Abstract
Methylation of the O6-methylguanine-DNA methyltransferase (MGMT) gene is a predictive and prognostic marker in newly diagnosed glioblastoma patients treated with temozolomide but how MGMT methylation should be assessed to ensure optimal detection accuracy is debated. We developed a novel quantitative methylation-specific PCR (qMSP) MGMT assay capable of providing allelic methylation data and analyzed 151 glioblastomas from patients receiving standard of care treatment (Stupp protocol). The samples were also analyzed by immunohistochemistry (IHC), standard bisulfite pyrosequencing, and genotyped for the rs1690252 MGMT promoter single nucleotide polymorphism. Monoallelic methylation was observed more frequently than biallelic methylation, and some cases with monoallelic methylation expressed the MGMT protein whereas others did not. The presence of MGMT methylation was associated with better overall survival (p = 0.006; qMSP and p = 0.002; standard pyrosequencing), and the presence of the protein was associated with worse overall survival (p = 0.009). Combined analyses of qMSP and standard pyrosequencing or IHC identified additional patients who benefited from temozolomide treatment. Finally, low methylation levels were also associated with better overall survival (p = 0.061; qMSP and p = 0.02; standard pyrosequencing). These data support the use of both MGMT methylation and MGMT IHC but not allelic methylation data as prognostic markers in patients with temozolomide-treated glioblastoma.
Keywords: Alkylating agents, Allele-specific, Glioblastoma, Immunohistochemistry, Methylation, MGMT, Temozolomide.
INTRODUCTION
Silencing by promoter methylation of the O6-methylguanine-DNA methyltransferase (MGMT) gene is an early event in several human cancers due to its central role in DNA repair. MGMT removes alkyl adducts from the O6 position of guanine thereby preventing G>A mutations in the genome. Under normal circumstances, this is an important cellular function; however, since alkylating agents such as temozolomide introduce alkyl adducts in the genome, MGMT methylation may be a predictive marker of response to alkylating agents in patients with glioblastoma multiforme (GBM) (1–3). The use of MGMT methylation status for routine clinical decision-making has been hampered by the fact that most GBM patients receive a combination of radiotherapy and temozolomide regardless of MGMT methylation status because no other good treatment options are available. However, MGMT methylation testing is useful for selecting patients for clinical trials (4). Recent studies also suggest that treatment decisions in older patients, for whom monotherapy with either radiotherapy or temozolomide is favored, should take MGMT methylation into account (5, 6).
A functional role of MGMT methylation in GBM has been confirmed at the RNA and protein levels (7–9), but it is unclear how detection of MGMT methylation should be performed in clinical practice to ensure that as many patients as possible receive optimal treatment. We have previously shown that allelic methylation patterns as well as methylation levels influence MGMT protein expression in malignant pleural mesothelioma (10). If this were also observed in GBMs, allelic methylation patterns might also influence responses to temozolomide. In addition to MGMT methylation, the T-allele of the rs16906252 C>T single nucleotide polymorphism (SNP) located in the MGMT promoter region has been shown to have a negative influence on MGMT expression and is associated with longer survival in GBM patients treated with temozolomide (11, 12). This reduced activity may, in turn, predispose the allele to become methylated. In several malignant tumors, including GBM, the T-allele is associated with MGMT methylation (9–11, 13–16).
Both quantitative and qualitative methodologies have been employed for monitoring MGMT methylation (17–19). The analytical sensitivity varies substantially among different assays. In particular, assays based on methylation-specific PCR (MSP) primers are generally more sensitive than methods based on methylation-independent PCR primers (20, 21). Gel electrophoresis, pyrosequencing, Sanger sequencing, fluorescent probes, or high-resolution melting are most often used to determine the methylation level/status of the samples. In addition to the method of choice, other factors such as tumor content, intratumor heterogeneity, heterogeneous methylation, and allelic methylation status likely affect the results.
Bisulfite sequencing of single clones may provide allelic methylation information if a heterozygous SNP is located in between the primers, but this may suffer from PCR and cloning bias, and takes several days to perform (17). In addition, it would require sequencing hundreds of clones to determine allelic methylation patterns with high reliability in samples containing low methylation levels (14). Low methylation levels in GBM likely occur because of contamination with inflammatory and stromal cells and intratumoral heterogeneity. It is also still debated whether detection of the MGMT protein by immunohistochemistry (IHC) should be part of the standard assessment of newly diagnosed glioblastoma (22).
Here, we report the development of a novel quantitative methylation-specific PCR (qMSP) MGMT assay that also provides allelic methylation information for individuals who are heterozygous for the rs16906252 SNP. The qMSP is performed using a biotinylated PCR primer to allow subsequent pyrosequencing of the PCR amplicon, which is used to determine allelic methylation status of the samples. The analyses can be completed within a day and allow for allelic methylation analysis even in samples containing low methylation levels. The methylation data obtained by qMSP were compared to data obtained by a standard bisulfite pyrosequencing assay based on methylation-independent PCR primers (Therascreen (R) MGMT Pyro (R) Kit, Qiagen, Hilden, Germany) and MGMT IHC data. In total, 151 GBM tumor biopsies from patients who received standard of care treatment (Stupp protocol) were analyzed. Allelic methylation patterns were compared with IHC data and survival analyses were performed according to methylation data obtained by each method, rs16906252 genotypes, and MGMT IHC data. In addition, we performed combined survival analyses of qMSP and standard pyrosequencing as well as combined methylation and IHC analyses.
MATERIALS AND METHODS
Patient Samples
This retrospective study examined material from 151 patients treated at Rigshospitalet, Denmark, who had been diagnosed with GBM (World Health Organization [WHO] grade IV) according to the WHO 2000/2007 guidelines. Patient inclusion criteria were that tumor DNA was available or could be obtained from fresh frozen tumor samples and further that patients had received primary standard GBM therapy according to the Stupp regimen (ie, concomitant radiation and temozolomide therapy followed by up to 6 courses of adjuvant temozolomide therapy).
The patients were diagnosed from 2005 to 2010, and the median duration of observation from the day patients first received radiation/temozolomide therapy to the project cutoff day (March 18, 2015) was 94 months (range, 53–123 months). Detailed descriptions of the treatments and patient evaluations have been described elsewhere (23, 24); patient demographics and clinical characteristics are shown in Supplementary Data 1.
Ethics Statement
This study was performed according to the Declaration of Helsinki and Danish legislation. Permissions were given from the Danish Data Protection Agency (2006-41-6979) and the ethical committee for the Capital Region of Denmark (H-C-2008-095).
DNA Extraction
Genomic DNA was purified from approximately 50 mg fresh frozen homogenized tumor tissue by the Proteinase K-Phenol/Chloroform protocol as described (25).
rs16906252 SNP Genotyping
Genotyping was performed using pyrosequencing using previously published primer sequences as described (14). To validate the results, SNP genotyping was also performed using high-resolution melting analysis on 137 of the samples. Nine samples with uncertain pyrosequencing and/or high-resolution melting results were Sanger sequenced and this result was used in subsequent data analyses. Additional details can be found in the online Supplementary Data.
Quantitative and Allelic Methylation Analyses by qMSP Followed by Pyrosequencing
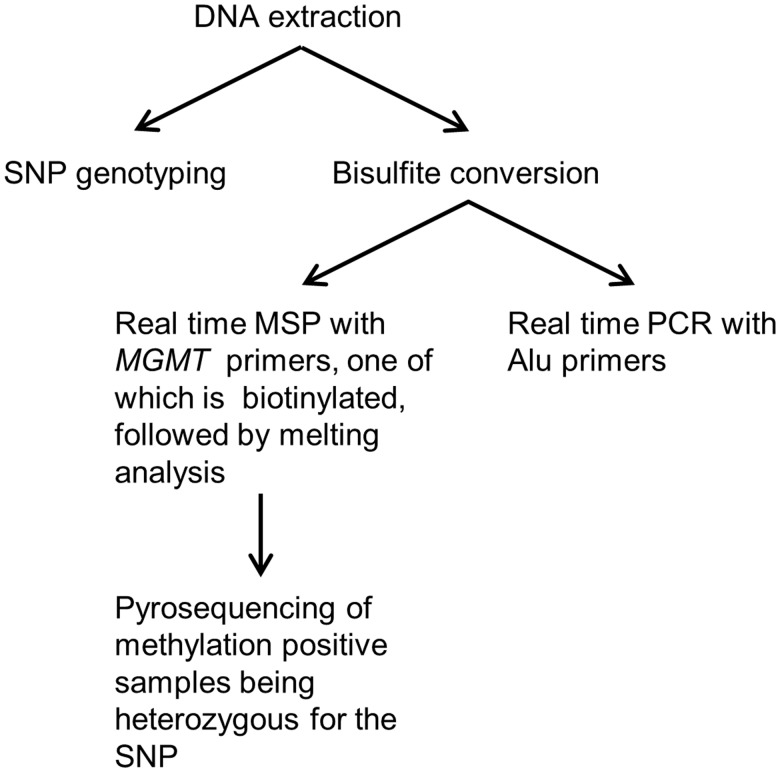
Quantitative and allelic methylation analyses were performed using qMSP and melting analyses followed by pyrosequencing of methylation-positive samples being heterozygous for the rs16906252 SNP (Fig. 1). The genomic location of the primers is shown in Supplementary Data 2. For normalization purposes, we used an assay based on Alu sequences depleted of CpG sites by evolutionary deamination (26). This assay is less susceptible to normalization errors caused by aneuploidy and copy number changes often observed in cancer cells. Additional details are in the Supplementary Data.
FIGURE 1.
Overview of the quantitative methylation-specific PCR (qMSP)-pyrosequencing workflow. First, genomic DNA is used for genotyping the single nucleotide polymorphism (SNP) in the region between the MSP primers. Then, for the methylation analysis, genomic DNA is bisulfite converted. Next, 2 separate real-time PCRs are performed; one with MSP primers (one of which is biotinylated) targeting the region of interest, and one with primers capable of amplifying both methylated and unmethylated templates targeting a control region (Alu sequences depleted of CpG sites). The data obtained are used to calculate the level of methylation in the samples relative to in vitro methylated DNA. Melting analysis is performed as an integrated part of the real-time PCR runs to confirm that the observed amplification is specific. Subsequently, amplification products are pyrosequenced to allow allelic methylation analysis for the patients being heterozygous for the SNP found between the MSP primers.
Quantification of Methylation Levels Using qMSP
Methylation levels of the samples were estimated relative to universal methylated DNA (Chemicon, Millipore, Billerica, MA) using the 2−ΔΔCt quantification approach (27), in which ΔΔCt = glioblastoma sample (Cttarget gene – CtALU control) − universal methylated DNA (Cttarget gene − CtALU control) (28). This approach implies that the assays have approximately the same PCR efficiency (E) (27). For the MGMT assay, E = 2.0; for the Alu assay, E = 1.9. Samples were analyzed in duplicate and the average Ct values were used in the calculations. Only samples for which both replicates amplified in the MGMT assay having a melting profile resembling the melting profile of universal methylated DNA were considered methylation positive. Samples with a mean Ct value above 17 in the Alu assay were bisulfite converted again and repeated. Samples with methylation levels below 0.1% were analyzed as unmethylated. This technical cutoff was defined following an evaluation of a serial dilution series of methylated DNA into unmethylated. Samples with a methylation level below 5% were scored as low-level methylation; samples with a methylation level between 5% and 20% as medium; and samples with a methylation level above 20% as high. No template control and unmethylated DNA, prepared as described (28), were included as negative controls.
Standard Bisulfite Pyrosequencing
PCR and pyrosequencing were performed using the Therascreen (R) MGMT Pyro (R) kit according to the manufacturer’s instructions with slight modifications (Supplementary Data). The CpG sites analyzed by the standard pyrosequencing assay are shown in Supplementary Data 2. Samples with a mean methylation level below 10% were considered unmethylated. Samples with a mean methylation level between 10% and 25% were scored as low-level methylation; samples with a mean methylation level between 25% and 50% as medium; and samples with a mean methylation level above 50% as high.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections were deparaffinized in xylene and rehydrated in decreasing concentrations of ethanol. After blocking of endogenous peroxidase with 3% H2O2, the sections were pretreated in a microwave oven with a Tris-EGTA buffer and immunostained on a DAKO Cytomation autostainer (DAKO, Glostrup, Denmark), using monoclonal mouse anti-human antibody against MGMT (MAB16200, 1:200, Millipore). Immunoreactivity was visualized with DAB + (DAKO K3468) as chromogen. The immunohistochemical reactions were semiquantitatively evaluated according to the number of tumor cells stained: 0 = 0; 1%–10% = 1+; 11%–25% = 2+; 26%–50% = 3+; and >50% = 4+. For MGMT evaluation, positive endothelial cells, lymphocytes, and microglia served as positive internal controls.
Statistical Analyses
Chi-square tests were used to compute p values for 2x2 contingency tables when all expected values were above 5. When at least 1 expected value was below 5, Fisher exact tests were used. The results of different MGMT analyses were compared for systematic differences using the McNemar test. Survival probabilities were estimated by the Kaplan-Meier method. Cox regression model was used for univariate as well as multivariate analysis of time to event, the latter for combined analysis of different MGMT detection methods. P values less than 0.05 were considered significant. Statistical calculations were performed using SPSS (version 19) and SAS (v9.3, SAS Institute, Cary, NC).
RESULTS
Quantitative Methylation Analyses
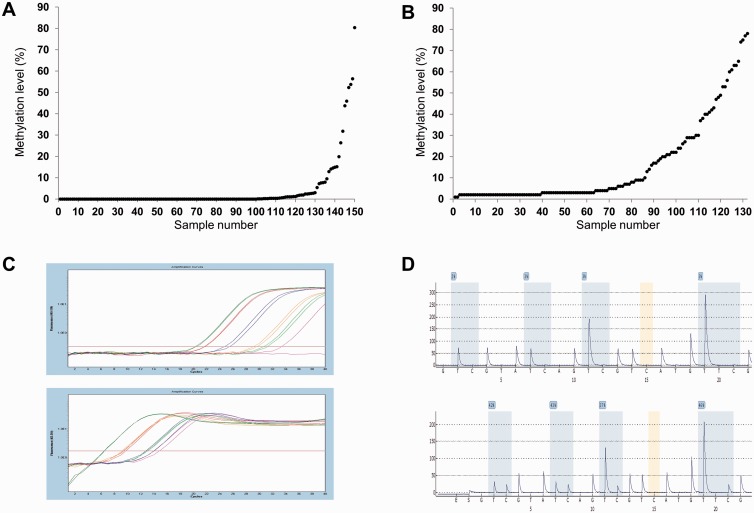
In total, 51 of 151 patient samples (34%) were methylation positive by qMSP. The mean methylation level of all samples was 5.1%. When considering only the 51 methylation-positive samples, the mean and median methylation levels were 15.2% and 2.4%, respectively. In total, 47 of 132 patient samples (36%) were methylation positive by standard pyrosequencing. The mean methylation level of all samples was 15.5%. When considering only the 47 methylation-positive samples (methylation levels above 9%), the mean and median methylation levels were 37.3% and 30%, respectively.
The quantitative methylation results are plotted for each of the samples for the 2 methods (Fig. 2A, B). Examples of individual results for each method are shown (Fig. 2C, D).
FIGURE 2.
The quantitative methylation results using quantitative methylation-specific PCR (qMSP) and standard pyrosequencing. (A) Methylation levels obtained by qMSP plotted for each of the samples. (B) Methylation levels obtained by standard pyrosequencing plotted for each of the samples. (C) Examples of individual results from the MGMT qMSP assay and the Alu assay. The red curves represent the in vitro methylated DNA; the other curves represent individual samples that were analyzed in duplicate. (D) Examples of individual results from the standard pyrosequencing assay that interrogates 4 individual CpG sites.
Comparison of qMSP and Standard Pyrosequencing
There was a strong association between the data obtained by the 2 methods when analyzing methylation status as “methylation positive” and “methylation negative” (p < 0.001); no systematic difference between the 2 methods was observed (p = 0.81) (Table 1). Next, samples that were methylation positive by both methods were divided into 2 groups according to whether or not the methylation level of each sample was below the median methylation level or above or equal to the median methylation level for each of the 2 methods, respectively. There was a significant association between the methylation levels (p = 0.002), and no systematic difference between the 2 methods was observed (p = 0.74) (Supplementary Data 3). When comparing the quantitative data directly for both methods, there was a relatively weak correlation (Supplementary Data 4). In total, 14 samples were methylated in the so-called “gray zone” as assessed by standard pyrosequencing (6%–13%). Twelve samples methylated below the cutoff (6%–9%) were negative as assessed by qMSP with the exception of one sample, which had a methylation level of 0.3% as assessed by qMSP and 9% by standard pyrosequencing. On the other hand, 2 samples that were methylated at 10% and 13% were both positive as assessed by qMSP with levels of 2.4% and 1%, respectively.
TABLE 1.
Comparison of Quantitative Methylation-Specific PCR and Standard Pyrosequencing
| Methylation Positive (Standard Pyrosequencing) | Methylation Negative (Standard Pyrosequencing) | Total | |
|---|---|---|---|
| Methylation positive (qMSP) | 38 | 8 | 46 |
| Methylation negative (qMSP) | 9 | 77 | 86 |
| Total | 47 | 85 | 132 |
qMSP, quantitative methylation-specific PCR.
Allelic Methylation Analyses by qMSP Followed by Pyrosequencing
Information on the methylation status of individual alleles could be obtained for samples heterozygous for the rs16906252 MGMT promoter SNP by pyrosequencing of the qMSP products. Eight of the heterozygous samples were methylated. Five of these were methylated only at the T-allele; 2 samples were methylated only at the C-allele; and the remaining sample was methylated at both alleles. Only 2 of these samples were methylated at low levels. Examples are shown in Supplementary Data 5. Some samples with the T-allele methylated did not express the protein whereas others did. This was also true for samples with monoallelic methylation of the C-allele (Table 2). The sample with biallelic methylation had low-level protein expression. This sample was methylated at medium level (19.9%), indicating that a minority of the cells had both alleles methylated.
TABLE 2.
Comparison of Quantitative- and Allelic Methylation Data With Immunohistochemistry Data
| Sample No. | Genotype | Methylation Level (qMSP) | Methylated Allele(s) | Protein Expression | Methylation Level (Standard Pyrosequencing) |
|---|---|---|---|---|---|
| 1050 | CT | Medium | C | Not expressed | No data |
| 1107 | CT | Medium | T | Not expressed | High |
| 1155 | CT | High | T | Not expressed | High |
| 1439 | CT | Medium | C and T | Low | Medium |
| 1518 | CT | High | T | High | High |
| 1650 | CT | Low | C | Low | Low |
| 20 | CT | Low | T | Not expressed | Low |
| 72 | CT | Low | T | High | No methylation |
| 1037 | CC | Low | C | Medium | No methylation |
| 1521 | CC | Low | C | Low | Low |
| 1664 | CC | Low | C | Not expressed | Medium |
| 55 | TT | High | T | Not expressed | Medium |
qMSP, quantitative methylation-specific PCR.
Four homozygous samples (3 CC and 1 TT) were also analyzed as controls (Table 2). The 2 CpG sites in between the primers were on average methylated above 85% in all samples except one, which had one site 96% methylated and the other site 47% methylated (100% and 37% when repeated). None of the samples showed incomplete bisulfite conversion. The experiments (bisulfite conversion and qMSP-pyrosequencing) were repeated for 7 of the samples, for which there was sufficient material for another bisulfite conversion, and the same allelic methylation results were observed again.
Comparison of DNA Methylation Data and IHC Data
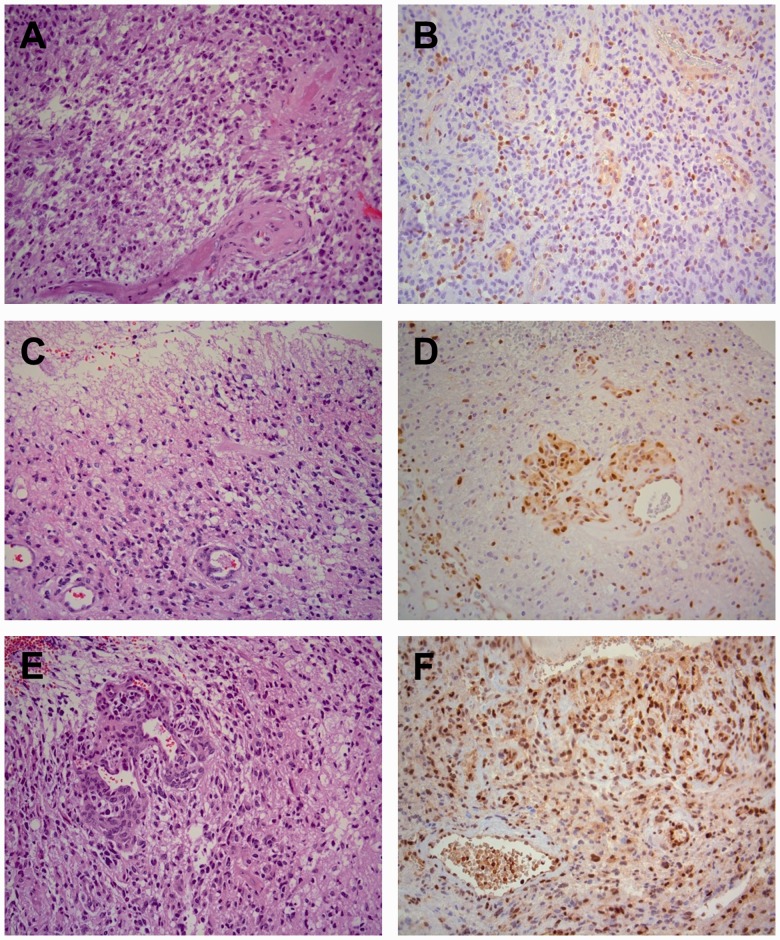
IHC was performed on 148 of the samples. Representative examples are shown (Fig. 3). The data are shown according to the quantitative methylation data by qMSP and standard pyrosequencing in Supplementary Data 6 and 7, respectively. When scoring samples as either positive or negative, there was a strong association between methylation status as determined by qMSP and IHC data (p = 0.009, Chi-square test and p = 0.74, McNemar test). This was also the case when comparing methylation status as determined by standard pyrosequencing and IHC data (p < 0.001, Chi-square test and p = 0.82, McNemar test). Absence of the MGMT protein was frequently observed in both methylation-negative and -positive samples; the frequency of unmethylated samples with protein expression was 43.9% and 48.8% for qMSP and standard pyrosequencing, respectively. On the other hand, expression of the protein was rare in methylation-positive samples; the frequency of methylated samples without protein expression was 78% and 84.4% for qMSP and standard pyrosequencing, respectively. Finally, it was observed that more samples with low-level methylation expressed the protein as compared to samples with medium and high methylation levels. However, this was not statistically significant for any of the methods.
FIGURE 3.
Representative examples of MGMT immunohistochemistry (IHC) on glioblastomas (World Health Organization [WHO], grade IV). (A, C, E) There are characteristic pleomorphic glial tumor cells, necrosis with pseudopalisading and endothelial cell proliferation. Hematoxylin and eosin stain. (B, C, F) Variable amounts of staining by IHC for MGMT of endothelial cells, lymphocytes, microglia, and tumor cells. (B) Only a minority of tumor cells is positive (0%–10%). (D) There is clear staining of endothelial cells in tumor vessels and diffusely in tumor cells (11%–25%). (F) Many endothelial cells and tumor cells are positive (26%–50%). The staining of tumor cells is heterogeneous, varying from area to area in the tissue. Magnification: x200 for all panels.
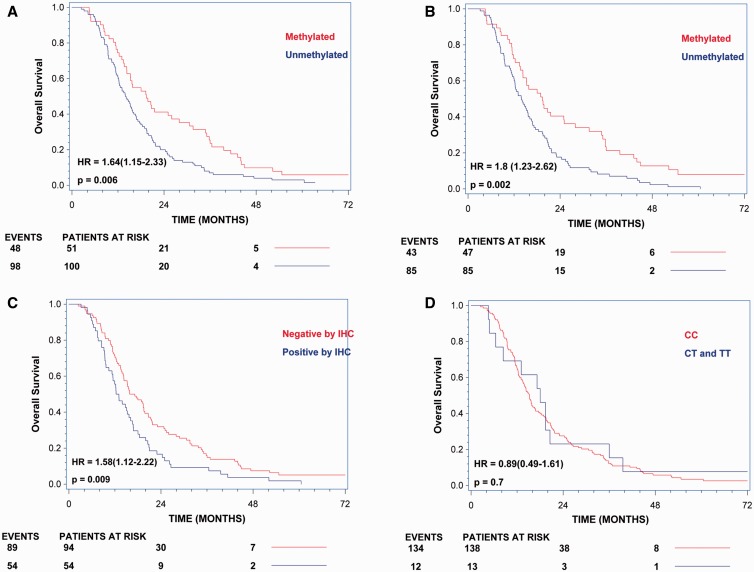
Overall Survival Analyses According to MGMT Methylation Status, IHC Data, and rs16906252 Genotypes
A total of 151 and 132 patients were evaluated for overall survival according to MGMT methylation status assessed by qMSP and standard pyrosequencing, respectively. The hazard ratio for death in the unmethylated group was 1.64 (95% confidence interval [CI], 1.15 to 2.33; p = 0.006) when assessed by qMSP and 1.8 (95% CI, 1.23 to 2.62; p = 0.002) when assessed by standard pyrosequencing (Fig. 4). A total of 148 patients were evaluated for overall survival according to MGMT protein expression assessed by IHC. The hazard ratio for death in the group expressing the protein was 1.58 (95% CI, 1.12 to 2.22; p = 0.009) (Fig. 4). Finally, a total of 151 patients were evaluated for overall survival according to rs16906252 genotypes. The hazard ratio for death in the group carrying the T-allele was 0.89 (95% CI, 0.49 to 1.61; p = 0.70) (Fig. 4).
FIGURE 4.
Overall survival analyses according to MGMT methylation status, immunohistochemistry (IHC) data, and rs16906252 genotypes. (A) Overall survival according to methylation status assessed by quantitative methylation-specific PCR. (B) Overall survival according to methylation status assessed by standard pyrosequencing. (C) Overall survival according to MGMT protein expression assessed by IHC. (D) Overall survival according to rs16906252 genotypes.
Overall Survival Analyses According to MGMT Methylation Levels
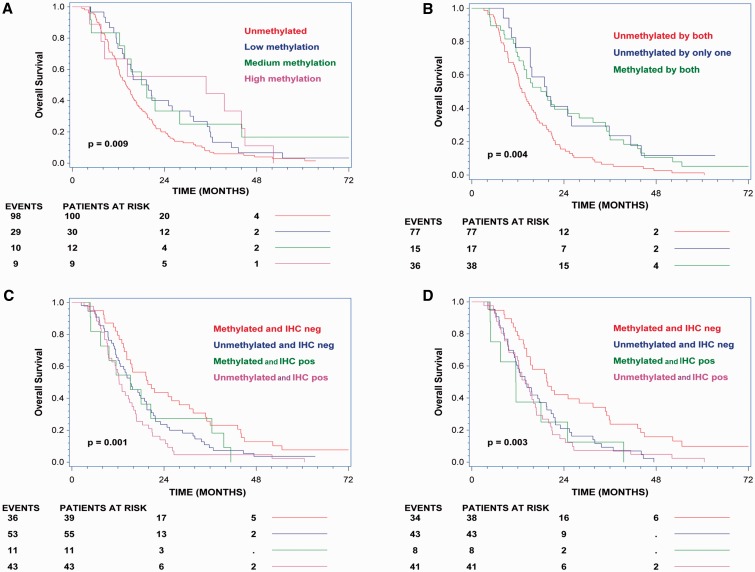
The patients were divided into subgroups according to methylation levels as described in “Materials and Methods.” A total of 151 and 132 patients were evaluated for overall survival according to MGMT methylation levels assessed by qMSP and standard pyrosequencing, respectively. The hazard ratio for death in the low methylated group versus the unmethylated group was 0.67 (95% CI, 0.44 to 1.02; p = 0.061) when assessed by qMSP and 0.43 (95% CI, 0.25 to 0.75; p = 0.02) when assessed by standard pyrosequencing. There was a trend towards a better overall survival in patients with higher methylation levels compared to patients with lower methylation levels when analyzed by qMSP (Fig. 5A). However, there was no difference in overall survival when comparing patients with medium methylation levels with patients with low methylation levels.
FIGURE 5.
Overall survival analyses according to MGMT methylation levels and combined methylation and immunohistochemistry (IHC) analyses. (A) Overall survival according to methylation levels determined by quantitative methylation-specific PCR (qMSP). (B) Overall survival according to methylation status determined by qMSP and standard pyrosequencing. (C) Overall survival according to methylation status determined by qMSP and IHC data. (D) Overall survival according to methylation status determined by standard pyrosequencing and IHC data.
Overall Survival Analyses According to Combined qMSP and Standard Pyrosequencing
A total of 132 patients were evaluated for overall survival according to MGMT methylation status assessed by both qMSP and standard pyrosequencing. It was observed that patients with only one positive test result did better than patients scored as methylation negative by both methods (Fig. 5B). It was also observed that patients with a positive methylation result assessed by qMSP and a negative methylation result by standard pyrosequencing performed equally well as patients with a positive methylation result assessed by standard pyrosequencing and a negative methylation result by qMSP (Supplementary Data 8).
Overall Survival Analyses According to Combined Methylation and IHC Analyses
A total of 148 patients were evaluated for overall survival according to MGMT methylation status assessed by both qMSP and IHC. It was observed that patients with either a positive methylation test or a negative IHC result did better relative to patients with a negative methylation test and a positive IHC result (Fig. 5C). A total of 130 patients were evaluated for overall survival according to both MGMT methylation status assessed by standard pyrosequencing and IHC. It was observed that patients with a negative IHC result and a positive methylation result did better relative to patients with a negative methylation test and a negative IHC result (Fig. 5D).
Association Between MGMT Methylation and Genotypes
In total, 13 of the 151 patients carried the T-allele of the rs16906252 MGMT promoter SNP. Ten of these patients were methylation positive by qMSP. Individuals carrying the T-allele of the rs16906252 SNP were more likely to have detectable MGMT methylation by qMSP (p = 0.01). In total, 132 patients were analyzed by standard pyrosequencing, and 12 of these carried the T-allele of the rs16906252 MGMT promoter SNP. Seven of these patients were methylation positive by standard pyrosequencing. No statistically significant association between the T-allele of the rs16906252 SNP and MGMT methylation by standard pyrosequencing was observed (p = 0.11). A summary of the genotypes and methylation status as determined by the 2 methods is shown in Supplementary Data 9.
DISCUSSION
MGMT methylation is used as a predictive biomarker of response to alkylating agents in patients with GBM but the methylation status of a sample is not straightforward to analyze. This is mainly due to the fact that numerous CpG sites are located in the MGMT promoter. These sites may often be methylated at different levels in different cancer clones, and the cells may also have different allelic methylation patterns. Because of these factors, different methodologies for DNA methylation detection may provide different results (29). Standard bisulfite pyrosequencing has been widely adopted for evaluation of MGMT methylation for several reasons; the methodology is quantitative and relatively accurate (30), and a standardized commercial kit, which has been approved for clinical use, is available. However, standard pyrosequencing is based on a methylation-independent PCR, which amplifies a pool of bisulfite-treated DNA molecules with no possibility of tracking individual epialleles. The displayed methylation levels at each individual CpG site are, therefore, an average of the methylation found in the sample across cell types, cancer clones, and alleles. Because the tumor cell content and allelic methylation status may vary from sample to sample, the quantitative results of standard pyrosequencing should always be interpreted with caution.
We have previously shown that allelic methylation patterns and methylation levels influence MGMT protein expression in malignant pleural mesothelioma (10). There have not been previous studies of allelic MGMT methylation patterns in GBM. Therefore, we investigated how allelic methylation patterns and methylation levels influence MGMT protein expression in GBM. For this purpose, we developed a novel MGMT promoter methylation assay capable of providing quantitative and allelic methylation data. The assay is based on qMSP followed by pyrosequencing of positive samples. Because MSP is considered prone to false-positive results (28, 31–33), the use of pyrosequencing is advantageous because it allows assessment of the efficiency of the bisulfite conversion as well as the methylation status of CpG sites found in between the primers (14, 32, 34).
We found that the majority of the samples, which were informative with regard to allelic methylation patterns, were monoallelically methylated; however, several of these samples did not express the MGMT protein. This is likely explained by hemizygous deletion of the MGMT locus, which occurs in a large fraction of GBM (12, 35). On the other hand, several samples with monoallelic methylation did express the protein possibly from the unmethylated allele. Therefore, our data suggest that allelic methylation analysis may not contribute additional information that would be useful in clinical settings. However, because the T-allele frequency was relatively low in our cohort compared to other studies using American and Australian cohorts (11, 12), additional studies of allelic MGMT methylation patterns in relation to protein expression and survival of patients with GBM treated with temozolomide is warranted. In addition, because DNA extracted from tumor biopsies was used for SNP genotyping in our study, it is possible that heterozygous patients with a hemizygous deletion of the MGMT locus in their tumor cells would be scored as homozygous. Indeed, we found several samples with pyrosequencing and high-resolution melting results being difficult to score because they were in the gray zone between being homozygous CC and heterozygous. These were all homozygous when analyzed by Sanger sequencing, which has a relatively poor analytical sensitivity (36).
Overall, we observed that MGMT promoter methylation was associated with lack of protein expression. Also, we found more samples with low-level methylation expressing the protein as compared to samples with medium and high methylation levels, although this was not statistically significant. Unmethylated samples were often negative by IHC, indicating that mechanisms other than promoter methylation may be responsible for silencing of the MGMT gene. Gene body methylation, histone modifications, as well as aberrant expression of microRNAs have been shown to downregulate expression of the MGMT protein (4, 37, 38). Therefore, our results were not surprising, and we have also previously observed the same phenomenon when studying p16/CDKN2B in lung cancer (39). Another recent study including 418 patients with GBM also found correlations between MGMT methylation and expression of the protein assessed by IHC (8), whereas others have not (40).
Despite qMSP and standard pyrosequencing being inherently different methods, we observed strong correlation between the 2 methods when analyzing the data in a purely qualitative and semiquantitatively manner using predetermined cutoff values. However, when the data were analyzed quantitatively, there was a relatively weak correlation. This was expected because the 2 methods do not analyze the exact same CpG sites and because qMSP data are strongly affected by the annealing temperature of the PCR and the number and positions of CpG sites within the primer sequences (33).
When performing standard pyrosequencing analysis, it has been suggested that tumors with percentage values close to the technical cutoff (in the so-called “gray zone”) should be tested again by another method or not assigned to either the methylated or unmethylated groups. We observed that 11 out of 12 samples being methylated between 6% and 9% were negative as assessed by qMSP with the exception of one sample being methylated at a very low level. On the other hand, 2 samples being methylated between 10% and 13% were both positive as assessed by qMSP. Thus, our data support the use of the currently recommended technical cutoff point.
The presence of MGMT methylation by the qMSP assay was associated with better overall survival (p = 0.006). This was also observed for the standard pyrosequencing assay (p = 0.002), whereas the presence of the protein was associated with worse overall survival (p = 0.009). It has been suggested that IHC is not reliable for use as a biomarker in the clinics (41); however, in our hands, IHC performed nearly as well as the methylation analyses. It has also previously been shown that the rs16906252 SNP is predictive of response to temozolomide in patients with GBM (12). Because relatively few of our patients carried the T-allele of the SNP, we cannot make firm conclusions on this issue. Likewise, too few patients were informative with regard to allelic methylation patterns to justify the performance of survival analysis according to this criterion.
Because standard pyrosequencing and qMSP are quantitative methods, we also analyzed the data with respect to methylation levels. Interestingly, our analyses indicate that patients with relatively low methylation levels have a better overall survival compared to patients with unmethylated tumor biopsies. However, the data do not support a further stratification of the patients according to methylation levels, and it has previously been shown that increasing levels of methylation (as assessed by standard pyrosequencing) do not have much impact on overall survival of patients with astrocytomas and GBM (9). This is likely a result of methylation levels being an average of the methylation found in the sample across cell types, cancer clones, and alleles, and that the tumor cell content and allelic methylation status vary from sample to sample.
Finally, we performed a combined analysis of qMSP and standard pyrosequencing and observed that patients testing positive by only 1 of the 2 methods benefit from treatment with temozolomide. Thus, our data suggest that patients should be tested by both methylation-independent and MSP-based methods. Combined analysis of qMSP and IHC likewise seem to identify additional patients who benefit from temozolomide treatment. This is in accordance with the findings of Lalezari et al (8).
In conclusion, our data indicate that allelic MGMT methylation information may not be useful for predicting if the MGMT protein is expressed. However, our results provide strong evidence to support the use of MGMT methylation as a prognostic marker in patients with GBM treated with temozolomide and suggest that combined qMSP and standard pyrosequencing analyses as well as qMSP and IHC analyses identify additional patients who may benefit from temozolomide treatment.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Elena Projva for excellent technical assistance and the Danish Cancer Society and the University of Copenhagen for funding. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 2000;343:1350–4 [DOI] [PubMed] [Google Scholar]
- 2.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 2005;352:997–1003 [DOI] [PubMed] [Google Scholar]
- 3.Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res 2004;10:1871–4 [DOI] [PubMed] [Google Scholar]
- 4.Wick W, Weller M, van den Bent M, et al. MGMT testing–the challenges for biomarker-based glioma treatment. Nat Rev Neurol 2014;10:372–85 [DOI] [PubMed] [Google Scholar]
- 5.Wick W, Platten M, Meisner C, et al. Temozolomide chemotherapy alone versus radiotherapy alone for malignant astrocytoma in the elderly: The NOA-08 randomised, phase 3 trial. Lancet Oncol 2012;13:707–15 [DOI] [PubMed] [Google Scholar]
- 6.Malmstrom A, Gronberg BH, Marosi C, et al. Temozolomide versus standard 6-week radiotherapy versus hypofractionated radiotherapy in patients older than 60 years with glioblastoma: The Nordic randomised, phase 3 trial. Lancet Oncol 2012;13:916–26 [DOI] [PubMed] [Google Scholar]
- 7.Everhard S, Tost J, El Abdalaoui H, et al. Identification of regions correlating MGMT promoter methylation and gene expression in glioblastomas. Neuro Oncol 2009;11:348–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lalezari S, Chou AP, Tran A, et al. Combined analysis of O6-methylguanine-DNA methyltransferase protein expression and promoter methylation provides optimized prognostication of glioblastoma outcome. Neuro Oncol 2013;15:370–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oberstadt MC, Bien-Moller S, Weitmann K, et al. Epigenetic modulation of the drug resistance genes MGMT, ABCB1 and ABCG2 in glioblastoma multiforme. BMC Cancer 2013;13:617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kristensen LS, Nielsen HM, Hager H, et al. Methylation of MGMT in malignant pleural mesothelioma occurs in a subset of patients and is associated with the T allele of the rs16906252 MGMT promoter SNP. Lung Cancer 2011;71:130–6 [DOI] [PubMed] [Google Scholar]
- 11.McDonald KL, Rapkins RW, Olivier J, et al. The T genotype of the MGMT C>T (rs16906252) enhancer single-nucleotide polymorphism (SNP) is associated with promoter methylation and longer survival in glioblastoma patients. Eur J Cancer 2013;49:360–8 [DOI] [PubMed] [Google Scholar]
- 12.Rapkins RW, Wang F, Nguyen HN, et al. The MGMT promoter SNP rs16906252 is a risk factor for MGMT methylation in glioblastoma and is predictive of response to temozolomide. Neuro Oncol 2015;17:1589–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawkins NJ, Lee JH, Wong JJ, et al. MGMT methylation is associated primarily with the germline C>T SNP (rs16906252) in colorectal cancer and normal colonic mucosa. Mod Pathol 2009;22:1588–99 [DOI] [PubMed] [Google Scholar]
- 14.Kristensen LS, Treppendahl MB, Asmar F, et al. Investigation of MGMT and DAPK1 methylation patterns in diffuse large B-cell lymphoma using allelic MSP-pyrosequencing. Sci Rep 2013;3:2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leng S, Bernauer AM, Hong C, et al. The A/G allele of rs16906252 predicts for MGMT methylation and is selectively silenced in premalignant lesions from smokers and in lung adenocarcinomas. Clin Cancer Res 2011;17:2014–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogino S, Hazra A, Tranah GJ, et al. MGMT germline polymorphism is associated with somatic MGMT promoter methylation and gene silencing in colorectal cancer. Carcinogenesis 2007;28:1985–90 [DOI] [PubMed] [Google Scholar]
- 17.Mikeska T, Bock C, El-Maarri O, et al. Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis. J Mol Diagn 2007;9:368–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karayan-Tapon L, Quillien V, Guilhot J, et al. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol 2010;97:311–22 [DOI] [PubMed] [Google Scholar]
- 19.Quillien V, Lavenu A, Karayan-Tapon L, et al. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer 2012;118:4201–11 [DOI] [PubMed] [Google Scholar]
- 20.Kristensen LS, Hansen LL. PCR-based methods for detecting single-locus DNA methylation biomarkers in cancer diagnostics, prognostics, and response to treatment. Clin Chem 2009;55:1471–83 [DOI] [PubMed] [Google Scholar]
- 21.Kristensen LS, Treppendahl MB, Grønbæk K. Analysis of epigenetic modifications of DNA in human cells. Curr Protoc Hum Genet 2013;Chapter 20;Unit 20.2 [DOI] [PubMed] [Google Scholar]
- 22.Weller M. Assessing the MGMT status in glioblastoma: One step forward, two steps back? Neuro Oncol 2013;15:253–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michaelsen SR, Christensen IJ, Grunnet K, et al. Clinical variables serve as prognostic factors in a model for survival from glioblastoma multiforme: An observational study of a cohort of consecutive non-selected patients from a single institution. BMC Cancer 2013;13:402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hasselbalch B, Lassen U, Hansen S, et al. Cetuximab, bevacizumab, and irinotecan for patients with primary glioblastoma and progression after radiation therapy and temozolomide: A phase II trial. Neuro Oncol 2010;12:508–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab Press; 1989 [Google Scholar]
- 26.Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res 2005;33:6823–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 2001;25:402–8 [DOI] [PubMed] [Google Scholar]
- 28.Kristensen LS, Mikeska T, Krypuy M, et al. Sensitive Melting Analysis after Real Time-Methylation Specific PCR (SMART-MSP): High-throughput and probe-free quantitative DNA methylation detection. Nucleic Acids Res 2008;36:e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikeska T, Candiloro IL, Dobrovic A. The implications of heterogeneous DNA methylation for the accurate quantification of methylation. Epigenomics 2010;2:561–73 [DOI] [PubMed] [Google Scholar]
- 30.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc 2007;2:2265–75 [DOI] [PubMed] [Google Scholar]
- 31.Rand K, Qu W, Ho T, et al. Conversion-specific detection of DNA methylation using real-time polymerase chain reaction (ConLight-MSP) to avoid false positives. Methods 2002;27:114–20 [DOI] [PubMed] [Google Scholar]
- 32.Shaw RJ, Akufo-Tetteh EK, Risk JM, et al. Methylation enrichment pyrosequencing: Combining the specificity of MSP with validation by pyrosequencing. Nucleic Acids Res 2006;34:e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kristensen LS, Raynor MP, Candiloro I, et al. Methylation profiling of normal individuals reveals mosaic promoter methylation of cancer-associated genes. Oncotarget 2012;3:450–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kristensen LS, Asmar F, Dimopoulos K, et al. Hypermethylation of DAPK1 is an independent prognostic factor predicting survival in diffuse large B-cell lymphoma. Oncotarget 2014;5:9798–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collins VP, Ichimura K, Di Y, et al. Prognostic and predictive markers in recurrent high grade glioma; results from the BR12 randomised trial. Acta Neuropathol Commun 2014;2:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kristensen LS, Daugaard IL, Christensen M, et al. Increased sensitivity of KRAS mutation detection by high-resolution melting analysis of COLD-PCR products. Hum Mutat 2010;31:1366–73 [DOI] [PubMed] [Google Scholar]
- 37.Kushwaha D, Ramakrishnan V, Ng K, et al. A genome-wide miRNA screen revealed miR-603 as a MGMT-regulating miRNA in glioblastomas. Oncotarget 2014;5:4026–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Zhang J, Hoadley K, et al. miR-181d: A predictive glioblastoma biomarker that downregulates MGMT expression. Neuro Oncol 2012;14:712–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kristensen LS, Wojdacz TK, Thestrup BB, et al. Quality assessment of DNA derived from up to 30 years old formalin fixed paraffin embedded (FFPE) tissue for PCR-based methylation analysis using SMART-MSP and MS-HRM. BMC Cancer 2009;9:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mason S, McDonald K. MGMT testing for glioma in clinical laboratories: Discordance with methylation analyses prevents the implementation of routine immunohistochemistry. J Cancer Res Clin Oncol 2012;138:1789–97 [DOI] [PubMed] [Google Scholar]
- 41.Riemenschneider MJ, Hegi ME, Reifenberger G. MGMT promoter methylation in malignant gliomas. Target Oncol 2010;5:161–5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.