Abstract
The mammalian circadian timing system coordinates key molecular, cellular and physiological processes along the 24-h cycle. Accumulating evidence suggests that many clock-controlled processes display a sexual dimorphism. In mammals this is well exemplified by the difference between the male and female circadian patterns of glucocorticoid hormone secretion and clock gene expression. Here we show that the non-circadian nuclear receptor and metabolic sensor Liver X Receptor alpha (LXRα) which is known to regulate glucocorticoid production in mice modulates the sex specific circadian pattern of plasma corticosterone. Lxrα-/- males display a blunted corticosterone profile while females show higher amplitude as compared to wild type animals. Wild type males are significantly slower than females to resynchronize their locomotor activity rhythm after an 8 h phase advance but this difference is abrogated in Lxrα-/- males which display a female-like phenotype. We also show that circadian expression patterns of liver 11β-hydroxysteroid dehydrogenase type 1 (11β-HSD1) and Phosphoenolpyruvate carboxykinase (Pepck) differ between sexes and are differentially altered in Lxrα-/- animals. These changes are associated with a damped profile of plasma glucose oscillation in males but not in females. Sex specific alteration of the insulin and leptin circadian profiles were observed in Lxα-/- females and could be explained by the change in corticosterone profile. Together this data indicates that LXRα is a determinant of sexually dimorphic circadian patterns of key physiological parameters. The discovery of this unanticipated role for LXRα in circadian physiology underscores the importance of addressing sex differences in chronobiology studies and future LXRα targeted therapies.
Introduction
In mammals, many molecular, cellular, physiological and behavioural processes show circadian (~24 h) oscillations synchronised to the external light/dark cycle. These circadian rhythms are under the control of a self-sustained internal clock present in nearly every cell. At the organism level, these clocks are organized hierarchically with at the top a central pacemaker located in the suprachiasmatic nuclei (SCN) of the hypothalamus that receives photic cues and in turn coordinates local clocks in the periphery. Peripheral clocks are entrained by the SCN through internal systemic synchronizers such as glucocorticoid hormones and body temperature and most probably other signals that remain to be identified. Although peripheral clocks display self-sustained oscillations at the single-cell level, at the organ and systemic levels they require an intact SCN clock to remain in phase [1, 2]. At the molecular level, the core mechanism of all cellular clocks is governed by a genetic network that integrates multiple time delayed negative and positive feedbacks loops [3]. The primary loop involves the two bHLH-PAS transcription factors CLOCK and BMAL1 which upon dimerization trigger transcription of the Period (Per1-3) and Cryptochrome (Cry1/2) clock genes. PER and CRY proteins then translocate to the nucleus where they in turn repress the CLOCK-BMAL1 transactivation [4]. The core clock mechanism also involves the nuclear receptors ROR(α,β,γ) and REV-ERB(α,β) which are direct CLOCK:BMAL1 targets and compete to activate or repress the transcription of the Bmal1 and Clock genes respectively. This secondary loop provides robustness to the circadian oscillator and is critical for normal circadian behavior and physiology [5, 6]. In addition to these transcriptional mechanisms, the circadian molecular network is also extensively regulated post-translationally [7–9] as well as through chromatin remodelling [10, 11]. Oscillation of this increasingly complex network directs the rhythmic expression of downstream clock-controlled genes through transcriptional, post-transcriptional, translational and post-translational mechanisms [12, 13]. A recent meta-analysis of available genome wide circadian gene expression data has estimated that approximately 43% of mouse genes oscillate somewhere in the body yet with a significant tissue-specificity, thus highlighting the extent of circadian regulation in mammals [14].
Nuclear hormone receptors form a large family of proteins which function as ligand-inducible transcription factors involved in virtually all key biological processes and expectedly in many diseases. In addition to the REV-ERB and ROR receptors, a substantial number of these receptors have been implicated either in the core circadian clock mechanism or as transcriptional links between the clock gene network and clock-controlled processes [15]. The glucocorticoid receptor (GR) is for instance directly involved in the resetting of peripheral clocks by glucocorticoids [16]. The essential clock gene Bmal1 is regulated by peroxisome proliferator activated receptors (PPARs) α and γ in the liver and the cardiovascular system respectively [17, 18]. Other examples include the estrogen receptor β, constitutive androstane receptor, short heterodimer partner and the estrogen related receptor α [19–22]. Many of these receptors have been implicated in the circadian regulation of metabolism [23]. Liver X receptor (LXR) α is another important metabolic nuclear receptor regulating cholesterol, fatty acids and glucose homeostasis [24]. A critical function of LXRα is to activate bile acid formation through upregulation of cholesterol catabolism by the CYP7α1 enzyme encoding gene. LXRα was suggested to be responsible for the circadian expression of the Cyp7α1 gene through rhythmic activation by its endogenous ligands that include 22(R)-hydroxycholesterol, 24(S)-hydroxycholesterol, 27-hydroxycholesterol, and cholestenoic acid [25]. Interestingly, male Lxrα-/- mice display adrenomegaly and increased corticosterone (CORT) secretion [26]. Given the pivotal role of glucocorticoids (GCs) in the synchronization of peripheral clocks, this observation suggests that although LXRα is not clock-controlled, it may play an unanticipated role in circadian physiology [27]. Notably, the mammalian hypothalamo-pituitary-adrenal (HPA) axis exhibits a marked and well documented sexual dimorphism [28]. In particular, females show higher mean levels and amplitude of corticosterone as compared to males. Given the impact of the loss of LXRα in the adrenals, we hypothesized that LXRα could contribute to such sex difference. To address this issue we analyzed the circadian physiology of LXRα deficient animals and obtained evidence that known and newly identified sex differences in circadian regulation are indeed altered in these animals.
Results
The sexual dimorphism of the circadian corticosterone pattern is differentially changed in male and female Lxrα-/-mice
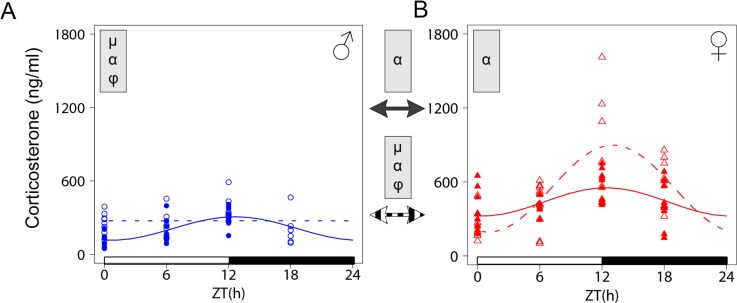
Plasma GCs levels show daily oscillations in mammals including mice with higher concentrations being observed at the end of the resting phase. Further, female mice display significantly higher mean CORT levels than males irrespective of the genetic background [29]. Of note a circadian pattern of CORT in females is observed in non estrus-synchronized females [29–31]. Based on the known implication of LXRα in adrenal corticosterone production, we measured plasma CORT levels at four time points around the clock in wild type (WT) and Lxrα-/- male and non estrus-synchronized female mice entrained to an LD12:12 cycle. Expectedly, plasma CORT showed a robust daily variation in WT mice with an acrophase at the beginning of the night (Fig 1 and Table 1). Mean levels were significantly higher in females than in male WT mice (Fig 1 and Table 1). Loss of LXRα resulted in a significant and opposite effect in males as compared to females. Indeed, male Lxrα-/- mice showed significantly higher mean CORT levels with no circadian variation as compared to WT mice (Fig 1 and Table 1). In sharp contrast, Lxrα-/- females had similar mean CORT levels as compared to WT females but they displayed a 3 times higher amplitude. We also confirmed that the expression of glucocorticoid receptor (GR) did not show circadian rhythmicity [27]. We found that mean level of GR mRNA was lower in Lxrα-/- males as compared to WT animals, consistent with the known downregulation of GR expression by its ligand [32]. We also found a small but significant difference between males and females (S1 Fig and S1 Table). We conclude from these observations that both the level and the circadian pattern of plasma CORT are differentially impacted by the LXRα mutation in males and females.
Fig 1. The sexually dimorphic circadian pattern of plasma CORT is differentially altered in male and female Lxrα-/- mice.
(A) Rhythm of plasma CORT was determined in wild type (plain line) and Lxrα-/- (dashed line) male mice entrained to a LD12:12 cycle. (B) Same as in (A) except that mice were females. Cosinor based non-linear regression was used for curve fitting. Plotted data were from 10 mice for each experimental group. The ZT0 time point is double plotted for visualization purpose. White and black bars represent the light and dark phases, respectively. Statistically significant differences in cosine fitting parameters (p<0.05) between wild type and Lxrα-/- mice or between male and female of the same genotype is indicated in the grey box at the top of the corresponding graph or between graphs (WT: plain arrow, Lxrα-/-: dashed arrow). µ, α and φ indicate a difference in mean level, amplitude and acrophase, respectively.
Table 1. Cosinor analysis of plasma CORT circadian pattern in WT and Lxrα -/- mice.
| Group | Mean level (ng/ml) | Amplitude | Acrophase (ZT:min) |
|---|---|---|---|
| ♂ WT | 206.7 (181.4 ; 231.2) | 94.9 (62.5 ; 129.0) | 12:16 (10:49 ; 13:47) |
| ♂ Lxrα -/- | 274.0 (235.8 ; 317.0)† | NSR | NSR |
| ♀ WT | 437.6 (388.7 ; 486.5)* | 114.7 (62.5 ; 179.4) | 12:24 (09:49 ; 14:52) |
| ♀ Lxrα -/- | 545.4 (458.3 ; 634.3)* | 356.6 (225.1 ; 492.1)*† | 13:03 (11:52 ; 14:23)* |
For each parameter measured, values are represented as median ± 95% bootstrap confidence intervals.
* indicates significant differences between females vs males
† indicates significant differences between Lxrα-/- vs WT mice.
Circadian rhythmicity was considered significant for a p-value < 0.05; NSR, not significantly rhythmic.
Time to reset after jetlag is decreased in Lxrα-/-males
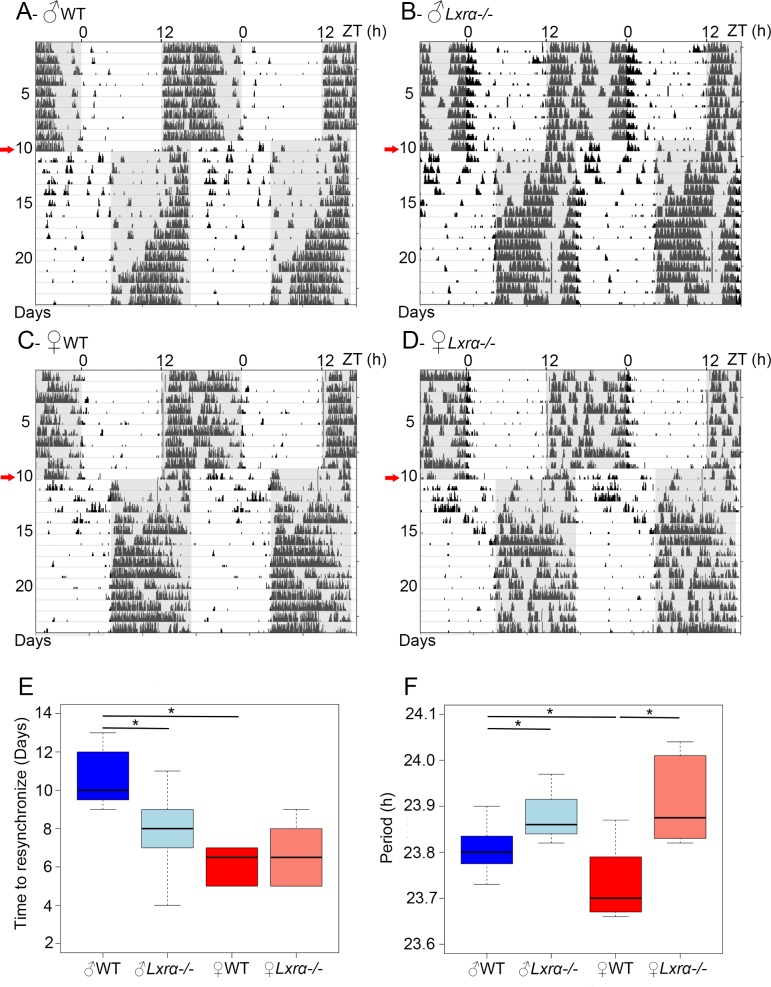
Glucocorticoid hormones are well known to play a role in the resetting of the central clock after a phase shift in mammals [33, 34]. As the CORT pattern was differentially changed in male and female Lxrα-/- mice, we investigated circadian pattern and resetting of the locomotor activity rhythm in these animals. Male and female Lxrα-/- mice entrained to an LD 12:12 cycle and then released in constant darkness showed a similar level of total wheel running activity and a small, although significant, increase of their free running period length of locomotor activity as compared to WT animals (Fig 2F and Table 2). After re-entrainment to an LD 12:12 cycle, the animals were subjected to an 8-hours phase advance of the LD cycle; we observed a significant sexual dimorphism in WT animals with females requiring approximately 6 days to re-entrain with the new phase as compared to 10 days for males (Fig 2A, 2C and 2E and Table 2). This difference was abolished in Lxrα-/- males which became significantly faster than their WT controls, and consequently were similar to females (Fig 2B, 2D and 2E, Table 2). Note that some Lxrα-/- females tended to re-entrain with a combination of advances (onset) and delays (offset) of their activity patterns, rather than simply advancing activity onset. These results suggest that LXRα does not play a major role in the control of the endogenous period by the central circadian pacemaker but in contrast dramatically impacts on its resetting properties in males.
Fig 2. Resetting after jetlag differs between male and female in wild type but not in Lxrα-/- mice.
Wheel running activity was measured in animals entrained to a LD12:12 and then submitted to an 8-hours jetlag (red arrow). Representative actograms for male (A) and female (B) wild type mice and male (C) and female (D) Lxrα-/- mice are shown. In A-D, the white and grey bars represent the light and dark phases respectively. Box and whiskers plots for resetting-time (E) and free-running period of wheel running activity (F) in male (n = 8) and female (n = 6) wild type mice and in male (n = 11) and female (n = 6) Lxrα-/- mice. * indicates statistically significant differences (p<0.05) between groups.
Table 2. Total daily activity, free-running period of locomotor activity and resynchronization time of WT and Lxrα-/- mice.
| Group | Daily activity (LD 12/12) | Period (DD, hours : min) | Resetting (days) |
|---|---|---|---|
| ♂ WT | 7736 (7048 ; 8294) | 23:48 (23:47 ; 23:50) | 10.00 (9.75 ; 12.00) |
| ♂ Lxrα -/- | 6638 (5849 ; 8474) | 23:52 (23:50; 23:55)† | 8.00 (7.00; 9.00)† |
| ♀ WT | 6732 (5555 ; 8612) | 23:42 (23:41 ; 23:46)* | 6.50 (5.25 ; 7.00)* |
| ♀ Lxrα -/- | 8097 (7384 ; 8681) | 23:53 (23:50 ; 23:59)† | 6.50 (5.25 ; 7.75) |
Values are represented as median ± 95% bootstrap confidence intervals.
* indicates significant differences between males vs females
† indicates significant differences between WT vs Lxrα-/- mice.
Loss of LXRα differentially alters circadian gene expression in males and females
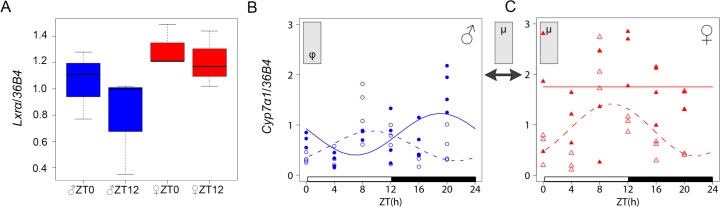
We first analysed the expression of Lxrα in liver and adrenals in WT mice and found no significant time of day- or sex- dependent variation confirming and extending previous data (Fig 3A, S2 Fig) [27]. Because Cyp7α1 is a well established direct target of LXRα, that displays a mRNA circadian variation, we compared its expression in male and female Lxrα-/- mice. Data shows that the Cyp7α1 mRNA oscillates in WT males but not in females. Upon deletion of LXRα this pattern is phase advanced by approximately 10-h in males and became rhythmic in females with a similar phase (Fig 3B and 3C, Table 3).
Fig 3. Analysis of Lxrα and Cyp7α1 in liver.
Lxrα mRNA expression determined at ZT0 and ZT 12 in WT males and females using qRT-PCR (A). Diurnal mRNA expression of liver Cyp7α1 was compared in males (B) and females (C) using qRT-PCR in WT (plain line) and Lxrα-/- mice (dashed line). For each time point, 3–4 mice were used. For the Cyp7α1 analysis, cosinor-based non-linear regression was used for curve fitting. The ZT0 time point is double plotted for visualization purposes. Expression data were normalized to the constitutively expressed 36B4 mRNA. The white and black bars represent the light and dark phases, respectively. Statistically significant differences in cosine fitting parameters (p<0.05) between wild type and Lxrα-/- mice or between male and female of the same genotype is indicated in the grey box at the top of the corresponding graph or between graphs (WT: plain arrow, Lxrα-/-: dashed arrow). µ, α and φ indicate a difference in mean level, amplitude and acrophase, respectively.
Table 3. Cosinor analysis of Cyp7α1 circadian expression in WT and Lxrα-/- mice.
| Gene | Group | Amplitude | Acrophase (ZT:min) | |
|---|---|---|---|---|
| Cyp7 α 1 | ♂ WT | 0.81 (0.64 ; 1.01) | 0.41 (0.18 ; 0.70) | 19:18 (16:22 ; 21:43) |
| ♂ Lxrα-/- | 0.58 (0.45 ; 0.74) | 0.30 (0.12; 0.52) | 9.21 (6:24; 12:35)† | |
| ♀ WT | 1.83 (1.53 ; 2.15)* | NSR | NSR | |
| ♀ Lxrα-/- | 0.90 (0.64 ; 1.18)† | 0.52 (0.20; 0.96) | 9.37 (6:29; 12:33) |
Values are represented as median ± 95% bootstrap confidence intervals.
* indicates significant differences between females vs males
† indicates significant differences between Lxrα-/- vs WT mice.
Circadian rhythmicity was considered significant for a p-value < 0.05; NSR, not significantly rhythmic.
Based on the changes observed in LXRα deficient mice regarding the plasma CORT pattern, we investigated the expression profiles of the Adreno-Cortico-Tropic-Hormone-receptor (MC2R) and Steroidogenic Acute Regulatory (StAR) mRNA, two key determinants of CORT synthesis in the adrenals. They both display a circadian expression at the mRNA level in males and the rhythmic CORT pattern has been linked to the circadian expression of StAR [35]. We observed higher amplitude of the MC2R mRNA profile in females as compared to males irrespective of the genotype, suggesting that LXRα is unlikely to significantly modulate the response of the adrenal to ACTH (Fig 4A and 4B and Table 4). We found that the acrophase of StAR mRNA was advanced by 4 to 6 hours in females as compared to males, independently of the genotype (Fig 4C and 4D and Table 4). This sex difference was potentiated in the knockout animals as Lxrα-/- females displayed an even earlier peak than their controls while Lxrα-/- and WT males were not different.
Fig 4. Analysis of circadian gene expression in the adrenals of WT and Lxrα-/- mice.
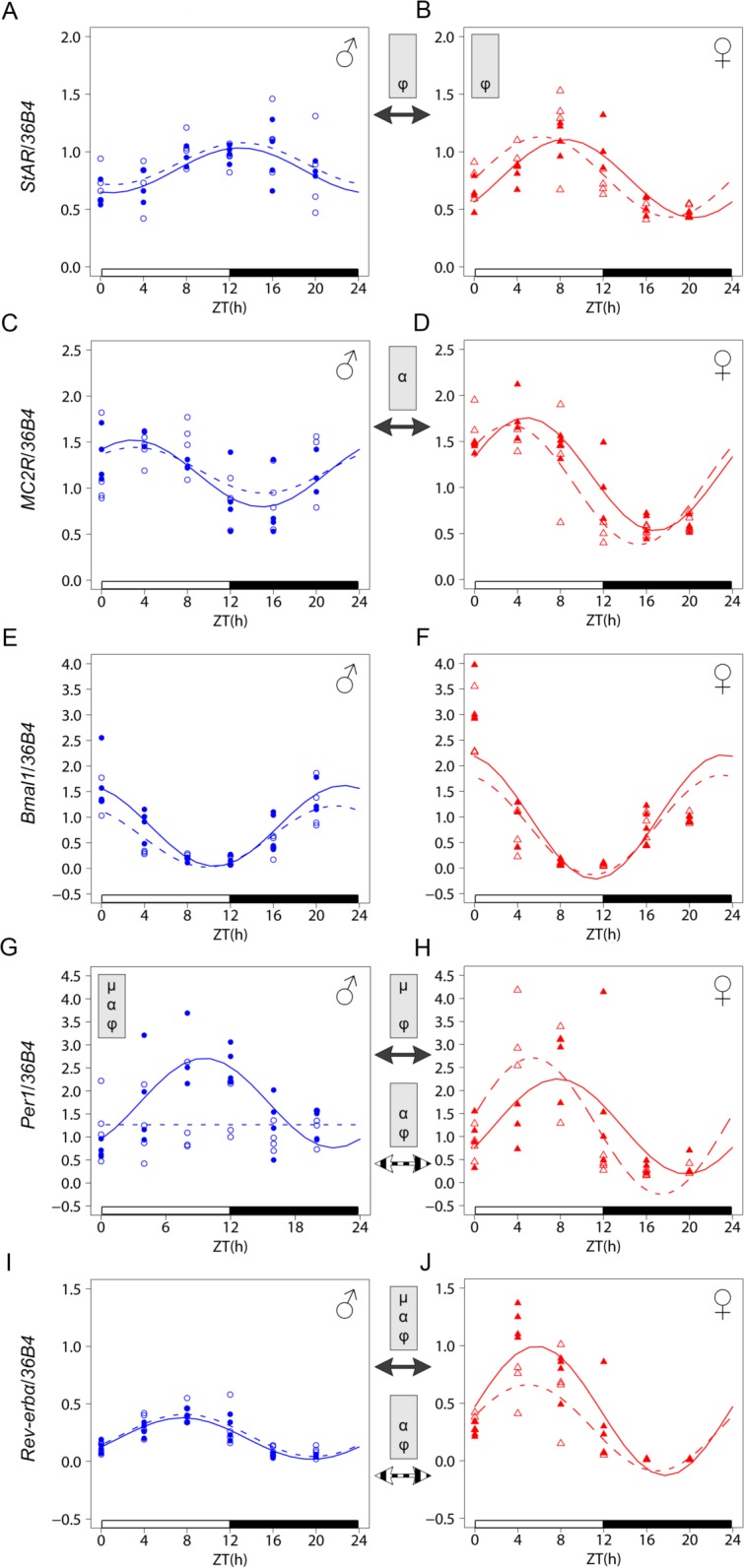
Diurnal mRNA expression of adrenal StAR (A, B), MC2R (C, D), Bmal1 (E, F), Per1 (G, H) and Rev-erbα (I, J) was compared using qRT-PCR in WT (plain line) and Lxrα-/- mice (dashed line). For each time point, 3–4 mice were used. Cosinor-based non-linear regression was used for curve fitting. The ZT0 time point is double plotted for visualization purposes. Expression data were normalized to the constitutively expressed 36B4 mRNA. The white and black bars represent the light and dark phases, respectively. Statistically significant differences in cosine fitting parameters (p<0.05) between wild type and Lxrα-/- mice or between male and female of the same genotype is indicated in the grey box at the top of the corresponding graph or between graphs (WT: plain arrow, Lxrα-/-: dashed arrow). µ, α and φ indicate a difference in mean level, amplitude and acrophase, respectively.
Table 4. Cosinor analysis of circadian gene expression in the adrenals of WT and Lxrα-/- mice.
| Gene | Group | Mean level | Amplitude | Acrophase (ZT:min) |
|---|---|---|---|---|
| StAR | ♂ WT | 0.83 (0.78 ; 0.90) | 0.20 (0.13 ; 0.28) | 12:58 (11:17 ; 14:37) |
| ♂ Lxrα -/- | 0.90 (0.82 ; 0.98) | 0.18 (0.08 ; 0.32) | 13:09 (10:35 ; 15:58) | |
| ♀ WT | 0.77 (0.72 ; 0.82) | 0.34 (0.27 ; 0.42) | 8:26 (7:32 ; 9:16) * | |
| ♀ Lxrα -/- | 0.78 (0.71 ; 0.85) | 0.35 (0.26 ; 0.46) | 6:14 (5:02 ; 7:26) † | |
| MC2R | ♂ WT | 1.15 (1.06 ; 1.27) | 0.36 (0.23 ; 0.50) | 2:56 (1:14 ; 4:25) |
| ♂ Lxrα-/- | 1.19 (1.07 ; 1.32) | 0.25 (0.10 ; 0.45) | 3:08 (0:08 ; 6:17) | |
| ♀ WT | 1.14 (1.07 ; 1.22) | 0.61 (0.51 ; 0.73)* | 4:49 (4:05 ; 5:32) | |
| ♀ Lxrα-/- | 1.03 (0.92 ; 1.15) | 0.66 (0.50 ; 0.82)* | 3:18 (2:23 ; 4:16) | |
| Per1 | ♂ WT | 1.74 (1.51 ; 1.97) | 0.98 (0.64 ; 1.35) | 9:34 (8:17 ; 10:53) |
| ♂ Lxrα-/- | 1.27 (1.04 ; 1.49)† | NSR | NSR | |
| ♀ WT | 1.09 (0.86 ; 1.33)* | 0.92 (0.61 ; 1.28) | 6:53 (5:33 ; 8:10)* | |
| ♀ Lxrα-/- | 1.23 (0.95 ; 1.51) | 1.50 (1.11 ; 1.90) | 5:22 (4:23 ; 6:21) | |
| Bmal1 | ♂ WT | 0.83 (0.71 ; 0.97) | 0.79 (0.61 ; 0.97) | 22:32 (21:40 ; 23:25) |
| ♂ Lxrα-/- | 0.62 (0.51 ; 0.74) | 0.60 (0.46 ; 0.76) | 21:47 (20:46 ; 22:48) | |
| ♀ WT | 1.00 (0.72 ; 1.26) | 1.22 (0.84 ; 1.66) | 23:11 (21:56 ; 00:21) | |
| ♀ Lxrα-/- | 0.84 (0.63 ; 1.08) | 0.98 (0.67 ; 1.34) | 23:00 (21:41 ; 00:17) | |
| Rev-erbα | ♂ WT | 0.20 (0.17 ; 0.22) | 0.18 (0.15 ; 0.21) | 7:35 (6:51 ; 8:17) |
| ♂ Lxrα-/- | 0.23 (0.19 ; 2.66) | 0.18 (0.14 ; 0.24) | 7:48 (6:44 ; 8:54) | |
| ♀ WT | 0.43 (0.35 ; 0.52)* | 0.56 (0.44 ; 0.69)* | 5:45 (4:53 ; 6:35)* | |
| ♀ Lxrα-/- | 0.29 (0.22 ; 0.36) | 0.38 (0.28 ; 0.48)* | 4:55 (3:56 ; 5:54)* |
For each parameter measured, values are represented as median ± 95% bootstrap confidence intervals.
* indicates significant differences between females vs males
† indicates significant differences between Lxrα-/- vs WT mice.
Circadian rhythmicity was considered significant for a p-value < 0.05; NSR, not significantly rhythmic.
To extend our analysis of possible LXR dependent sexual dimorphism in circadian regulation, we also analyzed clock gene expression in both the adrenals and the liver where LXRα has a prominent role. Our analysis focused on Per1, Bmal1 and Rev-erbα, three key components of the molecular clock. Differences were observed in the adrenals between WT males and females for both Per1 and Rev-erbα mean level and acrophase, while Bmal1 pattern did not differ between sexes (Fig 4E–4J and Table 4). Strikingly, we found that LXRα deficiency resulted in an arrhythmic Per1 expression in males while females showed no significant changes. By contrast, a trend for a decreased Rev-erbα mean level was only seen in Lxrα-/- females, so that they were not different from Lxrα-/- males any more. The Per1, Bmal1 and Rev-erbα expression profiles were similar in the liver irrespective of sex or genotype (Fig 5E–5J, Table 5).
Fig 5. Analysis of circadian gene expression in the liver of WT and Lxrα-/- mice.
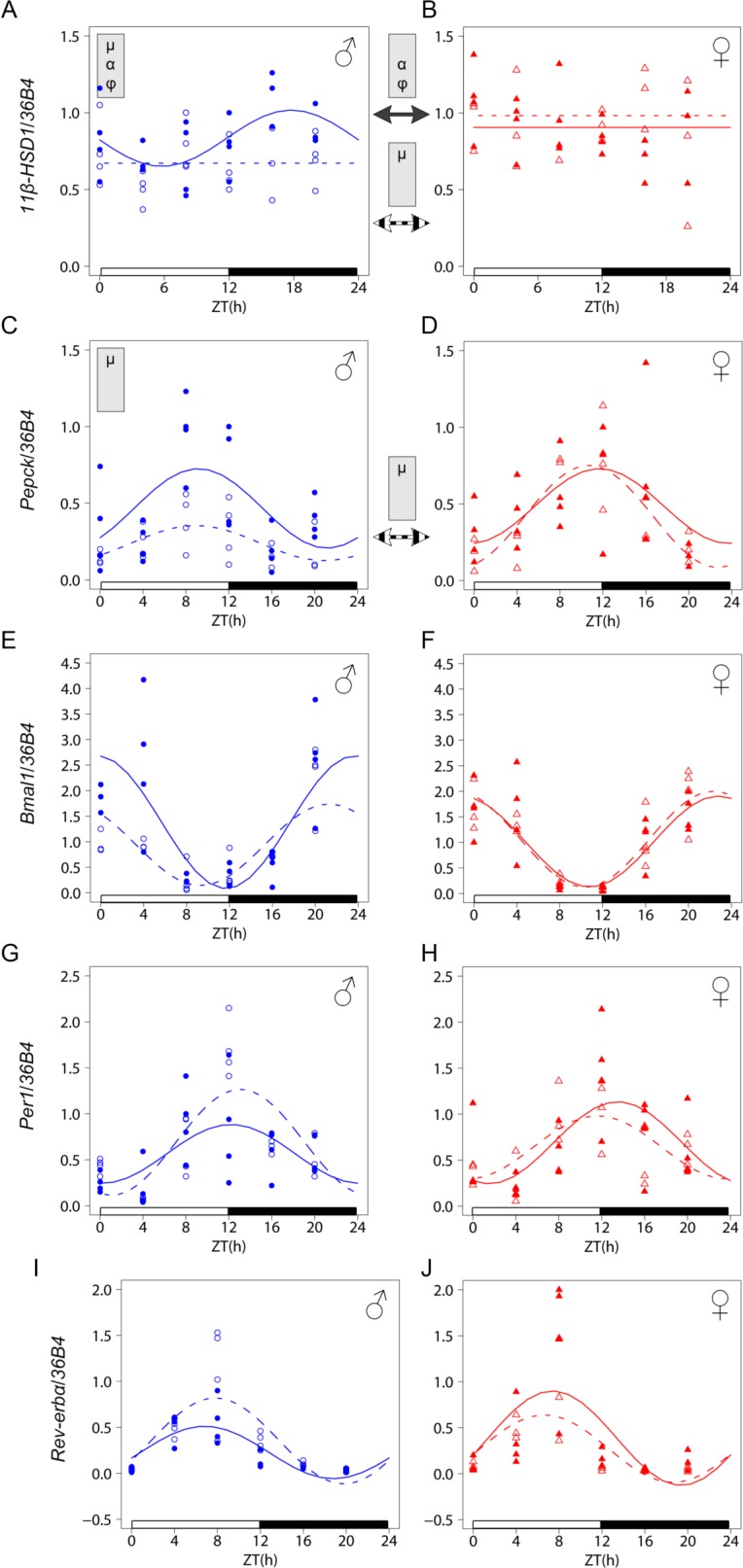
Diurnal mRNA expression of liver 11β-HSD1 (A, B), Pepck (C, D), Bmal1 (E, F), Per1 (G, H) and Rev-erbα (I, J) was compared using qRT-PCR in WT (plain line) and Lxrα-/- mice (dashed line). For each time point, 3–4 mice were used. Cosinor-based non-linear regression was used for curve fitting. The ZT0 time point is double plotted for visualization purpose. Expression data were normalized to the constitutively expressed 36B4 mRNA. The white and black bars represent the light and dark phases, respectively. Statistically significant differences in cosine fitting parameters (p<0.05) between wild type and Lxrα-/- mice or between male and female of the same genotype is indicated in the grey box at the top of the corresponding graph or between graphs (WT: plain arrow, Lxrα-/-: dashed arrow). μ, α and φ indicate a difference in mean level, amplitude and acrophase, respectively.
Table 5. Cosinor analysis of circadian gene expression in the liver of WT and Lxrα-/- mice.
| Gene | Group | Mean level | Amplitude | Acrophase (ZT:min) |
|---|---|---|---|---|
| 11β-HSD1 | ♂ WT | 0.83 (0.76 ; 0.91) | 0.18 (0.09 ; 0.29) | 17:27 (15:23 ; 20:01) † |
| ♂ Lxrα-/- | 0.67 (0.60 ; 0.74)† | NSR | NSR | |
| ♀ WT | 0.90 (0.83 ; 0.98) | NSR | NSR | |
| ♀ Lxrα-/- | 0.98 (0.84 ; 1.14) * | NSR | NSR | |
| Pepck | ♂ WT | 0.47 (0.36 ; 0.58) | 0.26 (0.12 ; 0.43) | 9:06 (6:21 ; 11:21) |
| ♂ Lxrα-/- | 0.24 (0.19 ; 0.30)† | 0.12 (0.04 ; 0.19) | 8:33 (6:12 ; 11:24) | |
| ♀ WT | 0.49 (0.38 ; 0.61) | 0.24 (0.10 ; 0.41) | 11:24 (8:33 ; 14:19) | |
| ♀ Lxrα-/- | 0.42 (0.34 ; 0.50)* | 0.34 (0.23 ; 0.45) | 10:25 (9:14 ; 12:01) | |
| Per1 | ♂ WT | 0.56 (0.43 ; 0.71) | 0.32 (0.15 ; 0.52) | 12:12 (9:38 ; 14:30) |
| ♂ Lxrα-/- | 0.69 (0.54 ; 0.85) | 0.58 (0.39 ; 0.79) | 13:01 (11:37 ; 14:29) | |
| ♀ WT | 0.69 (0.53 ; 0.85) | 0.45 (0.24 ; 0.68) | 13:19 (11:15 ; 15:19) | |
| ♀ Lxrα-/- | 0.64 (0.51 ; 0.76) | 0.34 (0.19 ; 0.53) | 11:22 (9:21 ; 13:21) | |
| Bmal1 | ♂ WT | 1.38 (1.05 ; 1.72) | 1.30 (0.86 ; 1.80) | 23:41 (22:20 ; 01:00) |
| ♂ Lxrα-/- | 0.94 (0.75 ; 1.15) | 0.80 (0.54 ; 1.10) | 21:17 (19:55 ; 22:42) | |
| ♀ WT | 1.02 (0.84 ; 1.21) | 0.89 (0.64 ; 1.17) | 22:47 (21:39 ; 23:56) | |
| ♀ Lxrα-/- | 1.06 (0.90 ; 1.23) | 0.95 (0.72 ; 1.21) | 22:20 (21:20 ; 23:17) | |
| Rev-erbα | ♂ WT | 0.23 (0.17 ; 0.29) | 0.29 (0.21 ; 0.37) | 6:29 (5:25 ; 7:33) |
| ♂ Lxrα-/- | 0.20 (0.10 ; 0.32) | 0.31 (0.17 ; 0.47) | 7:42 (5:45 ; 9:42) | |
| ♀ WT | 0.41 (0.24 ; 0.60) | 0.50 (0.26 ; 0 ;78) | 7:00 (4:48; 9:03) | |
| ♀ Lxrα-/- | 0.27 (0.17; 0.39) | 0.37 (0.22; 0.54) | 6:37 (4.50; 8:23) |
For each parameter measured, values are represented as median ± 95% bootstrap confidence intervals.
* indicates significant differences between females vs males
† indicates significant differences between Lxrα-/- vs WT mice.
Circadian rhythmicity was considered significant for a p-value < 0.05; NSR, not significantly rhythmic.
Because both the StAR and MC2R patterns were unlikely to explain the observed CORT pattern in LXRα deficient mice, while clock function was not globally altered in the periphery, we additionally explored the expression of hepatic type 1 11β-hydroxysteroid dehydrogenase (11β-HSD1) which catalyzes the reactivation of 11-dehydrocorticosterone in the periphery and thereby locally controls glucocorticoid signaling [36]. We found that 11β-HSD1 is rhythmically expressed with low amplitude in the liver from WT males but not in females (Fig 5A and 5B and Table 5). The 11β-HSD1 mRNA oscillation was abolished in Lxrα -/- males while remaining unchanged in females as compared to WT animals (Fig 5A and 5B and Table 5). We conclude from this gene expression profiling that loss of LXRα has a highly selective effect that is restricted to adrenal Per1 and hepatic 11β-HSD1in males and StAR in females. This data may point to a primary defect both in liver and adrenal in males whereas only adrenal steroidogenesis seems to be affected in females.
Circadian rhythms of metabolic parameters are differentially impacted in LXR α -/- male and female mice
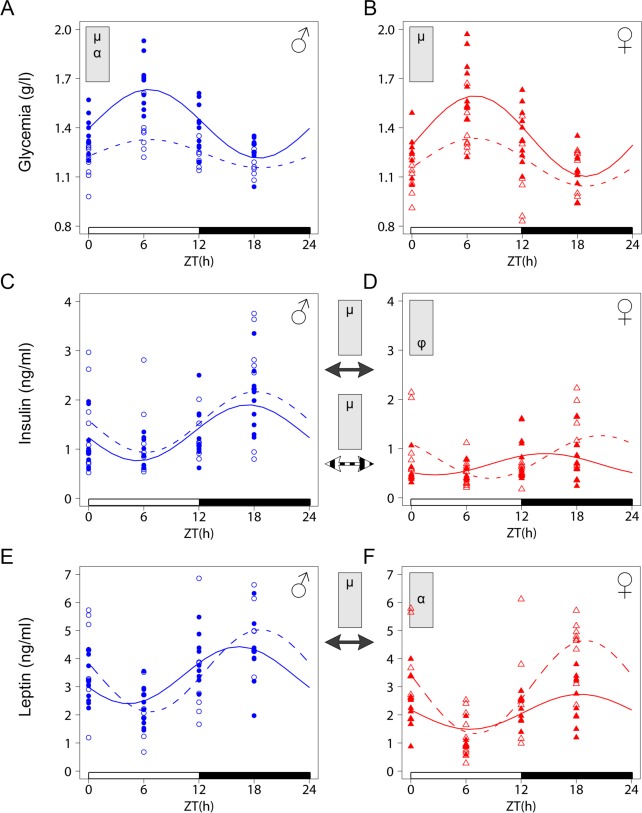
Glucocorticoid hormones play a crucial role in metabolic homeostasis and LXRα has been established as an important metabolic sensor in mammals. We therefore sought to link the observed selective changes in CORT levels and circadian gene expression to physiological parameters related to glucose homeostasis. We first analyzed plasma glucose levels at 4 time points along the 24 h cycle and observed an approximately two-fold decrease of amplitude in Lxrα-/- males as compared to their controls, leading to hypoglycemia during all the resting phase (Fig 6A and 6B, Table 6). Lxrα-/- females were also affected but to a lesser extent. Hepatic glycogen content measured at ZT0 and ZT12 varied as expected but revealed no effect of sex or genotype (S3A and S3B Fig). However, we noticed that the sex difference in food intake was abolished in Lxrα -/- mice (S3C Fig).
Fig 6. Analysis of circadian variation of glucose, insulin and leptin in WT and Lxrα-/- mice.
Plasma glucose (A, B), insulin (C, D) and leptin (E, F) were determined in WT and Lxrα-/- mice entrained to a LD12:12 cycle. Cosinor-based non-linear regression was used for curve fitting. The ZT0 time point is double plotted for visualization purposes. The white and black bars represent the light and dark phases, respectively. Statistically significant differences in cosine fitting parameters (p<0.05) between wild type and Lxrα-/- mice or between male and female of the same genotype is indicated in the grey box at the top of the corresponding graph or between graphs (WT: plain arrow, Lxrα-/-: dashed arrow). μ, α and φ indicate a difference in mean level, amplitude and acrophase, respectively.
Table 6. Cosinor analysis of circadian variation of glucose, insulin and leptin in WT and Lxrα-/- mice.
| Parameter | Group | Mean level | Amplitude | Acrophase (ZT:min) |
|---|---|---|---|---|
| Glucose | ♂ WT | 1.42 (1.38 ; 1.46) | 0.21 (0.16 ; 0.27) | 6:53 (5:27 ; 7:36) |
| ♂ Lxrα-/- | 1.24 (1.21 ; 1.27)† | 0.09 (0.04 ; 0.14)† | 6:40 (4:23 ; 8:49) | |
| ♀ WT | 1.35 (1.30 ; 1.40) | 0.25 (0.18 ; 0.32) | 6:53 (5:46 ; 7:59) | |
| ♀ Lxrα-/- | 1.19 (1.13 ; 1.24)† | 0.15 (0.08 ; 0.22) | 6:51 (4:34 ; 8:54) | |
| Insulin | ♂ WT | 1.33 (1.18 ; 1.49) | 0.57 (0.36 ; 0.81) | 17:20 (15:50 ; 18:49) |
| ♂ Lxrα-/- | 1.55 (1.27 ; 1.85) | 0.62 (0.27 ; 1.07) | 18:12 (15:26 ; 20:54) | |
| ♀ WT | 0.68 (0.57 ; 0.80)* | 0.22 (0.08 ; 0.39) | 14:34 (10:43 ; 17:32) | |
| ♀ Lxrα-/- | 0.83 (0.68 ; 0.98)* | 0.44 (0.25 ; 0.67) | 20:34 (18:16 ; 22:27)† | |
| Leptin | ♂ WT | 3.41 (3.14 ; 3.70) | 1.01 (0.67 ; 1.40) | 16:19 (14:54 ; 17:44) |
| ♂ Lxrα-/- | 3.57 (3.14 ; 4.03) | 1.48 (0.86 ; 2.14) | 18:43 (17:09 ; 20:23) | |
| ♀ WT | 2.10 (1.86 ; 2.36)* | 0.62 (0.30 ; 1.03) | 18:25 (16:15 ; 20:35) | |
| ♀ Lxrα-/- | 2.99 (2.61 ; 3.39) | 1.67 (1.19 ; 2.21)† | 19:01 (17:34 ; 20:20) |
For each parameter measured, values are represented as median ± 95% bootstrap confidence intervals.
* indicates significant differences between females vs males
† indicates significant differences between Lxrα-/- vs WT mice.
Circadian rhythmicity was considered significant for a p-value < 0.05; NSR, not significantly rhythmic.
To determine whether changes in glycemia could be caused by a differential hepatic glucose production, we monitored the expression of the phosphoenolpyruvate carboxykinase (Pepck) gene, which encodes a rate limiting step in the hepatic gluconeogenesis pathway. Results showed a significant decrease in the amplitude of Pepck mRNA oscillation in Lxrα -/- males as compared to controls while females from both genotypes exhibited a similar circadian expression profile. (Fig 5C and 5D and Table 5). At the physiological level, plasma insulin, oscillated in both males and females but with higher amplitude in males. This profile was not altered in Lxrα -/-males while females displayed a 6 hours phase delay as compared to WT animals (Fig 6C and 6D and Table 6). Because leptin is known to regulate glucose metabolism through its action in the brain, we also investigated its circadian pattern in the Lxrα -/- model. We found that WT females displayed lower mean levels than WT males. In contrast, this sexual dimorphism was not observed in Lxrα -/- animals because Lxrα -/- females exhibited significantly higher amplitude of the leptin rhythm thus resembling males (Fig 6E and 6F and Table 6). These data collectively indicate that the circadian regulation of metabolic parameters is altered in a sex-specific manner in LXRα deficient mice.
Discussion
LXRα is an essential metabolic sensor which upon binding to oxysterols regulates the transcription of genes critical for cholesterol, lipid, and glucose homeostasis [24]. In this report we identified a novel and unanticipated role for LXRα as a determinant of the sexual dimorphism of circadian physiology. Lxrα-/- male mice have previously been reported to display adrenomegaly and increased plasma CORT levels, although the circadian pattern was not investigated [26]. We and others reported significant sex specific differences in the circadian rhythm of plasma CORT which show higher levels and amplitude in females than in males [29, 37, 38]. The observation that CORT was arrhythmic in Lxrα-/- males while females displayed higher amplitude is of importance because there is compelling evidence from the literature that glucocorticoid hormones are potent synchronizers of peripheral circadian clocks in addition to their multiple physiological roles [16, 39–41]. For instance, phase-shifting the CORT pattern using metyrapone changed the speed of behavioral re-entrainment after a jetlag and transplanting clock-deficient adrenals in adrenalectomized WT hosts caused a faster re-entrainment after a 6-hour phase advance [33]. Furthermore it was shown in rat that CORT rhythmic secretion is critical for normal resetting behavior [34]. We hypothesize that the faster re-entrainment seen in Lxrα-/- males may result at least in part from the altered CORT pattern. The circadian rhythm of CORT secretion by the adrenal cortex is regulated both at the central level by the SCN and locally through the HPA axis. Because ACTH levels are unchanged in Lxrα-/- mice while adrenals from these animals secrete more CORT in vitro, Cummins et al excluded a dysregulation of the HPA axis in these animals [26]. At the adrenal level, the rhythmic secretion pattern of GCs is determined by both the sensitivity to ACTH and GCs biosynthesis rate. Accordingly, the adrenal responsiveness to ACTH follows a diurnal rhythm, with a higher sensitivity during the activity phase in rodents [42, 43]. The StAR gene that encodes a rate-limiting enzyme of the steroidogenic pathway is the only component showing a robust circadian expression in the adrenal [44]. Expression of the MC2R and StAR mRNA was circadian and sexually dimorphic in our study, but this was not changed in Lxrα -/- mice suggesting that the damped CORT pattern observed in Lxrα -/- males may primarily result from the adrenomegaly reported by Cummins et al [26].
Notably, the biological effects of GCs involve to a large extent the 11β-HSD1 enzyme that catalyzes the reduction of plasma 11-dehydrocorticosterone to CORT. This reactivation pathway plays a major role for local production of CORT and can also indirectly impact adrenal CORT secretion [36, 45, 46]. The liver, which is the main site of LXRα expression, also contains the highest concentration of 11β-HSD1. Mice lacking 11β-HSD1 display compensatory adrenomegaly and elevated morning (diurnal nadir) plasma CORT concentrations but similar peak levels [45]. Intriguingly, this phenotype is highly reminiscent to that observed in Lxrα-/- male mice and prompted us to investigate hepatic 11β-HSD1expression around the clock. We found that the circadian pattern of 11β-HSD1 mRNA expression was totally damped in Lxrα-/- males. An earlier study failed to detect this change possibly because only one time point in the early light phase was analyzed [47]. Yet this previous paper reported a decreased hepatic expression of 11β-HSD1 upon treatment with the LXR agonist TO901317 [47]. The similar effect of both the agonist treatment and the lack of LXRα on 11β-HSD1 expression can be explained by the fact that the apoLXRα is a repressor which, upon deletion or activation, induces an intermediary repressor of the 11β -HSD1 gene [47]. Altogether, this data strongly suggest that LXRα regulates the circadian rhythm of GCs production in males both at the systemic and local levels through its action on the adrenals and 11β -HSD1 respectively. Presumably, females which exhibit higher CORT levels and amplitude than males do not require further circadian time dependent increase of the local regeneration of CORT. This may explain why 11β -HSD1 is neither rhythmic nor regulated by LXRα in females. Interestingly, mice lacking 11β -HSD1 are hypoglycemic and a have decreased Pepck response to fasting hypoglycemia [45]. Thus the lower amplitude of plasma glucose oscillation seen in Lxα-/-males may result, at least in part, from an altered stimulation of the Pepck gene by locally produced GCs. This demonstrates that LXRα is required not only for normoglycemia but also for normal daily oscillation of glucose levels. We do not exclude that this role in the interaction between glucose homeostasis and circadian timing could also involve LXRα outside the liver. Indeed insulin sensitivity is known to be under control of the SCN clock and consequently displays daily variations [48]. Activation of LXRs using the GW3965 agonist improves glucose tolerance in a mouse model of diet-induced obesity and insulin resistance [49]. These effects are attributable to the role of LXRα in insulin secretion and glucose uptake. It is therefore plausible that the damped plasma glucose profile in Lxrα-/- males also originates from a time specific increase (ZT0-ZT12) of peripheral sensitivity to insulin resulting from the loss of LXRα [49, 50]. Although female LXRα mice did not show significant changes in their plasma glucose profile, there was also a trend toward decreased levels. Additionaly, they displayed a dramatic phase delay of their insulin profile (6 hours) that can be interpreted as a consequence of the significant increase in the amplitude of leptin, an adipokine showing a circadian variation in plasma and regulated by GCs[51, 52]. The dramatic increase in the amplitude of CORT in females could therefore explain the gender specific increase in the amplitude of leptin rhythmicity, resulting in a higher demand for insulin secretion.
LXRα not only influences the production of corticosteroids but also that of sex steroids. Lxrα-/- female mice display elevated levels of 17β-estradiol and an ovarian hyperstimulation phenotype [53]. Unexpectedly, expression of Cyp19 which aromatizes androgens was found to be decreased while that of Cyp11a1 was increased in Lxrα-/- female mice, suggesting that they may also produce more testosterone than WT animals. Males lacking LXRα show a contrasting situation as they display decreased levels of testicular testosterone associated to an increased apoptosis rate of germ cells [54]. These effects of LXRα on sex steroid production could therefore also contribute the sexual dimorphism of physiological parameters including those under circadian variation. Importantly, there is evidence that the gonadotropic axis and glucocorticoid signaling interfere at different levels. GCs are known to directly inhibit gonadotropin-stimulated testosterone production [55], and serum corticosterone and testosterone levels are inversely correlated [56]. Consistently, chronic corticotherapy reduces serum testosterone levels In human patients [57]. Crosstalk between the estrogen, GCs and LXR pathways have been described. [58]. [59, 60]. Collectively, this and our data suggests that loss of LXRα compromises at the organismal level the fine tuning of the sex-specific balance between glucocorticoid, estrogen and androgen production and action.
The LXRα dependent co-regulation of clock outputs is another mechanism that could also underlie the observed phenotype. For instance LXRα shares many targets with the circadian nuclear receptor PPARα and the formation of LXRα-PPARα heterodimer has been shown to downregulate Cyp7α1 expression [61, 62]. LXRα is also positively regulated by the deacetylase SIRT1, an enzyme displaying a circadian activity in liver [63, 64]. Thus, LXRα activity could be modulated posttranslationally by the circadian clock through rhythmic deacetylation.
We excluded a direct role of LXRα in the transcriptional regulation of the clock network because Per1 was the only core clock gene found to be changed in Lxrα-/- animals and this was restricted male adrenals. This suggests that Per1 rhythmic expression in the adrenal is more likely to be driven by a systemic cue such as CORT, the production of which is altered in Lxrα-/- males (see below). This hypothesis is supported by the observation that Per1 is an hyper-responsive gene to GCs [65]. In addition two previous genome wide studies failed to identify core clock components among genes regulated by the LXRα agonist T0901317 or directly bound by LXRα [61, 66]. Finally the marginal effects of the LXRα mutation on the free-running period of locomotor activity also supports that LXRα does not play a significant role in core clock mechanism.
Despite a considerable male bias in most animal studies [67], sexual dimorphism in metabolic and circadian physiology has been recognized in mammals [68, 69]. Reproductive factors including sex hormones are considered as major determinants of such differences but non-reproductive factors including glucocorticoid hormones also appear to play a significant role [70]. Compelling evidences have recently linked metabolic homeostasis and circadian timing and involved numerous nuclear hormone receptors. We extend this concept by linking the metabolic nuclear receptor LXRα to the circadian and sex-dependent regulation of physiology. These findings are of importance in the context of pharmacological studies or future personalized therapies targeting LXRα.
Materials and Methods
Animals
Mutant Lxrα-/- breeders in the C57BL/6j background were obtained from Taconic and subsequently crossed in our facility. Control C57BL/6j animals were from Charles River (France) and adapted to the facility environment for at least 6 weeks before the experiments to prevent any bias related to housing conditions. Mice were housed in a temperature-controlled room with a 12-hours light (325 lux)/12-hours dark (LD 12:12) cycle and fed ad libitum. All experiments were performed with 3 months-old animals. Animal experiment procedures were carried out in accordance with the CNRS and INSERM institutional guidelines. The local ethical committee (Comité Institutionnel d'Éthique Pour l'Animal de Laboratoire CIEPAL-AZUR PEA N° NCE 2011–26) specifically approved this study.
Plasma metabolic parameters measurements
Blood samples were collected from the retro-orbital venous plexus in heparin containing tubes, and plasma was separated by centrifugation for 20 min at 3,000 rpm. For each mouse, blood was collected only twice (two different ZT times), at 2 weeks intervals, to avoid stress effects. Plasma glucose was determined using an Accu-Check glucometer (Roche Diagnostics, France). Serum insulin, leptin and corticosterone concentrations were measured with commercial enzyme-linked immunosorbent assays from respectively Mercodia (Uppsala, Sweden) R&D (MOB00) and Molecular Devices companies with an enzyme standard instrument for serum corticosterone levels, according to the manufacturer’s recommendations.
RNA extraction and quantitative real-time (qRT)-PCR
Total RNA was extracted using the single step method described by Chomczynski andmRNA levels were measured by real-time (RT)–qPCR using a Light Cycler 1.5 (RocheApplied Science) and SYBR green I dye detection according to the manufacturer's recommendation. cDNA, synthesized from 2 to 5 μg of total RNA using random primers and Superscript II (Invitrogen), was added to a reaction mixture (Faststart DNA SYBR green I; Roche Diagnostics) with appropriate primers at 0.5 mM each (Lxrα: forward 5’-CTGATGTTTCTCCTGATTCTGC-3’ and reverse 5’-CTTTTTCCGCTTTTGTGGAC-3’, Cyp7α1: forward 5’-TACTTCTGCGAAGGCATTTGG-3’ and reverse 5’-TACTTCTGCGAAGGCATTTGG-3’, StAR: forward 5’-AGGAAAGCCAGCAGGAGAAC-3’ and reverse 5’-TGATGACCGTGTCTTTTCCA-3’, 11β-HSD1: forward 5’-GGCGGGAAAGCTCATGG-3’ and reverse 5’-AAGGAGGAGATGACGGCAAT-3’, MC2R: forward 5’-GCCCTTCTAAGCCAGATC-C-3’ and reverse 5’-ATTTCTTGCGGTGTCATTGG-3’, Per1: forward 5’-GAAGTTTGAGCTCCCGAAGT-3’ and reverse 5’-TGAGAGCAGCAAGAGTACAAAC-3’, Rev-erbα: forward 5’-AACCTCCAGTTTGTGTCAAGGT-3’ and reverse 5’-GATGACGATGATGCAGAAGAAG-3’ Bmal1: forward 5’-CTCATTGATGCCAAGACTGG-3’ and reverse 5’-GGTGGCCAGCTTTTCAAATA-3’, Pepck: forward 5’-TTTGATGCCCAAGGCAACTT-3’ and reverse 5’-ATCGATGCCTTCCCAGTAAA-3’, 36B4: forward 5’-GCTGATGGGCAAGAACACCA-3’ and reverse 5’-CCCAAAGCCTGGAAGAAGGA-3’). The relative mRNA abundance was calculated using a standard-curve method. Expression levels were normalized to the levels of the constitutively expressed 36B4 ribosomal protein mRNA.
Running wheel activity measurement
Mice were individually housed within light-controlled isolation chambers in cages containing monitored activity wheels, and they were allowed ad libitum access to food and water. Animals were weighed weekly and their daily food intake in each condition was estimated by measuring the difference between the quantity of food provided and food remaining after 1 week / 7. Mice were entrained to an initial 12:12 LD cycle (Light phase from 7:00 to 19:00, ZT0 = lights on = 7:00). Daily as well as (subjective) day and night activity were quantified using Clocklab plugin for Matlab (Actimetrics). After 3 weeks of activity recording on this cycle, they were transferred to constant darkness (DD). Their free running period was assessed based on activity onset on the second week of DD using Clocklab. Mice were then returned to the initial 12:12 LD cycle for re-entrainment. After 3 weeks, animals were subjected to an 8h phase shift (Jetlag experiment) by advancing the light phase from 23:00 to 11:00. The time to re-entrain to the new lighting schedule was determined based on activity onset and acrophase using Clocklab.
Statistical analysis
Activity data are reported as median ± 95% bootstrap confidence intervals. For the jetlag experiment, results are represented as boxplots. Data were analysed using a Kruskal and Wallis rank sum test for multiple comparison between groups followed by non-parametric pairwise post-hoc test. For time series, data was modelled using a non-linear regression (Cosine fitting analysis: a + b * cos(2 * pi * (ZT—c)/24); where a = mean expression level, b = amplitude of the oscillation and c = acrophase). Cosine fitting analysis was followed by bootstrapping to compare confidence intervals on a, b and c parameters between time series. Significance level was set at p < 0.05. Statistical analysis was performed using the R software (version 2.15.2; The R Foundation for Statistical Computing).
Supporting Information
Diurnal mRNA expression of liver GR was compared in males (blue) and females (red) using qRT-PCR in WT and Lxrα-/- mice. For each time point, 3–4 mice were used. Cosine-based non-linear regression was used for curve fitting. The ZT0 time point is double plotted for visualization purposes. Expression data were normalized to the constitutively expressed 36B4 mRNA. The white and black bars represent the light and dark phases, respectively.Statistically significant differences in cosine fitting parameters (p<0.05) between wild type and Lxrα-/- mice or between male and female of the same genotype is indicated in the grey box at the top of the corresponding graph or between graphs (WT: plain arrow, Lxrα-/-: dashed arrow). μ, α and φ indicate a difference in mean level, amplitude and acrophase, respectively.
(TIF)
Diurnal mRNA expression of adrenal Lxrα was compared in males (blue) and females (red) using qRT-PCR in WT mice. For each time point, 3–4 mice were used. Cosine-based non-linear regression was used for curve fitting. The ZT0 time point is double plotted for visualization purposes. Expression data were normalized to the constitutively expressed 36B4 mRNA. The white and black bars represent the light and dark phases, respectively. Statistically significant differences in cosine fitting parameters (p<0.05) between wild type and Lxrα-/- mice or between male and female of the same genotype is indicated in the grey box at the top of the corresponding graph or between graphs (WT: plain arrow, Lxrα-/-: dashed arrow). μ, α and φ indicate a difference in mean level, amplitude and acrophase, respectively.
(TIF)
(A) Mean daily food intake (g) in male and female WT and Lxrα-/- mice. Note there is a significant difference between WT males and WT females (p>0.05). (B-C) Glucose content liberated from glycogen in liver pieces at ZT0 and ZT12. (B) Female WT vs Lxrα-/- mice (n = 4) and (C) male WT vs Lxrα-/- mice (n = 4). There is a significant difference in liver glycogen content between ZT0 and ZT12 (p>0.0001) but no influence of the LXRα mutation.
(TIF)
Liver pieces collected at ZT0 and ZT12, from 4 animals of each group were used in this experiment. For each sample, 30 mg of liver was lysed in KOH 0.5M at 95°C. Na2SO4 6% (25 μl) and 750 μl methanol were then added. Glycogen was precipitated at -80°C in 2 separate aliquots for each sample. After centrifugation, glycogen was either resuspended in 200 μl amyloglucosidase 2 mg/ml (Sigma-Aldrich) or in 200 μl sodium acetate, to assay total glucose and free glucose respectively. Suspensions were incubated for 1-h at 37°C. Free/total glucose content was measured on 5 μL of supernatant in 300 μl of reagent using a glucose hexokinase assay kit (Sigma-Aldrich) according to manufacturer’s protocol. Glucose was expressed in μmol/g wet liver. Glucose coming from glycogen was determined as (total glucose)-(free glucose) in each sample.
(DOCX)
Values are represented as median ± 95% bootstrap confidence intervals. * indicates significant differences between females vs males, † indicates significant differences between Lxrα-/- vs WT mice. Circadian rhythmicity was considered significant for a p-value < 0.05; NSR, not significantly rhythmic.
(DOCX)
Acknowledgments
The authors thank the staff of the Institute of Biology Valrose animal facility for their technical help.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by University Nice Sophia Antipolis France http://unice.fr/ (MT, FD); Centre National de la Recherche Scientifique http://www.cnrs.fr/ (MT, FD); Institut National de la Santé et de la Recherche Médicale http://www.inserm.fr/ (MT, FD); European Commission grants FP6 CRESCENDO LSHM-CT-2005-01865 (FD); Agence Nationale de la Recherche http://www.agence-nationale-recherche.fr/; CHRONOMET project ANR-12-BSV1- 0014-01 (FD); Agence Nationale de la Recherche http://www.agence-nationale-recherche.fr/ LABEX SIGNALIFE program ANR-11-LABX-0028-01 (FD); The Robert A Welch Foundation http://www.welch1.org/ (E-0004 to JÅG).
References
- 1.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101(15):5339–46. 10.1073/pnas.0308709101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tahara Y, Kuroda H, Saito K, Nakajima Y, Kubo Y, Ohnishi N, et al. In vivo monitoring of peripheral circadian clocks in the mouse. Curr Biol. 2012;22(11):1029–34. 10.1016/j.cub.2012.04.009 . [DOI] [PubMed] [Google Scholar]
- 3.Buhr ED, Takahashi JS. Molecular components of the Mammalian circadian clock. Handb Exp Pharmacol. 2013;(217):3–27. Epub 2013/04/23. 10.1007/978-3-642-25950-0_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15 Spec No 2:R271–7. 10.1093/hmg/ddl207 . [DOI] [PubMed] [Google Scholar]
- 5.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485(7396):123–7. Epub 2012/03/31. 10.1038/nature11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Fang B, Emmett MJ, Damle M, Sun Z, Feng D, et al. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science. 2015;348(6242):1488–92. 10.1126/science.aab3021 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanselow K, Kramer A. Role of phosphorylation in the mammalian circadian clock. Cold Spring Harbor symposia on quantitative biology. 2007;72:167–76. Epub 2008/04/19. 10.1101/sqb.2007.72.036 . [DOI] [PubMed] [Google Scholar]
- 8.Virshup DM, Eide EJ, Forger DB, Gallego M, Harnish EV. Reversible protein phosphorylation regulates circadian rhythms. Cold Spring Harbor symposia on quantitative biology. 2007;72:413–20. Epub 2008/04/19. 10.1101/sqb.2007.72.048 . [DOI] [PubMed] [Google Scholar]
- 9.Reischl S, Kramer A. Kinases and phosphatases in the mammalian circadian clock. FEBS letters. 2011;585(10):1393–9. Epub 2011/03/08. 10.1016/j.febslet.2011.02.038 . [DOI] [PubMed] [Google Scholar]
- 10.Feng D, Lazar MA. Clocks, metabolism, and the epigenome. Mol Cell. 2012;47(2):158–67. Epub 2012/07/31. 10.1016/j.molcel.2012.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sahar S, Sassone-Corsi P. The epigenetic language of circadian clocks. Handb Exp Pharmacol. 2013;(217):29–44. Epub 2013/04/23. 10.1007/978-3-642-25950-0_2 . [DOI] [PubMed] [Google Scholar]
- 12.Lipton JO, Yuan ED, Boyle LM, Ebrahimi-Fakhari D, Kwiatkowski E, Nathan A, et al. The Circadian Protein BMAL1 Regulates Translation in Response to S6K1-Mediated Phosphorylation. Cell. 2015;161(5):1138–51. 10.1016/j.cell.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, et al. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science. 2012;338(6105):349–54. Epub 2012/09/01. 10.1126/science.1226339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A. 2014;111(45):16219–24. 10.1073/pnas.1408886111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teboul M, Guillaumond F, Gréchez-Cassiau A, Delaunay F. Minireview: the nuclear hormone receptor family round the clock. Molecular Endocrinology. 2008;22(12):2573–82. 10.1210/me.2007-0521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, et al. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289(5488):2344–7. . [DOI] [PubMed] [Google Scholar]
- 17.Canaple L, Rambaud J, Dkhissi-Benyahya O, Rayet B, Tan NS, Michalik L, et al. Reciprocal regulation of brain and muscle Arnt-like protein 1 and peroxisome proliferator-activated receptor α defines a novel positive feedback loop in the rodent liver circadian clock. Molecular Endocrinology. 2006;20(8):1715–27. [DOI] [PubMed] [Google Scholar]
- 18.Wang N, Yang G, Jia Z, Zhang H, Aoyagi T, Soodvilai S, et al. Vascular PPARgamma controls circadian variation in blood pressure and heart rate through Bmal1. Cell Metab. 2008;8(6):482–91. 10.1016/j.cmet.2008.10.009 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai W, Rambaud J, Teboul M, Masse I, Benoit G, Gustafsson J- Å, et al. Expression levels of estrogen receptor β are modulated by components of the molecular clock. Molecular and cellular biology. 2008;28(2):784–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanno Y, Otsuka S, Hiromasa T, Nakahama T, Inouye Y. Diurnal difference in CAR mRNA expression. Nucl Recept. 2004;2(1):6 10.1186/1478-1336-2-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dufour CR, Levasseur MP, Pham NH, Eichner LJ, Wilson BJ, Charest-Marcotte A, et al. Genomic convergence among ERRα, PROX1, and BMAL1 in the control of metabolic clock outputs. PLoS Genet. 2011;7(6):e1002143 10.1371/journal.pgen.1002143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan X, Zhang Y, Wang L, Hussain MM. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 2010;12(2):174–86. 10.1016/j.cmet.2010.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong SH, Ahmadian M, Yu RT, Atkins AR, Downes M, Evans RM. Nuclear receptors and metabolism: from feast to famine. Diabetologia. 2014;57(5):860–7. 10.1007/s00125-014-3209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calkin AC, Tontonoz P. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat Rev Mol Cell Biol. 2012;13(4):213–24. 10.1038/nrm3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Lo Sasso G, et al. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7(9):e1000181 Epub 2009/09/02. 10.1371/journal.pbio.1000181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cummins CL, Volle DH, Zhang Y, McDonald JG, Sion B, Lefrançois-Martinez AM, et al. Liver X receptors regulate adrenal cholesterol balance. J Clin Invest. 2006;116(7):1902–12. 10.1172/JCI28400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126(4):801–10. 10.1016/j.cell.2006.06.050 . [DOI] [PubMed] [Google Scholar]
- 28.Goel N, Workman JL, Lee TT, Innala L, Viau V. Sex differences in the HPA axis. Compr Physiol. 2014;4(3):1121–55. 10.1002/cphy.c130054 . [DOI] [PubMed] [Google Scholar]
- 29.Ahowesso C, Li X-M, Zampera S, Peteri-Brunbäck B, Dulong S, Beau J, et al. Sex and dosing-time dependencies in irinotecan-induced circadian disruption. Chronobiology international. 2011;28(5):458–70. 10.3109/07420528.2011.569043 [DOI] [PubMed] [Google Scholar]
- 30.Muglia LJ, Jacobson L, Weninger SC, Luedke CE, Bae DS, Jeong KH, et al. Impaired diurnal adrenal rhythmicity restored by constant infusion of corticotropin-releasing hormone in corticotropin-releasing hormone-deficient mice. J Clin Invest. 1997;99(12):2923–9. 10.1172/JCI119487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGinnis R, Walker J, Margules D, Aird F, Redei E. Dysregulation of the Hypothalamus-Pituitary-Adrenal Axis in Male and Female, Genetically Obese (ob/ob) Mice. J Neuroendocrinol. 1992;4(6):765–71. 10.1111/j.1365-2826.1992.tb00229.x . [DOI] [PubMed] [Google Scholar]
- 32.Dong Y, Poellinger L, Gustafsson JA, Okret S. Regulation of glucocorticoid receptor expression: evidence for transcriptional and posttranslational mechanisms. Mol Endocrinol. 1988;2(12):1256–64. 10.1210/mend-2-12-1256 . [DOI] [PubMed] [Google Scholar]
- 33.Kiessling S, Eichele G, Oster H. Adrenal glucocorticoids have a key role in circadian resynchronization in a mouse model of jet lag. J Clin Invest. 2010;120(7):2600–9. 10.1172/JCI41192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sage D, Ganem J, Guillaumond F, Laforge-Anglade G, François-Bellan AM, Bosler O, et al. Influence of the corticosterone rhythm on photic entrainment of locomotor activity in rats. J Biol Rhythms. 2004;19(2):144–56. 10.1177/0748730403261894 . [DOI] [PubMed] [Google Scholar]
- 35.Park SY, Walker JJ, Johnson NW, Zhao Z, Lightman SL, Spiga F. Constant light disrupts the circadian rhythm of steroidogenic proteins in the rat adrenal gland. Mol Cell Endocrinol. 2013;371(1–2):114–23. 10.1016/j.mce.2012.11.010 . [DOI] [PubMed] [Google Scholar]
- 36.Seckl JR, Walker BR. Minireview: 11beta-hydroxysteroid dehydrogenase type 1- a tissue-specific amplifier of glucocorticoid action. Endocrinology. 2001;142(4):1371–6. 10.1210/endo.142.4.8114 . [DOI] [PubMed] [Google Scholar]
- 37.Griffin AC, Whitacre CC. Sex and strain differences in the circadian rhythm fluctuation of endocrine and immune function in the rat: implications for rodent models of autoimmune disease. J Neuroimmunol. 1991;35(1–3):53–64. . [DOI] [PubMed] [Google Scholar]
- 38.Chmielarz P, Kreiner G, Nalepa I. Selective ablation of glucocorticoid receptors in the noradrenergic system affects evening corticosterone levels in a sex-dependent manner. Pharmacol Rep. 2015;67(6):1201–3. 10.1016/j.pharep.2015.05.013 . [DOI] [PubMed] [Google Scholar]
- 39.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119(5):693–705. 10.1016/j.cell.2004.11.015 . [DOI] [PubMed] [Google Scholar]
- 40.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, et al. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288(5466):682–5. Epub 2000/04/28. . [DOI] [PubMed] [Google Scholar]
- 41.Le Minh N, Damiola F, Tronche F, Schütz G, Schibler U. Glucocorticoid hormones inhibit food-induced phase-shifting of peripheral circadian oscillators. EMBO J. 2001;20(24):7128–36. 10.1093/emboj/20.24.7128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kaneko M, Kaneko K, Shinsako J, Dallman MF. Adrenal sensitivity to adrenocorticotropin varies diurnally. Endocrinology. 1981;109(1):70–5. 10.1210/endo-109-1-70 . [DOI] [PubMed] [Google Scholar]
- 43.Dallman MF, Engeland WC, Rose JC, Wilkinson CW, Shinsako J, Siedenburg F. Nycthemeral rhythm in adrenal responsiveness to ACTH. Am J Physiol. 1978;235(5):R210–8. . [DOI] [PubMed] [Google Scholar]
- 44.Stocco DM. Steroidogenic acute regulatory (StAR) protein: what's new? Bioessays. 1999;21(9):768–75. . [DOI] [PubMed] [Google Scholar]
- 45.Kotelevtsev Y, Holmes MC, Burchell A, Houston PM, Schmoll D, Jamieson P, et al. 11beta-hydroxysteroid dehydrogenase type 1 knockout mice show attenuated glucocorticoid-inducible responses and resist hyperglycemia on obesity or stress. Proc Natl Acad Sci U S A. 1997;94(26):14924–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eckel-Mahan KL, Patel VR, Mohney RP, Vignola KS, Baldi P, Sassone-Corsi P. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci U S A. 2012;109(14):5541–6. Epub 2012/03/21. 10.1073/pnas.1118726109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stulnig TM, Oppermann U, Steffensen KR, Schuster GU, Gustafsson JA. Liver X receptors downregulate 11beta-hydroxysteroid dehydrogenase type 1 expression and activity. Diabetes. 2002;51(8):2426–33. . [DOI] [PubMed] [Google Scholar]
- 48.la Fleur SE, Kalsbeek A, Wortel J, Fekkes ML, Buijs RM. A daily rhythm in glucose tolerance: a role for the suprachiasmatic nucleus. Diabetes. 2001;50(6):1237–43. . [DOI] [PubMed] [Google Scholar]
- 49.Laffitte BA, Chao LC, Li J, Walczak R, Hummasti S, Joseph SB, et al. Activation of liver X receptor improves glucose tolerance through coordinate regulation of glucose metabolism in liver and adipose tissue. Proc Natl Acad Sci U S A. 2003;100(9):5419–24. 10.1073/pnas.0830671100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beaven SW, Matveyenko A, Wroblewski K, Chao L, Wilpitz D, Hsu TW, et al. Reciprocal regulation of hepatic and adipose lipogenesis by liver X receptors in obesity and insulin resistance. Cell Metab. 2013;18(1):106–17. 10.1016/j.cmet.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Vos P, Saladin R, Auwerx J, Staels B. Induction of ob gene expression by corticosteroids is accompanied by body weight loss and reduced food intake. J Biol Chem. 1995;270(27):15958–61. . [DOI] [PubMed] [Google Scholar]
- 52.Kalsbeek A, Fliers E, Romijn JA, La Fleur SE, Wortel J, Bakker O, et al. The suprachiasmatic nucleus generates the diurnal changes in plasma leptin levels. Endocrinology. 2001;142(6):2677–85. 10.1210/endo.142.6.8197 . [DOI] [PubMed] [Google Scholar]
- 53.Mouzat K, Volat F, Baron S, Alves G, Pommier AJ, Volle DH, et al. Absence of nuclear receptors for oxysterols liver X receptor induces ovarian hyperstimulation syndrome in mice. Endocrinology. 2009;150(7):3369–75. 10.1210/en.2008-1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Volle DH, Mouzat K, Duggavathi R, Siddeek B, Déchelotte P, Sion B, et al. Multiple roles of the nuclear receptors for oxysterols liver X receptor to maintain male fertility. Mol Endocrinol. 2007;21(5):1014–27. 10.1210/me.2006-0277 . [DOI] [PubMed] [Google Scholar]
- 55.Welsh TH, Bambino TH, Hsueh AJ. Mechanism of glucocorticoid-induced suppression of testicular androgen biosynthesis in vitro. Biol Reprod. 1982;27(5):1138–46. . [DOI] [PubMed] [Google Scholar]
- 56.Cumming DC, Quigley ME, Yen SS. Acute suppression of circulating testosterone levels by cortisol in men. J Clin Endocrinol Metab. 1983;57(3):671–3. 10.1210/jcem-57-3-671 . [DOI] [PubMed] [Google Scholar]
- 57.MacAdams MR, White RH, Chipps BE. Reduction of serum testosterone levels during chronic glucocorticoid therapy. Ann Intern Med. 1986;104(5):648–51. . [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y, Leung DY, Nordeen SK, Goleva E. Estrogen inhibits glucocorticoid action via protein phosphatase 5 (PP5)-mediated glucocorticoid receptor dephosphorylation. J Biol Chem. 2009;284(36):24542–52. 10.1074/jbc.M109.021469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krycer JR, Brown AJ. Cross-talk between the androgen receptor and the liver X receptor: implications for cholesterol homeostasis. J Biol Chem. 2011;286(23):20637–47. 10.1074/jbc.M111.227082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nader N, Ng SS, Wang Y, Abel BS, Chrousos GP, Kino T. Liver x receptors regulate the transcriptional activity of the glucocorticoid receptor: implications for the carbohydrate metabolism. PLoS One. 2012;7(3):e26751 10.1371/journal.pone.0026751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boergesen M, Pedersen T, Gross B, van Heeringen SJ, Hagenbeek D, Bindesbøll C, et al. Genome-wide profiling of liver X receptor, retinoid X receptor, and peroxisome proliferator-activated receptor α in mouse liver reveals extensive sharing of binding sites. Mol Cell Biol. 2012;32(4):852–67. 10.1128/MCB.06175-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gbaguidi GF, Agellon LB. The inhibition of the human cholesterol 7alpha-hydroxylase gene (CYP7A1) promoter by fibrates in cultured cells is mediated via the liver x receptor alpha and peroxisome proliferator-activated receptor alpha heterodimer. Nucleic Acids Res. 2004;32(3):1113–21. 10.1093/nar/gkh260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28(1):91–106. 10.1016/j.molcel.2007.07.032 . [DOI] [PubMed] [Google Scholar]
- 64.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–40. 10.1016/j.cell.2008.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reddy TE, Gertz J, Crawford GE, Garabedian MJ, Myers RM. The hypersensitive glucocorticoid response specifically regulates period 1 and expression of circadian genes. Mol Cell Biol. 2012;32(18):3756–67. 10.1128/MCB.00062-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steffensen KR, Neo SY, Stulnig TM, Vega VB, Rahman SS, Schuster GU, et al. Genome-wide expression profiling; a panel of mouse tissues discloses novel biological functions of liver X receptors in adrenals. J Mol Endocrinol. 2004;33(3):609–22. 10.1677/jme.1.01508 . [DOI] [PubMed] [Google Scholar]
- 67.Zucker I, Beery AK. Males still dominate animal studies. Nature. 2010;465(7299):690 10.1038/465690a . [DOI] [PubMed] [Google Scholar]
- 68.Hedrington MS, Davis SN. Sexual Dimorphism in Glucose and Lipid Metabolism during Fasting, Hypoglycemia, and Exercise. Front Endocrinol (Lausanne). 2015;6:61 10.3389/fendo.2015.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mong JA, Baker FC, Mahoney MM, Paul KN, Schwartz MD, Semba K, et al. Sleep, rhythms, and the endocrine brain: influence of sex and gonadal hormones. J Neurosci. 2011;31(45):16107–16. 10.1523/JNEUROSCI.4175-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Quinn M, Ramamoorthy S, Cidlowski JA. Sexually dimorphic actions of glucocorticoids: beyond chromosomes and sex hormones. Ann N Y Acad Sci. 2014;1317:1–6. 10.1111/nyas.12425 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Diurnal mRNA expression of liver GR was compared in males (blue) and females (red) using qRT-PCR in WT and Lxrα-/- mice. For each time point, 3–4 mice were used. Cosine-based non-linear regression was used for curve fitting. The ZT0 time point is double plotted for visualization purposes. Expression data were normalized to the constitutively expressed 36B4 mRNA. The white and black bars represent the light and dark phases, respectively.Statistically significant differences in cosine fitting parameters (p<0.05) between wild type and Lxrα-/- mice or between male and female of the same genotype is indicated in the grey box at the top of the corresponding graph or between graphs (WT: plain arrow, Lxrα-/-: dashed arrow). μ, α and φ indicate a difference in mean level, amplitude and acrophase, respectively.
(TIF)
Diurnal mRNA expression of adrenal Lxrα was compared in males (blue) and females (red) using qRT-PCR in WT mice. For each time point, 3–4 mice were used. Cosine-based non-linear regression was used for curve fitting. The ZT0 time point is double plotted for visualization purposes. Expression data were normalized to the constitutively expressed 36B4 mRNA. The white and black bars represent the light and dark phases, respectively. Statistically significant differences in cosine fitting parameters (p<0.05) between wild type and Lxrα-/- mice or between male and female of the same genotype is indicated in the grey box at the top of the corresponding graph or between graphs (WT: plain arrow, Lxrα-/-: dashed arrow). μ, α and φ indicate a difference in mean level, amplitude and acrophase, respectively.
(TIF)
(A) Mean daily food intake (g) in male and female WT and Lxrα-/- mice. Note there is a significant difference between WT males and WT females (p>0.05). (B-C) Glucose content liberated from glycogen in liver pieces at ZT0 and ZT12. (B) Female WT vs Lxrα-/- mice (n = 4) and (C) male WT vs Lxrα-/- mice (n = 4). There is a significant difference in liver glycogen content between ZT0 and ZT12 (p>0.0001) but no influence of the LXRα mutation.
(TIF)
Liver pieces collected at ZT0 and ZT12, from 4 animals of each group were used in this experiment. For each sample, 30 mg of liver was lysed in KOH 0.5M at 95°C. Na2SO4 6% (25 μl) and 750 μl methanol were then added. Glycogen was precipitated at -80°C in 2 separate aliquots for each sample. After centrifugation, glycogen was either resuspended in 200 μl amyloglucosidase 2 mg/ml (Sigma-Aldrich) or in 200 μl sodium acetate, to assay total glucose and free glucose respectively. Suspensions were incubated for 1-h at 37°C. Free/total glucose content was measured on 5 μL of supernatant in 300 μl of reagent using a glucose hexokinase assay kit (Sigma-Aldrich) according to manufacturer’s protocol. Glucose was expressed in μmol/g wet liver. Glucose coming from glycogen was determined as (total glucose)-(free glucose) in each sample.
(DOCX)
Values are represented as median ± 95% bootstrap confidence intervals. * indicates significant differences between females vs males, † indicates significant differences between Lxrα-/- vs WT mice. Circadian rhythmicity was considered significant for a p-value < 0.05; NSR, not significantly rhythmic.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.