Abstract
Concanavalin A (ConA), a T-cell mitogen that induces acute autoimmune hepatitis, is widely used to model pathophysiological processes of human acute autoimmune liver disease. Although autophagy has been extensively studied in the past decade, little is known about its molecular mechanism underlying the regulation of ConA-induced acute hepatitis. In this study, we used a Cre-conditional atg7 KO mouse to investigate the effects of Atg7-associated autophagy on ConA-induced murine hepatitis. Our results demonstrated that atg7 deficiency in mice enhanced macrophage activation and increased pro-inflammatory cytokines upon ConA stimulation. Atg7 silencing resulted in accumulation of dysfunctional mitochondria, disruption of reactive oxygen species (ROS) degradation, and increase in pro-inflammatory cytokines in Raw264.7 cells. p38/MAPK and NF-κB levels were increased upon ConA induction due to Atg7 deficiency. Blocking ROS production inhibited ConA-induced p38/IκB phosphorylation and subsequent intracellular inflammatory responses. Hence, this study demonstrated that atg7 knockout in mice or Atg7 knockdown in cell culture augmented ConA-induced acute hepatitis and related cellular malfunction, indicating protective effects of Atg7 on regulating mitochondrial ROS via a p38/MAPK-mediated pathway. Collectively, our findings reveal that autophagy may attenuate macrophage-mediated inflammatory response to ConA and may be the potential therapeutic targets for acute liver injury.
Introduction
Concanavalin A (ConA), the most widely used Jack bean seed lectin, is a T-cell mitogen that can induce acute autoimmune hepatitis [1,2]. ConA-induced hepatitis, which exhibits pathological changes of acute hepatic inflammation, has long been regarded as an appropriate model for studying the pathogenic mechanisms of human acute autoimmune liver disease [3,4]. It is well recognized that ConA-mediated T lymphocyte activation, followed by increased inflammatory cytokines, is the main cause of the rapid and severe inflammatory response exclusively seen in the liver. During this process, CD4+ T lymphocyte-mediated activation of macrophage function serves as an crucial pathogenic parameter [5]. Furthermore, a study unveiled that macrophages residing in the hepatic sinusoid were activated prior to the migration of splenic lymphocytes into the liver [6]. Kupffer cells are dedicated hepatic macrophages and functions as an essential component in liver homeostasis as well as in the acute or chronic responses to chemical compounds [7]. Activation of Kupffer cells augments the production of an array of inflammatory mediators, growth factors, and reactive oxygen species (ROS), whereas inhibition of Kupffer cell activation or depletion of Kupffer cells appears to protect against liver injury induced by immunostimulant ConA [4,8].
Autophagy is an evolutionarily conserved lysosomal pathway critical to cytoplasmic homeostasis to control the turnover of long-lived proteins and excess or dysfunctional organelles [9,10]. It occurs at basal levels, but can be induced by nutritional cues and stress signals. During the autophagic process, cellular contents including organelles are sequestered in double-membrane vesicles called autophagosomes, then the autophagosomes fuse with lysosomes where hydrolysis occurs, supplying amino acids and macromolecular precursor for cells [11,12]. Among various autophagy related genes (Atgs), Atg7, an E1-like enzyme, is essential for the autophagy conjugation system, the formation of autophagosomes and starvation-induced degradation of proteins and organelles in mammalian cells. [13,14].
To date, autophagy has been implicated in several human hepatic pathological conditions, such as hepatocellular carcinoma, HCV-associated chronic liver disease, and liver cirrhosis [15–18]. Although ConA has been demonstrated to be a potent autophagy inducer via a mitochondria-mediated pathway, the role of autophagy in ConA-induced fulminant hepatitis has been incompletely understood [19].
In this study, we utilized a Cre-conditional atg7 KO mouse model to investigate the effect of autophagy in ConA-induced murine autoimmune hepatitis, explored the mechanisms and indicated possible ways to prevent or treat ConA-induced liver injury.
Material and Methods
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the University of North Dakota, School of Medicine (Assurance Number: A3917-01). Dissections and injections were performed under anesthesia that was induced and maintained with ketamine hydrochloride (40 mg/kg) and xylazine, and all efforts were made to minimize animal suffering.
Reagents
N-Acetyl-L-Cysteine (NAC) is clinically used as an ROS scavenger to attenuate liver injury [20]. In our study, NAC (Cat #: A7250, Sigma Aldrich, St Louis, MO) was used to inhibit ROS production. NF-κB inhibitor Sodium 4-aminosalicylate (S4073), p38 mitogen-activated protein kinase (MAPK) inhibitor SB202190 (S1077) and mTOR inhibitor rapamycin (S1039) were purchased from Selleck Company. The mitochondria toxicant Rotenone was bought from Sigma Aldrich.
Animal experiments and ConA treatment
To investigate the physiological roles of autophagy in hepatic injury, Atg7 conditional knockout mice were employed in this study. atg7 KO mice (atg7-/-) on a C57BL/6J background were graciously provided by Dr. Youwen He at Duke University. The atg7-/- mice were originally generated by Masaaki Komatsu at Tokyo Metropolitan Institute of Medical Science. To conditionally delete the target gene, atg7-/- mice were bred with estrogen receptor (ER) cre mice and were injected with 0.1 mg/kg of tamoxifen (T5648, Sigma Aldrich) daily for 6 days before experiments. Mouse genotyping for Atg7 was performed by polymerase chain reaction (PCR) analysis of tail DNA, essentially as described previously [21]. Six to eight week-old wild-type (WT) mice (C57BL/6J) were obtained from the Jackson Laboratory (Bar Harbor, ME) [22]. After acclimation for 6–7 days, mice were randomly divided into a control group and a treatment group, each containing 6 mice. Body weight was assessed at baseline.
Concanavalin A (ConA) (C2272, Sigma Aldrich) was freshly dissolved before each experiment, in pyrogen-free phosphate-buffered saline (PBS) at 2 mg/ml. ConA was injected intravenously via the tail vein of mice in the ConA model group, with equivalent volume of PBS given to the control group. For survival experiment, ConA was dosed at 40 mg/kg and was then adjusted to 20 mg/kg for modelling. Based on the results from previous reports [5] and our preliminary experiments (data not shown), we identified eighteen hours after ConA or PBS injection as an appropriate time point to assess the pathophysiology in this mouse model. Mice were executed by cervical dislocation after ketamine anesthesia (100 mg/kg body weight). Blood and liver samples were collected aseptically.
Measurements of serum liver enzymes
Whole blood was collected by orbital sinus puncture. After centrifugation at 1, 600 × g for 10 min at room temperature, sera samples were stored at −80°C until analysis. The serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were determined by ALT activity assay kit (MAK052, Sigma Aldrich) and AST activity assay kit (MAK055, Sigma Aldrich), respectively, to evaluate the specific hepatic injury and hepatic parenchymal damage.
Histopathology and immunohistochemically staining
Liver tissues from median and left lobes were collected, incubated in 4% paraformaldehyde (MPX00553, Thermofisher Scientific, Rockford, IL) and embedded in paraffin wax according to the traditional method. Sections were made at 4 μm thickness and stained with hematoxylin-eosin (H&E) for observation under a light microscope.
To perform immunohistochemistry staining, liver sections were prepared by heating at 70°C for 30 min. Having dewaxed in dimethylbenzene for 10 min, we dehydrated the sections in a graded series of alcohol. Antigen retrieval was then performed by microwaving the samples in citrate buffer for four cycles. Endogenous peroxidase activity was blocked by treatment with 3% H2O2 for 15 min at room temperature. Nonspecific proteins were blocked with 5% bovine serum albumin (BSA) (BP9706100, Thermofisher Scientific) for 30 min. The sections were incubated with an F4/80 (ab74383, 1:200, Abcam Company, Cambridge, MA) antibody at 4°C overnight. The sections were then washed with phosphate-buffered saline (PBS) and treated immediately with a corresponding secondary antibody at 1:500 in PBS (sc2027, Santa Cruz Biotechnology) for another 30 min. The positive reactions were finally visualized using a diaminobenzidine (DAB) kit (ab64238, Abcam) and the stained tissue was then observed under a light microscope.
Cell viability assay
Cell viability was assessed by an MTT assay kit (M6494, Thermofisher Scientific) according to the manufacturer’s instruction. Briefly, Raw264.7 cells were seeded in a 96-well plate (21377203, Thermofisher Scientific). After ConA treatment, MTT was added to each well at a final concentration of 250 μg/ml. After 2–4 hours, the medium was removed and the cells were dissolved in DMSO (BP231100, Thermofisher Scientific) and kept overnight for complete dissolution at room temperature. Absorbance at 590nm was determined for each well using SpectraMax M5 (Molecular Devices) [23]. After subtracting the background absorbance, the A590 of the cells was divided by that of the control groups to obtain the percentage of the viable cells.
Intracellular ROS measurement
Intracellular ROS were analyzed using hydro-dichlorofluorescein diacetate (H2DCF-DA) (C369, Life technologies) as probes. H2DCF-DA, a non-fluorescent compound, is deacetylated by viable cells to 2, 7-dichlorofluorescein (DCF) by hydrogen peroxide. Post-treatment, Raw264.7 cells were collected and incubated with 100 mM Mito-tracker Red FM/ H2DCF-DA (dissolved in DMSO) for 30 min at 37°C. Cells were then washed three times with PBS and the intracellular accumulation of fluorescent DCF-DA was measured by LSM 510 Meta confocal laser scanning microscope (CLSM) (Carl Zeiss MicroImaging, Thornwood, NY). The results were expressed as the intensity of DCF fluorescence.
Mitochondrial membrane potential Assay and Superoxide Dismutase (SOD) Enzyme Activity Assay
The mitochondrial membrane potential was measured by JC-1 staining (T3168, Life technologies) bearing a delocalized positive charge that can enter the mitochondrial matrix due to the negative charge established by the intact mitochondrial membrane potential. After incubation with JC-1 dye, fluorescence at 485 nm was measured by a fluorometer (BioTek, Winooski, VT). The level of SOD in Raw264.7 cells were analyzed by commercially available SOD kits (19160, Sigma Aldrich) according to the manufacturer’s instruction.
ELISA
The levels of TNF-α, IL-1β, IFN-γ and IL-6 in murine plasma or Raw264.7 cell suspensions were analyzed by ELISA using commercially available kits (eBioscience, San Diego, CA) according to the manufacturer’s instruction.
Cell transfection and confocal microscopy
Raw264.7 cells were respectively transfected with Atg7 siRNA (S20651, Invitrogen, Grand Island, NY) or GFP-LC3 plasmid, using LipofectAmine 2000 reagent (11668019, Invitrogen) in serum-free RPMI 1640 medium (MT15040CV, Thermofisher Scientific) per the manufacturer’s instructions. For immunostaining, Raw264.7 cells were grown in glass-bottomed dishes (MatTek, Ashland, MA) and fixed in 4% paraformaldehyde, permeabilized with 0.2% Triton X-100 in PBS and incubated with blocking buffer for 30 min, then cell nuclei were stained with 2-(4-amidinophenyl)-6-indolecarbamidine dihydrochloride (DAPI) (1:1000 dilution of stock in PBS) for 5 min [24]. The cells were incubated with a p65 antibody (sc7151, Santa Cruz Biotechnology) at 1:500 dilution in blocking buffer overnight and washed three times with wash buffer. Images were captured after incubation with an appropriate secondary antibody containing FITC using a 510 Meta confocal microscope (Carl Zeiss MicroImaging), and processed using the software provided by the manufacturer [25].
Western blotting
Protein concentrations were determined by the BCA Protein Assay Kit (Bio-Rad, Hercules, CA) [26]. Total proteins were extracted from the tissues and cells using a RIPA lysis buffer method and stored at −80°C until western blotting analysis. After boiled at 100°C for 5 min, an equal amount of protein samples was loaded for sodium dodecyl sulfate polyacrylamide gel for electrophoresis (SDS-PAGE), and then transferred to nitrocellulose membranes, blocked with 5% skim milk for 1 h at room temperature, and then incubated with primary antibodies at 4°C overnight. After the samples were incubated with a secondary antibody for 120 min at room temperature, the signals were detected with the chemiluminescence (34096, SuperSignal West Femto Maximum Sensitivity Substrate, Thermofisher Scientific).
Electrophoretic Mobility Shift Assay
Nuclear extracts were isolated following the instruction of NE-PER Nuclear and Cytoplasmic Extraction Reagents Kit (78833, Pierce-Thermofisher Scientific). After preparation of nuclear extracts, the biotin end-labeled DNA was subjected to gel electrophoresis on a native polyacrylamide gel and transferred to a nylon membrane, then detected using a LightShift Chemiluminescent EMSA kit (20148, Pierce-Thermofisher Scientific) as previously described [27]. The NF-κB p65 probe used for this experiment was purchased by Viagene Biotech.
Statistical analysis
All experiments were performed in triplicate and repeated at least three times. Data were presented as ratios or percentage changes compared with mean+SD from the three independent experiments. Parametric data were statistically analyzed by the Student’s t test or one-way ANOVA followed by post hoc tests when appropriate. Differences in Non-parametric data were evaluated by the Mann-Whitney U test. Survival curves were analyzed using Kaplan-Meier test. p<0.05 was used to indicate a statistically significant difference. The calculations were performed with the SPSS software, version 11.0 (SPSS, Inc., Chicago, IL).
Results
atg7 deficiency augmented ConA-induced murine acute hepatic injury
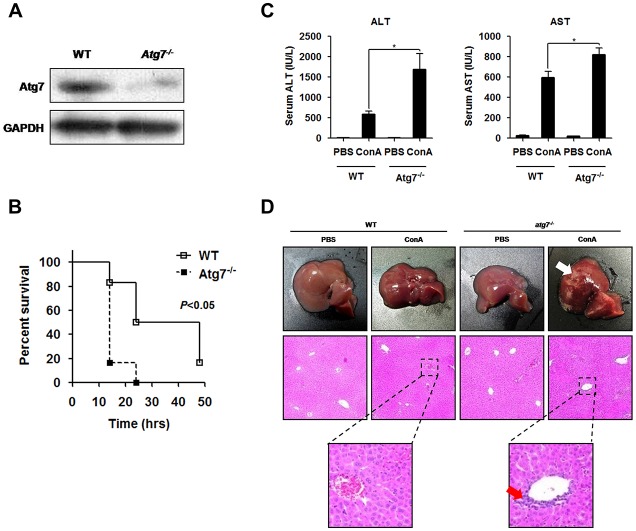
To examine the role of atg7-depleted autophagy in the pathogenesis of ConA-induced autoimmune hepatitis, tamoxifen-induced Cre-mediated atg7 mutant (atg7-/-) mice and its littermate controls were employed in this study. Atg7 protein was expressed at background levels in the liver of atg7-/- mice (Fig 1A). When mice were challenged with 40 mg/kg of ConA, a lethal dose reported by Tiegs G [28], a robust statistically significant difference in survival rates was observed between the two groups. atg7-/- mice manifested worse outcomes compared with control mice (Fig 1B). Based on the results from previous study [5] and preliminary experiments (data not shown), we utilized ConA at a lower dose of 20 mg/kg to construct acute hepatitis model in our study. Serum aminotransferase levels were markedly elevated in atg7-/- mice compared to the littermate controls (Fig 1C). Histopathological changes were consistent with the liver enzyme results. Though hepatic injury, including hepatocytic swelling, degeneration vacuolization and neutrophil infiltration, was apparently observed in both groups, liver damage was more serious in atg7-/- mice (Fig 1D). These results delineated that depletion of atg7 increased susceptibility to ConA hepatitis in mouse models.
Fig 1. atg7 deficiency augmented ConA-induced murine acute hepatic injury.
A. Western blotting of Atg7 protein expression in the liver of atg7-/- mice and the littermate controls. B. Survival curve of the two groups following ConA injection. Mice were challenged with 40 mg/kg ConA and then observed every 2 h until 48 h. C. Serum ALT and AST levels after ConA injection. D. Histopathological staining of liver sections from atg7-/- mice and the littermate controls. Representative results were from 3 independent experiments. Data are presented as mean±SD (n = 6), * p<0.05 vs. littermate controls.
atg7 deficiency increased the production of pro-inflammatory cytokines following ConA injection
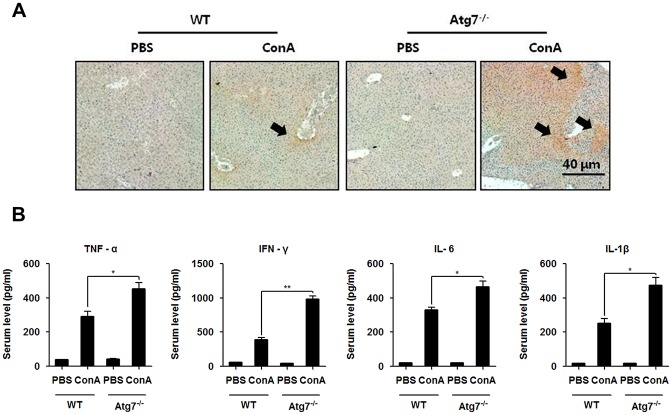
Pro-inflammatory cytokines are essential in the pathogenesis of ConA hepatitis. To investigate whether and how resident hepatic macrophages, Kupffer cells, were activated in this setting, mouse macrophage-specific marker F4/80 mAb was used to visualize the phenotypic characterization of mature macrophages in the liver. Indeed, we observed an increase in the F4/80 positive cells in atg7-/- mice than the littermate controls (Fig 2A). To further explore the mechanism of atg7 deficiency in intra-hepatic inflammation, serum pro-inflammatory mediators were assayed. Lacking of Atg7 in mice led to an increased production of circulatory TNF-α, IFN-γ, IL-6 and IL-1β as measured by ELISA (Fig 2B). Thus, it is suggested that worsened hepatic injury upon atg7 deficiency in ConA hepatitis may be resulted from over activation of macrophages and excessive production of pro-inflammatory cytokines.
Fig 2. atg7 deficiency increased the production of pro-inflammatory cytokines following ConA injection.
A. The immunochemistry micrographs stained by F4/80 mAb of liver tissues from atg7-/- mice and the littermate controls at 18 h following ConA injection (Arrows showing inflammatory regions). B. ELISA analysis of serum TNF-α, IFN-γ, IL-6 and IL-1β 18 h after ConA injection. Representative results were from 3 independent experiments. Data are presented as mean±SD (n = 6), * p<0.05 and ** p<0.01 vs. littermate controls.
Inhibition of autophagy via Atg7 silencing resulted in accumulation of dysfunctional mitochondria and abolished ROS degradation
To validate the observations derived from animal experiments, we used immortalized murine macrophages, Raw264.7 cells, as an in vitro model to investigate the role of Atg7-related autophagy in ConA-induced macrophage activation. ConA has been shown to be a potent autophagic inducer [29–32]. In preliminary experiments, we confirmed that autophagy was induced in Raw264.7 cells when incubated in the complete media supplemented with ConA and was inhibited when Atg7 was knocked down by siRNA silencing (S1A Fig and Fig 1B).
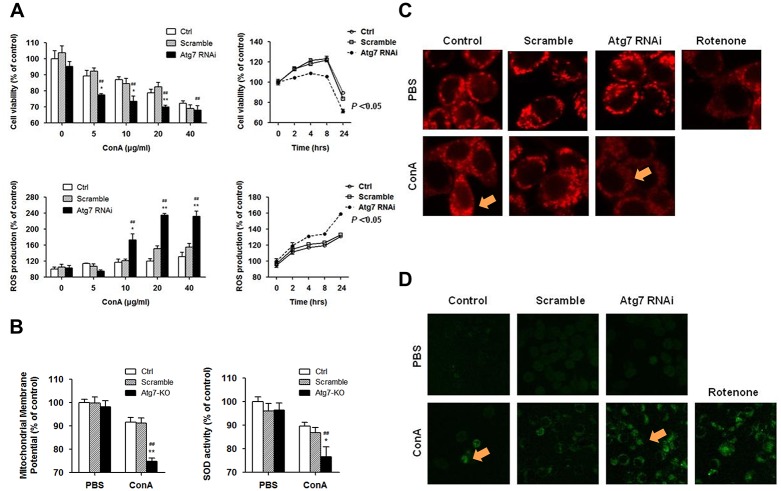
Raw264.7 cells were then stimulated with ConA at different concentrations and time points. We found that ConA increased intra-cellular ROS accumulation and decreased cell viability in a concentration-dependent manner, whereas these changes were more significant upon Atg7 depletion. For the time-dependent tests, cell viability slightly increased and peaked at earlier times of ConA exposure and decreased to a lower level 24 h post-treatment in Atg7-silenced cells than in controls. By contrast, intra-cellular ROS levels rose over time and reached a higher level in Atg7-silenced cells. Loss of autophagy by atg7 silencing exhibited significant decreases in cell viability and increases ROS production 24 h post-treatment (Fig 3A).
Fig 3. Atg7 silencing resulted in accumulation of dysfunctional mitochondria and abolished ROS degradation in Raw264.7 cells.
A. Cell viability measured by an MTT assay and ROS production by mean fluorescence intensity of H2DCF-DA at different concentrations and time points for ConA exposure in Raw264.7 cells lacking Atg7. B. Measurement of mitochondria membrane potential and SOD activity after ConA stimulation. Raw264.7 cells were transfected with Atg7 siRNA or control siRNA and treated with 10 μg/ml ConA for 24 h. Representative results were from 3 independent experiments. Data are presented as mean±SD; * p<0.05 and ** p<0.01 vs. controls, # p<0.05 and ## p<0.01 vs. pre-treatments. C and D. Fluorescence microscopy of mitochondrial membrane by Mito-tracker Red FM (red) and ROS production by H2DCF-DA (green) after ConA stimulation (Arrows showing fluorescence staining regions). Raw264.7 cells were transfected with Atg7 siRNA or control siRNA and treated with 10 μg/ml ConA for 4 h. Magnifications: 1000X. Representative results were from 3 independent experiments.
We then evaluated whether these ConA-induced alterations were mitochondria-associated. ConA 10 μg/ml for 24 h was chosen for a detailed mechanistic study. Mitochondrial membrane potential and SOD activity were detected pre- and post-ConA treatment, respectively. Exposure to ConA decreased mitochondrial membrane potential and SOD activity, which was further reduced in Atg7-silenced Raw264.7 cells (Fig 3B).
To further validate our observation, fluorescence microscopy was used to visualize the phenotypic characterization of cultured cells lacking Atg7. Exposure to ConA greatly impaired the mitochondrial membrane as reflected by reduction in the red fluorescent intensity of Mito-tracker Red FM. The alteration was more significant in Atg7-silenced cells when compared to controls (Fig 3C). The contents of ROS in Raw264.7 cells were similarly determined with H2DCF-DA. The green fluorescence started to increase upon ConA exposure and appeared to be stronger in Atg7-silenced cells when compared to control cells (Fig 3D). These findings confirmed that ConA-induced mitochondrial dysfunction and viscous ROS accumulation in Raw264.7 cells, which were worsened by Atg7 knockdown.
Atg7 knockdown extravagantly activated p38/MAPK and NF-κB signaling in Raw264.7 macrophages
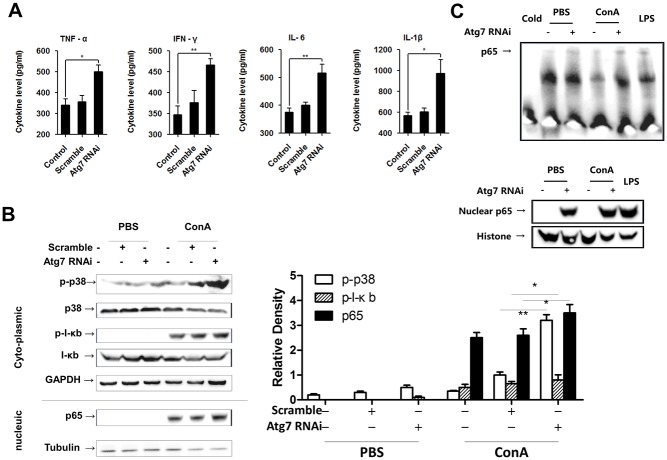
Based on the finding that an Atg7-dependent process was specifically involved in the ROS-associated cell function and death in macrophages, we thus switched our focus onto the interface between autophagy and ConA-induced inflammatory signaling pathway in this setting. We first measured the levels of pro-inflammatory cytokines in the supernatant of cell cultures. As expected, TNF-α, IFN-γ, IL-6 and IL-1β were all significantly elevated in Raw264.7 cells lacking Atg7 (Fig 4A).
Fig 4. Atg7 knockdown extravagantly activated p38 and NF-κB signaling in Raw264.7 macrophages.
A. ELISA analysis of pro-inflammatory cytokines in the supernatant of cell cultures upon ConA challenge. B. Western blotting analysis of p-p38, p-IκB and nuclear p65 protein expression in cultured cells upon ConA challenge. Differences in p38 and IκB phosphorylation were quantitated using densitometry. * p<0.05 and ** p<0.01 vs. controls. C. EMSA results of NF-κB p65 activation and translocation upon ConA challenge. Raw264.7 cells were transfected with Atg7 siRNA or control siRNA and treated with 10 μg/ml ConA for 2 h. Representative results were from 3 independent experiments. Data are presented as mean±SD; * p<0.05 and ** p<0.01 vs. controls.
It is recognized that the p38/NF-κB signaling pathway plays important roles in regulating hepatic inflammatory responses [20,33,34]. In our initial experiments, the kinetics of p-p38/p38, rather than p-JNK/JNK and p-ERK/ERK, was found to be consistent with inflammatory changes in Raw264.7 cells after ConA challenging. By immunofluorescence staining, we confirmed the translocation of NF-κB p65 into the nucleus of Raw264.7 cells, irrespective of Atg7 (S2 Fig). When the expression levels of cytosolic p-p38/p-IκB and nucleic NF-κB p65 were measured by western blotting, we found that p-p38 and p-IκB were surprisingly up-regulated by ConA stimulation. These changes were more significant in Atg7-silenced cells (Fig 4B). We next performed EMSA to examine the conjugation of NF-κB p65 with the promoter gene in the nucleus. In line with previous results, EMSA also exhibited an augmented NF-κB p65 activation, nuclear translocation and combination with the target gene in immortalized macrophages lacking Atg7 (Fig 4C).
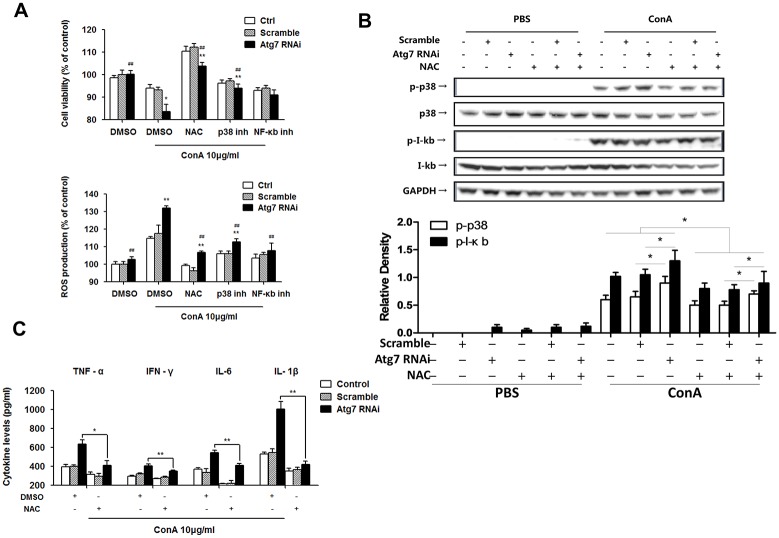
Blocking ROS production reduced ConA-induced p38/IκB phosphorylation and subsequent pro-inflammatory responses in Atg7-silenced macrophages
To verify the effect of ROS in mediating p38/NF-κB pathway in macrophages, we employed chemical inhibitors (ROS inhibitor NAC, p38 inhibitor SB202190 and NF-κB inhibitor Sodium 4-aminosalicylate). Cell viability and ROS production upon ConA stimulation were measured in Raw264.7 cells lacking Atg7. Although siRNA-mediated knockdown of Atg7 led to an increase in ROS production and decrease in cell viability, NAC exhibited the strongest inhibition in ROS among the three differently pre-treated cell groups (Fig 5A). Western blotting shows that addition of NAC reversed over-phosphorylation of p38 and IκB (Fig 5B). The significant alterations in phosphorylation of p38 and IκB were quantitated using densitometry (Fig 5C). Furthermore, NAC also reduced subsequent overproduction of pro-inflammatory cytokines, including TNF-α, IFN-γ, IL-6 and IL-1β upon ConA stimulation in Raw264.7 cells (Fig 5D).
Fig 5. Blocking ROS reduced ConA-induced p38/IκB phosphorylation and subsequent pro-inflammatory responses in Atg7-silenced macrophages.
A. Raw264.7 cells were pretreated with NAC (1 mM), SB202190 (10 μM) and Sodium 4-aminosalicylate (10 μM), respectively, for 30 min prior to 10 μg/ml ConA treatment for 24 h. Cell viability was measured by an MTT assay and ROS production detected by H2DCF-DA. B. Western blotting on p-p38 and p-IκB phosphorylation levels in cultured cells. Densitometry of quantification of p-p38 and p-IκB phosphorylation. * p<0.05 and ** p<0.01 vs. controls, # p<0.05 vs. non NAC-treatments. C. ELISA of TNF-α, IFN-γ, IL-6 and IL-1β levels in the supernatant of cultured cells. Raw264.7 cells were transfected with Atg7 siRNA or scrambles siRNA, pretreated with 1 mM NAC for 30 min and then treated with 10 μg/ml ConA for 24 h. Representative results were from 3 independent experiments. Data are presented as mean±SD; * p<0.05 and ** p<0.01 vs. control, # p<0.05 and ## p<0.01 vs. ConA alone.
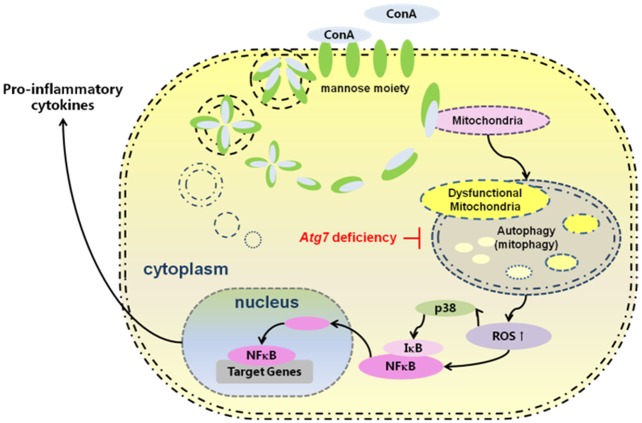
These findings indicate that ConA-induced ROS are associated with p38/IκB phosphorylation in macrophages, which is responsible for pro-inflammatory responses. We put forward to a mechanistic model by which Atg7 effects in the center stage in controlling proinflammatory response in liver hepatitis induced by ConA (Fig 6).
Fig 6. Putative mechanism in which Atg7 knockdown augments ConA-induced hepatic injury through an ROS-mediated p38/MAPK pathway in intra-hepatic macrophages.
Discussion
The current understanding of mammalian autophagy in the liver stems mainly from studies of cancer [29,32] and ischemia/reperfusion injury [35–37], whereas the autophagic mechanism in the pathogenesis of acute autoimmune hepatitis has barely been investigated. Although the regulatory effect of autophagy in liver injury remains elusive, a growing amount of researches have demonstrated that autophagy plays a protective role in hepatocytes under stress conditions. Complete activation of autophagic process attenuates liver injury and improves survival in septic mice [38,39]. In this study, using a tamoxifen-induced conditional atg7-/- mice, we investigated the role of Atg7 in the pathogenesis of ConA-induced autoimmune hepatitis. Thus, we uncovered that autophagy disruption by atg7 depletion increased mortality and enhanced liver damage in mice models.
It is well established that ConA-induced liver injury is initiated by specific activation of CD4+ lymphocytes, which is followed by activation of macrophage function and release of inflammatory cytokines, resulting in necrosis and apoptosis of hepatocytes [5,40,41]. However, a recent study unveils that Kupffer cell plays an important role in T cell activation-induced liver injury by impacting TNF-α production. Liver damage was restricted and TNF-α levels were unaltered when Kupffer cells were depleted by clodronate liposome [8]. In our study, the intensity of immuno-histochemistry staining of macrophage-specific marker F4/80 in liver samples were consistent with the severity of liver injury, indicating that macrophages may serve as a critical inflammatory and immune responder in ConA-induced hepatitis, whose cooperative signaling is indispensable for this pathogenic process. Meanwhile, atg7 deficiency promoted activation of macrophages, which is likely to be responsible for exacerbated systematic inflammatory response and liver injury in atg7-/- mice. Based on the in vivo results, we hypothesize that autophagy is, at least partially, involved in the pathophysiology of ConA hepatitis in Kupffer cells.
In order to further investigate the regulatory role of autophagy in Kupffer cells in ConA-induced autoimmune hepatitis, immortalized macrophage cell line, Raw264.7 cells were used. We found that ConA treatment increased intra-cellular ROS generation and decreased cell viability in a time- and concentration-dependent manner in cultured cells. Spectrophotometric analysis on mitochondrial membrane potential and SOD activity showed an exaggerated mitochondrial damage with Atg7 knockdown. Similar results about ROS quantity and mitochondrial morphological alteration were obtained using fluorescence microscopy analysis.
It has been previously reported that aberrant mitochondrial function and disrupted ROS clearance activate the p38 MAPK pathway [42]. In addition, the p38 MAPK pathway acts as a key regulator in immune and inflammatory responses in ROS-mediated autophagy [43] and apoptosis [34]. Here, we confirmed that inhibition of autophagy augmented ConA-induced inflammatory response via a ROS-mediated pathway, demonstrated by up-regulated activation of p38/IκB, which in turn led to global inflammatory responses and cell death in cultured cells. Given that ConA-induced hepatitis can be ameliorated by antioxidants [44–46], we used ROS, p38 and NF-κB inhibitors to investigate the importance of ROS-mediated signaling in macrophages upon ConA challenging. The administration of NAC, a ROS inhibitor, appeared to robustly correct defects in ROS degradation, leading to improved cell viability, downregulated p38/IκB phosphorylation, and, ultimately, reduced pro-inflammatory cytokine secretion. We thus concluded that enhanced ConA signaling might have resulted from the accumulation of dysfunctional mitochondria and elevated ROS levels, which can be blocked by NAC. Together, these results suggest that Atg7-associated autophagy indeed plays a protective role in response to ConA stimulation by interfering with the p38 MAPK pathway to control the overzealous pro-inflammatory response in the liver.
Intriguingly, although p38 and NF-κB inhibitors also blocked ROS production, such inhibition could not confer capacity to suppress ROS in Atg7-silenced cells to the levels found in control counterparts, indicating that significant part of dysfunctional mitochondria and altered ROS generation are constitutively degraded in the lysosome via an autophagic pathway. In addition, NAC did not substantially inhibit the phosphorylation of p38/ IκB, implying that the activation of the ROS-mediated p38 MAPK pathway was significantly but not exclusively involved in ConA-induced inflammatory response in macrophages. ROS is a group of highly reactive molecular forms of oxygen that are continuously produced as a byproduct of the mitochondrial respiratory chain [47–49]. It is usually maintained at tolerable levels in normal conditions. Imbalance due to either increased ROS production or decreased ROS degradation can cause excessive ROS accumulation, which evokes damage to cell structures and compromising cellular function [50–53]. ROS are recognized as the crossroads of mitochondria, inflammation and autophagy signaling pathways [53]. Autophagy is one of the major intra-cellular pathways by which dysfunctional mitochondria are eliminated [54]. Inhibition of autophagy promoted mitochondrial ROS accumulation, which further aggravated mitochondrial damage. These changes represent a vicious cycle that accelerated the process of cell death and inflammatory pathway activation.
Hence our study demonstrates that atg7 deficiency augmented ConA-induced murine acute hepatitis, indicating a protective effect of Atg7 on regulating mitochondrial ROS degradation via a p38 MAPK pathway. These findings may indicate novel therapeutic targets for fulminant hepatitis. Given that a variety of mechanisms may be involved in the pathogenesis of autoimmune hepatitis, further studies are required to elucidate the mechanisms of atg7 deficiency relating to T cells upon ConA stimulation.
Supporting Information
A. Western blotting analysis of Atg7, p63 and LC3 protein expression in Raw264.7 cells after ConA (10 μg/ml) treatment for 2 h. Disrupted consumption of p62 and interrupted conversion of LC3-II to LC3-I were observed in Atg7 siRNA transfected cells. B. Raw264.7 cells transfected with GFP-LC3 plasmid were treated with ConA (10 μg/ml) for 4 h (Arrows showing GFP-LC3 punctate regions). Rapamycin (50 nM) treatment for 24 h was used as positive control. GFP-LC3 punctate, the marker of autophagosome formation, was scarce to none in Atg7-silenced cells. Magnifications 1000X. Representative results were from 3 independent experiments. Data are presented as mean+SD; * p<0.05 vs. controls.
(TIF)
Immunofluorescence staining shows no significant difference in translocation of NF-κB p65 between Atg7-silenced cells or controls.
(TIF)
Acknowledgments
The authors thank Sarah Rolling of university of North Dakota imaging core for technical support.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by grants from the US National Institute of Health (AI109317-01A1) to M. W., and also supported by the National Natural Science Foundation of China (81171569, 81300316), Shanghai Science and Technology Committee (12XD1403600), The National Key Programs on Infectious Diseases of China (2012ZX10002003-003, 2012ZX10002007-002) to QX. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gupta D, Oscarson S, Raju TS, Stanley P, Toone EJ, Brewer CF. (1996) A Comparison of the Fine Saccharide-Binding Specificity of Dioclea grandiflora Lectin and Concanavalin A. European Journal of Biochemistry 242: 320–326. [DOI] [PubMed] [Google Scholar]
- 2.Ju Y, Shang X, Liu Z, Zhang J, Li Y, Shen Y, et al. (2014) The Tim-3/galectin-9 pathway involves in the homeostasis of hepatic Tregs in a mouse model of concanavalin A-induced hepatitis. Molecular Immunology 58: 85–91. 10.1016/j.molimm.2013.11.001 [DOI] [PubMed] [Google Scholar]
- 3.Hoffmann F, Sass G, Zillies J, Zahler S, Tiegs G, Hartkorn A, et al. (2009) A novel technique for selective NF-kappaB inhibition in Kupffer cells: contrary effects in fulminant hepatitis and ischaemia-reperfusion. Gut 58: 1670–1678. 10.1136/gut.2008.165647 [DOI] [PubMed] [Google Scholar]
- 4.Erhardt A, Biburger M, Papadopoulos T, Tiegs G (2007) IL-10, regulatory T cells, and Kupffer cells mediate tolerance in concanavalin A-induced liver injury in mice. Hepatology 45: 475–485. [DOI] [PubMed] [Google Scholar]
- 5.Tiegs G, Hentschel J, Wendel A (1992) A T cell-dependent experimental liver injury in mice inducible by concanavalin A. J Clin Invest 90: 196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Z, Otsuka H, Yamaguchi K, Kuroishi T, Sasano T, Hartkorn A, et al. (2011) Roles of platelets and macrophages in the protective effects of lipopolysaccharide against concanavalin A-induced murine hepatitis. Biochimica et Biophysica Acta (BBA)—Molecular Basis of Disease 1812: 1069–1079. [DOI] [PubMed] [Google Scholar]
- 7.Dixon LJ, Barnes M, Tang H, Pritchard MT, Nagy LE (2013) Kupffer Cells in the Liver. Comprehensive Physiology: John Wiley & Sons, Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schumann J, Wolf D, Pahl A, Brune K, Papadopoulos T, van Rooijen N, et al. (2000) Importance of Kupffer cells for T-cell-dependent liver injury in mice. Am J Pathol 157: 1671–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikeda Y, Sciarretta S, Nagarajan N, Rubattu S, Volpe M, Frati G, et al. (2014) New Insights into the Role of Mitochondrial Dynamics and Autophagy during Oxidative Stress and Aging in the Heart. Oxidative Medicine and Cellular Longevity 2014: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levine B, Kroemer G (2008) Autophagy in the pathogenesis of disease. Cell 132: 27–42. 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine B, Klionsky DJ (2004) Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 6: 463–477. [DOI] [PubMed] [Google Scholar]
- 12.Klionsky DJ (2007) Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol 8: 931–937. [DOI] [PubMed] [Google Scholar]
- 13.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, et al. (2000) A ubiquitin-like system mediates protein lipidation. Nature 408: 488–492. [DOI] [PubMed] [Google Scholar]
- 14.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19: 5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perlmutter DH (2002) Liver injury in alpha1-antitrypsin deficiency: an aggregated protein induces mitochondrial injury. J Clin Invest 110: 1579–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N, Ohsumi Y, Yoshimori T (2002) Autophagosome formation in mammalian cells. Cell Struct Funct 27: 421–429. [DOI] [PubMed] [Google Scholar]
- 17.Lu G-D, Ang YH, Zhou J, Tamilarasi J, Yan B, Lim YC, et al. (2014) C/EBPalpha predicts poorer prognosis and prevents energy starvation–induced cell death in hepatocellular carcinoma. Hepatology: n/a-n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandra PK, Gunduz F, Hazari S, Kurt R, Panigrahi R, Poat B, et al. (2014) Impaired Expression of Type I and Type II Interferon Receptors in HCV-Associated Chronic Liver Disease and Liver Cirrhosis. PLoS ONE 9: e108616 10.1371/journal.pone.0108616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W-w, Yu J-y, Xu H-l, Bao J-k (2011) Concanavalin A: A potential anti-neoplastic agent targeting apoptosis, autophagy and anti-angiogenesis for cancer therapeutics. Biochemical and Biophysical Research Communications 414: 282–286. 10.1016/j.bbrc.2011.09.072 [DOI] [PubMed] [Google Scholar]
- 20.Kamo N, Ke B, Ghaffari AA, Shen X-d, Busuttil RW, Cheng G, et al. (2013) ASC/caspase-1/IL-1β signaling triggers inflammatory responses by promoting HMGB1 induction in liver ischemia/reperfusion injury. Hepatology 58: 351–362. 10.1002/hep.26320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, et al. (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. The Journal of Cell Biology 169: 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Goldman S, Baerga R, Zhao Y, Komatsu M, Jin S. (2009) Adipose-specific deletion of autophagy-related gene 7 (atg7) in mice reveals a role in adipogenesis. Proceedings of the National Academy of Sciences 106: 19860–19865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, He S, Zhou X, Ye Y, Tan S, Zhang S, et al. (2016) Lyn Delivers Bacteria to Lysosomes for Eradication through TLR2-Initiated Autophagy Related Phagocytosis. PLoS Pathog 12: e1005363 10.1371/journal.ppat.1005363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D, Wu M, Nelson DE, Pasula R, Martin WJ II, (0000) Alpha-1-antitrypsin expression in the lung is increased by airway delivery of gene-transfected macrophages. Gene Ther 10: 2148–2152. [DOI] [PubMed] [Google Scholar]
- 25.Yuan K, Huang C, Fox J, Laturnus D, Carlson E, Zhang B, et al. (2012) Autophagy plays an essential role in the clearance of Pseudomonas aeruginosa by alveolar macrophages. Journal of Cell Science 125: 507–515. 10.1242/jcs.094573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kannan S, Pang H, Foster DC, Rao Z, Wu M (2005) Human 8-oxoguanine DNA glycosylase increases resistance to hyperoxic cytotoxicity in lung epithelial cells and involvement with altered MAPK activity. Cell Death Differ 13: 311–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Budunova IV, Perez P, Vaden VR, Spiegelman VS, Slaga TJ, Jorcano JL. (1999) Increased expression of p50-NF-kappaB and constitutive activation of NF-kappaB transcription factors during mouse skin carcinogenesis. Oncogene 18: 7423–7431. [DOI] [PubMed] [Google Scholar]
- 28.Tiegs G, Hentschel J, Wendel A (1992) A T cell-dependent experimental liver injury in mice inducible by concanavalin A. The Journal of Clinical Investigation 90: 196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang CP, Yang MC, Liu HS, Lin YS, Lei HY (2007) Concanavalin A induces autophagy in hepatoma cells and has a therapeutic effect in a murine in situ hepatoma model. Hepatology 45: 286–296. [DOI] [PubMed] [Google Scholar]
- 30.Lei HY, Chang CP (2007) Induction of autophagy by concanavalin A and its application in anti-tumor therapy. Autophagy 3: 402–404. [DOI] [PubMed] [Google Scholar]
- 31.Chang CP, Lei HY (2008) Autophagy induction in T cell-independent acute hepatitis induced by concanavalin A in SCID/NOD mice. Int J Immunopathol Pharmacol 21: 817–826. [DOI] [PubMed] [Google Scholar]
- 32.Chang CP, Yang MC, Lei HY (2011) Concanavalin A/IFN-gamma triggers autophagy-related necrotic hepatocyte death through IRGM1-mediated lysosomal membrane disruption. PLoS One 6: e28323 10.1371/journal.pone.0028323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu F-C, Yu H-P, Hwang T-L, Tsai Y-F (2012) Protective Effect of Tropisetron on Rodent Hepatic Injury after Trauma-Hemorrhagic Shock through P38 MAPK-Dependent Hemeoxygenase-1 Expression. PLoS ONE 7: e53203 10.1371/journal.pone.0053203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng Y, Qiu F, Ye YC, Guo ZM, Tashiro S, Onodera S, et al. (2009) Autophagy inhibits reactive oxygen species-mediated apoptosis via activating p38-nuclear factor-kappa B survival pathways in oridonin-treated murine fibrosarcoma L929 cells. FEBS J 276: 1291–1306. 10.1111/j.1742-4658.2008.06864.x [DOI] [PubMed] [Google Scholar]
- 35.Cursio R, Colosetti P, Gugenheim J (2015) Autophagy and liver ischemia-reperfusion injury. Biomed Res Int 2015: 417590 10.1155/2015/417590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gupta NA, Kolachala VL, Jiang R, Abramowsky C, Shenoi A, Kosters A, et al. (2014) Mitigation of autophagy ameliorates hepatocellular damage following ischemia-reperfusion injury in murine steatotic liver. Am J Physiol Gastrointest Liver Physiol 307: G1088–1099. 10.1152/ajpgi.00210.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Czaja MJ, Ding WX, Donohue TM Jr., Friedman SL, Kim JS, Komatsu M, et al. (2013) Functions of autophagy in normal and diseased liver. Autophagy 9: 1131–1158. 10.4161/auto.25063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chien WS, Chen YH, Chiang PC, Hsiao HW, Chuang SM, Lue SI, et al. (2011) Suppression of autophagy in rat liver at late stage of polymicrobial sepsis. Shock 35: 506–511. 10.1097/SHK.0b013e31820b2f05 [DOI] [PubMed] [Google Scholar]
- 39.Lin CW, Lo S, Perng DS, Wu DB, Lee PH, Chang YF, et al. (2014) Complete activation of autophagic process attenuates liver injury and improves survival in septic mice. Shock 41: 241–249. 10.1097/SHK.0000000000000111 [DOI] [PubMed] [Google Scholar]
- 40.Tiegs G (2007) Cellular and cytokine-mediated mechanisms of inflammation and its modulation in immune-mediated liver injury. Z Gastroenterol 45: 63–70. [DOI] [PubMed] [Google Scholar]
- 41.Tiegs G (1997) Experimental hepatitis and role of cytokines. Acta Gastroenterol Belg 60: 176–179. [PubMed] [Google Scholar]
- 42.Bode JG, Ehlting C, Haussinger D (2012) The macrophage response towards LPS and its control through the p38(MAPK)-STAT3 axis. Cell Signal 24: 1185–1194. 10.1016/j.cellsig.2012.01.018 [DOI] [PubMed] [Google Scholar]
- 43.Duan WJ, Li QS, Xia MY, Tashiro S, Onodera S, et al. (2011) Silibinin activated ROS-p38-NF-kappaB positive feedback and induced autophagic death in human fibrosarcoma HT1080 cells. J Asian Nat Prod Res 13: 27–35. 10.1080/10286020.2010.540757 [DOI] [PubMed] [Google Scholar]
- 44.Wang C, Xia Y, Zheng Y, Dai W, Wang F, Chen K, et al. (2015) Protective effects of N-acetylcysteine in concanavalin A-induced hepatitis in mice. Mediators Inflamm 2015: 189785 10.1155/2015/189785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Xia Y, Liu T, Wang J, Dai W, Wang F, et al. (2015) Protective effects of astaxanthin on ConA-induced autoimmune hepatitis by the JNK/p-JNK pathway-mediated inhibition of autophagy and apoptosis. PLoS One 10: e0120440 10.1371/journal.pone.0120440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kusters S, Gantner F, Kunstle G, Tiegs G (1996) Interferon gamma plays a critical role in T cell-dependent liver injury in mice initiated by concanavalin A. Gastroenterology 111: 462–471. [DOI] [PubMed] [Google Scholar]
- 47.Lin WJ, Kuang HY (2014) Oxidative stress induces autophagy in response to multiple noxious stimuli in retinal ganglion cells. Autophagy 10: 1692–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Halestrap AP, Richardson AP (2014) The mitochondrial permeability transition: A current perspective on its identity and role in ischaemia/reperfusion injury. J Mol Cell Cardiol. [DOI] [PubMed] [Google Scholar]
- 49.Shaughnessy DT, McAllister K, Worth L, Haugen AC, Meyer JN, Domann FE, et al. (2014) Mitochondria, Energetics, Epigenetics, and Cellular Responses to Stress. Environ Health Perspect. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ott M, Gogvadze V, Orrenius S, Zhivotovsky B (2007) Mitochondria, oxidative stress and cell death. Apoptosis 12: 913–922. [DOI] [PubMed] [Google Scholar]
- 51.Gutterman DD (2005) Mitochondria and reactive oxygen species: an evolution in function. Circ Res 97: 302–304. [DOI] [PubMed] [Google Scholar]
- 52.Sun Q, Gao W, Loughran P, Shapiro R, Fan J, Billiar TR, et al. (2013) Caspase 1 activation is protective against hepatocyte cell death by up-regulating beclin 1 protein and mitochondrial autophagy in the setting of redox stress. J Biol Chem 288: 15947–15958. 10.1074/jbc.M112.426791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perrotta I, Carito V, Russo E, Tripepi S, Aquila S, Donato G. (2011) Macrophage autophagy and oxidative stress: an ultrastructural and immunoelectron microscopical study. Oxid Med Cell Longev 2011: 282739 10.1155/2011/282739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim I, Rodriguez-Enriquez S, Lemasters JJ (2007) Selective degradation of mitochondria by mitophagy. Arch Biochem Biophys 462: 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Western blotting analysis of Atg7, p63 and LC3 protein expression in Raw264.7 cells after ConA (10 μg/ml) treatment for 2 h. Disrupted consumption of p62 and interrupted conversion of LC3-II to LC3-I were observed in Atg7 siRNA transfected cells. B. Raw264.7 cells transfected with GFP-LC3 plasmid were treated with ConA (10 μg/ml) for 4 h (Arrows showing GFP-LC3 punctate regions). Rapamycin (50 nM) treatment for 24 h was used as positive control. GFP-LC3 punctate, the marker of autophagosome formation, was scarce to none in Atg7-silenced cells. Magnifications 1000X. Representative results were from 3 independent experiments. Data are presented as mean+SD; * p<0.05 vs. controls.
(TIF)
Immunofluorescence staining shows no significant difference in translocation of NF-κB p65 between Atg7-silenced cells or controls.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.