Abstract
Introduction
Placental function is an important determinant of fetal growth, and fetal growth influences obesity risk in childhood and adult life. Here we investigated how FTO and MC4R gene variants linked with obesity relate to patterns of fetal growth and to placental FTO expression.
Methods
Southampton Women's Survey children (n = 1990) with measurements of fetal growth from 11 to 34 weeks gestation were genotyped for common gene variants in FTO (rs9939609, rs1421085) and MC4R (rs17782313). Linear mixed-effect models were used to analyse relations of gene variants with fetal growth.
Results
Fetuses with the rs9939609 A:A FTO genotype had faster biparietal diameter and head circumference growth velocities between 11 and 34 weeks gestation (by 0.012 (95% CI 0.005 to 0.019) and 0.008 (0.002–0.015) standard deviations per week, respectively) compared to fetuses with the T:T FTO genotype; abdominal circumference growth velocity did not differ between genotypes. FTO genotype was not associated with placental FTO expression, but higher placental FTO expression was independently associated with larger fetal size and higher placental ASCT2, EAAT2 and y + LAT2 amino acid transporter expression. Findings were similar for FTO rs1421085, and the MC4R gene variant was associated with the fetal growth velocity of head circumference.
Discussion
FTO gene variants are known to associate with obesity but this is the first time that the risk alleles and placental FTO expression have been linked with fetal growth trajectories. The lack of an association between FTO genotype and placental FTO expression adds to emerging evidence of complex biology underlying the association between FTO genotype and obesity.
Keywords: Fetal growth trajectories, FTO genotype, MC4R genotype, Placental amino acid transporter expression, Placental FTO expression
Highlights
-
•
Variants in the FTO gene are previously known to be associated with obesity.
-
•
discovered novel associations between FTO variants and growth trajectory of fetal head measures.
-
•
also found novel associations between placental FTO expression and fetal size.
1. Introduction
Obesity is a major public health problem as it increases the risk of coronary heart disease, stroke, metabolic syndrome, some cancers and a wide range of other diseases. Children who are obese are likely to continue to be obese when they become adults, highlighting the need for research into early determinants of obesity [1]. In order to develop effective interventions it is important to understand the genetic and environmental determinants of obesity. There is evidence suggesting critical windows during fetal and early postnatal life, within which altered development may predispose the individual to obesity in later life [2], [3].
Genome wide association studies (GWAS) have been instrumental in the discovery of genes associated with obesity, including the fat mass and obesity-associated (FTO) gene [4]. A common variant in the FTO gene, rs9939609 has been identified that predisposes to type 2 diabetes through an effect on body mass index (BMI) [4], [5], [6]. FTO variants, including rs9939609 and rs1421085, have been reported to be associated with BMI from age 5.5 years [4], [7], [8], [9], [10], [11] and throughout adult life [4], [5], [6], [12], although associations are less consistent in childhood. In addition, one study has shown that FTO genotype is associated with fat mass in the first 2 weeks of life [13].
Associations have also been observed between the genotype of a Single Nucleotide Polymorphism (SNP), rs17782313, near the MC4R gene [14], [15] and adult obesity. This SNP is also associated with obesity in children [14], [16], [17] and one study reported an association with changes in BMI over the first two weeks of life [18]. MC4R genotype is thought to act through an influence on satiety and it is therefore interesting to investigate the effect of this SNP on the fetus where conventional appetite mechanisms cannot play a part.
FTO is an RNA demethylase and could act through regulation of mRNA stability [19]. However, no direct connection between obesity associated variants and FTO expression and function has been made. A recent study has demonstrated that obesity-associated non-coding variants in FTO affect the expression of the gene IRX3 in humans, mice and zebrafish [20]. Obesity associated SNPs in FTO were found to be associated with the expression of IRX3, but not FTO, in the cerebellum of the human brain. IRX3 is a homeobox gene involved in pattern formation in the early embryo and is expressed at much lower levels in later life [21]. This raises the possibility that the effects of FTO genotype on fetal growth and postnatal obesity may have originated through IRX3 expression in embryonic life.
Recent research has shown that FTO plays a key role in the cellular sensing of amino acids and the regulation of cell growth and global mRNA translation through the mTORC1 pathway [22]. This could explain why carriers of the obesity predisposing SNPs in FTO not only consume more calories during test meals but also show an alteration in nutrient preference [23] and a higher dietary protein intake [24]. In utero, fetal growth is regulated by placental amino acid transporters as they control the supply of nutrients to the fetus. Thus FTO could potentially regulate fetal growth through a mechanism of altered placental amino acid transport.
The primary aim of this study was to investigate the association of obesity related SNPs (two FTO SNPs and one SNP near MC4R) with fetal growth throughout gestation in children of the Southampton Women's Survey (SWS) [25]. The secondary aim of this study was to investigate associations between FTO genotype, the expression of this gene in the placenta, a fetal tissue, and placental amino acid transporter expression.
2. Methods
Offspring of participants in the Southampton Women's Survey were studied [25]. Between 1998 and 2007, 3158 babies were born and data gathered on these babies both during the pregnancy via ultrasound scans and after the birth.
Gestational age of the SWS babies was determined using an algorithm based on last menstrual period, or where this was not available early ultrasound data. Using Acuson 128 XP, Aspen and Sequoia ultrasound machines calibrated to 1540 m/s; experienced research ultrasonographers used standardised anatomical landmarks to measure fetal head, abdominal circumference and biparietal diameter at 11, 19 and 34 weeks gestation.
The method of Royston [26] was used to calculate measures of fetal size, correcting for exact gestational age at measurement.
Maternal smoking during pregnancy was assessed by questionnaire in early and late pregnancy.
DNA was extracted from the cord blood of SWS babies by the salting out method and stored at −80 °C. 1990 children from the SWS cohort with DNA available were genotyped for the following polymorphisms: rs9939609 and rs1421085 located in the FTO gene on chromosome 16, and rs17782313 located near the MC4R gene on chromosome 18. Genotyping was performed by Kbiosciences (Hoddesdon, Herts, UK) using a custom SNP panel and an in-house calling algorithm. All three SNPs were found to be in HWE (chi squared p value ≥ 0.05); call rate was ≥ 97%; 5% duplicates were included with an error rate of 0%.
We analysed 99 SWS placentas based on availability of neonatal data and Caucasian ethnicity and collection within 30 min of delivery. Five villous tissue samples were selected using a stratified random sampling method and stored at −80 °C and powdered in a frozen tissue press. Total RNA was extracted from 30 mg tissue using the Rneasy fibrous tissue RNA isolation mini kit (Qiagen, UK) according to the manufacturer's instructions. The integrity of total RNA was confirmed by agarose gel electrophoresis. Total RNA (0.2 μg) was reverse transcribed with 0.5 μg random hexamer primer, 200 units M-MLV reverse transcriptase, 25 units recombinant RNasin ribonuclease inhibitor and 0.5 mM each of dATP, dCTP, dGTP and dTTP in a final reaction volume of 25 μl in 1 x MMLV reaction buffer (Promega, Wisconsin, USA). All 99 samples were produced in one batch to reduce variation.
Oligonucleotide probes and primers for FTO and the amino acid transporter genes were designed using the Roche (West Sussex, UK) ProbeFinder version 2.45 for human. Probes were supplied by Roche from the human universal probe library and primers were synthesised by Eurogentec (Seraing, Belgium). FTO (NM_001080432.2): Forward 5′- cacaacctcggtttagttcca -3′, Reverse 5′- aaatataatccaaggttcctgttgag -3′, probe #53. ASCT2 (NM_005628.2): Forward 5′-gaggaatatcaccggaacca-3′, Reverse 5′-aggatgttcatcccctcca-3′, probe 43. EAAT2 (NM_004171.3): Forward 5′-aaaatgctcattctccctctaatc-3′, Reverse 5′-gccactagccttagcatcca-3′, probe 78. y+LAT2 (NM_001076785.1): Forward 5′-gctgtgatcccccatacct-3′, Reverse 5′-ggcacagttcacaaatgtcag-3′, probe 66. Control genes were selected using the geNormTM human Housekeeping Gene Selection Kit (Primer Design Limited, Southampton, UK).
FTO and amino acid transporter mRNA expression levels were quantified by real-time reverse transcriptase PCR using a Roche LightCycler 480. For Roche universal probe library probes the cycle parameters were 95 °C for 5 min, followed by 45 cycles of 95 °C for 10 s, 60 °C for 30 s and 72 °C for 1 s. For primer design Perfect Probes (control genes) the cycle parameters were 95 °C for 10 min, followed by 40–50 cycles of 95 °C for 15 s, 50 °C for 30 s and 72 °C for 15 s. Intra-assay CV's for each gene were 5–8%. Each of the 99 samples was run on the same plate in triplicate. All mRNA levels are presented relative to the geometric mean of the three control genes, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide (YWHAZ), ubiquitin C (UBC) and topoisomerase (TOP1) [27].
2.1. Statistical analysis
Statistical procedures were performed in Stata version 12 (StataCorp, Texas, USA) and SPSS version 21 (IBM, Armonk, New York). Babies with congenital growth abnormalities and babies of mothers listed as having non-white ethnicity were excluded from analysis.
Anthropometric variables were checked for normality and then standardised to z-scores. For cross-sectional analysis genotypes were coded according to the additive model (0, 1 or 2 copies of the risk allele). The risk allele was considered to be A for FTO rs9939609, C for FTO rs1421085 and C for MC4R rs17782313. Univariate linear regressions were run with each genotype separately as predictor variable for each outcome (CRL, BPD, HC and AC), adjusting for sex, maternal smoking during pregnancy, pregnancy weight gain category (IOM (2009)) and pre-pregnancy maternal BMI. Results were expressed in SDs of change in outcome per unit increase in number of risk alleles.
Linear mixed-effect models [28] were used to conduct the longitudinal analyses. Intercept and gestational age were entered into the model as random effects and a genotype x gestational age interaction was also included to assess growth velocity in the measurements over time. Sex, maternal smoking, pregnancy weight gain category and pre-pregnancy maternal BMI were included as covariates. Genotype was included as a categorical covariate because exploratory analysis (see Fig. 1) indicated that this was more appropriate.
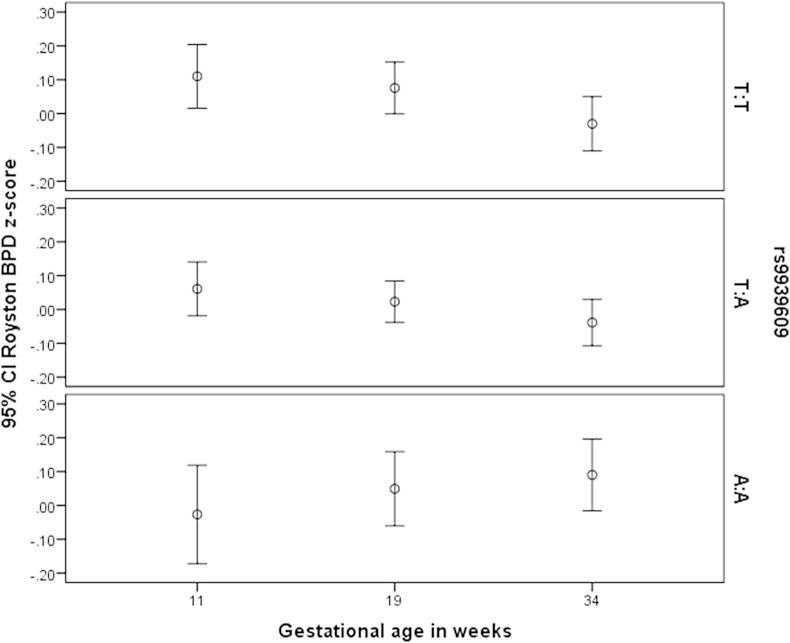
Fig. 1.
The means and 95% CIs of Royston biparietal diameter z-score at all three available gestational ages for each rs9939609 genotype separately. For the T:T and T:A genotypes the biparietal diameter z-score decreases with increasing gestational age, however for the A:A genotype biparietal diameter z-score increases with gestational age. This shows that the biparietal diameter for the A:A genotype has a significantly different fetal growth trajectory, with fetal biparietal diameter growing faster than the T:T genotype. This was confirmed by linear mixed-effect modelling (p = 0.0009).
Partial correlations of FTO gene expression in the placenta and anthropometric variables, were calculated, controlling for sex. Neonatal variables were additionally adjusted for gestational age and mode of delivery (Caesarean or vaginal).
Bonferroni correction was applied to account for multiple testing for each of the main hypotheses tested.
3. Results
Analysis of fetal growth, was based on 1990 SWS singleton births with available fetal measurements and FTO or MC4R genotype or placental gene RNA data. Supplementary Figure 1 shows the numbers of SWS babies with available DNA and genotype. Table 1 shows summary statistics for the SWS cohort divided into groups according to whether genotype was available or not. There does not appear to be much difference between these groups.
The minor allele frequency for the FTO rs9939609 genotype in the SWS was A = 0.41, similar to the minor allele frequency quoted in HapMap [29] for Caucasians (CEU: A = 0.45).
3.1. Cross-sectional analysis
Table 2 shows there were a number of potentially significant associations at the 5% level between FTO and MC4R SNPs and fetal anthropometric measurements. While none of these associations remained significant at p < 0.05 after a strict Bonferroni correction, many of the fetal measures are correlated, so a Bonferroni correction is likely to be overly conservative. A retrospective power calculation indicated that to find a difference in means of fetal measure z-scores between genotype groups of 0.18, 1446 SWS children would be required to achieve a power of 90% at a 5% significance level.
Fig. 1 shows the means and 95% confidence intervals for biparietal diameter z-score at 11, 19 and 34 weeks for the FTO rs9939609 genotypes. At 11 weeks gestation, fetuses with the A:A genotype tended to have a lower biparietal diameter z-score than either the T:A genotype or the T:T genotype. At 19 weeks, biparietal diameter z-score was similar in the three genotypes. However, at 34 weeks gestation, fetuses with the A:A genotype had a higher biparietal diameter z-score than either of the other two genotypes. This implies a different fetal biparietal diameter growth trajectory in fetuses with the A:A genotype compared to the other two genotypes, with a smaller 11 week size followed by a faster growth velocity for the A:A genotype.
3.2. Linear mixed-effect models
Maternal weight gain category and sex were significantly associated with biparietal diameter, head circumference and abdominal circumference when included in linear mixed-effect models (p < 0.0001), with males having a larger measurement than females. Maternal smoking in pregnancy was associated with biparietal diameter and head circumference (p ≤ 0.05), with the fetuses of mothers who did not smoke in pregnancy having a larger head size. Maternal smoking in pregnancy was not associated with abdominal circumference (p > 0.05). Maternal pre-pregnancy BMI was significantly associated with head circumference and abdominal circumference (p ≤ 0.007) but not with biparietal diameter.
Linear mixed-effect models for two measures of head size, namely biparietal diameter and head circumference z-score, measured at 11, 19 and 34 weeks gestation, showed that FTO genotype rs9939609 was strongly associated with the growth velocity of head size, as there was a significant interaction between genotype and gestational age (p = 0.0009 for biparietal diameter and p = 0.014 for head circumference). This demonstrates that FTO genotypes have different growth trajectories. When abdominal circumference z-score was used as the dependent variable, there was no association between the growth velocity of abdominal circumference and FTO genotype. The results from linear mixed-effect models analysing the rs1421085 genotype were very similar to those using the rs9939609 genotype due to the high LD between them (data not shown). When the MC4R genotype rs17782313 was used as a predictor in the longitudinal models there were no significant associations between this SNP and the growth velocity of biparietal diameter and abdominal circumference, but there was an association between MC4R and head circumference growth velocity (p = 0.013) across the three gestational ages of 11, 19 and 34 weeks.
Interactions between FTO genotype and sex, and between FTO genotype and maternal smoking were investigated but no significant interactions were found (p > 0.05).
3.3. Placental FTO relative RNA level
Placental FTO relative RNA level was available for 99 offspring (mean 1.23, sd 0.27).
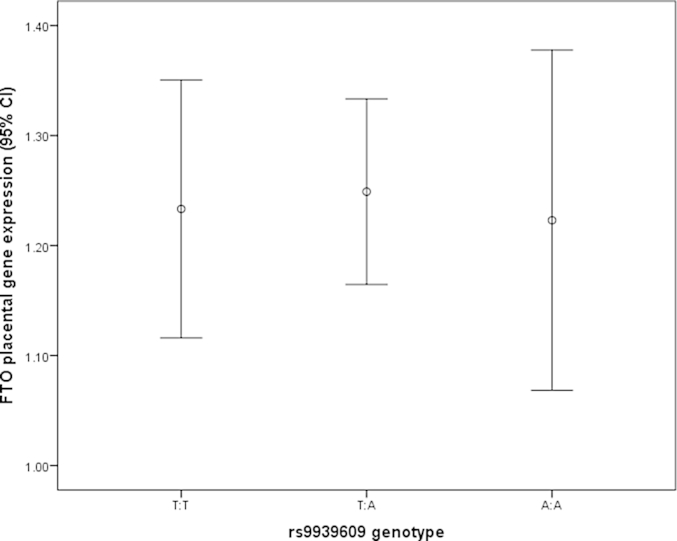
No association was observed between FTO genotype and relative RNA levels of FTO in the placenta (assessed by ANOVA, p value = 0.914, Fig. 2).
Fig. 2.
There is no association between FTO genotype and expression of FTO in the placenta (ANOVA p value = 0.914).
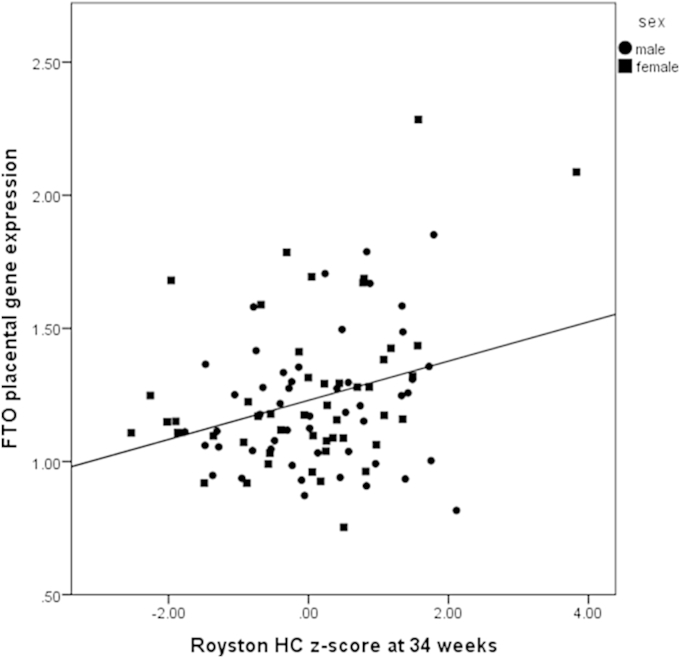
Table 3 shows partial correlations, controlling for sex, of fetal anthropometric measurements with placental FTO relative gene expression. Neonatal measures were adjusted for sex, gestational age and mode of delivery. Several fetal anthropometric measurements appeared highly correlated with the FTO expression in the placenta (Table 3, Fig. 3). Head circumference at 34 weeks was significantly associated with placental FTO relative gene expression after Bonferroni correction and neonatal head circumference showed a borderline association.
Fig. 3.
The correlation between FTO relative RNA level in the placenta and 34 week Royston head circumference z-score (r = 0.324, p = 0.001, n = 98). Males were plotted using circles and females using squares.
We also attempted to measure the levels of IRX3 in the term human placenta, but found that levels were too low to measure reliably.
Associations were also found between FTO mRNA expression and the expression of placental amino acid transporters. FTO expression was correlated with ASCT2 (rho = 0.29, p = 0.004), EAAT2 (rho = 0.22, p = 0.027) and y+LAT2 (rho = 0.37, p = 0.0002) mRNA expression.
4. Discussion
We have shown novel associations between FTO genotypes rs9939609 and rs1421085 and two measures of fetal head growth. From 11 to 34 weeks gestation fetuses with the homozygous FTO risk genotype (A:A or C:C) showed a higher velocity of biparietal diameter and head circumference growth in comparison with the other genotypes (T:A or T:C and T:T). Results were also in accordance with previous publications showing that maternal smoking is significantly associated with smaller fetal head size [30]. Trends were also identified in our cross-sectional analyses between the FTO SNPs and a SNP near MC4R and head circumference and crown-rump length, although these did not survive Bonferroni correction. For the above associations, the effect was in accordance with published literature for adults and babies [14], [15], [18] as the addition of each risk allele increased the measurements. We did not observe associations between the MC4R SNP and fetal growth trajectories of biparietal diameter and abdominal circumference, or between FTO SNPs and the fetal abdominal circumference growth trajectory.
Few authors have investigated the possible effect of FTO SNPs on fetal growth. Marsh et al. [31] reported that there was no evidence of any difference in fetal growth between the sexes and between rs9930609 genotype up to 28 weeks gestation. After 28 weeks gestation they observed a significant association between the rs9939609 A:A genotype and fetal growth restriction in non-smoking Australian mothers and fetal growth enhancement for mothers who smoked. This differs from our results as we did not observe a significant interaction between maternal smoking and genotype. However, the two sets of results are difficult to compare as our measurements were made by research staff following strict measurement protocols and we have considered the growth trajectory over all available gestational ages in the Southampton Women's Survey cohort; in contrast Marsh et al. used routinely collected data and analysed each trimester of pregnancy separately.
Placental function is an important determinant of fetal growth and may predispose to obesity later in life [32]. Our observations that FTO gene expression was correlated with birth weight are consistent with those of previous studies [33], [34], [35]. In addition we found novel associations between FTO gene expression and fetal growth parameters. However, placental FTO expression was not associated with either of the FTO genotypes in our SWS cohort, consistent with a previous report [34]. It is now thought that mutations in Intron 1 of the FTO gene control the expression of IRX3 [20]. Our observation that FTO expression was independent of its genotype is consistent with the suggestion that the associations between FTO genotype and fetal growth are not mediated via FTO expression. If FTO is not regulated by genotype the question arises as to what is regulating FTO expression. FTO may be involved in nutrient sensing and regulate the mTOR pathway and, in the placenta, FTO gene expression may be related to maternal nutritional status. We observed that FTO gene expression was related to the expression of specific amino acid transporters in the placenta. As amino acid transfer is essential for fetal development this provides a potential mechanism by which FTO expression within the placenta may affect placental function and therefore fetal growth [36].
Consistent with the observation that FTO genotype is associated with fetal head growth; in humans a ‘loss of function’ mutation in the FTO gene has been found to induce postnatal growth retardation and malformations in the head and brain [37] Other recent studies have reported that carriers of common FTO gene polymorphisms show both a reduction in frontal lobe volume of the brain [38] and an impaired verbal fluency performance [39], It was interesting that FTO genotype (thought to be controlling IRX3 gene expression) and FTO gene expression (which is independent of FTO genotype rs9939609) were both associated with fetal head growth in our study, however it is not clear whether there is a biological relationship underlying this. FTO knockout mice have altered head growth [40], but as knocking out the FTO gene also knocks out the FTO SNPs regulating IRX3 it is not clear whether the effects of FTO knockout is due to the loss of FTO gene expression or the loss of regulation of the IRX3 gene.
Fetal growth is a complex trait which is likely to be determined by multiple genetic [41] and environmental [42] factors. The cumulative effect of multiple variants could produce clinically relevant differences in growth. The detection of novel genetic variants associated with fetal growth has the potential to identify molecular mechanisms connected with growth and can yield insights of biological importance.
Author contributions
SJB contributed to the design of the study, performed the statistical analysis and interpretation of the results and drafted the article. SRC contributed to the statistical analysis, acquisition of data and interpretation of the results. MM contributed to acquisition of data, analysis and interpretation of data. JKC contributed to the design and interpretation of data. ASF contributed to the statistical analysis and interpretation of the data. CC contributed to the design, acquisition and interpretation of data. HMI contributed to acquisition of data, interpretation of data, revision and final approval of paper. JWH contributed to design, acquisition of data and interpretation of the data. RML contributed to sample collection, design and interpretation of the data. KMG contributed to design, analysis and interpretation of data. All authors reviewed and approved the final version of the article.
Conflict of interest
Keith Godfrey has received reimbursement for speaking at conferences sponsored by companies selling nutritional and pharmaceutical products. One of the research groups involved in this work are part of an academic consortium that has received funding from Abbott Nutrition, Nestec and Danone.
Ethical approval
The study was conducted according to the guidelines in the Declaration of Helsinki, and the Southampton and South West Hampshire Research Ethics Committee approved all procedures (276/97, 307/97).
Informed consent
Written informed consent was obtained from all participating women and by parents or guardians with parental responsibility on behalf of their children.
Table 1.
Summary statistics for the Southampton Women’s Survey Cohort, showing subgroups defined by availability of genotype
| Variable | Genotype available |
Genotype not available |
||||
|---|---|---|---|---|---|---|
| N | Mean | Std. Dev. | N | Mean | Std. Dev. | |
| 11 week scan: Crown rump length (mm) | 1593 | 53.48 | 9.07 | 808 | 53.32 | 8.88 |
| 11 week scan: Biparietal diameter (mm) | 1247 | 18.72 | 2.59 | 609 | 18.66 | 2.52 |
| 19 week scan: Biparietal diameter (mm) | 1912 | 45.5 | 2.54 | 961 | 45.75 | 2.49 |
| 34 week scan: Biparietal diameter (mm) | 1869 | 86.98 | 3.6 | 896 | 87.09 | 3.51 |
| 11 week scan: Head circumference (mm) | 1223 | 70.46 | 9.41 | 602 | 69.99 | 8.94 |
| 19 week scan: Head circumference (mm) | 1912 | 168.43 | 8.53 | 960 | 168.79 | 8.53 |
| 34 week scan: Head circumference (mm) | 1869 | 317.94 | 10.98 | 902 | 317.42 | 11.02 |
| Birth: Head circumference (mm) | 1955 | 350.33 | 13.47 | 848 | 348.74 | 14.87 |
| 11 week scan: Abdominal circumference (mm) | 1152 | 55.99 | 7.91 | 555 | 55.96 | 7.42 |
| 19 week scan: Abdominal circumference (mm) | 1902 | 146.46 | 8.95 | 960 | 146.97 | 8.75 |
| 34 week scan: Abdominal circumference (mm) | 1958 | 307.87 | 15.42 | 940 | 307.22 | 15.02 |
| Birth: Abdominal circumference (mm) | 1953 | 318 | 20.18 | 846 | 315.22 | 21.86 |
| Birth: Birthweight (g) | 1976 | 3485.49 | 506.91 | 984 | 3373.68 | 623.77 |
| Maternal pre-pregnancy BMI (kg/m2) | 1964 | 25.35 | 4.77 | 999 | 25 | 4.78 |
| Sex | 1982 | 51.6% male | 1008 | 52.5% male | ||
| IOM maternal weight gain | 1744 | 28.5% adequate | 600 | 31.7% adequate | ||
| maternal smoking | 1944 | 16.9% yes | 887 | 16.1% yes | ||
| maternal pre-eclampsia | 1982 | 2.8% yes | 1008 | 3.3% yes | ||
| maternal gestational diabetes | 1982 | 1.1% yes | 1008 | 1.0% yes | ||
| SGA (WHO UK z-score ) | 1976 | 7.5% yes | 984 | 8.8% yes | ||
Table 2.
Regression results for cross-sectional analysis of fetal measurements, controlling for sex, maternal smoking during pregnancy, gestational weight gain and maternal BMI. Beta values represent change in fetal measure per risk allele, p values are uncorrected.
| Variable | FTO rs9939609 |
FTO rs1421085 |
MC4R rs17782313 |
||||||
|---|---|---|---|---|---|---|---|---|---|
| beta | se | p value | beta | se | p value | beta | se | p value | |
| 11 week scan: Royston CRL z-score | -0.066 | 0.04 | 0.099 | -0.081 | 0.039 | 0.04 | 0.088 | 0.046 | 0.056 |
| 11 week scan: Royston BPD z-score | -0.074 | 0.043 | 0.083 | -0.076 | 0.042 | 0.073 | -0.026 | 0.049 | 0.592 |
| 19 week scan: Royston BPD z-score | -0.023 | 0.032 | 0.472 | -0.033 | 0.032 | 0.299 | 0.025 | 0.038 | 0.509 |
| 34 week scan: Royston BPD z-score | 0.034 | 0.035 | 0.33 | 0.022 | 0.034 | 0.526 | 0.05 | 0.04 | 0.211 |
| 11 week scan: Royston HC z-score | -0.11 | 0.042 | 0.013 | -0.1 | 0.042 | 0.016 | -0.061 | 0.049 | 0.22 |
| 19 week scan: Royston HC z-score | -0.031 | 0.032 | 0.332 | -0.035 | 0.031 | 0.266 | 0.03 | 0.037 | 0.41 |
| 34 week scan: Royston HC z-score | -0.011 | 0.034 | 0.74 | -0.016 | 0.034 | 0.644 | 0.059 | 0.04 | 0.136 |
| 11 week scan: Royston AC z-score | -0.071 | 0.047 | 0.128 | -0.083 | 0.047 | 0.076 | -0.048 | 0.053 | 0.373 |
| 19 week scan: Royston AC z-score | -0.027 | 0.032 | 0.396 | -0.03 | 0.031 | 0.332 | 0.047 | 0.036 | 0.201 |
| 34 week scan: Royston AC z-score | -0.022 | 0.033 | 0.51 | -0.025 | 0.033 | 0.446 | 0.054 | 0.039 | 0.16 |
| Birth weight z-score (also controlling for GA) | 0.004 | 0.03 | 0.888 | 0.012 | 0.03 | 0.678 | 0.052 | 0.035 | 0.142 |
Table 3.
Partial correlations, controlling for sex, of anthropometric measurements and placental FTO relative RNA level
| Variable | Correlation | Significance (2-tailed) | N |
|---|---|---|---|
| 11 week scan: Royston CRL z-score | 0.289 | 0.020 | 66 |
| 11 week scan: Royston BPD z-score | 0.359 | 0.014 | 47 |
| 19 week scan: Royston BPD z-score | 0.212 | 0.045 | 91 |
| 34 week scan: Royston BPD z-score | 0.241 | 0.018 | 97 |
| 11 week scan: Royston HC z-score | 0.205 | 0.172 | 47 |
| 19 week scan: Royston HC z-score | 0.180 | 0.090 | 91 |
| 34 week scan: Royston HC z-score | 0.324 | 0.001 | 98 |
| 11 week scan: Royston AC z-score | 0.345 | 0.023 | 44 |
| 19 week scan: Royston AC z-score | 0.179 | 0.091 | 91 |
| 34 week scan: Royston AC z-score | 0.242 | 0.017 | 98 |
| Birth weight (controlling for sex, GA and mode of delivery) | 0.204 | 0.046 | 99 |
| Neonatal HC (controlling for sex, GA and mode of delivery) | 0.276 | 0.006 | 99 |
| Neonatal AC (controlling for sex, GA and mode of delivery) | 0.165 | 0.107 | 99 |
Acknowledgements
This work was supported by grants from the UK Medical Research Council (MC_U147585827); British Heart Foundation; Arthritis Research UK; National Osteoporosis Society; International Osteoporosis Foundation; Cohen Trust; National Institute for Health Research Musculoskeletal Biomedical Research Unit, University of Oxford; National Institute for Health Research Southampton Biomedical Research Centre, University of Southampton and University Hospital Southampton National Health Service Foundation Trust; and by the European Union's Seventh Framework Programme (FP7/2007–2013), project EarlyNutrition under grant agreement n°289346. The funders had no role in study design, data collection and analysis, decision to publish or preparation of manuscript.
We thank the mothers of the Southampton Women's Survey who gave us their time and acknowledge the work of the Southampton Women's Survey Study Group, and the team of dedicated research nurses and ancillary staff for their assistance.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.placenta.2015.12.015.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Organization WH . 2014. Childhood Overweight and Obesity.http://www.who.int/dietphysicalactivity/childhood/en/ Available from: [Google Scholar]
- 2.Field C.J. Early risk determinants and later health outcomes: implications for research prioritization and the food supply. Summary of the workshop. Am. J. Clin. Nutr. 2009;89(5):1533s–1539s. doi: 10.3945/ajcn.2009.27113H. [DOI] [PubMed] [Google Scholar]
- 3.Okubo H., Crozier S.R., Harvey N.C., Godfrey K.M., Inskip H.M., Cooper C. Maternal dietary glycemic index and glycemic load in early pregnancy are associated with offspring adiposity in childhood: the Southampton Women's survey. Am. J. Clin. Nutr. 2014;100(2):676–683. doi: 10.3945/ajcn.114.084905. [DOI] [PubMed] [Google Scholar]
- 4.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dina C., Meyre D., Gallina S., Durand E., Korner A., Jacobson P. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat. Genet. 2007;39(6):724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 6.Scuteri A., Sanna S., Chen W.M., Uda M., Albai G., Strait J. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouchard C. Childhood obesity: are genetic differences involved? Am. J. Clin. Nutr. 2009;89(5):1494s–1501s. doi: 10.3945/ajcn.2009.27113C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouchard C. Defining the genetic architecture of the predisposition to obesity: a challenging but not insurmountable task. Am. J. Clin. Nutr. 2010;91(1):5–6. doi: 10.3945/ajcn.2009.28933. [DOI] [PubMed] [Google Scholar]
- 9.Haworth C.M., Carnell S., Meaburn E.L., Davis O.S., Plomin R., Wardle J. Increasing heritability of BMI and stronger associations with the FTO gene over childhood. Obes. (Silver Spring, Md) 2008;16(12):2663–2668. doi: 10.1038/oby.2008.434. [DOI] [PubMed] [Google Scholar]
- 10.Sovio U., Mook-Kanamori D.O., Warrington N.M., Lawrence R., Briollais L., Palmer C.N. Association between common variation at the FTO locus and changes in body mass index from infancy to late childhood: the complex nature of genetic association through growth and development. PLoS Genet. 2011;7(2):e1001307. doi: 10.1371/journal.pgen.1001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wardle J., Carnell S., Haworth C.M., Farooqi I.S., O'Rahilly S., Plomin R. Obesity associated genetic variation in FTO is associated with diminished satiety. J. Clin. Endocrinol. metabolism. 2008;93(9):3640–3643. doi: 10.1210/jc.2008-0472. [DOI] [PubMed] [Google Scholar]
- 12.Jess T., Zimmermann E., Kring S.I., Berentzen T., Holst C., Toubro S. Impact on weight dynamics and general growth of the common FTO rs9939609: a longitudinal Danish cohort study. Int. J. Obes. 2008;32(9):1388–1394. doi: 10.1038/ijo.2008.110. [DOI] [PubMed] [Google Scholar]
- 13.Lopez-Bermejo A., Petry C.J., Diaz M., Sebastiani G., de Zegher F., Dunger D.B. The association between the FTO gene and fat mass in humans develops by the postnatal age of two weeks. J. Clin. Endocrinol. metabolism. 2008;93(4):1501–1505. doi: 10.1210/jc.2007-2343. [DOI] [PubMed] [Google Scholar]
- 14.Hardy R., Wills A.K., Wong A., Elks C.E., Wareham N.J., Loos R.J. Life course variations in the associations between FTO and MC4R gene variants and body size. Hum. Mol. Genet. 2010;19(3):545–552. doi: 10.1093/hmg/ddp504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qi L., Kraft P., Hunter D.J., Hu F.B. The common obesity variant near MC4R gene is associated with higher intakes of total energy and dietary fat, weight change and diabetes risk in women. Hum. Mol. Genet. 2008;17(22):3502–3508. doi: 10.1093/hmg/ddn242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Timpson N.J., Sayers A., Davey-Smith G., Tobias J.H. How does body fat influence bone mass in childhood? A mendelian randomization approach. JBone MinerRes. 2009;24(3):522–533. doi: 10.1359/jbmr.081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stutzmann F., Cauchi S., Durand E., Calvacanti-Proenca C., Pigeyre M., Hartikainen A.L. Common genetic variation near MC4R is associated with eating behaviour patterns in European populations. Int. J. Obes. 2009;33(3):373–378. doi: 10.1038/ijo.2008.279. [DOI] [PubMed] [Google Scholar]
- 18.Petry C.J., Lopez-Bermejo A., Diaz M., Sebastiani G., Ong K.K., de Z.F. Association between a common variant near MC4R and change in body mass index develops by two weeks of age. Horm. Res. Paediatr. 2010;73(4):275–280. doi: 10.1159/000284392. [DOI] [PubMed] [Google Scholar]
- 19.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7(12):885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smemo S., Tena J.J., Kim K.H., Gamazon E.R., Sakabe N.J., Gomez-Marin C. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertshaw E., Matsumoto K., Lumsden A., Kiecker C. Irx3 and Pax6 establish differential competence for Shh-mediated induction of GABAergic and glutamatergic neurons of the thalamus. Proc. Natl. Acad. Sci. U. S. A. 2013;110(41):E3919–E3926. doi: 10.1073/pnas.1304311110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gulati P., Cheung M.K., Antrobus R., Church C.D., Harding H.P., Tung Y.C. Role for the obesity-related FTO gene in the cellular sensing of amino acids. Proc. Natl. Acad. Sci. U. S. A. 2013;110(7):2557–2562. doi: 10.1073/pnas.1222796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cecil J.E., Tavendale R., Watt P., Hetherington M.M., Palmer C.N. An obesity-associated FTO gene variant and increased energy intake in children. NEnglJMed. 2008;359(24):2558–2566. doi: 10.1056/NEJMoa0803839. [DOI] [PubMed] [Google Scholar]
- 24.Qi Q., Kilpelainen T.O., Downer M.K., Tanaka T., Smith C.E., Sluijs I. FTO genetic variants, dietary intake and body mass index: insights from 177 330 individuals. Hum. Mol. Genet. 2014;23(25):6961–6972. doi: 10.1093/hmg/ddu411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inskip H.M., Godfrey K.M., Robinson S.M., Law C.M., Barker D.J., Cooper C. Cohort profile: The Southampton Women's survey. IntJEpidemiol. 2006;35(1):42–48. doi: 10.1093/ije/dyi202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Royston P. Calculation of unconditional and conditional reference intervals for foetal size and growth from longitudinal measurements. StatMed. 1995;14(13):1417–1436. doi: 10.1002/sim.4780141303. [DOI] [PubMed] [Google Scholar]
- 27.Cleal J.K., Day P., Hanson M.A., Lewis R.M. Measurement of housekeeping genes in human placenta. Placenta. 2009;30(11):1002–1003. doi: 10.1016/j.placenta.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Laird N.M., Ware J.H. Random-effects models for longitudinal data. Biometrics. 1982;38(4):963–974. [PubMed] [Google Scholar]
- 29.Frazer K.A., Ballinger D.G., Cox D.R., Hinds D.A., Stuve L.L., Gibbs R.A. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dobson R. 2007-03-08. Smoking in Pregnancy Slows Growth of Baby's Head2007. 23:00:52. 499- p. [Google Scholar]
- 31.Marsh J.A., Pennell C.E., Warrington N.M., Mook-Kanamori D., Briollais L., Lye S.J. Fat mass and obesity-associated obesity-risk genotype is associated with lower foetal growth: an effect that is reversed in the offspring of smoking mothers. J. Dev. Orig. Health Dis. 2012;3(1):10–20. doi: 10.1017/S2040174411000638. [DOI] [PubMed] [Google Scholar]
- 32.Lewis R.M., Demmelmair H., Gaillard R., Godfrey K.M., Hauguel-de Mouzon S., Huppertz B. The placental exposome: placental determinants of fetal adiposity and postnatal body composition. Ann. Nutr. metabolism. 2013;63(3):208–215. doi: 10.1159/000355222. [DOI] [PubMed] [Google Scholar]
- 33.Mayeur S., Cisse O., Gabory A., Barbaux S., Vaiman D., Vambergue A. Placental expression of the obesity-associated gene FTO is reduced by fetal growth restriction but not by macrosomia in rats and humans. J. Dev. Orig. Health Dis. 2013;4(2):134–138. doi: 10.1017/S2040174412000748. [DOI] [PubMed] [Google Scholar]
- 34.Bassols J., Prats-Puig A., Vazquez-Ruiz M., Garcia-Gonzalez M.M., Martinez-Pascual M., Avelli P. Placental FTO expression relates to fetal growth. Int. J. Obes. 2010;34(9):1365–1370. doi: 10.1038/ijo.2010.62. [DOI] [PubMed] [Google Scholar]
- 35.Sebert S.P., Hyatt M.A., Chan L.L., Yiallourides M., Fainberg H.P., Patel N. Influence of prenatal nutrition and obesity on tissue specific fat mass and obesity-associated (FTO) gene expression. Reproduction. 2010;139(1):265–274. doi: 10.1530/REP-09-0173. [DOI] [PubMed] [Google Scholar]
- 36.Paolini C.L., Marconi A.M., Ronzoni S., Di Noio M., Fennessey P.V., Pardi G. Placental transport of leucine, phenylalanine, glycine, and proline in intrauterine growth-restricted pregnancies. J. Clin. Endocrinol. metabolism. 2001;86(11):5427–5432. doi: 10.1210/jcem.86.11.8036. [DOI] [PubMed] [Google Scholar]
- 37.Boissel S., Reish O., Proulx K., Kawagoe-Takaki H., Sedgwick B., Yeo G.S. Loss-of-function mutation in the dioxygenase-encoding FTO gene causes severe growth retardation and multiple malformations. Am. J. Hum. Genet. 2009;85(1):106–111. doi: 10.1016/j.ajhg.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ho A.J., Stein J.L., Hua X., Lee S., Hibar D.P., Leow A.D. A commonly carried allele of the obesity-related FTO gene is associated with reduced brain volume in the healthy elderly. Proc. Natl. Acad. Sci. U. S. A. 2010;107(18):8404–8409. doi: 10.1073/pnas.0910878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benedict C., Jacobsson J.A., Ronnemaa E., Sallman-Almen M., Brooks S., Schultes B. The fat mass and obesity gene is linked to reduced verbal fluency in overweight and obese elderly men. Neurobiol. aging. 2011;32(6):1159. doi: 10.1016/j.neurobiolaging.2011.02.006. e1-5. [DOI] [PubMed] [Google Scholar]
- 40.Gao X., Shin Y.H., Li M., Wang F., Tong Q., Zhang P. The fat mass and obesity associated gene FTO functions in the brain to regulate postnatal growth in mice. PloS one. 2010;5(11):e14005. doi: 10.1371/journal.pone.0014005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horikoshi M., Yaghootkar H., Mook-Kanamori D.O., Sovio U., Taal H.R., Hennig B.J. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat. Genet. 2013;45(1):76–82. doi: 10.1038/ng.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanson M., Godfrey K.M., Lillycrop K.A., Burdge G.C., Gluckman P.D. Developmental plasticity and developmental origins of non-communicable disease: theoretical considerations and epigenetic mechanisms. Prog. biophys Mol. Biol. 2011;106(1):272–280. doi: 10.1016/j.pbiomolbio.2010.12.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.