Many microbial opsin genes encode proteins that, upon absorption of a photon, move ions across the cell membrane. The resulting ion flow can activate, inhibit, or modulate cells depending on the type, direction, and quantity of the ion being conducted (1). For optogenetic experiments, expressing these proteins has been useful for providing activity patterns to targeted cells (1, 2). On page 647 of this issue, Govorunova et al. (3) report a potent new opsin from the microbe Guillardia theta that can inhibit target cells. The discovery of this channelrhodopsin punctuates the long search for a naturally occurring, light-activated ion channel with utility for inhibition in optogenetic studies.
Most channelrhodopsin proteins allow cations in the cellular milieu to flow down electrochemical gradients across the membrane in response to light (4). This translates into an excitatory stimulus for the opsin-expressing cell. In many cases, however, inhibiting the targeted cells is also of scientific value, for which light-activated potassium or chloride ion channels would be well-suited. However, for more than 12 years after the initial description of ion conductances of channelrhodopsins (4, 5), and for more than 7 years after initial reports of channel engineering and genomic screening (6) to create or identify channelrhodopsins with new properties—and despite intense efforts on both the genomic and channel engineering fronts—no potassium- or chloride-selective channelrhodopsins had emerged (7).
To overcome this obstacle, microbial opsins encoding chloride or proton pumps have been used, giving rise to many discoveries on the neural circuit control of behavior. However, these are less efficient than channels, moving only one ion per photon instead of the hundreds that channels can allow. The crystal structure of channelrhodopsin (8) allowed rational modification of the channel pore (9) and mutagenesis of amino acids involved in the photocycle (10), to generate inhibitory chloride channels. Both engineered channelrhodopsins [inhibitory C1C2 (iC1C2) and slow chlorideconducting channelrhodopsin (SloChloC), respectively] exclude sodium and potassium ions, but conduct chloride, thus effectively inhibiting action potentials in cultured neurons (9, 10).
Genomic studies had identified and characterized microbial opsins from Guillardia (7), but the new family members reported by Govorunova et al. [called Guillardia theta anion channelrhodopsin 1 (GtACR1) and GtACR2] show markedly reduced primarysequence homology and unusual chloride selectivity. Like the engineered chloride channels (9, 10), GtACR2 allows light-induced blockade of action potentials in cultured neurons. Two other properties of the GtACRs deserve mention: large photocurrents and high light sensitivity.
The photocurrents arising from GtACR2 are among the largest that have been reported with any microbial opsin (many nanoamperes of current in single mammalian cells). This implies especially robust expression (many channels per cell) and perhaps especially strong single-channel conductance as well. Although GtACR2 in neurons was challenged with relatively moderate action potential–inducing stimulation, its large inhibitory photocurrent amplitude may present a major upside for preventing action potentials driven by very strong synaptic input.
Also of interest is the low light intensity that can be employed. Govorunova et al. used an irradiance value of 0.026 mW/mm2 to inhibit action potentials in neurons, which is about two orders of magnitude weaker than might be used to saturate widely used channelrhodopsins (11). In optogenetics, the operational light sensitivity of targeted cells can be decomposed into several contributing factors (11), ranging from quantum efficiency of the photosensitive protein molecule itself (the likelihood of successful absorption of a photon arriving within the protein’s spatial cross-section, leading to photocycle initiation), to the density of proteins on the cell membrane (which is itself related to expression level and trafficking efficiency), to the kinetics of deactivation (slowed deactivation kinetics allow accumulation of proteins in the active state within a cell, and therefore longer light pulses can be effective at orders-of-magnitude lower irradiance values for slow-deactivating opsins). All of these factors could be operative for GtACR2. However, quantum efficiencies are already thought to be fairly high for wild-type channelrhodopsins (~0.5, leaving little room for orders-of-magnitude improvement). In addition, GtACR2 protein density in the membrane is presumably high, as reflected in the large photocurrents, but again perhaps not alone high enough to explain the sensitivity. Also, deactivation kinetics of GtACR2 (>40 ms) are somewhat slower than fast inhibitory pumps (~4 ms exhibited by cells expressing the chloride pump halorhodopsin eNpHR3.0) (11, 12) or inhibitory channels (~10 ms for the dominant fast component of iC1C2) (9). It may be that high expression is the dominant contributor to the effective light sensitivity reported by Govorunova et al., which will be fascinating to explore further but is, in principle, of high utility regardless of mechanism.
“The discovery of this channelrhodopsin punctuates the long search for a naturally occurring, light-activated ion channel with utility for inhibition in optogenetic studies.”
Are the slower off-kinetics problematic in other ways? For hypothetical experiments involving, for example, the deletion of single action potentials (spikes) from within >25-Hz trains, deactivation kinetics of >40 ms could be problematic. But most experiments with inhibition do not involve deleting a single spike within a high-frequency train (for which fast pumps or channels like eNpHR3.0 and iC1C2 would be used), but rather involve more sustained inhibition. The photocurrents of GtACR2 show suitable properties for such longer-term experiments, including temporal stationarity and large amplitude. Other factors could guide selection (for use in optogenetics) of a chloride pump over a chloride channel—for example, in cases where chloride gradients might be inverted as in developmental or pathological situations, and certain cell types or subcellular compartments. In these cases, even perfect chloride conductance and strong currents will not lead to better inhibition. Instead, excitation will result, indicating that chloride pumps would be a better choice. Pumps have their own challenges and can cause membrane instability if hyperpolarization that is too strong is achieved (11). It will be interesting to observe the extent of hyperpolarization elicited by the robust photocurrents of GtACR2 in typical application settings.
What are the next steps for the field? Individual optogenetic tools have frequently shown promise in vitro, but unpredictably encountered fundamental problems in the longer-term in vivo expression setting. It will be essential to actually test, for optogenetics, all of the new chloride channels (both engineered and naturally occurring) in intact tissue, in different cell types, and under different chloride concentrations. A large upside is possible for GtACR2, as many opportunities have not yet been leveraged, including adding the mutations from step-function opsins [for higher light sensitivity (11, 13)], adding surface-membrane and neurite trafficking sequences for higher expression levels (11– 13), and adding mutations for accelerating kinetics further (11, 13).
Knowing that channelrhodopsins can be chloride-selective to an extent that allows action potential inhibition (2, 9, 10) has been useful for understanding the proteins themselves, and now poses additional intriguing biophysical and structure-function questions. Solving the crystal structures of both the engineered and naturally occurring chloride channels will be necessary, to understand if shared or distinct pore configurations and related selectivity mechanisms are at work.
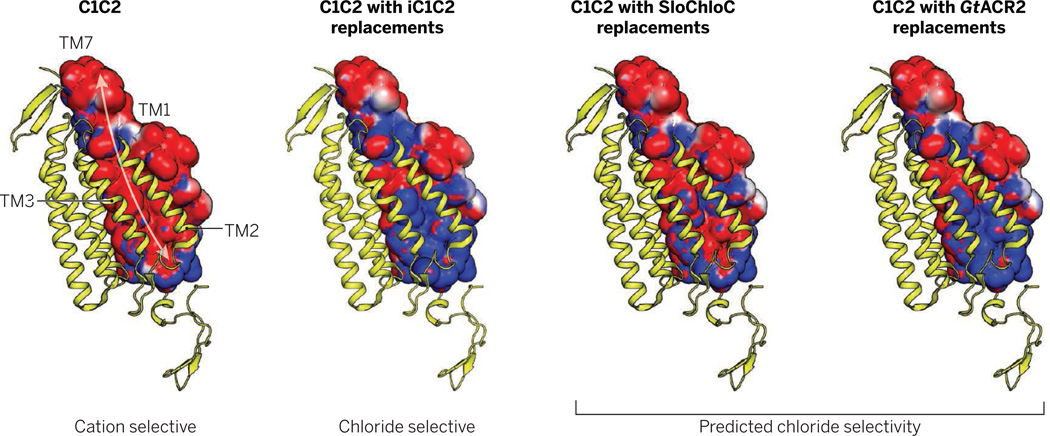
In particular, the crystal structure of channelrhodopsin (8)—with a large relatively disordered pore, absence of bound ions, and conduction pathway lined with residues expected to be negatively charged or polar—suggested an electrostatic model for cation-selective pore function that was empirically tested, and successfully led to creation of anion selectivity (9). Obtaining the structure of GtACR2 will be interesting in this regard; although GtACR2 is chloride selective while retaining a glutamate residue (E90) (2), which iC1C2 (9) and SloChloC (10) lack, certain pore similarities between iC1C2 and GtACR2 support the net electrostatic model (see the figure) with numerous replacements of pore-facing glutamate residues corresponding to the original C1C2 channelrhodopsin with noncharged residues in GtACR2 (including GtACR2 serine 57, threonine 67, alanine 71, and serine 93). Interestingly, the serine 93 site corresponds to a similar threonine in the chloride pump halorhodopsin from Halobacterium salinarum, where this residue is near the chloride ion binding site.
Figure. Conductivity in channelrhodopsins.
The wild-type channel C1C2 [Protein Data Bank: 3UG9 (8)] does not conduct chloride (9). The ion-conducting pore is formed by four transmembrane helices (TM 1, 2, 3, 7). In the modified structures shown, residues facing the ion-conducting pore in C1C2 are replaced to demonstrate the putative impact on the electrostatic surface potential of helices 1 and 7; side-chain positioning (except for the shown closed state of C1C2) (8) and electrostatics are not known and could vary in the open state as well. Structures were generated with PyMOL 1.7 and surface potentials were calculated with the APBS Tool 2.1 (14) assuming full deprotonation of acidic and full protonation of basic residues. Red and blue represent putative electrostatic potential of −1 kT/e and +1 kT/e, respectively. k, Boltzmann’s constant; T, temperature; e, elementary charge.
Structural information and molecular dynamics studies on GtACR2 will also enable understanding of the central and cytosolic pore gates of the closed-state channelrhodopsin structure, especially because tyrosine 109 of the C1C2 cytosolic gate is represented by methionine in GtACR2, while another contributing residue to this site in C1C2 (histidine 173) is replaced by tryptophan in GtACR2. These presumptive gates might still operate in GtACR2, but replacement of the cytosolic gate tyrosine by methionine suggests a substantial disturbance of that pore site, which based on the closed-state structure could contribute considerably to enhanced conductance. Complementation of crystal structures with spectroscopic studies and molecular dynamics simulations will be of value for deeper understanding of intermediate states. These and many other basic biophysical questions will go hand in hand with exploring the opportunities for optogenetics—both avenues representing intriguing directions arising from identification of GtACRs.
Acknowledgments
We thank S. Y. Lee, H. Kato, Y. S. Kim, and C. Ramakrishnan for helpful comments.
REFERENCES AND NOTES
- 1.Deisseroth K. Nature. 2014;505:309. doi: 10.1038/nature12982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grosenick L, Marshel JH, Deisseroth K. Neuron. 2015;86:106. doi: 10.1016/j.neuron.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Govorunova EG, Sineshchekov OA, Janz R, Liu X, Spudich JL. Science. 2015;349:647. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagel G, et al. Science. 2002;296:2395. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- 5.Nagel G, et al. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13940. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang F, et al. Nat. Neurosci. 2008;11:631. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang F, et al. Cell. 2011;147:1446. doi: 10.1016/j.cell.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato HE, et al. Nature. 2012;482:369. doi: 10.1038/nature10870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berndt A, Lee SY, Ramakrishnan C, Deisseroth K. Science. 2014;344:420. doi: 10.1126/science.1252367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wietek J, et al. Science. 2014;344:409. doi: 10.1126/science.1249375. [DOI] [PubMed] [Google Scholar]
- 11.Mattis J, et al. Nat. Methods. 2012;9:159. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gradinaru V, et al. Cell. 2010;141:154. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yizhar O, et al. Nature. 2011;477:171. doi: 10.1038/nature10360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10037. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]