Abstract
Protein-ligand docking is a commonly used method for lead identification and refinement. While traditional structure-based docking methods represent the receptor as a rigid body, recent developments have been moving toward the inclusion of protein flexibility. Proteins exist in an inter-converting ensemble of conformational states, but effectively and efficiently searching the conformational space available to both the receptor and ligand remains a well-appreciated computational challenge. To this end, we have developed the Flexible CDOCKER method as an extension of the family of complete docking solutions available within CHARMM. This method integrates atomically detailed side chain flexibility with grid-based docking methods, maintaining efficiency while allowing the protein and ligand configurations to explore their conformational space simultaneously. This is in contrast to existing approaches that use induced-fit like sampling, such as Glide or Autodock, where the protein or the ligand space is sampled independently in an iterative fashion. Presented here are developments to the CHARMM docking methodology to incorporate receptor flexibility and improvements to the sampling protocol as demonstrated with re-docking trials on a subset of the CCDC/Astex set. These developments within CDOCKER achieve docking accuracy competitive with or exceeding the performance of other widely utilized docking programs.
Keywords: protein-ligand, sampling, in silico screening
Introduction
Structure-based protein-ligand docking is an important methodology in the repertoire of tools for drug discovery and design. This approach is often used to rapidly predict ligand orientation and affinity for virtual screening in lead identification or optimization applications.1,2 A plethora of docking software has become available over the years, beginning with DOCK and including other programs such as Autodock, Glide and ICM.3–6 There have also been developments toward improving the ability to predict affinities of possible lead molecules through improved knowledge-based scoring functions such as Drugscore, IT-Score, and KECSA.7–10
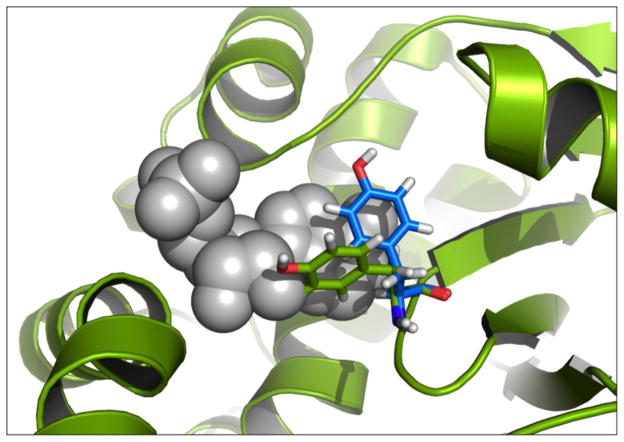
Traditionally, structure-based docking uses a rigid approximation of the receptor to reduce the number of degrees of freedom to be sampled and thus maintain computational efficiency. Speed in these calculations is desirable for lead discovery through virtual screening to achieve feasibility in screening the immense chemical space of small molecules. For example, the ZINC Database contains approximately 34 million purchasable compounds.11 Additionally, this type of rigid receptor model represents a reasonable approach in the context of the lock and key type understanding of ligand binding as is important in many protein-ligand binding mechanisms.12 However, with ever increasing structural and biophysical data available, the importance of protein flexibility has become more apparent.13–17 Proteins exist as an inter-converting ensemble of states, and the highest populated state in the apo protein is often not the dominant state when a ligand is present. This has been captured experimentally, where throughout a series of crystallographic structures of the same protein side chain conformations can vary substantially.17–20 For example, in alpha-Momorcharin (alpha-MMC) (Figure 1) there are different rotameric states for many of the side chains depending on the ligand in complex. Predicting the ligand conformation consistent with the experimental holo structure would not be successful when docking to the experimental apo structure with a rigid receptor docking method due to steric clashes.
Figure 1.
The importance of receptor flexibility has become more obvious with the increase of structural data. For instance, pictured here is an overlay of an apo (PDBID: 1AHC in green) and holo (PDBID: 1AHB in blue and grey) structure of alpha-Momorcharin. There is a steric clash between the ligand and the apo structure that would prevent rigid docking methods from predicted the crystallographic conformation.
Incorporation of protein flexibility has become more feasible due to advancements in computational resources as well as recent software development towards incorporating flexibility in docking.1,16,21,22 Existing methods try to balance computational cost and the enormous size of the configurational space that needs to be sampled through a variety of approaches. These approaches include sampling the ligand conformations relative to an ensemble of protein conformers, or representing the protein as a single averaged grid.23–25 Others take advantage of rotamer libraries to optimize the receptor after sampling the ligand on a rigid representation of the receptor, or use a soft representation of the protein, where terms for steric clash penalties are reduced.6,26–29 Though advances have been made in these areas, incorporation of receptor flexibility into docking remains at the forefront of challenges in the field.1,23,24
In this paper we present improvements to the sampling in the CHARMM based docking method to include receptor flexibility. Previously, CHARMM docking, or CDOCKER, achieved ligand sampling via simulated annealing on a rigid representation of the receptor, although genetic algorithm methods have also been explored in this context.29,30 In this setup, the protein is represented as a series of non-bonded grids, where the non-bonded interactions are softened to accommodate small differences in protein structure when the ligand is bound to facilitate ligand sampling.29,31 This representation works well for rigid pockets but may not be sufficient for a case like that portrayed in Figure 1. Nevertheless, the CDOCKER approach has served as a standard in the field for a number of years.28–30,32 To extend the capabilities and scope of CDOCKER to accommodate a wider variety of receptors, we have incorporated receptor flexibility by including explicit side chains while maintaining the rest of the receptor interactions using a grid representation. This allows the protein and ligand to sample configurational space simultaneously while having a minimal impact on the computational cost (SI Figure 1), which scales linearly with the number of explicit receptor atoms included in the sampling (SI Figure 2). In addition to incorporating receptor flexibility we have implemented a docking protocol that employs enhanced sampling molecular dynamics followed by minimization (MD+Minimization) utilizing self-guided Langevin dynamics, which further improves CDOCKER docking accuracy.33,34 These improvements to the CHARMM docking methods lead to docking accuracy that is competitive with other highly successful docking programs, such as Glide and Autodock.5,6
Methods
Setup of Protein-Ligand Test Set
For the docking trials described here a subset of 161 protein-ligand complexes from the CCDC/ASTEX set was used.35 All structures were obtained directly from the Protein Data Bank (see SI Table 1 for PDBIDs).36 The set was selected for direct comparison against results presented recently in publications from the Gohlke research group.37 From the full set examined by Gohlke and co-workers, the entries in complex with heme or those for which parameters were not available within the CHARMM CGENFF36 parameter library were removed.38–40 Single complexes were extracted from the downloaded PDBs using the MMTSB Toolset and all crystallographic waters and small molecules not representing the target ligand were removed.41
Only ions found in the region of the binding site were retained as part of the receptor.41 The hydrogen atoms for the receptors and ligands were prepared independently of one another. The protonation states of the titratable residues in the protein were predicted using PROPKA version 3.1.42 The hydrogen atoms were built using Babel version 2.3.2 for both the protein and ligand of each entry.43 Ligand parameters were obtained using MATCH, which assigns atom types, charges and force field parameters through a chemical pattern-matching engine.44 Symmetric models of the ligand, to assure uniquely named atoms that are of the same type do not contribute to higher RMSD, were generated using an in-house developed Perl script, mapPDB. This script is now in the MMTSB Toolset for use in root mean square deviation (RMSD) calculations.41
Docking Protocol: Initialization of ligands and receptor grids
Prior to docking the small molecule into the receptor the system must be initialized by generating parameters, building the grid and generating initial configurations of the ligand. We developed a Perl script as an automated tool to interface user input with CHARMM for docking initialization. During initialization the receptor representation may be set up as either rigid or flexible. The ligand parameters are generated using MATCH at the beginning of docking setup. The grid representation of the protein employs a set of van der Waals and electrostatic grid-based potentials that are used in the sampling and ranking steps of our docking protocol. The van der Waals grids are built from a set of test particles with 20 different van der Waals radii that are centered on the most highly populated radii found in the CHARMM general force-field (CGenFF).40 This grid is calculated with a mesh spacing of 1Å spanning 30Å centered at the specified center of mass for the target-binding pocket. In these trials the center of mass of the crystallographic conformation of the ligand was used. This grid size is sufficiently large to allow for a ligand to sample different rigid body rotations at the target binding site without a high energetic penalty. The “softness” of the grid can be varied for each non-bonded interaction: van der Waals, electrostatic attraction and repulsion, and follow the general form as given in equation 1.
| (1) |
Where is the energy of the regular non-bonded potential at a given distance rij for either electrostatic or van der Waals.28 Outlined in Table 1 are three different CDOCKER sampling protocols, each utilizing alternative grid representations with differing Emax values. The a and b coefficients are extracted from equations that express the potential and forces at the switching distance. The switching to the soft core potential occurs at the Rcut cutoff distance and is defined in equation 2.
Table 1.
The parameters that determine the softness of the grid potential, all Emax values are in units of kcal/mol. The methods outlined here use a either a single grid of different sampling methods, simulated annealing (SA-1 Grid) or molecular dynamics followed by minimization (MD+Minimization), or a two grids with a simulated annealing protocol as outlined by Wu, et. al.28
| Protocol | Emax (vdW) | Emax (attractive) | Emax (repulsive) |
|---|---|---|---|
| SA-1 Grid | 15.0 | −120.0 | −2.0 |
| SA-2 Grids | 0.6 | −0.4 | 8.0 |
| 3.0 | −20.0 | 40.0 | |
| MD+Minimization | 15.0 | −120.0 | −2.0 |
| (2) |
The coefficients “a” and “b” are chosen so that the energy and force terms agree with Rcut. For all other components, the usual interactions are used. Grids were generated with a distance dependent dielectric with the relative dielectric constant, ε, equal to 3 as previously outlined by Vieth et al.29 The grid soft-core potentials for the simulated annealing protocol with two grids were previously outlined in Wu, et. al. and are called here “SA-2 Grids”.28 The protocol named here as “SA-1 Grid” follows the same docking protocol as its two grid counterpart but on a single, harder grid. Our newly implemented molecular dynamics (MD) followed by minimization protocol, “MD+Minimization”, which is described later in this section, also uses a single receptor grid.
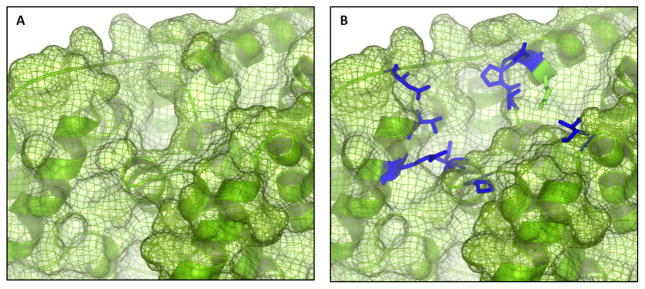
In previous versions of CDOCKER, the grid was generated with the entire protein held fixed, creating a rigid receptor, and will be referred to as rigid-docking (Figure 2A). To incorporate receptor flexibility, side chains selected around the target binding-site are removed prior to grid generation. These residues are kept as explicit side chains during the sampling step and they are not present in the grid representation of the receptor (Figure 2B). The backbone atoms of these selected explicit residues are held fixed during all sampling. However, the side chains of the explicit residues are allowed to sample configuration space simultaneously with the ligand, and both ligand and side chains interact with the grid representation of the receptor. The residues that remain flexible can be user specified or the residues may be selected to be within a specified cutoff distance from the reference small-molecule, 3Å in these docking trials. These residues allow for changes in the surface of the binding site to better accommodate the small molecule. Incorporating explicit side chains allows for receptor flexibility while adding a minimal number of degrees of freedom to the sampling calculations. Additionally, side chain conformational sampling should provide sufficient receptor flexibility to overcome the clashes between the apo receptor structure and the small molecule crystallographic conformation, as illustrated in Figure 1. The targeting of large-scale conformational changes involving the receptor backbone that occur upon ligand binding in some situations would need to be addressed in some other manner, such as ensemble docking, and is beyond the scope of this flexible receptor docking approach.22,25,45–47
Figure 2.
Rigid CDOCKER represents the receptor as a series of non-bonded grids (A). To include receptor flexibility, residues around a target binding site are kept as explicit side chains (B) allowing them to sample conformational space with the ligand simultaneously in the presence of the grids.
The initial configurations of the small molecule are generated through rigid body rotations and random rotations around the rotatable bonds. The energy for each configuration is calculated on the grid and all configurations below an energy cutoff of 1,000 kcal/mol are clustered. This removes the extremely high-energy conformations and also yields a structurally diverse set of starting conformations for docking. The clustering tool within the MMTSB toolset was used for the k-means based clustering of the configurations. The cutoff RMSD for the clustering was determined so that at least 50 clusters were generated. The lowest energy conformation from each of the lowest energy clusters was selected for the sampling steps. This protocol provides a diverse set of ligand conformations and also represents an ensemble of relatively low energy starting configurations from which to initialize sampling. Each docking trial generates 50 ligand conformations starting from 50 unique, low energy conformations of the small molecule.
Docking Protocol: Sampling
Flexible CDOCKER was implemented with two different sampling schemes; one uses a simulated annealing protocol while the other uses a series of short molecular dynamics simulations followed by minimization (MD+Minimization). Both implementations also utilize self-guided Langevin dynamics (SGLD) to accelerate the sampling.33,34 SGLD uses an average of local velocities to calculate a guiding force to accelerate dynamics of lower frequency motions. There are two key parameters that determine how aggressive the acceleration to the system is: the average time and the guiding factor. The larger the average time, 0.2ps in these simulations, the slower the motion that is enhanced. The guiding factor is related to the guiding force and governed by the target self-guiding temperature, 700K for these simulations. The larger the guiding factor the larger the energy barriers that can be overcome.
For the re-docking trials using SA the protocol as previously outlined by Wu, et al. was used.28 For details on the temperatures and lengths of phases for the SA protocols see SI Table 2. SGLD is a feature available for use with all the sampling protocols. However, for these trials was not used in the simulated annealing protocol, as it tends to cause the ligand to move out of the pocket because the sampling becomes too aggressive. SGLD was used only in the MD+Minimization protocol for the docking trials. Each small-molecule starting configuration was subjected to five repetitions of the simulated annealing protocol, where the end result of one step provided the starting configuration for the next. This protocol allows the ligand, and in the case of flexible receptor docking the side chains, to sample different conformations and rotameric states.
The MD+Miniminization protocol was inspired by the successful ICM implementation of Monte Carlo steps followed by minimization, following the concept to guide the ligand down the energy landscape towards a low energy state corresponding to the crystallographic configuration.4 Each ligand configuration from the clustering results is subjected to 20 rounds of 500 steps of molecular dynamics at 300K with SGLD, followed by 500 steps of Adopted Basis Newton-Raphson (ABNR) minimization. Before the start of each round an energy comparison between the current and the previous ligand configuration is made, and the lowest energy configuration is selected to continue the sampling, driving the system towards lower energy states. Each MD step is 2fs of enhanced sampling on the grid representation of the receptor. SGLD sampling can be quite aggressive, and when the receptor is in a grid representation the ligand can leave the target binding site region. We found that in order to prevent the ligand from being ejected from the binding site fewer than 2,000 MD steps must be used. Additionally, to achieve high acceptance rates between rounds of sampling the number of MD steps fewer than 1,000 steps should be used.
Each docking trial for the MD+Minimization and the simulated annealing protocol was limited to 2.6 × 106 energy evaluations to allow the fairest comparison with the benchmark studies by Gohlke and co-workers.37 The docking trials were repeated 10 times, with the same input structures for the protein and ligand. However, each trial generated a new set of random orientations and conformations of the small molecule followed by sampling on the grid representation of the receptor.
Docking Protocol: Ranking and RMSD calculation
As the focus of this work is to develop a new docking search strategy, the energy evaluation on the grid representation of the receptor is used for sorting the resulting configurations from the sampling step. The RMSD between the docked configurations and their respective crystallographic configurations was calculated with consideration for the symmetry of the small molecule. A configuration of the ligand is considered “docked” when it has an RMSD less than 2Å. This metric, as is the convention, will be used to assess docking accuracy.48
Molecular dynamics simulations for investigating side chain sampling
To investigate the ability of side chains to sample alternative rotameric states we carried out explicit solvent simulations of a few selected receptors as well as performed sampling in the presence of the grid. Simulations of two structures from the MMC alpha protein (PDBIDs: 1AHC and 1AHB) were run for comparison. The explicit solvent MD simulations were carried out using a GPU capable CHARMM version c39a2 with an OpenMM interface using OpenMM version 5.2.49–51 The simulations are 5ns in length using a 2fs time step and were run at constant pressure and temperature, 300K, with the CHARMM27 force field.38,39 Langevin dynamics was used to provide the thermal heat bath with a friction coefficient of 10 ps−1. Particle mesh Ewald with a non-bonded cutoff of 9Å was used with a FFT grid size of 72Å. SHAKE was applied to the hydrogen atoms and the parameters for the small molecules were obtained with MATCH using CGenFF.40,44 The simulations run on the grid were also 5ns in duration with flexible residues selected within a 3Å cutoff of the ligand. This is the same as the cutoff as used for the docking trials. The grid parameters used were identical to those described in the MD+Minimization protocol outlined in Table 1, as this is the “hardest” grid and would be the most constraining. To investigate side chain sampling, both the explicit solvent and grid-based simulations were run in the absence of the ligand. Analysis of the χ angles was performed using CHARMM.
Results & Discussion
Grid-embedded explicit side chains sample well the side chain rotameric states
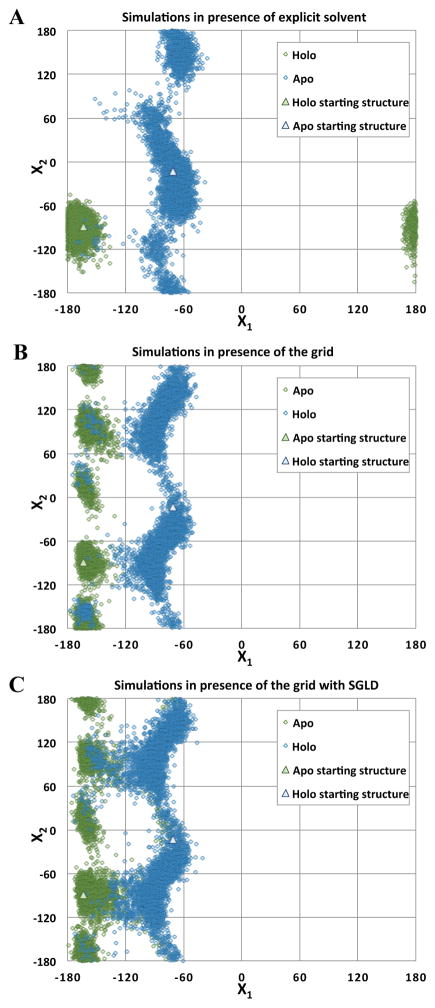
To investigate the side chain sampling in the presence of the grid we compared the sampling of the side chains to that of an explicit solvent simulation. It was important to ensure that the side chains were able to sample alternate conformations and not be constrained by the surrounding grid. As has been noted in Figure 1, with the alpha MMC protein, a clash would occur between the tyrosine and the ligand in a cross-docking trial, and it is thus critical to ensure that both side chain conformers are attainable during the course of a simulation on the grid-embedded side chain representation of the receptor. In Figure 3, we illustrate the χ1 and χ2 angles sampled in both explicit solvent and grid based simulations for the essential tyrosine residue noted in Figure 1. From the displayed configurational history maps we see more sampling on the grid than in the all-atom simulation. The triangles represent the angles observed in the starting structures. Indeed, independent of the starting conformation of the side chain, both conformations observed in the apo and holo-crystal structures are sampled. The grid-based simulations are able to sample the various conformations of the protein side chains without the enhanced sampling that is implemented in the docking protocol. As to be expected, even more sampling of alternate side chain rotameric states occurs with the SGLD enhanced sampling. This sampling is important to consider because most applications of the docking methodology will be in cross-docking experiments one is trying to either improve the potency of a lead compound or identify a new lead compound.
Figure 3.
Sampling of χ angles during molecular dynamics simulation of a single tyrosine of alpha-Momorcharin protein in the presence of explicit solvent (A) or the grid (B) with enhanced sampling (C). The simulations started from the apo structure (PDBID: 1AHC) begin at the green triangle and sample the rotameric states shown in green circles. Alternatively, the blue triangle was the starting rotameric state for the holo structure (PDBID: 1AHB) and the states sampled are shown in blue circles.
Incorporation of SGLD MD and minimization in sampling protocol improves docking accuracy
CDOCKER originally used a simulated annealing protocol, where the ligand was heated in the presence of the grid, allowed to sample at the higher temperature and then cooled. This would push the ligand out of minima and, ideally, into new ones, occasionally sampling the desired low-energy, docked conformation. While such an approach has worked well in many situations, for a single docking trial, a native-like pose (closer than 2Å) was sampled in less than 60% of the complexes, and this is without scoring the conformations (see Table 2). To improve this step we implemented a MD and minimization approach, where a single starting ligand conformer is subjected to a short round of enhanced sampling SGLD molecular dynamics simulation and is followed up with a round of minimization. Each subsequent round of MD and minimization is started from either the conformation at the beginning or end of the previous round of sampling; whichever conformation is the lowest in energy. This protocol aims to reduce the time spent in high energy, irrelevant states that may be sampled during a simulated annealing procedure. This method improves docking accuracy for a single docking trial on a subset of 161 protein-ligand complexes from the CCDC/ASTEX set by about 16% when flexible side chain sampling is employed (Table 2).
Table 2.
Results from a single docking trial of 161 complexes from the CCDC/ASTEX set.
| Rigid CDOCKER percent found | Flexible CDOCKER percent found | |
|---|---|---|
| Simulated Annealing (SA - 1 Grid) | 54.50% | 56.10% |
| MD + Minimization | 56.10% | 72.40% |
Docking to flexible receptor greatly improves re-docking accuracy
Re-docking trials are commonly used to assess the quality of a docking method. Re-docking takes a ligand and docks it into the same receptor structure with which it was originally crystallized. Presented here are re-docking trials of a subset of the CCDC/Astex set to compare various implementations of sampling and receptor flexibility within our novel extension of the CHARMM docking methodology.
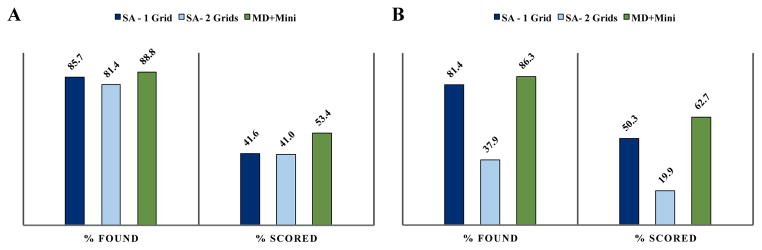
The docking accuracy results for the re-docking trials are presented in Figure 4, and include a percentage found and percentage scored. When a conformation that is “docked”, less than 2Å RMSD from the crystallographic conformation in the heavy atoms of the ligand, exists in the docking output it is considered “found”. If a docked conformation is chosen to be the lowest energy conformation it is considered “scored”, where the energy is that of the grid interacting with the ligand and, in the case of flexible docking, explicit receptor side chains. Results are presented for 10 docking trials. To be included in the “found” percentage, at least one structure for a given complex in at least a single trial must be identified as docked. However, to be “scored” it must be the lowest predicted energy of all of the trials. For the rigid receptor docking, finding at least one docked conformation across all of the sampling methods occurred with high frequency, all greater than 81%. The docked conformation is selected out of the results at approximately half the frequency that the docked conformation is found. The MD and minimization (MD+Minimization) protocol improves docking accuracy in both finding and identifying a docked conformation with 41% scored with simulated annealing methods and 53.4% when MD+Minimization is used. For the single grid, the incorporation of flexibility in both simulated annealing and the MD and minimization protocols improves the scoring step by 8.7 and 9.3 percentage points respectively. This is likely due to some reorganization of the protein side chains to better accommodate the ligand and improve the overall energy. The two grid simulated annealing (SA-2 Grids) protocol suffers greatly from the incorporation of the flexible side chains with a loss in the docked conformations found of 43.5 percentage points, which is also reflected in the scoring of 19.9%. Both of the grids in this SA-2 Grids method are much softer than those used in the other protocols. While this allows for sampling of side chain and ligand conformations without large energetic penalties, it also causes loss of sensitivity in flexible receptor docking. It should be noted that this reduction of docking accuracy with the inclusion of flexible side chains is not unlikely for a re-docking trial, where additional receptor degrees of freedom added allow for sampling away from the crystallographic conformation predefined in the rigid docking case. However, the loss of docking accuracy seen in the SA-2 Grids case was not observed in the MD+Minimization or the SA-1 Grid protocol, which have “harder” grids and larger penalties for high energy conformations. In general, the addition of the flexible receptor further improves the percent “scored” and does not diminish the percent “found” for the single grid sampling protocols. The MD+Minimization protocol improves re-docking accuracy over the simulated annealing protocols, an improvement of 11.5 and 12.4 percentage points for the rigid and flexible docking respectively.
Figure 4.
Re-docking trial results by metrics of finding at least one conformation in the ten trials that is less than 2Å RMSD from crystallographic pose (found) and the lowest energy conformation is docked (scored). Rigid CDOCKER method results are shown in panel A and flexible CDOCKER results are in panel B.
Flexible CDOCKER performs comparably with current docking software
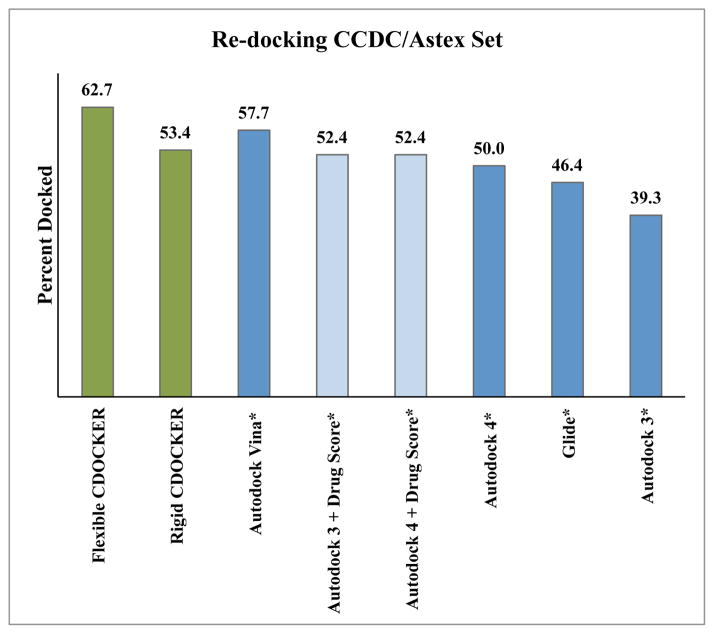
Numerous docking methodologies have been developed in recent years and many useful comparisons of the re-docking ability of these software packages have been published.37,52–59 Recently, the Gohlke research group published a comparative study of current docking programs for re-docking of ligand-receptor pairs in the CCDC/Astex dataset.35 Taking the results reported by Krueger, et al. for the subset of complexes from the CCDC/Astex set reported above, comparisons between the popular docking methods and CDOCKER can be made (Figure 5).37 In addition to reporting the Autodock results, these researchers reported Autodock results that were rescored using a knowledge based scoring function, DrugScore.37 Comparing the best performing CDOCKER, the flexible receptor with the MD+minimization sampling, and the other docking methods that were reported, CDOCKER is very competitive with the other methods. Rigid CDOCKER with the MD+Minimization protocol is competitive, with 53.4% of the complexes scored, which is on par with Autodock 3 and Autodock 4 rescored with DrugScore, which show 52.4%, docked. Only Autodock Vina and Flexible CDOCKER outperform Rigid CDOCKER with this subset of CCDC/Astex set. Flexible receptor CDOCKER outperforms the other docking programs, selecting the docked conformation for 62.7% of the complexes, while the next most successful docking software reported, Autodock Vina, has a success rate of 57.7%.
Figure 5.
Docking accuracy comparison of widely used docking programs and Flexible CDOCKER and CDOCKER for a subset of the CCDC/Astex. Docked is defined as the lowest predicted energy conformation is within 2Å RMSD of the crystallographic conformation. Results for the non-CDOCKER based methods are taken from the published results of Krueger et al, and are marked with an asterix.37
Flexible CDOCKER improves cross-docking accuracy: a case study
The primary motivation for incorporating receptor flexibility into docking was to target a receptor structure that requires side chain conformational flexibility to accommodate its ligand (Figure 1). As a case study, we performed re-docking and cross-docking experiments using the structures from Figure 1, utilizing the MD+Minimization implementation of CDOCKER. For proper comparison identical starting conformations of the small molecule were used for the sampling step of the docking trial with either rigid or flexible receptor representations and for both re-docking and cross-docking trials. Using the same small molecule conformations allows for investigation of the benefits of the sampling without concern for the variability in initial conditions. The re-docking trial docked the small-molecule to the receptor conformation from which it was derived (PDBID: 1AHB). The cross-docking trial docked the small-molecule to the experimentally determined apo receptor conformation (PDBID: 1AHC). The results for these trials are presented in Table 3, and show the percentage of the resulting conformations, from the ten docking trials, that have a RMSD of less than or equal to 2Å, as well as the RMSD of the lowest predicted free energy conformation. The percentages docked shown in Table 3 are different from the percentages shown in Figure 5, which shows the percent of complexes that the lowest predicted energy conformation is docked. Consistent with the results observed for the CCDC/Astex set, rigid and flexible CDOCKER sample docked conformations with similar frequency, 10.6% and 10.2% of the resulting conformations (500 total) docked for rigid and flexible CDOCKER respectively for the re-docking trial. Flexible CDOCKER is superior in identifying the docked conformation; selecting a conformation with a 1.69Å RMSD while the rigid version’s low energy conformation had an RMSD of 6.99Å. As expected, there is a drop in the frequency of sampling native-like poses for the cross-docking trial. We observed docked conformations with a frequency of 5.6% for rigid CDOCKER and 7.2% for flexible CDOCKER of 500 total conformations. Interestingly, in the cross-docking trial rigid CDOCKER was able to sample a docked conformation of the small-molecule 5.6% of the time. This suggests the softness of the grid mimics some aspects of receptor flexibility and enables the sampling of docked conformations. However, for scoring flexible CDOCKER was superior, identifying a docked conformation of 1.42Å RMSD compared to 5.0Å RMSD for the rigid implementation. Upon visual inspection of the predicted lowest energy conformation identified by Flexible CDOCKER, the dihedral angle for the tyrosine shown in Figure 1 remains closer to that of the apo crystallographic structure compared to the holo structure (SI Figure 1). This suggests that a small shift in dihedral angle is needed to achieve a low energy docked conformation and it is possible that this is why the rigid grid with a soft-core potential is capable of sampling docked conformations. Flexible CDOCKER, in this test case, has no loss in sampling for the re-docking trial despite an increase in the number of degrees of freedom. It also shows improved sampling compared to the rigid implementation for the cross-docking trial. Flexible CDOCKER is superior to the rigid implementation in ranking docked conformations, leading to improved docking accuracy.
Table 3.
| Docking Method | Percent of all conformations docked | Low Energy RMSD(Å) | |
|---|---|---|---|
| Re-docking | Rigid CDOCKER | 10.6 | 6.99 |
| Flexible CDOCKER | 10.2 | 1.69 | |
|
| |||
| Cross-docking | Rigid CDOCKER | 5.6 | 5.00 |
| Flexible CDOCKER | 7.2 | 1.42 | |
Flexible CDOCKER cross-docking HIV Reverse Transcriptase
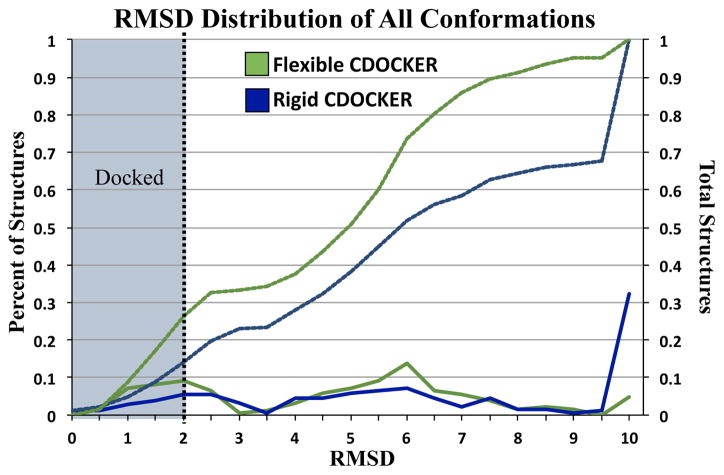
Flexible CDOCKER is capable sampling and scoring a docked conformation in a cross-docking test case of a single receptor. Cross-docking trials are more challenging than re-docking trials, as the receptor is not in the optimal configuration to interact with the small molecule. Results from the CSAR Benchmark Exercise 2011–2012, hosted by the Carlson research group, demonstrated a wide range of docking accuracy for different targets.53 The overall docking accuracy for the different receptors found that 33% of the top scoring results were docked, however some complexes were as low as 5.4% scored.53 As a challenge for the newly implemented CDOCKER methodology we performed a cross-docking trial of 131 unique complexes of HIV reverse transcriptase (SI Table 3). Consistent with previous results, the flexible and rigid receptor implementations had similar ability to sample docked conformations with 47.3% and 48.1% of the complexes docked respectively. Again, Flexible CDOCKER outperformed the rigid implementation in its ability to score the docked conformation as lowest in energy, with 19.1% docked compared to 10.7% for rigid. These docking percentages are low compared to redocking trials, which is to be expected, as the receptor structure is not in an optimal conformation for ligand binding. Histograms of the low energy conformations demonstrate the improvement of docking success with Flexible CDOCKER over rigid CDOCKER, as illustrated in Figure 6 left-side y-axis. It is especially apparent that the inclusion of flexible side chains improves the sampling of docked conformations when considering the total structures docked, which is plotted in dashed lines along the right-side y-axis of Figure 6. For Flexible CDOCKER, 26.2% of all the conformations sampled were docked while the rigid implementation only achieved 14.2% docked.
Figure 6.
Histogram of RMSD (solid lines) of all low energy conformations from the HIV reverse transcriptase for both rigid (blue) and flexible (green) CDOCKER. The cumulative sums (dashed lines) of the number of conformations for rigid (blue) and flexible (green) CDOCKER for a given RMSD show flexible side chains inclusion leads to a greater number of docked conformations sampled.
While adding the receptor flexibility improves the docking accuracy, it is still fairly low. Recent results published for this crossdocking set using DOCK 6 show 66.7% of the 61 overlapping complexes were scored (SI Table 4).6 The initial placement of the small molecule for CDOCKER is somewhat naïve, generating many random ligand conformations targeting a specific binding site, neglecting much information concerning the receptor surface, and perhaps improvements to the initial placement would lead to increased docking accuracy. DOCK 6 for example, uses pre-computed site points for excluded volume and chemical matching for initial ligand placement. There was an overlap of 61 complexes, while the remaining 70 complexes were considered non-viable pairings by Allen et al. A non-viable pairing was defined as a ligand minimized in a receptor structure that had an RMSD of greater than 2Å.6 For the complexes not attempted by Allen et al, Flexible CDOCKER had about twice the docking accuracy as the rigid implementation with 14.3% and 7.1%, respectively, scored and 32.9% of the complexes were found by both methods (SI Table 4). CDOCKER was able to find docked conformations of these more challenging complexes. However, many of the docked conformations that are being found by the docking protocol are not being scored as low energy conformations. This suggests a need for improved methods of ranking ligand conformations.
Conclusions
Incorporating receptor flexibility into docking methods is an essential step in more accurately modeling protein-ligand interactions, however, maintaining efficiency requires this to be done in a limited manner. Flexible CDOCKER represents the majority of the receptor as a rigid entity, representing side chains around a target binding pocket explicitly to minimize computational costs (Figure 1). When integrated with a grid representation of the remaining receptor that is softened with respect to its interactions one finds that the sampling of flexible side chains is maintained similarly to what is observed with conventional solvated molecular dynamics. Compared to the previously reported rigid receptor CDOCKER method, the inclusion of flexibility with a new MD+Minimization sampling protocol leads to improvements in docking accuracy, particularly in the scoring step of the docking process.28 This approach to incorporating receptor flexibility does not lead to loss of docking accuracy in re-docking trials over the rigid implementation, as may occur when including additional degrees of freedom. The computational cost of including the receptor flexibility increases linearly with the number of atoms (SI Figure 2). In a test case of cross-docking, the flexible receptor version of CDOCKER was superior to the rigid implementation in both sampling and scoring of docked conformations of the receptor’s paired small-molecule. Finally, comparing our new docking protocols to results reported by the Gohlke research group shows that Flexible CDOCKER outperforms other widely used docking methodologies in re-docking trials of the CCDC/Astex clean set.37
Supplementary Material
Acknowledgments
We wish to acknowledge H. Gohlke for sharing data from their published work. Support for this work is from the NIH through GM037554.
Contributor Information
Jessica K. Gagnon, Email: jcceditor@umich.edu, Department of Chemistry, University of Michigan, Ann Arbor, Michigan
Sean M. Law, Department of Chemistry, University of Michigan, Ann Arbor, Michigan
Charles L. Brooks, III, Department of Chemistry, University of Michigan, Ann Arbor, Michigan, Fax: 734-647-1604.
References
- 1.Borhani DW, Shaw DE. J Comp-Aided Mol Des. 2012;26(1):15–26. doi: 10.1007/s10822-011-9517-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jorgensen WL. Science. 2004;303(5665):1813–1818. doi: 10.1126/science.1096361. [DOI] [PubMed] [Google Scholar]
- 3.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. J Med Chem. 2004;47(7):1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 4.Neves MAC, Totrov M, Abagyan R. J Comp-Aided Mol Des. 2012;26(6):675–686. doi: 10.1007/s10822-012-9547-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trott O, Olson AJ. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allen WJ, Balius TE, Mukherjee S, Brozell SR, Moustakas DT, Lang PT, Case DA, Kuntz ID, Rizzo RC. J Comput Chem. 2015;36(15):1132–1156. doi: 10.1002/jcc.23905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang SY, Zou X. J Comput Chem. 2006;27(15):1866–1875. doi: 10.1002/jcc.20504. [DOI] [PubMed] [Google Scholar]
- 8.Liu J, Wang R. J Chem Inf Modl. 2015;55(3):475–482. doi: 10.1021/ci500731a. [DOI] [PubMed] [Google Scholar]
- 9.Velec HFG, Gohlke H, Klebe G. J Med Chem. 2005;48(20):6296–6303. doi: 10.1021/jm050436v. [DOI] [PubMed] [Google Scholar]
- 10.Zheng Z, Merz KM., Jr J Chem Inf Modl. 2013;53(5):1073–1083. doi: 10.1021/ci300619x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irwin JJ, Sterling T, Mysinger MM, Bolstad ES, Coleman RG. J Chem Inf Modl. 2012;52(7):1757–1768. doi: 10.1021/ci3001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jorgensen WL. Science. 1991;254(5034):954–955. doi: 10.1126/science.1719636. [DOI] [PubMed] [Google Scholar]
- 13.Alvarez-Garcia D, Barril X. J Chem Theor Comp. 2014;10(6):2608–2614. doi: 10.1021/ct500182z. [DOI] [PubMed] [Google Scholar]
- 14.Baron R, McCammon JA. Ann Rev Phys Chem. 2013;64:151–175. doi: 10.1146/annurev-physchem-040412-110047. [DOI] [PubMed] [Google Scholar]
- 15.Carlson HA. Curr Opin Chem Biol. 2002;6(4):447–452. doi: 10.1016/s1367-5931(02)00341-1. [DOI] [PubMed] [Google Scholar]
- 16.Cozzini P, Kellogg GE, Spyrakis F, Abraham DJ, Costantino G, Emerson A, Fanelli F, Gohlke H, Kuhn LA, Morris GM, Orozco M, Pertinhez TA, Rizzi M, Sotriffer CA. J Med Chem. 2008;51(20):6237–6255. doi: 10.1021/jm800562d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zavodszky MI, Kuhn LA. Prot Sci. 2005;14(4):1104–1114. doi: 10.1110/ps.041153605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bowman GR, Geissler PL. J Phys Chem B. 2014;118(24):6417–6423. doi: 10.1021/jp4105823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teague SJ. Nature Rev Drug Disc. 2003;2(7):527–541. doi: 10.1038/nrd1129. [DOI] [PubMed] [Google Scholar]
- 20.Kuhn LA. Strength in flexibility: Modeling side-chain conformational change in docking and screening. 2008 [Google Scholar]
- 21.Durrant JD, McCammon JA. Curr Opin Pharm. 2010;10(6):770–774. doi: 10.1016/j.coph.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lexa KW, Carlson HA. Quart Rev Biophys. 2012;45(3):301–343. doi: 10.1017/S0033583512000066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carlson HA, McCammon JA. Molec Pharmaco. 2000;57(2):213–218. [PubMed] [Google Scholar]
- 24.Sousa SF, Fernandes PA, Ramos MJ. Proteins. 2006;65(1):15–26. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- 25.Miranker A, Karplus M. Proteins. 1991;11(1):29–34. doi: 10.1002/prot.340110104. [DOI] [PubMed] [Google Scholar]
- 26.Armen RS, Chen J, Brooks CL., III J Chem Theor Comp. 2009;5(10):2909–2923. doi: 10.1021/ct900262t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meiler J, Baker D. Proteins. 2006;65(3):538–548. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- 28.Wu GS, Robertson DH, Brooks CL, III, Vieth M. J Comput Chem. 2003;24(13):1549–1562. doi: 10.1002/jcc.10306. [DOI] [PubMed] [Google Scholar]
- 29.Vieth M, Hirst JD, Dominy BN, Daigler H, Brooks CL., III J Comput Chem. 1998;19(14):1623–1631. [Google Scholar]
- 30.Vieth M, Hirst JD, Kolinski A, Brooks CL., III J Comput Chem. 1998;19(14):1612–1622. [Google Scholar]
- 31.Jiang F, Kim SH. J Molec Biol. 1991;219(1):79–102. doi: 10.1016/0022-2836(91)90859-5. [DOI] [PubMed] [Google Scholar]
- 32.Roche O, Kiyama R, Brooks CL., III J Med Chem. 2001;44(22):3592–3598. doi: 10.1021/jm000467k. [DOI] [PubMed] [Google Scholar]
- 33.Wu X, Brooks BR. J Chem Phys. 2011;134(13) doi: 10.1063/1.3574397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu XW, Brooks BR. Chem Phys Lett. 2003;381(3–4):512–518. [Google Scholar]
- 35.Nissink JWM, Murray C, Hartshorn M, Verdonk ML, Cole JC, Taylor R. Proteins. 2002;49(4):457–471. doi: 10.1002/prot.10232. [DOI] [PubMed] [Google Scholar]
- 36.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucl Acid Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krueger DM, Jessen G, Gohlke H. J Chem Inf Modl. 2012;52(11):2807–2811. doi: 10.1021/ci3003599. [DOI] [PubMed] [Google Scholar]
- 38.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 39.Mackerell AD, Feig M, Brooks CL., III J Comput Chem. 2004;25(11):1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 40.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, MacKerell AD., Jr J Comput Chem. 2010;31(4):671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feig M, Karanicolas J, Brooks CL., III J Molec Graph Modl. 2004;22(5):377–395. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Olsson MHM, Sondergaard CR, Rostkowski M, Jensen JH. J Chem Theor Comp. 2011;7(2):525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 43.O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. J Cheminfo. 2011:3. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yesselman JD, Price DJ, Knight JL, Brooks CL., III J Comput Chem. 2012;33(2):189–202. doi: 10.1002/jcc.21963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bowman AL, Nikolovska-Coleska Z, Zhong H, Wang S, Carlson HA. J Amer Chem Soc. 2007;129(42):12809–12814. doi: 10.1021/ja073687x. [DOI] [PubMed] [Google Scholar]
- 46.Lorber DM, Shoichet BK. Prot Sci. 1998;7(4):938–950. doi: 10.1002/pro.5560070411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zavodszky MI, Ming L, Thorpe MF, Day AR, Kuhn LA. Proteins. 2004;57(2):243–261. doi: 10.1002/prot.20179. [DOI] [PubMed] [Google Scholar]
- 48.Fleischman SH, Brooks CL., III Proteins. 1990;7(1):52–61. doi: 10.1002/prot.340070106. [DOI] [PubMed] [Google Scholar]
- 49.Brooks BR, Brooks CL, III, Mackerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4(2):187–217. [Google Scholar]
- 51.Eastman P, Friedrichs MS, Chodera JD, Radmer RJ, Bruns CM, Ku JP, Beauchamp KA, Lane TJ, Wang LP, Shukla D, Tye T, Houston M, Stich T, Klein C, Shirts MR, Pande VS. J Chem Theor Comp. 2013;9(1):461–469. doi: 10.1021/ct300857j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bursulaya BD, Totrov M, Abagyan R, Brooks CL., III J Comp-Aided Mol Des. 2003;17(11):755–763. doi: 10.1023/b:jcam.0000017496.76572.6f. [DOI] [PubMed] [Google Scholar]
- 53.Damm-Ganamet KL, Smith RD, Dunbar JB, Jr, Stuckey JA, Carlson HA. J Chem Inf Modl. 2013;53(8):1853–1870. doi: 10.1021/ci400025f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunbar JB, Jr, Smith RD, Yang CY, Ung PMU, Lexa KW, Khazanov NA, Stuckey JA, Wang S, Carlson HA. J Chem Inf Modl. 2011;51(9):2036–2046. doi: 10.1021/ci200082t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guthrie JP. J Phys Chem B. 2009;113(14):4501–4507. doi: 10.1021/jp806724u. [DOI] [PubMed] [Google Scholar]
- 56.Kim R, Skolnick J. J Comput Chem. 2008;29(8):1316–1331. doi: 10.1002/jcc.20893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skillman AG, Geballe MT, Nicholls A. J Comp-Aided Mol Des. 2010;24(4):257–258. doi: 10.1007/s10822-010-9358-0. [DOI] [PubMed] [Google Scholar]
- 58.Smith RD, Dunbar JB, Jr, Ung PMU, Esposito EX, Yang CY, Wang S, Carlson HA. J Chem Inf Modl. 2011;51(9):2115–2131. doi: 10.1021/ci200269q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taufer M, Crowley M, Price DJ, Chien AA, Brooks CL., III Concurr Comp-Pract E. 2005;17(14):1627–1641. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.