Abstract
Genomes of invertebrates and vertebrates exhibit highly divergent patterns of DNA methylation. Invertebrate genomes tend to be sparsely methylated, and DNA methylation is mostly targeted to a subset of transcription units (gene bodies). In a drastic contrast, vertebrate genomes are generally globally and heavily methylated, punctuated by the limited local hypo-methylation of putative regulatory regions such as promoters. These genomic differences also translate into functional differences in DNA methylation and gene regulation. Although promoter DNA methylation is an important regulatory component of vertebrate gene expression, its role in invertebrate gene regulation has been little explored. Instead, gene body DNA methylation is associated with expression of invertebrate genes. However, the evolutionary steps leading to the differentiation of invertebrate and vertebrate genomic DNA methylation remain unresolved. Here we analyzed experimentally determined DNA methylation maps of several species across the invertebrate–vertebrate boundary, to elucidate how vertebrate gene methylation has evolved. We show that, in contrast to the prevailing idea, a substantial number of promoters in an invertebrate basal chordate Ciona intestinalis are methylated. Moreover, gene expression data indicate significant, epigenomic context-dependent associations between promoter methylation and expression in C. intestinalis. However, there is no evidence that promoter methylation in invertebrate chordate has been evolutionarily maintained across the invertebrate–vertebrate boundary. Rather, body-methylated invertebrate genes preferentially obtain hypo-methylated promoters among vertebrates. Conversely, promoter methylation is preferentially found in lineage- and tissue-specific vertebrate genes. These results provide important insights into the evolutionary origin of epigenetic regulation of vertebrate gene expression.
Keywords: DNA methylation, chordate evolution, promoter DNA methylation, gene expression, epigenetic regulation.
Introduction
DNA methylation is a crucial epigenetic mechanism in mammalian genomes, yet it is evolutionarily highly labile (Suzuki and Bird 2008; Mendizabal et al. 2014). For instance, vertebrate and invertebrate genomes exhibit a remarkable contrast with respect to the patterns of genomic DNA methylation. Vertebrate genomes, in particular mammalian genomes, are generally heavily methylated at most CpG sites (“global” DNA methylation), although some cell types/tissues exhibit significantly reduced DNA methylation (e.g., placenta and sperm are markedly lowly methylated compared with other tissues and cell types [Ehrlich et al. 1982; Schroeder et al. 2013; Zeng et al. 2014]). Only a small number of CpGs, localized to short genomic regions, are lowly methylated (hypomethylation), and these regions often encode promoters and enhancers (Mendizabal and Yi 2016; Schultz et al. 2015).
In contrast, invertebrate genomes typically are only sparsely methylated compared with vertebrate genomes (Suzuki and Bird 2008; Zemach et al. 2010). Targets of DNA methylation in invertebrate genomes are concentrated in CpGs at exons and introns of certain genes, or “gene bodies” (Feng et al. 2010; Zemach et al. 2010). For example, in Ciona intestinalis, which is one of the closest invertebrate outgroups to vertebrates DNA methylation is targeted to approximately 60% of gene bodies in different cell types (Suzuki et al. 2013). Other more distant invertebrate genomes, such as those of the honey bee Apis mellifera, are even more sparsely methylated (Lyko et al. 2010; Galbraith et al. 2015). The “global” patterns of DNA methylation observed in vertebrates are a derived feature, originating during early vertebrate evolution (Tweedie et al. 1997; Zhang Z, Liu G, Zhou Y, Lloyd JPB, McCauley DW, Li W, Gu X, Su Z, unpublished data).
The function of DNA methylation varies according to their genomic targets. In the well-studied human genome, DNA methylation of regulatory regions such as promoters and enhancers is typically linked to silencing of downstream gene expression, although this effect is not absolute (Lou et al. 2014; Mendizabal and Yi 2016). DNA methylation of transposable elements (TEs) is also linked to silencing effect (Yoder et al. 1997). In contrast, DNA methylation of gene bodies is associated with active transcription of genes. Actively transcribed gene bodies are often substantially methylated (Feng et al. 2010; Zemach et al. 2010; Jjingo et al. 2012). Methylated gene bodies also show less expression variability compared with other genes, which may be related to the effect of DNA methylation to regulate spurious transcription (Bird 1995; Bird et al. 1995; Zemach et al. 2010; Huh et al. 2013). Overall, it is clear that gene body DNA methylation and promoter DNA methylation both regulate gene expression (Jones 2012; Park et al. 2012; Lou et al. 2014).
As gene body methylation is the pronounced form of genomic DNA methylation in invertebrates, it may play roles in regulation of gene expression. Indeed, invertebrate gene bodies are clearly separated to heavily and sparsely methylated genes, the former highly expressed and the latter lowly expressed (Suzuki et al. 2007; Sarda et al. 2012). In contrast, nearly all gene bodies of vertebrates are heavily methylated with relatively little variation among genes (e.g., Jjingo et al. 2012). Instead, vertebrate promoters exhibit a clear bimodal pattern of lowly and heavily methylated promoters, the former typically found near broadly expressed housekeeping genes and the latter adjacent to tissue specific genes (Antequera 2003; Saxonov et al. 2006; Elango and Yi 2008; Lou et al. 2014; Mendizabal and Yi 2016).
One pressing open question then is, how did the difference in the genomic DNA methylation and expression consequence between vertebrates and invertebrates arise? Given the intriguing relationships to tissue-specific patterns of gene expression, we have previously proposed a biased acquisition of promoter methylation during vertebrate evolution (Elango and Yi 2008). Specifically, it was proposed that methylated gene bodies in the genome of invertebrate ancestor, which encoded broadly expressed genes, preferentially obtained unmethylated promoters in vertebrates. On the other hand, genes used for tissue-specific functions in vertebrate body plan may have preferentially acquired methylated promoters (Elango and Yi 2008).
We examined genome sequence and DNA methylation data to investigate the origin and evolution of promoter and gene body methylation across the invertebrate–vertebrate boundary. We found that, as proposed, orthologs of methylated gene bodies in invertebrates are preferentially associated with unmethylated promoters in vertebrates. Moreover, based on bisulfite sequencing data, we identify genes whose promoters exhibit clear DNA methylation in an early chordate species. Our analyses reveal a complex DNA methylation landscape of an invertebrate chordate genome, and illuminate the evolutionary emergence of hypomethylated vertebrate promoters.
Results
Whole-Genome Bisulfite Sequencing Maps Reveal Pervasive Bimodality of Promoter and Gene Body DNA Methylation in Chordates
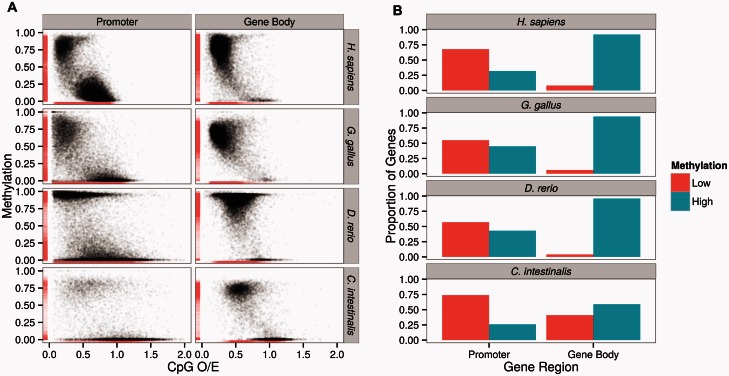
Previous studies often used CpG O/E as a proxy for DNA methylation (Elango and Yi 2008; Yi and Goodisman 2009; Gavery and Roberts 2010; Okamura et al. 2010). To directly analyze the patterns of DNA methylation, we used experimentally determined DNA methylation maps from four diverse chordates (Homo sapiens, Gallus gallus, Danio rerio, and C. intestinalis; data shown in table 1). These are generated by the whole-genome sequencing of bisulfite-converted genomic DNA (referred to as “WGBS”). As expected (Gavery and Roberts 2010; Park et al. 2011; Sarda et al. 2012), CpG O/E and DNA methylation are significantly negatively correlated, for both promoters and gene bodies in all species (fig. 1A).
Table 1.
WGBS Data and RNA-seq Gene Expression Data Used in This Study.
| Species | Genome Build | WGBS Data |
Gene Expression Data |
||
|---|---|---|---|---|---|
| Source | Accession No. | Source | Accession No. | ||
| Homo sapiens | hg19 | Psoas muscle | GSM1010986 | Muscle | Human Bodymap 2.0 project |
| Gallus gallus | galgal4 | Embryo | SRR942840 | — | — |
| Danio rerio | zv9 | 1,000-Cell zygote | GSM1133397 | 1,000-Cell embryo | GSM1085061 |
| Ciona intestinalis | JGI 2.0 | Muscle | GSM497251 | Muscle | GSM497252 |
Fig. 1.
(A) Distributions of promoter and gene body DNA methylation from whole-genome methylation maps in four species across the chordate phylogeny. Both promoters and gene bodies are composed of lowly (mean fractional methylation level < 0.3) and highly methylated (mean fractional methylation level > 0.5) subsets in these species, and show good correspondence with the CpG O/E values. (B) Using empirical cutoff values, the proportions of lowly and highly methylated promoters and gene bodies of the study species are depicted.
We find a clear “bimodal” pattern of DNA methylation from the invertebrate chordate C. intestinalis, of lowly and highly methylated gene bodies (fig. 1A, also in Suzuki et al. 2007; Elango and Yi 2008; Sarda et al. 2012; Suzuki et al. 2013). In contrast, the majority of vertebrate gene bodies is heavily methylated (fig. 1A). Interestingly, we observe that a small number of vertebrate genes remain sparsely methylated, even though at much lower frequencies than in the invertebrate outgroup C. intestinalis (e.g., 3.2% of all genes in H. sapiens can be classified as lowly methylated, whereas 34% of C. intestinalis genes are lowly methylated; fig. 1B).
It is well established that vertebrate promoters exhibit a clear “bimodality” of lowly and highly methylated promoters (Saxonov et al. 2006; Elango and Yi 2008). Indeed, we observe the expected pattern (fig. 1A). Moreover, we find that some promoters in the invertebrate outgroup C. intestinalis exhibit substantial levels of DNA methylation (fig. 1A).
Based upon the observed distribution of DNA methylation (fig. 1A), we can classify promoter and gene body methylation values into binary “highly” (mean fractional methylation levels > 0.5) and “lowly” methylated (mean fractional methylation levels < 0.3) groups for some analyses, given their generally bimodal nature (as in Sarda et al. 2012). This criterion retained between 80% and 95% genes per species (supplementary table S1, Supplementary Material online). Figure 1B represents the distributions of lowly and highly methylated promoters and gene bodies in the analyzed species, based upon this criterion. These analyses confirm the previous findings that gene body methylation and promoter bimodality are dominant patterns of genic DNA methylation in invertebrates and vertebrates, respectively (Suzuki et al. 2007; Elango and Yi 2008; Gavery and Roberts 2010; Sarda et al. 2012; Suzuki et al. 2013). At the same time however, we show that both promoter and gene body methylation can be classified into low and high methylation across the invertebrate–vertebrate boundary.
Methylated Promoters in C. intestinalis Affect Gene Expression
The role of promoter methylation on regulation of gene expression in invertebrate genomes has been little explored (however, see Olson and Roberts 2014; Saint-Carlier and Riviere 2015, also in discussion). The observed promoter DNA methylation in C. intestinalis might be of functional consequence, or merely a result of noisy methylation surrounding the region of functional importance, such as gene body. According to the latter hypothesis, promoter DNA methylation should be confined to those adjacent to heavily methylated gene bodies.
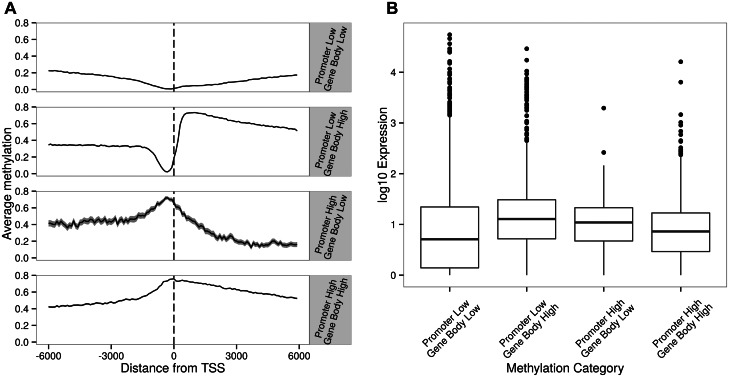
However, this is not the case. A detailed examination of DNA methylation near transcription start sites (TSSs) illustrates that we can identify four classes of C. intestinalis genes, with promoters and gene bodies exhibiting low and high DNA methylation, respectively (fig. 2A). The majority (71.4%) of genes is devoid of promoter DNA methylation (first and second categories in fig. 2A, 3,387 and 2,633 genes, respectively). Most of the remaining genes (fourth category in fig. 2A, 2,252 genes) exhibit heavy gene body DNA methylation as well as promoter DNA methylation. Notably, for approximately 2% of genes, promoter DNA methylation is present in the absence of gene body DNA methylation (third category in fig. 2A, 163 genes). The lists of genes belonging to these four categories are shown in supplementary table S2, Supplementary Material online.
Fig. 2.
(A) Sea squirt (Ciona intestinalis) genes with low and high levels of DNA methylation in promoters and gene bodies. Figures drawn using values calculated from sliding windows that are 100 bases wide with a 10-base step size. Genes in this species can be classified into four groups, where promoter and gene bodies are lowly and highly methylated, respectively. The numbers of genes in each group are described in the main text. (B) Variation of gene expression levels of these four methylation categories.
We then examined whether such distinctive DNA methylation patterns across TSS harbor functional significance. We approached this by investigating functional annotation, genome sequence analyses, and gene expression data. Gene ontology analyses found no significant enrichment of genes in any of the above categories. Nevertheless, we found that genes harboring HOX domain are enriched in the first category in figure 2A (3-fold enrichment according to the INTERPRO protein families database; Mitchell et al. 2015). Next, we examined gene expression data (RNA-seq data of the same muscle tissue where WGBS maps are from Zemach et al. [2010]). Consistent with previous findings (Zemach et al. 2010; Zeng and Yi 2010; Sarda et al. 2012; Gavery and Roberts 2013), highly methylated gene bodies in C. intestinalis exhibited significantly higher expression levels than lowly methylated gene bodies (t-test, P < 10−15). Interestingly, among genes with high gene body DNA methylation, those with high promoter methylation exhibit lower level of gene expression than those with low promoter methylation, although not significantly so (fig. 2B). Interestingly, when gene body methylation is low or absent, promoter methylated genes are more highly expressed than those without promoter methylation (t-test, P = 0.026, fig. 2B).
We have previously shown that methylated and non-methylated genes in some invertebrates, such as C. intestinalis, are associated with different gene lengths parameters (Zeng and Yi 2010; Sarda et al. 2012). As gene lengths and gene expression are correlated (e.g., Park et al. 2012), we examined whether the pattern we observe could be confounded by the difference in gene lengths. Indeed, the four categories of genes show different gene lengths (supplementary fig. S1, Supplementary Material online). Notably, genes with high promoter methylation in the near absence of gene body DNA methylation are the longest among all genes, which is due to unusually long introns of these genes (supplementary fig. S1, Supplementary Material online).
To assess the impact of promoter DNA methylation on gene expression while avoiding the confounding effect of gene lengths and sequence composition, we used a partial correlation analysis (Materials and Methods, table 2). The results from this analysis indicate that the qualitative findings from the figure 2B hold true when the effects of other variables are controlled. Specifically, promoter DNA methylation is consistently positively correlated with gene expression when gene body methylation is low, although not significantly so. We note that the sample size is much smaller for lowly methylated gene body data. In contrast, promoter DNA methylation is significantly negatively correlated with gene expression when gene body methylation is high. We have repeated the same analyses after limiting to those with ≥5CpGs and found similar results to table 2 (supplementary table S3, Supplementary Material online).
Table 2.
Partial Correlation between Promoter Methylation and Gene Expression in Ciona intestinalis.
| Gene Body Methylation | Conditioned Variables | Partial Correlation | P Value |
|---|---|---|---|
| Low | GC | 0.04 | 0.07 |
| Low | CDS length | 0.04 | 0.1 |
| Low | Gene length | 0.03 | 0.2 |
| Low | GC + CDS length | 0.05 | 0.046 |
| Low | GC + gene length | 0.04 | 0.1 |
| High | GC | −0.21 | <2.2e-16 |
| High | CDS length | −0.23 | <2.2e-16 |
| High | Gene length | −0.22 | <2.2e-16 |
| High | GC + CDS length | −0.22 | <2.2e-16 |
| High | GC + gene length | −0.21 | <2.2e-16 |
Note.—Analyses are performed separately for low and high gene body methylated genes. Data are from WGBS and RNA-seq data from muscle tissue in C. intestinalis (Zemach et al. 2010). Gene length variables are log-transformed to improve normality.
TE DNA Methylation Does Not Associate with Promoter Methylation in C. intestinalis
We analyzed several aspects of C. intestinalis genome to shed lights on the origin of promoter methylation in this species. We first examined the relationship between promoter DNA methylation and TE DNA methylation. Silencing of TEs may be a potential primary force driving the global DNA methylation of vertebrate genomes (Yoder et al. 1997). If indeed promoter methylation in Ciona is largely due to the methylation of TEs, most of methylated promoters in C. intestinalis should include TE-derived sequences.
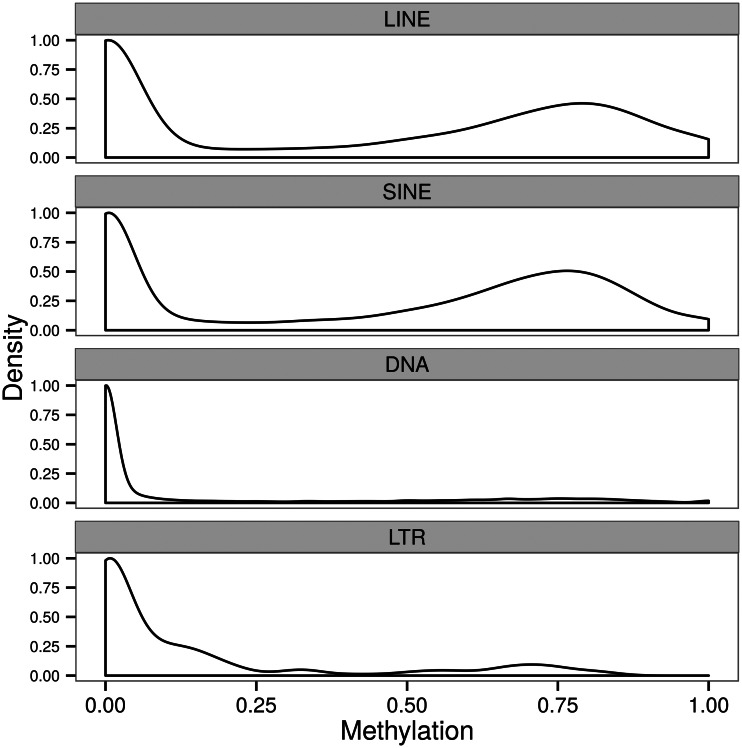
There are several classes of TEs in the genome of C. intestinalis (e.g., Sela et al. 2010). Unlike in the genomes of vertebrates, many TEs are not methylated in Ciona (e.g., Simmen et al. 1999; fig. 3). Specifically, we show that only subsets of long interspersed elements (LINEs) and short interspersed elements (SINEs) in Ciona exhibit substantial DNA methylation (fig. 3). Among approximately 36,000 SINEs and LINEs in C. intestinalis with WGBS data coverage, approximately half of them can be classified as highly methylated (e.g., mean fractional methylation ≥ 0.5; fig. 3).
Fig. 3.
Distribution of DNA methylation in different TE classes of Ciona intestinalis. Only LINEs and SINEs have substantially methylated subsets.
However, methylated SINEs and LINEs are not targeted to methylated promoters. In fact, they are found less frequently than expected in promoters (chi-square test, P < 0.0001; supplementary table S4, Supplementary Material online). Among the total SINEs and LINEs, only 4% of them overlap with promoter regions. In fact, most (84%) of promoters in this genome do not contain TEs of any type. Nevertheless, promoters harboring LINEs or SINEs exhibit slightly yet significantly higher level of DNA methylation than those without TEs (0.25 vs. 0.22, P = 0.0001, t-test). Thus, although there is no evidence of LINEs and SINEs specifically targeting promoters, some promoters harboring these elements exhibit slightly higher DNA methylation levels than those without TEs. We also examined what effect TEs might have for gene body DNA methylation. The overall levels of gene body methylation were very similar between those harboring LINEs/SINEs versus those without (average methylation 0.45 vs. 0.44, P = 0.09, t-test), suggesting that the presence of LINEs or SINEs does not contribute meaningfully to overall methylation level within a gene body.
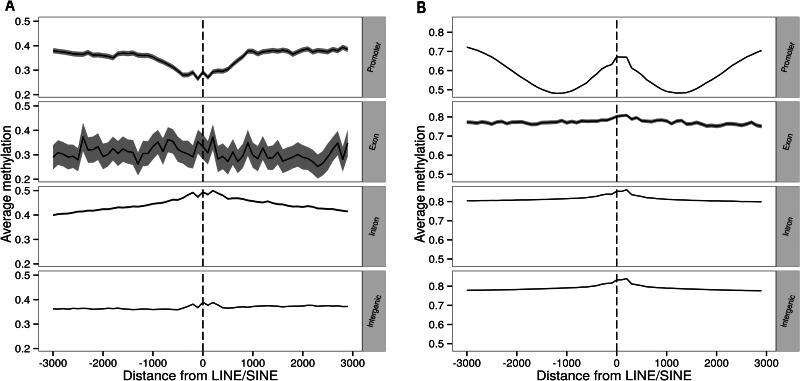
We further examined the association between specific genomic context and methylation of TEs. We divided TEs into groups depending on whether they were present in promoter, exon, intron, or intergenic regions and compared methylation in the upstream and downstream sequences. In C. intestinalis, only introns have a slight methylation peak near TEs (fig. 4A). In contrast, D. rerio genome is generally heavily methylated, and significant increases of DNA methylation near TEs are observed in promoters (fig. 4B). Other vertebrate genomes display similar methylation patterns (supplementary fig. S2, Supplementary Material online).
Fig. 4.
Variation of DNA methylation in different genomic regions as a function of the distance from LINEs/SINEs in (A) Ciona intestinalis and (B) Danio rerio. Although a peak of DNA methylation in the promoters of D. rerio is apparent in (B), promoters of C. intestinalis do not show such a pattern. Other vertebrates have similar methylation patterns (supplementary fig. S2, Supplementary Material online). Figures drawn using values calculated from sliding windows that are 100 bases wide with a 10-base step size.
Methylated Ciona Promoters Are Not Conserved during Vertebrate Evolution
We investigated whether the methylated promoters in C. intestinalis tend to maintain its methylated status throughout vertebrate evolution. Specifically, using the WGBS data, we examined the promoter methylation status of orthologs among the four chordates. We found that in all pairwise comparisons, heavily methylated promoters in Ciona do not preferentially maintain methylation in the vertebrate genomes analyzed. In the Ciona genome, the ratio of lowly versus highly methylated promoters is 6,020:2,415. This ratio in the orthologs is 508:203, not statistically different from that in the Ciona genome. In all pairwise comparisons with the other three species, there is no tendency for methylated promoters in Ciona to preferentially maintain their methylation status in other vertebrates (Odds ratio 1.09∼1.49, not significant in all cases, table 3). We conclude that DNA methylation of promoters in C. intestinalis does not directly translate to preferentially methylated vertebrate promoters.
Table 3.
No Evidence that Methylated Promoters in the Invertebrate Ciona intestinalis Are Preferentially Remained as Methylated along Vertebrate Evolution, Using Orthologs across These Species.
| Promoter Methylation in Vertebrates |
||||||
|---|---|---|---|---|---|---|
| Promoter Methylation in Ciona intestinalis |
Danio rerio |
Gallus gallus |
Homo sapiens |
|||
| Low | High | Low | High | Low | High | |
| Low | 440 | 68 | 439 | 42 | 377 | 110 |
| High | 165 | 38 | 172 | 18 | 143 | 56 |
Note.—Classification of promoters to low or high is as depicted in figure 1 and Materials and Methods.
Evolutionarily Conserved, Broadly Expressed Vertebrate Genes Avoid Promoter Methylation
As seen in table 3, promoters of most orthologs in the vertebrate genomes are lowly methylated, regardless of whether they were methylated or not in the outgroup C. intestinalis genome. Indeed, we have previously shown that housekeeping genes, which are more evolutionarily conserved than tissue-specific genes, harbor lowly methylated promoters (Elango and Yi 2008). We further proposed that body-methylated genes in invertebrate outgroups avoid promoter DNA methylation during vertebrate evolution (Elango and Yi 2008).
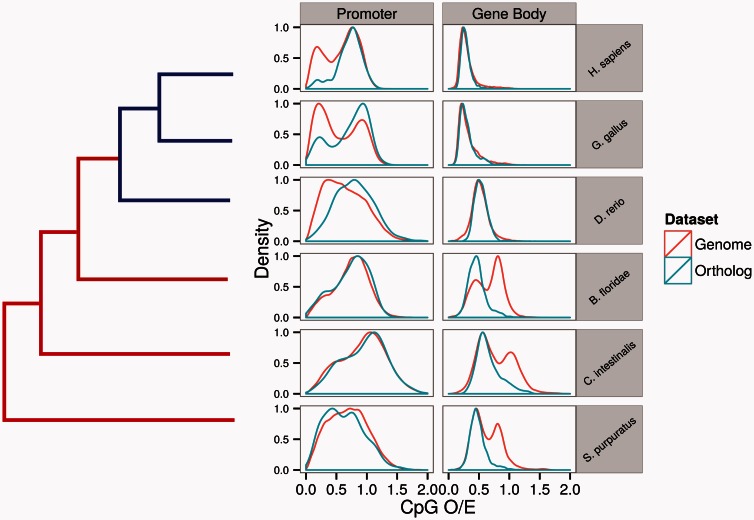
To directly test this hypothesis, we examined DNA methylation patterns of single-copy orthologs across six species straddling the invertebrate–vertebrate boundaries (Materials and Methods). We first examined CpG O/E profiles of single-copy orthologs. Comparison of CpG O/E profiles of whole genome versus orthologs in these species demonstrates a clear and intriguing pattern of bias for promoter and gene body DNA methylation (fig. 5). Specifically, orthologs are preferentially found in genes exhibiting low gene body CpG O/E (high methylation) in invertebrates, and in high CpG O/E promoters (low methylation) among vertebrates. WGBS data of four species (C. intestinalis, D. rerio, G. gallus, and H. sapiens) demonstrate the same pattern. For example, when we examined 1,807 pairwise orthologs between humans and C. intestinalis, the majority of gene pairs (75%) had low human promoter methylation and high C. intestinalis gene body methylation. Therefore, computational and experimental data confirm that ancestrally methylated gene body genes are preferentially found in lowly methylated promoter genes of vertebrates.
Fig. 5.
Contrasting distributions of CpG O/E in all genes versus orthologs across the invertebrate–vertebrate boundary.
We further examined another data set where nonmethylated regions of the genome were experimentally characterized. Specifically, Long et al. (2013) have identified, using biotinylated CAPCxxC affinity purification (Bio-CAP) sequencing method, regions harboring little DNA methylation (referred to as “nonmethylated islands [NMIs]”) from several vertebrate genomes. We investigated whether these NMIs are preferentially found near promoters of orthologous genes compared with the genomic background. Indeed, NMIs are highly biased toward the promoters of conserved, orthologous genes compared with the entire genome (supplementary fig. S3, Supplementary Material online, Fisher’s exact test <10−15 for all comparisons). The NMI genes area also enriched in housekeeping functions such as translation and RNA-processing (supplementary table S5, Supplementary Material online), consistent with the idea that lowly methylated promoters encode broadly expressed genes (Elango and Yi 2008).
Discussion
Whole-genome DNA methylation maps of several invertebrate species are now available, illuminating the evolutionary history of genomic DNA methylation in animals. The most pronounced difference between invertebrate whole-genome methylation maps and the well-characterized mammalian DNA methylation maps is that the latter is nearly ubiquitously methylated, whereas invertebrate genomes are sparsely or moderately methylated with most of DNA methylation found in gene bodies. DNA methylation of gene bodies in invertebrates has a clear consequence on gene expression, where methylated and unmethylated gene bodies represent highly and broadly expressed genes versus those that are lowly expressed, and when examined, in a more tissue- or phenotype (such as different castes in social insects)-specific manner (Elango et al. 2009; Foret et al. 2009; Gavery and Roberts 2010; Lyko et al. 2010; Zeng and Yi 2010; Patalano et al. 2012; Sarda et al. 2012; Hunt et al. 2013; Wang et al. 2013; Olson and Roberts 2014). DNA methylation is also associated with alternative splicing of genes in invertebrates (Park et al. 2011; Flores et al. 2012). The strong associations between gene body methylation and gene expression in invertebrate genomes bring intriguing questions with respect to the evolutionary contrast between vertebrate and invertebrate genomes. Unlike in invertebrate genomes, promoter DNA methylation is tightly linked to gene expression in mammals, where methylated promoters are often associated with suppression of transcription (Antequera 2003; Weber et al. 2007; Lou et al. 2014; Mendizabal and Yi 2016).
To shed lights on the evolutionary trajectories of leading to such a contrast between patterns of invertebrate and vertebrate DNA methylation, we examined whole-genome DNA methylation maps of several species straddling the invertebrate–vertebrate boundary. Unexpectedly, we found that a substantial number of promoters in the invertebrate chordate C. intestinalis were methylated (fig. 1). The level of methylation in the promoters is comparable to those of methylated gene bodies. Moreover, analyses of the genome-wide expression indicate that methylation of C. intestinalis promoters is significantly associated with levels of gene expression, independent of the effect of gene body DNA methylation and other genomic variables (fig. 2 and table 2).
Is promoter DNA methylation present in other invertebrate genomes, and if so, does it affect gene expression? So far the only other known examples are found in the Pacific oyster Crassostrea gigas, another moderately methylated species where approximately 15% of CpGs are methylated (Olson and Roberts 2014). It was shown that expression of homeobox genes in Cr. gigas was affected by differential promoter DNA methylation (Riviere et al. 2013; Saint-Carlier and Riviere 2015). In these studies, promoter DNA methylation suppressed the expression of homeobox genes (Saint-Carlier and Riviere 2015). In C. intestinalis however, homeobox genes tend to be devoid of DNA methylation, and promoter methylation in lowly methylated epigenomic context on average facilitates gene expression (table 2). The contrast between these two systems needs to be resolved. It is possible that the homeobox promoters may undergo cell type-specific DNA methylation during the C. intestinalis development, and our muscle data have failed to capture it. Another study, however, proposed that promoter methylation and gene expression were positively correlated in Cr. gigas (Olson and Roberts 2014), although it was not clear whether the effect of promoter methylation was independent of gene body DNA methylation. Whether the promoter methylation of C. intestinalis and Cr. gigas represent an ancestral origin of invertebrate promoter methylation or were independently acquired, and what functional roles promoter methylation in these species might play, remains to be determined.
Interestingly, in our analyses, the effect of promoter DNA methylation in C. intestinalis is dependent on the epigenomic context of nearby regions. When adjacent to methylated gene bodies (thereby resembling the epigenomic context of vertebrate promoters), methylation of promoters is negatively correlated with gene expression levels (table 2). On the other hand, when adjacent to unmethylated gene bodies, promoter methylation has negligible overall effect on gene expression (table 2). Given that global DNA methylation of genomes originated early in the vertebrate evolution (Tweedie et al. 1997; Zhang Z, Liu G, Zhou Y, Lloyd JPB, McCauley DW, Li W, Gu X, Su Z, unpublished data), and that DNA methylation typically suppresses transcription in vertebrate genomes, it is tempting to hypothesize that the silencing effect of DNA methylation in the methylated epigenomic context (e.g., adjacent to methylated gene bodies) may have been co-opted genome-wide early in the vertebrate evolution.
It was proposed that the invasion of TEs of vertebrate genomes provided a selective advantage toward massive DNA methylation and silencing of the genome (Yoder et al. 1997). Alternatively, with the increase of organismal complexity, global silencing of gene expression by DNA methylation offered a selective advantage for a more efficient regulation of tissue-specific genes (Bird 1995; Bird et al. 1995). To test whether promoter DNA methylation of C. intestinalis can be attributed to either of these evolutionary processes, we examined genomic, epigenomic, and expression contexts of methylated C. intestinalis promoters. If promoter methylation in this species was largely associated with the need to silence TEs, methylated promoters should harbor significantly more TEs than unmethylated promoters. It should be noted that not all TEs in C. intestinalis are methylated (Simmen et al. 1999), as we show that approximately half of TEs in this genome are not methylated (fig. 3). Defying the expectation of the TE-origin hypothesis, methylated TEs are not preferentially found in promoters. Nevertheless, promoters harboring LINEs and SINEs were slightly more methylated than other promoters. These results suggest that preferential methylation of at least some TEs may predate the invertebrate–vertebrate split. The presence of TEs cannot, however, explain the majority of methylated promoters in C. intestinalis.
With respect to the second hypothesis, data on tissue-specific gene expression and DNA methylation are currently lacking in C. intestinalis. Nevertheless, it is notable that in two tissues examined (testis and muscle), nearly identical sets of genes were methylated (Suzuki et al. 2013). More data are needed to test whether promoter DNA methylation regulates tissue- or cell type-specific gene expression in C. intestinalis.
Importantly, promoter methylation in C. intestinalis per se has not been maintained in orthologous vertebrate genes. Previous studies have shown that vertebrate genes harboring methylated promoters are tissue-specific (Antequera 2003; Weber et al. 2007; Elango and Yi 2008). Consequently, it was proposed that highly methylated gene bodies in invertebrates preferentially obtained lowly methylated promoters during vertebrate evolution (Elango and Yi 2008). Indeed, we show that orthologous genes are predominantly those with the combination of highly methylated gene bodies in C. intestinalis and lowly methylated vertebrate promoters (table 3). This observation supports the idea that vertebrate promoter DNA methylation evolved primarily as a regulatory mechanism to suppress expression of tissue-specific genes (Bird 1995; Bird et al. 1995; Elango and Yi 2008), even though the silencing effect of DNA methylation may have existed in the genome of ancestral chordates. The study of evolutionary transitions of patterns and functions of genomic DNA methylation during chordate evolution can potentially illuminate the mechanistic differences between suppressions and facilitations of gene expression through DNA methylation.
Materials and Methods
Genomic Data
We downloaded genomic regions (promoters and gene bodies) of four different chordate lineages (H. sapiens, G. gallus, D. rerio, and C. intestinalis) from Ensembl v. 65 (Cunningham et al. 2014). Single-copy orthologs among these four species were also obtained from the Ensembl database. In addition, we obtained data for Strongylocentrotus purpuratus from Ensembl Metazoa and the Branchiostoma floridae JGI2 genome build (Putnam et al. 2008). Putative orthologs between these two species and the other four chordates were identified using a reciprocal best-hit approach. We used the BLAT software with default options, using protein sequences as queries, using E = 10−10 as cutoff.
Experimentally Generated Genomic Methylation Maps
Data on DNA methylation and gene expression are summarized in table 1. WGBS data for a 1,000-cell zygote D. rerio and muscle tissue of C. intestinalis were obtained from accession numbers GSM497251 and GSM497252 (Zemach et al. 2010). We also obtained WGBS methylation map of the psoas muscle from human from the NIH Roadmap project (GSM1010986, http://nihroadmap.nih.gov/epigenomics/, last accessed December 2015). We chose psoas muscle to provide some consistency with the tissue used for WGBS in C. intestinalis. Raw reads for a WGBS methylome of a G. gallus embryo were downloaded from the Short Read Archive (SRR942840). These reads were adaptor trimmed using Cutadapt and mapped to the galgal2 genome using Bismark (Krueger and Andrews 2011) using default values. Reads that mapped to the same start and end position were defined as duplicate reads, and were removed using the default option in Bismark (Krueger and Andrews 2011).
Following these procedures the fractional methylation levels at each CpG site in the genome were estimated. Mean fractional methylation levels of specific genomic regions were then computed from all CpGs within that region, such as gene bodies and promoters (as in Lister et al. 2009; Sarda et al. 2012). These values are referred to as simply “methylation levels” in the manuscript. For some analyses, we classified promoter and gene body methylation values into binary “high” or “low” methylation values given their generally bimodal nature. Visual analysis of promoter and gene body methylation indicated that values of less than 0.3 to classify “low” and more than 0.5 to classify “high” would capture the majority of data (fig. 1). Indeed, retaining only genes that met these criteria resulted in at least 80% of genes and up to 95% of genes that were kept for further analysis (supplementary table S1, Supplementary Material online).
CpG O/E Calculation
We calculated the observed versus expected proportion of CpG dinucleotides as CpG O/E = (N*tCpG)/(tC*tG), where N is the length of the genomic region, tCpG is the number of CpG sites in the regions, and tC and tG are the numbers of cytosines and guanines in the region, respectively (Yi and Goodisman 2009).
Annotation
Promoter regions were defined as 500 bp upstream of the TSSs in all nonhuman species. The relatively poor correlation between CpG O/E and methylation in humans led us to consider expanding the putative “promoter” region. It has been shown that nucleotide composition upstream of TSS is different from those of the genomic background and that the length of this nucleotide composition deviation is longer in the human genome compared with other species (Aerts et al. 2004). Considering this observation it may be worthwhile to analyze longer regions upstream of TSS. Indeed some studies consider 2,000 bp upstream as promoters (Chen et al. 2011), we used 1,500 bases upstream to 500 bases downstream of TSS as promoters in the human genome. The correlation between CpG O/E and methylation is much higher with this extended promoter region, so we use it in humans for all subsequent analyses. RepeatMasker TE annotations for each genome were downloaded from the UCSC genome browser (Smit et al. 2013–2015).
Expression Data
We obtained RNA-seq expression data for C. intestinalis muscle tissue from Zemach et al. (2010) (GSM497252) and aligned it to the transcript models of the JGI2 build from Ensembl v65 using Tophat2 (Kim et al. 2013) with default options. RNA-seq data for H. sapiens muscle and D. rerio 1,000-cell embryo were obtained from the Human Bodymap 2.0 project (http://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-513/, last accessed December 2015) and SRR748490 (Jiang et al. 2013), respectively.
Analyses
All statistical analyses were carried out using the R programming language (R Core Team 2014). To compute partial correlations, we used the ppcor R package (Kim and Yi 2007). Functional annotation analyses are performed using the DAVID functional annotation tools (Huang et al. 2009), using ENSEMBL gene IDs as inputs. R code used for figure 2 is available upon request.
Supplementary Material
Supplementary figures S1–S3 and tables S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Elizabeth Smithgall Watts endowment and the Georgia Tech School of Biology. The authors thank the comments from the Yi lab members on the previous versions of the manuscript.
References
- Aerts S, Thijs G, Dabrowski M, Moreau Y, De Moor B. 2004. Comprehensive analysis of the base composition around the transcription start site in Metazoa. BMC Genomics 5:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antequera F. 2003. Structure, function and evolution of CpG island promoters. Cell Mol Life Sci. 60:1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. 1995. Gene number, noise reduction and biological complexity. Trends Genet. 11:94–100. [DOI] [PubMed] [Google Scholar]
- Bird A, Tate P, Nan X, Campoy J, Meehan J, Cross S, Tweedie S, Charlton J, Macleod D. 1995. Studies of DNA methylation in animals. J Cell Sci Suppl. 19:37–39. [DOI] [PubMed] [Google Scholar]
- Chen CH, Lin HY, Pan CL, Chen FC. 2011. The genomic features that affect the lengths of 5′ untranslated regions in multicellular eukaryotes. BMC Bioinformatics 12(Suppl 9):S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. 2014. Ensembl 2015. Nucleic Acids Res. 43:D662–D669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Gama-Sosa MA, Huang L-H, Midgett RM, Kuo KC, McCune RA, Gehrke C. 1982. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues or cells. Nucleic Acids Res. 10:2709–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango N, Hunt BG, Goodisman MAD, Yi SV. 2009. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Natl Acad Sci U S A. 106:11206–11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango N, Yi SV. 2008. DNA methylation and structural and functional bimodality of vertebrate promoters. Mol Biol Evol. 25:1602–1608. [DOI] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen P-Y, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. 2010. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 107:8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores K, Wolschin F, Corneveaux J, Allen A, Huentelman M, Amdam G. 2012. Genome-wide association between DNA methylation and alternative splicing in an invertebrate. BMC Genomics 13:480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foret S, Kucharski R, Pittelkow Y, Lockett G, Maleszka R. 2009. Epigenetic regulation of the honey bee transcriptome: unravelling the nature of methylated genes. BMC Genomics 10:472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DA, Yang X, Niño EL, Yi S, Grozinger C. 2015. Parallel epigenomic and transcriptomic responses to viral infection in honey bees (Apis mellifera). PLoS Pathog. 11:e1004713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavery MR, Roberts SB. 2010. DNA methylation patterns provide insight into epigenetic regulation in the Pacific oyster (Crassostrea gigas). BMC Genomics 11:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavery MR, Roberts SB. 2013. Predominant intragenic methylation is associated with gene expression characteristics in a bivalve mollusc. PeerJ 1:e215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Huh I, Zeng J, Park T, Yi S. 2013. DNA methylation and transcriptional noise. Epigenetics Chromatin 6:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt BG, Glastad K, Yi SV, Goodisman MAD. 2013. Patterning and regulatory associations of DNA methylation are mirrored by histone modifications in insects. Genome Biol Evol. 5:591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Zhang J, Wang JJ, Wang L, Zhang L, Li G, Yang X, Ma X, Sun X, Cai J, et al. 2013. Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell 153:773–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jjingo D, Conley AB, Yi SV, Lunyak VV, Jordan IK. 2012. On the presence and role of human gene-body DNA methylation. Oncotarget 3(4):462–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA. 2012. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 13:484–492. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Yi SV. 2007. Understanding relationship between sequence and functional evolution in yeast proteins. Genetica 131:151–156. [DOI] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2011. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo Q-M, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, David S, Andreas H, Neil PB, Claudia K, Megan LW, Frank GT, Duncan TO, Roger P, Chris PP, et al. 2013. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. eLife 2:e00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou S, Lee H-M, Qin H, Li J-W, Gao Z, Liu X, Chan L, Kl Lam V, So W-Y, Wang Y, et al. 2014. Whole-genome bisulfite sequencing of multiple individuals reveals complementary roles of promoter and gene body methylation in transcriptional regulation. Genome Biol. 15:408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyko F, Foret S, Kucharski R, Wolf S, Falckenhayn C, Maleszka R. 2010. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 8:e1000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendizabal I, Keller TE, Zeng J, Yi SV. 2014. Epigenetics and evolution. Int Comp Biol. 54:31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendizabal I, Yi SV. 2016. Whole-genome bisulfite sequencing maps from multiple human tissues reveal novel CpG island associated with tissue-specific regulation. Hum Mol Genet. 25:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A, Chang H-Y, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, et al. 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43:D213–D221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Matsumoto K, Nakai K. 2010. Gradual transition from mosaic to global DNA methylation patterns during deuterostome evolution. BMC Bioinformatics 11:S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson CE, Roberts SB. 2014. Genome-wide profiling of DNA methylation and gene expression in Crassostrea gigas male gametes. Front Physiol. 5:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Peng Z, Zeng J, Elango N, Park T, Wheeler D, Werren JH, Yi SV. 2011. Comparative analyses of DNA methylation and sequence evolution using Nasonia genomes. Mol Biol Evol. 28:3345–3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Xu K, Park T, Yi SV. 2012. What are the determinants of gene expression levels and breadths in the human genome? Hum Mol Genet. 21:46–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patalano S, Hore TA, Reik W, Sumner S. 2012. Shifting behaviour: epigenetic reprogramming in eusocial insects. Curr Opin Cell Biol. 24:367–373. [DOI] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453:1064–1071. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2014. R: a language and environment for statistical computing. Vienna (Austria: ): R Foundation for Statistical Computing. [Google Scholar]
- Riviere G, Wu G-C, Fellous A, Goux D, Sourdaine P, Favrel P. 2013. DNA methylation is crucial for the early development in the oyster C. gigas. Mar Biotechnol. 15:739–753. [DOI] [PubMed] [Google Scholar]
- Saint-Carlier E, Riviere G. 2015. Regulation of Hox orthologues in the oyster Crassostrea gigas evidences a functional role for promoter DNA methylation in an invertebrate. FEBS Lett. 589:1459–1466. [DOI] [PubMed] [Google Scholar]
- Sarda S, Zeng J, Hunt BG, Yi SV. 2012. The evolution of invertebrate gene body methylation. Mol Biol Evol. 29:1907–1916. [DOI] [PubMed] [Google Scholar]
- Saxonov S, Berg P, Brutlag DL. 2006. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 103:1412–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder DI, Blair JD, Lott P, Yu HOK, Hong D, Crary F, Ashwood P, Walker C, Korf I, Robinson WP, et al. 2013. The human placenta methylome. Proc Natl Acad Sci U S A. 110:6037–6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz MD, He Y, Whitaker JW, Hariharan M, Mukamel EA, Leung D, Rajagopal N, Nery JR, Urich MA, Chen H, et al. 2015. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 523:212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela N, Kim E, Ast G. 2010. The role of transposable elements in the evolution of non-mammalian vertebrates and invertebrates. Genome Biol. 11:R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen MW, Leitgeb S, Charlton J, Jones SJM, Harris BR, Clark VH, Bird A. 1999. Nonmethylated transposable elements and methylated genes in a chordate genome. Science 283:1164–1167. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2013–2015. RepeatMasker Open-4.0 Available from: http://www.repeatmasker.org.
- Suzuki M, Yoshinari A, Obara M, Takuno S, Shigenobu S, Sasakura Y, Kerr A, Webb S, Bird A, Nakayama A. 2013. Identical sets of methylated and nonmethylated genes in Ciona intestinalis sperm and muscle cells. Epigenetics Chromatin 6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. 2008. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 9:465–476. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Kerr AR, De Sousa D, Bird A. 2007. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res. 17:625–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tweedie S, Charlton J, Clark V, Bird A. 1997. Methylation of genomes and genes at the invertebrate-vertebrate boundary. Mol Cell Biol. 17:1469–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wheeler D, Avery A, Rago A, Choi J-H, Colbourne JK, Clark AG, Werren JH. 2013. Function and evolution of DNA methylation in Nasonia vitripennis. PLoS Genet. 9:e1003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D. 2007. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 39:457–466. [DOI] [PubMed] [Google Scholar]
- Yi SV, Goodisman MA. 2009. Computational approaches for understanding the evolution of DNA methylation in animals. Epigenetics 4:551–556. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335–340. [DOI] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. 2010. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328:916–919. [DOI] [PubMed] [Google Scholar]
- Zeng J, Nagrajan HK, Yi SV. 2014. Fundamental diversity of human CpG islands at multiple biological levels. Epigenetics 9:483–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Yi S. 2010. DNA methylation and genome evolution in honeybee: gene length, expression, functional enrichment co-vary with the evolutionary signature of DNA methylation. Genome Biol Evol. 2:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.