Abstract
Lampreys, one of the two surviving groups of ancient vertebrates, have become important models for study in diverse fields of biology. Lampreys (of which there are approximately 40 species) are being studied, for example, (a) to control pest sea lamprey in the North American Great Lakes and to restore declining populations of native species elsewhere; (b) in biomedical research, focusing particularly on the regenerative capability of lampreys; and (c) by developmental biologists studying the evolution of key vertebrate characters. Although a lack of genetic resources has hindered research on the mechanisms regulating many aspects of lamprey life history and development, formerly intractable questions are now amenable to investigation following the recent publication of the sea lamprey genome. Here, we provide an overview of the ways in which genomic tools are currently being deployed to tackle diverse research questions and suggest several areas that may benefit from the availability of the sea lamprey genome.
Keywords: lamprey, genomics, evolution, molecular mechanisms, control
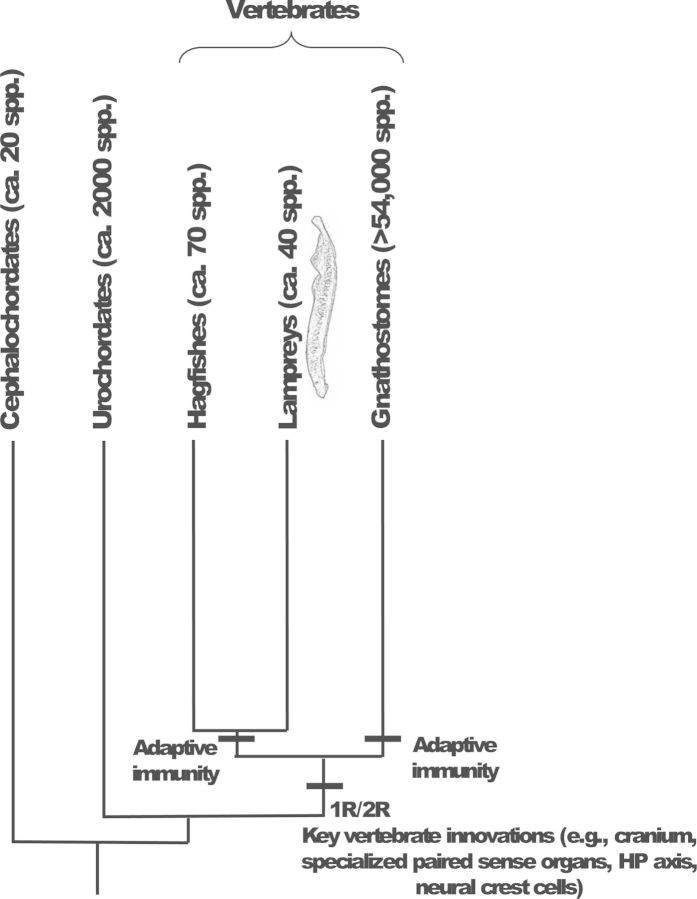
Lampreys are primitive vertebrates that, together with hagfishes, constitute the remaining members of a mostly extinct group of jawless fishes known as the Agnatha (figure 1; Potter et al. 2015). Although comparatively little is known about the biology of hagfishes, the anatomy, physiology, and embryology of lampreys have been subjects of research for more than 150 years (Schultze 1856). Lampreys are positioned at the base of vertebrate phylogeny and have long been recognized for their importance in understanding many aspects of vertebrate evolution (Docker et al. 2015). Although authors have debated the relationship of hagfishes and lampreys to other vertebrates for more than 200 years (Dumeril 1806), modern molecular analyses suggest that the jawless lampreys and hagfishes are monophyletic and together are the closest living relatives of the jawed vertebrates, the Gnathostomata (Heimberg et al. 2010). The invasion of the parasitic sea lamprey (Petromyzon marinus) from their native range in the Atlantic Ocean into the Great Lakes Basin brought about a very practical need to understand lamprey biology (Applegate 1950). Sea lamprey were first noted in Lake Ontario in the mid-1800s, although it is still not clear whether they colonized the Great Lakes Basin below Niagara Falls via canals in historical times or naturally in postglacial times (see the review by Eshenroder 2014). Nevertheless, the sea lamprey made its way into the popular press in North America in the early twentieth century, when, following improvements to the Welland Canal around Niagara Falls, sea lamprey soon began to appear in Lake Erie and subsequently throughout the upper Great Lakes (Applegate and Moffett 1955). Because of a parasitic lifestyle in which they feed on the blood and fluids of other fish (figure 2), sea lamprey soon negatively affected the fishing industry on the Great Lakes. This led to efforts to control their numbers that depended on gaining understanding into the ecological, physiological, and developmental aspects of their biology (Applegate 1950, Piavis 1961). Meanwhile, it was also recognized that among the vertebrates, the lampreys are unparalleled in their ability to recover behaviorally following transection of the spinal cord (Hibbard 1963), leading to their extensive use as a model for spinal cord repair (Hibbard 1963) and highlighting their importance among diverse fields of research.
Figure 1.
The interrelationships among the vertebrate lineages (jawless hagfishes and lampreys, as well as jawed vertebrates) and the nonvertebrate chordates (cephalochordates and urochordates) inferred from molecular data. Most evidence supports two rounds of genome duplication (1R, 2R) prior to the divergence of the agnathan and gnathostome lineages (but see the text), potentially permitting the origin of the many key innovations that characterize the vertebrates; adaptive immunity, however, likely arose independently in the agnathan and gnathostome lineages. Abbreviation: HP, hypothalamic–pituitary.
Figure 2.
Top: A salmon with a sea lamprey wound, caught in Lake Huron at the Rogers City, Michigan, Salmon Derby. Bottom: A lake trout with a parasitic sea lamprey attached just posterior to the gill. Wounding by sea lamprey is regularly observed in this location. According to the Great Lakes Fishery Commission, a single lamprey is capable of killing up to 40 pounds (18 kilograms) of fish during its 12- to 18-month feeding period. Photographs: Marc Gaden, Great Lakes Fishery Commission.
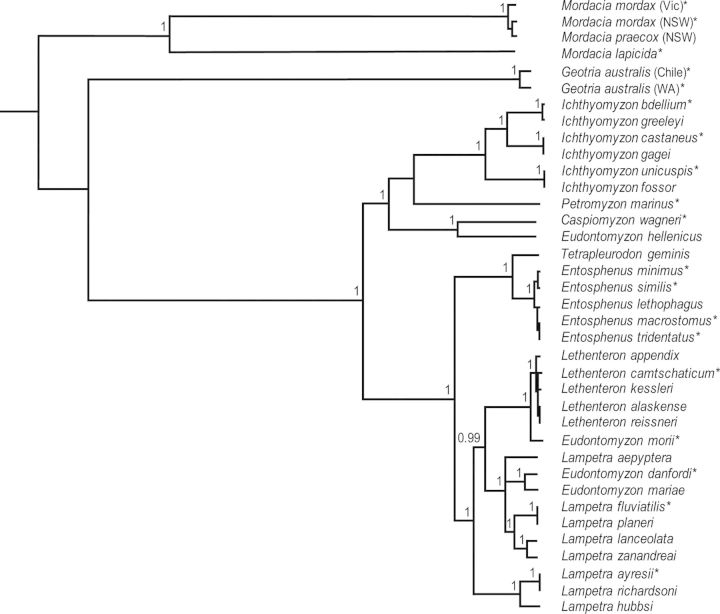
New knowledge on the mechanisms that control various aspects of lamprey life history, from embryogenesis up through the single spawning season that precedes their death, will be of cross-disciplinary interest to researchers in evolutionary developmental biology, immunology, and endocrinology, as well as in research for developing new lamprey management strategies. Although the invasion of sea lamprey into the Great Lakes has had considerable negative economic impact on the commercial and recreational fishing industries, populations of this same species are in decline in parts of its European range (Renaud 2011). The Pacific lamprey (Entosphenus tridentatus), found along the Pacific Rim from the North American West Coast to Asia, is likewise in decline in parts of its range, particularly in the Columbia River Basin, where dams have limited its upstream migration to former spawning habitats (Close et al. 2002). Worldwide, at least 16 of the approximately 40 extant lamprey species (figure 3) now receive legal protection at a national level, and others are protected at subnational levels (Maitland et al. 2015). For these reasons, management biologists (for both control and conservation), as well as basic researchers, stand to gain from having a greater understanding of lamprey biology.
Figure 3.
Phylogenetic relationships among 35 of the 41 lamprey species recognized by Potter and colleagues (2015), derived from cytochrome b sequence data. Bayesian posterior probabilities are given for those nodes where values are more than 0.95; asterisks designate parasitic species. Geotria and Mordacia occur in the Southern Hemisphere (Vic, Victoria; NSW, New South Wales; WA, Western Australia); all other genera are found in the Northern Hemisphere. At least 16 lamprey species are of conservation concern and receive legal protection at a national level—including four anadromous species (European populations of sea lamprey, Petromyzon marinus; European river lamprey, Lampetra fluviatilis; Caspian lamprey, Caspiomyzon wagneri; and Pacific lamprey, Entosphenus tridentatus) and three freshwater-resident parasitic and nine freshwater nonparasitic “brook lamprey” species (Maitland et al. 2015). Source: Reprinted from Potter and colleagues (2015) with kind permission from Springer Science+Business Media.
Historically, genetics and molecular biology have not played major roles in either basic research or management strategies because of the scarce availability of functional tools that could be adapted for research or management practice. Whereas biochemical and histological approaches have long been used to understand aspects, for example, of the neuroendocrine system in lampreys (Kavanaugh et al. 2008), molecular approaches to management practices are recent, beginning with the isolation of larval and adult pheromones that show promise as new management tools for both control (Li et al. 2002, Twohey et al. 2003) and possibly conservation efforts as a potential tool for the reintroduction of Pacific lampreys to spawning habitats (Maitland et al. 2015).
Importantly, limitations on the availability of resource material, a limited spawning season, and a life history that precludes the establishment and maintenance of germline mutants have limited the appeal of the lamprey as a genetic model. Because of their protected status, many lamprey species are simply not available to researchers; adult sea lamprey are most accessible during their spring migration, and embryos are available only during the spawning season in late spring to early summer. Furthermore, the life cycle of the sea lamprey may last as long as 5 to 7 years and contains an extended larval period (several years) during which they exist as filter feeding ammocoetes larvae, followed by a 1- to 2-year parasitic phase, and ending with a single short spawning period just prior to death (Potter et al. 2015). Establishing germline mutants would therefore require a long-term investment in rearing ammocoetes larvae over multiple years, through metamorphosis to the parasitic phase, and finally to sexual maturation, with F2 mutants available only after many years.
In spite of the limitations discussed above, the development of molecular genetic methods (e.g., reverse transcription polymerase chain reaction, RT-PCR, for the detection of gene expression) and functional “reverse” genetic tools (which investigate the phenotypes that result from particular—e.g., mutated, downregulated, or upregulated—gene sequences) has begun to enable investigators to explore the biology of lampreys using modern molecular techniques. Molecular genetics has been used to describe the expression patterns of many genes required for lamprey development (e.g., Takio et al. 2007), whereas functional genetic techniques have been applied to understand the mechanisms that regulate the basic biology of lamprey embryogenesis (e.g., McCauley and Bronner-Fraser 2006). Continued development and application of new genetic tools will open up new basic research avenues. The advent of next-generation sequencing technologies, which can generate tremendous amounts of DNA (e.g., genome) or RNA (e.g., transcriptome) sequence data, is also enabling considerable advances (Ellegren 2014). The publication of the sea lamprey genome (Smith et al. 2013) is a watershed event that is expected to lead to a wealth of new knowledge of the genes and gene networks that control many aspects of lamprey biology. These developments have the potential to lead to transformative control strategies based on unique aspects of sea lamprey biology, as well as new management practices and strategies to conserve populations of threatened lamprey species (Hess et al. 2013, Mateus et al. 2013). Here, we outline some areas of potential focus for adapting currently available genetic and molecular techniques to investigate the biology of lampreys. These tools will be of broad interest to the research community and may help to “cross-pollinate” the research activities of both basic and applied biologists seeking to understand the biology of these interesting animals.
We recognize that lampreys serve as important models for research in other areas that we have not covered in this overview (e.g., neural control of behavior), and other animal models, such as hagfishes, are equally positioned to inform our understanding of topics such as gene duplication and vertebrate evolution (see Docker et al. 2015). However, we have chosen to limit our focus to studies that have predominantly used genomics technologies in order to show how, going forward, advances in genomics may be used synergistically to gain an integrated understanding of lamprey biology that crosses disciplinary boundaries in both basic and applied research.
Genomics and transcriptomics
Genomics and transcriptomics have begun to revolutionize the study of biology as it has become possible to use systems-biology approaches to address formerly intractable problems. The sequencing of genomes, especially among nonmodel organisms, has begun to reveal lineage-specific adaptations that are likely to underlie evolutionary novelties (Ellegren 2014). A systems approach to lamprey biology has the potential to open new avenues for investigation. The sea lamprey genome, freely available as a public resource (Smith et al. 2013), contains some 26,000 genes spread over 84 pairs of chromosomes. A somatic genome was assembled from DNA obtained from the liver of a single sea lamprey. However, genome rearrangement is now known to occur during sea lamprey embryogenesis, in which approximately 20% of the germline genome is lost from somatic lineages (Smith et al. 2009), demonstrating the need for a germline genome resource (Smith et al. 2010a). Programmed rearrangement and loss may provide a biological strategy to ensure that pluripotential functions are segregated to the germline, thereby preventing the possibility of germline transmission in the somatic lineage (Smith et al. 2009, Smith et al. 2012). Comparison of somatic and germline genomic resources may provide important insights for understanding the role of genome rearrangement in the biology of lampreys (Smith et al. 2010a) and may represent an important taxon-specific trait to explore as a potential pest management tool.
The evolution of early vertebrates. Analysis of lamprey genomes has already begun to advance our understanding of early vertebrate evolution (Smith et al. 2010b), but the timing of whole genome duplications, considered key in the evolution of vertebrates, remains unsettled. A major finding that emerged from the sequencing of the sea lamprey genome is that two rounds of genome duplication (1R, 2R) likely occurred prior to the divergence of agnathan and gnathostome vertebrates (Barucchi et al. 2013, Smith et al. 2013). This interpretation is supported by analysis of neuroendocrine-associated genes in the lamprey, in which syntenic analysis of the gonadotropin-releasing hormone (GnRH) genes present in the sea lamprey supports two genome duplication events prior to agnathan–gnathostome divergence (Ohno 1970, Decatur et al. 2013). However, recently constructed high-density meiotic and comparative genomic maps for the sea lamprey do not support a second duplication event, suggesting instead several evolutionarily independent segmental duplications (Smith and Keinath 2015). Interestingly, sequencing and analysis of the Arctic lamprey (Lethenteron camtschaticum) genome suggest the possibility that two rounds of genome duplication in vertebrates might have occurred independently in lamprey and gnathostome lineages (Mehta et al. 2013). Comparison of the Arctic lamprey genome with the annotated sea lamprey genome indicates the existence of six Hox clusters in lampreys, suggesting that after 1R and 2R duplication events, lampreys might have subsequently undergone a third round of genome duplication. This finding has implications for understanding morphological diversity among vertebrates because the role of Hox genes for anteroposterior patterning is well established (Krumlauf 1994, Mehta et al. 2013).
Lampreys possess an adaptive immune system that is fundamentally different from the immunoglobulin-based B-cell and T-cell receptors found in the jawed vertebrates. Instead, lampreys have evolved a novel strategy for adaptive immunity that relies on leucine rich repeat (LRR)–based receptors to antigens, which are known as variable lymphocyte receptors (VLRs) (Pancer et al. 2004). VLRs, present only in lampreys and hagfishes, are expressed in cells that resemble gnathostome T-cells and B-cells and are encoded at VLRA and VLRB loci (Pancer et al. 2004, 2005). A VLRC locus has also been described in lamprey (Kasamatsu et al. 2010) and is expressed by a second lamprey-specific lineage of T cell–like lymphocytes (Hirano et al. 2013). Analysis of the sea lamprey draft genome has revealed the genomic composition of the VLRC locus and has enabled analysis of the mechanism for VLRC assembly (Smith et al. 2013). Such analyses have increased our understanding of unique aspects of acquired immunity in lampreys and may provide a target-rich environment for developing sea lamprey control strategies that would minimize adverse effects on nontarget species.
Paired species and the loss of the parasitic feeding phase
Another aspect of lampreys that is crucial to both their management and the basic understanding of their biology is the phenomenon of “paired species” (Docker 2009). The majority of extant lamprey species exist as parasitic–nonparasitic species pairs (figure 3). Both members of a pair spend several years as filter-feeding larvae, but following metamorphosis, one species passes through a sexually immature parasitic phase (for a few months to several years) before eventually spawning and dying. The nonparasitic member of the pair becomes sexually mature immediately following metamorphosis, bypasses the adult feeding stage, and spawns and dies at a small size within 6–10 months of metamorphosis. Evidence suggests that, independently within each pair, the nonparasitic species has arisen from the parasitic species (Docker 2009). The parasitic sea lamprey does not have a nonparasitic counterpart, but efforts to control invasive sea lamprey populations have adversely affected populations of the paired parasitic silver (Ichthyomyzon unicuspis) and nonparasitic northern brook (Ichthyomyzon fossor) lampreys that are found in the Great Lakes Basin; these native lamprey species are also vulnerable to the nitrophenol compound (3-trifluoromethyl-4-nitrophenol, TFM) that is used in tributary streams to kill larval sea lamprey (Schuldt and Goold 1980). Sea lamprey control efforts appear to negatively affect silver lamprey more than northern brook lamprey, possibly related to the in-stream distribution of the silver lamprey overlapping more with that of the sea lamprey (Schuldt and Goold 1980). Mitochondrial and microsatellite DNA marker analyses, however, suggest that silver and northern brook lampreys may represent a single species with ongoing gene flow when they co-occur, suggesting that less affected northern brook lamprey populations could potentially be used to help rehabilitate affected silver lamprey populations (Docker et al. 2012). Genomic analyses of paired species will be vital for understanding the evolution of lampreys and for identifying species differences. The paired European river (Lampetra fluviatilis) and brook (Lampetra planeri) lampreys similarly share mitochondrial haplotypes when sympatric, likewise suggesting a very recent divergence or ongoing gene flow between these species (Espanhol et al. 2007). However, genomic analysis of single nucleotide polymorphisms (SNPs) of this species pair in the Tagus River Basin of Portugal revealed 166 fixed genetic differences between the species (Mateus et al. 2013). Using the sea lamprey genome, these authors were able to link 12 of these fixed differences to genes likely related to adaptation from an anadromous to freshwater-resident lifestyle (Mateus et al. 2013) that, in this species pair, accompanied the transition from parasitism to nonparasitism. Further genomic study is required, especially in those pairs, such as silver and northern brook lampreys, in which both parasitic and nonparasitic members are freshwater resident. From a practical point of view, this will be important for understanding whether paired species of conservation concern should be managed as single or separate gene pools and for understanding the genetic mechanisms responsible for the loss of the parasitic feeding phase.
Identification of potential new targets for sea lamprey control
A key component to annotation of the sea lamprey genome was transcriptome analysis of different tissues and developmental stages. Whereas the genome is the entire DNA sequence of the organism, the transcriptome represents only the RNA that has been transcribed (e.g., the genes that have been expressed) in that particular tissue, stage of development, and physiological state. This enables researchers to make inferences about a gene's function on the basis of when and where it is expressed and under what conditions. Other molecular methods (e.g., quantitative RT-PCR, microarrays) are able to measure the expression of a relatively small number of targeted genes (those for which some prior sequence knowledge is available), but transcriptome analysis using next-generation sequencing allows the entire collection of RNAs in a sample to be analyzed (and, depending on the method, quantified). Comparisons of variation in global gene expression levels through life history, across tissue type, and among species can therefore be accomplished with relative ease. A global view of transcriptomes and their regulation throughout life history will help define the molecular programs that endow the species-specific or lineage-specific adaptation in lampreys. A transcriptomic approach, for example, was used recently to sequence and compare the relative abundances of genes expressed in the gonads of nonparasitic northern brook lampreys and parasitic chestnut lampreys (Ichthyomyzon castaneus) before, during, and after ovarian differentiation (Spice 2013; see the “Reproductive biology and the development of genetic control strategies” section below).
Transcriptomics might also be used to better understand the process of metamorphosis and to develop species-specific lampricides. Using nontranscriptomic approaches, empirical experiments on lampreys during embryogenesis have implicated molecular mechanisms related to important vertebrate innovations (McCauley and Bronner-Fraser 2006, Lakiza et al. 2011). Lampreys are one of the few vertebrate animals that go through a “true” metamorphosis, and decades of research on the radical changes that occur during this process have been a treasure trove for mining physiological and molecular information regarding organogenesis and developmental plasticity (Manzon et al. 2015). Further study using transcriptomics will no doubt now prove fruitful in yielding additional insights into vertebrate evolution and development and may help identify the basis for unique aspects of sea lamprey biology (e.g., metamorphosis) that could be exploited for management purposes. Similarly, the lampricide TFM that is used to kill larval sea lamprey for population management appears to be toxic specifically to lampreys because of their unique characteristics of metabolism of xenobiotics. Relative to other fishes, lampreys have a reduced capacity to biotransform TFM to TFM-glucuronide via the process of glucuronidation (Birceanu et al. 2014). However, although TFM is largely lamprey specific, it is not sea lamprey specific and can have negative effects on native lamprey species in the Great Lakes Basin (Schuldt and Goold 1980); transcriptomic approaches may help increase species specificity. Although functional studies will always be desired—and often needed—to validate predictions from sequence analyses, the depth of information afforded by the new sequencing technologies and the comprehensive analyses thereof should allow insightful and significant inference on lineage-specific mechanistic processes. In short, comprehensive analyses of transcriptomes will facilitate and accelerate research efforts to further define biochemical, genetic, metabolic, and physiological pathways in lampreys and eventually to impart novel or additional strategies for conservation and control.
Gene knockdown
Although forward genetic tools (in which genotypes that are responsible for specific phenotypes are identified) have been applied in model species (e.g., by generating mutant phenotypes and performing cross-breeding experiments to map the gene responsible onto its chromosome), lampreys (e.g., given their 5- to 7-year generation time and singular reproductive event) are not amenable to these approaches. However, the introduction of reverse genetics techniques, in which gene function can be determined by analyzing the phenotypic effects of a specific gene sequence, coupled with molecular analytical tools has resulted in an increase in information on the molecular mechanisms of lamprey biology. Gene knockdown or “silencing” has been used to investigate the basic mechanisms of development in the sea lamprey. Two approaches that have been used are morpholinos and RNA interference (RNAi). Morpholinos are synthetic oligonucleotides that can target the expression of specific RNA transcripts to prevent the expression and activity of a specific protein. Coupled with in situ hybridization to analyze changes in gene expression as a result of morpholino-induced down regulation, this technique has provided important insights for understanding the development of the lamprey craniofacial skeleton and also the evolution of the vertebrate neural crest under the regulation of a gene regulatory network (GRN; McCauley and Bronner-Fraser 2006, Sauka-Spengler et al. 2007, Lakiza et al. 2011). RNAi is an endogenous post-transcriptional regulatory mechanism to silence specific messenger RNA transcripts and plays significant roles in regulating development as well as cellular defense against foreign DNA. RNAi can be adapted as a tool to investigate the roles of specific genes, has provided important insight into developmental mechanisms and disease (Alvarez-Garcia and Miska 2005), and has recently been shown to be functional during lamprey development (Heath et al. 2014). Morpholino delivery has been accomplished by microinjection into individual embryos (McCauley and Bronner-Fraser 2006, Sauka-Spengler et al. 2007, Lakiza et al. 2011), whereas small interfering (si)RNA uptake has also been accomplished through feeding directly to larval lampreys (Heath et al. 2014).
A recent advance in the ability to target genes for knockdown has been the adaptation of CRISPR/Cas technology to delete gene sequences of interest from diverse model organisms (Cong et al. 2013). The CRISPR/Cas system, an adaptive defense mechanism to direct RNA-guided site-specific DNA cleavage, evolved in bacteria and archaea to provide acquired resistance to invading viruses and plasmids (Marraffini and Sontheimer 2010). The use of this technology to target the knockdown of genes of interest has begun to revolutionize genome editing, making it possible to easily modify genomes across a wide range of model organisms (Cong et al. 2013). Studies are underway to determine the potential for CRISPR/Cas technology to facilitate the understanding of gene function in lampreys (Romasek et al. 2015). CRISPR/Cas technology may also provide a new avenue for developing strategies to control pest lampreys.
A powerful tool in the molecular biologists’ toolbox has been the ability to misexpress either foreign or endogenous genes of interest under controlled conditions. The ability to express a reporter gene (green fluorescent protein, GFP) under the regulation of either a ubiquitous promoter or a tissue-specific promoter was demonstrated in the Arctic lamprey, in which GFP was expressed under the control of either a cytomegalovirus (CMV) promoter or a muscle actin promoter to drive the muscle-specific expression of GFP (Kusakabe et al. 2003). This study demonstrated the feasibility of tissue-specific expression as a tool for understanding lamprey development. With the availability of genomic sequence and analytical tools, it is now possible to isolate specific conserved noncoding cis regulatory elements (CNEs) and test their functional conservation across vertebrates. Recent studies have demonstrated the utility of this approach and have shown the viability of generating transgenic embryos in order to investigate the gene regulatory architecture in lampreys by characterizing the tissue-specific function of enhancer elements (Parker et al. 2014a, 2014b).
A variety of molecular techniques that may be used for investigating different aspects of lamprey biology are therefore becoming available. These techniques will be useful for exploring the importance of specific gene activities relative to the evolution of molecular mechanisms in basal vertebrates. Such tools may also prove beneficial in the search for lamprey-specific (and particularly sea lamprey–specific) mechanisms to target for management purposes. For example, although the genes targeted in the study by Heath and colleagues (2014) were highly conserved and were used only to test the efficacy of siRNAs in lampreys, siRNAs (given the sequence specificity of RNAi) have potential for use as sea lamprey–specific lampricides in which DNA sequences unique to the sea lamprey can be targeted for the knockdown of essential genes, resulting in a lethal loss-of-function phenotype specific to the sea lamprey without affecting nontarget species.
Reproductive biology and the development of genetic control strategies
One of the best-studied aspects of lamprey reproduction is its control by the hypothalamic–pituitary (HP) system. The HP system is considered to be a key innovation that emerged prior to or during the evolution of vertebrates (Sower 2015). Within this hierarchically organized endocrine system, gonadotropin-releasing hormone (GnRH) produced in the hypothalamus (and released in response to external and internal cues) stimulates the synthesis and release of the gonadotropin(s) from the pituitary gland, which in turn act at the gonads to regulate steroidogenesis and gametogenesis. Gnathostomes generally have one or two GnRHs, two pituitary gonadotropins, and one gonadal receptor for each of these gonadotropins; lampreys have three GnRHs but only one pituitary gonadotropin-type hormone and one gonadal receptor for the gonadotropin (Sower 2015). Elucidation of the HP system in lampreys has been crucial for understanding its evolution in vertebrates and may lead to possible innovative control strategies.
Other aspects of the genetic and molecular control of lamprey reproduction are considerably less well understood. These include the mechanisms of sex determination (the genetic and/or environmental process that establishes the gender of an organism) and the factors involved in sex differentiation (the processes that transform an undifferentiated gonad into a testis or an ovary; Piferrer et al. 2012). With regards to sex determination, extensive research has revealed that fishes exploit a wide range of mechanisms, from genetic sex determination (GSD) to environmental sex determination (ESD), and, in species with GSD, the genes involved in sex determination are highly variable among and within taxa (Piferrer et al. 2012). The sex-determination mechanism employed by lampreys remains unknown. Mateus and colleagues (2013) found no evidence of genomic differentiation between male and female European brook lamprey (L. planeri), suggesting that if sex determination is genetic, the underlying system evolved without major chromosome divergence. Alternatively, ESD may play a role in this species, as has been suggested in other lamprey species. The sex ratios of adult and larval sea lamprey in the Great Lakes have varied widely with abundance, ranging from male-biased sex ratios when abundance was high during their initial invasion to female-biased ratios following population declines after the initiation of sea lamprey control (Purvis 1979). The lampricide TFM appears not to be differentially toxic to males and females (Purvis 1979), indicating that the skewed sex ratios are not the result of sex-specific differences in mortality, and similar correlations between sex ratio and abundance have been observed in other lamprey species that have never been exposed to TFM (e.g., Docker and Beamish 1994). It is interesting to note, however, that TFM has been shown to bind estrogen receptors and was able to induce the formation of vitellogenin in other fish species (Hewitt et al. 1998), and the potential feminizing effect of TFM on lampreys should be examined.
Compared with sex determination, the process of sex differentiation in lampreys is somewhat better understood. For many years, it has been recognized that the lamprey gonad remains histologically undifferentiated well into the larval phase and that testicular differentiation (which does not occur until metamorphosis, at ages of approximately 4 to 6 years) is delayed relative to ovarian differentiation (Hardisty 1971). Primordial germ cells are first observed during embryogenesis following gastrulation. Mitotic cell divisions occur in the gonad prior to differentiation, which is accompanied in both sexes by the appearance of growing oocytes. Ovarian differentiation (which yields a larger number of oocytes, few or no residual undifferentiated germ cells, and a relatively large gonad in presumptive females) occurs in the larval stage (e.g., at approximately 1 year in nonparasitic species and 3 years in anadromous sea lamprey; Hardisty 1971). In presumptive males, oocytes (which are relatively small in size and number) appear to regress, and residual undifferentiated germ cells that escape degeneration may give rise to the male germ line in small cell nests and ultimately to spermatocytes at metamorphosis (Hardisty 1971). Interestingly, the presence of oocytes within the testes of adult lampreys has been noted, and it is thought that their presence represents a likely failure to fully differentiate into males at metamorphosis (Hardisty 1971, Clemens et al. 2012), but the importance of this observation for understanding sex-differentiation mechanisms (and the fertility of these males) is not clear.
Research showing the molecular mechanism(s) that regulate sex differentiation in lampreys will provide important insights for understanding the evolution of this process in vertebrates. The genetic basis of sex differentiation is reasonably well understood in birds (Chue and Smith 2011) and mammals (Wilhelm et al. 2007), but it is less well understood in the “lower vertebrates”—although studies in model fish species have identified some conserved genes involved in sex differentiation in teleost fishes (Piferrer et al. 2012). Recently, Spice and colleagues (2014) used quantitative RT-PCR to examine potential differences in the expression of eight target genes before, during, and after ovarian differentiation in chestnut and northern brook lampreys. These eight genes were chosen because of their involvement in sex differentiation in a variety of other vertebrates (e.g., 17β-hydroxysteroid dehydrogenase, germ cell-less, daz-associated protein 1) or because they were found to be differentially expressed in the gonadal transcriptomes of a small number of chestnut and northern brook lampreys (e.g., cytochrome c oxidase subunit III, dehydrocholesterol reductase 7; Spice 2013). Expression patterns suggested that cytochrome c oxidase subunit III and insulin-like growth factor 1 receptor may be involved in apoptosis and oocyte growth, respectively; Wilms’ tumor suppressor protein 1 may be involved in the undifferentiated gonad and/or later testicular development; and 17β-hydroxysteroid dehydrogenase and daz-associated protein 1 may be involved in female development (Spice et al. 2014). This study was the first to identify genes that may be involved in sex differentiation in lampreys, but follow-up research is clearly required.
Knowledge about the genetic basis of sex determination and differentiation in lampreys may also have potential management applications. The manipulation of sea lamprey reproduction is an untapped but potentially effective strategy to control spawning populations in the Great Lakes region. Genetic control technologies are being developed for insect pests, and it may be possible to adapt some of the technologies to invasive fishes (Thresher et al. 2014). As an instructive example, a prototype sex ratio–distorting (“daughterless”) technology is being tested in common carp (Cyprinus carpio) using a strategy that attempts to manipulate sex ratios with a bias toward males by targeting a gene that is required to convert testosterone into estrogens. Although adaptation of this strategy could have potential as an effective management tool to control sea lamprey in the Great Lakes, much greater knowledge regarding the hormonal control of sex determination and differentiation in lampreys is first required. Aromatase activity, required for the biosynthesis of estrogens, has been found in lampreys (Callard et al. 1980), but its role has yet to be established, and several studies have shown that lampreys appear to use a mixture of both classical (e.g., estradiol, progesterone) and nonclassical (e.g., 15-hydroxylated) sex steroids (Bryan et al. 2008). Furthermore, although hormonal control of sex differentiation has been accomplished in many teleost fishes (Hunter and Donaldson 1983), treatment with sex steroids has been unsuccessful in altering sex differentiation in larval sea lamprey (Docker 1992).
The sterilization and re-release of spawning male sea lamprey is another strategy that has been tried. Captured adult males were treated with the male selective chemosterilant, bisazir, and then released to spawning streams that had recently been treated with lampricide (Hanson 1981). The goal of this program was to reduce larval populations that result from residual spawning female lampreys in streams following lampricide treatment (Bergstedt and Twohey 2007). This program was discontinued in 2011, in part because of equivocal results and partially as a result of the cost of the chemosterilant and its potential human health hazards that limited the number of males that could be sterilized to compete with fertile males. However, the development of species-specific sterilization tools may still prove useful for reducing invasive sea lamprey populations.
Conclusions
Here, we have highlighted how modern genetic and genomic tools are being used to understand the biology of lampreys. The application of these new tools may provide important new avenues both for understanding the evolution and biology (ecology, physiology, development) of lampreys and also for developing management tools for conservation and control. Our desire is to stimulate research interests and partnerships that bridge the gulf that often separates basic and applied biology. The new tools that have recently become available (i.e., sea lamprey genome, molecular techniques) can have a synergistic effect on the ability of researchers to integrate new findings across disciplinary boundaries with the potential to provide important new insights into the biology of lampreys. For example, reverse genetic tools developed for understanding the basic biology of lampreys are also among the most promising for adoption as new control strategies. Conversely, in order to explore sex ratio–biasing technologies to control lamprey populations, as developed for other invasive species, a greater understanding of the basic molecular mechanisms regulating sex determination in lampreys will be required—knowledge that will also be useful to researchers interested in understanding the evolution of sex-determining mechanisms. In addition, potential synergies may also already exist within the lamprey community, in which research on new control methods that depend on the ability to culture large numbers of lamprey embryos and larvae may result in powerful new methods that are useful for the conservation of threatened lampreys. Going forward, we expect that new insights into lamprey biology will affect the economic development related to the conservation and control of lampreys, whereas basic research findings may affect our understanding of vertebrate evolution as well as the development of new biomedical research tools.
Acknowledgments
This contribution emerged from activities of the Sea Lamprey Research Board of the Great Lakes Fishery Commission intended to highlight the science behind sea lamprey control. Photographs in figure 2 are provided by Marc Gaden, courtesy of the Great Lakes Fishery Commission. David McCauley wrote the introduction and contributed to all sections of the manuscript. Margaret Docker provided input on the general biology of lampreys and contributed to the section on reproductive biology. Steven Whyard contributed to the section on gene knockdown. Weiming Li contributed to the section on transcriptomics. Andrew Muir, Steve Cooke, and three anonymous reviewers provided insightful comments to improve this manuscript.
References cited
- Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- Applegate VC. Natural History of the Sea Lamprey, Petromyzon marinus, in Michigan. US Fish and Wildlife Services Special Scientific Report no. 55; 1950. [Google Scholar]
- Applegate VC, Moffett JW. The sea lamprey. Scientific American. 1955;192:36–41. [Google Scholar]
- Barucchi VC, Giovannotti M, Cerioni PN, Splendiani A. Genome duplication in early vertebrates: Insights from agnathan cytogenetics. Cytogenetic and Genome Research. 2013;141:80–89. doi: 10.1159/000354098. [DOI] [PubMed] [Google Scholar]
- Bergstedt RA, Twohey MB. Research to support sterile-male-release and genetic alteration techniques for sea lamprey control. Journal of Great Lakes Research. 2007;33:48–69. [Google Scholar]
- Birceanu O, Sorensen LA, Henry M., McClelland GB, Wang YS, Wilkie MP. The effects of the lampricide 3-trifluoromethyl-4-nitrophenol (TFM) on fuel stores and ion balance in a non-target fish, the rainbow trout (Oncorhynchus mykiss) Comparative Biochemistry and Physiology. 2014;160:30–41. doi: 10.1016/j.cbpc.2013.10.002. [DOI] [PubMed] [Google Scholar]
- Bryan MB, Scott AP, Li W. Sex steroids and their receptors in lampreys. Steroids. 2008;73:1–12. doi: 10.1016/j.steroids.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Callard GV, Petro Z, Ryan KJ. Aromatization and 5-α-reduction in brain and nonneural tissues of a cyclostome, Petromyzon marinus. General and Comparative Endocrinology. 1980;42:155–159. doi: 10.1016/0016-6480(80)90181-1. [DOI] [PubMed] [Google Scholar]
- Chue J, Smith CA. Sex determination and sexual differentiation in the avian model. Federation of European Biochemical Societies Journal. 2011;278:1027–1034. doi: 10.1111/j.1742-4658.2011.08032.x. [DOI] [PubMed] [Google Scholar]
- Clemens BJ, Sower SA, van de Wetering S, Schreck CB. Incidence of male intersex in adult Pacific lamprey (Entosphenus tridentatus), with a brief discussion of intersex versus hermaphroditism in lampreys (Petromyzontiformes) Canadian Journal of Zoology. 2012;90:1201–1206. [Google Scholar]
- Close DA, Fitzpatrick MS, Li HW. The ecological and cultural importance of a species at risk of extinction, Pacific Lamprey. Fisheries. 2002;27:19–25. [Google Scholar]
- Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decatur WA, Hall JA, Smith JJ, Li W, Sower SA. Insight from the lamprey genome: Glimpsing early vertebrate development via neuroendocrine-associated genes and shared synteny of gonadotropin-releasing hormone (GnRH) General and Comparative Endocrinology. 2013;192:237–245. doi: 10.1016/j.ygcen.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docker MF. PhD dissertation. Guelph, Canada: University of Guelph; 1992. Labile Sex Determination in Lampreys: The Effect of Larval Density and Sex Steroids on Gonadal Differentiation. [Google Scholar]
- Docker MF. A review of the evolution of nonparasitism in lampreys and an update of the paired species concept. In: Brown LR, Chase SD, Mesa MG, Beamish RJ, Moyle PB, editors. Biology, Management, and Conservation of Lampreys in North America. American Fisheries Society; 2009. pp. 71–114. [Google Scholar]
- Docker MF, Beamish FWH. Age, growth, and sex ratio among populations of least brook lamprey, Lampetra aepyptera, larvae: An argument for environmental sex determination. Environmental Biology of Fishes. 1994;41:191–205. [Google Scholar]
- Docker MF, Mandrak NE, Heath DD. Contemporary gene flow between “paired” silver (Ichthyomyzon unicuspis) and northern brook (I. fossor) lampreys: Implications for conservation. Conservation Genetics. 2012;13:823–835. [Google Scholar]
- Docker MF, Hume JB, Clemens BJ. Introduction: A surfeit of lampreys. In: Docker MF, editor. Lampreys: Biology, Conservation, and Control. Vol. 1. Springer; 2015. pp. 1–34. [Google Scholar]
- Dumeril AMC. Zoologie Analytique, ou Methods Naturelle de Classification des Animaux. Didot; 1806. [Google Scholar]
- Ellegren H. Genome sequencing and population genomics in non-model organisms. Trends in Ecology and Evolution. 2014;29:51–63. doi: 10.1016/j.tree.2013.09.008. [DOI] [PubMed] [Google Scholar]
- Eshenroder RL. The role of the Champlain Canal and Erie Canal as putative corridors for colonization of Lake Champlain and Lake Ontario by sea lampreys. Transactions of the American Fisheries Society. 2014;143:634–649. [Google Scholar]
- Espanhol R, Almeida PR, Alves MJ. Evolutionary history of lamprey paired species Lampetra fluviatilis (L.) and Lampetra planeri (Bloch) as inferred from mitochondrial DNA variation. Molecular Ecology. 2007;16:1909–1924. doi: 10.1111/j.1365-294X.2007.03279.x. [DOI] [PubMed] [Google Scholar]
- Hanson LH. Sterilization of sea lampreys (petromyzon-marinus) by immersion in an aqueous-solution of bisazir. Canadian Journal of Fisheries and Aquatic Sciences. 1981;38:1285–1289. [Google Scholar]
- Hardisty MW. Gonadogenesis, sex differentiation, and gametogenesis. In: Hardisty MW, Potter IC, editors. The Biology of Lampreys, vol. 1. Academic Press; 1971. pp. 295–359. [Google Scholar]
- Heath G, Childs D, Docker MF, McCauley DW, Whyard S. RNA interference technology to control pest sea lampreys: A proof-of-concept. PLOS ONE. 2014;9 doi: 10.1371/journal.pone.0088387. (art. e88387). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimberg AM, Cowper-Sal-lari R, Semon M, Donoghue PC, Peterson KJ. MicroRNAs reveal the interrelationships of hagfish, lampreys, and gnathostomes and the nature of the ancestral vertebrate. Proceedings of the National Academy of Sciences. 2010;107:19379–19383. doi: 10.1073/pnas.1010350107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess JE, Campbell NR, Close DA, Docker MF, Narum SR. Population genomics of Pacific lamprey: Adaptive variation in a highly dispersive species. Molecular Ecology. 2013;22:2898–2916. doi: 10.1111/mec.12150. [DOI] [PubMed] [Google Scholar]
- Hewitt LM, Tremblay L, Van der Kraak GJ, Solomon KR, Servos MR. Identification of the lampricide 3-trifluoromethyl-4-nitrophenol as an agonist for the rainbow trout estrogen receptor. Environmental Toxicology and Chemistry. 1998;17:425–432. [Google Scholar]
- Hibbard E. Regeneration in the severed spinal cord of chordate larvae of Petromyzon marinus. Experimental Neurology. 1963;7:175–185. [Google Scholar]
- Hirano M, Guo P, McCurley N, Schorpp M, Das S, Boehm T, Cooper MD. Evolutionary implications of a third lymphocyte lineage in lampreys. Nature. 2013;501:435–438. doi: 10.1038/nature12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter GA, Donaldson EM. Hormonal sex control and its application to fish culture. Fish Physiology. 1983;9:223–303. [Google Scholar]
- Kasamatsu J, Sutoh Y, Fugo K, Otsuka N, Iwabuchi K, Kasahara M. Identification of a third variable lymphocyte receptor in the lamprey. Proceedings of the National Academy of Sciences. 2010;107:14304–14308. doi: 10.1073/pnas.1001910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavanaugh SI, Nozaki M, Sower SA. Origins of gonadotropin-releasing hormone (GnRH) in vertebrates: Identification of a novel GnRH in a basal vertebrate, the sea lamprey. Endocrinology. 2008;149:3860–3869. doi: 10.1210/en.2008-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Kusakabe R, Tochinai S, Kuratani S. Expression of foreign genes in lamprey embryos: An approach to study evolutionary changes in gene regulation. Journal of Experimental Zoology: Molecular Development and Evolution. 2003;296:87–97. doi: 10.1002/jez.b.11. [DOI] [PubMed] [Google Scholar]
- Lakiza O, Miller S, Bunce A, Lee EM, McCauley DW. SoxE gene duplication and development of the lamprey branchial skeleton: Insights into development and evolution of the neural crest. Developmental Biology. 2011;359:149–161. doi: 10.1016/j.ydbio.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Li WM, Scott AP, Siefkes MJ, Yan HG, Liu Q, Yun SS, Gage DA. Bile acid secreted by male sea lamprey that acts as a sex pheromone. Science. 2002;296:138–141. doi: 10.1126/science.1067797. [DOI] [PubMed] [Google Scholar]
- Maitland PS, Renaud CB, Quintella BR, Close DA, Docker MF. Conservation of native lampreys. In: Docker MF, editor. Lampreys: Biology, Conservation, and Control. Vol. 1. Springer; 2015. pp. 375–428. [Google Scholar]
- Manzon RG, Youson JH, Holmes JA. Lamprey metamorphosis. In: Docker MF, editor. Lampreys: Biology, Conservation, and Control. Vol. 1. Springer; 2015. pp. 139–214. [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference: RNA-directed adaptive immunity in bacteria and archaea. Nature Reviews Genetics. 2010;11:181–190. doi: 10.1038/nrg2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus CS, Stange M, Berner D, Roesti M, Quintella BR, Alves MJ, Almeida PR, Salzburger W. Strong genome-wide divergence between sympatric European river and brook lampreys. Current Biology. 2013;23:R649–R650. doi: 10.1016/j.cub.2013.06.026. [DOI] [PubMed] [Google Scholar]
- McCauley DW, Bronner-Fraser M. Importance of SoxE in neural crest development and the evolution of the pharynx. Nature. 2006;441:750–752. doi: 10.1038/nature04691. [DOI] [PubMed] [Google Scholar]
- Mehta TK, et al. Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum) Proceedings of the National Academy of Sciences. 2013;110:16044–16049. doi: 10.1073/pnas.1315760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Evolution by Gene Duplication. Springer; 1970. [Google Scholar]
- Pancer Z, Amemiya CT, Ehrhardt GRA, Ceitlin J, Gartland GL, Cooper MD. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature. 2004;430:174–180. [Google Scholar]
- Pancer Z, Saha NR, Kasamatsu J, Suzuki T, Amemiya CT, Kasahara M, Cooper MD. Variable lymphocyte receptors in hagfish. Proceedings of the National Academy of Sciences. 2005;102:9224–9229. doi: 10.1073/pnas.0503792102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HJ, Bronner ME, Krumlauf R. A Hox regulatory network of hindbrain segmentation is conserved to the base of vertebrates. Nature. 2014a;514:490–493. doi: 10.1038/nature13723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker HJ, Sauka-Spengler T, Bronner M, Elgar G. A reporter assay in lamprey embryos reveals both functional conservation and elaboration of vertebrate enhancers. PLOS ONE. 2014b;9 doi: 10.1371/journal.pone.0085492. (art. e85492). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piavis GW. Embryological stages in the sea lamprey and effect of temperature on development. US Fish and Wildlife Service Fishery Bulletin. 1961;61:111–143. [Google Scholar]
- Piferrer F, Ribas L, Díaz N. Genomic approaches to study genetic and environmental influences on fish sex determination and differentiation. Marine Biotechnology. 2012;14:591–604. doi: 10.1007/s10126-012-9445-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter IC, Gill HS, Renaud CB, Haoucher D. The taxonomy, phylogeny, and distribution of lampreys. In: Docker MF, editor. Lampreys: Biology, Conservation, and Control. Vol. 1. Springer; 2015. pp. 35–73. [Google Scholar]
- Purvis HA. Variations in Growth, Age at Transformation, and Sex Ratio of Sea Lampreys Reestablished in Chemically Treated Tributaries of the Upper Great Lakes. Great Lakes Fishery Commission; 1979. Great Lakes Fishery Commission Technical Report no. 35. [Google Scholar]
- Renaud CB. Lampreys of the World: An Annotated and Illustrated Catalogue of Lamprey Species Known to Date. Food and Agriculture Organization of the United Nations; 2011. [Google Scholar]
- Romasek M, Square T, Jandzik D, Medeiros DM. CRISPR/Cas system in the sea lamprey: A tool for understanding ancestral gene functions in vertebrates. Integrative and Comparative Biology. 2015;55:E321–E321. doi: 10.1242/dev.125609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauka-Spengler T, Meulemans D, Jones M, Bronner-Fraser M. Ancient evolutionary origin of the neural crest gene regulatory network. Developmental Cell. 2007;13:405–420. doi: 10.1016/j.devcel.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Schuldt RJ, Goold R. Changes in the distribution of native lampreys in Lake Superior tributaries in response to sea lamprey (Petromyzon marinus) control, 1953–77. Canadian Journal of Fisheries and Aquatic Sciences. 1980;37:1872–1855. [Google Scholar]
- Schultze MS. Die Entwickelungs-geschichte von Petromyzon planeri. Natuurkundige Verhandelingen van de Hollandsche Maatschappy der Wetenschappen te Haarlem 2: 1–51; 1856. [Google Scholar]
- Smith JJ, Keinath MC. The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Research. 2015;25:1081–1090. doi: 10.1101/gr.184135.114. doi:10.1101/gr.184135.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proceedings of the National Academy of Sciences. 2009;106:11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Stuart AB, Sauka-Spengler T, Clifton SW, Amemiya CT. Development and analysis of a germline BAC resource for the sea lamprey, a vertebrate that undergoes substantial chromatin diminution. Chromosoma. 2010a;119:381–389. doi: 10.1007/s00412-010-0263-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Saha NR, Amemiya CT. Genome biology of the cyclostomes and insights into the evolutionary biology of vertebrate genomes. Integrative and Comparative Biology. 2010b;50:130–137. doi: 10.1093/icb/icq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Baker C, Eichler EE, Amemiya CT. Genetic consequences of programmed genome rearrangement. Current Biology. 2012;22:1524–1529. doi: 10.1016/j.cub.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, et al. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nature Genetics. 2013;45:415–421. doi: 10.1038/ng.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sower SA. The reproductive hypothalamic–pituitary axis in lampreys. In: Docker MF, editor. Lampreys: Biology, Conservation, and Control. Vol. 1. Springer; 2015. pp. 305–373. [Google Scholar]
- Spice EK. Master's thesis. Canada: University of Manitoba, Winnipeg; 2013. Ovarian Differentiation in an Ancient Vertebrate: Timing, Candidate Gene Expression, and Global Gene Expression in Parasitic and Non-Parasitic Lampreys. [Google Scholar]
- Spice EK, Whyard S, Docker MF. Gene expression during ovarian differentiation in parasitic and non-parasitic lampreys: Implications for fecundity and life history types. General and Comparative Endocrinology. 2014;208:116–125. doi: 10.1016/j.ygcen.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Takio Y, Kuraku S, Murakami Y, Pasqualetti M, Rijli FM, Narita Y, Kuratani S, Kusakabe R. Hox gene expression patterns in Lethenteron japonicum embryos: Insights into the evolution of the vertebrate Hox code. Developmental Biology. 2007;308:606–620. doi: 10.1016/j.ydbio.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Thresher RE, Hayes K, Bax NJ, Teem J., Benfey TJ, Gould F. Genetic control of invasive fish: technological options. Biological Invasions. 2014;16:1201–1216. [Google Scholar]
- Twohey MB, Sorensen PW, Li WM. Possible applications of pheromones in an integrated sea lamprey management program. Journal of Great Lakes Research. 2003;29:794–800. [Google Scholar]
- Wilhelm D, Palmer S, Koopman P. Sex determination and gonadal development in mammals. Physiological Reviews. 2007;87:1–28. doi: 10.1152/physrev.00009.2006. [DOI] [PubMed] [Google Scholar]