Abstract
Gastric cancer is one of the most prevalent and aggressive cancers worldwide, and its molecular mechanism remains largely elusive. Here we report the genomic landscape in primary gastric adenocarcinoma of human, based on the complete genome sequences of five pairs of cancer and matching normal samples. In total, 103,464 somatic point mutations, including 407 nonsynonymous ones, were identified and the most recurrent mutations were harbored by Mucins (MUC3A and MUC12) and transcription factors (ZNF717, ZNF595 and TP53). 679 genomic rearrangements were detected, which affect 355 protein-coding genes; and 76 genes show copy number changes. Through mapping the boundaries of the rearranged regions to the folded three-dimensional structure of human chromosomes, we determined that 79.6% of the chromosomal rearrangements happen among DNA fragments in close spatial proximity, especially when two endpoints stay in a similar replication phase. We demonstrated evidences that microhomology-mediated break-induced replication was utilized as a mechanism in inducing ~40.9% of the identified genomic changes in gastric tumor. Our data analyses revealed potential integrations of Helicobacter pylori DNA into the gastric cancer genomes. Overall a large set of novel genomic variations were detected in these gastric cancer genomes, which may be essential to the study of the genetic basis and molecular mechanism of the gastric tumorigenesis.
Keywords: gastric cancer, next-generation sequencing, bioinformatics, genomic variations, cancer mutations
Gastric cancer is the second leading cause of cancer-associated death worldwide. While the invasion by Helicobacter pylori is believed to be tightly associated with the occurrence of the cancer, the molecular mechanisms of the cancer’s formation and progression remain largely elusive. The high-throughput sequencing technology could help to reveal new insights about the genomic predisposition for the disease and cancer-related genomic changes induced by H. pylori or other causes, providing genomic level information about the mechanism of the cancer. A number of large-scale genomic analyses have been published on gastric cancer, including the most recent whole-genome and exome sequencing studies identifying recurrent RHOA mutations in diffuse-type gastric cancer1,2 and novel mutations in chromatin remodeling gene ARID1A,3 a genome-wide association study that reported two suspicious loci associated with non-cardia gastric cancers,4 and others.5–7 These published data revealed a high degree of heterogeneity among the considered gastric cancer of different subtypes and very little consistent genomic alterations were observed across different individuals. The most exciting discovery so far is the recurrent RHOA mutation observed in 25.3% of the tested diffuse-type gastric cancers.1 Instead of focusing on the identification of recurrent mutations in gastric cancer, our goal here is to uncover the comprehensive genomic landscape of gastric adenocarcinoma, particularly novel structural variations (SV) through in-depth sequencing data analysis and investigate the possible causes for these genomic alterations through several association analyses. Specifically, we performed a whole-genome analysis on five pairs of gastric adenocarcinoma and matching controls and attempted to elucidate how the genomic changes may have arisen in the tumors and their possible roles in cancer progression, particularly through studying the associations between the identified genomic changes and cancer-causing factors relevant to impaired DNA repair, H. pylori integration and functional selection.
Materials and Methods
A full description of the methods is provided in Supporting Information Methods while a brief synopsis follows.
Samples preparation and whole genome sequencing
High-molecular weight genomic DNA was extracted from surgically resected gastric cancer tissues and the matching blood samples, and was purified using (Qiagen, MD, USA) according to the manufacturer’s instructions. Whole genome DNA sequencing was performed by “unchained combinatorial probe anchor ligation sequencing”, as described8. The resulting mate-paired reads were mapped to the human reference genome (NCBI Build 37) with the average coverage greater than 60× (Supporting Information Table S2). Overall, 97% of the human reference genome was fully called.
Mutation detection
Variant calls for each sample genome with respect to the reference were made as described8. Somatic mutations between tumor and control genomes were obtained by comparing their variant calls using calldiff-1.3 (http://cgatools.sourceforge.net) with somatic scores. After pooling the data from all five patients, an empirical threshold score of 0.1 was used to obtain mutations of high confidence at ~90 sensitivity and FDR at 0.8%. Mutations were then annotated in term of their effects on transcripts using the variant effect predictor tool.9 Gene annotation was done against the RefSeq transcripts, protein functional domains from Pfam,10 noncoding RNA annotations from literature and database search and microRNA from miRBase.11 In order to calculate genome-wide mutation rates, we define a base as covered if there are at least 14 and 8 reads that overlap the position in the tumor and control sample, respectively, and consider only mutations called at the covered positions. As a result, we estimated the average genome-wide mutation rate to be 8.7 per Mb. For each gene, the mutation rate is adjusted by the expected mutation rate according to the gene length. We normalized the mutation frequency in each samples against the gene length and ranked them in the decreasing order of the frequency divided by the expected frequency (Supporting Information Table S12).
SVs and copy number variations detection
As structural variants will create anomalous junctions, i.e., the fusion points that are not linked in the reference genome, the key step for this are to identify all junctions based on the discordant reads. First, all uniquely mapped reads from the whole genome sequence were used to estimate if the mate-pairs reads fall into the normal range of pair-span (500 bps) and the penalties for mismatches and indels. The discordant mate-pairs were defined as either (i) mate-span beyond normal range or (ii) with a discordant orientation. Adjacent discordant reads (within 500 bps) with the same orientation were merged to make a discordant reads cluster, representing a junction, as illustrated in Supporting Information Figure S4. Subsequently, cgatools was used to rationalize set of junctions into events (details in Supporting Information Methods). For copy number variations (CNVs) detection, DNA reads were binned at 2 kbps and 100 kbps intervals along the control and tumor genomes, respectively. The ratio of counts per bin in the tumor and its control, log2 transformed, was calculated as the raw measure of copy number changes and then was corrected with respect to the GC content. The adjusted values were segmented into discrete blocks of uniform copy numbers using the CBS algorithm from the Bioconductor package DNAcopy. Segments with a log2 ratio ≤−0.15 were considered as regions of copy loss whereas segments with a log2 ratio ≥0.15 were defined as copy gain regions.
In addition, major mutations and SVs discussed in the main text were validated by PCR followed by Sanger resequencing, in conjunction with gene expression data we previously collected on 80 pairs of gastric cancer samples along with their adjacent noncancerous control12 (GEO database under accession number GSE27342). More details can be found in Supporting Information Methods.
Spatial features of structural rearrangements in the chromosomes
Several sets of Hi-C data were analyzed to derive pairwise distance information in the folded human chromosomes on different cell types, including lymphoblastoid cell, erythroleukemia cell, human embryonic stem cells and human IMR90 fibroblasts. Pairwise contact frequencies were measured by the Hi-C to represent the distance between two DNA segments in the chromosomes, at 10 Mbps and 40 kbps resolutions for different data sets. Pearson correlation was used to assess the spatial proximity among DNA fragments based on the idea that the neighboring DNA fragments should in general share similar neighbors. The detailed analysis procedure is given in Supporting Information Methods.
Detection of bacterial/viral integrations into the host genome
H. pylori (98–10) and EBV are included in our study because of their relevance to gastric cancer in the Asian population, with two genomes, Escherichia coli (NC_000913) and Arcobacter nitrofigilis (NC_014166), as control (Supporting Information Table S14). We utilized the paired-ends of the sequence reads and searched for human-pathogen chimeric reads, with one end of the reads mapped to the human genome and the other mapped to a reference genome. Adjacent or overlapping chimeric reads (within 50 bps) aligning to the human and reference genomes were merged into clusters, considered as potential integration events. Then sequentially adjacent clusters within 500 bps of each other on the human genome were considered as the same integration event; and the cluster with the highest number of reads was retained as a representative for that event (n = 134 in total in five tumors per H. pylori analysis, Supporting Information Table S11). We did the same analysis on both tumor and normal samples separately. Besides integration, double-strand DNA breakpoints were also detected, which is explained in details in Supporting Information Methods.
Results
The landscape of genomic alterations
DNA from primary adenocarcinoma samples of five gastric cancer patients and their paired noncancerous samples were extracted for whole-genome sequencing analysis at Complete Genomics, INC (Mountain View, CA) with 60 coverage (Supporting Information Table S1), which yields over 360 million reads per genome and an average of fully called genome faction at 97.3% (detailed statistics in Supporting Information Table S2). We identified a median of 9,700 putative somatic point mutations [or single-nucleotide variations (SNVs)] per tumor, ranging from 8,553 to 57,789 across the five samples (Table 1) and the genome with the highest number of mutations is from a patient with some 20 years of smoking history. Among all, 37,861 SNVs have a somatic score cutoff at 0.1 or higher, which are used as high confidence SNVs for further analyses (Supporting Information Table S3). The mean mutation rate (including indels and substitutions; Supporting Information Table S4) was 8.7 per megabases, at the same level as small cell lung cancer and melanoma and approximately tenfold higher than the rates for acute myeloid leukemia, breast cancer and prostate cancer as reported in the literature. The difference in the mutation rates may imply the different carcinogens the cells were exposed to, e.g., from smoking and exposure to UV light, or different DNA repair mechanisms were used. Overall, 407 nonsynonymous somatic point mutations, ranging from 41 to 231 per genome across the five cancer genomes, were found in 365 protein-coding genes (Supporting Information Table S5), of which 315 genes have not been reported previously in other cancer genomes.5,13–18 Of these genes, twenty-five harbor nonsynonymous mutations in more than two of the five tumors and eight genes harbor mutations in at least three of the five tumors, namely two mucins (MUC3A and MUC12), three transcription factors (ZNF595, ZNF717 and TP53), one cell cycle gene (CDC27), one trafficking factor TRAK2 and one filaggrin (FLG2). Of these genes, the recurrent mutations in MUC3A and ZNF717 are the most significant, as they occur in four of the five tumors. The nonsynonymous mutations in MUC3A, particularly in its SEA domain, highly likely change the extracellular structure or the structure of the glycosylation site of this protein, thus affecting its efficacy as a barrier against bacterial infection (such as by H. pylori) and increasing the risk of infection and possibly development of gastric cancer. All five tumors show nonsilent mutations in ZNF717, including both deletion and substitution. These mutations may have implications to misregulation by the gene relevant to cancer growth, for example, through its interaction with proapoptosis gene ZAK that mediates gamma radiation signaling and regulates the cell cycle arrest.19,20
Table 1.
Summary of genomic statistics in each of the five cancer genomes
| Patient ID | GC-S01 | GC-S02 | GC-S03 | GC-S04 | GC-S05 |
|---|---|---|---|---|---|
| Gender | M | M | M | M | M |
| Age | 57 | 70 | 63 | 65 | 64 |
| Stage | I | II | II | III | IV |
| Smoke | No | Yes | No | No | No |
| No. of somatic SNVs | 8,553 | 57,789 | 20,101 | 9,341 | 9,700 |
| No. of indels/substitutions | 11,170 | 92,415 | 20,165 | 13,514 | 10,405 |
| Mutations per Mb of DNA | 6.8 | 51.5 | 13.8 | 7.8 | 6.9 |
| No. of nonsynonymous mutations | |||||
| In CDs | 151 | 267 | 124 | 104 | 98 |
| # of genes | 73 | 266 | 62 | 57 | 46 |
| Ka/Ks ratio | 0.0163 | 0.0153 | 0.0066 | 0.0101 | 0.0109 |
| No. of genomic rearrangements | 94 | 307 | 156 | 54 | 68 |
| # of genes | 56 | 160 | 80 | 27 | 31 |
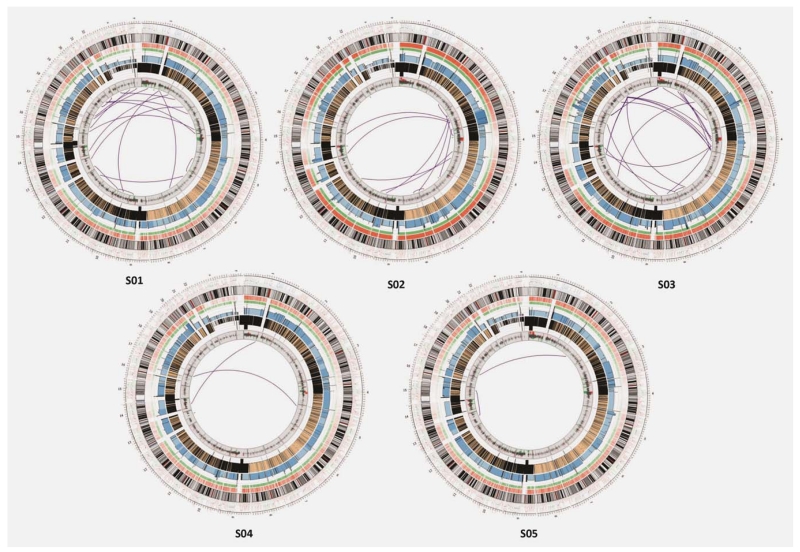
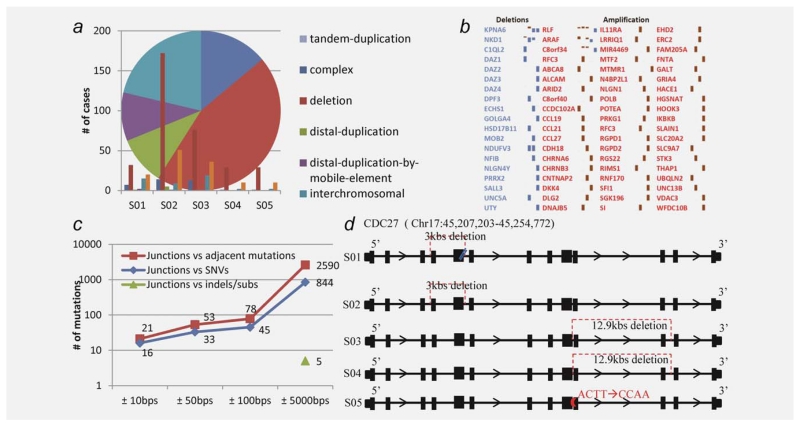
A total of 679 genomic rearrangements were identified in the five cancer genomes (Supporting Information Table S6), ranging from 54 to 307 per genome, determined based on 747 junction sites identified (Fig. 1). These junctions represent highly confident recombination regions that are supported by at least ten unique mate-pairs and satisfying a few stringent criteria (see Methods). Across the five tumors, approximately 50% of the genomic rearrangements are chromosomal deletions, ~40% are inversions and tandem/distal duplications and ~10% are interchromosomal translocations including 22 gene fusion events (Fig. 2). The number of rearrangements is comparable to that reported in prostate cancer.13 Among all, 379 rearrangements corrupted 355 protein-coding genes (Table 1), and the recurrent ones are involved in (i) cell cycle gene CDC27 and tumor suppressor gene FHIT, both harboring intragenic breakpoints in four of the five cancer genomes, (ii) two tumor suppressor genes DACH2 and WWOX, oncogene gene epidermal growth factor receptor (EGFR) and mucin gene MUC20, which are disrupted by deletions in three of five tumors and (iii) trafficking gene TRAK2, which contains a deletion in one tumor and harbors nonsynonymous mutations in three other tumors. Notably, most of these changes are deletions in genes, suggesting loss-of-function possibly selected by tumorigenesis. The vast majority of these genomic rearrangements represent novel discoveries in gastric cancer, except for a few such as deletions in FHIT and EGFR that have been reported in other cancers.21,22 The truncated proteins resulting from these altered genes may contribute to the oncogenic process (more discussion in the following section).
Figure 1.
The landscape of the genomic alterations in five gastric cancer genomes. Each circos plot depicts the genomic location in the outmost ring, and SNV (orange), indel (green), chromosomal copy number (blue for tumor and brown for control) and gene expression (red for upregulation and green for downregulation) in the next four rings from outside in. The purple lines in the innermost circle represent the chromosomal structural rearrangements.
Figure 2.
Structural rearrangements and associations between rearrangement breakpoints and point mutations. (a) The pie chart depicts different structural variations observed in the five tumors. (b) Copy-number changes observed in 76 genes (red, amplification; blue, copy loss, in five genomes). (c) The distribution of the junctions associated with point mutations in the surrounding region. 21 somatic mutations were found to be within ~10 bps to the rearrangement breakpoints, which involve 16 junctions. We suspect that these mutations may together contribute to the formation of the rearrangements to corrupt the genes. (d) An illustration of disruptions in gene CDC27 in the five tumors (red for deletion; blue for point mutations and purple for substitution). The missense mutations and substitutions in CDC27 were observed in GC-S01, S04 and S05, respectively, while deletions were found in GC-S01, S02, S03 and S04. We did not find any significant homology within 100 bps flanking region of the junction sites of CDC27. Instead, potential microhomologies of 2–5 bps were observed at its boundaries, such as CT in GC-S01, AT in GC-S03 and CAAT in GC-S04.
A translocation complex forms when multiple breakpoints from different chromosomal regions are involved, and this may lead to “balanced” DNA fusions.13 We clustered the junctions of all the genomic rearrangements based on chromosomal distance, allowing a 50-bps window size. Five 2-gene fusions out of the 22 total fusion events were identified with the balanced pattern (Supporting Information Table S7). The five fused genes include ELOVL6/ANKRD18B, FRMPD1/MICAL2, GARNL4/DOC2B in GC-S02, CNTFR/DNAI1 and TSS-UPSTREAM (APTX)/C10orf68 in GC-S03, possibly affecting the functions of DNA breakage repair (APTX), alternative splicing (C10orf68), fatty acid metabolism (ELOVL6), cell proliferation regulation (CNTFR), vesicle-mediated transport (DOC2B) and cytoskeletons that are involved in multiple cellular functions such as cell projection organization (DNAI1), G-protein receptor stabilization (FRMPD1) and oxidation–reduction (MICAL2). Most of these processes are cancer relevant so the fusion events may cause alterations in some cellular processes that favor cancer growth and thus were selected by the proliferating cells. In addition, 76 genes are detected with somatic CNVs (SCNVs), which show consistent changes (57 amplifications and 19 deletions) in at least two of the five tumors (Supporting Information Table S8). The most significant amplification is in a rearranged L-myc fusion (RLF) gene across all three early stage tumors (both stages I and II). According to our expression data, RLF is moderately overexpressed in gastric cancer.
Characterization of genomic rearrangements
The structural rearrangements and CNVs in a genome arise by several different DNA repair mechanisms.23,24 To sort out different types of genomic rearrangements, we first separated the ones that cause SCNVs, which involve deletions, amplifications and insertions, from those that do not result in chromosomal losses or gains such as inversions and “balanced” translocations (exchange of breakpoint arms13); and then examined the endpoints of all the rearrangements. Generally, deletions and amplifications are formed through two different mechanisms: homologous recombination (HR) between dispersed DNA repeats, or a type of nonhomologous recombination (NHR), called microhomologous recombination, between sequences sharing only few base-pairs of homology.24 Among the 747 identified junctions, 22.4% (167/747) show strong homology (>50 bps and > 98% sequence identity) between the flanking sequences of the impacted regions and additional 22.2% (166) have lower level but detectable homology (>50 bps with BLAST e-value <1.00E–06). We did the same analysis on the remaining 414 junctions and found 73.9% have only microhomologies involving a few base pairs (mostly 2–3 bps). Compared to HR as major repair mechanism for double-strand DNA breakage (DSB) in normal mammalian cells, NHRs are clearly being heavily used in gastric cancer. Interestingly different cancers may have used different mechanisms of NHRs as distinct recombination patterns have been observed. For example, most prostate cancers seem to have used the precise join mechanism for NHR13 while breast cancer tends to use microhomology in gene fusion events.25
Now a question is what other factors may have contributed to the creation of the genomic rearrangements in the five cancer genomes. We observed nonsense mutations in genes of the MLH1 or MSH2, suggesting the possible failure of the DNA repair system that may explain the observed genomic instability. In addition, we suspect that the deficiency of RecA or RecA-orthologous proteins such as RAD51 (suppressed expression was seen) may be another reason for the observed instability since the HR process uses them to lead the “homology search” in the genome for a single-stranded homologous DNA to pair up during the initial DNA synapsis stage of HR. We evaluated the statistical significance of the number of mutations neighboring to the structural variants within distance up to 200 kbps (detailed statistics in Supporting Information Table S9). The calculated p values show that significant number of small point mutations tend to be close to the structure variations, say, within 10 kbps in 7 chromosomes and within 100 kbps in 12 chromosomes (Supporting Information Table S9). We note the extensive indels and short repeats in the cancer genomes will increase the chances for NHR recombination since they may generate abundant microhomologies or disrupt the coding regions of the repair enzymes that assure the fidelity of DNA replication, potentially explaining the observed high prevalence of NHR recombination.
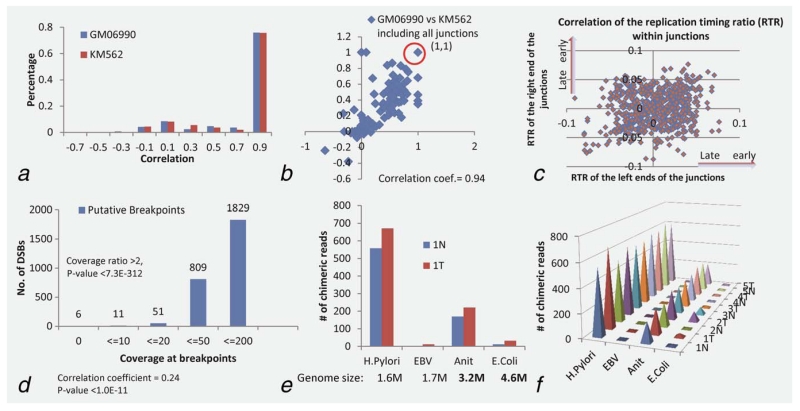
Two factors may affect why two specific DNA segments join in recombination instead of others. One is that the two segments are in close spatial proximity in the folded three-dimensional chromosomal structure. To determine if this is indeed the case for the observed recombination, we estimated the distance between two endpoints of any identified junction based on the genome-wide contact data derived using Hi-C experiments,26 where regions with high Hi-C reads, highly frequent contacts, represent spatially close regions in the folded chromosomes. The observed signals for each pair of DNA loci were extracted and the co-localization was evaluated by using Pearson correlation. 79.6% of the paired endpoints of the rearrangements are mapped to high Hi-C reads, achieving an overall correlation coefficient ≥0.7 with p value <0.001 (Fig. 3a). Plus, these endpoints of the rearrangements are highly enriched with regions having high numbers of Hi-C reads in contrast to the general background in the contact matrix (Supporting Information Fig. S1a) with low Hi-C reads, hence ensuring a high confidence of our speculation that DNA recombination tends to happen among the DNA segments in close spatial proximity. Considering that folded chromosomal structures may vary to some degree in different cell types under different conditions, we have carried out the same study using Hi-C data of other cell types, specifically of the human lymphoblastoid cell and the erythroleukemia cell at the 1 Mbp resolution,26 and human embryonic stem (ES) cells and human IMR90 fibroblasts with a higher Hi-C resolution, 40 Kb27 (details in Methods). The analysis results confirmed our general speculation that while chromosomal structures may vary across cell types and conditions, the pairwise spatial proximity remains unchanged in regions that involve translocations. Specifically, we observed a high correlation between the two sets of pairwise spatial proximity of the two cell types from the same experiment (Fig. 3b).
Figure 3.
Spatial proximity between both endpoints of the structural rearrangements in the 3D human chromosomal structure, identified double strand breakpoints and DNA integrations (a) Distribution of the correlation coefficients between two endpoints of each rearrangement. Hi-C matrix (Supporting Information Fig. S1a) representing the intrachromosomal interactions on Chromosome 1. Each pixel represents interaction correlation between two specific 1-Mb loci on Chromosome 1 (Methods). (b) Similarity between the correlation profiles of two cell systems human, lymphoblastoid cell (GM06990) and erythroleukemia cell (KM562), the coefficient as 0.94 for all 747 cases and 0.67 for those not staying in the same 1 Mb region. (c) Correlation between the replication timing profiles of both endpoints of each rearrangement. Replication timing profiles of both ends of the rearrangements. Supporting Information Figure S2 shows the replication timing profiles of Chromosome 1. The X-axis represents the locus along the chromosome and the y axis represents the replication timing ratio. (d) The putative DSBs detected in the cancer genomes with statistical confidence. Supporting Information Figure S3a shows an illustration of putative double-strand break sites (DSBs) in tumor versus control chromosomes [theoretically, there is no (or very few) reads over the DSB sites] as well as the DBS distribution versus gene lengths. (e) Putative integrations in human genome from H. pylori and EBV with two control genomes, Anit and E. coli. The histogram shows the number of chimeric reads (two mates are from different genomes) identified in the tumor and control samples of GC-S01, associated with each of the four viral/bacterial genomes; (f) shows consistent observations across the five pair of samples. The y-axis represents the number of chimeric reads that are involved in integration.
Another factor that affects if the neighboring DNA segments can join is if they replicate simultaneously, making their single-strand DNA accessible for possible recombination.28 We used the replication profiling data from one ENCODE study26 to assess the timing of replication on different DNA segments of the cancer genomes. The same rule used in Ref. [26 has been applied to the cancer genomes to classify all genomic rearrangements into early and late replicating regions. The early replicated genes include those encoding for cytoskeletons (ACTN4, DCLK1 and SPT), cell cycle protein (CDC27) and transporters (COG5 and PACS1), enzyme (NEDD4L), sperm-associated antigen (SPAG16) and transmembrane EGFR pathway regulator (TMEM8B); and late replicated genes include transcriptional regulator (ZNF57) and catenin (CTNNA3). We found that the fusion points of both ends across the 747 rearrangements are generally consistent with their replication timing, achieving a correlation coefficient cc = 0.24 and 74% of the junction pairs staying within replication timing window less than 0.01 (Fig. 3c), supporting our hypothesis of simultaneous timing holds.
Putative DSBs that are unfixed in gastric cancer genomes
Most of the DSBs are fixed by emergency DNA repair mechanisms in cancers such as microhomology-mediated break-induced replication and nonhomologous end joining,23,24 but some cannot. We have designed an algorithm (see Methods) to examine if some chromosomes may have open arms, resulting from unfixed chromosomal fragments after DSBs happen. Our examination results (Fig. 3d, details in Supporting Information Table S10) show 1,829 potential DSB sites, among which 51 are deemed to be highly confident as we restrict the heterogeneity level to no more than 20 reads covering breaking site and the ratio of the reads depths between breaking site and flanking region on any side as less than 0.5. We noted that this set of loci coincide with common fragile sites (CFSs),29 mostly within long genes such as CNTNAP2 (2.3 Mb, with know CFS FRA7I), DLG2 (2.2 Mb, FRA11F), ERBB4 (1.3 Mb, FRA2I) and WWOX (1.1 Mb, FRA16D). These breakpoints add to the whole picture of genomic instability in addition to CNVs and genomic rearrangements.
Infection-induced genomic integration in gastric cancer
Knowing that H. pylori and EBV infection could cause DSBs in the human genome and possibly induce gastric cancer,30,31 we have examined the sequence data to determine if the infection also causes the fusion of the viral/bacterial DNA with the cancer genomes. The challenge in identifying such fusion events is in identification of the possible integration sites through identifying chimeric reads with one end from the human genome and the other paired end from another organism, after clearing the possibility of contamination. We have developed an algorithm (see Methods) for detecting such sites involving H. pylori DNA while search against the EBV genome did not lead to significant hits (Figs. 3e and 3f). The observations are consistent across all the samples with cancer genomes showing more integrations than the matching controls. The integration sites are highly clustered on the human genome regions into 134 loci which cover 36 genes (Supporting Information Table S11). The most consistent integration shows that a 30–33 bps DNA from H. pylori integrated into the gene PREX2 in chromosome 18. Recent studies show that PREX2 (phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2) interacts with PTEN and negatively regulates this tumor suppressor in breast cancer32; and it was found to have high frequency mutations in melanoma, which accelerates melanoma formation in vivo.33 Future study will be carried out to validate if the integration-induced mutation in PREX2 may have a similar impact on gastric cancer development.
Recurrent mutations disrupt FHIT, CDC27, MUC20, EGFR and WWOX genes
We have examined the functional implications by the corrupted genes identified in the cancer genomes. Deletions in two tumor suppressor genes, the fragile histidine triad (FHIT) and WW domain-containing oxidoreductase (WWOX), were observed in four (GC-S01, S02, S03 and S05) and three (GC-S02, S03 and S05) of the five tumors, respectively. Both genes contain CFSs FRA3B and FRA16D, respectively, as observed in other cancers.29 The deleted regions ranging between 37 kbps and 700 kbps are all in the center of the CFSs while the recombination sites vary in each of the five tumors. Both genes are not expressed (or minimally expressed) in tumors harboring the deletions but show differential expression in other tumors with the intact gene sequence. This suggests that there may be a selection pressure for cancer to silence the functions of these two genes either through functional repression or deletion of the genes, knowing that these genes are involved in suppression of intracellular signaling, DNA damage response,34 and signaling to apoptotic pathway.35
Cell cycle gene CDC27, a major component of anaphase-promoting complex (APC), also has deletions (>3 kbps) in four tumors except for the Stage 4 tumor (GC-S05) where it shows a substitution (ACTT->CCAA) instead. A phosphorylated CDC27 can activate the anaphase-promoting complex (APC/cyclosome) in response to the TGF-beta signaling,36 which inhibits growth of epithelial cells, so a depleted CDC27 function would diminish the inhibition of cell growth, thus permitting cell growth. A study on the down-regulation of CDC27 in breast cancer revealed the role of this gene as a tumor suppressor37; and its downregulation or protein corruption in the TPR (tetratricopeptide repeat) interaction region observed in gastric cancers suggest another possible cause of cell cycle deregulation. In addition, EGFR, an oncogene, has a deletion (<1 kbps) in three tumors in a region close to the centromere, which consists of multiple tandem repeats. The elevated expression of EGFR in most gastric cancers versus their controls, reported in other cancers as well,38 may indicate the mutation (small deletion) yield an active and transforming receptor as reported in Ref. [37.
One of the most interesting findings is about the genetic changes in the mucin genes. These glycoproteins with one epidermal growth factor (EGF)-like domain are either membrane associated or secreted by epithelial cells. Gene MUC20 has deletions in three of the five tumors while two other mucin genes, MUC3A and MUC12, are highly mutated in four and three tumors, respectively, as discussed earlier. Normally, these proteins are involved in the formation of a mucus barrier to protect the cell from microorganism invasion, such as H. pylori. But the aberrant expression of mucin genes through epigenetic regulation has been used as an indicator for cancer prognosis and detection,39 and binding of H. pylori to some mucin proteins may trigger signaling transduction for cell growth of epithelial cells.40 From another perspective, their downregulation or functional corruption due to mutations or deletions observed in gastric cancers may also enable the cancer cells to break cell–cell contacts and migrate to other sites, i.e., metastasis, or make the relevant cells more accessible to H. pylori.
Sixteen other genes are also found to harbor somatic changes in at least two gastric tumors but only ten are considered as highly mutated genes according to the calculated mutation rate (see Methods and Supporting Information Table S12). In addition to those discussed above, this list also includes filaggrin (ELG2), melanoma antigen (MAGEC1), tumor protein (TP53), methyl-CpG binding domain protein (MBD6), olfactory receptors (OR5M11 and OR8U1) and zinc finger proteins (ZNF595, ZNF676 and ZNF717), most of which have not been reported to be cancer relevant.
Functional disruption in multiple pathways, particularly in signaling and energetic metabolisms
We have examined the functional impact by the 936 mutated genes at the pathway level, specifically the cancer hallmark related pathways.41 These genes are participating in pathways related to cell cycle, MAPK signaling, p53 signaling pathway, GnRH signaling pathway, cell adhesion molecules, focal adhesion, gap junction and purine metabolism and these pathway may form the core of gastric tumorigenesis. In addition, pathways relevant to cervical cancer, prostate cancer, nonsmall cell lung cancer and pancreatic cancer are highly enriched by these genes, suggesting a close relationship among these different types of cancer.
We also utilized gene-expression data of gastric cancer versus control to expand the pathway analysis. In total, 26 pathways are enriched by genes altered by genetic mutations and abnormal regulation consistently across more than one cancer genome (Supporting Information Table S13). Our main finding here is that the network expands the above pathway list to gastric acid secretion, protein digestion and absorption, DNA replication, ECM-receptor interaction, pyrimidine metabolism, T-cell receptor signaling pathway, oxidative phosphorylation, purine metabolism, tight junction, ribosome biogenesis, apoptosis and mTOR signaling pathway. We can see clearly a more thorough signaling and metabolic network involving not only major cancer hallmark pathways but also gastric cancer-specific pathways. Mutated pathways introduced42 such as TGF-beta (SMAD2 and SMAD3) and b-catenin (APC and CTNNB1) were observed in this study with relatively less significance, while no frequent mutations were found on RHO signaling, which may reflect the heterogeneity of this disease.
Like APC in colon tumors, cancer driver mutations often function through inducing aberrant downstream regulations and accumulation of other mutations. In another word, pathways altered by mere gene regulation are inclined to act as downstream process, “passengers,” compared to the initial drivers. For example, the dysregulation of glutathione metabolism and protein digestion and absorption are observed in early stage tumors where none of the mutations were detected, which in turn direct the study for more upstream inducers. Such integrated analysis of genetic mutations and functional regulation may lead to improved understanding about the early drivers of gastric cancer.
Discussion
Here we reported an in-depth genome analysis of five gastric adenocarcinomas, which has identified very complicated mutation patterns with most recurrent mutations in protein coding genes such as FHIT, TP53, Mucins (MUC3A and MUC12) and CDC27. No significant recurrent changes were identified in chromatin remodeling gene ARID1A as reported in other gastric cancer sequencing study of gastric cancer,5,42 reflecting the heterogeneity of the disease. Overall, we identified in the gastric cancers myriad genomic changes such as SCNVs (particular long deletions) and observed the utilization of microhomology mediated DNA repair during gastric cancer evolution. The integration of gene expression information complements our analysis toward discriminating the more upstream driving mutations among all somatic variations in a pathway context.
Other than the genomic changes intrinsic to the disease development, such in-depth analysis enables us to identify possible infection-induced genomic changes. Identification of the DNA integration from H. pylori first uncovers the bacterial invasion to the host genomes. Current knowledge relevant to this issue is that H. pylori can cause the DSBs after prolonged active infection,31 may be due to the action of reactive oxygen species (ROS) caused by chronic inflammation.43 The putative integrations from H. pylori may reflect a new mechanism about how H. pylori induce cancer development: the infection potentially induces the co-localized cancer gene with DNA pieces from the infectious agent by allowing DSBs to facilitate gene fusions. This discovery could lead to new and improved understanding about H. pylori infection as the key contributing factor to the onset of the disease.
The SCNVs and structural rearrangements detected in this study indicate that the affected genomic regions are particularly sensitive to DNA damages caused by replication stress or external stimulus and thus rearranged in cancers. In most cases, the boundaries of SCNVs are well correlated with rearrangement junctions. However, the reflection of underlying relationships between these groups can be diluted by inclusion of other factors, for instance, unfixed DSBs or cellular heterogeneity due to the complex lineages generated during the cancer evolution, which results in a mixture of changes irrelevant to tumorigenesis due to various DNA repair mechanisms and explains in part the observed insignificant correlation between two variations in the genomes. We have found that the spatial proximity and replication timing may be two key factors that make specific rearrangements possible, which are created directly by the emergency DNA repair mechanism, employed under hypoxic condition in the cancer cells,24 upon DNA breakage.
In addition to the to-be-determined causal relationships between the observed mutations in cancer genomes and cancer development, increased research attention has been put on identification of the microenvironment factors such as hypoxia or ROS stress that may trigger and select mutations in early disease stages.44 A widely accepted view is that cancer is the result of complex and continuous interplays among the changing microenvironment, responses at the metabolic levels and adaptation of the cellular system through functional regulation and selection for genomic changes. We presented in this study one piece of the puzzle related to gastric tumorigenesis. To gain a full understanding about the disease, we may need to study the genomic changes in the context of metabolic shifts in response to the evolving microenvironment.
Supplementary Material
What’s new?
Gastric cancer is the second leading cause of cancer deaths world-wide. Here, the authors performed whole-genome sequencing on five gastric adenocarcinomas and adjacent noncancerous tissue. More than 100,000 somatic point mutations were identified, the most recurrent ones located in genes encoding for mucins and transcription factors. The authors also report numerous genomic rearrangements including potential insertion of H. pylori, a bacterium associated with gastric cancerogenesis, into the tumor genome. These findings form the basis for interesting new hypotheses to be tested in future experiments.
Acknowledgements
The authors would like to thank all the individuals who have participated in this study. In particular, we thank Dr. Ming Hu at Harvard University and Ms. Yaping Li and Ms. Huan Xiong at UGA for their helpful discussions and technical assistance.
Grant sponsor: NIH; Grant numbers: DK033973, DK069711 and GM075331; Grant sponsor: NSF; Grant numbers: DBI-0354771, ITR-IIS-0407204, CCF-0621700 and DBI-0542119; Grant sponsor: Kuang-Cheng Wang Educational Foundation of Chinese Academy of Sciences
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Kakiuchi M, Nishizawa T, Ueda H, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46:583–7. doi: 10.1038/ng.2984. [DOI] [PubMed] [Google Scholar]
- 2.Wang K, Yuen ST, Xu J, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–82. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 3.Wang K, Kan J, Yuen ST, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–23. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y, Hu Z, Wu C, et al. A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet. 2011;43:1215–8. doi: 10.1038/ng.978. [DOI] [PubMed] [Google Scholar]
- 5.Zang ZJ, Cutcutache I, Poon SL, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012 doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 6.Nagarajan N, Bertrand D, Hillmer AM, et al. Whole-genome reconstruction and mutational signatures in gastric cancer. Genome Biol. 2012;13:R115. doi: 10.1186/gb-2012-13-12-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hillmer AM, Yao F, Inaki K, et al. Comprehensive long-span paired-end-tag mapping reveals characteristic patterns of structural variations in epithelial cancer genomes. Genome Res. 2011;21:665–75. doi: 10.1101/gr.113555.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drmanac R, Sparks AB, Callow MJ, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 9.McLaren W, Pritchard B, Rios D, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.UCSC http://hgdownload.cse.ucsc.edu/goldenPath/hg19/gc5Base/hg19.gc5Base.txt.gz.
- 11.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–63. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 12.Cui J, Chen Y, Chou WC, et al. An integrated transcriptomic and computational analysis for biomarker identification in gastric cancer. Nucleic Acids Res. 2011;39:1197–207. doi: 10.1093/nar/gkq960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berger MF, Lawrence MS, Demichelis F, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kan Z, Jaiswal BS, Stinson J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–73. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 15.Pleasance ED, Stephens PJ, O’Meara S, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–90. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pleasance ED, Cheetham RK, Stephens PJ, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenman C, Stephens P, Smith R, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–8. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bamford S, Dawson E, Forbes S, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91:355–8. doi: 10.1038/sj.bjc.6601894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang JJ. A novel zinc finger protein, ZZaPK, interacts with ZAK and stimulates the ZAK-expressing cells re-entering the cell cycle. Biochem Biophys Res Commun. 2003;301:71–7. doi: 10.1016/s0006-291x(02)02980-7. [DOI] [PubMed] [Google Scholar]
- 20.Yang JJ, Lee YJ, Hung HH, et al. ZAK inhibits human lung cancer cell growth via ERK and JNK activation in an AP-1-dependent manner. Cancer Sci. 2010;101:1374–81. doi: 10.1111/j.1349-7006.2010.01537.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Ramakrishnan R, Tang Z, et al. Quantifying EGFR alterations in the lung cancer genome with nanofluidic digital PCR arrays. Clin Chem. 2010;56:623–32. doi: 10.1373/clinchem.2009.134973. [DOI] [PubMed] [Google Scholar]
- 22.Muller CY, O’Boyle JD, Fong KM, et al. Abnormalities of fragile histidine triad genomic and complementary DNAs in cervical cancer: associa-tion with human papillomavirus type. J Natl Cancer Inst. 1998;90:433–9. doi: 10.1093/jnci/90.6.433. [DOI] [PubMed] [Google Scholar]
- 23.Hastings PJ, Lupski JR, Rosenberg SM, et al. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–64. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hastings PJ, Ira G, Lupski JR. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 2009;5:e1000327. doi: 10.1371/journal.pgen.1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens PJ, McBride DJ, Lin ML, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–10. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hastings PJ, Rosenberg SM. Genomic rearrangement in three dimensions. Nat Biotechnol. 2011;29:1096–8. doi: 10.1038/nbt.2064. [DOI] [PubMed] [Google Scholar]
- 29.Durkin SG, Glover TW. Chromosome fragile sites. Annu Rev Genet. 2007;41:169–92. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 30.Glover TW. Common fragile sites. Cancer Lett. 2006;232:4–12. doi: 10.1016/j.canlet.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 31.Toller IM, Neelsen KJ, Steger M, et al. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc Natl Acad Sci USA. 2011;108:14944–9. doi: 10.1073/pnas.1100959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fine B, Hodakoski C, Koujak S, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325:1261–5. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berger MF, Hodis E, Heffernan TP, et al. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature. 2012;485:502–6. doi: 10.1038/nature11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pekarsky Y, Garrison PN, Palamarchuk A, et al. Fhit is a physiological target of the protein kinase Src. Proc Natl Acad Sci USA. 2004;101:3775–9. doi: 10.1073/pnas.0400481101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang NS, Doherty J, Ensign A. JNK1 physically interacts with WW domain-containing oxidoreductase (WOX1) and inhibits WOX1-mediated apoptosis. J Biol Chem. 2003;278:9195–202. doi: 10.1074/jbc.M208373200. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Fujita T, Wu G, et al. Phosphorylation of the anaphase-promoting complex/Cdc27 is involved in TGF-beta signaling. J Biol Chem. 2011;286:10041–50. doi: 10.1074/jbc.M110.205518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pawar SA, Sarkar TR, Balamurugan K, et al. C/EBP{delta} targets cyclin D1 for proteasome-mediated degradation via induction of CDC27/APC3 expression. Proc Natl Acad Sci USA. 2010;107:9210–5. doi: 10.1073/pnas.0913813107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rocha-Lima CM, Soares HP, Raez LE, et al. EGFR targeting of solid tumors. Cancer Control. 2007;14:295–304. doi: 10.1177/107327480701400313. [DOI] [PubMed] [Google Scholar]
- 39.Yonezawa S, Higashi M, Yamada N, et al. Significance of mucin expression in pancreatobiliary neoplasms. J Hepatobiliary Pancreat Sci. 2010;17:108–24. doi: 10.1007/s00534-009-0174-7. [DOI] [PubMed] [Google Scholar]
- 40.Linden S, Mahdavi J, Hedenbro J, et al. Effects of pH on Helicobacter pylori binding to human gastric mucins: identification of binding to non-MUC5AC mucins. Biochem J. 2004;384:263–70. doi: 10.1042/BJ20040402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 42.Cancer Genome Atlas Research N Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Obst B, Wagner S, Sewing KF, Beil W. Helicobacter pylori causes DNA damage in gastric epithelial cells. Carcinogenesis. 2000;21:1111–5. [PubMed] [Google Scholar]
- 44.Cui J, Mao X, Olman V, et al. Hypoxia and mis-coupling between reduced energy efficiency and signaling to cell proliferation drive cancer to grow increasingly faster. J Mol Cell Biol. 2012;4:174–6. doi: 10.1093/jmcb/mjs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.