Abstract
DNA promoter methylation of tumor suppressor genes and global DNA hypomethylation are common features of head and neck cancers. Our goal was to identify early DNA methylation changes in oral premalignant lesions (OPLs) that may serve as predictive markers of developing oral squamous cell carcinoma (OSCC). Using high-throughput DNA methylation profiles of 24 OPLs, we found that the top 86 genes differentially methylated between patients who did or did not develop OSCC were simultaneously hypermethylated, suggesting that a CpG island methylation phenotype may occur early during OSCC development. The vast majority of the 86 genes were non-methylated in normal tissues and hypermethylated in OSCC versus normal mucosa. We used pyrosequencing in a validation cohort of 44 patients to evaluate the degree of methylation of AGTR1, FOXI2, and PENK promoters CpG sites that were included in the top 86 genes, and of LINE1 repetitive element methylation, a surrogate of global DNA methylation. A Methylation Index (MI) was developed by averaging the percent methylation of AGTR1, FOXI2, and PENK promoters; patients with a high MI had a worse OCFS (P=0.0030). On the other hand, patients with low levels of LINE1 methylation had a significantly worse OCFS (P=0.0153). In conclusion, AGTR1, FOXI2 and PENK promoter methylation and LINE1 hypomethylation may be associated with an increased risk of OSCC development in patients with OPLs.
Keywords: oral leukoplakia, oral cancer, squamous cell carcinoma, DNA promoter methylation, DNA global hypomethylation, risk biomarker
Introduction
Oral squamous cell carcinoma (OSCC) remains frequent worldwide with an estimated 263,000 cases each year causing about 127,000 deaths (1). The treatment of OSCC with surgery and radiation often results in substantial morbidity. A comprehensive molecular characterization of OPLs may allow refining oral cancer (OC) risk assessment and identifying new potential targets for chemoprevention (2).
Besides the identification of individual biomarkers of risk to develop OSCC (3-6), we have previously performed whole transcriptome profiling of biopsies of OPLs that were prospectively collected in a chemoprevention trial (2). We found that gene expression profiles were strongly associated with the development of OSCC and multiple transcripts identified in our study tend to be differentially expressed between normal mucosa and head and neck squamous cell carcinoma (HNSCC). DNA methyltransferase 3B (DNMT3B) is a de novo DNA methyltransferase and its transcript was one of the most significant ones associated with OSCC risk and was included in a 29-transcript model predictive of OC development (2). Furthermore, polymorphisms of the DNMT3B gene have been associated with risk of developing HNSCC (7), and DNMT3B expression has been associated with the CpG island methylator phenotype (CIMP) (8, 9), which is defined by high levels of simultaneous gene promoter methylation. CIMP was originally described in colorectal cancer but subsequently reported in other solid tumors including HNSCC (10, 11). Both of those observations lead us to suggest that a CpG island methylation phenotype may be an early event during OSCC development.
Studies evaluating increased promoter methylation from normal mucosa to premalignant lesions and to HNSCC have included one or a handful number of genes including retinoic acid receptor (RAR)-beta, p16Ink4, p14ARF, endothelin receptor type B (EDNRB), E-cadherin (CDH1), deleted in lung and esophageal cancer 1 (DLCE1), NDRG family member 2 (NDRG2), and O-6-methylguanine-DNA methyltransferase (MGMT) (12, 13). More recently, genome-wide methylation studies have shown that overall patterns of epigenetic alteration can more reliably distinguish tumor from normal head and neck epithelial tissues than individual gene methylation events (14, 15). Global DNA hypomethylation, measured by the degree of methylation of repetitive sequences across the genome such as long-interspersed nuclear element-1 (LINE-1), is another important feature of malignant tumors and has been associated with genome instability (16).
In the current study we utilized high-throughput CpG island methylation profiles of OPLs for which clinical outcomes were available in order to determine the utility of methylation profiles as predictive markers of OPLs progression and the potential importance of early DNA methylation changes to the development of OSCC. We found that i-the top 86 genes differentially methylated between patients who did or did not develop OSCC were simultaneously hypermethylated in patients who develop OSCC, ii- the vast majority of those 86 genes were non-methylated in normal tissues and hypermethylated in HNSCC versus normal mucosa, iii-among them, angiotensin II receptor type 1 (AGTR1), forkhead box I2 (FOXI2) and proenkephalin (PENK) promoters methylation were associated with OSCC development, and iv- LINE-1 hypomethylation was also positively associated with OCSCC development. We conclude that AGTR1, FOXI2 and PENK promoter methylation and global hypomethylation are promising markers of risk of OPLs progression.
Material and methods
Patients and specimens
From 1992 to 2001, 162 randomized and eligible patients were enrolled in a randomized OSCC chemoprevention trial at The University of Texas MD Anderson Cancer Center (MDACC). The patients had been diagnosed with OPLs and randomly assigned to intervention with 13-cis-retinoic acid (13cRA) versus retinyl palmitate (RP) with or without β-carotene (BC). Detailed information has been previously described (17). A total of 153 frozen samples were available at baseline or 3 months after enrollment but before any event (defined as the diagnosis of OSCC). DNA was available for 148 cases, including 38 from patients who developed OSCC. All samples selected were collected at baseline. The study was approved by the institutional review board, and written informed consent was obtained from all patients. For the discovery part of the study, 24 samples were selected, including 12 samples from patients who did not develop OSCC (group A, median follow-up: 7.64 years), and 12 samples from patients who did develop OSCC (group B, median follow-up: 2.15 years).
Identification of genes differentially methylated in OPLs from patients who developed OSCC
DNA was extracted and purified from OCT-embedded tissue using the QIAamp DNA Mini Kit (Qiagen, Valencia, CA) including on-column RNase (Qiagen) digestion as described by the manufacturer's protocol. An H&E stained section of all the samples was available to check for the presence of oral stratified epithelium. After DNA quantification using a ND-1000 spectrophotometer (Nanodrop Technologies), all DNAs were serially diluted in DNase-free water to obtain a 300ng/μL stock solution. Aliquots were prepared and stored at −80°C. Each aliquot was used once.
High-throughput DNA methylation profiles were generated using a PCR-based method, methylated CpG island amplification (MCA) method described in detail elsewhere (18). All restriction enzymes were from New England Biolabs (Beverly, MA). In brief, 2 μg DNA was digested with SmaI (methylation sensitive), followed by digestion with XmaI (methylation insensitive). The fragmented DNA was precipitated with ethanol and ligated with RXMA polymerase chain reaction (PCR) adaptors RXMA24 (50-AGCACTCTCCAGCCTCTCACCGA C-30) and RXMA12 (50-CCGGGTCGGTGA-30). The XmaI-DNA fragments were ligated to RXMA adaptors using T4 DNA ligase (New England Biolabs). PCR amplification was performed using a thermal cycler. Samples were subjected to 20 cycles of amplification. PCR products were then resolved on a 1.5% agarose gel and observed under ultraviolet light after ethidium bromide staining. Successfully methylated CpG island amplification reactions resulted in amplicons smear ranging from 300 bp to 3 kb, with most amplicons at 1 kb. PCR products were purified using the PCR purification kit from Qiagen.
Amplified CpG islands were then labeled with deoxuridine and hybridized to CGH microarray slides. Incorporation of amino-allyl deoxyuridine triphosphate (dUTP; Sigma, St Louis, MO) into the DNA from PMLs was conducted using the Bioprime Array CGH-labeling system protocol (Life Technologies, Gaithersburg, MD). Cy3 and Cy5 fluorescent dyes were coupled to amino-allyl dUTP-labeled OL (oral leukoplakia) from group A or group B, respectively. Each group A OL was randomly cohybridized with one group B OL to Agilent 4x44k custom CGH microarray slides, allowing the analysis and comparison of 12 pairs of OL. The array included 44,674 probes corresponding to 8,369 unique genes. The probes were selected to recognize Sma I/Xma I restriction fragments from regions close to gene transcriptional start sites (19). Microarray protocols including the hybridization and post-hybridization procedures were performed as previously reported (20). Hybridized slides were scanned with the GenePix 4000A scanner (Axon Instruments, Union City, CA), and the acquired images were analyzed with the software GenePix Pro 3.0. A two-step global lowess normalization was performed using the background-subtracted median intensity of each spot and the resultant Cy5/Cy3 log2 (ratio) were obtained for the 12 pairs of OPLs. In order to identify genes differentially methylated in patients with OPLs who develop OSCC, we filtered out probes with no SmaI fragment ID, on chromosome X and Y, with no CpG island, and with an absolute minimal distance to transcription starting site > 1kb. After filtering out those probes to identify genes differentially methylated in OL with malignant transformation, it remained 17,098 probes corresponding to 4,441 unique genes. We then selected probes with an average log2 (ratio) ≥ 1 or ≤ −1 in a least 7 of the 12 pairs of OL studied, to determine our candidate genes.
In silico validation of candidate genes
For in silico validation of candidate genes, we used the following 3 assumptions: i-genes associated with OSCC development should be unmethylated in normal tissues, including normal head and neck mucosa, but methylated in HNSCC; ii-promoter methylation of genes associated with OSCC development may be downregulated in HNSCC when compared to normal mucosa; and iii-genes associated with OC development may also be methylated in lung cancer when compared to normal lung as oral cavity has been reported as a surrogate tissue for lung smoking-induced molecular alterations, i.e. field effect, in particular for promoter methylation markers (21).
We tested those assumptions using the following datasets: 1-10 oral tongue SCC, blood and normal mucosa from the same patients with available genome wide DNA methylation profiles by Digital Restriction Enzyme Analysis of Methylation (DREAM), a method based on next generation sequencing analysis of methylation-specific signatures created by sequential digestion of genomic DNA with SmaI and XmaI enzymes, that has the ability to absolutely quantitate methylation of 13,000 unique genes (30,000 CpG sites) at transcription start sites individual genes (22); 2-publicly available DNA methylation and gene expression profiles of the following studies were downloaded from Gene Expression Omnibus (GEO): GSE19434 (23), GSE25083 (24), GSE13601 (25), GSE27902 (26), GSE9844 (27), and GSE46802 (28). A detailed description of the samples and platforms used in those studies is provided in Supplementary Table 1. DNA methylation studies used the Illumina Methylation Assay and BeadChip technology (GSE19434, GSE25083 and GSE46802). Normalized data, presented as beta values, and representing the degree of methylation at each CpG site (with 0 being unmethylated and 1 being fully methylated), was used to evaluate the degree of methylation of our candidate genes overlapping with genes analyzed on the Illumina platform. All probes were preserved irrespective of the number of probes per gene symbol. For the gene expression study (GSE13601, GSE6791, and GSE9844), raw microarray data were processed using quantile normalization and robust multi-array average algorithm. Using an Affymetrix platform, a unique gene can be represented by more than one probesets that were all preserved.
Validation of 5 candidate genes and of global DNA methylation in 37 patients with OPLs, using pyrosequencing
The validation set consisted of 37 OPLs collected in patients from the previously mentioned chemoprevention trial, including 23 from patients who did not develop OSCC and 14 from patients who did develop OSCC. Comprehensive data including demographics, smoking and alcohol history, histology, OPLs anatomical site, treatment arm and loss of heterozygocity (LOH) at various sites (d9s171, d3s1285, tp53, d951747, d1751176 and d85254) were available. The degree of promoter methylation of AGTR1, FOXI2, homeobox A9 (HOXA9), PENK, and Zic family member 1 (ZIC1) and the degree of methylation of the repetitive sequence Long Interspersed Elements (LINE1) was evaluated using pyrosequencing-based methylation analysis in the validation set of OPLs. All primers for pyrosequencing were designed using the Pyrosequencing Assay Design 2.0 software (Qiagen/Biotage, Uppsala, Sweden). To validate our pyrosequencing assays, the degree of AGTR1, FOXI2, HOXA9, PENK, and ZIC1 promoter methylation obtained by pyrosequencing in 61 head and neck cell lines described in Supplementary Table 2 was correlated with the results obtained in the same set of cell lines and the appropriate controls [HCT116-DKO (colon cancer cell line double knockdown for DNMT1 and DNMT3B), C42 (primary skin fibroblast cell line), HBEC16 (normal bronchial epithelial cell line), and HMVEC (primary dermal lymphatic endothelial cell line] using the Illumina HumanMethylation27 Beadchip (Illumina, Inc., San Diego, CA). This platform analyzes 27,579 CpG sites around promoters of 14,475 consensus coding sequences.
Bisulfite treatment of OPLs genomic DNA from the validation set was performed using the EpiTect bisulfite kit (Qiagen) according to manufacturer’s instructions. Bisulfite-treated DNA was used for each PCR. After an initial hotstart at 95 ºC for 5 minutes, all PCR reactions were run at 95ºC for 30 seconds, annealed at various temperatures for 30 seconds, and subjected to an extension step at 72 ºC for 30 seconds and 50 cycles . All reactions were carried out with a nested PCR step during which a biotinylated universal primer (50-GGGACACCGCTGATCGTT TA-30) was added. After PCR, the biotinylated strand was captured on streptavidin-coated beads (Amersham Bioscience, Uppsala, Sweden) and incubated with sequencing primers. Pyrosequencing was performed with PSQ HS 96 Gold reagents on a PS QHS 96 pyrosequencer (Biotage) as published previously (29). Methylation quantification was performed using the provided Pyro Q-CpG software. The program calculates for each single CpG dinucleotide the ratio between its methylated and nonmethylated form, resulting in percentage of methylation. The methylation degree of each CpG island area was then determined by calculating the average of the methylation differences of all CpG sites analyzed in each gene region.
Statistical analysis
Data analysis was performed using GraphPad Prism version 5.00 (San Diego, CA). Summary statistics, including median and range values, were used to describe the distribution of candidate genes in different datasets. Cancer and normal samples were compared using a log2 fold-change and a paired or non-paired two-sided Student’s t-test. Correlations were computed using the Spearman rank correlation coefficient. The Pearson correlation coefficient and Ward linkage method were used for hierarchical clustering of samples. The Kaplan-Meier method was used to construct oral cancer-free survival curves, and the log-rank test was used to test the difference by covariate levels. Univariate Cox proportional hazards model was fitted for all available variables and multi-covariable Cox model analysis including smoking status and significant variables in univariate analysis were performed. We applied Firth correction in the multivariate analysis in order to obtain a reasonable estimate of HR for variable MI without adding any dummy observations (30). All statistical tests were two-sided, and p-values of 0.05 or less were considered to be statistically significant.
Results
Identification of 86 candidate genes simultaneously methylated in patients developing OSCC
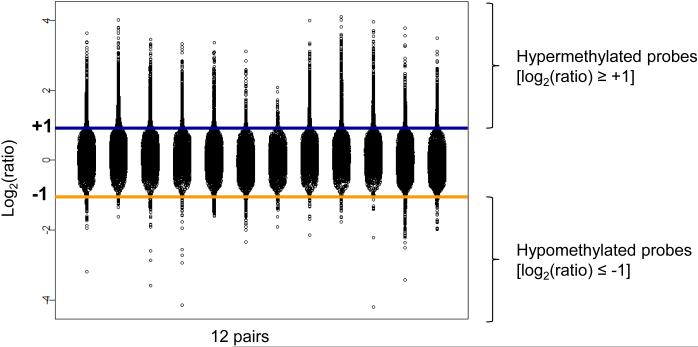
In each of the 12 pairs, the vast majority of the genes differentially methylated [log2(ratio) ≥ +1 or ≤ −1) were found to be hypermethylated [log2(ratio ≥ +1)] in OPLs from patients that subsequently went on to develop OSCC (Fig. 1). In order to select our candidate genes, we selected probes with a log2(ratio) ≥ +1 or ≤ −1 in a least 7 of the 12 pairs of OPLs (Supplementary Fig. 1). We identified 146 probes corresponding to 86 unique genes that were all hypermethylated in OPLs subsequently transforming to OSCC (Table 1). A total of 41/86 (48%) candidate genes were represented by at least 2 probes. Simultaneous methylation of these 86 genes in OPLs that subsequently underwent malignant transformation is consistent with CIMP being an early event in oral tumorigenesis (11).
Figure 1. Genes differentially methylated in oral premalignant lesions (OPL) undergoing malignant transformation are simultaneously methylated.
Distribution of the log2 ratio of 17,098 probes (4,441 unique genes) across 12 pairs of OPLs. In each pair, one OPL from a patient who developed OCSCC was labeled with Cy5 and cohybridized with one OPL from a patient who did not develop OCSCC labeled with Cy3. Probes on chromosome X and Y, with no SmaI fragment ID, with no CpG island, and with an absolute minimal distance to transcription starting site > 1 kb were removed.
Table 1.
Quantitative promoter methylation of 32/86 candidate genes in 10 oral squamous cell carcinoma (OSCC) and paired blood and normal tongue by Digital Restriction Enzyme Analysis of Methylation (DREAM) (22). Four of the 5 genes tested in the validation cohort were analyzed on DREAM and are in bold.
| Gene symbol | % methylation in OSCC |
Number ≥ 15% |
Number measured |
% methylation nal blood |
% methylation nal tongue |
|---|---|---|---|---|---|
| ZIC1 | 63.97 | 10 | 10 | 5.07 | 11.46 |
|
| |||||
| HOXA9 | 48.99 | 10 | 10 | 1.12 | 7.50 |
|
| |||||
| FOXI2 | 46.83 | 10 | 10 | 1.39 | 3.09 |
|
| |||||
| CLDN10 | 46.72 | 10 | 10 | 0.30 | 5.95 |
|
| |||||
| PENK | 40.82 | 10 | 10 | 9.09 | 7.41 |
|
| |||||
| GAD2 | 38.97 | 9 | 10 | 0.96 | 2.31 |
|
| |||||
| VSX2 | 34.52 | 10 | 10 | 7.63 | 5.39 |
|
| |||||
| KCNK12 | 33.53 | 8 | 10 | 1.19 | 1.79 |
|
| |||||
| TMEM132D | 33.24 | 10 | 10 | 2.86 | 4.16 |
|
| |||||
| SLC35F1 | 32.78 | 8 | 10 | 0.77 | 0.62 |
|
| |||||
| ST6GALNAC5 | 31.06 | 7 | 9 | 0.43 | 5.64 |
|
| |||||
| CNTN4 | 28.98 | 7 | 10 | 2.00 | 3.38 |
|
| |||||
| NXPH1 | 28.29 | 8 | 10 | 0.73 | 1.13 |
|
| |||||
| NETO1 | 27.70 | 9 | 10 | 1.58 | 4.13 |
|
| |||||
| STAC2 | 26.23 | 7 | 10 | 0.85 | 0.86 |
|
| |||||
| KCNA3 | 26.13 | 8 | 10 | 3.20 | 4.59 |
|
| |||||
| SALL1 | 24.31 | 5 | 10 | 0.82 | 1.97 |
|
| |||||
| IRF8 | 24.28 | 7 | 9 | 0.72 | 0.80 |
|
| |||||
| SNAP91 | 24.04 | 4 | 10 | 0.63 | 3.57 |
|
| |||||
| RYR2 | 22.43 | 7 | 10 | 0.65 | 1.32 |
|
| |||||
| NPY2R | 22.33 | 5 | 8 | 1.34 | 1.24 |
|
| |||||
| GRIK3 | 22.05 | 4 | 10 | 1.33 | 1.35 |
|
| |||||
| HS3ST2 | 21.50 | 5 | 10 | 0.59 | 2.53 |
|
| |||||
| ZNF529 | 20.90 | 5 | 10 | 0.37 | 0.65 |
|
| |||||
| RGS17 | 17.45 | 4 | 10 | 0.76 | 1.65 |
|
| |||||
| SOX14 | 14.10 | 4 | 10 | 1.30 | 1.10 |
|
| |||||
| ALDH1A2 | 12.53 | 3 | 7 | 0.00 | 0.33 |
|
| |||||
| WBSCR17 | 12.25 | 3 | 10 | 1.99 | 1.45 |
|
| |||||
| TMEFF2 | 11.03 | 1 | 10 | 0.49 | 0.60 |
|
| |||||
| ITGA4 | 9.18 | 2 | 10 | 0.33 | 0.33 |
|
| |||||
| PDE10A | 8.40 | 3 | 10 | 0.61 | 0.31 |
|
| |||||
| RUNDC3B | 4.18 | 1 | 8 | 0.63 | 0.10 |
In silico validation of candidate genes
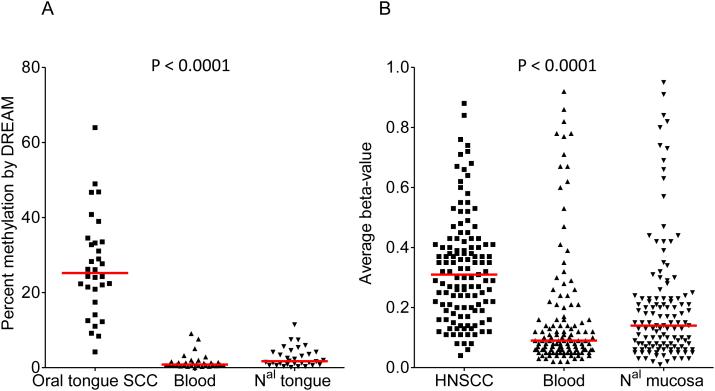
Our first assumption is that candidate genes should be unmethylated in normal tissues and frequently methylated in OSCC. Table 1 and Fig. 2A show the percent methylation of 32/86 (37%) candidate genes overlapping with those analyzed with DREAM in 10 oral tongue SCC, blood and normal oral mucosa. The 32 genes queried were found to have very low levels of methylation in the blood (median: 0.83%; range 0-9.9%) and normal mucosa (median: 1.72%; range 0.10-11.46%) as opposed to the degree of methylation found in oral tongue SCC (median: 25.22%; range: 4.18-63.97%). For further validation, we analyzed methylation data from a set of 146 blood samples, 18 head and neck normal mucosa, and 91 HNSCC that had been evaluated with the Illumina HumanMethylation27 platform which interrogates the promoter regions of 14,000 annotated genes, including 62/86 (72%) candidate genes (Supplementary Table 1) (24). With few exceptions, genes were found to have lower methylation levels in the blood (median: 9%; range: 2-92%) and normal head and neck mucosa (median: 14%; range: 1-95%), compared to HNSCC (median: 31%; range: 4-88%) (Fig. 2B). Details for each gene are provided in Supplementary Fig. 2. Consistent results were obtained using another set of 217 normal tissues (including among others blood, and head and neck mucosa) analyzed with an earlier platform that interrogates 1,505 CpG sites from the promoter region of 807 genes, including 13/86 (14%) candidate genes (23) (Supplementary Table 1 and Supplementary Fig. 3). Finally, in a cohort of 10 patients with paired normal, dysplastic lesions and carcinomas (in situ or invasive) profiled with the Illumina HumanMethylation27 platform (28), the methylation status of 62/86 candidate genes allowed to identify 2 clusters of samples: one included 7/10 (70%) carcinomas and 3/10 (30%) dysplastic samples, while the other one included 10/10 normal mucosa samples, 7/10 (70%) dysplastic mucosa and 3/10 (30%) carcinomas (Supplementary Fig. 4).
Figure 2. A large proportion of candidate genes are frequently methylated in head and neck cancer squamous cell carcinoma (HNSCC) and unmethylated in normal mucosa and blood.
(A) Average promoter methylation of 32/86 candidate genes in 10 oral squamous cell carcinoma and matched normal mucosa and blood by Digital Restriction Enzyme Analysis of Methylation (DREAM); (B) Average beta-values of 125 probes corresponding to 62/86 candidate genes in a set of 91 HNSCC, 146 blood samples, and 18 head and neck normal mucosa profiled with the Illumina HumanMethylation27 Beadchip (GSE25083) (24). ANOVA was used to compare both groups.
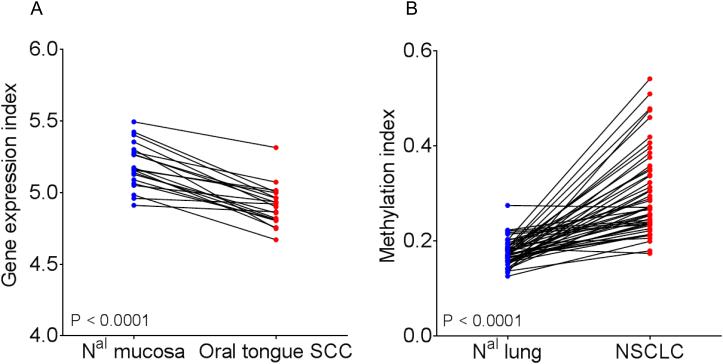
Our second assumption was that the expression of our candidate genes would be downregulated in OSCC compared to normal oral mucosa through promoter hypermethylation. We used gene expression profiles of 32 OSCC and 26 normal oral mucosa, including 20 matched samples (Supplementary Table 1) (25). A total of 62 probesets corresponding to 42/86 (49%) candidate genes were analyzed. An expression index was computed in 20 matched samples by the average expression levels of the 62 probesets. The expression index of our candidate genes was significantly lower in OSCC compared to normal oral mucosa (Fig. 3A). Hierarchical clustering of the 58 samples using this geneset identified 2 clusters of samples, the first one included 22/26 normal oral mucosa and 7/32 oral tongue SCC, and the second one including 4/26 normal oral mucosa and 25/32 oral tongue SCC (Fisher’s exact test P < 0.0001) (Supplementary Fig. 5A).
Figure 3. Candidate genes are often downregulated in oral squamous cell carcinoma (OCSCC) and methylated in non-small cell lung cancer (NSCLC).
(A) A gene expression index was computed in 20 matched OCSCC and normal oral mucosa by averaging the log2 expression values of 62 probesets corresponding to 42/86 candidate genes overlapping with the Affymetrix HG-U95av2 platform (GSE13601) (25). (B) A methylation index was computed in 47 matched non-small cell lung cancer (NSCLC) and normal lung by averaging the average beta-values of 29 probes corresponding to 12/86 candidate genes overlapping with the Illumina GoldenGate Methylation Cancer Panel I (GSE27902) (26). Statistical significance was determined by paired two-sided Student’s t-test.
Our third assumption was based on the fact that oral cavity has been reported as a surrogate tissue for lung smoking-induced molecular alterations. Genes methylated early during oral tumorigenesis may therefore be methylated as well in lung cancer when compared to normal lung. To test this, we computed a methylation index in 47 matched NSCLC and normal lung using 29 probes corresponding to 12/86 (14%) candidate genes overlapping with the platform used (GSE27902). Fig. 3B shows that the methylation index was significantly higher in NSCLC as compared to normal lung. Hierarchical clustering of the 94 samples showed 2 clusters of samples, the first one including 38/47 normal lung and 1/47 NSCLC, and the second one including 9/47 normal lung and 46/47 NSCLC (Fisher’s exact test P < 0.0001) (Supplementary Fig.5B).
Validation of 5 specific candidate genes and global DNA methylation using pyrosequencing in 37 other patients with OPLs
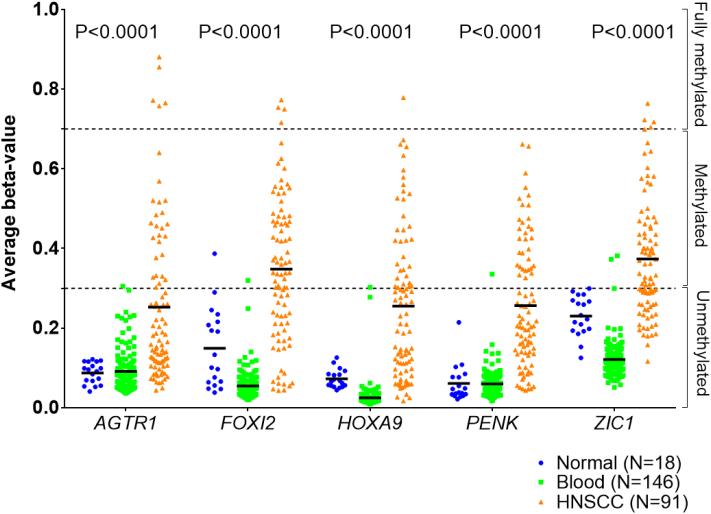
Among the 86 candidate genes methylated in OPLs that subsequently underwent malignant transformation, we selected AGTR1, FOXI2, HOXA9, PENK and ZIC1 for further validation using pyrosequencing. Their average beta-values in 146 blood samples, 18 head and neck normal mucosa, and 91 HNSCC are shown in Fig. 4. In order to validate our pyrosequencing assays, we tested 61 head and neck cell lines and found a significant correlation between the degree of methylation at the promoter of AGTR1, FOXI2, HOXA9, PENK and ZIC1 measured by pyrosequencing and their corresponding beta values extracted from methylation profiles using the Illumina HumanMethylation27 Beadchip, thus validating our pyrosequencing assays (Supplementary Fig. 6).
Figure 4. Promoter methylation of AGTR1, FOXI2, HOXA9, PENK and ZIC1 in head and neck squamous cell cancer (HNSCC), normal mucosa and blood.
Promoter methylation was given by the average beta values of AGTR1, FOXI2, HOXA9, PENK and ZIC1 in a set of 146 blood samples, 18 head and neck normal mucosa, and 91 HNSCC (GSE25083) (24). ANOVA was used to compare both groups.
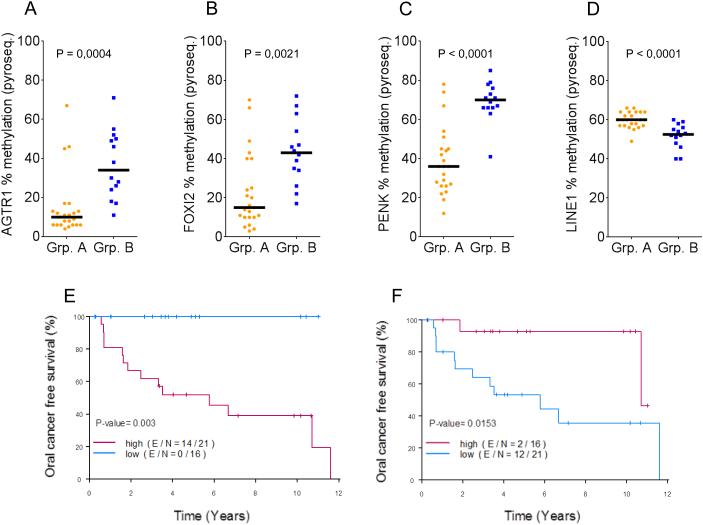
The percent methylation of AGTR1, FOXI2, HOXA9, PENK and ZIC1 was then evaluated in a validation set of 37 OPLs. Fig. 5A-C shows that the percent methylation of AGTR1, FOXI2 and PENK in patients who developed OSCC (group B) was significantly higher compared to patients who did not develop OC (group A). The difference between group A and B was not as pronounced for ZIC1 and was not significant for HOXA9 (data not shown). We then computed a Methylation Index (MI) by averaging the % methylation of AGTR1, FOXI2, and PENK. The group with high MI as defined by the median had a significantly worse oral cancer-free survival compared with the group with low MI (log-rank P=0.0031) (Fig. 5E).
Figure 5. Promoter methylation of AGTR1, FOXI2, and PENK, and global DNA hypomethylation are associated with oral cavity squamous cell carcinoma (OSCC) development.
Percentage methylation of AGTR1 (A), FOXI2 (B), and PENK (C) were obtained using pyrosequencing from 37 oral premalignant lesions (OPLs), including 23 who did not undergo malignant transformation (group A), and 14 who did develop OSCC (group B). (D) Global DNA methylation was measured by the percentage methylation of LINE1 by pyrosequencing and compared in patients from group A and group B. (E) Kaplan-Meier curve in patients with high versus low methylation index (MI) defined by the median of the average percentage methylation of AGTR1, FOXI2, and PENK. (F) Kaplan-Meier curve in patients with high versus low LINE1 percentage methylation by pyrosequencing. Statistical significance was determined by unpaired two-sided Student’s t-test (A-D), log-rank test (E, F).
We then tested whether the degree of methylation of LINE1, a group of genetic elements that composes 17% of the human genome and is being used as a surrogate of global DNA methylation, was associated with the development of OSCC. Using pyrosequencing, we measured the degree of methylation of LINE1 by pyrosequencing in 37 OPLs included in the validation set. LINE1 methylation was significantly lower in patients who developed OSCC (group B) as compared to patients who did not develop OSCC (group A) (Fig. 5D). Using the median to define patients with high versus low LINE1 methylation, this translated to a worse oral cancer-free survival in patients with low LINE1 methylation (log-rank P=0.0118) (Fig. 5F).
Histology (dysplasia versus hyperplasia) was the only other variable reaching statistical significance in a univariate Cox proportional hazards model (Supplementary Table 3). Due to the small sample size, multicovariate Cox models including histology, smoking status and MI or histology and LINE1 methylation status were marginally significant and only LINE1 methylation was significant at the 5% level with P=0.0368 (Supplementary Table 4).
Discussion
Promoter hypermethylation of specific genes have been already studied in oral premalignancy (31, 32), but description of early DNA methylation changes during oral carcinogenesis using genome-wide profiles is scarce. Towle et al. have reported early DNA promoter methylation alterations associated with histological changes during oral tumorigenesis (28). They confirmed that those alterations are early events during oral tumorigenesis and offer an opportunity for biomarker development (33). In our study, we used an alternative approach and identified candidate promoter methylation markers based on patients’ outcome rather than histological changes, because dysplastic lesions may progress to cancer, remain stable for years and/or may regress spontaneously or after tobacco/ alcohol cessation, and OPLs without dysplasia may still progress to cancer (34). We also report the value of global DNA methylation to identify patients at high risk to develop OSCC.
We identified a set of 86 candidate genes with high levels of simultaneous gene promoter methylation associated with the development of OSCC in patients with OPLs. A majority were consistently found to be methylated in OSCC but not in normal tissues. Although our approach may have been biased by the use of a restriction enzyme inherent to the MCA method and limited to 8,369 unique genes analyzed on the Agilent 4×44k custom CGH microarray, we validated our findings in multiple independent datasets either found in the public domain and using a different array-based technology (GEO), as well as in samples profiled by DREAM, a method based on next generation sequencing. We believe that consistent observations from independent studies using various platforms strengthen our conclusion.
Our observation of simultaneous gene promoter methylation in a large number of genes may be consistent with the emergence of a CIMP in the early steps of oral tumorigenesis. CIMP has been initially reported in colorectal cancer (11). Recent reports based on whole-genome methylation analysis support the concept of some tumors having a high propensity for CpG island DNA methylation (41). However, the absence of objective definition of CIMP may explain the challenge to determine its real clinical impact. In patients with breast cancer or glioblastoma, CIMP positive tumors have been associated with an improved outcome (42), although in lung adenocarcinoma and colorectal cancer CIMP positive tumors have been associated with worse outcome (43). The mechanism(s) associated with the onset of CIMP tumors remain(s) to be discovered.
In order to confirm the relevance of some of our 86 candidate genes, we focused on 5 specific ones: FOXI2, ZIC 1 and HOXA9 because they were on the top of the list, PENK because its aberrant promoter methylation has already been reported in various cancers and it has recently been reported as a potential blood-based diagnostic markers in colorectal cancer (37). Finally, AGTR1 was prioritized over CLDN10 due to intriguing reports that are discussed below, linking the use of angiotensin receptor blockers (ARBs) for the treatment of blood hypertension and the risk of cancer. We found that promoter methylation of FOXI2, PENK and AGTR1 were associated with the development of OSCC. FOXI2 plays a role during development, in particular during early craniofacial development (35). PENK plays a role in cell death and survival, and is downregulated by the proto-oncogenes FOS and JUN in the central nervous system. However, their role in cancer is not well understood (36).
Our finding of AGTR1 promoter methylation as a marker associated with OSCC development is consistent with a recent study of new DNA methylation markers for early diagnosis of colorectal cancer (37). This may also be consistent with provocative results of a recent meta-analysis of randomized controlled trials evaluating ARBs for the treatment of various medical conditions suggesting that ARBs may be associated with a modestly increased risk of new cancer diagnosis. Among specific solid organ cancers examined, only new lung-cancer occurrence was significantly higher in patients randomly assigned to receive ARBs than in those assigned to receive control (0.9% vs. 0.7%) (38). Together with our observation that AGTR1 promoter methylation is associated with OSCC development and is methylated as well in NSCLC, it is tempting to hypothesize that a loss of function of AGTR1 through pharmacologic intervention or DNA promoter methylation plays a role in lung and head and neck tumorigenesis. A complete understanding of the interaction between the two types of angiotensin receptors, ATR1 and ATR2 and functional studies in head and neck and lung preclinical models are warranted as those data contrast with other reports suggesting that AGTR1 may be a target for the treatment of gynecological malignancies (39).
Another aspect of DNA methylation aberrations in cancer is global DNA hypomethylation. In this report, we show that global DNA hypomethylation is an early event associated with the risk of developing OSCC in patients with OPLs. Global DNA hypomethylation has been associated with genome instability (16). Conflicting results have been reported between global DNA hypomethylation in peripheral blood and risk of cancer (40). The study of global DNA hypomethylation in the target tissue may be more relevant for cancer risk assessment.
The most robust and validated biomarker for risk prediction of OSCC development in patients with OPLs remains the LOH first reported by Mao et al. in 1996 (5). The value of LOH profiles to predict OCSS development was validated in multiple retrospective studies and more recently in a prospective Canadian cohort (32). The value of AGTR1, FOXI2 and PENK promoter methylation and LINE1 hypomethylation to predict OSCC development and whether it can improve the risk assessment obtained by the study of LOH needs to be validated in larger cohorts of patients. One of the advantages of methylation over LOH markers is that analyzing constitutional DNA may not be necessary. On the other hand, LOH status can be defined in paraffin-embedded tissues, which remains more difficult for DNA methylation markers that will require the development and evaluation of assays for tissues processed in every day practice.
In conclusion, AGTR1, FOXI2 and PENK promoter methylation may be associated with an increased risk of OSCC development in patients with OPLs. They may be associated with an early CIMP during oral tumorigenesis. LINE1 hypomethylation may also be associated with OSCC risk. Further studies are needed to validate these new biomarkers. The role of ARBs in head and neck and lung cancer warrants further investigation.
Supplementary Material
Acknowledgments
Grant Support: University of Texas SPORE in Head and Neck P50 CA97007, University of Texas SPORE in Head (Pickering CR, Papadimitrakopoulou V, William WN Jr, Mitchell FT, Wang J, Lang W, Feng L, Fan YH, Hong WK, El-Naggar AK, Lee JJ, Myers JN, Lippman SM, Mao L, Saintigny P) and Neck Career Development Award 2011-2012 (5 P50 CA097007 09) (Saintigny P), Fondation pour le Recherche Médicale (Foy JP), and Cancéropôle Lyon Auvergne Rhône-Alpes (CLARA) 201-2016 Structured Program - Grant N°CVPPRCAN000153 (International Head and Neck Prevention Act-IHNPACT) (Saintigny P)
Footnotes
Conflicts of interest: No
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Saintigny P, Zhang L, Fan YH, El-Naggar AK, Papadimitrakopoulou VA, Feng L, et al. Gene expression profiling predicts the development of oral cancer. Cancer Prev Res (Phila) 2011;4:218–29. doi: 10.1158/1940-6207.CAPR-10-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saintigny P, El-Naggar AK, Papadimitrakopoulou V, Ren H, Fan YH, Feng L, et al. DeltaNp63 overexpression, alone and in combination with other biomarkers, predicts the development of oral cancer in patients with leukoplakia. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:6284–91. doi: 10.1158/1078-0432.CCR-09-0498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taoudi Benchekroun M, Saintigny P, Thomas SM, El-Naggar AK, Papadimitrakopoulou V, Ren H, et al. Epidermal growth factor receptor expression and gene copy number in the risk of oral cancer. Cancer Prev Res (Phila) 2010;3:800–9. doi: 10.1158/1940-6207.CAPR-09-0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mao L, Lee JS, Fan YH, Ro JY, Batsakis JG, Lippman S, et al. Frequent microsatellite alterations at chromosomes 9p21 and 3p14 in oral premalignant lesions and their value in cancer risk assessment. Nat Med. 1996;2:682–5. doi: 10.1038/nm0696-682. [DOI] [PubMed] [Google Scholar]
- 6.Kawaguchi H, El-Naggar AK, Papadimitrakopoulou V, Ren H, Fan YH, Feng L, et al. Podoplanin: a novel marker for oral cancer risk in patients with oral premalignancy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:354–60. doi: 10.1200/JCO.2007.13.4072. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Wang L, Wang LE, Sturgis EM, Wei Q. Polymorphisms of the DNMT3B gene and risk of squamous cell carcinoma of the head and neck: a case-control study. Cancer letters. 2008;268:158–65. doi: 10.1016/j.canlet.2008.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nosho K, Shima K, Irahara N, Kure S, Baba Y, Kirkner GJ, et al. DNMT3B expression might contribute to CpG island methylator phenotype in colorectal cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2009;15:3663–71. doi: 10.1158/1078-0432.CCR-08-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibrahim AE, Arends MJ, Silva AL, Wyllie AH, Greger L, Ito Y, et al. Sequential DNA methylation changes are associated with DNMT3B overexpression in colorectal neoplastic progression. Gut. 2011;60:499–508. doi: 10.1136/gut.2010.223602. [DOI] [PubMed] [Google Scholar]
- 10.Marsit CJ, Houseman EA, Christensen BC, Eddy K, Bueno R, Sugarbaker DJ, et al. Examination of a CpG island methylator phenotype and implications of methylation profiles in solid tumors. Cancer research. 2006;66:10621–9. doi: 10.1158/0008-5472.CAN-06-1687. [DOI] [PubMed] [Google Scholar]
- 11.Taby R, Issa JP. Cancer epigenetics. CA Cancer J Clin. 2010;60:376–92. doi: 10.3322/caac.20085. [DOI] [PubMed] [Google Scholar]
- 12.Kresty LA, Mallery SR, Knobloch TJ, Song H, Lloyd M, Casto BC, et al. Alterations of p16(INK4a) and p14(ARF) in patients with severe oral epithelial dysplasia. Cancer research. 2002;62:5295–300. [PubMed] [Google Scholar]
- 13.Youssef EM, Lotan D, Issa JP, Wakasa K, Fan YH, Mao L, et al. Hypermethylation of the retinoic acid receptor-beta(2) gene in head and neck carcinogenesis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2004;10:1733–42. doi: 10.1158/1078-0432.ccr-0989-3. [DOI] [PubMed] [Google Scholar]
- 14.Guerrero-Preston R, Soudry E, Acero J, Orera M, Moreno-Lopez L, Macia-Colon G, et al. NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev Res (Phila) 2011;4:1061–72. doi: 10.1158/1940-6207.CAPR-11-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marsit CJ, Christensen BC, Houseman EA, Karagas MR, Wrensch MR, Yeh RF, et al. Epigenetic profiling reveals etiologically distinct patterns of DNA methylation in head and neck squamous cell carcinoma. Carcinogenesis. 2009;30:416–22. doi: 10.1093/carcin/bgp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 17.Papadimitrakopoulou VA, Lee JJ, William WN, Jr., Martin JW, Thomas M, Kim ES, et al. Randomized trial of 13-cis retinoic acid compared with retinyl palmitate with or without beta-carotene in oral premalignancy. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:599–604. doi: 10.1200/JCO.2008.17.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toyota M, Ho C, Ahuja N, Jair KW, Li Q, Ohe-Toyota M, et al. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer research. 1999;59:2307–12. [PubMed] [Google Scholar]
- 19.Estecio MR, Yan PS, Ibrahim AE, Tellez CS, Shen L, Huang TH, et al. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res. 2007;17:1529–36. doi: 10.1101/gr.6417007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–60. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 21.Bhutani M, Pathak AK, Fan YH, Liu DD, Lee JJ, Tang H, et al. Oral epithelium as a surrogate tissue for assessing smoking-induced molecular alterations in the lungs. Cancer Prev Res (Phila) 2008;1:39–44. doi: 10.1158/1940-6207.CAPR-08-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jelinek J, Liang S, Lu Y, He R, Ramagli LS, Shpall EJ, et al. Conserved DNA methylation patterns in healthy blood cells and extensive changes in leukemia measured by a new quantitative technique. Epigenetics. 2012:7. doi: 10.4161/epi.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet. 2009;5:e1000602. doi: 10.1371/journal.pgen.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poage GM, Houseman EA, Christensen BC, Butler RA, Avissar-Whiting M, McClean MD, et al. Global hypomethylation identifies Loci targeted for hypermethylation in head and neck cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011;17:3579–89. doi: 10.1158/1078-0432.CCR-11-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Estilo CL, P Oc, Talbot S, Socci ND, Carlson DL, Ghossein R, et al. Oral tongue cancer gene expression profiling: Identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9:11. doi: 10.1186/1471-2407-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson HH, Marsit CJ, Christensen BC, Houseman EA, Kontic M, Wiemels JL, et al. Key epigenetic changes associated with lung cancer development: results from dense methylation array profiling. Epigenetics. 2012;7:559–66. doi: 10.4161/epi.20219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye H, Yu T, Temam S, Ziober BL, Wang J, Schwartz JL, et al. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69. doi: 10.1186/1471-2164-9-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Towle R, Truong D, Hogg K, Robinson WP, Poh CF, Garnis C. Global analysis of DNA methylation changes during progression of oral cancer. Oral Oncol. 2013;49:1033–42. doi: 10.1016/j.oraloncology.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 29.Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–50. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- 30.Firth D. Bias reduction of maximum likelihood estimates. Biometrika. 1993;80:27–38. [Google Scholar]
- 31.Pattani KM, Zhang Z, Demokan S, Glazer C, Loyo M, Goodman S, et al. Endothelin receptor type B gene promoter hypermethylation in salivary rinses is independently associated with risk of oral cavity cancer and premalignancy. Cancer Prev Res (Phila) 2010;3:1093–103. doi: 10.1158/1940-6207.CAPR-10-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schussel J, Zhou XC, Zhang Z, Pattani K, Bermudez F, Jean-Charles G, et al. EDNRB and DCC salivary rinse hypermethylation has a similar performance as expert clinical examination in discrimination of oral cancer/dysplasia versus benign lesions. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:3268–75. doi: 10.1158/1078-0432.CCR-12-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arantes LM, de Carvalho AC, Melendez ME, Carvalho AL, Goloni-Bertollo EM. Methylation as a biomarker for head and neck cancer. Oral Oncol. 2014;50:587–92. doi: 10.1016/j.oraloncology.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 34.Warnakulasuriya S, Johnson NW, van der Waal I. Nomenclature and classification of potentially malignant disorders of the oral mucosa. J Oral Pathol Med. 2007;36:575–80. doi: 10.1111/j.1600-0714.2007.00582.x. [DOI] [PubMed] [Google Scholar]
- 35.Ohyama T, Groves AK. Expression of mouse Foxi class genes in early craniofacial development. Developmental dynamics : an official publication of the American Association of Anatomists. 2004;231:640–6. doi: 10.1002/dvdy.20160. [DOI] [PubMed] [Google Scholar]
- 36.Roperch JP, Incitti R, Forbin S, Bard F, Mansour H, Mesli F, et al. Aberrant methylation of NPY, PENK, and WIF1 as a promising marker for blood-based diagnosis of colorectal cancer. BMC Cancer. 2013;13:566. doi: 10.1186/1471-2407-13-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carmona FJ, Azuara D, Berenguer-Llergo A, Fernandez AF, Biondo S, de Oca J, et al. DNA methylation biomarkers for noninvasive diagnosis of colorectal cancer. Cancer Prev Res (Phila) 2013;6:656–65. doi: 10.1158/1940-6207.CAPR-12-0501. [DOI] [PubMed] [Google Scholar]
- 38.Sipahi I, Debanne SM, Rowland DY, Simon DI, Fang JC. Angiotensin-receptor blockade and risk of cancer: meta-analysis of randomised controlled trials. The lancet oncology. 2010;11:627–36. doi: 10.1016/S1470-2045(10)70106-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi CH, Park YA, Choi JJ, Song T, Song SY, Lee YY, et al. Angiotensin II type I receptor and miR-155 in endometrial cancers: synergistic antiproliferative effects of anti-miR-155 and losartan on endometrial cancer cells. Gynecologic oncology. 2012;126:124–31. doi: 10.1016/j.ygyno.2012.04.020. [DOI] [PubMed] [Google Scholar]
- 40.Brennan K, Flanagan JM. Is there a link between genome-wide hypomethylation in blood and cancer risk? Cancer Prev Res (Phila) 2012 doi: 10.1158/1940-6207.CAPR-12-0316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.