Abstract
Transcriptomic, proteomic, and metabolomic measurements are revolutionizing the way we model and predict cellular behavior, and multi-omic comparisons are being published with increased regularity. Some have expected a trivial and predictable correlation between mRNA and protein; however, the manifest complexity of biological regulation suggests a more nuanced relationship. Indeed, observing this lack of strict correlation provides clues for new research topics, and has the potential for transformative biological insight.
Keywords: bioinformatics, systems biology, proteomics, transcriptomics
Easy access to global -omic measurements
High-throughput transcriptomic, proteomic, and metabolomic measurements are revolutionizing the way we model and predict cellular behavior. Disruptive technologies are continually improving measurement speed, coverage, and accuracy of multi-omic data while simultaneously reducing costs. A popular avenue in the post-genomic era is the simultaneous interrogation of global abundance of protein and mRNA [1–3]. Because these studies are becoming more common and the results are being repeated and corroborated on multiple technology platforms, the goals and utilities of such comparisons now need to be evaluated. The purpose of this Forum article is not to exhaustively summarize the literature. Rather, the aim is to highlight the types of experiments that produce scientifically useful conclusions. It is clear from numerous reports that proteome and transcriptome abundances are not sufficiently correlated to act as proxies for each other. The majority of this difference is rooted in fundamental biological regulation, and not measurement bias or platform-specific error. Thus we should not wrestle with the differences, but rather leverage them to elucidate the underlying phenomena. With this emerging access to global measurements of multiple -omes, what new biology can be explored?
Biological processes necessarily lead to complexity in abundance measurements
One of the first comparisons of mRNA and protein was performed by the Aebersold group in 1999 using Saccharomyces cerevisiae growing at mid-log phase [4]. This study was limited compared to the technology of today, with only 106 genes being examined. Their major conclusion succinctly stated that ‘[they] found that the correlation between mRNA and protein levels was insufficient to predict protein expression levels from quantitative mRNA data.’ Over the past 15 years, technological innovation has drastically improved the breadth of both mRNA and protein measurements, but this fundamental observation continues to be widely, though not universally, supported [5–7]. An extensive review of the literature of protein/transcript comparisons can be found in Vogel and Marcotte [8].
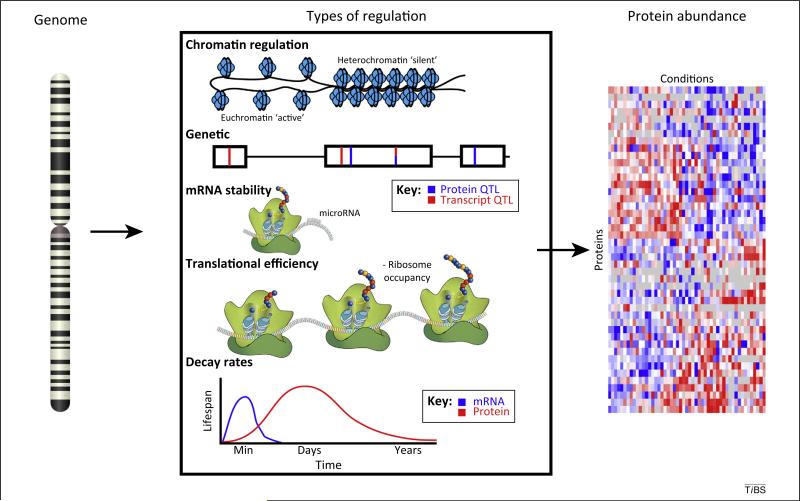
Fundamental biological processes control the information flow from genome to gene-product to functional output (Figure 1). It is now recognized that biological systems will regulate processes by modification, binding, concentration, and/or localization of nearly any biological molecule. In particular, protein abundance is regulated by a variety of complex mechanisms. By measuring mRNA abundance, only the early steps in a long chain of regulatory events are observed. The following exemplary studies have used multi-omic data to identify and characterize a variety of regulatory mechanisms. Using a cohort of 95 diverse individuals from the HapMap project to identify genetic variation that affects protein abundance, the Snyder group discovered that the loci controlling RNA expression (eQTLs) had only a 50% overlap with the loci controlling protein expression (pQTLs), highlighting distinct genetic regulatory sequences [9]. By coupling high-throughput sequencing of ribosome-protected transcripts to RNA-Seq experiments, Brar et al. teased apart the abundance of a transcript from the use of a transcript. In this genome-wide analysis they show that translational regulation is pervasive [10]. MicroRNAs are an additional specific mechanism used by cells to regulate protein synthesis. A global analysis found that micro-RNAs can affect protein abundance either through mRNA destabilization, which decreases mRNA abundance, or through translational repression, which does not alter mRNA abundance [11]. Another important factor in the differences between mRNA and protein abundance is the distinct synthesis and decay rates. Not only are these relative rates on different scales (the lifetime for an mRNA is minutes, the lifetime for a protein is hours to years), but the rates of synthesis or decay for mRNA and protein from a single gene are unrelated [12]. Finally, in a detailed study of the Prochlorococcus cell cycle, the Chisholm group characterized cycling proteins and transcripts by both phase and amplitude [13]. They concluded that ‘significant divergence between mRNA and protein levels in the relative timing and/or magnitude of abundance oscillations are the rule rather than the exception.’ Given these and numerous other regulatory mechanisms, we should not expect an easy correlation between protein and mRNA abundances.
Figure 1.
Diversity of regulation. The process of obtaining proteins from a genomic template is governed by many modalities of regulation, some of which are shown. Transcription can be regulated by the chromatin state of the DNA region containing the gene. Genetic regulation as inferred by quantitative trait locus (QTL) analysis identifies variable regions of a gene that affect the final abundance of the gene product. Regions that affect protein and transcript levels partially overlap but are not identical. mRNA stability can be affected by both intrinsic factors of the sequence itself, but also by extrinsic regulation such as through microRNAs. Translational efficiency denotes the amount of protein that is made from a transcript, and is affected by ribosome occupancy and other phenomena (e.g., codon usage). Decay rates for proteins are very different from those for mRNA, both as a global average and for a specific gene. Owing to the complexities of regulation, it is not currently possible to predict protein abundance from mRNA.
How to use multi-omic data for greatest insight?
As quantification technologies improve in coverage, accuracy, and cost it will become increasingly common to globally profile both protein and transcripts, which has great potential to elucidate novel biology. It is clear, however, that transcript measurements do not orthogonally validate proteome measurements and vice versa. In the utilitarian perspective, mRNA and protein abundance cannot proxy for one another, exactly as protein abundances of enzymes are not appropriate proxies for their enzymatic products. Given this knowledge and perspective, it is essential to consider the purpose for multi-omics experiments before one embarks.
For many experiments, collecting both data types will be valuable. In a generic experiment, one might ask what the effects of a stimulus are. To address this question, proteomics could be used to monitor phosphorylation signaling and dynamic cellular localization. Transcriptomics would elucidate the cohort of genes up or downregulated by activated transcription factors. Finally, proteomics would determine which transcripts become protein at which time, allowing researchers to see a multi-staged response to the stimulus, delineating between a rapid response and a long-term adaptation. Carefully considered hypotheses and experiments can use multi-omic data to drive biological insight. A large number of clinical and synthetic biology experiments are empowered by the recent decrease in cost of data generation and can now pursue genotype-to-phenotype hypotheses. Such experiments ask how changes in a gene (e.g., mutation or deletion) affect cellular function. By their nature, these are multi-omic questions. For clinical cohorts, these changes could be single-nucleotide polymorphisms or more complex genomic alterations. In synthetic biology, the focus is often on the insertion of new genes. The measurement of cellular function varies according to the hypothesis, but can be done with proteomics, metabolomics, or clinical meta-data.
A unique perspective on regulatory networks can be gained from multi-omic integration. Copy-number aberrations (CNA) measure the number of DNA segments corresponding to a particular gene in the genome. The normal copy number for diploid organisms is two; however, in a population there are typically some individuals with more or fewer copies of any given gene. As the copy number of a gene changes, it is expected that the abundance of its cognate protein changes accordingly. In genetics, this is termed a cis effect. This change in the abundance of the cognate protein may also affect the abundance of other interacting or downstream proteins, or a trans effect. In cancer, CNAs are often much more common and extreme. In a recent analysis of 90 colon tumors, Zhang et al. integrated proteomics data and genomic measurements of CNAs [14]. Through global correlation of these multi-omic data, they identified many CNAs affecting hundreds of proteins in trans. From this subset of CNAs they illuminated new driver mutations for colon cancer phenotypes. This integrative analysis identifies unique relationships in a regulatory network distinct from coexpression analysis, which finds groups of similarly behaving genes, but does not as clearly identify the drivers of the phenotype.
In systems biology, computational modeling methods can derive great insight from muti-omic integration. An exciting new pursuit is whole cell modeling, which accounts for all cellular processes and molecules [15]. This represents a significant expansion from initial metabolism-only models. To properly parameterize the model, proteome, metabolome, and transcriptome measurements are all utilized. Whole cell models offer incredible validation of predicted functions and relationships of molecules in all biological processes as well as presenting hypotheses for emergent properties of the cell. This level of characterization currently only exists for the most genome-reduced organisms. However, the recent increase in experimental data generation, particularly the elucidation of protein complexes and localization, will make such models feasible in the near future for many organisms.
As systems biology matures in its ability to both characterize and predict cellular functions, synthetic biology emerges as the tool to build and control cellular functions. In the myriad applications of synthetic biology it is essential to understand the complex regulatory relationships that govern molecular behavior. This understanding intrinsically comes from multi-omic analyses.
Finally, as computational biologists strive to integrate distinct data types, it is essential to develop a framework that understands and appreciates the differences between multi-omic data. This same appreciation is needed in the biological researchers who utilize the data. The needed task is not to determine which of the non-correlating data are ‘correct’; each is correctly measuring a different biomolecule. Rather, the task is to foster and utilize analytical paradigms that derive knowledge from multiple data types. We need a mindset that is truly integrative, and not simply correlative (Box 1).
Box 1. Outstanding questions.
What is data integration beyond correlation?
How do we educate new scientists to appreciate the diversity of -omics measurements?
How to determine which -omic data type is best to investigate a hypothesis?
When to generate multi-omic data?
Acknowledgments
The author thanks Karin Rodland, Josh Adkins, Jason McDermott, and Katrina Waters for critical reading of the manuscript, and Grant Fujimoto for assistance with images. S.H.P. is supported by an Early Career Award and the Pan-omics project from the US Department of Energy Genome Sciences Program and the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium (CPTAC, U24-CA-160019).
References
- 1.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 2.Ideker T, et al. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 3.Marguerat S, et al. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gygi SP, et al. Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Le Roch KG, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004;14:2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagaraj N, et al. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kislinger T, et al. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- 8.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu L, et al. Variation and genetic control of protein abundance in humans. Nature. 2013;499:79–82. doi: 10.1038/nature12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brar GA, et al. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 13.Waldbauer JR, et al. Transcriptome and proteome dynamics of a light–dark synchronized bacterial cell cycle. PloS ONE. 2012;7:e43432. doi: 10.1371/journal.pone.0043432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang B, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanghvi JC, et al. Accelerated discovery via a whole-cell model. Nat. Methods. 2013;10:1192–1195. doi: 10.1038/nmeth.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]