Abstract
Haldane’s Rule and Darwin’s Corollary to Haldane’s Rule are the observations that heterogametic F1 hybrids are frequently less fit than their homogametic siblings, and that asymmetric results are often obtained from reciprocal hybrid crosses. In Caenorhabditis, Haldane’s Rule and Darwin’s Corollary have been observed in several hybrid crosses, including crosses of Caenorhabditis briggsae and C. nigoni. Fertile F1 females are obtained from reciprocal crosses. However, F1 males obtained from C. nigoni mothers are sterile and F1 males obtained from C. briggsae die during embryogenesis. We have identified cbr-him-8 as a recessive maternal-effect suppressor of F1 hybrid male-specific lethality in this combination of species. This result implicates epigenetic meiotic silencing in the suppression of F1 male-specific lethality. It is also shown that F1 males bearing a C. briggsae X chromosome are fertile. When crossed to C. briggsae hermaphrodites or F1 females derived from C. briggsae hermaphrodites, viable F2 and backcross (B2) progeny were obtained. Sibling males that possessed a C. nigoni X chromosome were sterile. Therefore, the sterility of F1 males bearing a C. nigoni X chromosome must result from dysgenic interactions between the X chromosome of C. nigoni and the autosomes of C. briggsae. The fertility of F1 males bearing a C. briggsae X chromosome provides an opportunity to identify C. nigoni loci that prevent spermatogenesis, and hence hermaphroditic reproduction, in diplo-X hybrids.
Keywords: hybrid lethality, hybrid sterility, reproductive isolation, Haldane’s rule, Darwin’s corollary
Reproductive isolation refers collectively to all genetic mechanisms that prevent or limit gene flow between populations (Mayr 1963; Coyne and Orr 2004). These mechanisms can be divided into two discrete categories, prezygotic mechanisms that prevent mating or fertilization and postzygotic mechanisms that decrease the fitness hybrid progeny. Most genetic models of reproductive isolation invoke dysgenic interactions among two or more loci (Dobzhansky 1936; Muller 1940, 1942; Wu 2001; Lindtke and Buerkle 2015). Within populations, interactions among these genes are maintained. Between populations, interactions among these genes are disrupted. Genes involved in reproductive isolation “are ordinary genes that have normal functions within species” (Orr et al. 2004).
Postzygotic mechanisms of reproductive isolation include hybrid sterility and hybrid lethality. Genes involved in hybrid sterility and inviability include a receptor tyrosine kinase, transcription factors, nuclear pore proteins, and a histone H3 methyltransferase (Wittbrodt et al. 1989; Ting et al. 1998; Presgraves et al. 2003; Barbash et al. 2004; Tang and Presgraves 2009; Phadnis and Orr 2009; Mihola et al. 2009). While reproductive isolation may evolve through nonselective mechanisms (Mayr 1963), there is evidence that many of these and other ‘speciation genes’ are or have been under positive selection (Johnson 2010; Ting et al. 1998; Presgraves et al. 2003; Barbash et al. 2004; Tang and Presgraves 2009; Araripe et al. 2010; Hart et al. 2014). Therefore, speciation can result from adaptive evolution of normal cellular processes.
Two patterns frequently observed in postzygotic reproductive isolation are Haldane’s rule and Darwin’s corollary to Haldane’s rule. Haldane’s rule is the observation that when gender-specific differences are observed in hybrid fitness, it is generally the homogametic gender that is more fit (Haldane 1922; Laurie 1997; Coyne and Orr 2004). Darwin’s corollary to Haldane’s rule is the observation that reciprocal hybrid crosses often produce different results (Turelli and Moyle 2007). These patterns are of interest because of how they inform our understanding of speciation (Coyne and Orr 2004).
The primary explanation for Haldane’s rule is the dominance model (Wu and Davis 1993; Turelli and Orr 2000). The dominance model posits that most hybrid incompatibility genes are recessive. F1 female hybrids that are heterozygous for an X-linked hybrid incompatibility gene are viable. F1 male hybrids that are hemizygous for that gene are inviable. Support for this model in regard to hybrid lethality is especially strong (Wu and Davis 1993). The primary explanation for Darwin’s corollary is that F1 hybrids from reciprocal crosses have different mitochondria, different maternal contributions and F1 males have different X chromosomes (Turelli and Moyle 2007).
In the nematode genus Caenorhabditis, many species pairs are isolated by hybrid sterility and/or by hybrid lethality (Baird et al. 1992; Baird and Yen 2000; Woodruff et al. 2010; Kiontke et al. 2011; Kozlowska et al. 2011; Dey et al. 2012; Baird and Seibert 2013; Félix et al. 2014; Dey et al. 2014). Among these is the combination of Caenorhabditis briggsae and Caenorhabditis nigoni (Woodruff et al. 2010; Kozlowska et al. 2011). From crosses of C. briggsae males to C. nigoni females, fertile F1 adult females and sterile F1 adult males were obtained. Fertile adult females also are obtained from the reciprocal cross but all male hybrids die during embryogenesis. Therefore, both Haldane’s rule and Darwin’s corollary to Haldane’s rule are observed in crosses between C. briggsae and C. nigoni.
In this article, cbr-him-8 is identified as a maternal-effect suppressor of F1 male-specific lethality in crosses of C. nigoni males to C. briggsae hermaphrodites. It also is demonstrated that F1 males derived from cbr-him-8 mutant mothers that possess a C. briggsae X chromosome are fertile. Finally, it is shown that fertile adult progeny can be obtained from crosses of these C. briggsae-X-bearing F1 males to C. briggsae hermaphrodites or to F1 females derived from C. briggsae mothers.
Materials and Methods
Nematode strains and strain maintenance
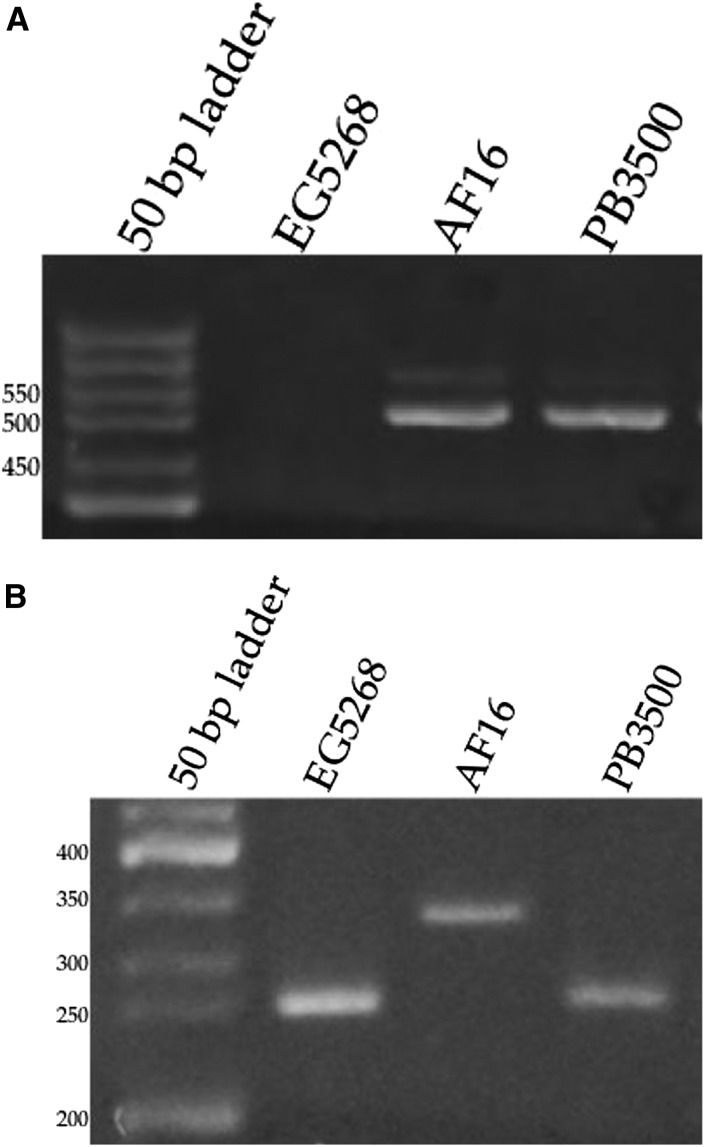
C. nigoni EG5268 (Kiontke et al. 2011; Félix et al. 2014) was provided by Marie-Anne Félix. C. briggsae AF16 (Fodor et al. 1983) was obtained from the Caenorhabditis Genetics Center. The C. briggsae AF16 derivatives RE980 [cbr-him-8(v188) I] (Wei et al. 2013) and RW20120 [stIs20120 (pmyo2::GFP) X] (Yan et al. 2012) were provided by Ron Ellis and Zhongying Zhao, respectively. PB192 [cbr-him-8(v188) I; stIs20120 X] was constructed from crosses of RE980 to RW20120. PB3500 was constructed from crosses of EG5268 males to AF16 hermaphrodites. Female progeny from this cross were backcrossed to EG5268 males for ten generations. Consistent with fixation of the C. nigoni nuclear genome, PB3500 had a female reproductive mode. Fixation of the C. briggsae AF16 mitotype and of the C. nigoni X chromosome in PB3500 was confirmed by amplification of species-specific mitochondrial and X chromosomal DNA products (Figure 1). Nematode strains were grown at 20° on lawns of Escherichia coli strain DA837. All strains used in this study are available from the Caenorhabditis Genetics Center.
Figure 1.
Confirmation of PB3500 cybrid genotypes. (A) Mitochondrial amplification products. Primers: cbr-nad-5 - AGCCAAACTCTAACACCACCT and cbr-nad-3 - TTCTTGGGGATTTTAGTTTCTGA. A 506 bp amplification product was expected from C. briggsae AF16 mitochondria. No product expected from C. nigoni EG5268 mitochondria. (B) Amplification products from the X-linked cbr-vab-3 and cni-vab-3 orthologs. Amplification products of 334 and 297 bp were expected from C. briggsae AF16 and C. nigoni EG5268, respectively. Primers: exon 4 - TGCACTCGGGCATACTGTAA and exon 6 - TGTACAACGGGCTCAGTCAG.
Crosses
Crosses always were of five males mated to three females or sperm-depleted hermaphrodites, and were conducted on freshly seeded mating plates (plates seeded with an approximately 1 cm spot of E. coli). Hermaphrodites were sperm-depleted by daily transfers for 4–5 d to fresh plates until egg laying ceased.
Microscopy
Crosses and routine microscopy were conducted using stereomicroscopes at magnifications of 25–50×. Pharyngeal GFP fluorescence was scored using an M2Bio fluorescence microscope (Kramer Scientific). Analyses of gonadal morphology were conducted using DIC optics at a magnification of 400× on a Zeiss Axiovert 35M microscope.
Reagents
All strains used in this study are available from the Caenorhabditis Genetics Center.
Data availability
Supplemental data on control crosses between C. nigoni EG5268 males and C. briggsae RW20120 hermaphrodites is available at figshare.com/articles/EG5268_x_RW20120_suppl_data_xlsx/2058864.
Results
F1 male-specific lethality is suppressed by cbr-him-8(v188)
Asymmetric results were observed in reciprocal crosses between the Caenorhabditis species C. nigoni and C. briggsae (Figure 2, A and B, and Table 1). Despite considerable embryonic lethality, viable and fertile F1 hybrid females were obtained from both cross directions. From C. nigoni mothers, some viable but sterile F1 hybrid males were obtained. However, from C. briggsae mothers, all F1 hybrid males died during embryogenesis. These results were consistent with results reported by Woodruff et al. (2010) and Kozlowska et al. (2011). Woodruff et al. (2010) reported no viable males from 186 F1s scored. Kozlowska et al. (2011) reported only seven viable males from 3705 F1s scored. Similarly, from 429 F1s scored in this study, no viable F1 males were observed (Table 1).
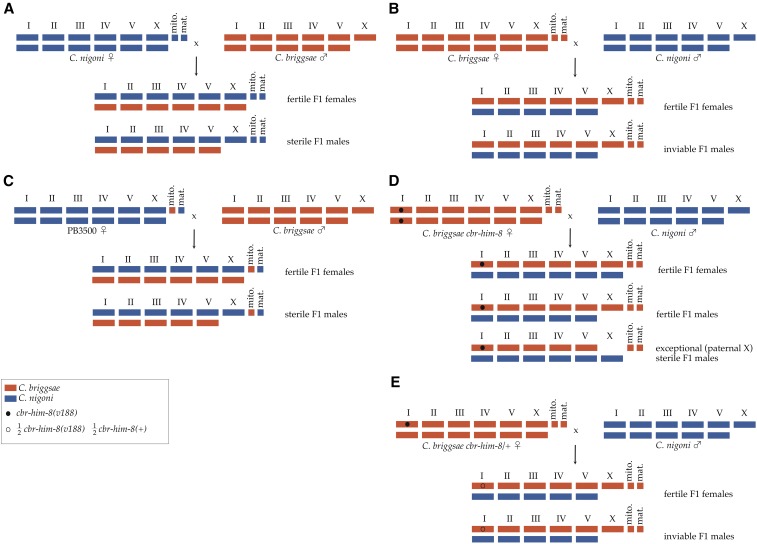
Figure 2.
Chromosome and mitochondrial segregation and maternal contributions in C. briggsae × C. nigoni hybrid crosses. In all panels, C. briggsae and C. nigoni genotypes are indicated in red and blue, respectively. In F1 hybrids, maternal chromosomes are shown above paternal chromosomes. In panels D and E, the v188 mutant allele of cbr-him-8 is indicated by a closed circle on chromosome I. In panel E, an open circle on chromosome I indicates that half of F1 hybrids were expected to be heterozygous for cbr-him-8(v188). Diagrammed are crosses between (A) C. nigoni females and C. briggsae males, (B) sperm-depleted C. briggsae hermaphrodites and C. nigoni males, (C) PB3500 cybrid females and C. briggsae males, (D) sperm-depleted C. briggsae cbr-him-8 mutant hermaphrodites and C. nigoni males, and (E) sperm-depleted C. briggsae cbr-him-8/+ heterozygous hermaphrodites and C. nigoni males.
Table 1. Frequency of F1 males derived from C. briggsae mothers.
| Cross | ♀♀ | ♂♂ | Fract. ♂ | ♂ Fract. XCbr (N) |
|---|---|---|---|---|
| C. briggsae AF16 ♂♂ × C. nigoni EG5268 ♀♀a | 293 | 32 | 0.098b | |
| C. nigoni EG5268 ♂♂ × C. briggsae AF16 ♀♀c | 429 | 0 | 0.000 | |
| C. briggsae AF16 ♂♂ × PB3500 cybrid ♀♀d | 383 | 39 | 0.092b | |
| C. nigoni EG5268 ♂♂ × C. briggsae RE980 ♀♀e | 330 | 68 | 0.171 | |
| C. nigoni EG5268 ♂♂ × C. briggsae PB192 ♀♀e | 634 | 142 | 0.183 | 0.60 (131)f |
| C. nigoni EG5268 ♂♂ × C. briggsae cbr-him-8(v188) ♀♀e,g | 964 | 210 | 0.179 |
AF16, C. briggsae wild-isolate; EG5268, C. nigoni wild-isolate; PB3500, EG5268 nuclear genome and AF16 mitochondria; RE980, C. briggsae cbr-him-8(v188) I; PB192, C. briggsae cbr-him-8(v188) I; stIs20120 [pmyo2::GFP] X (RE980 and PB192 are both AF16 derivatives).
These crosses are diagrammed in Figure 2, A, B, C, and D, respectively.
♂ frequencies not significantly different, P = 0.677 chi squared test, expected frequency = 0.098.
Sum of results from crosses using RE980 and PB192 ♀♀.
Pharyngeal expression of GFP observed in 79 of 131 F1 males scored.
F1 males from reciprocal crosses in Caenorhabditis differed in the source of their maternally derived X chromosome, their maternally derived mitochondria, and in maternal contributions to the oocyte prior to fertilization (Figure 2). These differences have been proposed as potential causes of asymmetric results in reciprocal crosses (Turelli and Moyle 2007; Dey et al. 2014). To test for dysgenic mitonuclear interactions, C. briggsae males were mated to females from the PB3500 cybrid strain. This strain possessed a C. nigoni nuclear genome and C. briggsae mitochondria (Figure 1). Viable F1 males were obtained from this cross. Frequencies of F1 males obtained from crosses of C. briggsae AF16 males to C. nigoni EG5268 and cybrid PB3500 mothers were identical (Figure 2C and Table 1). As mitochondria are maternally inherited, males derived from PB3500 mothers would have possessed C. briggsae mitochondria. The viability of these F1 males is not consistent with dysgenic mitonuclear interactions as a cause of F1 male-specific lethality of F1 males derived from C. briggsae mothers. This result was consistent with those of Bundus et al. (2015), who found that C. briggsae mitochondria did not have an impact on postzygotic reproductive isolation in crosses between C. briggsae and C. nigoni.
To discriminate between maternal-zygotic and X-autosomal interactions, C. nigoni males were mated to sperm-depleted C. briggsae cbr-him-8(v188) I hermaphrodites. The cbr-him-8(v188) mutation results in high rates of X chromosome nondisjunction and hence in high frequencies of XO males among self-progeny of mutant hermaphrodites (Wei et al. 2013). It was thought that this cross would produce exceptional males with a paternal C. nigoni X chromosome (XCni) through the fertilization of nullo-X oocytes by X-bearing sperm. Viability of these males would eliminate C. briggsae maternal-zygotic interactions as the cause of asymmetric F1 male-specific lethality. Viable F1 males were obtained from C. briggsae cbr-him-8(v188) mutant mothers (Figure 2D and Table 1). However, only 40% of these were the expected exceptional XCni males (Table 1). The rest of the viable F1 males possessed a maternally derived C. briggsae X (XCbr) chromosome. This was determined from crosses of C. nigoni males to hermaphrodites from the PB192 strain of C. briggsae. PB192 is an AF16 derivative that was mutant for cbr-him-8(v188) and that also included an X-linked insertion, stIs20120, of a cbr-myo2p::GFP transgene. Expression from stIs20120 results in pharyngeal GFP fluorescence (Yan et al. 2012). Frequencies of F1 males obtained from crosses that included or did not include stIs20120 were identical (Table 1). As PB192 is an AF16 derivative, the only difference between the viable XCbr F1 males derived from PB192 mothers and the inviable XCbr F1 males derived from wild-type AF16 C. briggsae mothers was the presence of cbr-him-8(v188) and stIs20120. In control crosses, stIs20120 was shown to have no effect on F1 male viability (not shown). For viable XCbr F1 males, cbr-him-8 was homozygous in the maternal genome and heterozygous in the zygotic genome. Hence, cbr-him-8(v188) was identified as a suppressor of the lethality of F1 XCbr males.
Suppression of hybrid by cbr-him-8(v188) is a maternal effect
In Caenorhabditis elegans, mutations in him-8 exhibit two distinct and separable phenotypes. Homozygosity of him-8 results in high rates of X chromosome nondisjunction (Hodgkin et al. 1979). This is caused by defects in X chromosome pairing during meiosis (Phillips et al. 2005). In somatic cells, him-8 mutations are dominant suppressors of missense mutations in transcription factor binding domains (Nelms and Hanna-Rose 2006; Sun et al. 2007). If C. briggsae cbr-him-8(v188) exhibits both of these phenotypes, then suppression of F1 male-specific lethality could be the result of maternal homozygosity or zygotic heterozygosity.
To distinguish between maternal and zygotic modes of suppression, C. nigoni males were crossed with cbr-him-8/+ C. briggsae heterozygotes. The X chromosome nondisjunction phenotype of cbr-him-8(v188) is recessive. If suppression results from X chromosome pairing defects during meiosis, then few if any F1 males would be expected from cbr-him-8 heterozygous mothers. Conversely, half of F1 male progeny from heterozygous mothers would inherit the mutant allele of cbr-him-8. These males would be genetically identical to F1 males derived from cbr-him-8 homozygotes. If suppression results from somatic suppression of transcription factor binding defects, then the abundance of viable F1 XCbr males derived from heterozygous mothers would be expected to be half of that observed from cbr-him-8 homozygotes. From crosses of C. nigoni males to C. briggsae cbr-him-8/+ hermaphrodites, a single F1 male was observed among 354 viable F1 progeny scored (Figure 2E and Table 2). This result excludes zygotic suppression but is consistent with maternal pairing defects as the cause of suppression of XCbr F1 male-specific lethality.
Table 2. Tests of zygotic and maternal suppression hypotheses.
| Observed | Females | Males | P valuea |
|---|---|---|---|
| C. nigoni × C. briggsae cbr-him-8/+b,c | 353 | 1 | |
| Expected | |||
| Zygotic suppressiond | 331.3 | 22.7 | 2.567 × 10−6 |
| Maternal suppressione | 353.3 | 0.7 | 0.685 |
P values from chi squared tests using the expected male frequencies for the zygotic and maternal suppression hypotheses described above.
C. nigoni EG5268 ♂♂ × C. briggsae cbr-him-8(v188)/+ I; stIs20120 [p-myo2::GFP] X, or C. nigoni EG5268 ♂♂ × C. briggsae cbr-him-8(v188)/+ I; stIs20120 [p-myo2::GFP]/+ X.
This cross is diagrammed in Figure 2E.
An expected male frequency of 6.4% was based on the expected 50% transmission rate of cbr-him-8(v188) from maternal heterozygotes and on the 12.8% frequency of viable adult XCbr males from cbr-him-8(v188) homozygous mothers.
An expected male frequency of 0.19% was based on the frequency of viable males obtained from crosses of C. nigoni males to wild-type C. briggsae hermaphrodites (Kozlowska et al. 2011).
F1 XCbr males are fertile
F1 XCbr males derived from crosses of C. nigoni males to C. briggsae cbr-him-8 mutant hermaphrodites had well-developed gonads and were fertile (Figure 3 and Table 3). When F1 XCbr males were crossed to C. nigoni females, fertilized embryos were observed. All of these embryos arrested prior to hatching. When F1 XCbr males were mated to C. briggsae hermaphrodites, viable F2 adult progeny were obtained. When F1 XCbr males were crossed to F1 females, the result varied depending upon the source of F1 females. When crossed to F1 females derived from C. nigoni mothers (F1Cni females), only arrested embryos were observed. When crossed to F1 females derived from C. briggsae mothers (F1Cbr females), viable F2 adults were obtained approximately a third of the time. Further crosses will be required to determine if these differences are significant.
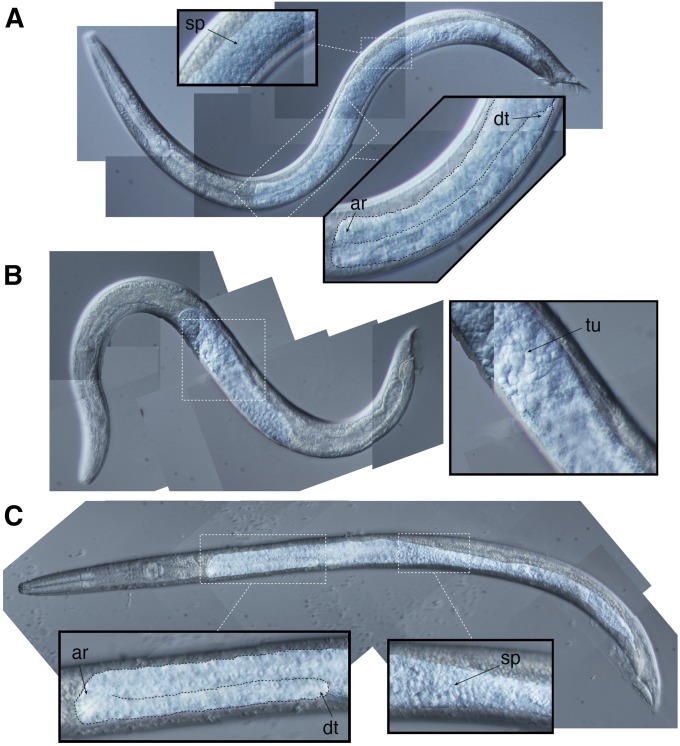
Figure 3.
Gonad morphology in F1 male hybrids. (A) C. nigoni EG5268, (B) F1 XCni, and (C) F1 XCbr males. Contrast of gonads enhanced in all panels. Boxes correspond to regions enlarged in insets. In panels A and C, the distal arm is outlined with a dashed line in the large insets to emphasize the tubular structure of the gonad. This tubular structure is absent in the F1 XCni male shown in panel C. Anterior reflex (ar), distal tip (dt), sperm (sp), and tumorous cells (tu) indicated in insets. The C. F1 XCni male was an ‘exceptional’ GFP– male obtained from crosses on C. nigoni EG5268 males to C. briggsae PB192 [cbr-him-8(v188) I; stIs20120 (pmyo2::GFP) X] hermaphrodites. The F1 XCbr male was a GFP+ male obtained from the same cross.
Table 3. Fertility of F1 XCbr males.
| Crossa | Resultb | Self-Fertile F2 | F2 Male |
|---|---|---|---|
| Female Fraction Nc | Fraction (N) | ||
| F1 XCbr ♂ × C. nigoni ♀ | Dead embryos (5) | ||
| No progeny (3) | |||
| F1 XCbr ♂ × C. briggsae ♀ | Viable adults (16) | 0.98 (48) | 0.20 (869) |
| F1 XCbr ♂ × F1Cni ♀ | Dead embryos (2) | ||
| F1 XCbr ♂ × F1Cbr ♀d | Dead embryos (6) | ||
| Viable adults (3) | ndf | ∼0.50e | |
| Viable adults (1) | 1.00 (30) | 0.005 (208) | |
| No progeny (1) |
F1 XCbr ♂ = GFP+ males derived from PB192 mothers, F1 ♀Cni = F1 females derived from C. nigoni mothers. F1 ♀Cbr = F1 females derived from C. briggsae mothers.
Number of crosses for each given result indicated in parentheses.
Fraction of anatomically female (i.e., XX) F2s that laid eggs. Number scored indicated in parentheses.
Includes results of full sib crosses as well as results of F1 XCbr males from PB192 mothers crossed to F1 females from AF16 mothers.
F2 males abundant but not counted. It is not clear why males were abundant in some crosses but not in others.
Not done.
Adult male, female and hermaphrodite progeny were obtained from crosses of F1 XCbr males to C. briggsae hermaphrodites and F1Cbr females (Table 3). However, the frequencies of these different progeny types were not consistent with expectations. Among cross progeny, haplo-X males were expected at a frequency of 0.50. From crosses to C. briggsae hermaphrodites, observed frequency of males, 0.20, was significantly lower than this expectation (P < 0.0001). From crosses to F1Cbr females, F2 males were sometimes, but not always, abundant. From both crosses, nearly all diplo-X progeny were self-fertile. Self-sterile (female) and self-fertile (hermaphrodite) diplo-X progeny were both expected from these crosses. However, Woodruff et al. (2010) demonstrated that self-sterility (female reproductive mode) was dominant and they observed very low frequencies (< 3%) of self-fertility among progeny of 2nd or 3rd generation hybrid males crossed to C. briggsae hermaphrodites. The high rates of self-fertility, ≥ 0.98, observed among diplo-X progeny was not consistent with this observation.
F1 XCni males are sterile regardless of cross direction
F1 XCni males derived from C. nigoni mothers have gonad defects and are sterile (Woodruff et al. 2010). In general, these F1 males were defective in gonadal outgrowth (Table 4). Gonad outgrowth in C. nigoni and C. briggsae is nearly identical to gonad outgrowth in C. elegans. Gonad outgrowth in C. elegans is regulated by the migration of the linker cell (Kimble and Hirsh 1979; Kato and Sternberg 2009). The linker cell initially migrates anteriorly along the ventral body wall until the L2 larval molt. It then migrates to the dorsal body wall where it turns and migrates posteriorly during the L3 and L4 larval stages. The result of these migrations is a thin tubular gonad with an anterior reflex (180° bend) near the posterior bulb of the pharynx. In some F1 XCni males, there is an apparent complete failure in gonad outgrowth. These males possess gonads that differ little from the gonad primordium present in L1 larvae at hatching. In other F1 XCni males, there is an apparent failure in the dorsal turn of the linker cell at the L2 molt. These males possess swollen, ovoid gonads that lack an anterior reflex (Table 4).
Table 4. Gonadal phenotypes of F1 XCni males.
| Cross | No Outgrowtha | Defective Outgrowthb | N |
|---|---|---|---|
| C. briggsae AF16 ♂♂ × C. nigoni EG5268 ♀♀ | 9 | 10 | 19 |
| C. briggsae PB192 ♂♂ × C. nigoni EG5268 ♀♀ | 6 | 15 | 21c |
| C. nigoni EG5268 ♂♂ × C. briggsae PB192 ♀♀ | 5 | 2 | 7c |
AF16, C. briggsae wild-isolate; EG5268, C. nigoni wild-isolate; PB192, C. briggsae cbr-him-8(v188) I.
Small ventral ovoidal masses of gonadal tissue, or degenerate vacuoles, located at midbody.
Larger masses of gonadal tissue extending anteriorly toward the pharynx but lacking the anterior reflex. Differentiated and/or tumorous cells often observed.
Distributions of gonadal phenotypes in XCni males derived from PB192 ♂♂ × EG5268 ♀♀ and EG5268 ♂♂ × PB192 ♀♀ do not differ significantly from the distribution of phenotypes derived from AF16 ♂♂ × EG5268 ♀♀. P values 0.084 and 0.20, respectively.
F1 XCni males obtained from C. briggsae cbr-him-8(v188) mutant mothers had the same gonadal outgrowth defects as those observed in F1 XCni males derived from C. nigoni mothers (Figure 3 and Table 4). The only genetic difference between these males and their F1 XCbr male siblings, which had well-developed functional gonads, was the X chromosome. Based on these results, the gonadal outgrowth defects observed in F1 XCni males must result from the presence of a hybrid sterile gene on the X chromosome of C. nigoni.
Discussion
In crosses between C. nigoni males and C. briggsae hermaphrodites, almost all F1 male hybrids die during embryogenesis. This F1 hybrid male-specific lethality was suppressed by the cbr-him-8(v188) mutation. This result was unexpected. There is evidence that F1 male-specific lethality results from dysgenic interactions between a C. briggsae X-linked locus and C. nigoni autosomal loci (Bi et al. 2015). cbr-him-8 does not correspond to this X-linked gene as it is located on chromosome I (www.wormbase.org). Rather, cbr-him-8 must be acting as a suppressor of this hybrid lethality gene.
In C. elegans, mutations in him-8 are pleiotropic. The HIM-8 protein binds to the pairing centers of the X chromosomes and is required for the meiotic pairing of X chromosomes (Phillips et al. 2005). Consequences of disrupted meiotic pairing include X-specific nondisjunction and an expansion of recombination distances on the X chromosome (Hodgkin et al. 1979; Broverman and Meneely 1994). The X-specific nondisjunction phenotype of cbr-him-8(v188) and the conservation of HIM-8 proteins in these species indicate that the role of HIM-8 in X chromosome pairing is conserved in C. briggsae (Phillips and Dernberg 2006; Wei et al. 2013). C. elegans him-8 mutations also act as dominant suppressors of missense mutations in the DNA-binding domains of transcription factors (Nelms and Hanna-Rose 2006; Sun et al. 2007). Conservation of this phenotype in C. briggsae has not been tested.
The suppression of F1 male-specific lethality by cbr-him-8 likely results from defects in X chromosome meiotic pairing during oogenesis in C. briggsae. This was evident from crosses of C. nigoni males to C. briggsae cbr-him-8/+ hermaphrodites. The X-nondisjunction phenotype, and hence the pairing defects, of cbr-him-8 are recessive. However, half the F1 hybrids derived from cbr-him-8/+ heterozygous mothers would also have been heterozygous for cbr-him-8. Thus, the absence of viable F1 male progeny from cbr-him-8/+ mothers demonstrates that zygotic heterozygosity of cbr-him-8(v188) is not sufficient to suppress male-specific lethality in F1 hybrids.
The suppression of F1 male-specific lethality by cbr-him-8 may result from meiotic silencing of the C. briggsae X chromosomes during oogenesis or from epigenetic suppression of X-linked gene expression during embryogenesis. In C. elegans, unpaired chromosomes are dimethylated on lysine 9 of histone H3 (H3K9me2) during meiosis (Bean et al. 2004; Bessler et al. 2010). H3K9me2 is a highly conserved epigenetic mark that is associated with transcriptional repression and meiotic silencing (Turner 2007; Kelly and Aramayo 2007; Kota and Feil 2010; Maine 2010; Mozzetta et al. 2015). Acquisition of H3K9me2 on unpaired X chromosomes in C. elegans her-1 XO hermaphrodites is associated with meiotic repression of transcription of X-linked genes (Bean et al. 2004). However, the repressive epigenetic imprint acquired by the X chromosome during spermatogenesis can also persist through the 14-cell stage of embryogenesis (Kelly et al. 2002; Bean et al. 2004). It should be possible to test for suppression by meiotic silencing by generating a mutation in cbr-met-2. In C. elegans, met-2 is required for dimethylation of H3K9 (Bessler et al. 2010). If suppression of F1 male-specific lethality results from H3K9me2 of X chromosomes in cbr-him-8 mutant hermaphrodites, then XCbr F1 males derived from cbr-him-8; cbr-met-2 doubly mutant hermaphrodites should die during embryogenesis.
Our results also demonstrated that the sterility of XCni F1 males was caused by dysgenic interactions between the X chromosome of C. nigoni and the autosomes of C. briggsae. From C. briggsae cbr-him-8 mothers, both XCni and XCbr F1 males were obtained. The XCbr F1 males had well-developed gonads and were fertile whereas their XCni siblings had defects in gonad development and were sterile. The XCbr and XCni males obtained from these crosses shared the same maternal and mitochondrial genotypes. They differed only in the identity of their X chromosomes. Moreover, unpaired X chromosomes in male spermatogenesis (i.e., XCni) were expected to share similar epigenetic modifications as unpaired X chromosomes in hermaphrodite oogenesis in cbr-him-8 mutant mothers (Bean et al. 2004). Thus, the cryptic asymmetry observed in F1 male fertility likely results from the divergence of one or more loci on the C. briggsae and C. nigoni X chromosomes.
Finally, the fertility of F1 XCbr males provides an opportunity to define the genetic requirements for hermaphroditic reproduction in C. briggsae. Woodruff et al. (2010) demonstrated that the hermaphroditic mode of reproduction was recessive to the female mode in diplo-X hybrids. We found females to be rare among diplo-X backcross progeny of XCbr males mated to C. briggsae hermaphrodites. Genotyping of these rare backcross females should allow for the identification of C. nigoni loci that suppress spermatogenesis in female hybrids.
Acknowledgments
We thank Labib Rouhana, Eric Haag, and two anonymous reviewers for their insightful comments and critical reading of the manuscript. We thank Ron Ellis, Zhonying Zhao, and Marie-Anne Félix for generously providing us with the RE980 and RW20120 strains of C. briggsae and the EG5268 strain of C. nigoni, respectively. Other strains were provided by the Caenorhabditis Genetic Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440).
Footnotes
Communicating editor: P. C. Phillips
Literature Cited
- Araripe L. O., Montenegro H., Lemos B., Hartl D. L., 2010. Fine-scale mapping of a hybrid sterility factor between Drosophila simulans and D. mauritiana: the varied and elusive functions of “speciation genes”. BMC Evol. Biol. 10: 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird S. E., Seibert S. R., 2013. Reproductive isolation in the Elegans-Group of Caenorhabditis. Nat. Sci. 5(4A): 18–25. [Google Scholar]
- Baird S. E., Yen W.-C., 2000. Reproductive isolation in Caenorhabditis: terminal phenotypes of hybrid embryos. Evol. Dev. 2: 9–15. [DOI] [PubMed] [Google Scholar]
- Baird S. E., Sutherlin M. E., Emmons S. W., 1992. Reproductive isolation in Rhabditidae (Nematoda: Secernentea); mechanisms that isolate six species of three genera. Evolution 46: 585–594. [DOI] [PubMed] [Google Scholar]
- Barbash D. A., Awadalla P., Tarone A. M., 2004. Functional divergence caused by ancient positive selection of a Drosophila hybrid incompatibility locus. PLoS Biol. 2(6): 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean C. J., Schaner C. E., Kelly W. G., 2004. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat. Genet. 36: 100–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessler J. B., Andersen E. C., Villeneuve A. M., 2010. Differential localization and independent acquisition of H3K9me2 and H3K9me3 chromatin modifications in the Caenorhabditis elegans adult germ line. PLoS Genet. 6(1): e1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi Y., Ren X., Yan C., Shao J., Xie D., et al. , 2015. A genome-wide hybrid incompatibility landscape between Caenorhabditis briggsae and C. nigoni. PLoS Genet. 11(2): e1004993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broverman S. A., Meneely P. M., 1994. Meiotic mutants that cause a polar decrease in recombination on the X chromosome in Caenorhabditis elegans. Genetics 136: 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundus J. D., Alaei R., Cutter A. D., 2015. Gametic selection, developmental trajectories, and extrinsic heterogeneity in Haldane’s rule. Evolution 69: 2005–2017. [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation, Sinauer Associates, Sunderland, MA. [Google Scholar]
- Dey A., Jeon Y., Wang G.-X., Cutter A. D., 2012. Global population genetic structure of Caenorhabditis remanei reveals incipient speciation. Genetics 191: 1257–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey A., Jin Q., Chen Y.-C., Cutter A. D., 2014. Gonad morphogenesis defects drive hybrid male sterility in asymmetric hybrid breakdown of Caenorhabditis nematodes. Evol. Dev. 16: 363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T., 1936. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila pseudoobscura hybrids. Genetics 21: 113–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix M.-A., Braendle C., Cutter A. D., 2014. A streamlined system for species diagnosis in Caenorhabditis (Nematoda: Rhabditidae) with name designations for 15 distinct biological species. PLoS One 9(4): e94723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor A., Riddle D. L., Nelson F. K., Golden J. W., 1983. Comparison of a new wild-type Caenorhabditis briggsae with laboratory strains of C. briggsae and C. elegans. Nematologica 29: 203–217. [Google Scholar]
- Haldane J. B. S., 1922. Sex ratio and unisexual sterility in hybrid animals. J. Genet. 12: 101–109. [Google Scholar]
- Hart M. W., Sunday J. M., Popovic I., Learning K. J., Konrad C. M., 2014. Incipient speciation of sea star populations by adaptive gamete recognition coevolution. Evolution 68: 1294–1305. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Horvitz H. R., Brenner S., 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91: 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson N. A., 2010. Hybrid incompatibility genes: remnants of a genomic battlefield? Trends Genet. 26: 317–325. [DOI] [PubMed] [Google Scholar]
- Kelly W. G., Aramayo R., 2007. Meiotic silencing and the epigenetics of sex. Chrom. Res. 15: 633–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly W. G., Schaner C. E., Dernburg A. F., Lee M.-H., Kim S. K., et al. , 2002. X-chromosome silencing in the germline of C. elegans. Development 129: 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Sternberg P. W., 2009. The C. elegans tailess/Tlx homolog nhr-67 regulates a stage-specific program of linker cell migration in male gonadogenesis. Development 136: 3907–3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., Hirsh D., 1979. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70: 396–417. [DOI] [PubMed] [Google Scholar]
- Kiontke K. C., Félix M.-A., Ailion M., Rockman M. V., Braendle C., et al. , 2011. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol. Biol. 11: 339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kota S. K., Feil R., 2010. Epigenetic transitions in germ cell development and meiosis. Dev. Cell 19: 675–686. [DOI] [PubMed] [Google Scholar]
- Kozlowska J. L., Ahmad A. R., Jahesh E., Cutter A. D., 2011. Genetic variation for postzygotic reproductive isolation between Caenorhabditis briggsae and Caenorhabditis sp. 9. Evol. 66: 1180–1195. [DOI] [PubMed] [Google Scholar]
- Laurie C. C., 1997. The weaker sex is heterogametic: 75 years of Haldane’s rule. Genetics 147: 937–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindtke D., Buerkle C. A., 2015. The genetic architecture of hybrid incompatibilities and their effect on barriers to introgression in secondary contact. Evolution 69: 1987–2004. [DOI] [PubMed] [Google Scholar]
- Maine E. M., 2010. Meiotic silencing in Caenorhabditis elegans, pp. 91–134 in International Review of Cell and Molecular Biology, v282, edited by Jeon K. W. Elsevier, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- Mayr E., 1963. Animal Species and Evolution, Harvard University Press, Cambridge, MA. [Google Scholar]
- Mihola O., Trachtulec Z., Vlcek C., Schimenti J. C., Forejt J., 2009. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323: 373–375. [DOI] [PubMed] [Google Scholar]
- Mozzetta C., Boyarchuk E., Pontis J., Ait-Si-Ali S., 2015. Sound of silence: the properties and functions of repressive Lys methyltransferases. Natl. Rev. 16: 499–513. [DOI] [PubMed] [Google Scholar]
- Muller H. J., 1940. Bearing of the Drosophila work on systematics, pp. 185–268 in The New Systematics, edited by Huxley J. S. Clarendon Press, Oxford. [Google Scholar]
- Muller H. J., 1942. Isolating mechanisms, evolution and temperature. Biol. Symp. 6: 71–125. [Google Scholar]
- Nelms B. L., Hanna-Rose W., 2006. C. elegans HIM-8 functions outside of meiosis to antagonize EGL-13 Sox protein function. Dev. Biol. 293: 392–402. [DOI] [PubMed] [Google Scholar]
- Orr H. A., Masly J. P., Presgraves D. C., 2004. Speciation genes. Curr. Opin. Genet. Dev. 14: 675–679. [DOI] [PubMed] [Google Scholar]
- Phadnis N., Orr H. A., 2009. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323: 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. M., Dernberg A. F., 2006. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell 11: 817–829. [DOI] [PubMed] [Google Scholar]
- Phillips C. M., Wong C., Bhalla N., Carlton P. M., Weiser P., et al. , 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123: 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., Balagopalan L., Abmayr S. M., Orr H. A., 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423: 715–719. [DOI] [PubMed] [Google Scholar]
- Sun H., Nelms B. L., Sleiman S. F., Chamberlin H. M., Hanna-Rose W., 2007. Modulation of Caenorhabditis elegans transcription factor activity by HIM-8 and the related zinc-finger ZIM proteins. Genetics 177: 1221–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Presgraves D. C., 2009. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science 323: 779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting C.-T., Tsaur S.-C., Wu M.-L., Wu C.-I., 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282: 1501–1504. [DOI] [PubMed] [Google Scholar]
- Turelli M., Orr H. A., 2000. Dominance, epistasis and the genetics of postzygotic isolation. Genetics 154: 1663–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M., Moyle L. C., 2007. Asymmetric postmating isolation: Darwin’s Corollary to Haldane’s Rule. Genetics 176: 1059–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J. M. A., 2007. Meiotic sex chromosome inactivation. Development 134: 1823–1831. [DOI] [PubMed] [Google Scholar]
- Wei Q., Shen Y., Chen X., Shifman Y., Ellis R. E., 2013. Rapid creation of forward-genetics tools for C. briggsae TALENs: lessons for nonmodel organisms. Mol. Biol. Evol. 31: 468–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittbrodt J., Adam D., Malitschek B., Mäueler W., Raulf F., et al. , 1989. Novel putative receptor tyrosine kinase encoded by the melanoma-inducing Tu locus in Xiphophorus. Nature 341: 415–421. [DOI] [PubMed] [Google Scholar]
- Woodruff G. C., Eke O., Baird S. E., Félix M.-A., Haag E. S., 2010. Insights into species divergence and the evolution of hermaphroditism from fertile interspecies hybrids of Caenorhabditis nematodes. Genetics 186: 997–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C.-I., 2001. The genic view of the process of speciation. J. Evol. Biol. 14: 851–865. [Google Scholar]
- Wu C.-I., Davis A. W., 1993. Evolution of postmating reproductive isolation: the composite nature of Haldane’s rule and its genetic basis. Am. Nat. 142: 187–212. [DOI] [PubMed] [Google Scholar]
- Yan C., Bi Y., Yin D., Zhao Z., 2012. A method for rapid and simultaneous mapping of genetic loci and introgression sizes in nematode species. PLoS One 7(8): e43770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Supplemental data on control crosses between C. nigoni EG5268 males and C. briggsae RW20120 hermaphrodites is available at figshare.com/articles/EG5268_x_RW20120_suppl_data_xlsx/2058864.