Abstract
The release of extracellular vesicles (EVs), including exosomes and microvesicles, is a phenomenon shared by many cell types as a means of communicating with other cells and also potentially removing cell contents. The cargo of EVs includes the proteins, lipids, nucleic acids, and membrane receptors of the cells from which they originate. EVs released into the extracellular space can enter body fluids and potentially reach distant tissues. Once taken up by neighboring and/or distal cells, EVs can transfer functional cargo that may alter the status of recipient cells, thereby contributing to both physiological and pathological processes. In this article, we will focus on EV composition, mechanisms of uptake, and their biological effects on recipient cells. We will also discuss established and recently developed methods used to study EVs, including isolation, quantification, labeling and imaging protocols, as well as RNA analysis.
Keywords: extracellular vesicles, exosomes, microvesicles, intercellular communication, methods
Definitions and subtypes
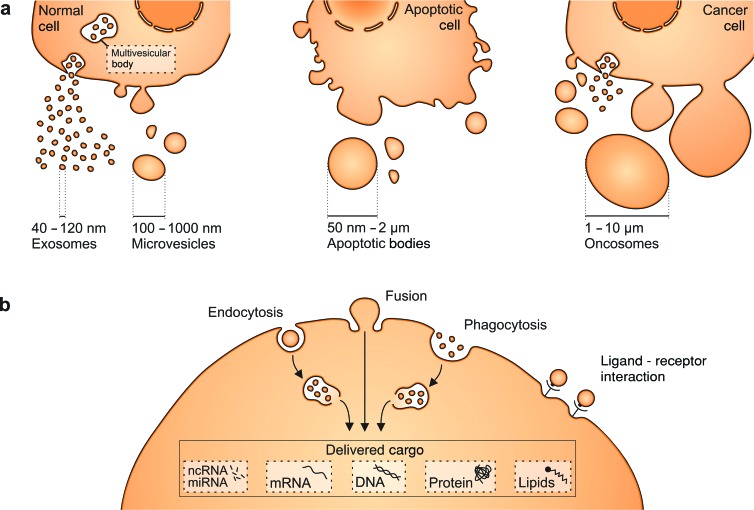
Extracellular vesicles (EVs) are nanosized, membrane-bound vesicles released from cells that can transport cargo—including DNA, RNA, and proteins—between cells as a form of intercellular communication. Different EV types, including microvesicles (MVs), exosomes, oncosomes, and apoptotic bodies, have been characterized on the basis of their biogenesis or release pathways (figure 1a). Microvesicles bud directly from the plasma membrane, are 100 nanometers (nm) to 1 micrometer (μm) in size, and contain cytoplasmic cargo (Heijnen et al. 1999). Another EV subtype, exosomes, is formed by the fusion between multivesicular bodies and the plasma membrane, in which multivesicular bodies release smaller vesicles (exosomes) whose diameters range from 40 to 120 nm (El Andaloussi et al. 2013, Cocucci and Meldolesi 2015). Dying cells, however, release vesicular apoptotic bodies (50 nm–2 μm) that can be more abundant than exosomes or MVs under specific conditions and can vary in content between biofluids (Thery et al. 2001, El Andaloussi et al. 2013). Membrane protrusions can also give rise to large EVs, termed oncosomes (1–10 μm), which are produced primarily by malignant cells in contrast to their nontransformed counterparts (Di Vizio et al. 2012, Morello et al. 2013). Because EV isolation methods to date only enable enrichment but not distinct separation of these EV subpopulations, the current article collectively refers to all vesicles released by cells as EVs unless otherwise stated by the cited studies.
Figure 1.
Cells produce different types of extracellular vesicles (EVs) that vary in size. (a) Exosomes and microvesicles (MVs) are produced by normal and diseased cells. Apoptosis triggers the release of apoptotic bodies. In addition, some cancer cells were reported to generate large EVs, termed oncosomes. (b) EVs can be taken up via different mechanisms, including endocytosis, membrane fusion, or phagocytosis. They deliver nucleic acids, proteins, and lipids that can be functional in recipient cells. Ligand–receptor interactions on the cell surface can also result in biological effects and help to target vesicles to specific cell types. Abbreviations: nm, nanometers; μm, micrometers.
EV structure and composition
The content of EVs includes lipids, nucleic acids, and proteins from donor cells.
Lipid content and membrane features
EV membranes consist of a lipid bilayer similar to that of cell plasma membrane, in contrast to the single-layered high- and low-density lipoprotein (HDL and LDL) found in body fluids (Laulagnier et al. 2004). Exosomes are enriched in sphingomyelin, gangliosides, and disaturated lipids, and their phosphatidylcholine and diacylglycerol proportion are decreased relative to the membranes of their cells of origin (Laulagnier et al. 2004). Some studies also describe an increased fraction of cholesterol in exosomes compared with that in cellular membranes (Llorente et al. 2013). In contrast to cellular membranes, exosomes contain more phosphatidylserine in the outer leaflet, which may facilitate their internalization by recipient cells (Fitzner et al. 2011). A comparison of banked red blood cells and MVs derived from them revealed a high similarity in lipid composition, with the exception of polyunsaturated glycerophosphoserine (38:4), which was enriched in MVs (Bicalho et al. 2013). These differences are consistent with the distinctive biogenesis of exosomes and MVs, because the latter stem directly from the plasma membrane.
The increased content of sphingomyelin and disaturated lipids implies a higher rigidity of the exosome lipid bilayer compared with that of cell membranes. Indeed, studying the anisotropy of a hydrophobic probe demonstrated that exosomes exhibit greater rigidity than cell plasma membranes, which was confirmed using Laurdan fluorescence spectroscopy (Laulagnier et al. 2004, Parolini et al. 2009). Interestingly, exosome membrane rigidity has been suggested to be pH dependent. For example, EVs derived from basophils (RBL-2H3) and treated with acidic solution became less rigid, more nearly matching the rigidity of the cell plasma membrane, which remained unchanged under the acidic pH (Laulagnier et al. 2004). The pH dependence may be linked to the origin of exosomes, because the pH in multivesicular bodies is lower than in the cytoplasm (Laulagnier et al. 2004). This is also consistent with the observation that a lower pH in the tumor microenvironment increases the cellular uptake of EVs (Parolini et al. 2009). The greater acidity renders the fluidity of EV membranes more similar to that of the cell plasma membrane, thereby promoting fusion (Laulagnier et al. 2004, Record et al. 2014). This concept, however, requires further study, because melanoma cells cultured under an acidic condition release EVs with more rigid membranes (Parolini et al. 2009). The discrepancy may be attributed to different cell types and methods used—the former study analyzed EVs isolated under normal conditions followed by acidic pH treatment, whereas the latter investigated EVs isolated from cells grown in an acidic environment. Transmembrane flip-flop lipid movements, which facilitate the exchange between the outer and inner leaflet of the membrane, are higher in EV membranes than in the cell plasma membrane (Laulagnier et al. 2004). The difference in biophysical properties between EVs and cell membranes may arise from their variation in lipid as well as protein composition. Finally, the greater rigidity of EVs as a result of their high sphingomyelin, disaturated-lipid, and cholesterol content may contribute to their resistance to degradation and therefore their stability as carriers of various biomolecules (Huang et al. 2013, Ridder et al. 2014).
EVs contain enzymes involved in lipid metabolism, including phospholipases D and A2, whose activities are triggered by the addition of GTP (Subra et al. 2010). Adipocytes under hypoxic conditions secrete EVs with increased levels of enzymes needed for de novo lipogenesis, including fatty acid synthase (Sano et al. 2014). Normal adipocytes treated with these EVs tended to accumulate more lipids, possibly via the EV-transferred fatty acid synthase. EVs from RBL-2H3 basophil cells also transport bioactive lipids, such as arachidonic acid (AA) and prostaglandin E2 (PGE2; Subra et al. 2010). When compared with the parental cancer cell line of origin, EVs showed an enrichment in ceramide and phosphatidic acid that have the capacity to influence the recipient cell status (Llorente et al. 2013). In addition, the high proportion of cholesterol and sphingomyelin in EVs may contribute to the apoptosis of recipient cells, because artificial nanoparticles mimicking this lipid composition of EVs reduced pancreatic tumor cell survival (Beloribi et al. 2012). Taken together, lipids and lipid metabolic enzymes transferred by EVs can participate in changing the status of recipient cells.
RNA
RNA transported by EVs (EV-RNA) is predominantly shorter in size (less than 200 nucleotides [nt]) than the average cellular fraction, although the observation is based on bioanalyzer data which can be affected by sample purity (table 1; Huang et al. 2013, Eirin et al. 2014). The sequencing of total RNA from serum-derived EVs suggested that microRNAs (miRNA) and transfer RNA (tRNAs) constitute about 15% of EV-RNA (Bellingham et al. 2012). The presence of miRNAs as well as long noncoding RNAs was also identified in the fraction of large oncosomes (Di Vizio et al. 2012, Morello et al. 2013). In the pool of long transcripts (more than 200 nt), both coding and noncoding RNAs were detected (Bellingham et al. 2012, Huang et al. 2013). The amount of RNA in EVs varies depending on the cell type of origin. Some cancer-derived EVs contain more total RNA than those derived from normal cells (Balaj et al. 2011). Interestingly, some profiles of EV-RNA do not mirror those of cellular RNA (Skog et al. 2008). Although they contain many transcripts in common, some RNAs were systematically enriched in the released EVs (Valadi et al. 2007, Skog et al. 2008, Eirin et al. 2014). The sequencing of RNA derived from mesenchymal stem cell EVs, for example, demonstrated an increased proportion of mRNA encoding transcription factors and proteins involved in alternative splicing and Golgi apparatus components, with relative depletion of transcripts encoding proteins for mitochondria, the cytoskeleton, or calcium signaling (Eirin et al. 2014). A study applying whole human-genome microarrays revealed high levels of retrotransposon sequences—such as human endogenous retroviruses (HERVs), Alu, or L1—in glioblastoma-derived EVs when compared with those in the donor cells (Balaj et al. 2011). The fraction of RNA repeat regions—long interspersed elements and short interspersed elements—has been estimated by RNA sequencing to constitute up to 50% of EV-RNA (Bellingham et al. 2012). Although various RNA types have been identified in EVs, it remains to be determined to what extent EV-RNAs are full length and/or mainly fragments of the transcripts and to what extent they are functional in recipient cells.
Table 1.
RNA species detected in EV-RNA in different biological samples.
| Study | Biological material | Method | RNA species |
|---|---|---|---|
| Valadi et al. 2007 | Mast-cell line (MC/9), primary bone marrow–derived mast cells (BMMC), human mast-cell line (HMC-1) | Affymetrix microarray miRCURY™ LNA Array | mRNA (including polyadenylated fraction) miRNA |
| Skog et al. 2008 | Primary glioblastoma multiforme cells Matched glioblastoma tumor and serum | Agilent whole human genome microarray, 4 ×44K Nested PCR | mRNA (including EGFRvIII mutant) miRNA |
| Balaj et al. 2011 | Primary glioblastoma cell lines 20/3 and 11/5 | Agilent whole human genome microarray, 4 ×44K | mRNA (including c-Myc amplification) human endogenous retroviruses (HERVs) |
| Primary medulloblastoma cell lines D458, D384 and D425 | |||
| Rhabdoid tumor cell line NS224 | |||
| Melanoma cell line, Yumel 0106 | |||
| Epidermoid carcinoma cell line, A431 | |||
| Human fibroblast lines, HF19 and HF27, Serum from xenograft mouse model of medulloblastoma | |||
| Bellingham et al. 2012 | Mouse hypothalamic neuronal (GT1–7) cell lines GT1–7 cells infected with human Fukuoka-1 prion strain | Small RNA deep sequencing | mRNA, RNA repeats, rRNA, small RNA (Transfer RNA (tRNA), small interfering (siRNA), small nucleolar RNA (snoRNA), small cytoplasmic RNA (scRNA), small nuclear RNA (snRNA), miRNA |
| Huang et al. 2013 | Human plasma | Small RNA deep sequencing | miRNA, lncRNA, tRNA, snoRNA, snRNA, piwi-interacting RNA (piRNA), mRNA |
| Eirin et al. 2014 | Porcine adipose tissue–derived mesenchymal stromal/stem cells | RNA deep sequencing | mRNA, rRNA, miRNA |
Because several RNA species are enriched in EVs, it raises the question as to whether certain mechanisms are responsible for their selective packaging. A defined sequence motif (CUGCC) in the three prime (3′) untranslated region in the presence of a binding site for miR-1289 has been shown to promote mRNA uploading into EVs (Bolukbasi et al. 2012). Another proposed mechanism involves sequence motifs in miRNAs recognized by the ribonucleoprotein hnRNPA2B1, which may promote miRNA packaging into EVs (Villarroya-Beltri et al. 2013). This process seems to be controlled by the posttranslational modification of hnRNPA2B1 in the form of SUMOylation that regulates protein stability and cellular trafficking (Villarroya-Beltri et al. 2013). The sorting of miRNAs to exosomes may also be driven by 3′ end posttranscriptional modifications, because 3′ adenylated miRNAs are prevalent in cells, whereas 3′ uridylated miRNAs are characteristic of exosomal miRNAs (Koppers-Lalic et al. 2014). Mechanisms that increase the likelihood of enrichment in EVs support the view that the phenomenon of RNA export into EVs is biologically relevant.
Several studies have shown that RNA can be transferred to recipient cells by EVs (Valadi et al. 2007, Skog et al. 2008, Mittelbrunn et al. 2011). Therefore, the next interesting question is to what extent is the transferred RNA functional. The fraction of polyadenylated mRNAs suggests its translation potential after uptake, which has been confirmed in recipient cells by translation assays (Valadi et al. 2007, Skog et al. 2008, Lai et al. 2015). miRNAs transported between cells by EVs may regulate the translation of target mRNAs in recipient cells (for more examples, see Mittelbrunn et al. 2011, Redzic et al. 2014). Examples include EVs released by T cells that can transfer specific miRNAs (e.g., miR-335) to recipient antigen-presenting cells (Mittelbrunn et al. 2011). The uptake of EVs containing Epstein Barr virus (EBV)—specific miRNAs by immune cells is discussed in detail in box 1. Recently, functionally relevant RNA transfer has also been shown in vivo. The authors studied transgenic mice expressing Cre recombinase under a promoter specific for hematopoietic cells and detected the enzyme activity in Purkinje cells in the cerebellum without evidence of cell fusion (Ridder et al. 2014). EVs isolated from peripheral blood did not contain detectable Cre protein, whereas both smaller and larger EVs carried Cre mRNA. The intracranial injection of EVs resulted in flox recombination detectable in Purkinje cells and in other nervous tissue cells. Furthermore, systemic inflammation induced by either a lung tumor or peritonitis increased the number of these recombination events in Purkinje cells (Ridder et al. 2014). Taken together, these observations support the concept that EV-RNA can be transferred between cells via EVs not only in a paracrine fashion but also distally, where it maintains its function.
Box 1. Extracellular vesicles in viral infection.
Extracelluar vesicles (EVs) also play a role in viral infection. EVs from Epstein Barr Virus (EBV)–transformed lymphoblastoid B cells (LCL) are taken up by dendritic cells and transfer EBV-specific BamHI A rightward transcript (BART) miRNAs, resulting in the repression of the immunostimulatory gene IFN-inducible T-cell attracting chemokine (CXCL11; Pegtel et al. 2010). The effect was specific to the sequence of BART miRNA because a firefly luciferase reporter construct with the 3′ untranslated region of CXCL11 (which contains the BART miRNA binding site) demonstrated a reduction in reporter activity. This suppressive effect was less pronounced when the reporter was tagged with mutated BART miRNA binding sites. The release of viral miRNA from EVs capable of repressing genes responsible of immune activation may therefore enhance infection efficiency.
The proteins transferred by EVs may also precondition recipient cells to be more susceptible to viral infection. HIV 1–infected cells produce EVs harboring Nef viral protein (Arenaccio et al. 2014). Nef protein was shown to make EV-recipient cells more permissive to subsequent HIV-1 replication (Arenaccio et al. 2014). Moreover, EVs provide a direct route for hepatitis C virus (HCV) transmission. EVs released from HCV-infected Huh7.5.1 hepatoma cells contain viral core protein and entire RNA viral genome (Ramakrishnaiah et al. 2013). Naive Huh7.5.1 cells incubated with EVs derived from infected cells were capable of inducing infection to the same extent as pure viral particles (Ramakrishnaiah et al. 2013). EV-pretreated cells were capable of inducing secondary infection in naive cells, supporting the notion that EVs mediate viral particle spread (Ramakrishnaiah et al. 2013). Furthermore, HCV-specific immunoglobulins derived from 3 out of 10 patients had a stronger inhibitory effect on free virus– than on EV-transmitted infection (Ramakrishnaiah et al. 2013). It suggests that EVs may provide viral particles with protection against antiviral immune response and therefore facilitate viral propagation. In addition, herpes simplex virus 1 (HSV-1)–infected cells release EVs containing stimulator of IFN genes (STING), which hinders the infection of intracellular pathogens, prior to the release of active virus particles (Kalamvoki and Roizman 2014)
DNA
EVs can transport DNA that is referred to as EV-DNA (Guescini et al. 2010, Balaj et al. 2011, Kahlert et al. 2014, Thakur et al. 2014). DNA ranging in size from 100 base pairs (bp) to 2.5 kilobase pairs (kB) can be enclosed within EVs (Thakur et al. 2014). A comparison of DNA extracted from intact EVs and EVs pretreated with DNase demonstrated a decrease in double-stranded DNA (dsDNA) longer than 2.5 kB in the fraction subject to enzymatic cleavage (Thakur et al. 2014). This finding suggests that these long dsDNA fragments are present in the EV pellet but not enclosed within the EV membrane that would protect them from DNase activity (Thakur et al. 2014). The total pool of EV-DNA studied by sequencing in exosomes derived from murine B16-F10 melanoma cells and from serum in pancreatic cancer patients spans sequences across all chromosomes of genomic DNA (gDNA; Kahlert et al. 2014, Thakur et al. 2014). Probing of gDNA in cells and EV-DNA in the extracellular medium conditioned by B16-F10 melanoma cells with anti-5′ methylcytosine antibody demonstrated similar staining signatures, suggesting comparable overall methylation status (Thakur et al. 2014), although it remains unclear whether the same regions are equally methylated in gDNA and EV-DNA. In some instances, EV-DNA has been shown to reflect the parental cell gDNA, because cancer cell mutations in BRAF, epithelial growth factor receptor (EGFR), KRAS and p53 were successfully detected in EV-DNA derived from melanoma and pancreatic cancer cells (Kahlert et al. 2014, Thakur et al. 2014). EV-DNA also corresponds to the gDNA quantitatively: c-Myc amplification was identified in EV-DNA and gDNA from medulloblastoma cells (Balaj et al. 2011). These findings were confirmed in serum EVs in mouse tumor models and human samples (Balaj et al. 2011, Kahlert et al. 2014, Thakur et al. 2014). Although there is compelling evidence for the presence of EV-DNA in EVs, the functional significance of this cargo remains unknown.
Protein
The protein composition of EVs is, in some instances, related to the cell type and mode of biogenesis. Exosomes that originate from the endolysosomal compartment tend to be more enriched in major histocompatibility complex class II (MHC class II) and tetraspanins CD37, CD53, CD63, CD81, and CD82 (Heijnen et al. 1999, Tauro et al. 2012). The endosomal sorting complex required for transport (ESCRT) is a group of proteins indispensable for internal membrane budding in the formation of multivesicular bodies (Morita et al. 2007). ESCRT pathway function also requires accessory proteins, including Alix and tumor susceptibility gene protein 101 (TSG101) (Morita et al. 2007). Multivesicular bodies can direct proteins to lysosomes for degradation. Consequently, exosomes contain ESCRT proteins, Alix, TSG101, and chaperones, such as Hcs70 and Hsp90, irrespective of cell type (Thery et al. 2001). Some studies indicate that compared with cells, exosomes are enriched in glycoproteins and transmembrane proteins (Escrevente et al. 2011, Sinha et al. 2014). Owing to their plasma membrane origin, MVs tend to be enriched in a different repertoire of proteins as compared with those of exosomes, including integrins, glycoprotein Ib (GPIb), and P-selectin (Heijnen et al. 1999). One pathway of plasma membrane budding involves arrestin containing protein 1 (AARDC1) and, similarly to exosomes, TSG101 (Nabhan et al. 2012). MVs carry more proteins with posttranslational modifications, such as glycoproteins or phosphoproteins, when compared with exosomes (Palmisano et al. 2012). Another class of EVs, apoptotic bodies, contains DNA-binding histones and is depleted in glycoproteins, which is in direct contrast to exosomes (Thery et al. 2001, Escrevente et al. 2011). The GTP-binding protein ARF6, which regulates cytoskeleton remodeling, promotes the shedding of oncosomes (Di Vizio et al. 2012) and was found to be incorporated into both MVs and large oncosomes (Di Vizio et al. 2012, Morello et al. 2013).
Many of the above-mentioned proteins—especially MHC II, tetraspanins, ESCRT proteins, Alix, TSG101, and heat-shock chaperones—are commonly found in EVs, irrespective of cell of origin, and can consequently be used as general EV markers (Thery et al. 2001, Tauro et al. 2012). In contrast, proteins contained within mitochondria (e.g., aconitase), the Golgi apparatus (e.g., GM130), the endoplasmic reticulum (e.g., calreticulin), and some cytoplasmic proteins (e.g., α-tubulin) have been reported to be depleted in EVs isolated by differential centrifugation (Christianson et al. 2013, Sinha et al. 2014). Therefore, their absence may serve as an additional confirmation of the purity of EV preparations as long as there was no cell stress and/or death. The protein composition of different EV subtypes shows a substantial overlap, although some proteins are more enriched in one than in other EV subtypes (Palmisano et al. 2012). It is unclear whether the overlap is, at least partially, contributed by applied isolation techniques, which currently do not allow the complete separation of EV subtypes and protein aggregates. However, the separation of EV fractions by sucrose gradient centrifugation supports, for instance, reduced content—but not the complete absence—of glycoprotein 1b alpha in exosomes when compared with MVs (Heijnen et al. 1999). Specific markers to identify individual EV subtypes remain to be determined.
Some EV proteins can be grouped in functional classes. Mass spectrometric analysis of EVs derived from ovarian cancer cell lines indicated enrichment in proteins that undergo phosphorylation and acetylation (Sinha et al. 2014). Among the most enriched were phosphatidylinositol-3-kinase, mitogen-activated protein kinase (MAPK), and ErbB family members (Liang et al. 2013, Sinha et al. 2014). This could partially explain the EVs potent biological effects on recipient cells, with kinases often serving as key signaling molecules. In accordance with abundant RNA and DNA transferred by EVs, nucleic acid binding is the most common function among proteins extracted from EVs (Sinha et al. 2014). Catalytic activity was also a well represented function (Liang et al. 2013). The surface protein repertoire in EVs may reflect the biologic status of parental cells. Indeed, the expression of EpCam, CD24, Ca-125, CA19–9, EGFR, and cloudin 3 was highly consistent between ovarian cancer cells and their released EVs (Runz et al. 2007, Im et al. 2014). The antigen profile was specific to ovarian cancer cells and their derived EVs. This characteristic enables the distinction between EVs derived from cancer and those derived from nonmalignant ovarian epithelium cells (TIOSE6; Im et al. 2014).
The proteins released in association with EVs—including receptors, transcription factors, and enzymes—can be functional and drive phenotypic changes in recipient cells. This form of transfer may contribute to the spread of the aggressive phenotype of malignant subpopulations of cancer cells within heterogeneous tumors. Glioblastoma cells, for instance, were shown to release the oncogenic mutant form of epidermal growth factor (EGFRvIII) in EVs (Al-Nedawi et al. 2008). The receptor successfully transferred to other glioma cells activates the MAPK/Akt cascade and increases their proliferation rate. In nasopharyngeal cancer, EBV latent membrane protein 1 (LMP1) up-regulated hypoxia-inducible factor 1-alpha (HIF-1α) in exosomes (Aga et al. 2014). Interestingly, HIF-1α was detected in multivesicular bodies of donor cells, which participate in exosome biogenesis. The overexpression of wild-type HIF-1α or a mutated HIF-1α (with suppressive transcriptional activity) resulted in equal packaging into exosomes. The subsequent addition of exosomes to recipient cells revealed increased transcriptional activity in response to wild-type HIF-1α but not to mutant-containing exosomes (Aga et al. 2014). In support of this finding, exosomal HIF-1α was shown capable of binding the DNA sequence of the hypoxia transcriptional response element (Aga et al. 2014). Moreover, treatment with wild-type HIF-1α decreased levels of E-cadherin and triggered up-regulation of N-cadherin in nasopharyngeal cancer cells, with the opposite effect caused by the mutant form of HIF-1α protein (Aga et al. 2014). Proteins released by EVs can also alter the extracellular space. EVs, including large oncosomes, derived from tumor cells contain matrix metalloproteinases (MMPs) that can digest the extracellular matrix (Di Vizio et al. 2012, Shimoda and Khokha 2013), which enhances the invasiveness of cancer cells. Metalloproteinase ADAM10 transferred via EVs from cancer-associated fibroblasts also promotes the motility of breast cancer cells (Shimoda et al. 2014). In summary, EV protein cargo can convey signaling messages to recipient cells in the immediate and distal environments.
EVs in cell communication
The number of EVs released and taken up likely depends on donor and recipient cell types, their physiological state, and the conditions in the microenvironment (Parolini et al. 2009, Mittelbrunn et al. 2011, EL Andaloussi et al. 2013). Exosome formation is promoted by Staphylococcus enterotoxin superantigen-E in T cells (Mittelbrunn et al. 2011). The production of EVs can also be increased in cancer cells under hypoxic conditions, which is mediated by HIF-1α (King Michael and Gleadle 2012). Similarly, it was observed that an acidic microenvironment augments EV release in melanoma cells (Parolini et al. 2009). The induction of cell stress and the use of calcium ionophores can also increase the release of vesicles (Valadi et al. 2007, El Andaloussi et al. 2013).
In parallel, variability is also found between cell types in vesicle internalization: ovarian cancer–derived EVs, for instance, are taken up avidly by natural-killer cells but very little by T cells (Keller et al. 2009). These differences raise important questions about the mechanisms of the interactions between EVs and a recipient cell and its specificity, as well as factors perturbing it (figure 1b). EV uptake is suppressed in low temperature (4 degrees Celsius [°C]), suggesting energy dependence (Morelli et al. 2004, Escrevente et al. 2011, Christianson et al. 2013). EVs can be either internalized and/or adhere to the surface of a recipient cell (Feng et al. 2010). With the use of fluorescently labeled EVs, the internalization can be observed within approximately 30 minutes and becomes most prominent after 4–6 hours using fluorescence-activated cell sorting (Feng et al. 2010, Escrevente et al. 2011, Christianson et al. 2013). Punctate fluorescence may be seen in recipient cells for as long as 24 hours after EV exposure. Studying this phenomenon with the use of a lipid fluorescent probe, R18, suggests that the fusion between EV and cell membranes can be a mechanism of uptake (Parolini et al. 2009). A growing body of evidence, however, supports endocytosis as a primary means of the internalization of EVs (Svensson et al. 2013). Indeed, cytochalasin D, which interferes with actin polymerization and endocytosis, significantly reduces the uptake of EVs (Morelli et al. 2004, Escrevente et al. 2011, Svensson et al. 2013). Similarly, the inhibition or knockout of dynamin, a GTPase that is responsible for the formation of new endosomal vesicles, significantly suppressed EV uptake (Nanbo et al. 2013). Similar effects are exerted by nocodazol, which interferes with microtubules and therefore perturbs endosomal trafficking (Svensson et al. 2013). Tracking of EV fate after the uptake revealed that they are taken up into endosomes first and then move along microtubules (Svensson et al. 2013). It is still under debate as to which type(s) of endocytosis drives this process. In one study, the inhibition of clathrin-dependent endocytosis by chlorpromazine reduced EV internalization (Escrevente et al. 2011), whereas the knockdown of clathrin had no effect in another study (Svensson et al. 2013). The discrepancy may be attributed to the low specificity of chlorpromazine toward the clathrin-mediated pathway. However, EVs secreted by EBV-infected B cells were primarily taken up by nasopharyngeal carcinoma CNE-1 cell via caveola-dependent endocytosis (Nanbo et al. 2013). By contrast, the knockdown of caveolin increased the internalization of glioblastoma-derived EVs into mouse embryonic fibroblasts (Svensson et al. 2013). There is also compelling evidence that lipid raft–dependent endocytosis plays a major role in EVs’ uptake. EVs co-localize with lipid-raft markers, flotillin-1, and cholera toxin subunit B, which binds to GM1 gangliosides on the cell surface (Svensson et al. 2013). The depletion of cholesterol that is enriched in these areas of the plasma membrane—either by methyl-beta-cyclodextrin or by simvastatin—reduced EV uptake (Escrevente et al. 2011, Svensson et al. 2013). Disturbing cell membrane composition by binding filipin to cholesterol exerted the same effect (Parolini et al. 2009). This is consistent with the increased cholesterol level in EVs that may make them exhibit a higher affinity toward the lipid rafts of cell membranes (Llorente et al. 2013). Indeed, liposomes, the synthetic counterpart of EVs, demonstrated an enhanced uptake with increasing cholesterol content (Smyth et al. 2014).
Phagocytosis may also play a role in EV uptake, because cells with a phagocytic phenotype (RAW 264.7, U937, J774A.1 macrophages) internalized EVs that were subsequently identified in phagosomes (Feng et al. 2010). To corroborate this, the phagocytosis inhibitor wortmanin suppressed this EV uptake (Feng et al. 2010). The blockade of macropinocytosis by amiloride also reduced EV uptake in ovarian cancer cells SKOV3 (Escrevente et al. 2011). The distinct or even contradictory results obtained by different studies suggest that the mechanism of EV uptake may occur by multiple mechanisms even in the same cell and is also influenced both by the cell of origin, the type of EVs, the nature of the recipient cells, and the specificity of inhibitory molecules, as well as potentially by the methods of EV labeling.
The efficacy of EV exchange between cells probably also depends on their surface antigen repertoires. The digestion of membrane proteins exposed on the EVs with proteinase K can decrease the uptake by 32 ± 8% in recipient cells (Escrevente et al. 2011). The blockage of integrins (CD51, CD61) or tetraspanins (CD9, CD81) with monoclonal antibodies also had suppressive effects on EV internalization by dendritic cells (ranging from 15% and 20%) (Morelli et al. 2004). A recent study revealed that internalization is dependent on heparan sulfate proteoglycans (HSPGs) expressed on the surface of recipient cells, with EVs co-localizing with HSPG on the plasma membrane of recipient cells (Christianson et al. 2013). The addition of heparin to U87 glioblastoma cells diminished EV uptake (Atai et al. 2013), with the effect being more potent when HSPG contained more negative sulphate groups (Christianson et al. 2013). The charge of heparan sulfate (HS) appears to be primarily responsible for this inhibition, because the addition of calcium ions reduced the inhibitory activity of HS on EV uptake (Christianson et al. 2013). Consistently, cells with mutation in xylosyl transferase and therefore a lower expression of HSPG revealed lower uptake (Christianson et al. 2013). In accordance with reduced uptake, the migration of glioblastoma cells induced by glioblastoma-derived EVs was inhibited by the addition of heparin (Christianson et al. 2013). The level of expression of HSPGs may partly account for the intercellular variability in the uptake efficiency or the mechanisms of uptake of EVs. Overall, the interaction between antigens expressed at the surfaces of recipient cells and on EVs, at least in part, determines the extent of internalization.
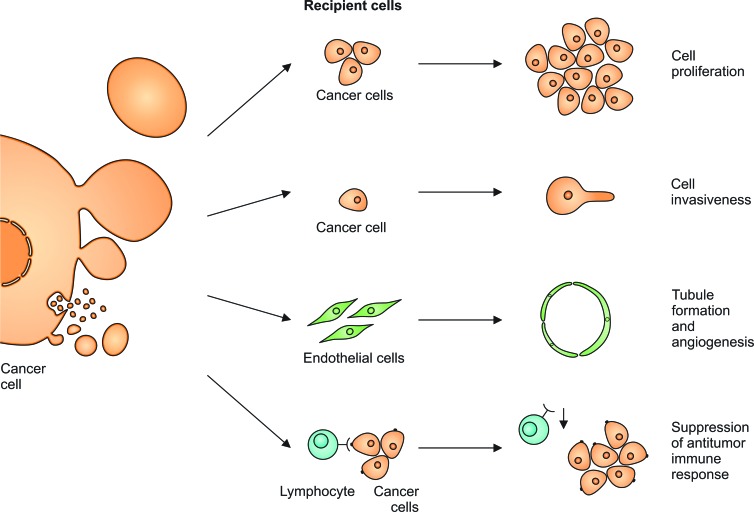
The biological effects of EVs
Owing to their rich composition and capacity to interact with other cells, EVs play a functional role in many biological processes (figure 2). EVs are readily exchanged between cancer cell populations, which can promote proliferation (Al-Nedawi et al. 2008, Skog et al. 2008, Keller et al. 2009). They can also enhance the migration of cancer cells, as well as the invasion and metastases of tumors via the release of EVs enriched with MMP 2 and 9, which digest the extracellular matrix (Di Vizio et al. 2012, Aga et al. 2014). The induction of epithelial-to-mesenchymal transition (EMT) upon interaction with EVs was confirmed by showing EV-induced reduction of E-cadherin expression by cancer cells (Aga et al. 2014), with mesenchymal cancers being more malignant.
Figure 2.
Extracellular vesicles (EVs) released from transformed cells can exert biological effects. They increase the proliferation rate and invasiveness of other cancer cells. They stimulate endothelial cells to form tubules that support tumor angiogenesis. Tumor-derived EVs render cytotoxic T lymphocytes less reactive, which results in the suppression of antitumor immune response.
Neoplastic cells can also release EVs that modify the phenotype of host cells to facilitate tumor growth. EVs released from ovarian cancer cells contain CD147, which promotes the expression of MMP-1, -2, and -9 in endothelial cells (Millimaggi et al. 2007). EVs promote angiogenesis by stimulating the migration and tubule formation of endothelial cells (Skog et al. 2008, Svensson et al. 2011, Di Vizio et al. 2012). Interestingly, this effect was also exerted by EVs derived from renal cancer stem cell populations (CD105 positive cells with the confirmed ability to form spheres and initiate tumors; Grange et al. 2011) and therefore may be a common property of cancer cell–derived EVs. The angiogenic activity was stronger if EV-producing cells were cultured under hypoxic conditions (Kucharzewska et al. 2013). EGFR transferred to endothelial cells from cancer cells via EVs induced the autocrine release of vascular endothelial growth factor to support angiogenesis (Al-Nedawi et al. 2009). Oncosomes were also shown to trigger the migration of cancer-associated fibroblasts (CAFs; Morello et al. 2013). Interestingly, CAFs, in turn, shed EVs with a high content of miR-409, which contributed to the EMT transition and high cancer stem cell phenotypes (Josson et al. 2014). EVs released by ovarian cancer cells contributed to the expansion and higher functional competence of regulatory T lymphocytes and the apoptosis of cytotoxic T lymphocytes that results in the suppression of antitumor immune responses (Szajnik et al. 2010). The incubation of breast cancer– or glioma-released EVs with fibroblasts and epithelial cells resulted in the increased anchorage-independent growth and survival of host cells, suggesting features of transformation in nonneoplastic host cells (Antonyak et al. 2011). Taken together, the exchange of EVs between cancer and normal cells in the tumor microenvironment can result in the promotion of tumor growth through multiple mechanisms.
The influence of EVs is not confined to paracrine activity but is also potent toward remote organs. Intravenously injected EVs accumulate in the spleen, liver, lungs, and kidneys and reside for at least 6 hours postinjection (Lai et al. 2014). The pro-angiogenic and pro-invasive properties of EVs were also confirmed by the intravenous injection of cancer stem cell–derived EVs that increased the number of remote pulmonary metastases of malignant renal tumors (Grange et al. 2011). Consistent with this, melanoma-released EVs administered into the tail vein of naive mice were found in the lungs, spleen, bone marrow, and liver—setting the stage for common metastatic sites for melanoma patients (Peinado et al. 2012). These EVs increased the leakiness of vessels within these organs, as well as the number and the distribution of metastases (Peinado et al. 2012). Melanoma EVs were also found to recruit bone marrow–derived cells to the primary location of the tumor, which had a supportive effect on neoplastic growth (Peinado et al. 2012). Subcutaneously injected melanoma EVs accumulated in lymph nodes, and subsequently implanted neoplastic cells were recruited to the same location as a model for melanoma metastases commonly found in lymphatic tissue (Hood et al. 2011). A recent study demonstrated that EVs released from a metastatic breast cancer cell line (4T1E) contained high levels of miR-200, which promotes mesenchymal-to-epithelial transition and facilitates the colonization of distant tissues (Le et al. 2014). Nonmetastatic breast cancer cells (4TO7V) pretreated with 4T1E-derived EVs formed more lung metastases following tail injection (Le et al. 2014). The inhibition of miR-200 in 4T1E cells reduced this pro-metastatic effect of EVs (Le et al. 2014). These studies demonstrate that cancer-derived EVs can have a systemic effect by preconditioning remote tissues and other neoplastic cell subpopulations to facilitate tumor dissemination. It should be noted, however, that these studies investigated the effects of EVs generated ex vivo on tumor growth in vivo. Therefore, further research is required to elucidate the potential of endogenously released tumor vesicles in mediating the same tumor-promoting processes.
Methods and challenges in studying EVs
Choosing a suitable method based on the EV subtypes is important in studying EVs. Below, we discuss the strategies of isolation, quantification, and RNA analysis and ways to image EVs.
Methods of isolation
EVs are typically isolated by following the major types of methods: differential centrifugation culminating in ultrafiltration, density gradient/cushion centrifugation, and immunoaffinity-based capture. The most widely used method of EV isolation is based on differential centrifugation. It enables enrichment—but not complete separation—of different EV fractions (table 2). Ultracentrifugation consists of an initial low-speed centrifugation step to remove cells (300 × gravitational force [g], 10 minutes, 4°C) and debris (2000 × g, 10 minutes, 4°C). In most protocols, this is followed by filtration through a 0.2 μm–0.8 μm filter or a 10,000–20,000 × g centrifugation to separate small and large EV subpopulations. Applying a wide-pore filter (2 μm) or skipping the filtration step or the 10,000–20,000 × g centrifugation step results in the pooling of smaller and larger EV subtypes. The major step to pellet EVs requires centrifugation at 100,000 × g for 1.5–2 hours using a 70Ti rotor (k factor 44; Beckman Coulter). Longer centrifugation increases the yield of pelleted EVs; however, spinning over 4 hours results in significant contamination with soluble protein (Cvjetkovic et al. 2014). The additional purification of EVs can be achieved by multiple washings with PBS and centrifugations at 100,000 × g. The isolation of EVs from body fluids (e.g., serum, ascites fluid) that have higher viscosity and numerous protein aggregates can be improved by increasing centrifugation force and time (e.g., 2 hours), increasing dilutions in PBS, or performing sucrose gradient density ultracentrifugation (Rani et al. 2011). The main drawback of the differential centrifugation method is the copelleting of high molecular mass protein complexes such as 26S proteasome, HSPG, fatty acid synthase, lipoproteins, and viral particles (Vickers et al. 2011, Tauro et al. 2012). It is also time consuming and requires an ultracentrifuge.
Table 2.
Isolation methods of different extracellular vesicles subtypes.
| Exosomes | Microvesicles | Apoptotic bodies | Oncosomes | |
|---|---|---|---|---|
| Size | 40–120 nm | 100 nm–1 μm | 50 nm–2 μm | 1–10 μm |
| Differential centrifugation and filtration steps | 1. 300 × g, 10 min | 1. 300 × g, 10 min | 1. 300 × g, 10 min | 1. 2,800 × g, 10 min |
| 2. 2,000 × g, 10 min | 2. 2,000 × g, 10 min | 2. 2,000 × g, 20 min | 2. 10,000 × g, 30 min, | |
| 3. Filtration (0.1, 0.22 or 0.8 μm) | 3. Filtration 0.8; 1.0 μm (optional) | Comment: Apoptotic bodies are also present in 10,000 and 100,000 × g fractions | Comment: Alternatively filtration through 0.2 μm with centrifugation 30 s, 8000 × g – oncosomes are captured by the filter | |
| 4. 100,000–120,000 × g, 1.5–2 hours | 4. 10–20,000 × g, 30 min | (Thery et al. 2001, Crescitelli et al. 2013) | (Morello et al. 2013) | |
| (Théry et al. 2009, Rani et al. 2011, Tauro et al. 2012) | (Heijnen et al. 1999, Palmisano et al. 2012) | |||
| Additional purification | 1. Multiple dilutions in PBS and centrifugations 100,000 × g, 70 min | |||
| 2. sucrose cushion (Rani et al. 2011) | ||||
| Fraction in density gradient | 1.07–1.18 g/ml | Unspecified | 1.24–1.28 g/ml | Unspecified |
| (Heijnen et al. 1999, Thery et al. 2001, Keller et al. 2009, Tauro et al. 2012) | (Thery et al. 2001) |
Density gradient centrifugation enables the increased purification of EVs and the partial separation of different subpopulations (Heijnen et al. 1999, Thery et al. 2001). This method improves the removal of high molecular weight proteins and is especially applicable in the characterization of EVs extracted from body fluids, which contain high levels of protein aggregates (Runz et al. 2007, Tauro et al. 2012). Although sucrose solutions are the most widely used for this purpose, iodixanol gradients also appear useful in the separation of exosomes from viral particles that are also likely to copellet using the ultracentrifugation method (Cantin et al. 2008).
An important disadvantage of the ultracentrifugation method is the isolation of all EVs regardless of their subtypes and cells of origin. Given that body fluids contain EVs from multiple cell types, it would be valuable to obtain EVs derived from specific cell types to study their contents and biological effects. Immunoaffinity-based methods can be useful for this purpose. A33 antibody–coated beads, for instance, were used to purify EVs from colorectal cancer cell–conditioned medium (Mathivanan et al. 2010). Magnetic microbeads coupled to anti-EpCAM antibody enabled the enrichment of ovarian cancer–derived EVs from serum for analysis of miRNAs content (Taylor and Gercel-Taylor 2008). The superparamagnetic microbeads coated with anti-L1CAM antibodies were used to demonstrate an increased level of α-synuclein in plasma exosomes in Parkinson's disease patients (Shi et al. 2014). The recently designed nano-plasmonic assay (nPLEX) detects exosomes on the basis of the wavelength shift in light spectrum that appears as a result of EV binding to specific antibodies (Im et al. 2014). This method was used to quantitate ovarian cancer exosomes bearing specific antigens in ascites fluid from ovarian cancer patients (Im et al. 2014). Antibodies that react with antigens common to many cell types can also be used as a method of purification, including CD63 (Chen et al. 2010, Kanwar et al. 2014) or peptides that bind to heat-shock proteins (Ghosh et al. 2014).
A comparison of all three procedures performed on conditioned media–derived EVs revealed that ultracentrifugation yielded the highest protein content (Tauro et al. 2012). Mass spectrometry analysis, however, demonstrated that high molecular weight protein complexes constituted a substantial fraction of the isolated EVs. Although the immunoaffinity method (antibodies coupled with magnetic beads) extracted lower overall protein, it enabled significant enrichment in EV-specific proteins and provided more homogenous EV profiles by electron microscopic evaluation (Tauro et al. 2012). In addition to the above-mentioned methods, EVs can also be isolated by size-exclusion chromatography; the strategy remains to be compared with other methods (Szajnik et al. 2010). An increasing number of commercial kits based on various proprietary technologies for EV isolation are also becoming available and remain to be validated.
RNA analysis
Because the amount of RNA recovered from EVs is low, it is important to use the most efficient protocol for isolation. The RNA enclosed in EVs is protected from RNase activity (Skog et al. 2008). The addition of RNase to EV pellets reduced the RNA content by less than 7% (Skog et al. 2008, Huang et al. 2013). However, the treatment of EVs with detergent before RNase addition resulted in the removal of mRNA coding for Cre recombinase as evaluated by reverse transcription polymerase chain reaction (RT-PCR), supporting the notion that nucleic acids are protected inside EVs (Ridder et al. 2014). However, some RNAs have also been detected in non-EV fractions at significant levels (Wang et al. 2010, Chevillet et al. 2014). They were shown to be bound to ribonucleoproteins, such as nucleophosmin 1, that protected them from RNase activity (Wang et al. 2010). miRNAs in plasma are also found in Argonaute2 protein complexes and in association with lipoproteins (Arroyo et al. 2011, Vickers et al. 2011).
A few studies have demonstrated the feasibility of the sequencing of EV-RNA derived from conditioned media and serum (Bellingham et al. 2012, Huang et al. 2013, Eirin et al. 2014). A comparison of library construction methods for EV-RNA sequencing revealed significant differences between protocols (Huang et al. 2013). With 2 nanograms of input EV-RNA and 15 polymerase chain reaction (PCR) amplification cycles, the NEBNext multiplex small RNA library preparation kit yielded the most 140–160 bp fragments and the highest percentage of mappable reads (62.72%; Huang et al. 2013). Studies are underway to compare the protocols for RNA sequencing from EVs through the National Institutes of Health's Extracellular RNA Common Fund Consortium (exrna.org), and the results should be online in June 2015.
The detection of defined mutations in EV-RNA constitutes an attractive biomarker approach. Quantitative RT-PCR assay, however, may not be sensitive enough given the low yield of nucleic acid. Increased sensitivity can be achieved using droplet digital PCR, in which the sample is split into picoliter-sized droplets that serve as separate PCR reaction compartments (Takahashi et al. 2014). Amplification in every droplet produces a fluorescent signal, and the quantification of positive droplets provides an estimate of copy numbers of the starting molecules. This technique has been used in detecting rare mutant copies, such as IDH1 mRNA in cerebrospinal fluid of glioblastoma patients, which may be used for the diagnosis and monitoring of disease progression (Chen et al. 2013)
Quantification
The quantification of EVs remains a major challenge (table 3). Earlier studies measured total protein content to estimate EV amount. However, the number is often overestimated by contamination with high molecular weight proteins that copurify with isolated EVs. This approach also does not take into consideration the notion that protein content per vesicle may differ between EV subtypes. Nanoparticle tracking analysis (NTA) is based on the detection of light scatter of particles in suspension and their Brownian motion to estimate the number and volume distribution of EVs both in conditioned media and body fluids (Dragovic et al. 2011, Soo et al. 2012). This instrument, however, measures moving particles as a point of light, and it can underestimate the number of larger EVs (i.e., those more than 500 nm). The precision of the quantification can be affected by the heterogeneity of EV size and optical parameters (Maas et al. 2014). It should also be noted that the analysis of EV-containing serum is disturbed by chylomicrons and very low–density lipoproteins that similarly scatter light (Dragovic et al. 2011). This effect can be reduced by the ultracentrifugation of serum samples (Dragovic et al. 2011). The NTA also enables dye-labeled vesicle detection under fluorescent mode (Dragovic et al. 2011). The fluorescence correlation spectroscopy that analyzes fluctuations of fluorescence signal enables studying EV size distribution, as well as the expression of membrane proteins (Wyss et al. 2014).
Table 3.
Methods of extracellular vesicles quantification.
| Technique | Feature |
|---|---|
| Protein Concentration Assay | Biased because of copelleted protein aggregates |
| Nanoparticle tracking analysis (NTA) | Can be used for small EVs (i.e., less than 500 nm) |
| Conventional flow cytometry | Suitable for large EVs (more than 500 nm), however can be biased by immune complexes from peripheral blood |
| Modified flow cytometry with membrane dye | Applicable for EVs larger than 100 nm |
Most conventional flow cytometry is not suitable for smaller-size (less than 300 nm) particles (Dragovic et al. 2011). Flow cytometry can be used to count EVs of more than 500 nm (Orozco and Lewis 2010), including oncosomes (Di Vizio et al. 2012, Morello et al. 2013). Smaller EVs can be analyzed if bound to beads coated with antibodies against surface antigens, such as MHC class II. Subsequent labeling by fluorophore-conjugated antibodies enables semi-quantitative evaluation by flow cytometry (Théry et al. 2006). Flow cytometry analysis of EVs from body fluids, however, can be disturbed by immune complexes that have similar biophysical properties as EVs (György et al. 2011). The use of fluorescent dyes that intercalate into membranes (e.g., PKH67) and instrument modifications adjusted for nanoparticle detection enable the quantification of EVs as small as 100 nm by flow cytometry (Nolte-'t Hoen et al. 2012, van der Vlist et al. 2012). Therefore, this method allows the simultaneous detection of EVs of heterogeneous size. Another technique to quantify EVs is tunable resistive pulse sensing (tRPS). tRPS is based on the disruption of ionic flow as particles pass through a single nanopore separating two fluidic cells. The rate and magnitude of the disruptions can be used to calculate the concentration and volume of EVs, respectively (Maas et al. 2014). A thorough comparison of NTA, tRPS, and flow cytometry demonstrated significant differences among the instruments, which illustrates that the absolute quantification of EVs remains challenging (Maas et al. 2014).
Imaging
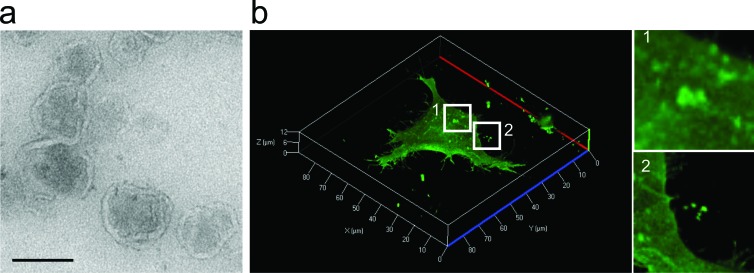
All types of EVs can be visualized by electron microscopy with the additional confirmatory immunolabelling of vesicular proteins (figure 3a; Heijnen et al. 1999). Cup-shaped structures once considered characteristic of EVs likely result from the chemical fixation step during sample preparation because frozen EVs analyzed in cryo-electron microscopy are round shaped (Raposo and Stoorvogel 2013).
Figure 3.
A visualization of extracellular vesicles. (a) A transmission electron micrograph showing extracellular vesicles (EVs) isolated from HEK293T cells. Note the lipid bilayer–enclosed vesicle characteristic of EVs. Bar, 100 nanometers. (b) Live-cell confocal microscopy of human embryonic kidney (HEK) 293T cells expressing palmitoylated GFP (PalmGFP). Plasma membranes are labeled with PalmGFP, allowing the observation of bud-like structures on the cell surface (subpanel 1), suggesting their subsequent release as EVs. Released PalmGFP-EVs were readily observed around the microenvironment of 293T-PalmGFP cells (subpanel 2).
Studying the biological functions of EVs requires tools to trace their fate in recipient cells and tissues. Various membrane-specific dyes have been in use for this purpose, including PKH67 (Morelli et al. 2004), PKH26 (Grange et al. 2011), DiI, and DiR (Hood et al. 2011). The use of the above-mentioned dyes has two main limitations: they label all EVs regardless of their origin, and their long half-life makes them less suitable for longitudinal studies because of potential dye deposition in cells and organs, which could be falsely interpreted as EV accumulation. To overcome these caveats, bioluminescent reporters such as Gaussia luciferase fused to a transmembrane domain was developed to label EVs for sensitive spatiotemporal studies in vivo (Lai et al. 2014). The overexpression of membrane-bound fluorescent proteins, such as green fluorescent protein (GFP) or tdTomato fused to palmitoylation signal peptide at the N-terminus, results in the production of labeled EVs only from transduced cells (Lai et al. 2015). Fluorescent proteins can be also fused to antigens found in EVs (e.g., CD63 and red fluorescent protein fusion; Le et al. 2014). By the latter techniques, or the above-mentioned membrane-specific dyes, the transfer of EVs between cells can be visualized by confocal microscopy (figure 3b). To distinguish that EVs are not simply attached to the cell surface but are indeed internalized, trypsinization can be applied to wash away EVs that have not entered the cells (Feng et al. 2010). The flow cytometric quantification of recipient cells positive for EV-coupled dyes reflects the extent of transfer (Escrevente et al. 2011). The above-mentioned techniques help in the accurate measurement of EV uptake by cells in order to normalize the biological effects assessed in recipient cells.
Box 2. Current challenges.
Current extracellular vesicle (EV) isolation techniques do not allow precise EV subpopulation enrichment.
Most analyses of the transfer of RNA transported by EVs (EV-RNA) are based on overexpression and detection in recipient cells. Although EV-RNA transfer is possible, more proof is needed as to when and whether this mechanism is biologically functional and significant, especially in vivo.
EVs have been shown to play many physiological and pathological roles; however, many of these observations were made in vitro or in vivo, with EVs that were generated in vitro administered into animals. Future studies with labeled EV-secreting cells in xenograft or transgenic models will reflect the physiological function of EVs in vivo more closely.
Experimental procedures used to block or enhance EV formation still lack specificity.
EV isolation protocols (and the subsequent extraction of EV contents) are not yet standardized and vary between studies, thereby making the comparison of results between studies challenging.
Because EVs contain a variety of proteins, nucleic acids, and lipids, it can be challenging to prove that EV-induced phenotype in recipient cells is attributed to the transfer of one or multiple EV-cargoes.
Biofluids contain EVs released from multiple cell types (e.g., cancer cells, immune cells, platelets). Although some studies successfully enrich for cancer-specific EVs in biofluids that contain EVs from platelets, immune cells, and tumor cells, the isolation of EVs with high cell-type specificity and abundance from other biofluids, such as plasma, remains a challenge.
Conclusions
We summarize the variety of cargos transported by EVs and their effects on biological functions. It should be noted that most of the conclusions were made based on in vitro experimental setups. It remains unclear as to what extent the isolation procedures and quantification of in vitro isolated EVs reflect physiological conditions. For these reasons, further in vivo models need to be developed to better elucidate the functional roles of EVs. Despite the many questions to be answered in the future (box 2), the intercellular exchange of EVs has emerged as a biologically relevant phenomenon under both physiological and pathological conditions. The use of EV contents in different biofluids as biomarkers for different disease states is a very active field.
Acknowledgments
We would like to thank Ms. Suzanne McDavitt for her supportive editorial assistance, Zofia Zaborowska for the graphical preparation of schematics, Sybren L.N. Maas for discussions on EV quantification methods, and Maria Ericsson for performing electron microscopy imaging. This work has been supported by NIH NCI P01CA069246 grant, NIH NCI U19 CA179563 supported by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director and the Richard Floor Biorepository Fund (MPZ, LB, XOB, CPL). LB is supported by the FDM Fellowship (ECOR MGH). Mikolaj Piotr Zaborowski is a recipient of a scholarship from the Kosciuszko Foundation and is thankful for support by the Poznan University of Medical Sciences.
References cited
- Aga M, Bentz GL, Raffa S, Torrisi MR, Kondo S, Wakisaka N, Yoshizaki T, Pagano JS, Shackelford J. Exosomal HIF1α supports invasive potential of nasopharyngeal carcinoma-associated LMP1-positive exosomes. Oncogene. 2014;33:4613–4622. doi: 10.1038/onc.2014.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Nedawi K, Meehan B, Kerbel RS, Allison AC, Rak J. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR; Proceedings of the National Academy of Sciences; 2009. pp. 3794–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nature Cell Biology. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- Antonyak MA, Li B, Boroughs LK, Johnson JL, Druso JE, Bryant KL, Holowka DA, Cerione RA. Cancer cell–derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells; Proceedings of the National Academy of Sciences; 2011. pp. 4852–4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenaccio C, Chiozzini C, Columba-Cabezas S, Manfredi F, Affabris E, Baur A, Federico M. Exosomes from human immunodeficiency virus type 1 (HIV-1)–infected cells license quiescent CD4+ T lymphocytes to replicate HIV-1 through a Nef- and ADAM17-dependent mechanism. Journal of Virology. 2014;88:11529–11539. doi: 10.1128/JVI.01712-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma; Proceedings of the National Academy of Sciences; 2011. pp. 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atai NA, Balaj L, van Veen H, Breakefield XO, Jarzyna PA, Van Noorden CJF, Skog J, Maguire CA. Heparin blocks transfer of extracellular vesicles between donor and recipient cells. Journal of Neuro-Oncology. 2013;115:343–351. doi: 10.1007/s11060-013-1235-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaj L, Lessard R, Dai L, Cho Y-J, Pomeroy SL, Breakefield XO, Skog J. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nature Communications. 2011;2 doi: 10.1038/ncomms1180. art. 180. doi:10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Research. 2012;40:10937–10949. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloribi S, Ristorcelli E, Breuzard G, Silvy F, Bertrand-Michel J, Beraud E, Verine A, Lombardo D. Exosomal lipids impact notch signaling and induce death of human pancreatic tumoral SOJ-6 cells. PLOS ONE. 2012;7 doi: 10.1371/journal.pone.0047480. art. e47480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicalho B, Holovati JL, Acker JP. Phospholipidomics reveals differences in glycerophosphoserine profiles of hypothermically stored red blood cells and microvesicles. Biochimica et Biophysica Acta. 2013;1828:317–326. doi: 10.1016/j.bbamem.2012.10.026. [DOI] [PubMed] [Google Scholar]
- Bolukbasi MF, Mizrak A, Ozdener GB, Madlener S, Ströbel T, Erkan EP, Fan J-B, Breakefield XO, Saydam O. miR-1289 and “Zipcode”-like Sequence Enrich mRNAs in Microvesicles. 2012. Molecular Therapy–Nucleic Acids 1, art. e10. [DOI] [PMC free article] [PubMed]
- Cantin R, Diou J, Bélanger D, Tremblay AM, Gilbert C. Discrimination between exosomes and HIV-1: Purification of both vesicles from cell-free supernatants. Journal of Immunological Methods. 2008;338:21–30. doi: 10.1016/j.jim.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Chen C, Skog J, Hsu C-H, Lessard RT, Balaj L, Wurdinger T, Carter BS, Breakefield XO, Toner M, Irimia D. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab on a Chip. 2010;10:505–511. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, et al. BEAMing and Droplet Digital PCR Analysis of Mutant IDH1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Molecular Therapy—Nucleic Acids. 2013;2 doi: 10.1038/mtna.2013.28. art. e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevillet JR, et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes; Proceedings of the National Academy of Sciences; 2014. pp. 14888–14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson HC, Svensson KJ, van Kuppevelt TH, Li J-P, Belting M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity; Proceedings of the National Academy of Sciences; 2013. pp. 17380–17385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocucci E, Meldolesi J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends in Cell Biology. 2015;25:364–372. doi: 10.1016/j.tcb.2015.01.004. doi:10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- Crescitelli R, Lässer C, Szabo TG, Kittel A, Eldh M, Dianzani I, Buzás EI, Lötvall J. Distinct RNA profiles in subpopulations of extracellular vesicles: Apoptotic bodies, microvesicles and exosomes. Journal of Extracellular Vesicles. 2013;2 doi: 10.3402/jev.v2i0.20677. art. 20677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvjetkovic A, Lötvall J, Lässer C. The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. Journal of Extracellular Vesicles. 2014;3 doi: 10.3402/jev.v3.23111. art. 23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragovic RA, et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine. 2011;7:780–788. doi: 10.1016/j.nano.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirin A, Riester SM, Zhu X-Y, Tang H, Evans JM, O'Brien D, van Wijnen AJ, Lerman LO. MicroRNA and mRNA cargo of extracellular vesicles from porcine adipose tissue–derived mesenchymal stem cells. Gene. 2014;551:55–64. doi: 10.1016/j.gene.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Andaloussi S, Mäger I, Breakefield XO, Wood MJA. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nature Reviews Drug Discovery. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- Escrevente C, Keller S, Altevogt P, Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011;11 doi: 10.1186/1471-2407-11-108. art. 108, doi:10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Zhao W-L, Ye Y-Y, Bai X-C, Liu R-Q, Chang L-F, Zhou Q, Sui S-F. Cellular internalization of exosomes occurs through phagocytosis. Traffic. 2010;11:675–687. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, Regen T, Hanisch U-K, Simons M. Selective transfer of exosomes from oligodendrocytes to microglia by macropinocytosis. Journal of Cell Science. 2011;124:447–458. doi: 10.1242/jcs.074088. [DOI] [PubMed] [Google Scholar]
- Ghosh A, et al. Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLOS ONE. 2014;9 doi: 10.1371/journal.pone.0110443. art. e110443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grange C, Tapparo M, Collino F, Vitillo L, Damasco C, Deregibus MC, Tetta C, Bussolati B, Camussi G. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Research. 2011;71:5346–5356. doi: 10.1158/0008-5472.CAN-11-0241. [DOI] [PubMed] [Google Scholar]
- Guescini M, Genedani S, Stocchi V, Agnati LF. Astrocytes and glioblastoma cells release exosomes carrying mtDNA. Journal of Neural Transmission. 2010;117:1–4. doi: 10.1007/s00702-009-0288-8. [DOI] [PubMed] [Google Scholar]
- György B, et al. Detection and isolation of cell-derived microparticles are compromised by protein complexes resulting from shared biophysical parameters. Blood. 2011;117:e39–e48. doi: 10.1182/blood-2010-09-307595. [DOI] [PubMed] [Google Scholar]
- Heijnen HF, Schiel AE, Fijnheer R, Geuze HJ, Sixma JJ. Activated platelets release two types of membrane vesicles: Microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–3799. [PubMed] [Google Scholar]
- Hood JL, San RS, Wickline SA. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Research. 2011;71:3792–3801. doi: 10.1158/0008-5472.CAN-10-4455. [DOI] [PubMed] [Google Scholar]
- Huang X, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im H, Shao H, Park Y. Il, Peterson VM, Castro CM, Weissleder R, Lee H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor Nature Biotechnology 2014. 32 490 495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josson S, et al. Stromal fibroblast–derived miR-409 promotes epithelial-to-mesenchymal transition and prostate tumorigenesis. Oncogene. 2014;34:2690–2699. doi: 10.1038/onc.2014.212. doi:10.1038/onc.2014.212. [DOI] [PubMed] [Google Scholar]
- Kahlert C, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. Journal of Biological Chemistry. 2014;289:3869–3875. doi: 10.1074/jbc.C113.532267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalamvoki M, Du T, Roizman B. Cells infected with herpes simplex virus 1 export to uninfected cells exosomes containing STING, viral mRNAs, and microRNAs; Proceedings of the National Academy of Sciences; 2014. pp. E4991–E4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S. Microfluidic device (ExoChip) for on-chip isolation, quantification, and characterization of circulating exosomes. Lab on a Chip. 2014;14:1891–1900. doi: 10.1039/c4lc00136b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S, König A-K, Marmé F, Runz S, Wolterink S, Koensgen D, Mustea A, Sehouli J, Altevogt P. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Letters. 2009;278:73–81. doi: 10.1016/j.canlet.2008.12.028. [DOI] [PubMed] [Google Scholar]
- King HW, Michael MZ, Gleadle JM. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer. 2012;12 doi: 10.1186/1471-2407-12-421. (art. 421). doi:10.1186/1471-2407-12-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppers-Lalic D, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Reports. 2014;8:1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- Kucharzewska P, Christianson HC, Welch JE, Svensson KJ, Fredlund E, Ringnér M, Mörgelin M, Bourseau-Guilmain E, Bengzon J, Belting M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development; Proceedings of the National Academy of Sciences; 2013. pp. 7312–7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CP, Mardini O, Ericsson M, Prabhakar S, Maguire CA, Chen JW, Tannous BA, Breakefield XO. Dynamic biodistribution of extracellular vesicles in vivo using a multimodal imaging reporter. ACS Nano. 2014;8:483–494. doi: 10.1021/nn404945r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CP, Kim EY, Badr CE, Weissleder R, Mempel TR, Tannous BA, Breakefield XO. Visualization and tracking of tumor extracellular vesicle delivery and RNA translation using multiplexed reporters. Nature Communications. 2015;6 doi: 10.1038/ncomms8029. (art. 7029). doi:10.1038/ncomms8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laulagnier K, et al. Mast cell– and dendritic cell–derived exosomes display a specific lipid composition and an unusual membrane organization. Biochemical Journal. 2004;380:161–171. doi: 10.1042/BJ20031594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MTN, Hamar P, Guo C, Basar E, Perdigão-Henriques R, Balaj L, Lieberman J. miR-200-containing extracellular vesicles promote breast cancer cell metastasis. Journal of Clinical Investigation. 2014;124:5109–5128. doi: 10.1172/JCI75695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, et al. Characterization and proteomic analysis of ovarian cancer–derived exosomes. Journal of Proteomics. 2013;80C:171–182. doi: 10.1016/j.jprot.2012.12.029. [DOI] [PubMed] [Google Scholar]
- Llorente A, Skotland T, Sylvänne T, Kauhanen D, Róg T, Orłowski A, Vattulainen I, Ekroos K, Sandvig K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochimica et Biophysica Acta. 2013;1831:1302–1309. doi: 10.1016/j.bbalip.2013.04.011. [DOI] [PubMed] [Google Scholar]
- Maas SLN, de Vrij J, van der Vlist EJ, Geragousian B, van Bloois L, Mastrobattista E, Schiffelers R, Wauben MHM, Broekman ML, Nolte-'t Hoen ENM. Possibilities and limitations of current technologies for quantification of biological extracellular vesicles and synthetic mimics. Journal of Controlled Release. 2014;200:87–96. doi: 10.1016/j.jconrel.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathivanan S, Lim JWE, Tauro BJ, Ji H, Moritz RL, Simpson RJ. Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Molecular and Cellular Proteomics. 2010;9:197–208. doi: 10.1074/mcp.M900152-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millimaggi D, Mari M, D'Ascenzo S, Carosa E, Jannini EA, Zucker S, Carta G, Pavan A, Dolo V. Tumor vesicle–associated CD147 modulates the angiogenic capability of endothelial cells. Neoplasia. 2007;9:349–357. doi: 10.1593/neo.07133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, Bernad A, Sánchez-Madrid F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells Nature Communications 2011. 2 (art. 282), doi:10.1038/ncomms1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morelli AE, et al. Endocytosis, intracellular sorting, and processing of exosomes by dendritic cells. Blood. 2004;104:3257–3266. doi: 10.1182/blood-2004-03-0824. [DOI] [PubMed] [Google Scholar]
- Morello M, et al. Large oncosomes mediate intercellular transfer of functional microRNA. Cell Cycle. 2013;12:3526–3536. doi: 10.4161/cc.26539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, Chung H-Y, Morham SG, Gygi SP, Rodesch CK, Sundquist WI. Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO Journal. 2007;26:4215–4227. doi: 10.1038/sj.emboj.7601850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan JF, Hu R, Oh RS, Cohen SN, Lu Q. Formation and release of arrestin domain-containing protein 1–mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein; Proceedings of the National Academy of Sciences; 2012. pp. 4146–4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A, Kawanishi E, Yoshida R, Yoshiyama H. Exosomes derived from Epstein Barr virus–infected cells are internalized via caveola-dependent endocytosis and promote phenotypic modulation in target cells. Journal of Virology. 2013;87:10334–10347. doi: 10.1128/JVI.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte-'t Hoen ENM, et al. Quantitative and qualitative flow cytometric analysis of nanosized cell-derived membrane vesicles. Nanomedicine. 2012;8:712–720. doi: 10.1016/j.nano.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco AF, Lewis DE. Flow cytometric analysis of circulating microparticles in plasma. Cytometry A. 2010;77:502–514. doi: 10.1002/cyto.a.20886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmisano G, Jensen SS, Le Bihan M-C, Lainé J, McGuire JN, Pociot F, Larsen MR. Characterization of membrane-shed microvesicles from cytokine-stimulated β-cells using proteomics strategies. Molecular and Cellular Proteomics. 2012;11:230–243. doi: 10.1074/mcp.M111.012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parolini I, et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. Journal of Biological Chemistry. 2009;284:34211–34222. doi: 10.1074/jbc.M109.041152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MAJ, Hopmans ES, Lindenberg JL, de Gruijl TD, Würdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes; Proceedings of the National Academy of Sciences; 2010. pp. 6328–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peinado H, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nature Medicine. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnaiah V, et al. Exosome-mediated transmission of hepatitis C virus between human hepatoma Huh7.5 cells; Proceedings of the National Academy of Sciences; 2013. pp. 13109–13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani S, O'Brien K, Kelleher FC, Corcoran C, Germano S, Radomski MW, Crown J, O'Driscoll L. Isolation of exosomes for subsequent mRNA, MicroRNA, and protein profiling. Methods in Molecular Biology. 2011;784:181–195. doi: 10.1007/978-1-61779-289-2_13. [DOI] [PubMed] [Google Scholar]
- Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. Journal of Cell Biology. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Record M, Carayon K, Poirot M, Silvente-Poirot S. Exosomes as new vesicular lipid transporters involved in cell–cell communication and various pathophysiologies. Biochimica et Biophysica Acta. 2014;1841:108–120. doi: 10.1016/j.bbalip.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Redzic JS, Balaj L, van der Vos KE, Breakefield XO. Extracellular RNA mediates and marks cancer progression. Seminars in Cancer Biology. 2014;28:14–23. doi: 10.1016/j.semcancer.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridder K, et al. Extracellular vesicle-mediated transfer of genetic information between the hematopoietic system and the brain in response to inflammation. PLOS Biology. 2014;12 doi: 10.1371/journal.pbio.1001874. art. e1001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runz S, Keller S, Rupp C, Stoeck A, Issa Y, Koensgen D, Mustea A, Sehouli J, Kristiansen G, Altevogt P. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecologic Oncology. 2007;107:563–571. doi: 10.1016/j.ygyno.2007.08.064. [DOI] [PubMed] [Google Scholar]
- Sano S, et al. Lipid synthesis is promoted by hypoxic adipocyte-derived exosomes in 3T3-L1 cells. Biochemical and Biophysical Research Communications. 2014;445:327–333. doi: 10.1016/j.bbrc.2014.01.183. [DOI] [PubMed] [Google Scholar]
- Shi M, et al. Plasma exosomal α-synuclein is likely CNS-derived and increased in Parkinson's disease. Acta Neuropathologica. 2014;128:639–650. doi: 10.1007/s00401-014-1314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda M, Khokha R. Proteolytic factors in exosomes. Proteomics. 2013;13:1624–1636. doi: 10.1002/pmic.201200458. [DOI] [PubMed] [Google Scholar]
- Shimoda M, et al. Loss of the Timp gene family is sufficient for the acquisition of the CAF-like cell state. Nature Cell Biology. 2014;16:889–901. doi: 10.1038/ncb3021. [DOI] [PubMed] [Google Scholar]
- Sinha A, Ignatchenko V, Ignatchenko A, Mejia-Guerrero S, Kislinger T. In-depth proteomic analyses of ovarian cancer cell line exosomes reveals differential enrichment of functional categories compared to the NCI 60 proteome. Biochemical and Biophysical Research Communications. 2014;445:694–701. doi: 10.1016/j.bbrc.2013.12.070. [DOI] [PubMed] [Google Scholar]
- Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nature Cell Biology. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth TJ, Redzic JS, Graner MW, Anchordoquy TJ. Examination of the specificity of tumor cell–derived exosomes with tumor cells in vitro. Biochimica et Biophysica Acta. 2014;1838:2954–2965. doi: 10.1016/j.bbamem.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo CY, Song Y, Zheng Y, Campbell EC, Riches AC, Gunn-Moore F, Powis SJ. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology. 2012;136:192–197. doi: 10.1111/j.1365-2567.2012.03569.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subra C, et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. Journal of Lipid Research. 2010;51:2105–2120. doi: 10.1194/jlr.M003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson KJ, Christianson HC, Wittrup A, Bourseau-Guilmain E, Lindqvist E, Svensson LM, Mörgelin M, Belting M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid raft–mediated endocytosis negatively regulated by caveolin-1. Journal of Biological Chemistry. 2013;288:17713–17724. doi: 10.1074/jbc.M112.445403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson KJ, Kucharzewska P, Christianson HC, Sköld S, Löfstedt T, Johansson MC, Mörgelin M, Bengzon J, Ruf W, Belting M. Hypoxia triggers a proangiogenic pathway involving cancer cell microvesicles and PAR-2-mediated heparin-binding EGF signaling in endothelial cells. Proceedings of the National Academy of Sciences. 2011;108:13147–13152. doi: 10.1073/pnas.1104261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajnik M, Czystowska M, Szczepanski MJ, Mandapathil M, Whiteside TL. Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg) PLOS ONE. 2010;5 doi: 10.1371/journal.pone.0011469. art. e11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yan IK, Kim C, Kim J, Patel T. Analysis of extracellular RNA by digital PCR. Frontiers in Oncology. 2014;4:129. doi: 10.3389/fonc.2014.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauro BJ, Greening DW, Mathias RA, Ji H, Mathivanan S, Scott AM, Simpson RJ. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecologic Oncology. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Thakur BK, et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Research. 2014;24:766–769. doi: 10.1038/cr.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. In: Bonifacino JS, Dasso M, Harford JB, Lippincott-Schwartz J, Yamada KM, editors. Current Protocols in Cell Biology. John Wiley and Sons; 2006. Chapter 3, unit 22 in. [DOI] [PubMed] [Google Scholar]
- Thery C, Boussac M, Veron P, Ricciardi-Castagnoli P, Raposo G, Garin J, Amigorena S. Proteomic analysis of dendritic cell–derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. Journal of Immunology. 2001;166:7309–7318. doi: 10.4049/jimmunol.166.12.7309. [DOI] [PubMed] [Google Scholar]
- Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nature Reviews Immunology. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nature Cell Biology. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M, Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nature Communications. 2013;4 doi: 10.1038/ncomms3980. (art. 2980), doi:10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Vizio D, et al. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. American Journal of Pathology. 2012;181:1573–1584. doi: 10.1016/j.ajpath.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Vlist EJ, Nolte-'t Hoen ENM, Stoorvogel W, Arkesteijn GJA, Wauben MHM. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nature Protocols. 2012;7:1311–1326. doi: 10.1038/nprot.2012.065. [DOI] [PubMed] [Google Scholar]
- Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Research. 2010;38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss R, Grasso L, Wolf C, Grosse W, Demurtas D, Vogel H. Molecular and dimensional profiling of highly purified extracellular vesicles by fluorescence fluctuation spectroscopy. Analytical Chemistry. 2014;86:7229–7233. doi: 10.1021/ac501801m. [DOI] [PubMed] [Google Scholar]