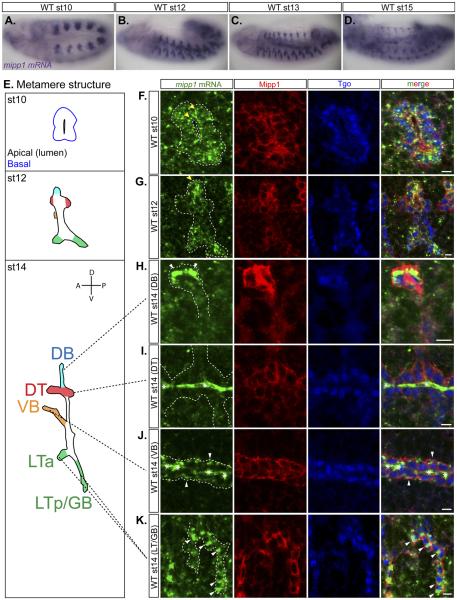
mipp1 expression was observed in all tracheal cells from embryonic st10 to st12 (Figures 2A-2B). From st13 to st15, mipp1 levels were diminished in the DT and maintained in the DB, VB, LT and GB - branches that undergo extensive migration and elongation (Figures 2C-2D). Fluorescent in situ hybridization revealed mipp1 accumulation in small puncta dispersed throughout the cytoplasm with some enrichment near the plasma membrane during st10 to st12 (Figures 2F-2G). During st13 to st15, mipp1 accumulated in a large focus localized to one side of the cell for DBs, VBs, and LTs/GBs. For DBs, which migrate dorsally, mipp1 mRNA foci localized to the dorsal sides of the two to three most distal tip cells (Figure 2H). For VBs, which migrate inwardly, mipp1 mRNA foci localized to the basal side of every cell (Figure 2J). For LTs/GBs, which migrate ventrally, mipp1 mRNA foci localized to the ventral side of each cell (Figure 2K). Similar large foci of mRNA accumulation were not observed with two other tracheal genes, sano and trh, which encode an apically-enriched cytosolic protein and a nuclear transcription factor, respectively (Figures S1Q-S1R). mipp1 mRNA expression/localization coincides with the onset and localization of filopodia formation in the trachea. Extensive filopodia were visible from st13 at the distal side of the most distal DB cells, on the basal side of VB cells, and the ventral side of LT/GB cells (Figure S1S). We conclude that mipp1 mRNA is maintained in migrating branches with enrichment at regions where extensive filopodia form.
Figure 2. mipp1 is dynamically expressed in the trachea with late transcripts concentrated in large foci on the distal side of each cell.
(A-D) in situ hybridization of mipp1 mRNA in st10 to st15 WT embryos (lateral views).
(E) Cartoon images showing tracheal metamere with labeled branches indicated by color at multiple embryonic stages. (Dorsal branches (DB) = blue; dorsal trunks (DT) = red; visceral branches (VB) = orange; lateral trunks/ganglionic branches (LT/GB) = green)
(F-K) Fluorescent in situ hybridization of st10 to st14 embryos detecting mipp1 mRNA (green) in combination with antibody staining for Mipp1 (red) and Tgo (blue), a tracheal nuclear marker. * = trapping of mRNA probes in st14 trachea, a frequent occurrence with in situ of later stage embryos; white arrowheads = large foci of mipp1 mRNA; yellow arrowheads = small puncta of mipp1 mRNA; white dash lines = outline of the trachea; scale bars = 5µm.
See also Figure S1.
Mipp1 protein localizes to the plasma membrane and to filopodia
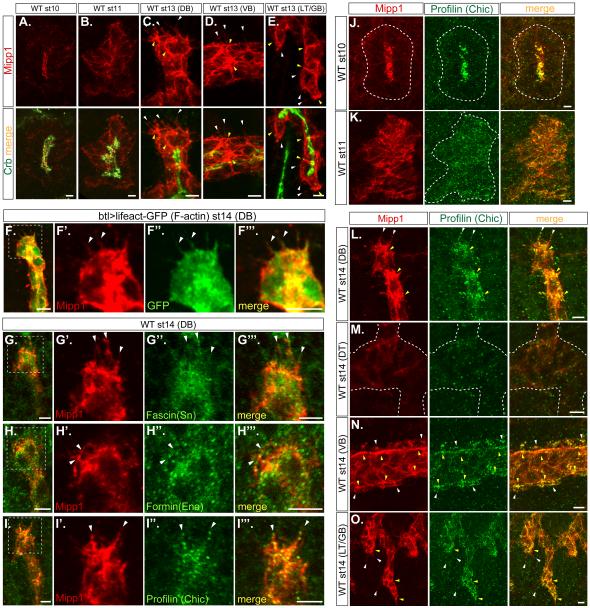
To learn the endogenous cellular localization of Drosophila Mipp1, Mipp1-specific antiserum was generated (Figures S3A-S3E). Mipp1 protein expression (Figures S3F-S3P) paralleled mipp1 mRNA expression (Figures S1A-S1L). Mipp1 localized to the plasma membrane, showing no overlap with the ER marker KDEL (Figure S3Q). During invagination (st10), Mipp1 preferentially localized to the apical surface (Figure 3A). During primary branching (st11), Mipp1 localization shifted to the basolateral domain (Figure 3B). At late stages (st13-15), Mipp1 was enriched at the distal ends of DB (Figure 3C) and LT/GB cells (Figure 3E), the basal side of VB cells (Figure 3D), in cell-cell junctions and in filopodia-like structures of these branches (Figures 3C-3E). Filopodial structures at the distal tip of the DB have been characterized by live in vivo imaging (Okenve-Ramos and Llimargas, 2014; Ribeiro et al., 2002); both F-actin and fascin (Singed) localize to these structures. We observed F-actin (Figure 3F-3F”’), fascin (Singed) (Figures 3G-3G”’) and other filopodia markers – formin (Enable/Vasp) (Figures 3H-3H”’) and profilin (Chickadee) (Figures 3I-3I”’) in the Mipp1-stained-filopodia-like structures, indicating that they are filopodia. Mipp1 overlapped with Chickadee at all stages (Figures 3J-3O); both proteins were highly enriched in the apical domain at st10 (Figure 3J), shifted to basolateral during st11 (Figure 3K), were enriched at cell-cell junctions and distal tips of the DB, VB, and LT/GB at st14 (Figures 3L, 3N, and 3O; yellow arrowheads), and localized to filopodia (Figures 3L, 3N, and 3O; white arrowheads). Similar Mipp1-stained-filopodia-like structures were observed when mipp1 was expressed in the embryonic salivary gland (another epithelial organ) (Figure S4A) and in S2 cells (Figures S4G-S4G”).
Figure 3. Mipp1 localizes to different plasma membrane domains and to filopodia.
(A-E) Mipp1 (red) and apical Crb (green) staining in st10 to st14 embryos (projection images).
(F-I) Mipp1 (red) staining overlaps with filopodial markers, including (F-F”’) lifeact-GFP, an F-actin marker, (G-G”’) Fascin, (H-H”’) Formin, and (I-I”’) Profilin.
(J-O) Mipp1 (red) staining overlaps with Profilin (green) staining in (J) apical domain at st 10, in (K) basolateral domain at st11, in filopodia and in tracheal branches at st14 – (L) DB, (N) VB and (O) LT/GB (projection images).
White arrowheads = filopodia; yellow arrowheads = Mipp1 enrichment; white dash lines = outline of the trachea; white dash boxes = zoom in area; scale bars = 5µm.
See also Figure S3.
FGF signaling regulates Mipp1 expression
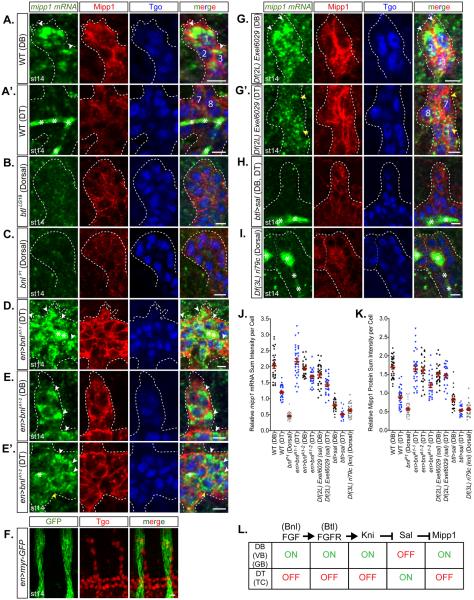
FGF signaling guides tracheal branch migration (Klambt et al., 1992; Ohshiro et al., 2002; Sutherland et al., 1996). The FGF receptor Breathless (Btl) is expressed in tracheal cells, and the FGF ligand Branchless (Bnl) is expressed in tissues towards which tracheal cells migrate. Activated FGF signaling at the tips of migrating branches induces filopodia formation (Ghabrial and Krasnow, 2006; Ribeiro et al., 2002). Since the mipp1 expression pattern is similar to the pattern of activated FGF signaling, we asked if mipp1 expression is downstream of FGF signaling. In WT late stage embryos, mipp1 mRNA was enriched at the distal tip of the DB and detected only at low levels in the DT (Figures 4A-4A’ and 4J-4K). In btlLG19 or bnlP1 mutants, in which branch migration is inhibited and only rudimentary branches form (Klambt et al., 1992; Sutherland et al., 1996), mipp1 mRNA and protein levels were reduced and the large foci of mipp1 accumulation were not observed (Figures 4B-4C). Ectopic overexpression of Bnl in epidermal stripes resulted in high levels of mipp1 in the DT and proximal DB, with large mRNA puncta on the side of cells close to the new source of Bnl (Figures 4D-4F). The high levels of Bnl expression were linked to failures in DB formation (Figure 4D) and elongation (Figure 4E). Knirps (Kni) and Knirps-related (Knrl) are transcription factors downstream of FGF signaling that are required for DB, VB, and LT/GB migration (Chen et al., 1998; Myat et al., 2005). mipp1 mRNA was decreased in Df(3L)ri79c (a deficiency removing both kni and knrl) (Figure 4I). In DB, VB, and LT/GB, Kni/Knrl is also responsible for suppressing Spalt (Sal), the transcription factor that promotes DB elongation and suppresses SCI in the DT (Kuhnlein and Schuh, 1996; Ribeiro et al., 2004). Suppression of Sal is required for DB, VB, and LT/GB elongation by SCI. In Df(2L)Exel6029 (a deficiency removing sal) – DT elongation was blocked, the large foci of mipp1 accumulation at the distal tip of the DB (Figure 4G) and small intense punctate mipp1 staining near the plasma membrane in the DT region (Figure 4G’) was observed. Correspondingly, overexpression of Sal throughout the trachea resulted in low levels of mipp1 mRNA and protein expression in all branches (Fig 4H). Thus, FGF signaling upregulates mipp1 in the DB, VB, and LT/GB, where extensive filopodia form to drive branch migration and elongation by SCI; Spalt suppresses mipp1 expression in the DT, where minimal filopodia form and branches elongate by other mechanisms (Figure 4L).
Figure 4. Expression of mipp1 is activated by FGF signaling and repressed by Spalt.
Fluorescent in situ hybridization of mipp1 mRNA (green) with immunostaining of Mipp1 protein (red) and tracheal nuclear Tgo (blue). The three DB cells located at most distal tip are labeled with 1, 2, and 3. The two DT cells located at the base of DB are labeled with 7 and 8.
(A-A’) WT st14 DB and DT.
(B-C) btl (FGF receptor null) (B) and bnl (FGF ligand null) (C) mutant st14 dorsal trachea. (D-E’) bnl overexpression st14 DB and DT.
(F) en-Gal4 driven myr-GFP expression relative to trachea. (G-G’) sal deficient st14 DB and DT.
(H) kni and knrl deficient st14 dorsal trachea.
(J-K) Quantification of the relative mipp1 mRNA and Mipp1 protein sum intensity per cell for DB and DT using Imaris. Error bars = SEM
(L) Model of the upstream pathway regulating the Mipp1 expression in trachea. * = trapping of green mipp1 signals in the tracheal lumen; white arrowheads = large foci of mipp1 mRNA; yellow arrowheads = small puncta of mipp1 mRNA; white dash lines = outline of the trachea; scale bars = 5µm.
The Mipp1 HP domain faces outside the cell and dephosphorylates extracellular IPs
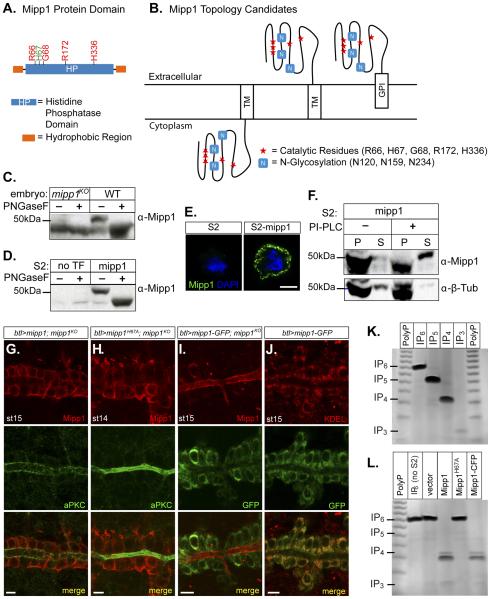
We asked if Mipp1 functions on the intracellular or extracellular side of the plasma membrane. Mipp1 contains an N-terminal signal sequence, a histidine phosphatase (HP) domain, and a putative C-terminal transmembrane (TM) domain (predicted by MemBrain) with a presumptive GPI anchor site (predicted by big-PI Predictor) (Figure 5A). Candidate Mipp1 topologies include a C-terminal TM-tethered protein with an intracellular HP domain and a C-terminal TM-tethered or GPI-anchored protein with an extracellular HP domain (Figure 5B). The Mipp1 HP domain contains three predicted N-glycosylation sites (Figure 5B). N-glycosylation occurs in the ER lumen, which is topologically equivalent to the extracellular side of the plasma membrane. Glycosidase treatment, which removes the N-glycan group, of extracts from WT embryos (Figure 5C) and S2 cells expressing Mipp1 (Figure 5D), revealed a mobility shift on Westerns, indicating that Mipp1 is glycosylated. Thus, the HP domain faces outside the cell. Mipp1 staining of unpermeabilized Mipp1-expressing S2 cells detected Mipp1 on the cell surface, confirming the extracellular localization of Mipp1 (Figure 5E).
Figure 5. Mipp1 is GPI anchored and dephosphorylates extracellular pools of IPs.
(A) Cartoon of Mipp1 protein showing histidine phosphatase (HP) domain and hydrophobic N-and C-termini.
(B) Candidate topologies for Mipp1, which is anchored in the plasma membrane by either its C-terminal hydrophobic span or GPI anchor.
(C-D) Glycosidase treatment of Mipp1 from (C) embryo extracts and from (D) S2 cell extracts.
(E) Extracellular Mipp1 can be detected in unpermeabilized S2 cells expressing Mipp1.
(F) PI-PLC cleavage releases GPI anchored Mipp1 into the extracellular media. P = pellet (cells); S = supernatant (extracellular media)
(G-H) Trachea overexpressing (G) WT mipp1 in a mipp1KO (btl>mipp1; mipp1KO) and (H) enzyme dead mipp1 in a mipp1KO (btl>mipp1H67A; mipp1KO) co-istained with Mipp1 (red) and aPKC (apical marker; green).
(I) Trachea overexpressing mipp1-GFP in a mipp1KO (btl>mipp1-GFP; mipp1KO) co-stained with (I) Mipp1 and GFP and with (J) GFP and KDEL (ER marker).
(J) Trachea overexpressing mipp1-GFP (btl>mipp1-GFP) co-stained with GFP and KDEL (ER marker).
(K) Toludine-blue stained PAGE gel of IP6, IP5, IP4, IP3, and polyphosphate (PolyP) as size indicator.
(L) The reaction products of IP6 added to the media of S2 cells expressing vector, Mipp1, enzyme-dead Mipp1H67A , or Mipp1-CFP for 30 mins.
See also Figure S4.
To learn if Mipp1 is GPI-anchored, we treated S2 cells expressing Mipp1 with PI-PLC, which cleaves the GPI-anchor and releases GPI-anchored proteins on the plasma membrane into the extracellular media. Western analysis revealed the appearance of the Mipp1 protein band in the supernatant fraction upon treatment with PI-PLC (Figure 5F). Thus, Mipp1 is GPI-anchored in S2 cells. To determine whether Mipp1 is also GPI-anchored in vivo, we generated a C-terminal GFP tagged Mipp1 (Mipp1-GFP) and observed localization of the GFP and Mipp1 in a mipp1 null background. GPI modification occurs in the lumen of the ER, where the C-terminal-TM domain of a target protein is cleaved and the remaining portion is transferred to the pre-assembled GPI moiety. If Mipp1 is GPI-anchored in the trachea, we expect the GFP to be in the ER and Mipp1 to be trafficked to the plasma membrane. We observed retention of GFP in the ER (Figures 5I-5J) with Mipp1 localizing to only the apical membrane (Figure 5I). Compared to untagged Mipp1 and untagged enzyme-dead Mipp1H67A, which localized to both apical and basolateral plasma membrane domains (Figures 5G-5H), Mipp1-GFP lacked the basolateral pool. Similar localization patterns were observed with expression of the same UAS constructs in the salivary gland, another polarized epithelial tubular organ that does not normally express mipp1 (Figures S4A-S4E). Deleting the GPI anchor site of Mipp1 also trapped Mipp1 in the ER (Figures S4F and S4K). An explanation for the lack of a basolateral pool of Mipp1-GFP is that the C-terminal-tagged-GFP allows GPI-anchored Mipp1 to be trafficked to the apical membrane but traps the Mipp1 that does not undergo the GPI modification in the ER. If this hypothesis is correct, then the apical pool of Mipp1 is GPI-anchored and the basolateral pool of Mipp1 is likely to be trans-membrane tethered. Alternative splicing can generate an apically localized GPI-anchored isoform and a basolaterally localized transmembrane-tethered isoform of the same protein [example: N-CAM (Powell et al., 1991)] or one protein isoform can either undergo GPI-anchor modification or remain as transmembrane tethered, regulated by an unknown mechanism [example:ULBP2 (Fernandez-Messina et al., 2011)]. Since the three alternative mipp1 mRNAs translate the identical protein sequence, the second scenario is likely for Mipp1.
We next tested if Mipp1 dephosphorylates extracellular IPs. IP6 was added to the media of S2 cells transfected with the vector, Mipp1, enzyme-dead Mipp1H67A, or C-terminal CFP-tagged Mipp1, which all trafficked to the cell surface (Figures S4G-S4K). After 30 min, media was collected and electrophoresed in high percentage polyacrylamide gels; that were stained with toluidine blue to detect the IP bands. Dephosphorylation of IP6 to lower order IPs was observed only in the media from S2-Mipp1 and S2-Mipp1-CFP (Figure 5L). We conclude that Mipp1 HP domain localizes to the extracellular side of the plasma membrane and dephosphorylates extracellular IPs.
Mipp1 affects tracheal morphogenesis
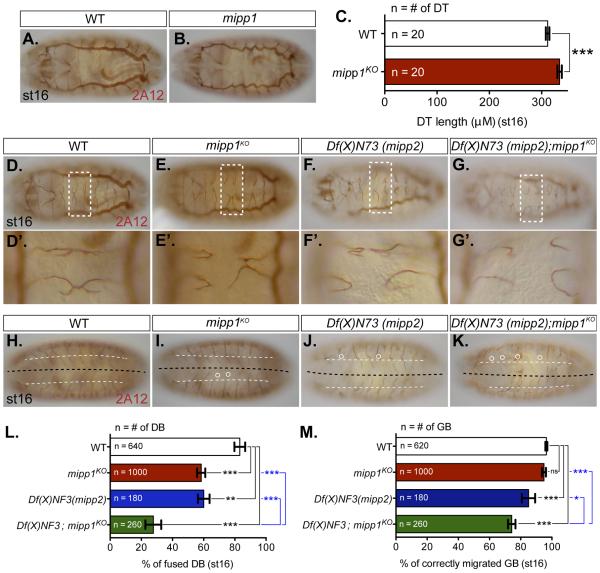
To elucidate the extracellular function of Mipp1 in trachea, we generated a mipp1KO by homologous recombination (Figure S5). Staining with the 2A12 lumenal marker revealed that tracheal morphology was largely normal in mipp1KO, with a few exceptions. In mipp1KO, the DTs were significantly elongated (Figures 6B-6C), the percentage of DBs that had fused with their contralateral partners was significantly decreased (Figures 6E-6E’ and 6L), and a small, statistically insignificant percentage of GBs failed to reach their targets in the CNS (Figures 6I and 6M). Similar tracheal phenotypes were observed in embryos deficient for mipp2 (Df(X)N73) (Figures 6F-F’, 6J, 6L and 6M). Since these embryos had significant DT breaks, we did not measure DT length. Embryos simultaneously deficient for mipp2 and null for mipp1 (Df(X)N73; mipp1KO) exhibited significantly increased severity of DB and GB defects (Figures 6G-6G’ and 6K-6M), suggesting that the two proteins could have redundant tracheal functions.
Figure 6. Loss of mipp perturbs DT tube length, DB fusion and GB migration to CNS.
Luminal morphology of trachea revealed by 2A12 staining (A-B) DT luminal morphology of (A) WT and (B) mipp1KO
(C) Measurement of DT length.
(D-G) DB fusion morphology of (D) WT and (E) mipp1KO, (F) mipp2 deficiency (Df(x)N73), (G) double mutant (Df(x)N73; mipp1KO).
(D’-G’) Zoomed in areas indicated by the white dash boxes.
(H-K) GB migration morphology of (H) WT, (I) mipp1KO, (J) mipp2 deficiency (Df(x)N73), (K) double mutant (Df(x)N73; mipp1KO). Black dashed line = the CNS midline; White dash line = ventral nerve cord (VNC) border; White circle = GB that either does not migrate beyond the VNC border or does not migrate close to the CNS midline and turn posteriorly.
(L-M) Quantification of the percentage of (L) fused DBs and (M) correctly migrated GBs. (error bars = SEM; ns = not significant; * = p-value <0.05; ** = p-value <0.005; *** = p-value <0.0001)
See also Figures S5 and S6.
The elongated DT phenotype has also been observed with mutations in septate junction proteins, in enzymes involved in chitin synthesis, secretion or deacetylation, in apical determinant proteins such as Crb, and in components of the PCP pathway (Chung et al., 2009; Laprise et al., 2010; Luschnig and Uv, 2014; Tonning et al., 2005; Wang et al., 2006). We examined the septate junction proteins Dlg (data not shown) and Coracle (Figures S6B), chitin (Figures S6D), chitin deacetylase (Verm) (Figures S6F), and Crb accumulation (Figures S6H) and did not observe abnormalities in mipp1KO. We also did not observe PCP defects in mipp1KO (data not shown) or with mipp1 overexpression in adult fly wing hair patterns (Figures S6J) or thoracic bristle orientation (data not shown). Thus, Mipp1 likely affects DT length by a novel mechanism.
Unfused DB phenotypes can be caused by a lack of fusion cell fate specification. We confirmed the presence of DB fusion cells at the appropriate position by staining with the fusion marker Dysfusion (Dys) (Figures S6L), ruling out the possibility that DB fusion defects are due to changes in fusion cell fates.
Mipp1 affects sister-cell-intercalation
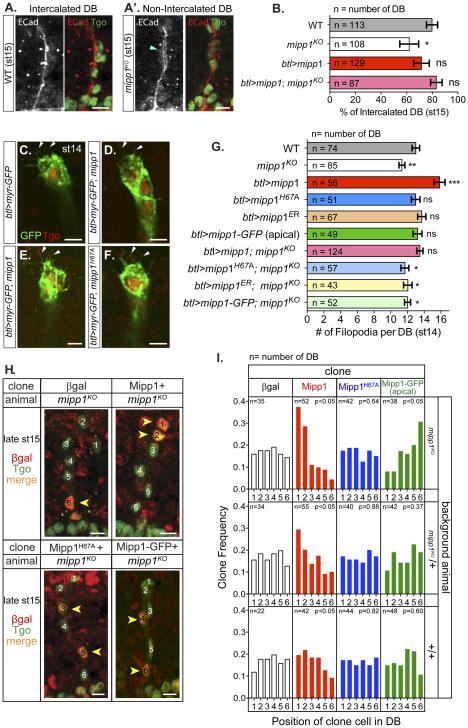
From the Mipp1 expression pattern and the mipp1KO phenotypes in the DB and GB, we speculate that Mipp1 facilitates tracheal branch migration/elongation. Since the DB defect was more pronounced and individual DB cells are easily visualized, we focused on the DB. The DB elongates by SCI, which transforms a multicellular tube to a unicellular tube by rearranging cells from a side-by-side to an end-to-end configuration (Figure S2B) (Ribeiro et al., 2004). During this cell rearrangement, adherens junctions (AJs) are remodeled from intercellular to autocellular. We visualized intercalation by the position of tracheal cells (Tgo) and AJ staining (ECad), which changes from an intercellular AJ staining pattern (ring-staining-pattern) (Figure S2C’) to an autocellular AJ staining pattern (line-staining-pattern) (Figure S2D’). We quantified the intercalation status of each DB at early st15, when the percentage of intercalated DBs in WT was ~80% (Figure 7B). Loss of mipp1 decreased the percentage of intercalated DBs to 62%, a phenotype rescued by expressing mipp1 in the trachea (Figure 7B); the decrease of percentage for successful SCI from 80% for wild type to 62% for mipp1KO closely matched the percentage change in DB fusion events. Overexpression of mipp1 insignificantly decreased the percentage of intercalated DBs, a phenotype likely due to the greater increase of filopodia number in the DB stalk compared to the tip (Figures S7B and S7E). Note that SCI was completely blocked in trachea where high filopodia numbers occurred throughout the DB, as observed with en-Gal4 driven UAS-Bnl expression (Figure S7C-S7D).
Figure 7. Mipp1 promotes SCI, filopodia formation and gives tracheal cells a migratory advantage.
(A-A’) Adherens junctions (ECad) and tracheal nuclei (Tgo) stainings of (A) fully intercalated DB and (A’) non-intercalated DB. Blue arrowhead = ring staining pattern of AJs for non-intercalated DB
(B) Quantification of percentage of intercalated DB for WT, mipp1KO, mipp1 overexpression (btl>mipp1), and mipp1KO rescue with mipp1 (btl>mipp1; mipp1KO).
(C-F) Trachea filopodia detected with btl-driven expression of Myr-GFP in (C) WT, (D) mipp1ko, (E) mipp1 overexpression, and (F) enzyme-dead mipp1 overexpression. White arrowheads = filopodia
(G) Quantification of filopodia number. (error bars = SEM; ns = not significant; * = p-value <0.05; ** = p-value <0.005; *** = p-value <0.0001) when compared to WT
(H) DB cell position of Mipp1-expressing clones. Clones = nuclear βgal (red); tracheal nuclei = Tgo (green) (projection images). One is the most distal and 6 is the most proximal tracheal cell. Yellow arrowheads = βgal marked clones in DB.
(I) Quantification of the frequency of clone distribution in positions 1-6 of the tracheal DB in the different genetic backgrounds.
Scale bars = 5µm.
See also Figures S2 and S7.
Mipp1 facilitates filopodia formation
SCI is induced by the pulling force created by migration of the distal tip of the branch and attachment of the proximal end of the branch to the DT (Figure S2) (Caussinus et al., 2008). Filopodia at the distal tip promote the migration of the DB leading cells (Ribeiro et al., 2002). DB cells are connected though cell-cell junctions, so trailing DB cells collectively migrate with the leading cells and rearrange to elongate the DB. mipp1 is highly enriched at the distal tip of the DB at the onset of filopodia formation (Figure 2H and Figure S1S). The extensive DB filopodia could also be detected by Mipp1 staining (Figure 3C). We determined filopodia number in both mipp1KO embryos and in embryos overexpressing tracheal-specific mipp1 using the membrane marker (Myr-GFP) at st14 (Figures 7C-7G). Filopodia number was decreased in mipp1KO and was increased with overexpression of WT Mipp1 but not overexpression of enzyme-dead Mipp1H67A, ER localized Mipp1ER or apically localized Mipp1-GFP (Figure 7G). Hence, both phosphatase activity and basal/filopodia localization of Mipp1 are required for Mipp1 function in filopodia formation. The reduced filopodia number in mipp1KO was rescued by specifically expressing mipp1 in the trachea (Figure 7G). Thus, Mipp1 is enriched at the distal tip of the DB to facilitate filopodia formation in a phosphatase activity dependent manner.
Mipp1 gives migrating cells the lead
DB migration and elongation occur with cell rearrangement. Since leading DB cells have the highest mipp1 expression, we asked if Mipp1 expression affects position of individual DB cells in the DB. We created βgal, Mipp1, enzyme-dead Mipp1H67A or apically localized Mipp1-GFP clones in a mipp1KO homozygous, heterozygous or WT background at st11, when primary branching initiates, and determined the position of cell clones in the DB at late st15, when DB migration and elongation are mostly complete. Cell clones were marked by nuclear βgal and tracheal cell nuclei were marked by Tango (Tgo) (Figure 7H). The most distal DB cell position was numbered 1 and the most proximal DB cell position was numbered 6. The βgal and mipp1H67A clones were evenly distributed from positions 1 to 6, whereas the Mipp1 clones preferentially localized to a distal position and Mipp1-GFP clones favored a proximal position (Figure 7I). Mipp1 normally localizes to the basolateral domain – the leading edge of migrating cells, whereas Mipp1-GFP localizes Mipp1 to the apical domain – the lagging edge of migrating cells; this switch of Mipp1 to the lagging edge appears to compromise either migration speed or direction. Thus, Mipp1 expression and proper basolateral localization is sufficient to advance migrating DB cells to the leading positions, although the advantage is less notable in trachea expressing the high wild-type levels of Mipp1 in the most distal tracheal cells.
DISCUSSION
Here, we show that mipp1 is highly and dynamically expressed in the developing trachea. Whereas expression occurs initially in all cells, mipp1 mRNA persists in migrating branches, especially in the distal-most cells. There, mipp1 mRNA forms large foci that localize to the side of the cell closest to the direction of migration and filopodia formation. We provide evidence that Drosophila Mipp1 sits on the cell surface where it dephosphorylates extracellular IP6 to IP3. During primary branching, Mipp1 localization shifts from being enriched in the apical membrane to the basolateral domain. During DB elongation and migration, Mipp1 localizes to filopodia and is enriched at the distal tip and cell-cell junctions. We show that Mipp1 facilitates filopodia formation and enhances cell competitiveness for the lead position of the DB in a phosphatase-activity dependent manner, suggesting either that local depletion of extracellular IP6 or local increases in lower order IP intermediates (IP5, IP4) or product (IP3) promote collective tracheal cell migration and branch elongation.
The extracellular localization of Drosophila Mipp1 raises the question of where Mipp proteins function in other organisms. In both mammals and Dictyostelium, Mipp has been suggested to affect intracellular IP profiles (Chi et al., 2000; King et al., 2010). In Dictyostelium, Li+ treatment reduces the speed and linear migration path of cAMP-dependent chemotaxis by lowering IP3 production, cytosolic Ca2+ levels and inositol synthesis (Harwood, 2011). Li+ is proposed to exert its effects through prolyl oligopeptidase (PO), since loss of this enzyme confers Li+ resistance. Because altering Mipp1 function increases sensitivity to Li+, it has been suggested that PO reduces cellular IP3 levels by blocking the activity of Mipp1. A model has been proposed suggesting a role for Mipp1-dependent increases in cellular IP3 levels in elevating transcription of enzymes required for inositol production (King et al., 2010). One problem with this model is that both loss and overexpression of Mipp1 makes Dictyostelium cells hypersensitive to Li+ treatment (King et al., 2010). Moreover, Dictyostelium Mipp1 contains a putative N-terminal signal peptide and C-terminal transmembrane domain, suggesting that it enters the secretory pathway and traffics to the plasma membrane where it faces the extracellular space, just like the Drosophila protein. We suggest that Mipp1 loss and overexpression may independently exacerbate the inhibitory effects of Li+ on cell migration by affecting the polarized distribution of Mipp1 substrates and/or reaction products in the immediate vicinity of migrating cells.
Recent studies in lung carcinoma cells suggest that mammalian Mipp1 also converts extracellular IP6 to IP3 (Windhorst et al., 2013). In these cells, Mipp1 localizes not only to the ER but also to Golgi/plasma membrane fractions and in the supernatant. Moreover, the extracellular/secreted Mipp1 could dephosphorylate extracellular IP6 to IP3 (Windhorst et al., 2013). The author suggested that the internalized IP6 was converted to IP5 in the lysosome, an organelle whose acidic environment is inconsistent with the optimum pH range of 7.0 to 7.8 for Mipp1 activity. It is thus likely that other enzymes are involved in the dephosphorylation of the internalized IP6. With RNAi knockdown of mipp1, extracellular conversion of IP6 to IP3 was more reduced compared to the intracellular dephosphorylation. Altogether, these findings suggest that extracellular Mipp activity exists in Drosophila, in Dictyostelium, and in mammalian systems.
IP6 is a key substrate of Mipp and is a well-known anti-tumor agent. High concentrations of extracellular IP6 (>1mM) inhibit migration and proliferation of tumor cells (Shamsuddin et al., 1997). Breast cancer cells treated with IP6 round up and exhibit decreased plating efficiency through down-regulation of integrin expression (Tantivejkul et al., 2003a, b). IP6 inhibition of invasive migration in vitro in matrigel or in vivo during metastasis has also been demonstrated (Tantivejkul et al., 2003a; Ullah and Shamsuddin, 1990; Vucenik et al., 1992). These findings suggest that a critical role for Mipp1 could be to remove local pools of IP6 to allow for attachment and cell migration.
A potential role for extracellular IP6 has been demonstrated in vitro (Morrison et al., 1994). Heparin association with FGF facilitates FGF binding to FGF receptor for signal activation (Spivak-Kroizman et al., 1994). IP6 can also bind FGF due to structural similarities between IP6 and the saccharide residues on heparin. IP6 binding to FGF blocks FGF association with heparin and inhibits FGF signal activation (Morrison et al., 1994). In the trachea, cells expressing the highest levels of FGF receptor outcompete other cells for the lead position in the migrating DB (Ghabrial and Krasnow, 2006). Here, we have shown that expression of Mipp1 similarly increases the competitiveness of DB cells for the leading position. Mipp1 may deplete local pools of extracellular IP6, thus enhancing local FGF activity in the distal tip cells of the migrating DB. Alternatively, Mipp1 at the tips of migrating cells may change the local extracellular IP profile and extracellular IPs may bind to other extracellular proteins or receptors to affect filopodia formation during cell migration.
Early Mipp1 at the apical domain might also promote filopodia formation. We observed significant overlap between Mipp1 and known filopodia markers in the apical domain during invagination. Our lab observed dynamic actin-rich filopodia-like extensions in the apical domain of the invaginating salivary gland (unpublished), so perhaps similar structures form in invaginating tracheal cells. Filopodia at the apical domain might sense the extracellular environment to control the size of the forming apical lumen, perhaps explaining the other tracheal phenotype we observe with mipp1 loss – over-elongation of the DT.
In conclusion, we show that Drosophila Mipp1 functions extracellularly to facilitate filopodia formation to drive tracheal tube elongation. This single target of FGF signaling provides the same advantage to migrating cells as having the highest levels of FGF signal. Importantly, Mipp1 localization to the external surface of the plasma membrane also solves the enigma of how Mipps access their IP substrates. We expect that our demonstration that extracellular pools of IPs affect tissue morphogenesis will fuel extensive research into how cells perceive and respond to such molecules.
EXPERIMENTAL PROCEDURES
Drosophila strains
btlLG19, bnlP1, Df(2L)Exel6029, Df(3L)ri79c, Df(X)N73, da-GAL4, en-GAL4, UAS-myr-GFP, UAS-lifeact-GFP, AyGAL-FRT; UAS-lacZ, and hsFLP were obtained from Bloomington. btl-GAL4 was provided by S. Hayashi. UAS bnlA1-1 and UAS bnlA1-1 were provided by M. Krasnow. sage-GAL4 was created by our lab (Chung et al., 2009).
Generation of mipp1KO and transgenic mipp1 flies
The mipp1 ORF was replaced by w+ by homologous recombination (Gong and Golic, 2003). mipp1KO was confirmed by PCR and in situ hybridization. UAS lines were created using the Drosophila Gateway Vector system (Carnegie). The mipp1 ORF was PCR amplified and cloned into pTW and pTWG to generate UAS-mipp1 and UAS-mipp1-GFP, respectively. Enzyme-dead Mipp1 was made by mutating the histidine at position 67 to an alanine and GPI-anchor-site-deleted Mipp1 was made by deleting amino acids 439 to 444, using the QuickChange II Site-Directed Mutagenesis Kit (Stratagene). All cloning details and primer sequences are available upon request.
Generation of Mipp1 antiserum
Antiserum was made against full length Mipp1 excluding the N-terminal 15 amino acids. The mipp1 ORF from nucleotide 46 to 1404 was cloned into pET28b vector (Novagen). Protein was purified from Mipp1-expressing BL21 cells by the side-strip method with electro-elution and lyophilization as described (Steiner, 1989). Guinea pig polyclonal Mipp1 antiserum was generated (Covance, Inc.).
in situ hybridization
Whole mount in situ hybridization and fluorescence in situ hybridization were performed as described (Knirr et al., 1999; Lehmann and Tautz, 1994) using an anti-sense digoxygenin-labeled mipp1 RNA probe generated from cDNA GM09242. sano and trh probes were generated in our lab (Chung et al., 2011; Chung et al., 2009).
Embryo Immunostaining
Embryos were formaldehyde fixed, and immunofluorescence and biotin-conjugated HRP immunohistochemistry were performed as described (Reuter et al., 1990). For ECad staining, embryos were fixed in formaldehyde-saturated heptane and devitellinized with EtOH. For Mipp1, Singed, Enable, and Chickadee staining, embryos were fixed in formaldehye or in Bouin’s solution (Sigma) and devitellinized with MeOH. Mipp1-stained filopodia could be detected with Bouin’s but not formaldehyde fixation. Mipp1 antiserum was used at 1:500 for immunofluorescence and 1:10,000 for HRP immunostaining. Primary antibodies used for immunostaining were mouse 2A12 (1:10, DHSB), mouse α-βgal (1:500, Promega), mouse α-Chic (1:10, DHSB), mouse α-Crb (1:10, DHSB), guinea pig α-Cor (1:2000, R. Fehon), mouse α-Dlg (1:500, DHSB), rabbit α-Dys (1:800, S. Crews), rat α-DE-Cad (1:10, DHSB), mouse α-Ena (1:20, DHSB), rabbit α-GFP (1:10,000, Invitrogen), rabbit α-aPKC (1:200, Santa Cruz), rabbit α-SAS (1:500, D. Cavener), mouse α-Sn (1:10, DHSB), rat α-PH4αSG1 (1:500, Abrams et al., 2006), guinea pig α-Verm (1:500, C. Samakovlis), WGA-488 (1:1000, Invitrogen). Fluorescently-labeled (Alexa 488, 555, 647) secondary antibodies were used at a dilution of 1:500 (Invitrogen). Images were obtained with LSM 510 confocal microscope (Carl Zeiss). Immunohistochemical images were taken with an Axiophot light microscope (Carl Zeiss) equipped with ProgRes C14PLUS image capture system. Statistical analysis were performed with unpaired t-test.
Generation of mutant cell clones
Fly lines used: 1) AyGAL-FRT, UAS-lacZ; mipp1KO, 2) hs-FLP; mipp1KO, 3) hs-FLP; mipp1KO, UAS mipp1, 4) hs-FLP; mipp1KO, UAS mipp1H67A, 5) hs-FLP; mipp1KO, UAS mipp1-GFP, 6) AyGAL-FRT, UAS-lacZ, 7) hs-FLP, 8) hs-FLP; UAS mipp1, 9) hs-FLP; UAS mipp1H67A, 10) hs-FLP; UAS mipp1-GFP. Embryos were collected for 2 hrs at 25 °C, aged for 5 hrs at 25 °C, heat-shocked for 45 min at 37 °C, and developed for 8 hrs at 25 °C.
Western Blots
Western blot for embryos and S2 cells were performed as described (Abrams et al., 2013; Ismat et al., 2013). Samples were separated on an 8% SDS-PAGE gel. Guinea pig α-Mipp1 (1:10,000) and mouse α-βtub (1:1000, DHSB) and HRP-conjugated secondary antibodies (1:10,000, Invitrogen) were used.
Transfection of S2 cells with mipp1 constructs
The mipp1, mipp1H67A, mipp1-CFP, mipp1ER constructs for S2 cell transfection were generated using the Drosophila Gateway Vector system (Carnegie) with the pAW or pAWC vector. Transient transfections of S2 were performed with Effectene Transfection Reagent (Sigma) following the manufacturer’s protocol.
S2 cell immunofluorescence
S2 cell immunofluorescence was performed as described (Rogers and Rogers, 2008), except PBS was used in the initial washes through primary antibody incubation to minimize cell permeability for better detection of membrane localized Mipp1. Guinea pig α-Mipp1 (1:1000), fluorescently-labeled (Alexa 488 or 555) secondary antibodies (1:1000, Invitrogen), phalloidin Alexa 546 (1:500 Invitrogen), and DAPI (1:1000) were used.
Mipp1 immunostaining of non-permeabilized cells
S2 cells were plated on ConA (0.5mg/ml) coverslips for 1hr at RT, then transferred to a 4°C cold room and kept on ice. Cells were blocked with 5% NGS:PBS for 5 min and incubated with primary antibodies in 5% NGS:PBS for 30 min on ice. Cells were fixed with 4% formaldehyde in PBS at RT for 10 min, washed with PBTriton at RT, incubated with secondary antibodies in 5% NGS:PBTriton for 1hr at RT and washed with PBTriton at RT.
Glycosidase Digestion
Embryo and S2 cell lysates were digested with PNGase-F (New England Biolabs) for 1hr at 37°C, according to the manufacturer’s protocol, and run on SDS-PAGE. Protein bands were detected by Westerns.
PI-PLC Treatment
S2 cells were incubated with PI-PLC (Sigma) at a concentration 0.2 UN/106 cell in 1X PBS at 37°C for 1hr. The cell pellet and supernatant were separated by centrifugation at 14000 rpm for 5 min. Both the cell pellet and supernatant samples were run on SDS-PAGE and analyzed by Westerns. All protein bands in the supernatant fraction were shifted slightly upward compared to proteins bands in the cell pellet fraction.
Extracellular dephosphorylation of IP6 by Mipp1
S2 cells were transfected with the pAW vector, mipp1-pAW, mipp1H67A-pAW or mipp1-pAWC. 200 μM IP6 was incubated with 5X 106/100µl of S2 cells in PBS for 30min at 25 °C. Cells were centrifuged at 14,000 rpm for 10 min and the supernatant was kept. IPs in the supernatant were separated on a 37% PAGE gel overnight at 4°C and stained with toluidine blue, as previously described (Losito et al., 2009). Standard IPs (I(1,3,4,5,6)P5, I(1,3,4,5)P4, and I(1,4,5)P3) were obtained commercially from Cayman Chemical.
Supplementary Material
Acknowledgements
We thank C. Machamer, J. Gagnon and the lab of S. Snyder for advice on Mipp1 topology, GPI anchor addition and activity assays. We thank S.Y. Chung, R.M. Fox, C.D. Hanlon, T. Inoue, R. Loganathan, G. Seydoux and M. Wells for critiquing the manuscript. This work was supported by NIH grants RO1 DE012873 (D.J.A.) and F31 DE021285 (Y.L.C.).
Footnotes
Author Contributions
Y.L.C. and D.J.A. conceived the experiments. Y.L.C. did the experiments. Y.L.C. and D.J.A. interpreted the experiments and wrote the paper.
References
- Abrams EW, Cheng YL, Andrew DJ. Drosophila KDEL Receptor Function in the Embryonic Salivary Gland and Epidermis. Plos One. 2013;8 doi: 10.1371/journal.pone.0077618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrams EW, Mihoulides WK, Andrew DJ. Fork head and Sage maintain a uniform and patent salivary gland lumen through regulation of two downstream target genes, PH4 alpha SG1 and PH4 alpha SG2. Development. 2006;133:3517–3527. doi: 10.1242/dev.02525. [DOI] [PubMed] [Google Scholar]
- Ali N, Craxton A, Shears SB. Hepatic Ins(1,3,4,5)P4 3-Phosphatase Is Compartmentalized inside Endoplasmic-Reticulum. Journal of Biological Chemistry. 1993;268:6161–6167. [PubMed] [Google Scholar]
- Caussinus E, Colombelli J, Affolter M. Tip-Cell Migration Controls Stalk-Cell Intercalation during Drosophila Tracheal Tube Elongation. Current Biology. 2008;18:1727–1734. doi: 10.1016/j.cub.2008.10.062. [DOI] [PubMed] [Google Scholar]
- Chen CK, Kuhnlein RP, Eulenberg KG, Vincent S, Affolter M, Schuh R. The transcription factors KNIRPS and KNIRPS RELATED control cell migration and branch morphogenesis during Drosophila tracheal development. Development. 1998;125:4959–4968. doi: 10.1242/dev.125.24.4959. [DOI] [PubMed] [Google Scholar]
- Chi HB, Yang XN, Kingsley PD, O'Keefe RJ, Puzas JE, Rosier RN, Shears SB, Reynolds PR. Targeted deletion of Minpp1 provides new insight into the activity of multiple inositol polyphosphate phosphatase in vivo. Mol Cell Biol. 2000;20:6496–6507. doi: 10.1128/mcb.20.17.6496-6507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Chavez C, Andrew DJ. Trachealess (Trh) regulates all tracheal genes during Drosophila embryogenesis. Dev Biol. 2011;360:160–172. doi: 10.1016/j.ydbio.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Vining MS, Bradley PL, Chan CC, Wharton KA, Jr., Andrew DJ. Serrano (sano) functions with the planar cell polarity genes to control tracheal tube length. Plos Genet. 2009;5:e1000746. doi: 10.1371/journal.pgen.1000746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craxton A, Ali N, Shears SB. Comparison of the activities of a multiple inositol polyphosphate phosphatase obtained from several sources: a search for heterogeneity in this enzyme. Biochem J. 1995;305(Pt 2):491–498. doi: 10.1042/bj3050491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craxton A, Caffrey JJ, Burkhart W, Safrany ST, Shears SB. Molecular cloning and expression of a rat hepatic multiple inositol polyphosphate phosphatase. Biochem J. 1997;328(Pt 1):75–81. doi: 10.1042/bj3280075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Messina L, Ashiru O, Aguera-Gonzalez S, Reyburn HT, Vales-Gomez M. The human NKG2D ligand ULBP2 can be expressed at the cell surface with or without a GPI anchor and both forms can activate NK cells. J Cell Sci. 2011;124:321–327. doi: 10.1242/jcs.076042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghabrial AS, Krasnow MA. Social interactions among epithelial cells during tracheal branching morphogenesis. Nature. 2006;441:746–749. doi: 10.1038/nature04829. [DOI] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosphila. P Natl Acad Sci USA. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grases F, Costa-Bauza A, Prieto RM. Intracellular and extracellular myo-inositol hexakisphosphate (InsP(6)), from rats to humans. Anticancer Res. 2005;25:2593–2597. [PubMed] [Google Scholar]
- Harwood AJ. Prolyl oligopeptidase, inositol phosphate signalling and lithium sensitivity. CNS & neurological disorders drug targets. 2011;10:333–339. doi: 10.2174/187152711794653779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvine RF, Schell MJ. Back in the water: The return of the inositol phosphates. Nat Rev Mol Cell Bio. 2001;2:327–338. doi: 10.1038/35073015. [DOI] [PubMed] [Google Scholar]
- Ismat A, Cheshire AM, Andrew DJ. The secreted AdamTS-A metalloprotease is required for collective cell migration. Development. 2013;140:1981–1993. doi: 10.1242/dev.087908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King J, Keim M, Teo R, Weening KE, Kapur M, McQuillan K, Ryves J, Rogers B, Dalton E, Williams RS, et al. Genetic control of lithium sensitivity and regulation of inositol biosynthetic genes. Plos One. 2010;5:e11151. doi: 10.1371/journal.pone.0011151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klambt C, Glazer L, Shilo BZ. breathless, a Drosophila FGF receptor homolog, is essential for migration of tracheal and specific midline glial cells. Gene Dev. 1992;6:1668–1678. doi: 10.1101/gad.6.9.1668. [DOI] [PubMed] [Google Scholar]
- Knirr S, Azpiazu N, Frasch M. The role of the NK-homeobox gene slouch (S59) in somatic muscle patterning. Development. 1999;126:4525–4535. doi: 10.1242/dev.126.20.4525. [DOI] [PubMed] [Google Scholar]
- Kuhnlein RP, Schuh R. Dual function of the region-specific homeotic gene spalt during Drosophila tracheal system development. Development. 1996;122:2215–2223. doi: 10.1242/dev.122.7.2215. [DOI] [PubMed] [Google Scholar]
- Kumar V, Sinha AK, Makkar HPS, Becker K. Dietary roles of phytate and phytase in human nutrition: A review. Food Chem. 2010;120:945–959. [Google Scholar]
- Laprise P, Paul SM, Boulanger J, Robbins RM, Beitel GJ, Tepass U. Epithelial polarity proteins regulate Drosophila tracheal tube size in parallel to the luminal matrix pathway. Current biology: CB. 2010;20:55–61. doi: 10.1016/j.cub.2009.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R, Tautz D. In-Situ Hybridization to Rna. Method Cell Biol. 1994;44:575–598. doi: 10.1016/s0091-679x(08)60933-4. [DOI] [PubMed] [Google Scholar]
- Losito O, Szijgyarto Z, Resnick AC, Saiardi A. Inositol Pyrophosphates and Their Unique Metabolic Complexity: Analysis by Gel Electrophoresis. Plos One. 2009;4 doi: 10.1371/journal.pone.0005580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luschnig S, Uv A. Luminal matrices: an inside view on organ morphogenesis. Exp Cell Res. 2014;321:64–70. doi: 10.1016/j.yexcr.2013.09.010. [DOI] [PubMed] [Google Scholar]
- Maruyama R, Andrew DJ. Drosophila as a model for epithelial tube formation. Dev Dyn. 2012;241:119–135. doi: 10.1002/dvdy.22775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison RS, Shi E, Kan M, Yamaguchi F, McKeehan W, Rudnicka-Nawrot M, Palczewski K. Inositolhexakisphosphate (InsP6): an antagonist of fibroblast growth factor receptor binding and activity. In vitro cellular & developmental biology Animal. 1994;30A:783–789. doi: 10.1007/BF02631302. [DOI] [PubMed] [Google Scholar]
- Myat MM, Lightfoot H, Wang P, Andrew DJ. A molecular link between FGF and Dpp signaling in branch-specific migration of the Drosophila trachea. Dev Biol. 2005;281:38–52. doi: 10.1016/j.ydbio.2005.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshiro T, Emori Y, Saigo K. Ligand-dependent activation of breathless FGF receptor gene in Drosophila developing trachea. Mechanisms of development. 2002;114:3–11. doi: 10.1016/s0925-4773(02)00042-4. [DOI] [PubMed] [Google Scholar]
- Okenve-Ramos P, Llimargas M. Fascin links Btl/FGFR signalling to the actin cytoskeleton during Drosophila tracheal morphogenesis. Development. 2014;141:929–939. doi: 10.1242/dev.103218. [DOI] [PubMed] [Google Scholar]
- Powell SK, Cunningham BA, Edelman GM, Rodriguezboulan E. Targeting of Transmembrane and Gpi-Anchored Forms of N-Cam to Opposite Domains of a Polarized Epithelial-Cell. Nature. 1991;353:76–77. doi: 10.1038/353076a0. [DOI] [PubMed] [Google Scholar]
- Reuter R, Panganiban GEF, Hoffmann FM, Scott MP. Homeotic Genes Regulate the Spatial Expression of Putative Growth-Factors in the Visceral Mesoderm of Drosophila Embryos. Development. 1990;110:1031–1040. doi: 10.1242/dev.110.4.1031. [DOI] [PubMed] [Google Scholar]
- Ribeiro C, Ebner A, Affolter M. In vivo Imaging reveals different cellular functions for FGF and Dpp signaling in tracheal branching morphogenesis. Developmental cell. 2002;2:677–683. doi: 10.1016/s1534-5807(02)00171-5. [DOI] [PubMed] [Google Scholar]
- Ribeiro C, Neumann M, Affolter M. Genetic control of cell intercalation during tracheal morphogenesis in Drosophila. Current Biology. 2004;14:2197–2207. doi: 10.1016/j.cub.2004.11.056. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC. Culture of Drosophila S2 cells and their use for RNAi-mediated loss-of-function studies and immunofluorescence microscopy. Nature protocols. 2008;3:606–611. doi: 10.1038/nprot.2008.18. [DOI] [PubMed] [Google Scholar]
- Sasakawa N, Sharif M, Hanley MR. Metabolism and biological activities of inositol pentakisphosphate and inositol hexakisphosphate. Biochemical pharmacology. 1995;50:137–146. doi: 10.1016/0006-2952(95)00059-9. [DOI] [PubMed] [Google Scholar]
- Shamsuddin AM, Vucenik I, Cole KE. IP6: a novel anti-cancer agent. Life sciences. 1997;61:343–354. doi: 10.1016/s0024-3205(97)00092-1. [DOI] [PubMed] [Google Scholar]
- Singh RP, Agarwal C, Agarwal R. Inositol hexaphosphate inhibits growth, and induces G1 arrest and apoptotic death of prostate carcinoma DU145 cells: modulation of CDKI-CDK-cyclin and pRb-related protein-E2F complexes. Carcinogenesis. 2003;24:555–563. doi: 10.1093/carcin/24.3.555. [DOI] [PubMed] [Google Scholar]
- Spivak-Kroizman T, Lemmon MA, Dikic I, Ladbury JE, Pinchasi D, Huang J, Jaye M, Crumley G, Schlessinger J, Lax I. Heparin-induced oligomerization of FGF molecules is responsible for FGF receptor dimerization, activation, and cell proliferation. Cell. 1994;79:1015–1024. doi: 10.1016/0092-8674(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Steiner L. Antibodies - a Laboratory Manual - Harlow,E, Lane,D. Nature. 1989;341:32–32. [Google Scholar]
- Sutherland D, Samakovlis C, Krasnow MA. Branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 1996;87:1091–1101. doi: 10.1016/s0092-8674(00)81803-6. [DOI] [PubMed] [Google Scholar]
- Tantivejkul K, Vucenik I, Shamsuddin AM. Inositol hexaphosphate (IP6) inhibits key events of cancer metastasis: I. In vitro studies of adhesion, migration and invasion of MDA-MB 231 human breast cancer cells. Anticancer Res. 2003a;23:3671–3679. [PubMed] [Google Scholar]
- Tantivejkul K, Vucenik I, Shamsuddin AM. Inositol hexaphosphate (IP6) inhibits key events of cancer metastasis: II. Effects on integrins and focal adhesions. Anticancer Res. 2003b;23:3681–3689. [PubMed] [Google Scholar]
- Tonning A, Hemphala J, Tang E, Nannmark U, Samakovlis C, Uv A. A transient luminal chitinous matrix is required to model epithelial tube diameter in the Drosophila trachea. Developmental cell. 2005;9:423–430. doi: 10.1016/j.devcel.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Ullah A, Shamsuddin AM. Dose-dependent inhibition of large intestinal cancer by inositol hexaphosphate in F344 rats. Carcinogenesis. 1990;11:2219–2222. doi: 10.1093/carcin/11.12.2219. [DOI] [PubMed] [Google Scholar]
- Verbsky J, Lavine K, Majerus PW. Disruption of the mouse inositol 1,3,4,5,6-pentakisphosphate 2-kinase gene, associated lethality, and tissue distribution of 2-kinase expression. Proc Natl Acad Sci U S A. 2005;102:8448–8453. doi: 10.1073/pnas.0503656102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucenik I, Shamsuddin AM. Cancer inhibition by inositol hexaphosphate (IP6) and inositol: From laboratory to clinic. J Nutr. 2003;133:3778S–3784S. doi: 10.1093/jn/133.11.3778S. [DOI] [PubMed] [Google Scholar]
- Vucenik I, Shamsuddin AM. Protection against cancer by dietary IP6 and inositol. Nutrition and cancer. 2006;55:109–125. doi: 10.1207/s15327914nc5502_1. [DOI] [PubMed] [Google Scholar]
- Vucenik I, Tomazic VJ, Fabian D, Shamsuddin AM. Antitumor activity of phytic acid (inositol hexaphosphate) in murine transplanted and metastatic fibrosarcoma, a pilot study. Cancer Lett. 1992;65:9–13. doi: 10.1016/0304-3835(92)90206-b. [DOI] [PubMed] [Google Scholar]
- Wang S, Jayaram SA, Hemphala J, Senti KA, Tsarouhas V, Jin H, Samakovlis C. Septate-junction-dependent luminal deposition of chitin deacetylases restricts tube elongation in the Drosophila trachea. Current biology : CB. 2006;16:180–185. doi: 10.1016/j.cub.2005.11.074. [DOI] [PubMed] [Google Scholar]
- Windhorst S, Lin H, Blechner C, Fanick W, Brandt L, Brehm MA, Mayr GW. Tumour cells can employ extracellular Ins(1,2,3,4,5,6)P(6) and multiple inositol-polyphosphate phosphatase 1 (MINPP1) dephosphorylation to improve their proliferation. Biochem J. 2013;450:115–125. doi: 10.1042/BJ20121524. [DOI] [PubMed] [Google Scholar]
- York JD. Regulation of nuclear processes by inositol polyphosphates. Bba-Mol Cell Biol L. 2006;1761:552–559. doi: 10.1016/j.bbalip.2006.04.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.