Abstract
ESSENCE refers to early symptomatic syndromes eliciting neurodevelopmental clinical examinations. It includes a broad range of early onset neurodevelopmental disorders affecting more than 10% of children before 5 years of age. ESSENCE includes among others attention deficit hyperactivity disorder (ADHD), intellectual disability (ID) and autism spectrum disorders (ASD). Some degree of disability is the rule rather than the exception. The causes are heterogeneous ranging from extreme social deprivation, pre- and perinatal risk factors, genetic and metabolic diseases, immune and infectious disorders, nutritional factors, physical trauma, and postnatal toxic and environmental factors (and combinations/interactions of some or several of these). Treatments often involve a combination of psychoeducational interventions, home- and school-based programmes, and medication. Here, I will first briefly review our main knowledge on the biological pathways associated with early onset neurodevelopmental disorders and will provide useful links to be informed of the progress in the field. Five main pathways are associated with ASD and ID: chromatin remodelling, cytoskeleton dynamics, mRNA translation, metabolism and synapse formation/function. I will then detail three propositions coming from institutions, researchers and/or communities of patients and families to foster research: 1) to use more dimensional and quantitative data than diagnostic categories; 2) to increase data sharing and research on genetic and brain diversity in human populations; 3) to involve patients and relatives as participants for research. Finally, I will provide examples of very stimulating initiatives towards a more inclusive world for individuals with ESSENCE.
Keywords: Autism, Genes, Synapses
Children with neurodevelopmental disorders can experience problems with language and speech, motor skills, behaviour, memory, learning, or other neurological functions. These difficulties are also frequently associated with co-morbidities such as sensory-motor, sleep, and gastrointestinal problems (1). To better tackle this heterogeneity Christopher Gillberg coined the acronym ESSENCE, referring to “early symptomatic syndromes eliciting neurodevelopmental clinical examinations”. It is a term to refer to the reality of children (and their parents) presenting in clinical settings with impairing child symptoms before age 3–5 years in the fields of 1) general development, 2) communication and language, 3) social inter-relatedness, 4) motor coordination, 5) attention, 6) activity, 7) behaviour, 8) mood, and/or 9) sleep. Symptoms of neurodevelopmental disorders often evolve and may improve as a child grows older, but many disabilities are permanent. Diagnosis and treatment of these disorders can be difficult; treatment often involves a combination of professional therapy, pharmaceuticals, and home- and school-based programmes. With progress in genetics and neurobiology, the causes of early onset neurodevelopmental disorders (or ESSENCE) are better understood. Here, I will summarize the current knowledge on the genetic causes. Then I will summarize propositions that were suggested to improve research in this field.
Definition and prevalence
Early onset neurodevelopmental disorders affect more than 10% of children (Table 1) often with consequences throughout their lives and with significant effects on their families (1–3). This grouping is diverse in terms of severity and pathophysiology: fetal alcohol syndrome (FAS), attention deficit hyperactivity disorder (ADHD), intellectual disability (ID), tic disorder, developmental coordination disorder, dyslexia, specific language disorders and autism spectrum disorders (ASD). Neuromuscular disorders such as Becker or Duchenne muscular dystrophies could also be included in neurodevelopmental disorders since they also affect cognition in a subset of patients, but such disorders are often considered as a separate cluster because of their predominant symptoms. Boys seem to be at elevated risk compared with girls for most neurodevelopmental disorders, suggesting gender-specific risk and protective factors.
Table 1. Prevalence and biological pathways associated with ESSENCE.
| Neurodevelopmental disorders | Prevalence% | Proteins or biological pathways |
|---|---|---|
| Learning disabilities | 2–4 | Chromatin remodelling Metabolism Actin skeleton organization Channels Synaptogenesis Neurotransmission |
| Dyslexia | 5–15 | Neuronal migration? |
| Attention deficit hyperactivity disorder | 1.7–9 | Synapses? Cortical maturation? |
| Autism spectrum disorders | 0.6–1.2 | Chromatin remodelling Metabolism Actin skeleton organization Channels Synapses |
| Epilepsy | 0.45–1 | Synapses Channels |
| Fetal alcohol syndrome | 0.1–5 | – |
The amount of funding and research dedicated to a disorder is often correlated to its prevalence and its severity (4). Thus, it is noteworthy that the amount of research on intellectual disability is below the predicted level (4). Causes of ESSENCE range from severe social deprivation, genetic risk factors, metabolic diseases, immune disorders, infectious diseases, nutritional factors, physical trauma, and toxic and environmental factors. Among these factors, we have recently gained better knowledge concerning genetic risk factors, which is, in turn, motivating new neurobiological research.
The genetics of ESSENCE
The growing list of genes that contribute to early onset developmental disorders includes hundreds of genes. However, the complexity is multiplied by the observation that each patient can carry a specific combination of alleles of large and small effect that occur de novo or inherited.
De novo mutations in ESSENCE
De novo mutations include single base mutations, amplification of trinucleotide repeats, copy-number variations (CNVs), large chromosomal rearrangements and chromosomal aneuploidy (5). Chromosomal aneuploidy (an abnormal number of chromosomes) is observed in syndromic forms of neurodevelopmental disorders such as Down, Klinefelter or Turner syndromes. Large chromosomal rearrangements and CNVs can be recurrent in some regions of the genome such as on chromosome 22q11 (velocardiofacial syndrome), 15q (Angelman and Prader-Willi syndromes), or 17p (Smith-Magenis syndrome). However, in most cases, CNVs are unique to each patient, affecting from one to hundreds of genes. A trinucleotide repeat expansion of CGG repeats is observed in fragile X syndrome. This expansion upstream of the FMR1 gene impedes its expression resulting in increased translation at the synapse. An additional example: single nucleotide mutations can affect X-linked genes such as MECP2 to cause Rett syndrome or autosomal genes such as CDH8 or SHANK3 to cause ASD.
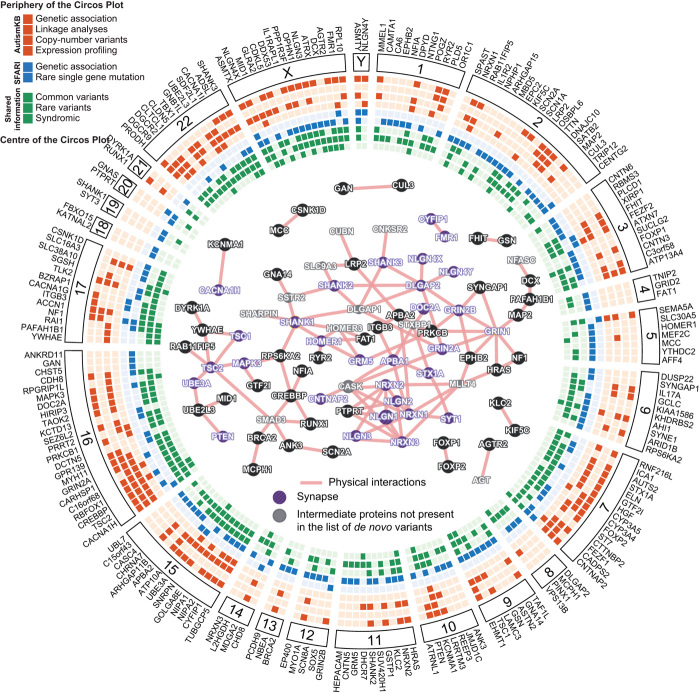
Highly penetrant de novo mutations probably account for a significant fraction (15–50%) of severe early onset developmental disorders (6, 7–13). This has been clearly demonstrated for ID (14) and ASD (11, 12, 15). The risk factors that increase the occurrence of de novo mutations, amplifications, deletions or duplications are better understood (16). For example, regions of the human genome flanked by large segmental duplications (such as on chromosome 15, 16p,) are more prone to be deleted/duplicated through illegitimate recombination. Increased paternal age was also shown to be a factor in de novo single base pair change. For ASD and ID, de novo chromosomal rearrangements and CNVs are more frequently observed in patients compared with controls. In contrast, patients and controls usually carry the same number of de novo single base mutations (on average 60–70 de novo mutations in each genome of 3 billion base pairs and one in each exome of 60 million base pairs). However, in patients, there is a significant increase, compared with controls, of damaging mutations (e.g. loss of function) in evolutionarily constrained genes expressed in the brain (Fig. 1) (11–13, 17).
Fig. 1. Circos plot of de novo mutations in ASD. All coding-sequence variants and copy-number variants present in AutismKB and SFARI Gene are shown. A GeneMANIA network analysis (centre) highlights proteins with synaptic function (36% of the proteins have at least one interaction with another protein, 61% are expressed in the brain, and 14% are known to be involved in synaptic function). From Huguet et al. 2013 (36).
The vast majority of mutations reported in patients with ASD were identified using DNA isolated from their blood (or from saliva in some projects). As a consequence, de novo somatic mutations occurring in specific brain cell lineage are missed (18, 19). Only studies using deep genomic sequencing and post-mortem brain tissues of the patients will be able to inform us as to whether somatic mutations in the brain are increased in early onset neurodevelopmental disorders.
Inherited monogenic and polygenic forms of ESSENCE
Among patients with early onset developmental disorders, inherited monogenic forms might account for a relatively significant fraction (> 10%) (20). In ASD it was estimated that 3–6% of patients are “homozygous knock-out” carriers of two loss of function mutations in the same gene (21, 22). In countries with higher consanguinity, the impact of recessive mutations is likely to be higher (23).
Multiple hits in different regions of the genome might also contribute to susceptibility to early onset neurodevelopmental disorders. Several studies have demonstrated the presence of more than one deleterious mutation in such patients (24–26). In a large-scale study of 2312 children known to carry a CNV associated with ID and congenital abnormalities, 10% carried a second large CNV in addition to the primary genetic lesion (25). Children who carried two large CNVs of unknown clinical significance were eight times more likely than controls to have developmental delay than controls. Among affected children, inherited CNVs tended to co-occur with a second-site large CNV. No parental bias was observed for the primary de novo or inherited site, but for the second-site, 72% of the second-site CNVs were inherited from the mother (25).
Other studies have supported a multiple-hits model in patients carrying a similar “first hit”. In 42 carriers of a 16p11.2 microdeletion, 10 carried an additional large copy-number variant, a significantly higher proportion when compared with controls conditional on a large first hit (10 of 42 cases, 21 of 471 controls; P = 0.000057, odds ratio = 6.6) (24). The clinical features of individuals with two mutations were distinct from and/or more severe than those of individuals carrying only the co-occurring mutation. Another study showed that three patients with ASD carrying a de novo SHANK2 deletion were also carriers of a second CNV at the 15q11 locus (26). Two were carrying CNVs including CHRNA7 and ARHGAP11B; the third was carrying a mutation that removed CYFIP1, NIPA1, NIPA2, and TUBGCP5. After this initial publication, another child with neurodevelopmental disorder carrying a SHANK2 translocation and a CHRNA7 duplication was reported (27).
Beside de novo and inherited rare mutations, one of the current challenges for geneticists is to identify the myriad of frequent alleles across the genome, which in an additive manner increase the risk of developing a disorder. Common variants could contribute to 17–60% and 25–30% of the heritability of ASD and ADHD, respectively (28–30). The same methodology was also used to estimate the contribution of genotyped single nucleotide polymorphisms (SNPs) in the heritability of the IQ (> 40%) (31, 32) and on the human brain anatomy (50%) estimates that common variants might contribute to such quantitative phenotypes (Toro et al. Molecular Psychiatry, in press. Given that these common variants have individually only a weak additive effect (33), genome-wide association studies (GWAS) to date have been significantly underpowered and identified very few if any replicated common sequence variants that contribute to risk of early onset neurodevelopmental disorders (34). Based on these results, even if this genetic information is difficult to translate into clinical diagnosis, the identification of low risk alleles represents an important goal for understanding the genetic architecture of early onset neurodevelopmental disorders (35). Moreover, even weak alleles shown with confidence to influence disease risk, point to genes and pathways involved in pathogenesis.
Database of genes associated with ESSENCE
Several genetic databases provide clinical and functional annotation of genes associated with early onset neurodevelopmental disorders. The Online Mendelian Inheritance in Man (OMIM) database catalogues more than 5000 human genetic diseases (http://www.omim.org/). Decipher (http://decipher.sanger.ac.uk/) and the Database of Genomic Variants (http://dgv.tcag.ca/dgv/app/home) are interactive Web-based databases which incorporate a suite of tools designed to aid the interpretation of genomic variants. Two databases of genes associated with ASD are updated regularly: AutismKB (http://autismkb.cbi.pku.edu.cn) and SFARI Gene (https://gene.sfari.org) (36). A total of 197 genes are included in both databases, and 481 are additionally included in either one or the other (255 in AutismKB and 226 in SFARI Gene). The main difference between the two databases concerns the selection of the genes. AutismKB usually selects genes from linkage analyses, copy-number variant studies, and GWAS, whereas SFARI Gene usually selects genes from copy-number variant studies, sequencing analyses of large cohorts, and case reports.
Biological pathways involved in ESSENCE
In the last 10 years, tremendous progress has been made in our comprehension of early onset developmental disorders. Animal models (37–47) as well as induced pluripotent stem cells (48, 49) have both contributed to better understanding of pathophysiology and to suggest new treatments. Understanding the symptoms and course for each individual, and the biology ranging from genetic and environmental risk factors to the neural circuits involved remains a substantial challenge for geneticists and neurobiologists (50–52).
Several pathway analyses have been performed using either genetic or transcriptome data to gain insight into the biological functions associated with ASD. Pinto et al. (53) recently analysed 2446 ASD-affected families and confirmed an excess of genic deletions and duplications in affected versus control groups (1.41-fold, p = 0.00001) and an increase in affected subjects carrying exonic pathogenic CNVs overlapping known loci associated with dominant or X-linked ASD and intellectual disability (odds ratio = 12.62, p = 02.7 × 10−15, ∼3% of ASD subjects). Consistent with hypothesized sex-specific modulators of risk, female patients with ASD were more likely to have highly penetrant CNVs (p = 0.017) and were also overrepresented among subjects with mutations in genes that encode fragile X syndrome protein targets (p = 0.02) suggesting that severe genetic lesions were required to overcome the lower liability to ASD in girls. Genes affected by de novo CNVs and/or loss-of-function single-nucleotide variants converged on networks related to neuronal signalling and development, synaptic function, and chromatin regulation. Voineagu and colleagues (54) analysed genes that are differentially expressed between two brain regions (frontal and temporal lobes) in patients with ASD and controls. Interestingly, the typical regional differences between the gene expression profiles of the frontal and temporal lobes were attenuated in patients. A first network module was related to interneurons and to genes involved in synaptic function, and was down-regulated in brains from patients compared with those from controls; a second module was enriched for genes related to immunity and microglial activation, and was up-regulated in brains from patients with ASD compared with those of controls.
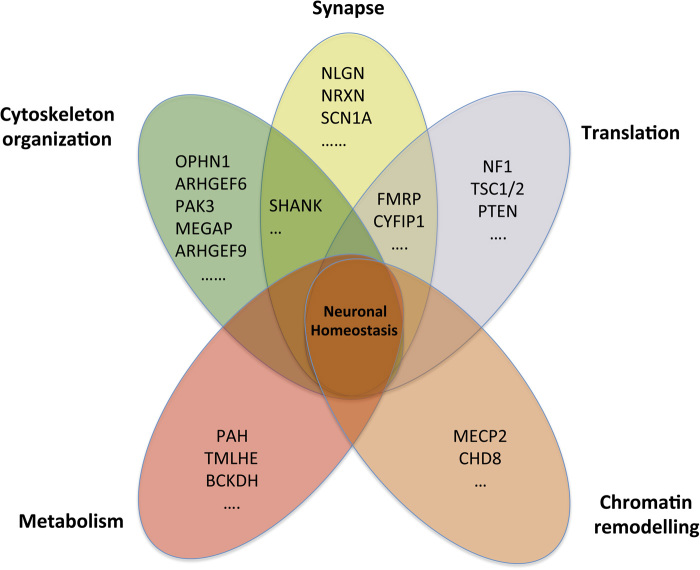
To date, five main pathways have been identified as candidates for early onset neurodevelopmental disorders (Fig. 2): chromatin remodelling, cytoskeleton dynamics, mRNA translation, metabolism and synapse formation/function. This list is, however, far from exhaustive.
Fig. 2. Five pathways are associated with early onset neurodevelopmental disorders. For each pathway, examples of mutated genes are indicated.
The first pathway concerns chromatin remodelling and was suggested by reports of mutations in genes such as MECP2 or CDH8 in Rett syndrome and ASD, respectively (11, 12, 55, 56). A second pathway is related to metabolism and includes mutations in genes such as PAH in phenylketonuria, BCKDH in disorders of branched-chain amino acids, TMLHE in carnitine deficiency or AGAT and GAMT in creatine deficiency syndromes. Interestingly, patients with mild forms of inborn errors of metabolism may present with predominantly autistic symptoms (22). Identifying such mutations is of clinical importance since treatments may already be available (57). A third pathway is related to aberrant translation of mRNA encoding synaptic proteins, (58) and includes mutations affecting several proteins that normally inhibit translation through the PI3K-mTOR signalling pathway (TSC1, TSC2, NF1, and PTEN) as well as mutations affecting proteins directly involved in inhibiting mRNA translation at the synapse (FMRP, CYFIP1, and EIF4E) (58, 59). A fourth pathway concerns the actin cytoskeleton organization and includes mutations affecting OPHN1, ARHGEF6, PAK3, MEGAP, ARHGEF9 and the regulation of the RhoGTPase, the Ras, the Rab, the Arf and the JNK pathways (60). While mutations affecting these pathways were mostly identified in patients with ID, they might account for a fraction of patients with ASD (53). Finally, a fifth pathway is involved in synapse formation and excitation/inhibition balance (17, 61). Several genes associated with ASD, such as NLGN3/4X, NRXN, and SHANK1-3, appear to be involved in the formation of excitatory and inhibitory synapses (62–64). In addition, genes associated with epilepsy, such as SCN1A, which encodes a voltage-gated sodium channel, were also found mutated in patients with ASD (7).
While different, these pathways most likely affect neuronal homeostasis at the end point (17, 65). Some suggest potential drug targets; indeed some early clinical trials are ongoing to determine whether targeting some such proteins could improve the symptoms of patients (see (66, 67) for reviews).
Three propositions to improve research in the field of ESSENCE
While tremendous progress has been made in the understanding of the causes of early onset neurodevelopmental disorders, several issues detailed below represent potential breaks for research in this field. Three propositions are listed below.
Proposition 1: fewer categories, more dimensions
The recent advances in genomics have demonstrated that an identical genetic variant may increase the risk for a wide range of diagnoses formerly thought of as distinct (29, 68, 69). These findings are contributing to an ongoing re- conceptualization of the current psychiatric nosology. The use of epidemiological samples, studies grouping individuals based first on genetic findings, and efforts at combining existing categorical schema with dimensional phenotypes and biomarkers, all promise to provide important new insights into the aetiology and classification of these disorders. DSM-5 now makes it easier to recognize overlap between different diagnostic categories, but in the main the existing narrow and rigid categories tend to disconnect researchers from the real phenotypes. Recently, several initiatives such as the ESSENCE from Christopher Gillberg were undertaken to improve phenotype characterization using more dimensional approaches. The Research Domain Criteria (RDoC) project has been launched by the US National Institute of Mental Health (NIMH), calling for the development, for research purposes, of new ways of classifying psychopathology based on dimensions of observable behaviour and neurobiological and genetic measures (http://www.nimh.nih.gov/research-priorities/rdoc/nimh-research-domain-criteria-rdoc.shtml). This effort is attempting to define basic dimensions of functioning related to known neural circuitry to be studied across multiple units of analysis, from genes to neural circuits to behaviours, cutting across disorders as traditionally defined.
In summary, it is most likely that progress in the comprehension of the risk factors for neurodevelopmental disorders will come from dimensional and quantitative data that goes well beyond current psychiatric classification. One first step would be to gather the information that is currently separated by DSM-5 diagnostic categories and dispersed in different laboratories that may fail to communicate. To achieve this, there is a need for more data sharing (see below).
Proposition 2: more research on genetic and brain diversity in human populations and more data sharing
Based on current case-control design, there is a tendency for researchers to know better the genotypes and phenotypes of the patients than those of the controls. Indeed, in the vast majority of genetic studies, controls are often not investigated at the phenotypic level, and in phenotypic studies, controls are very limited in their number and their cultural and socioeconomic status diversity (70). As a consequence, early onset developmental disorders are therefore considered as binary traits “affected” versus “non-affected” without taking into account the genetic and phenotypic diversity of both “affected” and “non-affected” individuals. The same is true for studies using transgenic mice; most of our knowledge is based on the effect of the mutations in C57BL6 mice. However, we know that mutations might produce a different phenotype in a different strain. The crucial role of the genetic background was very nicely illustrated in a recent paper showing the phenotypic consequence of the scalloped mutation in different strains of Drosophila melanogaster (71).
No progress could have been made in the genetics of neurodevelopmental disorders if thousands of genomes had not been sequenced to ascertain their genetic diversity. The same is true for human brains. The first initiatives of the Allen Brain Institute (http://www.brain-map.org) or the Sestan Laboratory (http://medicine.yale.edu/lab/sestan/index.aspx) are impressive in their description of human gene expression at very high resolution. However, if we want to better ascertain the links that exist between the variability of genomes and human brains, thousands of brains will need to be studied at the gene expression level as well as the functional level, even if this proves costly and difficult.
Integrating diversity into our experimental design will require an increase in the sample size of our study populations. Indeed, risk factors for early onset neurodevelopmental disorders are either rare with large effect or frequent but with a small effect (72). In both situations, robust genotype–phenotype relationships are difficult to ascertain in small samples. One opportunity to increase sample size is to foster data sharing. Many constraints reduce efficient data sharing (73). Hence, there is need 1) to agree on an ethical informed consent for research subjects that will allow data sharing; 2) to agree on standardized measures, 3) to change the reward system regarding publications, 4) to set up systems to make data sharing easy and secure.
There is an emerging community of researchers involved in data sharing. Specifically in neuroscience, initiatives such as the Neuroscience Information Framework (NIF) or the International Neuroinformatics Coordinating Facility (INCF) were launched recently. NIF (http://www.neuinfo.org) is a dynamic inventory of Web-based neuroscience resources: data, materials, and tools accessible via any computer connected to the Internet. This should advance neuroscience research by enabling discovery and access to public research data and tools worldwide through an open source, networked environment. INCF (http://www.incf.org/) develops collaborative neuroinformatics infrastructure and promotes the sharing of data and computing resources to the international research community. Neuroinformatics integrates information across all levels and scales of neuroscience to help understand the brain and treat disease. In addition to increasing sample size of the studies, these initiatives for more data sharing in the scientific community should also lead to a reduction of the important publication bias in the field of early onset neurodevelopmental disorders (74).
Proposition 3: patients and relatives as participants for research
Many aspects of the quality of life of patients and their relatives are not adequately taken into account by researchers. For example, in ASD, co-morbidities such as gastrointestinal and sensory problems are under-explored. The movement “no research about me, without me” is calling for patients and their relatives to be more involved in research designs. For example, the UK National health Service (NHS) initiative INVOLVE (http://www.invo.org.uk) is a national advisory group that supports greater public involvement in NHS, public health and social care research. There is also the James Lind Alliance (http://www.lindalliance.org), the Patient-Centered Outcomes Research Institute (PCORI) (http://www.pcori.org) and PatientsLikeMe (www.patientslikeme.org) initiatives for patients who want to monitor their own health and chronic illness. Using such frameworks, patients can propose and conduct their own studies among memberships – with some successes to report already. For example, a trial of lithium for amyotrophic lateral sclerosis (ALS) was completed faster than randomized control trials (RCTs) (75). In this study, PatientsLikeMe reached exactly the same conclusion as previous RCTs suggesting that data reported by patients over the Internet may be useful for accelerating clinical discovery and evaluating the effectiveness of drugs already in use. Another example is the initiative for cancer research at Sage Bionetworks (http://sagebase.org). Sage develops tools so that medical patients can keep their own data rather than storing it in particular medical institutions. The aim is to offer predictive, personalized, preventive, participatory (P4) cancer medicine (76). These types of initiatives require “the creation of new types of strategic partnerships – between patients, large clinical centers, consortia of clinical centers and patient-advocate groups. For some clinical trials it will be necessary to recruit very large numbers of patients – and one powerful approach to this challenge is the crowd-sourced recruitment of patients by bringing large clinical centers together with patient-advocacy groups. p. 184” (76)
Perspectives: towards a more inclusive world
For patients, the burden of neurodevelopmental disorders makes daily activities difficult and lowers the odds of living an independent life. Progress on the causes of neurodevelopmental disorders hopefully will provide knowledge-based treatments to improve quality of life for those affected. Nevertheless, in addition to improved medical care, innovative initiatives towards a more inclusive world point towards other important advances. For example, Aspiritech, a non-profit organization based in Highland Park, Illinois, USA, places people who have autism (mainly Asperger's syndrome) in jobs testing software (http://www.aspiritech.org). The Danish company Specialisterne has helped more than 170 individuals with autism obtain jobs since 2004. Its parent company, the Specialist People Foundation, aims to connect one million autistic people with meaningful work (http://www.specialistpeople.com). Laurent Mottron, a psychologist working in Montreal, has offered jobs for patients with ASD in his group and this new perspective has had a positive impact on his research on autism. As he said “The hallmark of an enlightened society is its inclusion of non-dominant behaviours and phenotypes, such as homosexuality, ethnic differences and disabilities. Governments have spent time and money to accommodate people with visual and hearing impairments, helping them to navigate public places and find employment, for instance – we should take the same steps for autistics” (77). As suggested by Waterhouse and Gillberg, it might be better to abandon the belief that there is a single defining ASD brain dysfunction (78). Instead, we should understand the diversity of ASD (or autismS). Considering autism not as a single entity, but as a continuum of human diversity and tackling this heterogeneity using information coming from different fields of research (including direct information from the affected individuals and their families (79)) should allow a better diagnostic, care and integration of individuals with autism (77).
Acknowledgements
I want to thank Steve Hyman and Roberto Toro for their helpful reading of the manuscript. This work was funded by the Institut Pasteur, the Bettencourt-Schueller Foundation, Centre National de la Recherche Scientifique, University Paris Diderot, Agence Nationale de la Recherche (ANR-13-SAMA-0006; SynDivAutism), the Conny-Maeva Charitable Foundation, the Cognacq Jay Foundation, the Orange Foundation, and the Fondation FondaMental.
Disclosure of interest: The author declares that there are no conflicts of interest. The author alone is responsible for the content and writing of the paper.
References
- Gillberg C. The ESSENCE in child psychiatry: early symptomatic syndromes eliciting neurodevelopmental clinical examinations. Res Dev Disabil. 2010;31:1543–51. doi: 10.1016/j.ridd.2010.06.002. [DOI] [PubMed] [Google Scholar]
- Developmental Disabilities Monitoring Network Surveillance Year Principal I. Prevalence of autism spectrum disorder among children aged 8 years – autism and developmental disabilities monitoring network, 11 sites, United States, 2010. MMWR Surveill Summ. 2014;63((Suppl2)):S1–21. [PubMed] [Google Scholar]
- Perou R, Bitsko RH, Blumberg SJ, Pastor P, Ghandour RM, Gfroerer JC, et al. Mental health surveillance among children – United States, 2005–2011. MMWR Surveill Summ. 2013;62((Suppl2)):S1–35. [PubMed] [Google Scholar]
- Bishop DV. Which neurodevelopmental disorders get researched and why? PLoS One. 2010;5:e15112. doi: 10.1371/journal.pone.0015112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoischen A, Krumm N, Eichler EE. Prioritization of neurodevelopmental disease genes by discovery of new mutations. Nat Neurosci. 2014;17:764–72. doi: 10.1038/nn.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Deriziotis P, Lee C, Vives L, Schwartz JJ, Girirajan S, et al. Exome sequencing in sporadic autism spectrum disorders identifies severe de novo mutations. Nat Genet. 2011;43:585–9. doi: 10.1038/ng.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–85. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–99. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488((7412)):471–5. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma'ayan A, Samocha KE, Sabo A, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485((7397)):242–5. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485((7397)):246–50. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485((7397)):237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilissen C, Hehir-Kwa JY, Thung DT, van de Vorst M, van Bon BW, Willemsen MH, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511((7509)):344–7. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485((7397)):237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell CD, Eichler EE. Properties and rates of germline mutations in humans. Trends Genet. 2013;29:575–84. doi: 10.1016/j.tig.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toro R, Konyukh M, Delorme R, Leblond C, Chaste P, Fauchereau F, et al. Key role for gene dosage and synaptic homeostasis in autism spectrum disorders. Trends Genet. 2010;26:363–72. doi: 10.1016/j.tig.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Frank SA. Somatic mosaicism and disease. Curr Biol. 2014;24:R577–81. doi: 10.1016/j.cub.2014.05.021. [DOI] [PubMed] [Google Scholar]
- Poduri A, Evrony GD, Cai X, Walsh CA. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341:1237758. doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Need AC, Petrovski S, Goldstein DB. One gene, many neuropsychiatric disorders: lessons from Mendelian diseases. Nat Neurosci. 2014;17:773–81. doi: 10.1038/nn.3713. [DOI] [PubMed] [Google Scholar]
- Lim ET, Raychaudhuri S, Sanders SJ, Stevens C, Sabo A, MacArthur DG, et al. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron. 2013;77:235–42. doi: 10.1016/j.neuron.2012.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu TW, Chahrour MH, Coulter ME, Jiralerspong S, Okamura-Ikeda K, Ataman B, et al. Using whole-exome sequencing to identify inherited causes of autism. Neuron. 2013;77:259–73. doi: 10.1016/j.neuron.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow EM, Yoo SY, Flavell SW, Kim TK, Lin Y, Hill RS, et al. Identifying autism loci and genes by tracing recent shared ancestry. Science. 2008;321:218–23. doi: 10.1126/science.1157657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Cooper GM, Antonacci F, Siswara P, Itsara A, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42:203–9. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S, Rosenfeld JA, Coe BP, Parikh S, Friedman N, Goldstein A, et al. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N Engl J Med. 2012;367:1321–31. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblond CS, Heinrich J, Delorme R, Proepper C, Betancur C, Huguet G, et al. Genetic and functional analyses of SHANK2 mutations suggest a multiple hit model of autism spectrum disorders. PLoS Genet. 2012;8:e1002521. doi: 10.1371/journal.pgen.1002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilian B, Abdollahpour H, Bierhals T, Haltrich I, Fekete G, Nagel I, et al. Dysfunction of SHANK2 and CHRNA7 in a patient with intellectual disability and language impairment supports genetic epistasis of the two loci. Clin Genet. 2013;84:560–5. doi: 10.1111/cge.12105. [DOI] [PubMed] [Google Scholar]
- Klei L, Sanders SJ, Murtha MT, Hus V, Lowe JK, Willsey AJ, et al. Common genetic variants, acting additively, are a major source of risk for autism. Mol Autism. 2012;3:9. doi: 10.1186/2040-2392-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium Lee SH, Ripke S, Neale BM, Faraone SV, Purcell SM, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–94. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaugler T, Klei L, Sanders SJ, Bodea CA, Goldberg AP, Lee AB, et al. Most genetic risk for autism resides with common variation. Nat Genet. 2014;46:881–5. doi: 10.1038/ng.3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deary IJ, Yang J, Davies G, Harris SE, Tenesa A, Liewald D, et al. Genetic contributions to stability and change in intelligence from childhood to old age. Nature. 2012;482((7384)):212–15. doi: 10.1038/nature10781. [DOI] [PubMed] [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Liewald D, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry. 2011;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–9. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anney R, Klei L, Pinto D, Almeida J, Bacchelli E, Baird G, et al. Individual common variants exert weak effects on the risk for autism spectrum disorderspi. Hum Mol Genet. 2012;21:4781–92. doi: 10.1093/hmg/dds301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratten J, Wray NR, Keller MC, Visscher PM. Large-scale genomics unveils the genetic architecture of psychiatric disorders. Nat Neurosci. 2014;17:782–90. doi: 10.1038/nn.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguet G, Ey E, Bourgeron T. The genetic landscapes of autism spectrum disorders. Annu Rev Genomics Hum Genet. 2013;14:191–213. doi: 10.1146/annurev-genom-091212-153431. [DOI] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472((7344)):437–42. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Mol Autism. 2010;1:15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, Lee H-R, Gee HY, Mah W, Kim J-I, Lee J, et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486((7402)):261–5. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]
- Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel AV, Kuebler A, et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486((7402)):256–60. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, et al. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–6. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamain S, Radyushkin K, Hammerschmidt K, Granon S, Boretius S, Varoqueaux F, et al. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proc Natl Acad Sci USA. 2008;105:1710–15. doi: 10.1073/pnas.0711555105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, et al. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–54. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Baudouin SJ, Gaudias J, Gerharz S, Hatstatt L, Zhou K, Punnakkal P, et al. Shared synaptic pathophysiology in syndromic and nonsyndromic rodent models of autism. Science. 2012;338:128–32. doi: 10.1126/science.1224159. [DOI] [PubMed] [Google Scholar]
- Han K, Holder JL, Jr, Schaaf CP, Lu H, Chen H, Kang H, et al. SHANK3 overexpression causes manic-like behaviour with unique pharmacogenetic properties. Nature. 2013;503((7474)):72–7. doi: 10.1038/nature12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ey E, Leblond CS, Bourgeron T. Behavioral profiles of mouse models for autism spectrum disorders. Autism Res. 2011;4:5–16. doi: 10.1002/aur.175. [DOI] [PubMed] [Google Scholar]
- Shcheglovitov A, Shcheglovitova O, Yazawa M, Portmann T, Shu R, Sebastiano V, et al. SHANK3 and IGF1 restore synaptic deficits in neurons from 22q13 deletion syndrome patients. Nature. 2013;503((7475)):267–71. doi: 10.1038/nature12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissart C, Poulet A, Georges P, Darville H, Julita E, Delorme R, et al. Differentiation from human pluripotent stem cells of cortical neurons of the superficial layers amenable to psychiatric disease modeling and high-throughput drug screening. Transl Psychiatry. 2013;3:e294. doi: 10.1038/tp.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Sanders SJ, Li M, Dong S, Tebbenkamp AT, Muhle RA, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155:997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, et al. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–21. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale A, Larimore J, Werner E, So L, Moreno-De-Luca A, Lese-Martin C, et al. Quantitative proteomic and genetic analyses of the schizophrenia susceptibility factor dysbindin identify novel roles of the biogenesis of lysosome-related organelles complex 1. J Neurosci. 2012;32:3697–711. doi: 10.1523/JNEUROSCI.5640-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466((7304)):368–72. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474((7351)):380–4. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–8. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novarino G, El-Fishawy P, Kayserili H, Meguid NA, Scott EM, Schroth J, et al. Mutations in BCKD-kinase lead to a potentially treatable form of autism with epilepsy. Science. 2012;338:394–7. doi: 10.1126/science.1224631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, III, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135:401–6. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Costa-Mattioli M, Monteggia LM. mTOR complexes in neurodevelopmental and neuropsychiatric disorders. Nat Neurosci. 2013;16:1537–43. doi: 10.1038/nn.3546. [DOI] [PubMed] [Google Scholar]
- Ba W, van der Raadt J, Nadif Kasri N. Rho GTPase signaling at the synapse: implications for intellectual disability. Exp Cell Res. 2013;319:2368–74. doi: 10.1016/j.yexcr.2013.05.033. [DOI] [PubMed] [Google Scholar]
- Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19:231–4. doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, et al. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–9. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–7. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari P, Paterson AD, Zwaigenbaum L, Roberts W, Brian J, Liu XQ, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–28. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramocki MB, Zoghbi HY. Failure of neuronal homeostasis results in common neuropsychiatric phenotypes. Nature. 2008;455((7215)):912–18. doi: 10.1038/nature07457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooren W, Lindemann L, Ghosh A, Santarelli L. Synapse dysfunction in autism: a molecular medicine approach to drug discovery in neurodevelopmental disorders. Trends Pharmacol Sci. 2012;33:669–84. doi: 10.1016/j.tips.2012.09.004. [DOI] [PubMed] [Google Scholar]
- Delorme R, Ey E, Toro R, Leboyer M, Gillberg C, Bourgeron T. Progress toward treatments for synaptic defects in autism. Nat Med. 2013;19:685–94. doi: 10.1038/nm.3193. [DOI] [PubMed] [Google Scholar]
- Moreno-De-Luca A, Myers SM, Challman TD, Moreno-De-Luca D, Evans DW, Ledbetter DH. Developmental brain dysfunction: revival and expansion of old concepts based on new genetic evidence. Lancet Neurol. 2013;12:406–14. doi: 10.1016/S1474-4422(13)70011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YS, State MW. Recent challenges to the psychiatric diagnostic nosology: a focus on the genetics and genomics of neurodevelopmental disorders. Int J Epidemiol. 2014;43:465–75. doi: 10.1093/ije/dyu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manly JJ. Critical issues in cultural neuropsychology: profit from diversity. Neuropsychol Rev. 2008;18:179–83. doi: 10.1007/s11065-008-9068-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari S, Dworkin I. The conditional nature of genetic interactions: the consequences of wild-type backgrounds on mutational interactions in a genome-wide modifier screen. PLoS Genet. 2013;9:e1003661. doi: 10.1371/journal.pgen.1003661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll SA, Feng G, Hyman SE. Genome-scale neurogenetics: methodology and meaning. Nat Neurosci. 2014;17:756–63. doi: 10.1038/nn.3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poline JB, Breeze JL, Ghosh S, Gorgolewski K, Halchenko YO, Hanke M, et al. Data sharing in neuroimaging research. Front Neuroinform. 2012;6:9. doi: 10.3389/fninf.2012.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joober R, Schmitz N, Annable L, Boksa P. Publication bias: what are the challenges and can they be overcome? J Psychiatry Neurosci. 2012;37:149–52. doi: 10.1503/jpn.120065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks P, Vaughan TE, Massagli MP, Heywood J. Accelerated clinical discovery using self-reported patient data collected online and a patient-matching algorithm. Nat Biotechnol. 2011;29:411–14. doi: 10.1038/nbt.1837. [DOI] [PubMed] [Google Scholar]
- Hood L, Friend SH. Predictive, personalized, preventive, participatory (P4) cancer medicine. Nat Rev Clin Oncol. 2011;8:184–7. doi: 10.1038/nrclinonc.2010.227. [DOI] [PubMed] [Google Scholar]
- Mottron L. Changing perceptions: The power of autism. Nature. 2011;479((7371)):33–5. doi: 10.1038/479033a. [DOI] [PubMed] [Google Scholar]
- Waterhouse L, Gillberg C. Why autism must be taken apart. J Autism Dev Disord. 2014;44:1788–92. doi: 10.1007/s10803-013-2030-5. [DOI] [PubMed] [Google Scholar]
- Kohane IS, Eran A. Can we measure autism? Sci Transl Med. 2013;5:209ed, 18. doi: 10.1126/scitranslmed.3007340. [DOI] [PubMed] [Google Scholar]