Abstract
The overproduction of inflammatory lipid mediators derived from arachidonic acid contributes to asthma and cardiovascular diseases, among other pathologies. Consequently, the enzyme that initiates the synthesis of pro-inflammatory leukotrienes, 5-lipoxygenase (5-LOX), is a target for drug design. The crystal structure of 5-LOX revealed a fully encapsulated active site, thus the point of substrate entry is not known. We asked whether a structural motif, a “cork” present in 5-LOX but absent in other mammalian lipoxygenases, might be ejected to allow substrate access. Our results indicate that reduction of cork volume facilitates access to the active site. However, if cork entry into the site is obstructed, enzyme activity is significantly compromised. The results support a model in which the “cork” that shields the active site in the absence of substrate serves as the active site portal, but the “corking” amino acid Phe-177 plays a critical role in providing a fully functional active site. Thus the more appropriate metaphor for this structural motif is a “twist-and-pour” cap. Additional mutagenesis data are consistent with a role for His-600, deep in the elongated cavity, in positioning the substrate for catalysis.
The enzyme 5-lipoxygenase (5-LOX) initiates biosynthesis of the eicosanoid lipid mediators known as leukotrienes (LT). Together with 5-Lipoxygenase-Activating Protein (FLAP), localized to the nuclear membrane, 5-LOX transforms arachidonic acid (AA) to LTA4 in a two-step reaction that proceeds via a hydroperoxy intermediate(1–3). Downstream metabolites of LTA4 are potent signaling molecules involved in diverse processes, some of which are mediated by G-protein coupled receptors. For example, LTD4 and LTC4 induce bronchoconstriction upon binding to their cognate G-protein coupled receptors in smooth muscle cells, while LTB4 is a potent leukocyte chemo attractant(4, 5). Due to its role in the production of inflammatory lipid mediators, 5-LOX is a target for the development of therapeutics for conditions as diverse as asthma, cardiovascular disease,(6, 7) pancreatic cancer(8) and traumatic brain injury(9).
Lipoxygenases are non-heme iron dioxygenases that catalyze the peroxidation of polyunsaturated fatty acids(10–12). Humans express six lipoxygenases(13), and each is named according to the site of peroxidation of AA. Thus, 5-LOX produces the intermediate 5-hydroperoxyiecosatetraenoic acid (5-HPETE), whereas, for example, a 15-LOX produces 15-HPETE. Thus, while animal LOXs share a common substrate, they differ in product specificities. The product generated is both regio-specific and stereo-specific, as either a specific R- or S-isomer is produced. Both the 5- and 15-enzymes produce the S-isomers of their respective products. The stereo-chemistries of these two products are consistent with two distinct modes of substrate binding (Fig. 1), corresponding to head-first (carboxyl innermost) or tail-first (hydrocarbon innermost) positioning in the active site(14–16). One of three AA pentadienes must be situated with its methylene carbon accessible to the catalytic machinery for H abstraction, and H abstraction generates a free radical that is accessed by O2 from the opposite face of the AA. The stereo-chemistry of the product is a consequence of this antarafacial relationship between the catalytic iron and the O2 access channel/pocket.
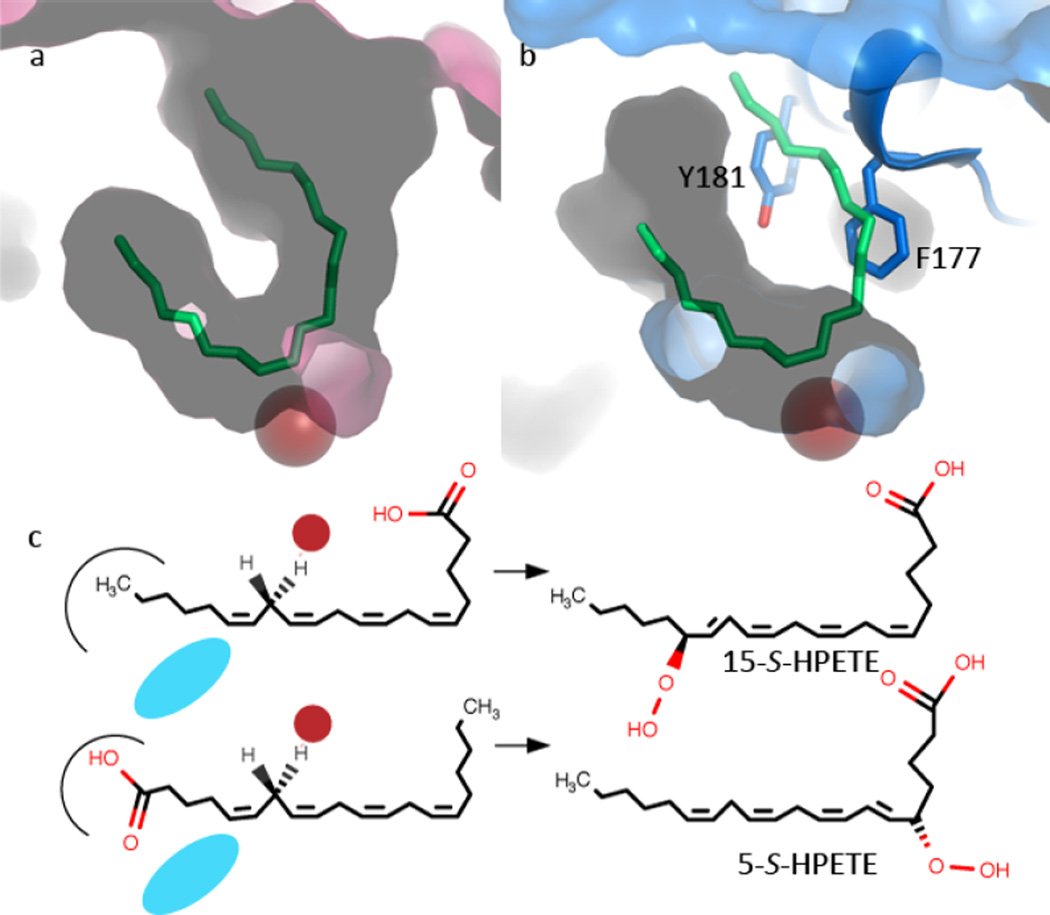
Fig. 1. LOX product stereo-chemistries are consistent with one of two orientations of AA in the active site.
(a) Contour of the active site of 15-LOX-2 (pink, 4NRE) as defined by a competitive inhibitor (green, stick). The red sphere marks the position of the catalytic iron. (b) The equivalent rendering of 5-LOX, including a cartoon and stick rendering of the St-5-LOX (3O8Y) “cork.” (c) For cavities of equal depth, inverse positioning of the substrate generates the 15-S-“tail-first” product or the 5-S “head-first” product. The catalytic iron is represented as a red sphere; the O2 as a blue ellipse.
Several LOX structures are available and combine to provide details of a consensus active site(17), but the 5-LOX isoform differs significantly from 15-LOX-1(18, 19), 15-LOX-2(20), 12-LOX(21) and 8R-LOX(22, 23) in three important ways: (1) The HPETE produced by 5-LOX is an intermediate; a second reaction in the same 5-LOX active site converts the intermediate to LTA4. (However, in vitro 15-LOX-1 can also utilize 15-HPETE as a substrate and generate a product analogous to LTA4(24)). (2) Only 5-LOX requires accessory proteins for maximal activity: an arachidonic acid-binding protein (FLAP) embedded in the nuclear membrane(25, 26) and a cytosolic coactosin-like protein(27, 28). (3) 5-LOX displays a distinctive structural variation on the canonical LOX fold(29). In general the relative placements of the roughly 20 α-helices that make up the catalytic domain are conserved among LOX isoforms. 5-LOX, as well as an 11R-LOX from Baltic coral(30), displays alternate placement of helix α2. This helix rims the U-shaped active site in other isoforms, but in 5-LOX (and 11R-LOX) the helix is “broken” to seal it off with insertion of aromatic amino acids into an otherwise fully accessible U-shaped active site. Thus the structure of a Stable-5-Lipoxygenase (St-5-LOX, so-called because of the presence of stabilizing mutations introduced distal to the active site) revealed a fully encapsulated catalytic machinery and neither entry into nor positioning of substrate in the active site are obvious in the reported structure(29).
We used site-directed mutagenesis, coupled with substrate and product analyses, to sample possible entry portals that may permit the substrate to gain access to the sheltered catalytic machinery and position itself in the active site. Our results support a model in which active site access appears to require repositioning of two aromatic amino acids (F177, Y181) that penetrate the active site cavity. Although paring down the “cork” volume appears to facilitate entry into the active site, obstruction of “cork” entry into the active site leads to loss of enzyme specificity, stability, and activity. In essence, our results suggest the “cork” metaphor, which conveys the idea that ejection of the cork is required for active site entry or exit, needs to be refined. The corking amino acid Phe-177, which is conserved in 5-LOX homologues with open active sites, appears to play a key role in product specificity in 5-LOX. However, Tyr-181 is dispensable for activity, but serves as a gatekeeper to the active site. Mutations designed to promote the open-access cavity conformations observed in homologous enzymes compromise both the reaction rate and specificity.
Experimental Procedures
Plasmid preparation
The St-5-LOX construct in a pET-14b (Novagen) plasmid was used as the starting point for mutation(29). The various mutations were introduced by site-directed mutagenesis. Primers were designed to contain a given mutation and then used to amplify the 5-LOX plasmid using whole plasmid PCR. The resultant products were gel purified and re-circularized. The primers also contained unique restriction sites as silent mutations to verify mutation. The presence of mutations was confirmed by DNA sequencing.
Protein expression and Purification
E. coli Rosetta2 (Novagen) cells were transformed with the mutated plasmids and grown in small scale cultures to check for protein expression and solubility via auto-induction(31). For large scale protein purification the cells were grown in Terrific Broth (Alpha Bioscience, MD) with 25 µg/ml chloramphenicol, 100 µg/ml ampicilin and 5mM MgSO4. The cultures were shaken at 250 rpm for 4 hours at 37 °C and then the temperature was reduced to 25 °C for an additional 25 hours to induce leaky expression. The cells were pelleted by centrifugation and frozen at −80 °C. For protein purification, the cells were suspended in 3ml/gm Bugbuster (Novagen) with 1 µM pepstatin, 1 µM leupeptin, 100 µM PMSF and 2 KU/gm DNAse I (all from Sigma). The mixture was stirred on ice for 30 min and lysed by passing through a French pressure cell. The lysate was centrifuged at 40000 g for 45 min. The resulting supernatant was loaded onto a HisTrap 5ml cobalt Sepaharose (GE Healthcare) column equilibrated in 50 mM Tris-HCl (pH 8.0), 500 mM NaCl, and 20 mM imidazole on an AKTA FPLC system (GE Healthcare). Subsequently, the column was washed with ten column volumes of the above buffer and then the protein was eluted by applying a linear gradient of 50 mM Tris-HCl (pH 8.0), 500 mM NaCl and 200 mM imidazole. The peak fractions were concentrated in an Amicon Ultra 30K (Millipore) filter to a final volume of 500 µl. Buffer exchange was performed twice with 15 ml of 20 mM Tris-HCl, pH8.0, 150 mM NaCl and 5 mM TCEP-HCl. Protein was concentrated to a final concentration of 1mg/ml to 5 mg/ml as detected by a NanoDrop spectrometer (Thermo Scientific). The protein was flash frozen in liquid N2 and stored at −80 °C until usage.
Kinetic Assays
The kinetic parameters for St-5-LOX and the different mutants were determined by monitoring the increase in absorbance at 238 nm in an Agilent 8453 Diode Array Spectrophotometer (Agilent Technologies, Santa Clara, CA, USA). The assays were performed with 500 nM enzyme in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl and 5 mM TCEP. Substrate concentration was varied from 5 to 50 µM. Substrate concentrations (5, 10, 20, 30, 40, 50 µM) were monitored in triplicate for each sample. Km and Vmax values were determined by non-linear regression analysis of a plot of velocity versus substrate concentration to equation 1. Vmax is the maximal velocity, S is the substrate concentration, Km is the Michaelis constant.
| Equation 1 |
Kinetic constants are reported in Tables 1 and 2 as kcat to facilitate the comparison of rates among samples which may differ by enzyme concentration.
Table 1.
Stability, kinetic parameters, and product analysis of mutants constructed to explore substrate access
| Mutant | St-5- LOX |
Y181A | F177A | F177A Y181A |
A603L | A603L Y181A |
G174N | D170S G174N |
W147L |
|---|---|---|---|---|---|---|---|---|---|
| TD (°C) | 53.1±0.6 | 52.9±0.1 | 51.7±0.1 | 54.8±0.5 | 49.1±0.3 | 55.8±0.6 | 54.5±0.2 | 52.5±0.4 | 49.9±0.1 |
| Kinetic parameters | |||||||||
| Km(µM) | 14±1.6 | 7.6±3.4 | 9.8±2.1 | 11±2 | 9.2±3.2 | 5.0±2.8 | 12±5.3 | 14±2.5 | 22±7 |
| kcat (s−1) | 0.8±0.03 | 1.2±0.2 | 0.7±0.04 | 0.7±0.03 | 0.05±0.01 | 0.7±0.1 | 0.1±0.002 | 0.20±0.01 | 0.01±0.01 |
| kcat/Km(µM−1s−1) | 0.06±0.007 | 0.16±0.07 | 0.07±0.02 | 0.06±0.006 | 0.005±0.002 | 0.14±0.08 | 0.008±0.003 | 0.002±0.002 | |
| Products (ng) | |||||||||
| 5-HETE | 840±41 | 2300±61 | 1200±61 | 630±82 | 27±20 | 1600±6 | 430±102 | nd | 350±12 |
| 11-HETE | nd | nd | nd | nd | nd | 116±4 | nd | 200±61 | nd |
| 8/12-HETE | nd | 55±8 | nd | nd | 45±8 | 390±12 | nd | nd | 120±10 |
| 15-HETE | nd | nd | nd | nd | nd | 135±11 | nd | 190±61 | 35±2 |
| “other” | 100±7 | 67±5 | nd | nd | nd | 680±80 | 103±20 | 130±30 | 42±20 |
| Products (ng) from analogs | |||||||||
| DGLA | nd | nd | nd | nd | 270±130 | 750±30 | 170±50 | 410±160 | -------- |
| AEA | nd | nd | 470±300 | 300±40 | 300±10 | 700±50 | 300±90 | 440±30 | -------- |
| NAGly | nd | 190±60 | 390±100 | 160±50 | 340±30 | 1500±100 | 270±100 | nd | -------- |
nd = not detected; ----- not determined
Table 2.
Stability, kinetic parameters and product analysis of His-600 mutants
| Mutant | St-5-LOX | H600V | H600V | F177A:Y181A:H600V |
|---|---|---|---|---|
| TD (°C) | 53.1 (0.1) | 56.4 (0.2) | 56.7(0.2) | 54.8 (0.1) |
| Kinetic parameters | ||||
| Km (µM) | 14±1.6 | nd | 27±0.7 | 5.4±0.5 |
| Kcat(s−1) | 0.8±0.03 | nd | <0.01 | 0.3±0.01 |
| kcat/Km(µM−1s−1) | 0.06±0.007 | nd | <0.001 | 0.06±0.005 |
| Products (ng) | ||||
| 5-HETE | 840±40 | nd | nd | nd |
| 8/12-HETE | nd | nd | nd | 700±5 |
| 15 HETE | nd | nd | nd | nd |
| “Other” | 100±7 | nd | nd | nd |
nd=not detected
Product Assays
The St-5-LOX and mutant enzymes (5µM) were incubated at 22 °C with 100 µM substrate or substrate analog (dihomo-γ-linolenic acid (DGLA), N-arachidonyl glycine (NaGly), anandamide (AEA), docosahexaenoic acid (DHA) all Cayman Chemical Company) for 10 min in 20 mM Tris-HCl. (pH 7.5), 150 mM NaCl, and 5 mM TCEP-HCl in a final volume of 200 µl for 5 min. Prostaglandin B1 (PGB1) (5 µl of 50 ng/µl, Cayman) was added after completion of the incubation as an internal standard. The products of the reaction were extracted by addition of 2 µl 1N HCl and 20 µl 1M K2HPO4 followed by 400 ul dichloromethane. The mixture was vigorously shaken for 10 sec and centrifuged briefly to separate the phases. The organic phase was collected and washed twice with 400 µl water, dried under a stream of N2 and stored at −20 °C. The products were resuspended in 30 µl of 50% methanol and reduced by addition of triphenylphosphine and loaded onto a Supelco Discovery C18 Column (5µm particle size, 25 × 0.46 cm, Sigma-Aldrich) for identification and quantitation using reversed phase HPLC with a diode array detector (Dionex). Peak areas were integrated and quantitated relative to that of the PGB1 internal standard. The mobile phase was acetonitrile/water/formic acid (60:40:0.01), and the flow rate 1ml/min. Four UV channels (220, 235, 270 and 280 nm) were monitored.
Thermal shift assays
Thermal induced unfolding assays were performed to determine the impact of the mutations on protein stability as described by others(32, 33). Briefly, samples were prepared with 2 µM enzyme in 20 mM Tris-HCl (pH 7.5), 150 mM NaCl and 5 mM TCEP and 1× SYPRO Orange stain (Invitrogen) in a final volume of 150 µL. Triplicate volumes of 40 µL each for every individual enzyme sample were aliquoted into a 96 well reaction plate. The plate was subjected to a temperature gradient of 5–94°C in 1° increments in a 7500 Fast Real-Time PCR system (Applied Biosystems). The thermal denaturation of the proteins was monitored by fluorescence of SYPRO Orange that occurs with protein binding. The resulting data were exported to SigmaPlot (v. 9) and averaged for each triplicate. The average sigmoidal part of each curve was then fit into a four parameter sigmoidal equation. The midpoints of denaturation (TD) values were measured in triplicate for each mutant.
Results
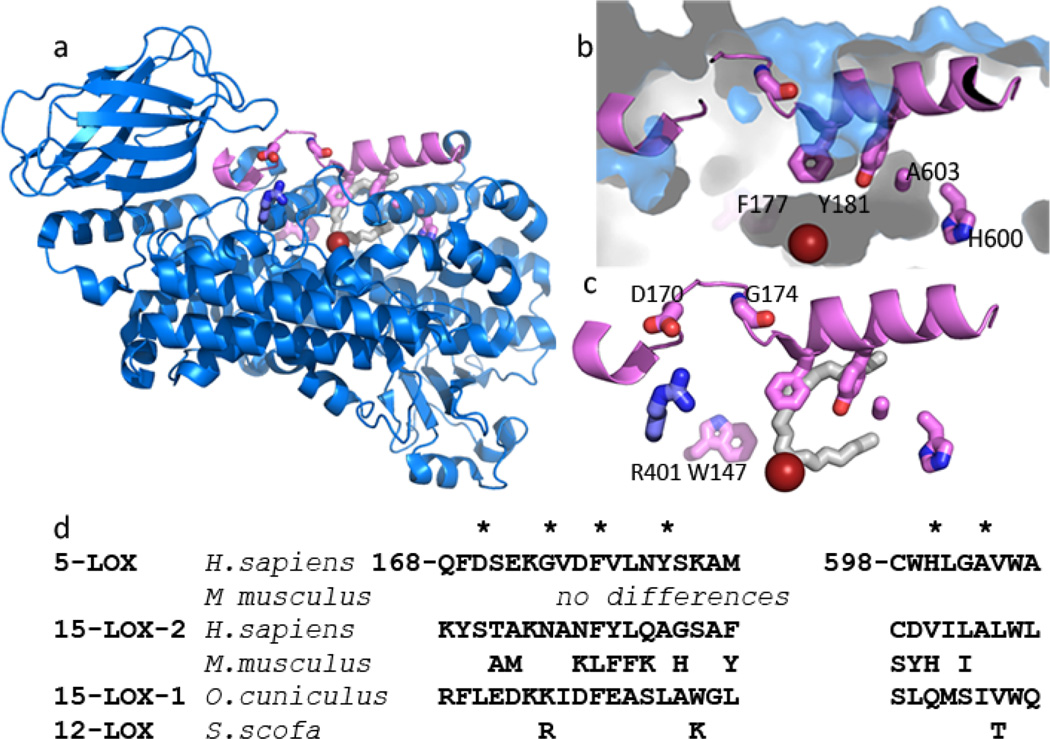
We sampled, via site-directed mutagenesis, several amino acids that line the enclosed active site cavity of 5-LOX (Fig 2) to determine what role, if any, they might play in allowing AA to gain access to the catalytic iron and position itself in the active site such that 5-HPETE and LTA4 can be generated. Mutations were made to Phe-177, Tyr-181, Ala-603, and Trp-147. The first three cluster to truncate one arm of what has been described as a U-shaped cavity in homologous enzymes with accessible catalytic sites(17) (15-LOX types-1(19) and -2(20), 12-LOX(21) and 8R-LOX(22, 23). However, the St-5-LOX structure revealed that Phe-177 and Tyr-181 form what has been described as an active site “cork” and the small size of the Ala at 603 appears to allow insertion of the “cork” into the active site. In contrast, Trp-147 appears to block access to the enclosed active site from an alternate approach(29).
Fig. 2. St-5-lipoxygenase has an encapsulated active site.
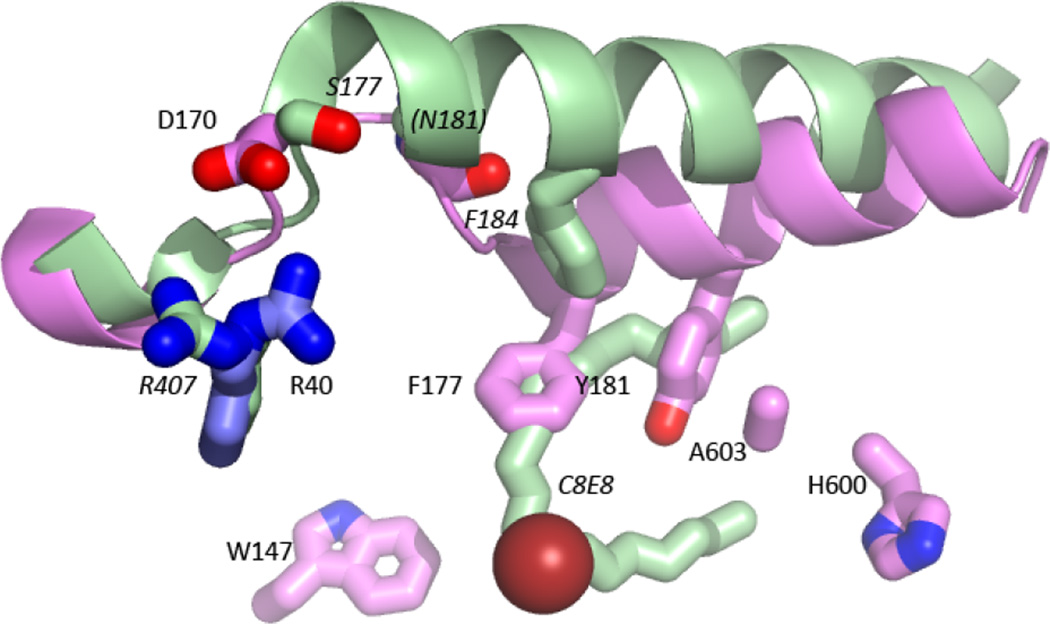
(a) Cartoon rendering of St-5-lipoxygenase (3O8Y). The region corresponding to helix α2 is in pink. The iron is shown as an orange sphere. The amino acids mutated in this study are in stick rendering (C pink, O red, N blue). The white stick rendering marks the position of the substrate mimic (C8E4) in the homologue 15-LOX-2. (b) Close-up of the internal cavity with the mutated amino acids in stick rendering. The proximity of A603 and Y181 is visible in this view. (c) D170 and G174 are 5-LOX specific amino acids that appear to confer its unique conformation of α2. D170 is positioned to participate in a salt-link with the conserved R401. The inhibitor (C8E4) in the 15-LOX-2 structure is superimposed in white stick rendering to indicate where the substrate mimic is bound in that enzyme. (d) Sequences of the mutated regions from related lipoxygenases. The positions mutated are marked with *.
Phe-177 is conserved in 5-LOX homologues with open active sites but in those enzymes it does not obstruct access to the site. Rather, it contributes to the contours of the tubular, U-shaped cavity. Therefore, in a parallel approach to prevent “corking” of the St-5-LOX active site, mutations were constructed to disfavor placement of helix α2 such that Phe-177 can plug the 5-LOX active site, a consequence of the anomalous placement of this element of secondary structure in 5-LOX. 5-LOX specific amino acids Asp-170 and Gly-174 appear to allow the unique placement of the helix and were substituted with their counterparts in 15-LOX-2, the closest sequence homologue of 5-LOX with an open active site.
Lastly, H600, positioned innermost in the tubular, U-shaped cavity that accommodates the methyl end of arachidonic acid in the 8R-LOX:AA complex structure(17, 23), was mutated to a hydrophobic amino acid to test whether it is important for “head first” binding.
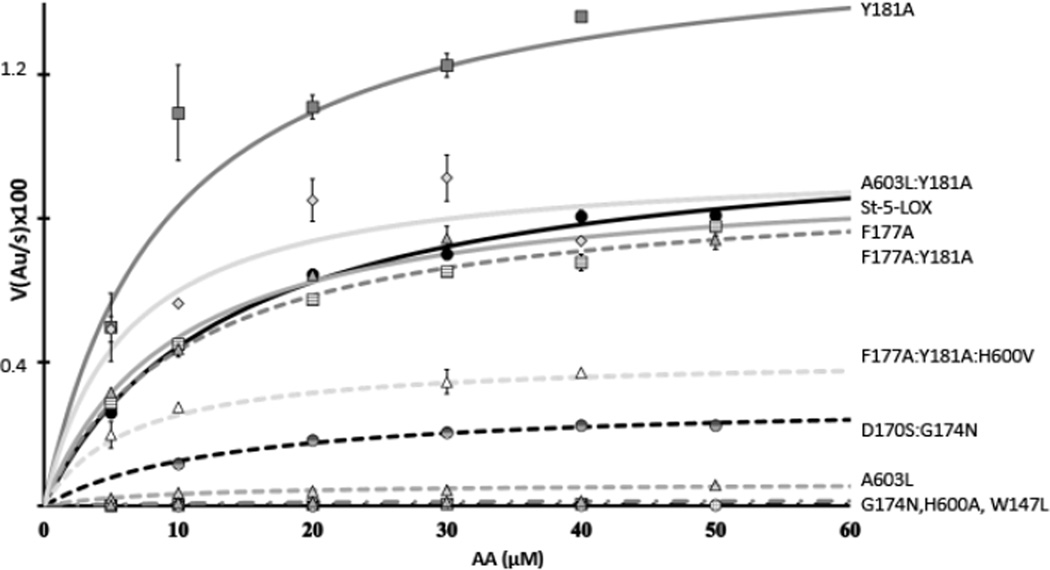
Each of the mutants was evaluated in terms of kinetic parameters and product profiles (Tables 1 and 2, Fig 3). While the in vivo product of the 5-LOX reaction is LTA4, the capacity of 5-LOX to convert the 5-HPETE intermediate to the leukotriene is significantly compromised in vitro, in part due to the absence of its helper protein FLAP(25, 34). Thus, kinetic parameters derived from initial reaction rates are reported for HPETE formation, monitored at 235 nm. (The hydroperoxy products are rapidly reduced to the corresponding hydroxyeicosatetraenoic acids (HETEs). For simplification, the products will henceforth be referred to as HETEs). Although wild-type 5-LOX activity is stimulated by Ca2+ and phosphotidylcholine(35), St-5-LOX lacks the Ca2+ binding sites and membrane binding loops. Kinetic analysis of the basal activity of the variants reveals the direct effects of the mutations on enzyme turnover.
Fig. 3. Kinetics traces for St-5-LOX and mutants.
Plots of the increase at 235 nm vs. substrate concentration for the 5-LOX variants studied. Data were fit to the Michaelis-Menten equation in Sigma Plot. St-5-LOX ( ), Y181A (
), Y181A ( ), F177A (
), F177A ( ), A603L:Y181A (
), A603L:Y181A ( ), F177A:Y181A (
), F177A:Y181A ( ), F177A:Y181A:H600V (
), F177A:Y181A:H600V ( ), D170S:G174N (
), D170S:G174N ( ), A603L (
), A603L ( ). Plots for H600A (
). Plots for H600A ( ), G174N (
), G174N ( ) and W147L (
) and W147L ( ) cluster at the X-axis and cannot be readily distinguished.
) cluster at the X-axis and cannot be readily distinguished.
LTA4 as well as di-HETEs that may form from 5-LOX recognition of HETEs as substrates have absorption maxima at 270–280 nm and UV spectra that clearly differentiate them from mono-HETEs. The time frame of the kinetic measurements minimizes signal interference from di-HETE formation, but these off-pathway products do accumulate in the course of the incubations. LTA4 breakdown products and di-HETEs, both formed by 5-LOX activity with a product HETE, are grouped together as “other” in the product analyses (Table 1, 2).
How does substrate enter the active site?
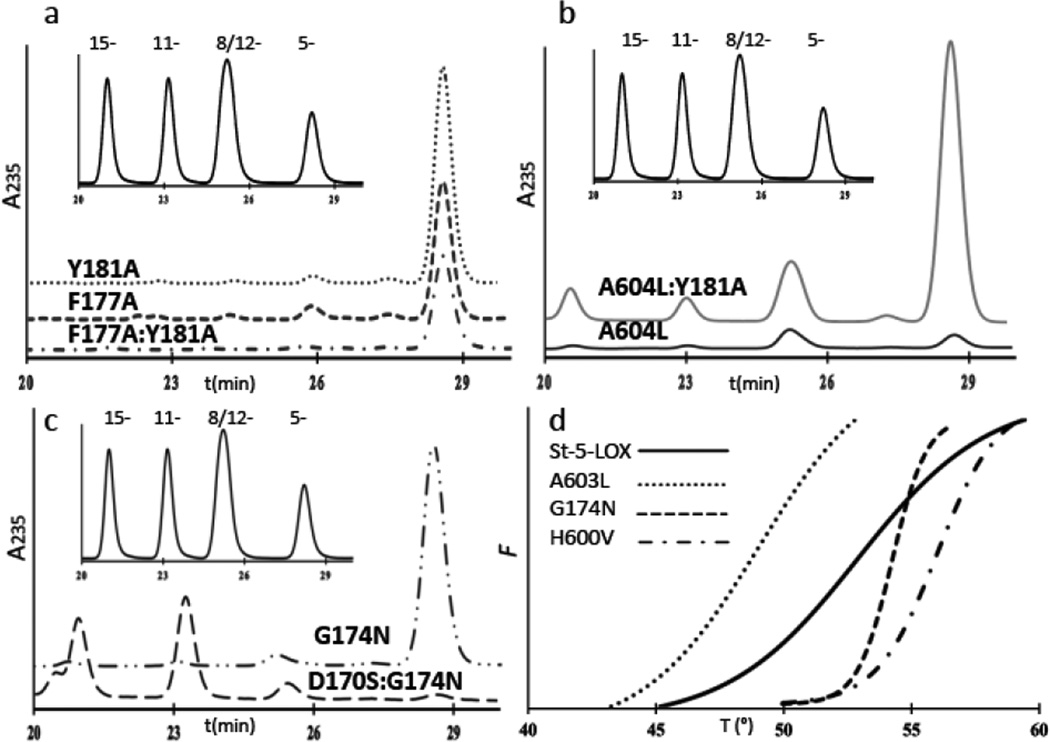
Mutations to the “cork” cluster clearly impacted enzyme activity. Trimming the cork by removal of either or both of the aromatic side chains (Y181A or F177A) increased 5-HETE yields from enzyme-substrate incubations ~3 and ~1.5-fold, respectively. However, the increased yield came at a cost to product specificity, as a small fraction of HETE other than 5-HETE was produced by Y181A. Sample HPLC traces of incubations mixtures are shown in Fig 4.
Fig. 4. Sample chromatograms from products analyses.
(a) HETE product profiles (absorbance at 235nm) from incubations of Y181A, F177A and F1771A:Y181A with substrate. The inset is the elution profile (A235) of HETE standards 15-HETE, 11-HETE, 12- and 8-HETEs, and 5-HETE. (b) Product profiles for A603L and A603L:Y181A. While the double mutant is active, its product specificity is compromised. The inset is the elution profile of HETE standards 15-HETE, 11-HETE, 12- and 8-HETEs, and 5-HETE. (c) HETE product profiles for G174N and the double mutant D170S:G174N. The inset is the elution profile of HETE standards 15-HETE, 11-HETE, 12- and 8-HETEs, and 5-HETE. (d) Representative sample thermal induced unfolding curves for St-5-LOX (—), and its destabilized (A603L, ●●●) and stabilized (G174 – – –, H600V–●–●–●) variants. The fluorescence of SYPRO orange (Y axis) increases as thermal denaturation exposes hydrophobic surfaces that bind fluorophore. Each curve represents the average of triplicate measurements.
The presence of F177 appears to be necessary for utilization of 5-HETE as a substrate, as none of the mutants in which F177 was replaced with Ala produced compounds that might be leukotrienes or di-HETEs, despite the fact that the F177A mutant generates 50% more 5-HETE than its progenitor. Y181A and F177A mutants were able to utilize AA analogs with bulky groups at the carboxyl end, arachidonyl-ethanolamine (AEA) and N-arachidonyl-glycine (NAGly), to some extent, but not dihomo-γ-linolenic acid (DGLA), which lacks the pentadiene centered at C7 for H abstraction by 5-LOX.
Neither the Y181A nor the F177A mutation appears to have an impact on enzyme stability, since thermal induced unfolding measurements revealed no significant shifts in TD (Table 1). Kinetic analyses indicate that, while the Km remains essentially unchanged, the turnover rate for Y181A is roughly 50% enhanced and the catalytic efficiency is improved ~3-fold with respect to St-5-LOX. Thus “removal” of the bulky Tyr is consistent with an enhanced entry and/or egress from the active site. (For comparison, the values of kcat for lipoxygenases with open-access sites can vary from ~1 s−1 to ~200 s−1 (15-LOX-2(20) and 8-R-LOX(23)). The exceptional activity of the 8R-enzyme is likely a consequence of an O2 access channel that is open to solvent.) The F177A:Y181A double mutant resembles the F177A single mutant with respect to reaction rate as well as substrate and product specificities. Notably, though, the double mutant has greater thermal stability than either of the single mutant forms.
We tested whether impeding “cork” insertion by mutation of A603 would have a similar impact to paring down the “cork,” as this would result in constitutive ejection of these resides from the cavity. A large amino acid side chain at this position, as is found in homologous LOXs with open cavities, should preclude cork insertion because of its proximity to Tyr-181. In contrast to the mutations which minimize cork volume, the A603L mutation resulted in significant destabilization of the protein (Table 1) with a 4° C drop in the TD. While the Km for AA is unchanged relative to St-5-LOX, the enzyme turnover is less than 10% of the parent enzyme. Moreover, the enzyme has significantly impaired substrate specificity and is able to oxygenate arachidonic acid analogs that are not substrates for St-5-LOX. Remarkably, we can recover a St-5-LOX-like kcat (0.7 vs. 0.8 s−1) by making a compensatory mutation: The substitution of Y181 with Ala to whittle down the cork and allow insertion of Phe-177 despite the added bulk at position 603. This double mutant (Y181A:A603L) has enhanced thermal stability. Consistent with the fact that the A603L mutants generate isomers other than 5-HETE, which requires attack at either C10 or C13, these variants can utilize DGLA as a substrate. In addition, A603L:Y181A is able to generate products from both AEA and NAGly, more so than any of the variants assayed.
In yet another approach to obstructing “cork” closure, we made mutations that might allow placement of α2 in the “standard” LOX position such that the “corking” amino acid side chains cannot reach the cavity to plug it(17). This approach avoids the substitution of a binding site amino acid (A603), and could provide an indirect strategy to engineer an open access cavity. A 5-LOX specific Asp (170) and Gly (174) appear to disfavor the formation of the longer α2 in active-site-open lipoxygenases by promoting an extended loop conformation at the amino-terminus of α2 (Fig 5). The G174N mutation showed an increase in thermal stability and no loss of product specificity, but like the A603L mutation, its enzymatic activity is significantly compromised (the kcat is reduced to <15% of that of St-5-LOX). The double mutant D170S:G174N had slightly more activity (kcat reduced to 25% of that of St-5-LOX) but was less stable. More importantly, this variant was no longer a 5-LOX, as it produced only 11- and 15-HETEs. Both D170 and G174, amino acids outside the cavity that affect positioning of the elements of secondary structure that provide the “cork,” are important for activity. Moreover, these mutants were able to process substrates that carry bulky head groups, suggestive of a larger volume of the active site. Consistent with the altered regio-specificity of D170S:G174N and the relatively high yield of Di-HETEs with G174N, these variants were able to transform DGLA, which only has pentadienes at C10 and C13.
Fig. 5. Helix α2 is “unraveled” in 5-LOX.
Helix α2 of 15-LOX-2 (light green), superimposed on 5-LOX (pink), displays the typical orientation for lipoxygenases with open-access active sites. Note that the counterpart of 5-LOX F177 (F184) in 15-LOX-2 does not occupy the same position as F177. The competitive inhibitor of 15-LOX-2 (C8E4) is shown in green stick. The 15-LOX-2 counterparts of 5-LOX D170, G174 and R401 are also included. N181 is on the backside of the helix and not visible from this perspective. 15-LOX-2 residues are labeled in italics.
Since an open “corked” portal would have a structure similar to what is observed in other LOX structure, it seemed the more likely entrance to the active site. However, we also tested a possible access portal obstructed by Trp-147. The substitution of Trp with the smaller Leu led to a significant reduction in enzyme stability. In addition, the enzyme had significantly reduced activity and the product specificity was compromised (Table 1). These characteristics are inconsistent with Trp-147 serving as a functional portal. Given the diminished activity and stability of the variant, we did not probe whether alternate substrates could be oxygenated by this mutant.
How is the substrate positioned in the active site?
A second question we addressed is whether AA is positioned with its carboxyl innermost, as has been inferred from the S-stereochemistry of the product. Extrapolating from the structure of 8R-LOX in complex with AA(23), His-600 is deep in the active site where the tubular cavity dead-ends (Fig 1, 2). We tested whether substitution of this charged/polar amino acid with a hydrophobic amino acid might impact 5-LOX activity with two mutations: H600A and H600V. Multiple substitutions of non-polar amino acids are important at this position, because the depth of the active site cavity determines whether the pentadiene to be attacked can be positioned productively at the catalytic iron. Both substitutions resulted in proteins which were expressed and purified at levels comparable to that of the parent enzyme, and had TDs that exceed that of the parent protein (Table 2). H600A displayed less than 10% of St-5-LOX activity and had reduced affinity for substrate. The low level of activity of the mutant precluded product profile analysis by HPLC. The H600V mutation resulted in lack of detectable catalytic activity. However, we were able to rule out a non-functional catalytic center with a “rescue” mutation that combined H600V with F177A:Y181A. This triple mutant produced HETEs, but not 5- or 15-HETE. Instead products consistent with attack at C10 were observed. Thus H600 appears to be essential to proper positioning of the substrate.
Discussion
Lipoxygenases are characterized by two domains(17, 18, 36): a β-barrel domain which is implicated in membrane binding and a much larger α-helical domain that harbors the catalytic iron. Invariant histidines, as well as the main chain carboxyl of the terminal invariant Ile position the catalytic iron. 5-LOX displays a distinct variation of the typical LOX fold. While the structures of 15-LOX-2, 15-LOX-1, 8R-LOX, and 12-LOX have revealed a solvent accessible active center at the base of a tubular, U-shaped cavity, access is firmly “corked” in 5-LOX by the insertion of the side chains of Phe-177 and Tyr-181. Thus the path of substrate entry is not apparent in the crystal structure. We asked whether 5-LOX may undergo a conformational change to “uncork” the active site by site-directed mutations that (1) reduce cork volume and/or (2) hinder “cork” insertion. Wild-type 5-LOX undergoes rapid auto-(37) and suicide-inactivation(38–40), thus a “stabilized” form of 5-LOX, which harbors two Cys mutations (C240A and C561A) that may make it less susceptible to product inactivation, was utilized. Despite this precaution, the caveat remains that some differences in product quantitation may be related to auto-inactivation.
Active site access
Our data are most consistent with a model in which the cork must reposition, but it is not fully ejected from the binding site. The cork is comprised of two aromatic amino acids (F177 and Y181), and while our data indicate that replacement of either or both with Ala results in catalytically active mutants, only Y181A has an increased catalytic efficiency. This observation suggests that Y181 is dispensable for the catalytic reaction, and in the native enzyme it may limit access to the active site. The increase in activity, reflected by a 50% increase in kcat, and 3-fold increase in kcat/Km, is consistent with facilitated entry/egress to/from the active site.
An increase in the rate of HPETE production by a 5-LOX mutant is not without precedent. Rakonjac et al. recently reported a similar observation for an R101D mutation. This substitution disrupts a salt-link between the tightly associated membrane-binding and catalytic domains mediated by Arg-101 and Asp-166(35). The proximity of the corking amino acids to this domain interface is consistent with their observation, in the sense that disruption of the tightly packed interface would loosen the restraining van der Waals contacts that favor the “corked” conformation of Phe-177 and Tyr-181.
While our data support repositioning of the “cork” for substrate entry, it is clear that the entire cork cannot be displaced, as a mutation that blocks cork insertion displays less than 10% of the activity observed for the parent enzyme. The mutation A603L, designed to interfere with cork insertion due to the proximity of Tyr-181 to Ala-603, leads to a nearly total loss of activity, indicating that a corking amino acid side chain is necessary during the catalytic cycle. A “rescue” mutation, in which substitution of Tyr-181 of the cork with Ala relieves the steric clash with the added bulk at position 603, renders a highly active enzyme. Thus, again we see that Tyr-181 is dispensable for catalytic activity. In contrast, the corking amino acid Phe-177 appears to be required for 5-LOX to process 5-HPETE, which it must do to complete the transformation of arachidonic acid to LTA4. No metabolites of 5-HPETE are observed in any of the mutant forms that lack Phe-177. In addition, a second approach to preventing the entry of the “corking” amino acids into the active site was taken. In the structures of LOX’s with open catalytic sites the positioning of α2 precludes “corking” (Fig 5). There are two distinct features in 5-LOX that appear to “unravel” 5-LOX helix α2 so that the “cork” can insert: a salt link between Asp-170 to a conserved Arg (R401) and a Gly (G174). Note that the counterpart for F177 in 15-LOX-2 (Phe-184) is not positioned for corking. We found that substitution of these amino acids with their counterparts in 15-LOX-2 significantly compromised enzyme activity and/or specificity. These observations are consistent with what we observed for the A603L mutations: substitutions that impact the placement of α2 (and by extension cork placement) have detrimental effects on enzyme activity.
Previous mutagenesis studies aimed at reducing the volume of the active site of mouse and human 5-LOX have explored single site mutants at Ala-603(41–43). The investigators observed that substitution with Ile was tolerated at this position, however for the human enzyme a reduction of product isolated from cell lysates (relative to that isolated for the bacterially expressed wild-type enzyme) was observed. In addition, loss of regio-specificity was reported. In contrast, substitution with Phe at this position led to a total loss of enzyme activity. These results can also be interpreted in the structural context we propose: that addition of bulk at position 603 likely interferes with the insertion of Phe-177 and Tyr-181 in the active site and can lead to significantly compromised enzyme activity.
Head first vs. tail first entry
Further experiments described herein suggest that His-600 is essential for positioning of the substrate, as mutations at this position abolish enzyme activity. This amino acid is deepest is an elongated cavity that must accommodate the 20 carbon substrate. A wealth of biochemical data are consistent with AA entering “head” (carboxyl) first into the 5-LOX active site, while in the homologous 15-LOX-2 (42% sequence identity), the “tail” (hydrocarbon) enters first. This “head-first” vs “tail-first” relationship is a crucial determinant of product stereo-chemistry as it places the carbon for peroxidation proximal to the O2 channel(16, 17). One can see from Fig. 1 that the 5-S and 15-S products have their peroxide groups on opposite sides of the product, consistent with the inverse relationship of arachidonic positioning in the active site. We observed that mutation of His-600 to either a Val or an Ala resulted in an enzyme that no longer generates 5-HPETE, presumably because the absence of a polar/charged amino acid deep in the cavity results in an enzyme that no longer favors “head-first” substrate entry.
The “head-first” entry LOX mouse 8S-LOX also has a histidine deepest in the cavity. However, in mutagenesis experiments with this enzyme, the substitution of the His with a hydrophobic amino acid at the deep end of the pocket resulted in the generation of product, but a product consistent with tail-first, rather than head-first, entry(44). So why is a “tail-first” product, which should be a 15-HETE according to Fig 1, not observed for the St-5-LOX H600A or H600V mutations? Cavity depth is an essential aspect of LOX structure as it allows the substrate to slide into the active site and position the pentadiene for attack. Cavity depth is clearly impacted when His is replaced by Ala or a Val, and one possible reason that no product is observed is that these mutations disallow productive alignment of a pentadiene at the catalytic center. Thus we combined the H600V mutant with mutations that permit “slack” in the binding site by opening the “corked” end. F177A:Y181A “rescues” the H600V mutation, recovering ~50% of the parent enzyme activity. However, no 5-HETE is produced; only products consistent with attack at carbon 10 are isolated from the incubations. Thus H600 does not play a catalytic role, but it is required for productive alignment of the substrate.
Activity with substrate analogs
The ability of a LOX to utilize an AA analog which carries a bulky group at the carboxyl has been used as a proxy for “tail-first” entry, as “head-first” entry would require that the additional bulk be accommodated deep in the active site. With tail first entry the extra cargo need not enter the active site(45). 5-LOX does not recognize AEA or NAGly as substrates, consistent with head-first entry. However, several of our mutants are able to recognize these substrates. How might that be possible? Note in Fig 5, 5-LOX A603 and Y181 are positioned between the tops of the U-shaped substrate mimic of 15-LOX-2 and mutations to Y181 could free up space and tolerate extra bulk at either end of a substrate analog. Thus, the ability to recognize AEA or NAGly as substrates is not indicative of the methyl end of the substrate situated innermost, but simply a consequence of a roomier cavity that allows “head-first” entry with added bulk. Similarly, mutations that do not allow the “corking” amino acids to enter the active site would be expected to utilize substrate analogs that carry extra cargo. Thus the A603L mutation, which prevents Y181 from positioning in the cavity, as well as the G174N and D170S:G174N mutations, also designed to promote an open conformation of the active site, are able to utilize the analogs. In contrast, the analog DGLA is simply AA without the C5-C6 double bond, making H abstraction at C7 by 5-LOX impossible. Variants that produce HETES other than the 5-isomer in appreciable amounts were able to utilize DGLA, as expected. The one exception to this trend was G174N. However, this variant produces high levels of di-HETES (relative to 5-HETE), and is clearly able to oxygenate multiple pentadienes.
Why is the active site of 5-LOX “corked”?
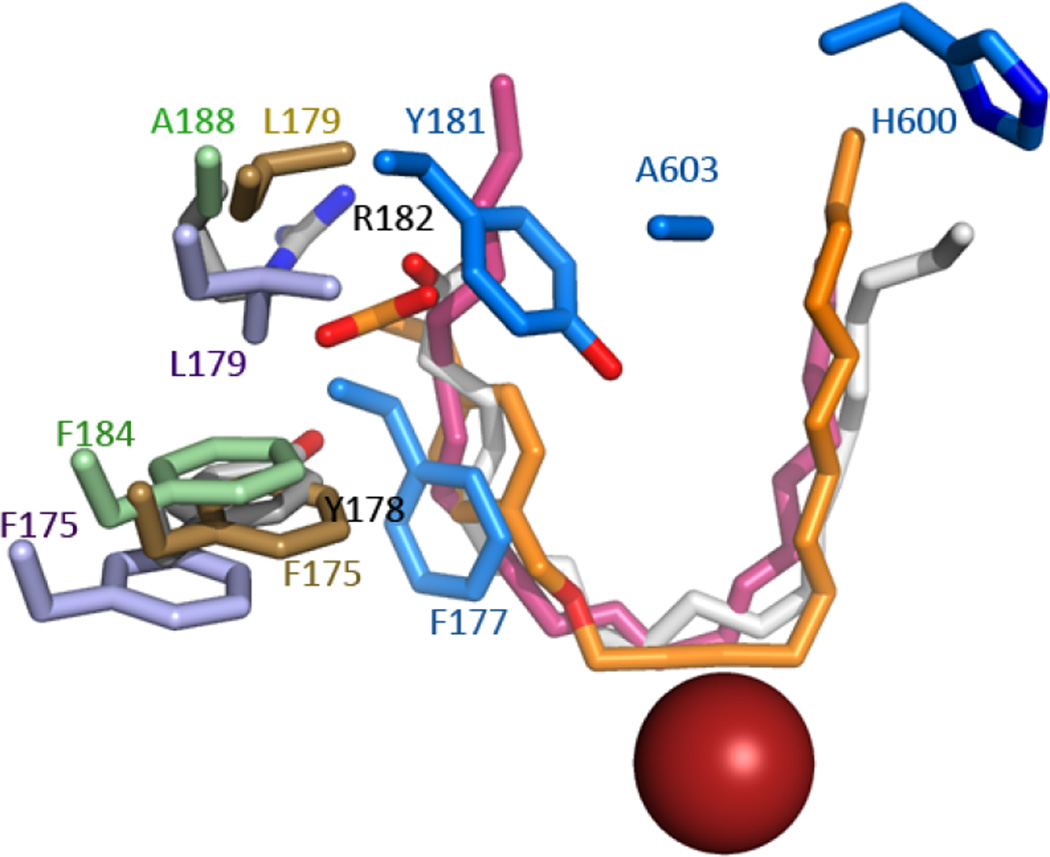
Recent structures of lipoxygenases have combined to provide a robust model for understanding product specificity in this family(17) and suggest a U-shaped cavity with a highly conserved core of amino acids that position the appropriate pentadiene for attack. What is not clear from the consensus model is why the active site of 5-LOX is “corked” since Phe-177 is a highly conserved amino acid whose counterparts in other arachidonate-utilizing LOX do not block access to the U-shaped channel. Instead they abut the active site cavity with the plane of the aromatic ring perpendicular to the fatty acid tail (Fig 6). (It can be noted here that a 12-LOX-specific inhibitor with an aromatic ring appears to mimic Phe-177: its placement in the 12-LOX crystal structure coincides with the position of the Phe-177 in 5-LOX i.e. the site of 12-LOX is “corked” in an intermolecular fashion by an inhibitor(17).) The structures of 15-LOX-2 and 5-LOX (~42% sequence identity) superimpose with an rms of 1.17 Å for 576 of 674 Cα’s, but the distance between the Cα’s of Phe-177 and its equivalent in 15-LOX-2 (Phe-184) is ~8 Å. This major shift in side chain placement is a consequence of an unraveling of α2 in 5-LOX so that the helical segment that contains Phe-177 and Tyr-181 is displaced in this isoform and these side chains can insert into the active site. Notably, substrate or inhibitors shield at least 50% of the accessible surface area of the Phe-177 counterparts in 8R-LOX, 12-LOX or 15-LOX-2, i.e they form part of the wall of the active site and even in their “open” conformations play critical roles in substrate recognition. In contrast, the counterparts of Tyr-181 are largely solvent-accessible amino acids at the entrance to the site. The 8R-LOX counterpart of Tyr-181 (Arg-182) contributes significantly to positioning of substrate in a productive conformation(23), presumably because it provides a counter charge to the substrate carboxylate that helps anchor the pentadiene to be attacked at the catalytic center. However, a charged amino acid is not conserved at this position in other tail-first LOX’s with open cavity access. Thus the amino acids at the periphery of the binding sites, specifically 5-LOX Phe-177 and Tyr-181 and their counterparts, can vary in placement and play distinct roles in binding site definition.
Fig. 6. Anomalous placement of the corking residues in 5-LOX.
5-LOX “corking” amino acids (bright blue, F177,Y181) are displaced relative to their counterparts F175, L179 in 15-LOX (lavender, 2P0M); F175, L179 in 12-LOX (gold, 3RDE); F184, A188 in 15-LOX-2 (green, 4NRE) and Y178, R182 in 8R-LOX (white). AA (white) and 12-LOX (orange) and 15-LOX-2 (pink) inhibitors are rendered in sticks. For reference, the catalytic iron (red sphere) and 5-LOX residues A603 and H600 are included.
In conclusion, our results are consistent with a model in which the substrate gains access to the 5-LOX active site via a “cork” which must be repositioned, but not ejected, to allow substrate access. Phe-177 is likely to form an integral part of the active site, and His-600 is required to position the substrate for 5-HETE production. In retrospect, “cork” was a poor choice of metaphor; the 5-LOX active site entry portal is more accurately described as a “twist-and-pour” closure.
Acknowledgments
Funding Statement
The work described was supported by National Institutes of Health Heart, Lung and Blood Institute R01 107887.
Abbreviations
- 5-LOX
5-Lipoxygenase
- AA
Arachidonic Acid
- AEA
Arachidonoyl Ethanolamide
- C8E4
Polyoxyethylene alkyl ether C8E4
- DGLA
Dihomo-γ-Linolenic Acid
- NAGly
N-arachidonyl Glycine
- FLAP
5-Lipoxygenase Activating Protein
- HETE
hydroxyeicosatetraenoic acid
- HPETE
hydroperoxyeicosatetraenoic acid
- LOX
lipoxygenase
- LT
leukotriene
- PGB1
Prostaglandin B1
- St-5-LOX
Stable 5-Lipoxygenase
- TCEP
tris (2-carboxyethyl) phosphine
References
- 1.Haeggstrom JZ, Funk CD. Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem Rev. 2011;111:5866–5898. doi: 10.1021/cr200246d. [DOI] [PubMed] [Google Scholar]
- 2.Radmark O, Werz O, Steinhilber D, Samuelsson B. 5-Lipoxygenase, a key enzyme for leukotriene biosynthesis in health and disease. Biochim Biophys Acta. 2015;1851:331–339. doi: 10.1016/j.bbalip.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 3.Shimizu T, Radmark O, Samuelsson B. Enzyme with dual lipoxygenase activities catalyzes leukotriene A4 synthesis from arachidonic acid. Proc Natl Acad Sci U S A. 1984;81:689–693. doi: 10.1073/pnas.81.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funk CD. Prostaglandins and leukotrienes: advances in eicosanoid biology. Science. 2001;294:1871–1875. doi: 10.1126/science.294.5548.1871. [DOI] [PubMed] [Google Scholar]
- 5.Hui Y, Funk CD. Cysteinyl leukotriene receptors. Biochem Pharmacol. 2002;64:1549–1557. doi: 10.1016/s0006-2952(02)01357-6. [DOI] [PubMed] [Google Scholar]
- 6.Steinhilber D, Hofmann B. Recent advances in the search for novel 5-lipoxygenase inhibitors. Basic & clinical pharmacology & toxicology. 2014;114:70–77. doi: 10.1111/bcpt.12114. [DOI] [PubMed] [Google Scholar]
- 7.Pergola C, Werz O. 5-Lipoxygenase inhibitors: a review of recent developments and patents. Expert opinion on therapeutic patents. 2010;20:355–375. doi: 10.1517/13543771003602012. [DOI] [PubMed] [Google Scholar]
- 8.Sarveswaran S, Chakraborty D, Chitale D, Sears R, Ghosh J. Inhibition of 5-lipoxygenase selectively triggers disruption of c-Myc signaling in prostate cancer cells. J Biol Chem. 2015;290:4994–5006. doi: 10.1074/jbc.M114.599035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corser-Jensen CE, Goodell DJ, Freund RK, Serbedzija P, Murphy RC, Farias SE, Dell'Acqua ML, Frey LC, Serkova N, Heidenreich KA. Blocking leukotriene synthesis attenuates the pathophysiology of traumatic brain injury and associated cognitive deficits. Experimental neurology. 2014;256:7–16. doi: 10.1016/j.expneurol.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brash AR. Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. J Biol Chem. 1999;274:23679–23682. doi: 10.1074/jbc.274.34.23679. [DOI] [PubMed] [Google Scholar]
- 11.Kuhn H, Thiele BJ. The diversity of the lipoxygenase family. Many sequence data but little information on biological significance. FEBS Lett. 1999;449:7–11. doi: 10.1016/s0014-5793(99)00396-8. [DOI] [PubMed] [Google Scholar]
- 12.Andreou A, Feussner I. Lipoxygenases - Structure and reaction mechanism. Phytochemistry. 2009;70:1504–1510. doi: 10.1016/j.phytochem.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Funk CD, Chen XS, Johnson EN, Zhao L. Lipoxygenase genes and their targeted disruption. Prostaglandins Other Lipid Mediat. 2002;68–69:303–312. doi: 10.1016/s0090-6980(02)00036-9. [DOI] [PubMed] [Google Scholar]
- 14.Egmond MR, Vliegenthart JF, Boldingh J. Stereospecificity of the hydrogen abstraction at carbon atom n-8 in the oxygenation of linoleic acid by lipoxygenases from corn germs and soya beans. Biochem Biophys Res Commun. 1972;48:1055–1060. doi: 10.1016/0006-291x(72)90815-7. [DOI] [PubMed] [Google Scholar]
- 15.Coffa G, Imber AN, Maguire BC, Laxmikanthan G, Schneider C, Gaffney BJ, Brash AR. On the relationships of substrate orientation, hydrogen abstraction, and product stereochemistry in single and double dioxygenations by soybean lipoxygenase-1 and its Ala542Gly mutant. J Biol Chem. 2005;280:38756–38766. doi: 10.1074/jbc.M504870200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider C, Pratt DA, Porter NA, Brash AR. Control of oxygenation in lipoxygenase and cyclooxygenase catalysis. Chem Biol. 2007;14:473–488. doi: 10.1016/j.chembiol.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newcomer ME, Brash AR. The structural basis for specificity in lipoxygenase catalysis. Protein Sci. 2015;24:298–309. doi: 10.1002/pro.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillmor SA, Villasenor A, Fletterick R, Sigal E, Browner MF. The structure of mammalian 15-lipoxygenase reveals similarity to the lipases and the determinants of substrate specificity [published erratum appears in Nat Struct Biol 1998 Mar;5(3)242] Nat Struct Biol. 1997;4:1003–1009. doi: 10.1038/nsb1297-1003. [DOI] [PubMed] [Google Scholar]
- 19.Choi J, Chon JK, Kim S, Shin W. Conformational flexibility in mammalian 15S-lipoxygenase: Reinterpretation of the crystallographic data. Proteins. 2008;70:1023–1032. doi: 10.1002/prot.21590. [DOI] [PubMed] [Google Scholar]
- 20.Kobe MJ, Neau DB, Mitchell CE, Bartlett SG, Newcomer ME. The structure of human 15-lipoxygenase-2 with a substrate mimic. J Biol Chem. 2014;289:8562–8569. doi: 10.1074/jbc.M113.543777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu S, Mueser TC, Marnett LJ, Funk MO., Jr Crystal structure of 12-lipoxygenase catalytic-domain-inhibitor complex identifies a substrate-binding channel for catalysis. Structure. 2012;20:1490–1497. doi: 10.1016/j.str.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oldham ML, Brash AR, Newcomer ME. Insights from the X-ray crystal structure of coral 8R-lipoxygenase: calcium activation via a C2-like domain and a structural basis of product chirality. J Biol Chem. 2005;280:39545–39552. doi: 10.1074/jbc.M506675200. [DOI] [PubMed] [Google Scholar]
- 23.Neau DB, Bender G, Boeglin WE, Bartlett SG, Brash AR, Newcomer ME. Crystal structure of a lipoxygenase in complex with substrate: the arachidonic acid-binding site of 8R-lipoxygenase. J Biol Chem. 2014;289:31905–31913. doi: 10.1074/jbc.M114.599662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bryant RW, Schewe T, Rapoport SM, Bailey JM. Leukotriene formation by a purified reticulocyte lipoxygenase enzyme. Conversion of arachidonic acid and 15-hydroperoxyeicosatetraenoic acid to 14, 15-leukotriene A4. J Biol Chem. 1985;260:3548–3555. [PubMed] [Google Scholar]
- 25.Evans JF, Ferguson AD, Mosley RT, Hutchinson JH. What's all the FLAP about?: 5-lipoxygenase-activating protein inhibitors for inflammatory diseases. Trends Pharmacol Sci. 2008;29:72–78. doi: 10.1016/j.tips.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 26.Dixon RA, Diehl RE, Opas E, Rands E, Vickers PJ, Evans JF, Gillard JW, Miller DK. Requirement of a 5-lipoxygenase-activating protein for leukotriene synthesis. Nature. 1990;343:282–284. doi: 10.1038/343282a0. [DOI] [PubMed] [Google Scholar]
- 27.Basavarajappa D, Wan M, Lukic A, Steinhilber D, Samuelsson B, Radmark O. Roles of coactosin-like protein (CLP) and 5-lipoxygenase-activating protein (FLAP) in cellular leukotriene biosynthesis. Proc Natl Acad Sci U S A. 2014;111:11371–11376. doi: 10.1073/pnas.1410983111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rakonjac M, Fischer L, Provost P, Werz O, Steinhilber D, Samuelsson B, Radmark O. Coactosin-like protein supports 5-lipoxygenase enzyme activity and up-regulates leukotriene A4 production. Proc Natl Acad Sci U S A. 2006;103:13150–13155. doi: 10.1073/pnas.0605150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gilbert NC, Bartlett SG, Waight MT, Neau DB, Boeglin WE, Brash AR, Newcomer ME. The structure of human 5-lipoxygenase. Science. 2011;331:217–219. doi: 10.1126/science.1197203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eek P, Jarving R, Jarving I, Gilbert NC, Newcomer ME, Samel N. Structure of a calcium-dependent 11R-lipoxygenase suggests a mechanism for Ca2+ regulation. J Biol Chem. 2012;287:22377–22386. doi: 10.1074/jbc.M112.343285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Vedadi M, Niesen FH, Allali-Hassani A, Fedorov OY, Finerty PJ, Jr, Wasney GA, Yeung R, Arrowsmith C, Ball LJ, Berglund H, Hui R, Marsden BD, Nordlund P, Sundstrom M, Weigelt J, Edwards AM. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci U S A. 2006;103:15835–15840. doi: 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ericsson UB, Hallberg BM, Detitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem. 2006;357:289–298. doi: 10.1016/j.ab.2006.07.027. [DOI] [PubMed] [Google Scholar]
- 34.Gerstmeier J, Weinigel C, Barz D, Werz O, Garscha U. An experimental cell-based model for studying the cell biology and molecular pharmacology of 5-lipoxygenase-activating protein in leukotriene biosynthesis. Biochim Biophys Acta. 2014;1840:2961–2969. doi: 10.1016/j.bbagen.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 35.Rakonjac Ryge M, Tanabe M, Provost P, Persson B, Chen X, Funk CD, Rinaldo-Matthis A, Hofmann B, Steinhilber D, Watanabe T, Samuelsson B, Radmark O. A mutation interfering with 5-lipoxygenase domain interaction leads to increased enzyme activity. Arch Biochem Biophys. 2014;545:179–185. doi: 10.1016/j.abb.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 36.Boyington JC, Gaffney BJ, Amzel LM. The three-dimensional structure of an arachidonic acid 15-lipoxygenase. Science. 1993;260:1482–1486. doi: 10.1126/science.8502991. [DOI] [PubMed] [Google Scholar]
- 37.Zhang YY, Hamberg M, Radmark O, Samuelsson B. Stabilization of purified human 5-lipoxygenase with glutathione peroxidase and superoxide dismutase. Anal Biochem. 1994;220:28–35. doi: 10.1006/abio.1994.1294. [DOI] [PubMed] [Google Scholar]
- 38.Rouzer CA, Shimizu T, Samuelsson B. On the nature of the 5-lipoxygenase reaction in human leukocytes: characterization of a membrane-associated stimulatory factor. Proc Natl Acad Sci U S A. 1985;82:7505–7509. doi: 10.1073/pnas.82.22.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aharony D, Redkar-Brown DG, Hubbs SJ, Stein RL. Kinetic studies on the inactivation of 5-lipoxygenase by 5(S)-hydroperoxyeicosatetraenoic acid. Prostaglandins. 1987;33:85–100. doi: 10.1016/0090-6980(87)90307-8. [DOI] [PubMed] [Google Scholar]
- 40.Lepley RA, Fitzpatrick FA. Irreversible inactivation of 5-lipoxygenase by leukotriene A4. Characterization of product inactivation with purified enzyme and intact leukocytes. J Biol Chem. 1994;269:2627–2631. [PubMed] [Google Scholar]
- 41.Schwarz K, Walther M, Anton M, Gerth C, Feussner I, Kuhn H. Structural basis for lipoxygenase specificity. Conversion of the human leukocyte 5-lipoxygenase to a 15-lipoxygenating enzyme species by site-directed mutagenesis. J Biol Chem. 2001;276:773–779. doi: 10.1074/jbc.M005114200. [DOI] [PubMed] [Google Scholar]
- 42.Schwarz K, Gerth C, Anton M, Kuhn H. Alterations in leukotriene synthase activity of the human 5-lipoxygenase by site-directed mutagenesis affecting its positional specificity. Biochemistry. 2000;39:14515–14521. doi: 10.1021/bi001447n. [DOI] [PubMed] [Google Scholar]
- 43.Hofheinz K, Kakularam KR, Adel S, Anton M, Polymarasetty A, Reddanna P, Kuhn H, Horn T. Conversion of pro-inflammatory murine Alox5 into an anti-inflammatory 15S-lipoxygenating enzyme by multiple mutations of sequence determinants. Arch Biochem Biophys. 2013;530:40–47. doi: 10.1016/j.abb.2012.11.015. [DOI] [PubMed] [Google Scholar]
- 44.Jisaka M, Kim RB, Boeglin WE, Brash AR. Identification of amino acid determinants of the positional specificity of mouse 8S-lipoxygenase and human 15S-lipoxygenase-2. J Biol Chem. 2000;275:1287–1293. doi: 10.1074/jbc.275.2.1287. [DOI] [PubMed] [Google Scholar]
- 45.Coffa G, Brash AR. A single active site residue directs oxygenation stereospecificity in lipoxygenases: stereocontrol is linked to the position of oxygenation. Proc Natl Acad Sci U S A. 2004;101:15579–15584. doi: 10.1073/pnas.0406727101. [DOI] [PMC free article] [PubMed] [Google Scholar]