Abstract
Candidate phylum TM6 is a major bacterial lineage recognized through culture-independent rRNA surveys to be low abundance members in a wide range of habitats; however, they are poorly characterized due to a lack of pure culture representatives. Two recent genomic studies of TM6 bacteria revealed small genomes and limited gene repertoire, consistent with known or inferred dependence on eukaryotic hosts for their metabolic needs. Here, we obtained additional near-complete genomes of TM6 populations from agricultural soil and upflow anaerobic sludge blanket reactor metagenomes which, together with the two publicly available TM6 genomes, represent seven distinct family level lineages in the TM6 phylum. Genome-based phylogenetic analysis confirms that TM6 is an independent phylum level lineage in the bacterial domain, possibly affiliated with the Patescibacteria superphylum. All seven genomes are small (1.0–1.5 Mb) and lack complete biosynthetic pathways for various essential cellular building blocks including amino acids, lipids, and nucleotides. These and other features identified in the TM6 genomes such as a degenerated cell envelope, ATP/ADP translocases for parasitizing host ATP pools, and protein motifs to facilitate eukaryotic host interactions indicate that parasitism is widespread in this phylum. Phylogenetic analysis of ATP/ADP translocase genes suggests that the ancestral TM6 lineage was also parasitic. We propose the name Dependentiae (phyl. nov.) to reflect dependence of TM6 bacteria on host organisms.
Keywords: candidate phylum TM6, parasitism, comparative genomics, upflow anaerobic sludge blanket, soil.
Introduction
Members of candidate phylum TM6 are widespread in the environment based on 16S rRNA gene (16S) surveys. TM6 16S sequences were first described in peat bogs (Rheims et al. 1996) and were subsequently found in diverse habitats such as hypersaline microbial mats (Sørensen et al. 2005), sulfur springs (Youssef et al. 2012), arsenic-rich sediments (Escudero et al. 2013) and also in biofilms collected from showerheads (Feazel et al. 2009), sinks (McLean et al. 2013), and drinking water supply systems (Henne et al. 2012). Current knowledge of the ecology and evolution of the TM6 phylum is limited to 16S sequences and two near-complete reference genomes. The first genome, TM6SC1, was recovered from a “mini-metagenome” of a presorted pool of cells collected from a hospital sink biofilm (McLean et al. 2013). Based on the habitat and similarities to other amoebal symbionts, the authors suggested that TM6SC1 and the TM6 phylum may more broadly be symbionts of amoeba (McLean et al. 2013). Consistent with this hypothesis, the second genome was recovered from a strain cocultured with Acanthamoeba, which the authors named Babela massiliensis and initially misclassified as a member of the Deltaproteobacteria (Cohen et al. 2011). The authors identified numerous features indicative of an obligate parasitic lifestyle including the inability to grow without amoebal hosts, a limited set of biosynthetic capabilities, and substantial degradation of its cell division machinery (Pagnier et al. 2015). A partial TM6 population genome (ACD64) was also recently reported as part of a larger metagenomic analysis of an aquifer metagenome, but was not discussed further in the context of that study (Wrighton et al. 2014).
Here, we obtained four near-complete genomes and one substantially complete genome from TM6 populations present in agricultural soil and a full-scale upflow anaerobic sludge blanket (UASB) reactor (Soo et al. 2014; Sekiguchi et al. 2015) through differential coverage binning (Imelfort et al. 2014). Differential coverage binning groups together anonymous metagenomic fragments (contigs) belonging to the same population based on the similarity of their sequencing coverage across multiple related metagenomes (Albertsen et al. 2013). Together with the two publicly available near-complete TM6 genomes, two class-level lineages are now represented in this phylum and concatenated gene analysis indicates that TM6 may be affiliated with the recently described Patescibacteria superphylum (Rinke et al. 2013). Shared characteristics of the TM6 genomes which include small genome size, limited cell envelope and cellular building block biosynthetic capacity, ability to parasitize external ATP pools, and presence of protein repeat domains that facilitate interaction with eukaryotes support a common parasitic lifestyle with eukaryotic host organisms. We propose the name Dependentiae (phyl. nov.) to reflect these phylum-level characteristics.
Results and Discussion
Recovering TM6 Genomes from Metagenomic Sequence Data
Two previously reported metagenomic data sets were mined for TM6 genomes. The first data set was obtained from four agricultural soil samples collected during a sugarcane field trial (NCBI SRA accession numbers: SRS881276, SRS881281, SRS881283, SRS881286; Yeoh et al. 2015) and the second from four UASB reactor samples (Soo et al. 2014; Sekiguchi et al. 2015). The agricultural soil data set was de novo assembled for the purposes of this study. Approximately a third of the soil data set (152.3 Gb) assembled into contigs greater than 500 bp long with an N50 of 1,055. Differential coverage binning (Albertsen et al. 2013; Imelfort et al. 2014) was used to obtain population genomes from the metagenomic assembly and the completeness and contamination of the recovered genomes was estimated by the presence/absence of 104 conserved single copy marker genes (Dupont et al. 2012; Parks et al. 2015). Six population genomes with greater than 65% completeness and less than 10% contamination (Parks et al. 2015) were recovered from this assembly despite the complexity of the resident soil microbial communities. Of these, two near-complete genomes (>90% complete) were identified (see section below) as belonging to candidate phylum TM6 (SOIL31 and SOIL82; table 1). The assembly and binning of the UASB data set have been previously reported (Soo et al. 2014; Sekiguchi et al. 2015) and resulted in 239 population genomes, including two near-complete (UASB124 and UASB340; table 1) and one substantially complete (>70% complete) TM6 genomes (UASB293; table 1). Including the two public TM6 genomes (TM6SC1 and Babela massiliensis) the GC content for this phylum ranges from 27.4% to 40.8%, comparable to other phyla of similar phylogenetic radiation (Lightfield et al., 2011), and the genomes are all uniformly small by bacterial standards (1.0–1.5 Mb in size). Four of the 104 marker genes were not identified in any of the TM6 genomes (OBG GTPase, Dephospho-CoA kinase, leucyl-tRNA synthetase, methionyl-tRNA formylytransferase; supplementary table S1, Supplementary Material online), suggesting that these genes are absent in the TM6 lineage. Sequence composition integrity analysis of each genome identified a small subset of contigs with atypical kmer signatures (supplementary fig. S1, Supplementary Material online) encoding mostly hypothetical genes (supplementary table S2, Supplementary Material online). However, some of these contigs were connected to other contigs with typical kmer profiles and likely represent integrated foreign elements such as viruses.
Table 1.
Summary Statistics for TM6 Population Genomes.
| Population Genome | Genome Origin | Number of Scaffolds | Genome Size (bp) | Longest Scaffold (bp) | N50 | %GC | 16S Copies (length) | % Completenessa | Completeness Categorya | % Contaminationa | Contamination Categorya | Original Study |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TM6SC1 | Hospital sink biofilm mini-metagenome | 7 | 1,074,689 | 464,047 | 273,590 | 36.4 | 1 (1,551 bp) | 98.0 | Near complete | 0.0 | Low | McLean et al. (2013) |
| Babela massiliensis | Amoeba co-culture | 1 | 1,118,422 | 1,118,422 | 1,118,422 | 27.4 | 1 (1,550 bp) | 98.0 | Near complete | 0.0 | Low | Pagnier et al. (2015) |
| SOIL31 | Soil metagenome | 69 | 1,460,402 | 183,769 | 43,767 | 37.9 | 1 (252 bp)b | 94.0 | Near complete | 0.0 | Low | Present study |
| SOIL82 | Soil metagenome | 13 | 1,001,943 | 242,158 | 92,171 | 40.8 | 1 (115 bp)b | 97.0 | Near complete | 0.0 | Low | Present study |
| UASB124c | UASB reactor metagenome | 4 | 1,289,444 | 806,336 | 806,336 | 37.5 | 1 (315 bp)b | 99.0 | Near complete | 4.0 | Low | Sekiguchi et al. (2015) |
| UASB293c | UASB reactor metagenome | 127 | 1,302,184 | 65,238 | 20,517 | 37.9 | 1 (185 bp)b | 77.0 | Substantially complete | 0.0 | Low | Sekiguchi et al. (2015) |
| UASB340c | UASB reactor metagenome | 36 | 1,078,523 | 162,571 | 95,078 | 28.7 | — | 94.0 | Near complete | 0.0 | Low | Sekiguchi et al. (2015) |
| ACD64d | Aquifer metagenome | 135 | 748,730 | 27,387 | 6,654 | 39.8 | — | 42.0 | Partial | 0.0 | Low | Wrighton et al. (2014) |
aGenome completeness and contamination were estimated using CheckM version 1.0.0 (Parks et al. 2015) with the bacterial root marker set. Four of the 104 root markers were missing in all TM6 genomes and were excluded from the completeness estimate as they were inferred to be genuinely absent in this phylum.
bHigh conservation and repeating nature of 16S rRNA genes in microbial genomes confound their assembly and binning, therefore presence/absence of 16S rRNA genes cannot be used as a measure of quality in population genomes.
cUASB124, 293, and 340 genomes were recovered from the reactor metagenome described in Sekiguchi et al. (2015), which is a comparative genomics study of two members of candidate bacterial phylum KSB3.
dThe ACD64 partial TM6 genome from Wrighton et al. (2014) was not directly included in the comparative analysis described in this study due to its low completeness estimate.
Phylogenetic Placement of Candidate Phylum TM6 Genomes
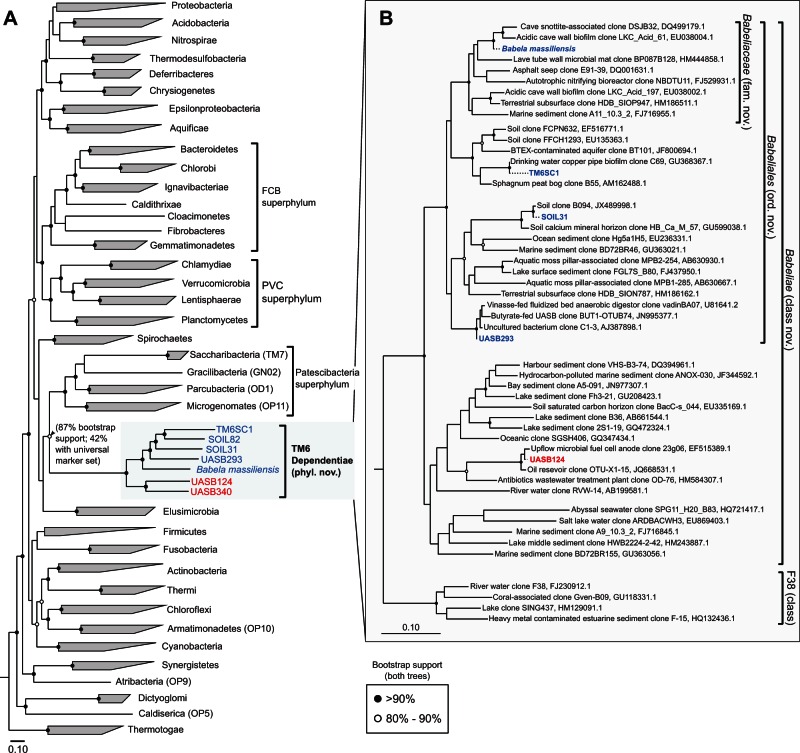
The population genomes were placed into genome trees comprising 2,350 reference genomes obtained from Integrated Microbial Genomes (IMG) database release 4.1 (Markowitz et al. 2012) to establish their relationship to one another and to other bacterial lineages. Two trees were constructed; one using an alignment of 38 concatenated universal marker genes from Phylosift (Darling et al. 2014) with an archaeal outgroup, and the other using 83 concatenated bacterial marker genes (supplementary table S3, Supplementary Material online; Soo et al. 2014) using Candidatus Acetothermus autotrophicum, previously inferred to be the most basal bacterial lineage (Takami et al. 2012), as the outgroup (fig. 1A). The seven TM6 genomes were reproducibly resolved as a monophyletic group with high bootstrap confidence (fig. 1A). In both trees, candidate phylum TM6 was affiliated with the recently proposed Patescibacteria superphylum with which it shares the features of small genome size (approximately 1 Mb) and low GC content (approximately 35%; Rinke et al. 2013). However, this relationship was only well supported in the 83 bacterial marker gene-based tree (87% bootstrap support), and is in contrast to the original placement of TM6SC1 and B. massiliensis, which branched with the Acidobacteria (McLean et al. 2013) and Deferribacteres (Pagnier et al. 2015), respectively. Therefore, specific affiliations (if any) of TM6 to other bacterial phyla remain an open question at this stage.
Fig. 1.
Phylogenetic relationships of candidate phylum TM6 bacteria. (A) Genome-based phylogeny of TM6 bacteria relative to other bacterial phyla. A maximum-likelihood tree was constructed from 2,175 bacterial reference genomes using an amino acid alignment of 83 phylogenetically informative bacterial marker genes. Candidatus Acetothermus autotrophicum (OP1) was used as the outgroup (Takami et al. 2012). (B) 16S rRNA gene-based phylogeny of TM6 bacteria using 16S sequences obtained from population genomes in this study (dotted lines) and representative sequences from the Greengenes database. The population genome labels for TM6 bacteria in both trees are highlighted in blue or red to reflect order-level relatedness. In both trees, black circles at nodes indicate >90% bootstrap support and open circles indicate 80–90% bootstrap support. The scale bars indicate 0.1 amino acid and nucleotide substitutions per site for panels (A) and (B), respectively.
Within the TM6 phylum, two major monophyletic groups could be resolved; one consisting of UASB124 and UASB340 and the other comprising B. massiliensis, TM6SC1, SOIL31, SOIL82, and UASB293 (denoted in red and blue, respectively, in fig. 1). To provide a broader phylogenetic context, we inferred the position of five of the seven population genomes based on partial 16S sequences identified in the bins (table 1), relative to publicly available clone TM6 sequences (fig. 1B; McDonald et al. 2012; Quast et al. 2013). We propose that all seven genomes belong to a single class, Babeliae, and that a separate class, F38, defined only by 16S sequences still lacks genome representation (fig. 1B). Average amino acid identity (AAI) values between pairs of TM6 genomes suggest that each of these genomes likely represents at least individual families (supplementary table S4, Supplementary Material online; 45–65%, Konstantinidis and Tiedje, 2005). Within the class Babeliae, we therefore propose that B. massiliensis, TM6SC1, SOIL31, SOIL82, and UASB293 are members of the order Babeliales and that UASB124 and UASB340 belong to one or possibly two separate orders (fig. 1B).
Overview of Genome Functionality
The seven TM6 genomes contain an average of 1,126 predicted open-reading frames (ORFs; maximum 1,462, minimum 955), of which approximately 60% had a predicted function. Assigning ORFs to cluster of orthologous group (COG) categories indicates that TM6 have significantly reduced functionality compared with the bacterial average in 15 of 22 categories (P < 0.05, Mann–Whitney test; supplementary fig. S2, Supplementary Material online). We compared the COG category profile of several endosymbiotic (host-beneficial), intracellular parasitic (host-detrimental), and free-living bacterial genera and found that the TM6 COG profile was most similar to that of parasitic intracellular bacterial genera (supplementary fig. S3, Supplementary Material online), such as Chlamydia, Wolbachia and Rickettsia. Specifically, the proportion of amino acid, coenzyme transport and metabolism-related genes in the TM6 genomes, like other parasitic intracellular bacteria, was smaller compared with endosymbiotic bacteria suggesting dependence on an external source for these essential compounds. In contrast, endosymbiotic bacteria generally have larger proportions of these genes, which reflect a symbiotic lifestyle of provisioning essential amino acid and coenzymes to their hosts (Merhej et al. 2009). Next, we examined the reciprocal best Basic Local Alignment Search Tool (BLAST) hits between TM6 genomes to identify the core set of orthologs shared by members of this phylum. Of 4,503 orthologous groups identified, 171 (3.8%) were common to all seven genomes, and a further 184 (4.1%) were common to any six of the seven genomes (supplementary table S5, Supplementary Material online). These orthologs predominantly encode core functions such as DNA/RNA synthesis and repair machinery, ribosomal proteins, cell division proteins, tRNAs, ATP synthase and also components for a type II secretion system and possibly a type IV pilus. Although the type IV pilus membrane pore pilQ is absent in TM6, two copies of the homologous type II secretion channel gene gspD were identified in all genomes (supplementary table S5, Supplementary Material online). It is possible that one gspD copy has functionally substituted pilQ in TM6 bacteria; however, this remains an open question as no pilus structures were observed in electron micrographs of B. massiliensis (Pagnier et al. 2015). Type IV pili help mediate interactions with other organisms including human cells (Craig et al. 2004), plants and fungi (Dörr et al. 1998) and, if present, may assist TM6 bacteria to attach to their host cells. Type IV pili genes are also found in representative genomes of the bacterial candidate phyla Parcubacteria (OD1), Microgenomates (OP11), WWE3, and Saccharibacteria (TM7) (Albertsen et al. 2013; Kantor et al. 2013; He et al. 2015) which may be phylogenetically related to TM6 (fig. 1). It has been proposed that the type-IV pili in these lineages mediate adhesion to larger host cells facilitating nutrient acquisition from the host cell (Luef et al. 2015).
Babela massiliensis and TM6SC1 have or are hypothesized to have amoebal hosts, respectively (Cohen et al. 2011; McLean et al. 2013; Pagnier et al. 2015), and a subset of their proteomes contain domains that facilitate interaction with eukaryotic hosts, similar to other amoeba-associated bacteria such as Candidatus Amoebophilus (McLean et al. 2013; Pagnier et al. 2015). In our sequence data, the coverage profile of an unbinned contig containing an 18S rRNA sequence identified as a ciliate protist (class Spirotrichea) tracked the coverage profiles of SOIL31 and SOIL82 TM6 population genomes (supplementary fig. S4, Supplementary Material online) suggesting a specific association between these organisms. Spirotrich ciliates are known to harbor endosymbiotic bacteria (Boscaro et al. 2012), thus we speculate that in addition to amoebae, some TM6 lineages may have ciliate hosts. We performed a principal component analysis (PCA) of the relative abundance of COGs identified in the TM6 genomes to determine the distribution of accessory functionalities within the TM6 phylum. A striking separation of B. massiliensis from the other genomes was observed due to a high number of ankyrin repeats (COG0666) in the B. massiliensis genome compared with the other TM6 genomes, although all seven genomes contained these repeats (supplementary fig. S5A, Supplementary Material online). Ankyrin repeats are amino acid motifs that commonly mediate protein–protein interactions of diverse cellular functions (Mosavi et al. 2004). Such motifs are uncommon in prokaryotes with the exception of a few facultative and obligate intracellular bacteria (Siozios et al. 2013), and are hypothesized to mediate eukaryote–bacterial interactions (Pan et al. 2008), which is consistent with the lifestyle of B. massiliensis as an intracellular parasite of amoebae (Cohen et al. 2011; Pagnier et al. 2015). Ankyrin repeat-containing proteins were also identified in the partial ACD64 TM6 genome from Wrighton et al. 2014 (data not shown), suggesting that this organism can interact with eukaryotic hosts. The higher number of ankyrin repeats in B. massiliensis compared with the other TM6 genomes suggests that B. massiliensis has the highest potential to interact with the cellular machinery of its host. When the ankyrin repeat family was removed from the PCA analysis, a separation of taxa by phylogeny was observed (supplementary fig. S5B, Supplementary Material online).
We also investigated the TM6 genomes for other protein domains known to facilitate and regulate protein–protein interactions in eukaryotes, including the WD40, F-box, leucine-rich repeat, and tetratricopeptide repeat. These domains were previously described in the genomes of TM6SC1 (McLean et al. 2013) and B. massiliensis (Pagnier et al. 2015), which the authors suggested allow TM6 bacteria to interact with cellular processes of their amoebal hosts and to modify the host intracellular environment, thereby facilitating survival of the bacterium. We identified numerous instances of genes containing these domains in all five additional SOIL and UASB TM6 genomes (supplementary table S6, Supplementary Material online).
Cell Envelope, Division and Morphology
The TM6 cell envelope is most likely Gram-negative based on cell envelope marker profiles (fig. 2), and also because the groEL gene sequences have an amino acid insertion characteristic of Gram-negative bacteria (supplementary fig. S6, Supplementary Material online; Gupta 1998). The five representatives of order Babeliales (fig. 1) lack most of the genes necessary to synthesize lipopolysaccharide (LPS), whereas the other two TM6 representatives (UASB124 and UASB340) likely are capable of producing LPS as part of their cell envelope (fig. 2). This distribution suggests differential Gram-negative cell envelope structures between the major TM6 lineages. However, even the most complete TM6 cell envelope in UASB124 and UASB340 is most likely basic (fig. 3) compared with typical Gram-negative bacteria due to a lack of multiple modification genes (fig. 2). LPS typically consists of three components: A core oligosaccharide (KDO), O antigen, and lipid A that is usually modified posttranslationally (Heinrichs et al. 1998). Both UASB124 and UASB340 lack the O antigen ligase gene (waaL) and downstream modification of the LPS molecule was variable between genomes (fig. 2). For example, addition of acyl groups by lipid-A acyltransferases should only be possible in UASB340, whereas both UASB124 and UASB340 have the potential to add heptose units to KDO–lipid A through heptosyl transferase encoded by waaC (Heinrichs et al. 1998). Taken together, these findings suggest that the TM6 bacteria outer membrane has undergone degeneration and part of its synthesis machinery has been lost, particularly in the order Babeliales (fig. 1B). Loss of LPS and bacterial outer membrane components is hypothesized to be an adaptation to host-associated lifestyles because LPS elicits immune responses from eukaryotic hosts (Bennett et al. 2014).
Fig. 2.
Distribution of cell envelope, division, and shape-determining genes in the studied TM6 population genomes indicating degenerated Gram-negative cell envelope and division apparatus. Reference bacteria were included for comparison of gene distributions: Bacillus subtilis and Streptomyces coelicolor as examples of Gram-positive bacteria, Escherichia coli and Bacteroides fragilis as examples of Gram-negative bacteria, and Buchnera aphidicola, Wigglesworthia glossinidia, Chlamydia trachomatis and Rickettsia prowazekii as examples of host-associated intracellular bacteria. Gene families were identified using protein HMMs and match probabilities are indicated by shading (Dark blue for e values <=0.001, light blue for e values >0.001, and gray for no positive matches). Cell envelope markers are from Soo et al. (2014).
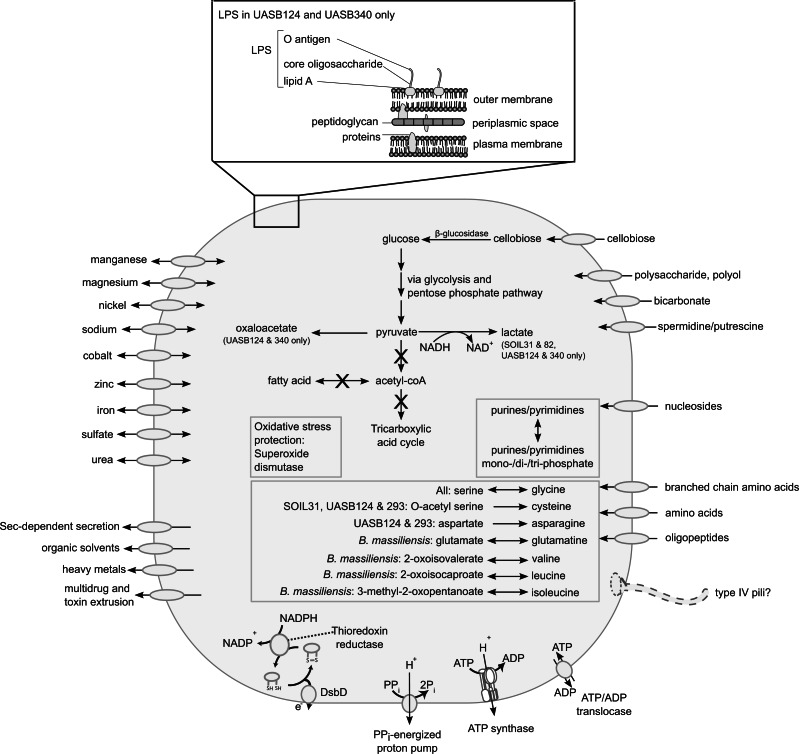
Fig. 3.
Predicted metabolic pathways of TM6 bacteria based on the seven population genomes studied. Import and/or export of compounds through transporters and permeases in the cell membrane are indicated by uni- or bidirectional arrows. β-glucosidase, pentose phosphate pathway, and partial glycolysis genes were identified indicating that TM6 bacteria are able to hydrolyze cellobiose for energy through substrate-level phosphorylation. No complete pathways for the TCA cycle, fatty acid, nucleotide and amino acid biosynthesis were identified, and only a limited number of amino acids can be interconverted or synthesized from intermediate metabolites. TM6 bacteria most likely rely on the uptake of nucleosides and amino acids from their surroundings, suggesting a host-associated lifestyle. Superoxide dismutase and thioredoxin reductase genes suggest potential for oxidative stress tolerance. A pyrophosphate-energized proton pump and ATP synthase potentially serve to maintain the transmembrane proton gradient. The presence of ATP/ADP translocases and a degenerated cell envelope structure indicate a parasitic lifestyle in TM6 bacteria.
The TM6 cell division machinery shows similar degeneration, particularly in B. massiliensis which has only three of nine key cell division genes (fig. 2). The TM6 cell division apparatus is atypical particularly in B. massiliensis, resulting in dense, amorphous bodies that separate into long bacillary forms as part of its developmental cycle (Pagnier et al. 2015). We found all genomes to lack ftsQ, ftsN, zapB and zipA and have varied distribution of other cell division genes (fig. 2), indicating that atypical cell division, such as that identified in B. massiliensis, is likely common in TM6 bacteria.
We also investigated the TM6 genomes for the presence of the mreBCD operon and the rodA and pbpA genes, which have been shown to be necessary for rod shape morphology in model organisms including Escherichia coli (Matsuzawa et al. 1989; Wachi et al. 1989) and Bacillus subtilis (Henriques et al. 1998; Jones et al. 2001; Leaver and Errington 2005). With the exception of B. massiliensis, the TM6 genomes encode the majority of shape-determining proteins only consistently lacking the mreD subunit (fig. 2). As a complete mreBCD complex is required for the formation of rods in E. coli (Kruse et al. 2005), TM6 cells may be spherical, although care should be taken in extrapolating results over such broad phylogenetic distances. It is likely, however, that B. massiliensis is not be able to form rods as it lacks all five shape-determining genes, as well as most genes for peptidoglycan synthesis. These findings suggest that, of the investigated TM6 bacteria, B. massiliensis is the most advanced in its adaptation to an intracellular lifestyle as exemplified by various host-associated intracellular bacteria that have undergone genome size and gene repertoire reduction (McCutcheon and Moran 2012).
Carbon Metabolism and Energy Production
All TM6 bacteria studied lack a tricarboxylic acid (TCA) cycle and electron transport chain, suggesting that they share a fermentative metabolism with lactate as the only potential fermentation end product (fig. 3). Superoxide dismutases and thioredoxin reductase were identified in all genomes indicating oxidative stress tolerance. All genomes encode an ATP synthase which in fermentative bacteria can function to maintain the transmembrane proton gradient by hydrolyzing ATP. A pyrophosphatase-driven proton pump is also present in all genomes for this purpose (fig. 3). To obtain ATP, TM6 bacteria may rely on ADP/ATP translocases to exchange ADP for their hosts’ ATP. These translocases, found in all studied TM6 genomes, are considered a feature of obligate intracellular parasites (Schmitz-Esser et al. 2004), thereby providing further evidence for TM6 bacteria having an intracellular parasitic lifestyle. In addition to parasitizing their hosts’ energy pool, TM6 bacteria may also rely on substrate level phosphorylation of cellobiose to gain ATP. All seven TM6 genomes encode β-glucosidase for hydrolyzing the β-glycosidic linkage in cellobiose to produce glucose monomers, and ATP is then generated from the conversion of glucose to pyruvate through a combination of glycolytic and pentose phosphate pathway (PPP) enzymes. In SOIL31, SOIL82, UASB124, and UASB340, pyruvate can then be converted to lactate through lactate dehydrogenase to regenerate NAD+. Pyruvate dehydrogenase and other downstream enzymes for synthesis of other fermentation products from acetyl-coA, including acetate, ethanol, butanediol, and propionate were absent. In this regard, the carbon metabolism of TM6 bacteria resembles candidate phylum Parcubacteria (OD1) whereby glucose is metabolized to pyruvate but genes for acetyl-coA synthesis and utilization were also absent (Kantor et al. 2013). In addition, like TM6 bacteria, Parcubacteria was hypothesized to also use lactate fermentation to recover NAD+.
Amino Acid, Nucleotide, Lipid and Cofactor Biosynthesis
No complete pathways for amino acid, nucleotide, lipid or cofactor biosynthesis were identified in any of the TM6 genomes (fig. 3). The absence of biosynthetic machinery for essential cellular building blocks is consistent with a host-associated lifestyle (Moran 2002). All seven TM6 bacteria have limited capacity to synthesize amino acids and most likely depend on uptake of oligopeptides and amino acids from their surroundings through a small set of transporters and permeases (fig. 3). They have some capacity to synthesize a limited set of amino acids from precursors with B. massiliensis being the most versatile in this regard and TM6SC1, SOIL82, and UASB340 only being able to interconvert serine and glycine (fig. 3). All TM6 bacteria also appear to lack the capacity for nucleoside biosynthesis. Purine (adenosine, guanosine, and inosine) and pyrimidine (uridine, cytidine, and thymidine) nucleosides are normally synthesized from ribose-5-phosphate produced through the PPP; however, all nucleoside biosynthesis genes downstream of ribose-5-phosphate are absent in TM6 bacteria. As with amino acids, these bacteria likely rely on nucleoside permeases and nucleotide transporters for the uptake of nucleosides from their local environment. Once inside the cell, nucleoside kinases and phosphatases are present to convert among mono-, di-, and tri-phosphate forms of purines and pyrimidines (fig. 3). TM6 bacteria lack genes for fatty acid biosynthesis and degradation. In the absence of these capabilities, we speculate that TM6 bacteria may exploit host-derived lipids, as suggested for Buchnera aphidicola where synthesis of the cell membrane is not possible without fatty acid biosynthesis genes (Bennett et al. 2014). Mechanisms by which host-associated bacteria obtain host lipids are mostly unknown. Chlamydia trachomatis can acquire lipids from host lipid droplets using Lda1 and IncA proteins (Kumar et al. 2006; Cocchiaro et al. 2008), but no homologs were identified in the TM6 genomes. The synthesis of cofactors is also not possible in TM6 bacteria, with the possible exception of UASB293 which has genes for a portion of the vitamin B12 pathway (from co-sirohydrochlorin to cobyrinate). A sodium/pantothenate symporter and a biotin transporter were only identified in UASB340. Taken together, these observations indicate that TM6 bacteria depend on external sources, most likely a host organism, for essential nutrients and building blocks to survive.
Evolution and Ecology of Inferred Parasitic Lifestyle
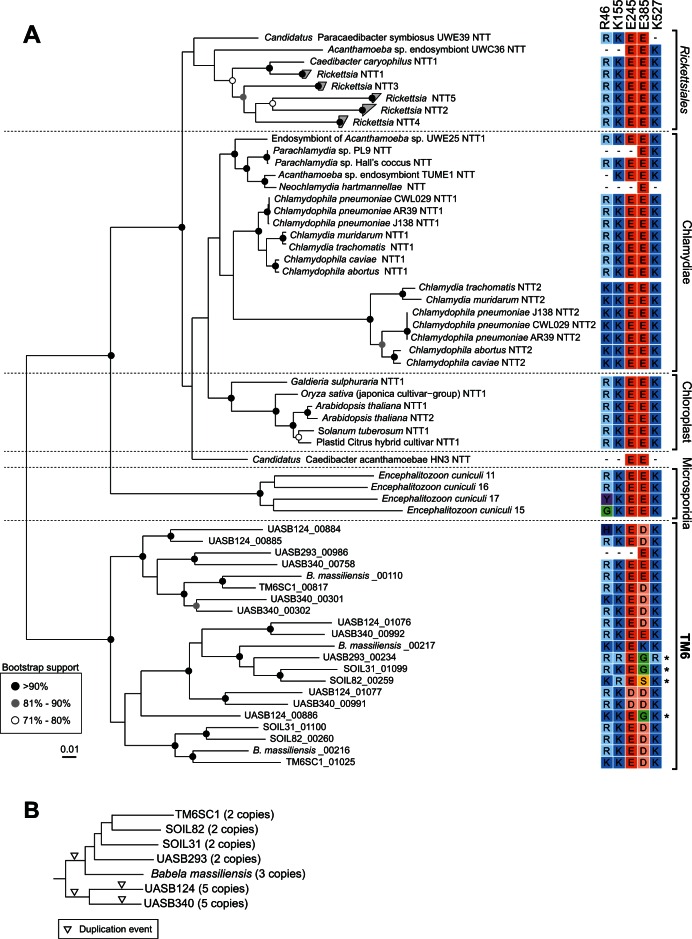
Host-associated lifestyles are well known in bacteria and are characteristic of multiple lineages such as the phylum Chlamydiae, order Rickettsiales, and the genera Buchnera and Wigglesworthia (Moran 2002; McCutcheon and Moran 2012). Candidate phylum TM6 is striking in that the entire phylum appears to be host associated, which has only been observed in one other phylum, the Chlamydiae, although genome representation from class F38 is required to confirm this inference (fig. 1B). The proposal of an amoeba-dependent lifestyle in TM6 bacteria was first inferred based on genome features of TM6SC1 (McLean et al. 2013). This proposal was subsequently supported by the isolation of B. massiliensis in coculture with Acanthamoeba castellanii confirming an obligate intracellular parasitic lifestyle for a member of the TM6 phylum (Pagnier et al. 2015). With the findings from McLean et al. (2013), Pagnier et al. (2015), and genomes of five additional TM6 bacteria described in this study, we provide further evidence that TM6 bacteria are intracellular parasites of eukaryotes based on several features in common with known parasitic bacteria: 1) Small genome size; 2) degenerated cell envelope; 3) lack of metabolic pathways for energy production and synthesis of essential building blocks including amino acids, coenzymes, nucleotides, and fatty acids; 4) proteins containing domains for interaction with cellular machinery of eukaryotic hosts; and 5) ATP/ADP translocases for scavenging ATP from host cells. As the TM6 and Chlamydiae phyla are not sister to each other (fig. 1A), these similarities likely arose from independent adaptations to parasitic lifestyles. Parasitism is thought to have evolved once in the Chlamydiae with the last common chlamydial ancestor having a protist host (Horn 2008). This raises the question as to whether the TM6 ancestor was also parasitic, or whether parasitism evolved independently on multiple occasions in this phylum. We inferred a phylogenetic tree from an alignment of the ATP/ADP translocase genes identified in the TM6, Chlamydiae, Rickettsiales, and chloroplast genomes. Consistent with a previous analysis, we found that the Chlamydiae translocases are monophyletic supporting the position that the chlamydial ancestor was parasitic (Schmitz-Esser et al. 2004). Similarly, the TM6 translocases are monophyletic (fig. 4A) indicating that at least the ancestor of the class Babeliae (fig. 1) was also parasitic. Genome representation from class F38 is necessary to determine whether these findings extend to the TM6 phylum ancestor. TM6 translocase genes were duplicated independently several times particularly in the UASB124/340 lineage (fig. 4B). Such duplications are also apparent in the Chlamydiae and particularly in the Rickettsiales (Schmitz-Esser et al. 2004), some of which have been shown to increase the substrate range of the enzyme to other nucleotides including GTP, UTP, CTP, and ATP (Tjaden et al. 1999; Audia and Winkler 2006). We note that several TM6 translocase paralogs have substitutions of essential amino acid residues (fig. 4A) suggesting neofunctionalization of these genes, possibly also to other substrates. We did not identify any ATP/ADP translocases in the ACD64 genome (Wrighton et al. 2014); however, these may have been missed since the genome is estimated to be only 42% complete (table 1).
Fig. 4.
Phylogenetic relationships of TM6 ATP/ADP translocase proteins. (A) Unrooted maximum-likelihood tree of aligned ATP/ADP transporter (nucleotide transporter; NTT) amino acid sequences identified in the TM6 genomes and representatives of the Chlamydiae, Rickettsiales, chloroplasts, and Encephalitozoon cuniculi (microsporidia; Schmitz-Esser et al. 2004). Paralogs are indicated as NTT1–NTT5. Five-digit numbers included in the TM6 bacteria labels represent sequential gene ordering (i.e., relative genomic location) in the respective population genomes as annotated by PROKKA. The scale bar indicates amino acid substitutions per site. Alignment of key ATP/ADP translocase amino acid residues for transport efficiency and substrate specificity is depicted in a colored matrix adjacent to the tree, in which conservative amino acid mutations are represented by shades of blue (H, K, and R) or orange (D and E). Asterisks indicate paralogs with substitutions in these key residues. Residue position R46 is based on the Rickettsia prowazekii ATP/ADP translocase (Alexeyev and Winkler 2000) whereas positions K155, E245, E385, and K527 are based on the Arabidopsis thaliana ATP/ADP translocase (Trentmann et al. 2000). (B) Genome-based tree of the TM6 phylum (inset from fig. 1A) showing inferred ATP/ADP translocase gene duplication events and number of paralogs identified in the respective population genomes. Inverted triangles indicate gene duplications that can be inferred from ATP/ADP translocase paralog count, genomic location, and gene phylogeny (fig. 4A).
An open question is host-specificity of TM6 bacteria. Recent studies indicate that B. massiliensis and TM6SC1 are intracellular parasites of amoebae (Cohen et al. 2011; McLean et al. 2013; Pagnier et al. 2015), and we predict that SOIL31 and SOIL82 are associated with ciliate hosts based on co-occurrence analysis (supplementary fig. S4, Supplementary Material online). Amoebae are thought to act as reservoirs for pathogenic bacterial genera such as Legionella and Chlamydia, and coevolution of the bacterium with its amoebal host has led to evolution of mechanisms for survival in higher eukaryotes (Molmeret et al. 2005). One such mechanism is the accumulation of proteins containing domains that enable interaction with eukaryotic hosts, of which two proteins with ankyrin repeats in Legionella pneumophila have been shown to be involved in modulating host GTPases (AnkX; Mukherjee et al. 2011) and intracellular proliferation in amoebae and macrophages (AnkB; Al-Khodor et al. 2008). As at least some members of the TM6 phylum have amoebal hosts and most possess numerous ankyrin repeats (supplementary table S6, Supplementary Material online), it is tempting to speculate that they may have the capacity to invade macrophages. Further exploration of the TM6 phylum through comparative genomics will shed light on this possibility and on host-specificity in general.
Based on genome features of TM6 bacteria, we propose the following names for the TM6 phylum, class, order, and family level lineage described in this study:
“Dependentiae phyl. nov.” L. fem. n. dependentia, dependent, L. fem. pl. n. Dependentiae to reflect dependence of TM6 bacteria on host organisms.
“Babeliae class nov.,” N.L. fem. pl. n. Babeliae, -ae ending to denote a class, the class-level lineage of genus Babela; derived from Babela massiliensis, the first named representative of the TM6 phylum (Pagnier et al. 2015).
“Babeliales ord. nov.,” N.L. fem. n. Babeliales, -ales ending to denote an order, the order-level lineage of genus Babela. Description is the same as the class Babeliae.
“Babeliaceae fam. nov.,” N.L. fem. n. Babeliaceae, -aceae ending to denote a family, the family-level lineage of genus Babela. Description is the same as the class Babeliae.
During review of this manuscript, 15 additional TM6 population genomes recovered from aquifer metagenomes became publicly available (Brown et al. 2015). A preliminary inspection of these genomes indicates that they represent members of the class Babeliae and have features consistent with parasitism including reduced genome size, ankyrin repeats, and ATP/ADP translocases, supporting our conclusions concerning this phylum.
Materials and Methods
Source of TM6 Population Genomes
Two previously reported metagenomic data sets were used for mining TM6 population genomes, one from a UASB reactor treating organic wastewater from a sugar manufacturing factory (Soo et al. 2014) and the other from sugarcane agricultural soil (Yeoh et al. 2015). Metagenomic assembly and recovery of population genomes from the UASB reactor metagenome are described in Soo et al. (2014) and Sekiguchi et al. (2015). For the agricultural soil, a de novo metagenome assembly was performed. Briefly, library adapter sequences were first removed and overlapping read pairs merged using SeqPrep (https://github.com/jstjohn/SeqPrep, last accessed December 10, 2015) with default settings. Nonoverlapping reads were then quality trimmed to a Phred quality score threshold of 20 using Nesoni 0.128 (https://github.com/Victorian-Bioinformatics-Consortium/nesoni, last accessed December 10, 2015). These sequences were then coassembled using the de novo assembly algorithm in CLC Genomics Workbench 6.5 (CLC bio). After assembly, reads from the respective samples were mapped to the scaffolds using BWA-MEM in BWA 0.7.10 (Li and Durbin 2009). Population genome binning using differential coverage was performed using GroopM version 0.2 (Imelfort et al. 2014). Scaffolds greater than 5,000 bp were used as seed sequences for recruitment of scaffolds that were greater than 4,000 bp.
Inferring Taxonomic Assignment of Recovered Population Genomes
Population genomes recovered from the UASB and soil metagenomes were classified by placement in a concatenated marker gene tree. Amino acid sequences of 38 universal (Darling et al. 2014) and 83 bacterial phylogenetically informative marker genes (Soo et al. 2014) were identified in the recovered population genomes, TM6SC1, Babela massiliensis and finished genomes in the IMG database (release 4.1; Markowitz et al. 2012). The amino acid sequences of these marker genes were aligned using HMMER version 3.1b1 (Eddy, 2011) and concatenated. Ambiguous alignment positions were masked using Gblocks version 0.91b (Talavera and Castresana 2007). Trees were inferred using FastTree v2.1.7 with default settings (JTT model, CAT approximation; Price et al. 2010) and visualized using ARB (Ludwig et al. 2004). Support values were determined using nonparametric bootstrapping (Felsenstein 1985). The final figure was edited in Inkscape version 0.48 for publication. Population genomes robustly clustering with TM6SC1 and B. massiliensis were classified as representatives of the TM6 phylum, and subsequently verified in some instances by the presence of partial or complete 16S rRNA gene sequences (see below).
Genome Completeness and Contamination Estimates
Completeness and contamination of the TM6 genomes were estimated by the presence/absence of bacterial single-copy marker genes as described in Rinke et al. (2013), Soo et al. (2014), Wrighton et al. (2014), and Sekiguchi et al. (2015). Completeness was reported as the percentage of 104 bacterial single-copy genes (modified from Dupont et al. 2012) present in each TM6 genome, whereas contamination was reported as the percentage of single-copy genes found in multiple copies indicating possible inclusion of genome sequences belonging to other populations (Albertsen et al. 2013; Soo et al. 2014; Sekiguchi et al. 2015). Of the 104 marker genes used, four were absent in all TM6 genomes and were inferred to be a lineage-specific loss. Completeness and contamination estimates were hence adjusted to be based on the remaining 100 marker genes. All completeness and contamination analyses were performed using CheckM version 1.0.0 (Parks et al. 2015).
In addition, GC content deviation and tetranucleotide frequencies of the five SOIL and UASB TM6 population genomes were determined to identify putative contaminating contigs that may lead to erroneous gene annotation and metabolic inferences. Contig %GC and tetranucleotide frequencies were plotted against contig size and compared with %GC and tetranucleotide frequency deviation windows calculated from IMG reference genomes. All TM6 contigs that were not within these windows were examined for gene content (see Annotation and Metabolic Reconstruction section for methods). These analyses were performed using RefineM version 0.0.6 (https://github.com/dparks1134/RefineM, last accessed December 10, 2015).
Average AAIs
The average AAI between pairs of genomes was calculated from orthologous genes identified through reciprocal best BLAST hits as implemented in CompareM version 0.0.5 (https://github.com/dparks1134/CompareM, last accessed December 10, 2015).
16S rRNA Gene Phylogeny
Representative phylum TM6 16S rRNA gene (16S) sequences (>1,200 nt) were exported from an ARB database of Greengenes 16S sequences (May 2013 release) with Lane masking, and then aligned de novo with 16S sequences identified in the TM6 population genomes using MUSCLE version 3.8.31 (Edgar 2004). Trees were inferred using FastTree v2.1.7 using default settings (JTT model, CAT approximation; Price et al. 2010) and visualized using ARB (Ludwig et al. 2004). Support values were determined using nonparametric bootstrapping (Felsenstein 1985).
Annotation and Metabolic Reconstruction
ORFs in TM6 population genomes were identified using Prodigal v2.60 and annotated using 1) PROKKA version 1.10 (Seemann 2014), 2) BLAST alignment against IMG finished genomes (Release 4.1) and Uniref90 (downloaded April 3, 2014), and 3) HMMER against Hidden Markov Models (HMMs) in Pfam-A (27.0) (Finn et al. 2014) and TIGRFAM (release 14) (Haft et al. 2003) databases. The genomes were also submitted to the IMG Expert Review (ER) and Rapid Annotation using Subsystem Technology (RAST) systems for annotation. Metabolic pathways were visualized using the Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al. 2014) pathway maps based on annotations provided by the KEGG Automatic Annotation Server (KAAS; Moriya et al. 2007). Gene assignments to COGs were identified by IMG ER, and COG profiles were visualized using PCA implemented in the vegan package for R version 3.0.2 (R Core Team 2014). Mann–Whitney tests in R were used to statistically compare differences in COG categories between genomes. Orthologous groups of amino acid sequences were identified using Proteinortho 5 (Lechner et al. 2011). Orthologous groups consisting of genes present in at least six of the seven genomes were considered to be core genes, whereas groups with lower representation across the phylum were considered to be accessory genes.
Protein domains that facilitate interaction with eukaryotic hosts in the TM6 bacteria genomes were identified using an HMMER search with the corresponding HMMs obtained from Pfam (Finn et al. 2014). Matching ORFs with e values less than 0.001 were then queried against the UNIREF90 (downloaded April 2014) database using BLAST to identify their best BLAST hits.
ATP/ADP Translocase Gene Phylogeny
An ARB database containing chlamydial, rickettsial, and chloroplast ATP/ADP translocase protein sequences was downloaded from http://www.microbial-ecology.net/download (last accessed December 10, 2015) (Schmitz-Esser et al. 2004) and aligned de novo together with ATP/ADP translocases identified in the TM6 genomes using MAFFT-linsi version 6.864b (Katoh and Toh 2010). Functional conserved amino acid residues were identified in the alignment by comparison to reference sequences. Phylogenetic trees were inferred using FastTree v2.1.7 with default settings (JTT model, CAT approximation; Price et al. 2010) and visualized using ARB (Ludwig et al. 2004). Support values were determined using nonparametric bootstrapping (Felsenstein 1985).
Supplementary Material
Supplementary figures S1–S6 and tables S1–S6 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Satoshi Hanada (National Institute of Advanced Industrial Science and Technology, Japan) for invaluable etymological advice. The SOIL and UASB TM6 genomes are deposited in GenBank as Whole Genome Shotgun projects with accessions LDIV00000000, LDIW00000000, LDIX00000000, LDIY00000000, and LDIZ00000000. This research was supported through access to facilities managed by Bioplatforms Australia and funded by the Australian Government National Collaborative Research Infrastructure Strategy and Education Investment Fund Super Science Initiative. It was also supported by a Queensland Smart State Co-investment fund awarded to Mark A. Ragan, Susanne Schmidt, and P.H. Y.K.Y. was supported by a University of Queensland International PhD scholarship; D.H.P and P.H. were supported by an Australian Research Council Discovery Outstanding Researcher Award (DP120103498) and an Australian Research Council Laureate Fellowship. The authors declare no competing interests.
References
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. 2013. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 31:533–538. [DOI] [PubMed] [Google Scholar]
- Al-Khodor S, Price CT, Habyarimana F, Kalia A, Abu Kwaik Y. 2008. A Dot/Icm-translocated ankyrin protein of Legionella pneumophila is required for intracellular proliferation within human macrophages and protozoa. Mol Biol. 70:908–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyev MF, Winkler HH. 2000. Complete replacement of basic amino acid residues with cysteines in Rickettsia prowazekii ATP/ADP translocase. Biochim Biophys Acta. 1565:136–142. [DOI] [PubMed] [Google Scholar]
- Audia JP, Winkler HH. 2006. Study of five Rickettsia prowazekii proteins annotated as ATP/ADP translocases (Tlc): only Tlc1 transport ATP/ADP, while Tlc4 and Tlc5 transport other ribonucleotides. J Bacteriol. 188:6261–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, McCutcheon JP, MacDonald BR, Romanovicz D, Moran NA. 2014. Differential genome evolution between companion symbionts in an insect-bacterial symbiosis. MBio 5:e01697–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boscaro V, Vannini C, Forkin SI, Verni F, Petroni G. 2012. Characterization of “Candidatus Nebulobacter yamunensis” from the cytoplasm of Euplotes aediculatus (Ciliophora, Spirotrichea) and emended description of the family Francisellaceae. Syst Appl Microbiol. 35:432–440. [DOI] [PubMed] [Google Scholar]
- Brown CT, Hug LA, Thomas BC, Sharon I, Castelle CJ, Singh A, Wilkins MJ, Wrighton KC, Williams KH, Banfield JF. 2015. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature 523:208–211. [DOI] [PubMed] [Google Scholar]
- Cocchiaro JL, Kumar Y, Fischer ER, Hackstadt T, Valdivia RH. 2008. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc Natl Acad Sci U S A. 105:9379–9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen G, Hoffart L, La Scola B, Raoult D, Drancourt M. 2011. Ameba-associated keratitis, France. Emerg Infect Dis. 17:1306–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig L, Pique ME, Tainer JA. 2004. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2:363–378. [DOI] [PubMed] [Google Scholar]
- Darling AE, Jospin G, Lowe E, Matsen FA, Bik HM, Eisen JA. 2014. Phylosift: phylogenetic analysis of genomes and metagenomes. PeerJ 2:e243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr J, Hurek T, Reinhold-Hurek B. 1998. Type IV pili are involved in plant–microbe and fungus–microbe interactions. Mol Microbiol. 30:7–17. [DOI] [PubMed] [Google Scholar]
- Dupont CL, Rusch DB, Yooseph S, Lombardo MJ, Ritcher RA, Valas R, Novotny M, Yee-Greenbaum J, Selengut JD, Haft DH, et al. 2012. Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. Isme J. 6:1186–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput Biol. 7:e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escudero LV, Casamayor EO, Chong G, Pedrós-Alió C, Demergasso C. 2013. Distribution of microbial arsenic reduction, oxidation and extrusion genes along a wide range of environmental arsenic concentrations. PLoS One 8:e78890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feazel LM, Baumgartner LK, Peterson KL, Frank DN, Harris JK, Pace NR. 2009. Opportunistic pathogens enriched in showerhead biofilms. Proc Natl Acad Sci U S A. 106:16393–16399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RS. 1998. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among Archaebacteria, Eubacteria, and Eukaryotes. Microbiol Mol Biol Rev. 62:1435–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut JD, White O. 2003. The TIGRFAM database of protein families. Nucleic Acids Res. 31:371–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, McLean JS, Edlund A, Yooseph S, Hall AP, Liu SY, Dorrestein PC, Esquenazi E, Hunter RC, Cheng G, et al. 2015. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci U S A. 112:244–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs DE, Yethon JA, Whitfield C. 1998. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol Microbiol. 30:221–232. [DOI] [PubMed] [Google Scholar]
- Henne K, Kahlisch L, Brettar I, Höfle MG. 2012. Analysis of structure and composition of bacterial core communities in mature drinking water biofilms and bulk water of a citywide network in Germany. Appl Environ Microbiol. 78:3530–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques AO, Glaser P, Piggot PJ, Moran CP. 1998. Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol Microbiol. 28:235–247. [DOI] [PubMed] [Google Scholar]
- Horn M. 2008. Chlamydiae as symbionts in eukaryotes. Annu Rev Microbiol. 62:113–131. [DOI] [PubMed] [Google Scholar]
- Imelfort M, Parks D, Woodcroft BJ, Dennis P, Hugenholtz P, Tyson GW. 2014. GroopM: an automated tool for the recovery of population genomes from related metagenomes. PeerJ 2:e603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LJF, Carballido-López R, Errington J. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913–922. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 42:D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor RS, Wrighton KC, Handley KM, Sharon I, Hug LA, Castelle CJ, Thomas BC, Banfield JF. 2013. Small genomes and sparse metabolisms of sediment-associated bacteria from four candidate phyla. MBio 4:e00708–e00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. 2010. Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26:1899–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. 2005. Towards a genome-based taxonomy for prokaryotes. J Bacteriol. 187:6258–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Bork–Jensen J, Gerdes K. 2005. The morphogenetic MreBCD proteins of Escherichia coli form an essential membrane-bound complex. Mol Microbiol. 55:78–89. [DOI] [PubMed] [Google Scholar]
- Kumar Y, Cocchiaro J, Valdivia RH. 2006. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr Biol. 16:1646–1651. [DOI] [PubMed] [Google Scholar]
- Leaver M, Errington J. 2005. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol Microbiol. 57:1196–1209. [DOI] [PubMed] [Google Scholar]
- Lechner M, Findeiß S, Steiner L, Marz M, Stadler PF, Prohaska SJ. 2011. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics 12:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfield J, Fram NR, Ely B. 2011. Across bacterial phyla, distantly-related genomes with similar genomic GC content have similar patterns of amino acid usage. PLoS One 6:e17677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 25:1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luef B, Frischkorn KR, Wrighton KC, Holman HYN, Birarda G, Thomas BC, Singh A, Williams KH, Siegerist CE, Tringe SG, et al. 2015. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun. 6:6372. [DOI] [PubMed] [Google Scholar]
- Markowitz VM, Chen AI, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, et al. 2012. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40:D115–D122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa H, Asoh S, Kunai K, Muraiso K, Takasuga A, Ohta T. 1989. Nucleotide sequence of the rodA gene, responsible for the rod shape of Escherichia coli: rodA and the pbpA gene, encoding penicillin-binding protein 2, constitute the rodA operon. J Bacteriol. 171:558–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10:13–26. [DOI] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. Isme J. 6:610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean JS, Lombardo MJ, Badger JH, Edlund A, Novotny M, Yee-Greenbaum J, Vyahhi N, Hall AP, Yang Y, Dupont CL, et al. 2013. Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci U S A. 110:E2390–E2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D. 2009. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol Direct. 4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. 2005. Amoeba as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol. 71:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583–586. [DOI] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35:W182–W185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. 2004. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 13:1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Liu X, Arasaki K, McDonough J, Galán JE, Roy CR. 2011. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature 477:103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnier I, Yutin N, Croce O, Makarova KS, Wolf YI, Benamar S, Raoult D, Koonin EV, La Scola B. 2015. Babela massiliensis, a representative of a widespread bacterial phylum with unusual adaptations to parasitism in amoebae. Biol Direct. 10:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Lührmann A, Satoh A, Laskowski-Arce MA, Roy CR. 2008. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320:1651–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25:1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 10:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41:D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. 2014. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; Available from: http://www.R-project.org/ [Google Scholar]
- Rheims H, Rainey FA, Stackebrandt E. 1996. A molecular approach to search for diversity among bacteria in the environment. J Ind Microbiol. 17:159–169. [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, et al. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. [DOI] [PubMed] [Google Scholar]
- Schmitz-Esser S, Linka N, Collingro A, Beier CL, Neuhaus HE, Wagner M, Horn M. 2004. ATP/ADP translocases: a common feature of obligate intracellular amoebal symbionts related to Chlamydiae and Rickettsiae. J Bacteriol. 186:683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. [DOI] [PubMed] [Google Scholar]
- Sekiguchi Y, Ohashi A, Parks DH, Yamauchi T, Tyson GW, Hugenholtz P. 2015. First genomic insights into members of a candidate bacterial phylum responsible for wastewater bulking. PeerJ 3:e740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siozios S, Loannidis P, Klasson L, Andersson SGE, Braig HK, Bourtzis K. 2013. The diversity and evolution of Wolbachia ankyrin repeat domain genes. PLoS One 8:e55390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo RM, Skennerton CT, Sekiguchi Y, Imerfort M, Paech SJ, Dennis PG, Steen JA, Parks DH, Tyson GW, Hugenholtz P. 2014. An expanded genomic representation of the phylum Cyanobacteria. Genome Biol Evol. 6:1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen KB, Canfield DE, Teske AP, Oren A. 2005. Community composition of a hypersaline endoevaporitic microbial mat. Appl Environ Microbiol. 71:7352–7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takami H, Noguchi H, Takaki Y, Uchiyama Y, Toyoda A, Nishi A, Chee GJ, Arai W, Nunoura T, Itoh T, et al. 2012. A deeply branching thermophilic bacterium with an ancient acetyl-coA pathway dominates a subsurface ecosystem. PLoS One 7:e30559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56:564–577. [DOI] [PubMed] [Google Scholar]
- Tjaden J, Winkler HH, Schwöppe C, Van Der Laan M, Möhlmann T, Neuhaus HE. 1999. Two nucleotide transport proteins in Chlamydia trachomatis, one for net nucleoside triphosphate uptake and the other for transport of energy. J Bacteriol. 181:1196–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trentmann O, Decker C, Winkler HH, Neuhaus HE. 2000. Charged amino-acid residues in transmembrane domains of the plastidic ATP/ADP transporter from Arabidopsis are important for transport efficiency, substrate specificity, and counter exchange properties. Eur J Biochem. 267:4098–4105. [DOI] [PubMed] [Google Scholar]
- Wachi M, Doi M, Okada Y, Matsuhashi M. 1989. New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J Bacteriol. 171:6511–6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton KC, Castelle CJ, Wilkins MJ, Hug LA, Sharon I, Thomas BC, Handley KM, Mullin SW, Nicora CD, Singh A, et al. 2014. Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer. Isme J. 8:1452–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoh YK, Paungfoo-Lonhienne C, Dennis PG, Robinson N, Ragan MA, Schmidt S, Hugenholtz P. 2015. The core root microbiome of sugarcanes cultivated under varying nitrogen fertiliser application. Environ Microbiol. Advance Access publication July 30, 2015; doi:10.1111/1462-2920.12925 [DOI] [PubMed] [Google Scholar]
- Youssef N, Steidley BL, Elshahed MS. 2012. Novel high-rank phylogenetic lineages within a sulfur spring (Zodletone spring, Oklahoma), revealed using a combined pyrosequencing-sanger approach. Appl Environ Microbiol. 78:2677–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.