Abstract
We present results of the first large-scale interlaboratory study carried out in synthetic biology, as part of the 2014 and 2015 International Genetically Engineered Machine (iGEM) competitions. Participants at 88 institutions around the world measured fluorescence from three engineered constitutive constructs in E. coli. Few participants were able to measure absolute fluorescence, so data was analyzed in terms of ratios. Precision was strongly related to fluorescent strength, ranging from 1.54-fold standard deviation for the ratio between strong promoters to 5.75-fold for the ratio between the strongest and weakest promoter, and while host strain did not affect expression ratios, choice of instrument did. This result shows that high quantitative precision and reproducibility of results is possible, while at the same time indicating areas needing improved laboratory practices.
Introduction
Rapid improvements in our ability to both understand and genetically engineer biological organisms offer the potential for revolutionary applications for medicine, manufacturing, agriculture, and the environment [1–5]. A major barrier to transition from principle to practice, however, is the frequent sensitivity of biological systems to small changes in their cellular or environmental context [6]. This makes it difficult to reproduce or build on prior results in the lab, let alone to ensure desirable behavior in a deployed application, and may play a part in significant concerns that have been raised with respect to the state of published biomedical literature [7–10].
Practitioners of synthetic biology face particularly strong challenges in this area, because the engineering approaches applied in this area are often particularly demanding of quantitative precision in models and measurements. At the same time, synthetic biology offers a unique opportunity for improving our understanding of biological systems, through insertion of artificial systems intended to operate relatively independently from the evolved systems of their host or else to apply precise interventions to that host. Due to their engineered nature, these systems are likely to be more tractable to study than natural systems, as well as to provide leverage for the study of natural systems.
An important step toward addressing these issues is to quantify the degree of variability exhibited by engineered genetic constructs across different laboratories. Toward this end, we present the results of the first large-scale interlaboratory study of reproducibility in synthetic biology, carried out by students at 88 institutions around the world as part of the 2014 and 2015 International Genetically Engineered Machine (iGEM) competitions. This study focused on one of the most widely used tools in genetic engineering: constitutive expression of fluorescent protein from an engineered plasmid (although there are a number of well-known disadvantages to assays based on fluorescent proteins, such as oxygen dependence and folding times, these techniques are readily accessible, widely used, and still one of the best and easiest ways to study many biological phenomena). In particular, each participating team measured three engineered constitutive constructs in E. coli grown under standardized conditions following a particular protocol. Previously, a small-scale interlaboratory study with similar design demonstrated that it is possible for several laboratories to obtain relative measurements accurate to within a 2-fold range using flow cytometry [11]. Our new study enhances these results with a much wider range of participants and instruments, as well as identifying likely sources of variation and key challenges to be addressed.
In particular, we find that strong fluorescent expression can be measured with a remarkable degree of precision—to a maximum of 1.54-fold standard deviation. Precision degrades significantly, however, for constructs with weaker expression. Surprisingly, we find no significant correlation between strain and observed behavior, suggesting that the constructs measured are not overly sensitive to host context, while choice of instrument does appear to affect results. These results are promising, in that they show that engineered genetic constructs can exhibit a high degree of consistency in behavior even in the face of significant variations in context. At the same time, the limits encountered in this study highlight the need for adoption of calibrated measurements producing standardized units (e.g., [12, 13]) and for protocol approaches that can reduce the impact of “cultural art” in laboratory methods (i.e., the undocumented or undocumentable differences in how an apparently identifical measurement is executed differently between two individuals or laboratories, and in how the data is handled to turn “raw” observations into reported values), which can be reduced by methods such as [14] and [15]. Finally, this study also demonstrates how critical issues in science and reproducibility can be addressed through focused undergraduate research challenges and “citizen science” involving students around the world.
Materials and Methods
The aim of this study is to establish a broadly relevant baseline for precision and replicability in engineered biological systems. As such, we selected the following experimental conditions as being both readily accessible to laboratories at varying levels of sophistication world-wide, and also representative of much work in biological engineering:
Host organism: E. coli (preferably a DH5-alpha strain)
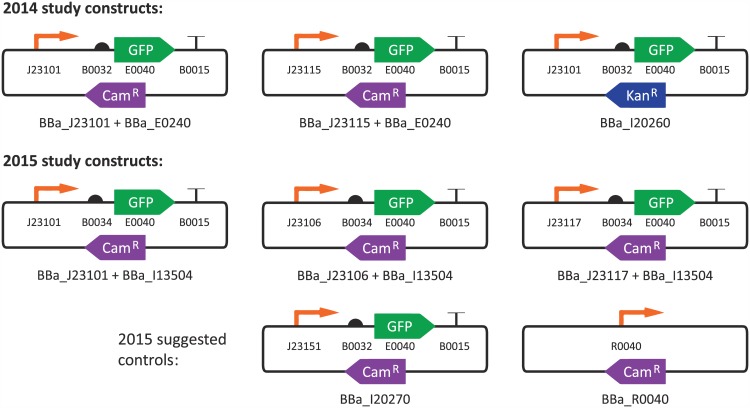
Engineered construct: constitutive expression of green fluorescent protein at three levels (“strong”, “medium”, and “weak”), driven by constitutive promoters from the Anderson promoter collection [16] (the same collection drawn from in [11]). Constructs are shown in Fig 1, with additional details provided in S1 File.
Molecular cloning: BioBricks Assembly [17] was recommended for building constructs, but not required; some teams substituted DNA synthesis or other assembly strategies.
Culture conditions: 16–18 hours of growth in LB broth plus appropriate antibiotic, at 37°C and shaking at 300 rpm.
Replication: triplicate
Measurement: green fluorescence measured as best capable, in absolute SI units if possible.
Fig 1. Constitutive fluorescence constructs measured in the 2014 and 2015 iGEM Interlab Studies.
In the 2014 iGEM Interlab Study, the protocol was intentionally left at this vague level of specification, for two reasons. The first goal was to determine what types of equipment were commonly accessible at participating institutions worldwide. The second goal was to determine how much variation was common in protocol execution and reporting, for both culturing and measurement.
Following analysis of the 2014 study, the protocol was adjusted with the aim of improving precision. In particular, the following key changes were made for the 2015 iGEM Study:
The set of constructs measured was changed to all use the same plasmid backbone (thus eliminating differences in growth and expression due to the use of different antibiotics) and to have a greater range of expression between constructs.
A set of defined positive and negative controls were added.
Standardized protocols were specified for construction, culturing, and measurement, along with checklists and forms for participating teams to fill out as they executed these protocols. These were split across two forms, attached as S2 and S3 Files.
When possible, teams were encouraged to provide data in both biological and technical triplicate (for a total of nine replicates). Standardized reporting of individual sample measurements also allowed elimination of anomalous samples: in cases where one sample differs from all the rest by at least 5-fold or one set of replicates differs by at least 3-fold (and values are not very close to zero), those samples were not used for analysis.
Both studies were advertised to all teams participating in iGEM through an information page on the iGEM website, as well as social media and iGEM newsletters. The information pages for the 2014 and 2015 studies are attached as S4 and S5 Files, respectively. Beginning from the initial advertisement, teams had several months in which to indicate that they were volunteering to collect data for the study, to execute the specified protocols and collect fluorescence data, and finally to return protocol records and fluorescence data via email or web forms. Finally, we analyzed the returned data and protocols and presented preliminary results and recognition of the contributing teams at each year’s iGEM Giant Jamboree.
For the 2014 study, 45 teams participated, and of these 36 teams were ultimately able to contribute data for inclusion in the study. For the 2015 study, 85 teams participated, and of these 67 teams were ultimately able to contribute data for inclusion in the study. There were 18 teams that participated in both studies, of which 15 contributed data to both studies and 2 contributed data to only one of the studies. The results presented in this paper thus represent experiments conducted at 88 institutions worldwide. These institutions are widely distributed across countries and continents: contributing teams hail from 27 different countries, with 42 teams from Europe, 19 teams from Asia, 19 teams from North America, 7 teams from Central and South America, and 1 team from Oceania. The vast majority of participants were undergraduate students, but many teams also included or were comprised of graduate students or high-school students. All members designated by a team as deserving co-authorship for their contributions are recognized as consortium authors in the Acknowledgments section.
Results
For the 2014 study, almost all contributing teams produced one usable set of measurements. Of the 36 teams contributing data, five teams measured samples with two instruments, but issues in data reporting means that all but two of those dual data sets had only one readily usable set of data. Accordingly, we analyze using only the best documented dataset for each team, a total of 36 datasets.
For the 2015 study, a number of teams decided to collect more than one dataset, using multiple instruments and/or multiple strains: all told, the 67 contributing teams produced a total of 95 datasets. This presents an additional opportunity for intra-team data analysis, which will be explored below in our examination of sources of variability.
All values in these datasets are reported in S1 and S2 Tables. In addition, data from the 2015 study protocol and measurement forms is included as S3 and S4 Tables.
Absolute vs. Relative Units
First and foremost in our analysis, we observed that almost none of the datasets include data in directly comparable SI units. Only two datasets from the 2014 study are reported in absolute units, and the units obtained for these datasets are not comparable. In the 2015 study, twelve teams reported at least one dataset in absolute units, generally calibrated to a standard fluorophore, most frequently fluorescein. Two groups of these datasets are directly comparable: two datasets are reported in MEFL (molecules of equivalent fluorescein), and four datasets are reported in ng/mL fluorescein.
In both of these groups of data, however, the ratios between constructs within the datasets are quite similar, while there are two or more magnitudes of difference across the absolute numbers reported. It thus appears likely that calibration techniques are being applied inconsistently. Highly inconsistent calibration is effectively no better than simply using arbitrary units to begin with. Thus, as is frequently the case in current biological research, we cannot directly compare measurements from one lab to another.
We can still, however, consider the precision and reproducibility of relative fluorescent expression. Accordingly, the remainder of our analysis will use normalized fluorescence, comparing the ratio of the constructs most similar between the two years (Strong14 and Strong15) to each other promoter measured. This gives us the following five sets of ratio data to analyze:
2014: Strong14/Medium14, Strong14/Weak14
2015: Strong15/Medium15, Strong15/Positive15, Strong15/Weak15
The exact construct for each device is noted in Fig 1 and S1 File. Note that for the 2015 study we include ratios with the suggested positive control, as teams used this construct for 75 data sets, producing a significant body of data to analyze. Negative controls, however, were highly variable in both identity and data handling, including many zero, negative, and near-zero numbers. This renders them a poor target for ratiometric analysis, and we thus do not include them in our analysis.
The remainder of our analysis focuses on the five expression ratios that we have identified here.
Replicability and Precision
In evaluating precision and replicability, we began with a hypothesis about the sources of variation that are likely to be encountered. In particular, we hypothesized that observed variation can be viewed as a mixture of three main sources of variation:
Mistakes and Failures: The most extreme variations will likely be driven by gross protocol failures, such as contamination of samples, incorrect assembly of constructs, mixing up samples, or mistakes in data interpretation and entry. The occurrence of such failures is often best addressed by replication and by improvement of controls and procedures, such that mistakes and failures can be more easily detected and an affected experiment repeated.
Communication-Based Variability: Some variations are driven by differences in how various people in various laboratories interpret a given experimental protocol. These types of variation can often be addressed by standardization and training, and by more precise communication of protocols.
Systemic Variability: Some variation is due to the nature of the system under observation, e.g., inherent variability of biological organisms, precision limits of instruments, variation in reagents, environmental differences (e.g., laboratory altitude). This type of variation is often the most difficult to address, as it is likely to involve fundamental, technological, or economic limits.
Single-cell fluorescence levels tend to vary following an approximately log-normal distribution (see e.g., [12, 18–21]). As such, we hypothesize that the ratios in the data-set will also vary following a log-normal distribution, plus some outliers caused by mistakes and failures (Note that it is for this reason that we report variability as x-fold rather than ±x: a plus/minus range on a log-scale translates to a multiply/divide range in the standand linear scale. Thus, for example, one 2-fold standard deviation around a mean of 10 is a range of 5 to 20, and two standard deviations is a range from 2.5 to 40).
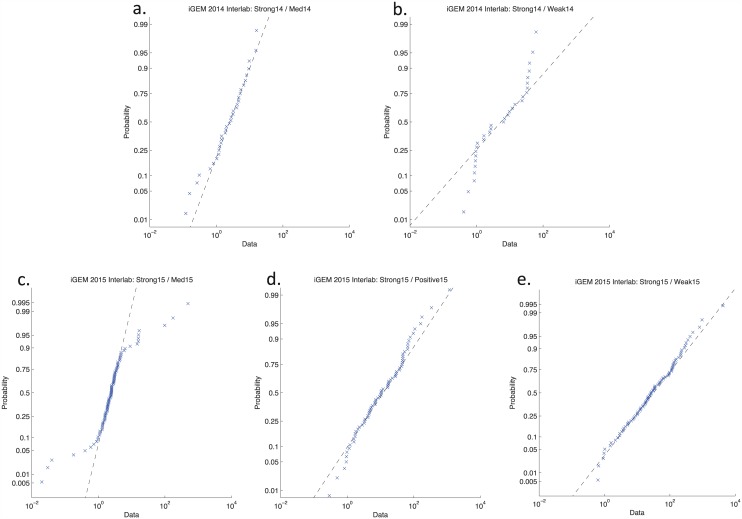
To evaluate this hypothesis, we constructed log-normal distribution diagrams for each fluorescence expression ratio, in which sorted data is plotted against the expected probability distribution (Fig 2). The mostly linear structure of these plots indicates that variation in expression does generally conform closely to log-normal distribution, with more vertical clustering indicating tighter distributions.
Fig 2. Log-Normal distribution diagrams for fluorescence expression ratios.
The mostly linear structure indicates that variation in expression does generally conform closely to log-normal distribution, with more vertical clustering indicating tighter distributions. Data points significantly off the line will be considered outliers and not used in further analysis of these distributions.
Data points significantly off of the linear distribution will be considered outliers: Table 1 gives the number of points that we are interpreting as outliers above and below the log-normal distribution of each data set. These we attribute to mistakes and failures and will not use in further analysis aimed at understanding the relationship between precision of communication and system variability. In total, 17% of all data points are outliers, and the rate of outliers in the 2015 data set is lower than in the 2014 data set. We hypothesize that the decreased rate of outliers is due to the use of more detailed protocol instructions and checklists, but there is not enough data to conclude that this difference is necessarily significant.
Table 1. Number of data points total for each ratio, along with number and fraction of data points that we interpret as outliers (i.e., not conforming to the log-normal distribution hypothesis).
| Expression Ratio | Samples | Non-Outliers | Outliers | ||
|---|---|---|---|---|---|
| High | Low | Fraction | |||
| Strong14/Medium14 | 34 | 28 | 1 | 5 | 0.18 |
| Strong14/Weak14 | 32 | 18 | 7 | 7 | 0.43 |
| Strong15/Medium15 | 94 | 77 | 9 | 8 | 0.18 |
| Strong15/Positive15 | 71 | 55 | 9 | 7 | 0.22 |
| Strong15/Weak15 | 90 | 89 | 0 | 1 | 0.01 |
| Total | 321 | 267 | 26 | 28 | 0.17 |
It is also possible that some outliers may be due to sequence variations induced by different assembly methods, since assembly scars are known to have a significant effect on promoter expression [22]. This appears to be unlikely, however: in 2015, when teams were required to report their assembly method, 10 data sets were produced with methods other than BioBricks. These account for only three Strong15/Medium14, three Strong15/Positive15, and no Strong15/Weak15 outliers, a proportion in line with the overall fraction of outliers.
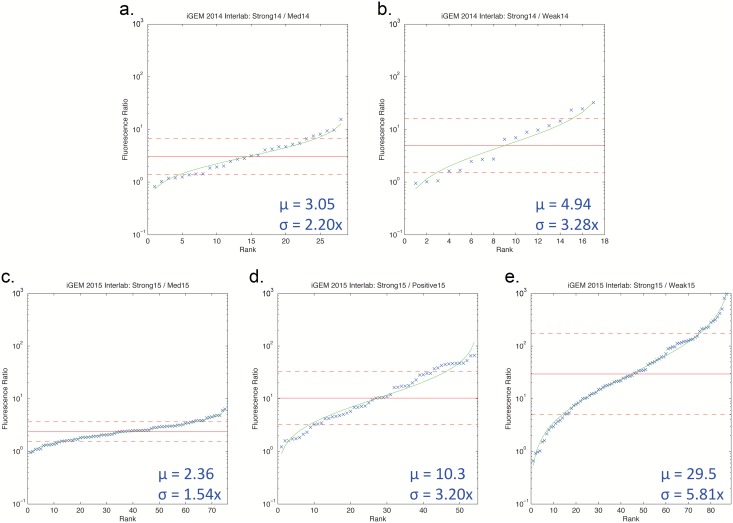
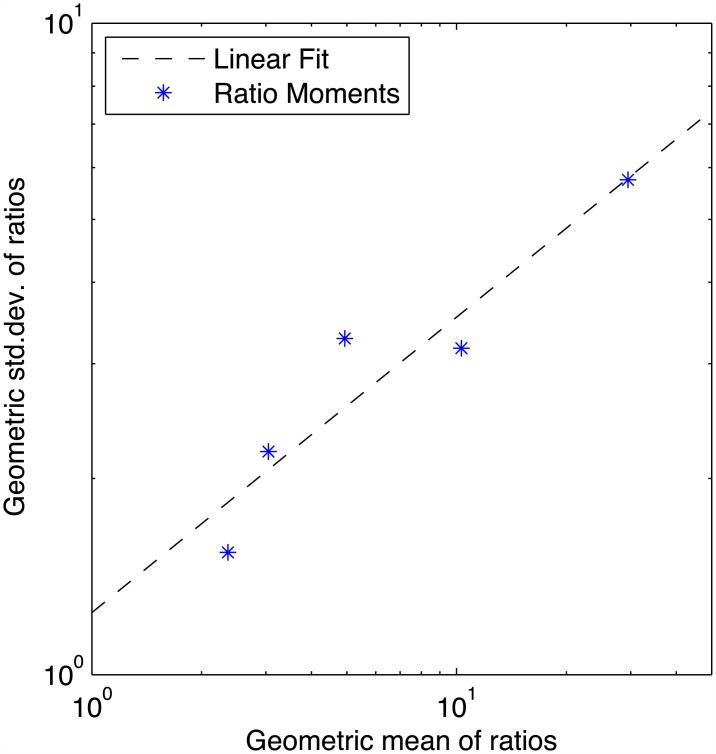
Turning to the non-outlier data, Fig 3 shows a plot of rank-sorted data against a log-normal distribution model, along with the mean (μ) and standard deviation (σ) for each distribution. Surprisingly, there is a great degree of variation in the precision of measurements. The lower a promoter’s expression is in relation to the Strong14 or Strong15 construct, the higher the variation in measurement. Fig 4 plots mean against standard deviation, showing that standard deviation grows approximately in proportion to the square root of the mean ratio. This might indicate either that measurement is less precise for weak fluorescence or that error grows as the two values grow farther apart. We can test which is more likely by considering the other four ratios: Medium14/Weak14, Medium15/Positive15, Medium15/Weak15, and Positive15/Weak15. S1 and S2 Figs show the results, which indicate that error appears to be controlled more strongly by the denominator, suggesting that the problem is in fact decreased precision in quantifying weak fluorescence. This conclusion is further supported by our analysis of instrument-linked variation in the next section.
Fig 3. Rank-sorted data for each ratio, omitting outliers.
Blue points are observed ratios, red lines show geometric mean (solid) and ±1 std.dev. (dashed), and green line shows log-normal distribution fit.
Fig 4. Relationship between mean ratio and precision.
The lower another promoter’s expression is in relation to the strong construct, the higher the variation in measurement: standard deviation grows approximately in proportion to the square root of the mean ratio.
Sources of Imprecision
To begin investigation of possible sources of variation, we identified the two largest and most systematic differences in protocol from team to team: the strain of E. coli used for culturing, and the instrument used to measure behavior. For this analysis, we coded strains into four categories, as shown in Table 2, and instruments into five categories, as shown in Table 3.
Table 2. Strains reported in 2014 and 2015 iGEM Interlab Studies.
| Strain | 2014 | 2015 |
|---|---|---|
| DH5-alpha | 16 | 50 |
| TOP10 | 7 | 16 |
| BL21 | 2 | 9 |
| Other | 6 | 20 |
| Not reported | 5 | — |
Table 3. Instruments reported in 2014 and 2015 iGEM Interlab Studies.
| Instrument | 2014 | 2015 |
|---|---|---|
| Plate Reader | 23 | 56 |
| Flow Cytometer | 6 | 25 |
| Microscope | 3 | 7 |
| Other Spectrofluorimeter | 2 | 5 |
| Other | 2 | 2 |
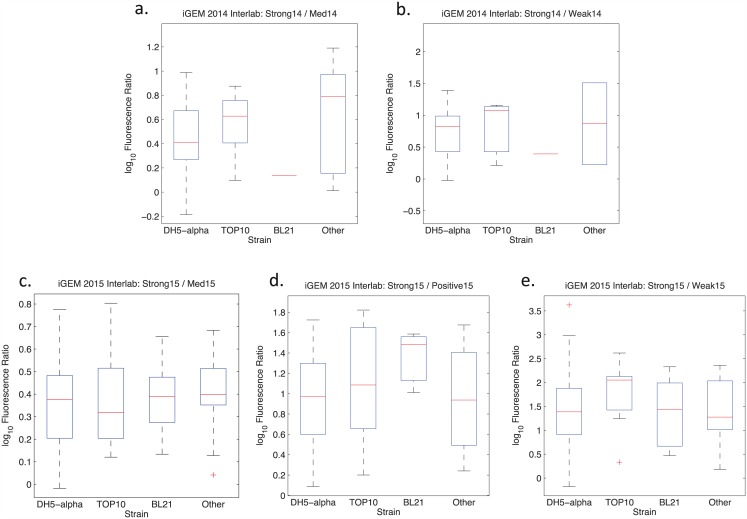
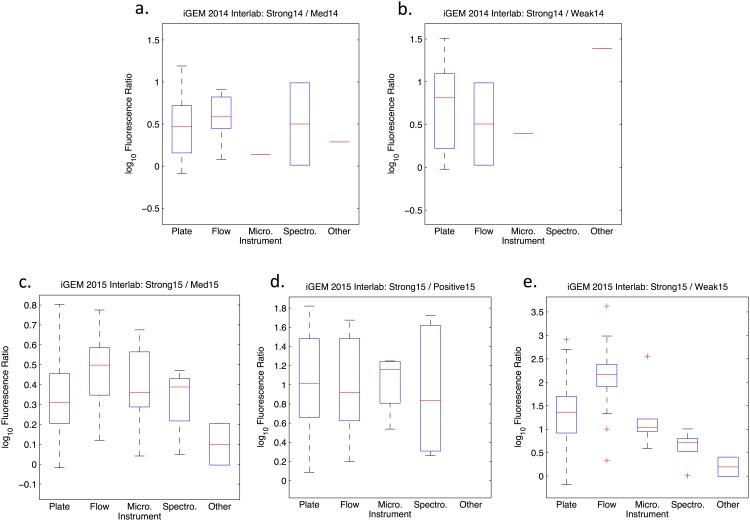
The results, shown in Figs 5 and 6, were unexpected. While we had expected to see significant variation by strain, the distributions were for the most part quite consistent from strain to strain. Between classes of instruments, however, there appears to be more variability. Most notably, there is more than a 6-fold difference between the mean values returned by flow cytometry and by other methods for the Strong15/Weak15 ratio. Such instrument-linked variability might be due to the instruments themselves, but could also be due to other causes, such as differences in how measurement and analysis is carried out on different instruments by different people.
Fig 5. Distributions of data partitioned by E. coli strain for each ratio, omitting outliers.
Note that the BL21 subset in 2014 has only a single non-outlier data point and may thus be effectively ignored.
Fig 6. Distributions of data partitioned by measuring instrument for each ratio, omitting outliers.
Note that some subsets have only one or zero non-outlier data points and may thus be effectively ignored.
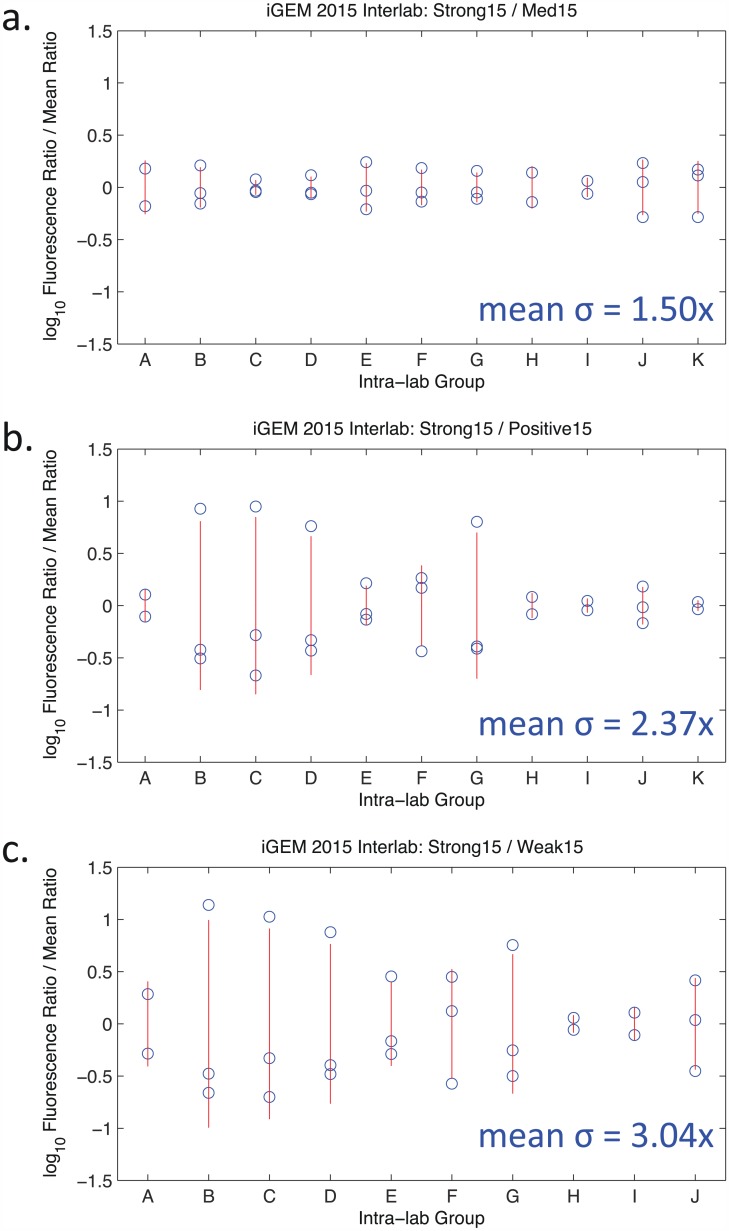
To further investigate the possibility that instrument-linked variation is a source of significant error, we take advantage of the fact that a number of the contributing teams for the 2015 study contributed multiple data sets per sample by measuring the same samples with different instruments. For the ratios with Medium15 and Positive15, there are 11 available data-sets, and for Weak15 there are 10 available data-sets. Most of these have three measurements, two of which are a plate reader and a flow cytometer. If instrument-linked variation is a driving force in imprecision, then the intra-team variation should be similar in scale to the team-to-team variation reported in Fig 3. This is in fact the case, as shown in Fig 7. Most importantly, note that instrument-linked variation is larger for ratios comparing with lower expression samples, just as it is for variation across data sets overall.
Fig 7. Instrument-to-instrument variation within a single team.
Each column represents a set of replicates measured by the same laboratory on different instruments. Blue circles are the ratios for individual instruments, normalized by mean ratio; the red line spans ±1 std.dev.
As a cross-check, we also compute the variation between replicates. This variation is much tighter: the geometric mean of geometric standard deviations across ratios for individual samples is only 1.24-fold, meaning that the fundamental measurement variation of individual instruments can account for at most approximately half of the tightest observed distribution.
Together, all of these various pieces of evidence support the hypothesis that measurement precision is poor for weak fluorescence, and that this is largely due to differences in how measurement and analysis is carried out by various laboratories, rather than systemic variability in either the biological organisms or the instruments themselves.
Discussion
Our results reveal a mixed picture of reproducibility and precision for engineered expression of fluorescence. On the positive side, the fraction of outliers observed is not too great, and the distributions of results conforms overall quite nicely with the prediction of log-normal distribution. Furthermore, we see that relative expression can be quite tightly quantified for pairs of strong fluorescent proteins.
On the negative side, however, the lack of standardized absolute units limits analysis to comparison of relative levels, which are much less useful than absolute units. Critically, we believe that the difficulty of performing “quality control” on a data set is a key contributor to both the number of outliers and to the variation between non-outlier measurements, since mistakes and failures cannot be readily excluded. Absolute unit measurements can allow control samples to be used for more stringent quality control of experiments by direct comparison of control measurements against expected standard values, thereby allowing detection of experimental or analytic problems and potentially greatly improving data quality (e.g., as described in [23]).
The difficulty of quantifying larger ratios is also quite concerning, as is the significant variation of values from instrument to instrument, even for the same samples within a single laboratory. We hypothesize that this phenomenon indicates general difficulties in quantifying weaker levels of fluorescent expression. It is not surprising that weak signals are more difficult to quantify than strong signals. What is surprising is that the dynamic range of effective quantification appears to be so narrow, since fluorescence is routinely quantified across much wider dynamic ranges than the mean strong/weak ratio observed in this study. Our suspicion is that this is not fundamentally due to the instruments (though flow cytometers do typically have a wider dynamic range than the other instruments used), but rather due to differences in how various laboratories deal with background fluorescence and with data interpretation and analysis.
One possible criticism of this study is the predominance of undergraduate researchers. We argue, however, that undergraduate researchers are more representative of the actual culture and practice of a laboratory than is perhaps comfortable to admit. There is a good deal of similarity between undergraduate researchers and the situation of a new graduate student or even a postdoctoral researcher entering a laboratory and learning the particulars of its protocols, equipment, and practices. This may be further intensified by the interdisciplinary nature of the field, meaning that even senior researchers often come from highly diverse backgrounds.
Overall, we think that the results of this study are largely good news for synthetic biology and for any other biological research that makes use of engineered expression of fluorescent proteins. First, the Anderson collection promoters seem to be fairly stable in their performance between different strains of E. coli, meaning that they are indeed generally a good starting point for tuning of gene expression. Second, we have established a clear baseline for precision and reproducibility. Since this has been established with the aid of undergraduates at a wide variety of institutions around the world, we argue that it should be considered as strong minimum standard for accuracy and reproducibility of published work.
Paradoxically, the measurement problems highlighted in this study may also be good news. Is is not surprising that this study shows problems in precision and reproducibility: challenges with variation in behavior and difficulty in reproducing and building upon results are commonly voiced concerns within the field. What is surprising and significant, however, is that they appear to be tied closely to the use of instruments and/or analysis of data. The instruments themselves appear to be quite reliable, as indicated by the tight replicate-to-replicate distribution. If the instruments are reliable, yet the same sets of samples produce markedly different relative values on different instruments, then this indicates that the problem is likely to be in how people are assaying cells with their instruments and the how they are analyzing the data produced by those instruments. This is likely to be compounded by differences in sensitivity and detection range: for example, flow cytometers typically have a wider range than plate readers, and thus a flow cytometer data set is likely to be less sensitive to how weak signals and background fluorescence are handled. In short, these simple biological systems appear to be much less variable than the ways in which we are studying them. This is good news because if the variability in precision and reproducibility was primarily systemic—i.e., due to the biological systems being studied, then it might be very difficult or even impossible to make significant improvements. Instead, however, much of the observed variability appears to be communication-based, i.e., closely tied to the ways in which people are studying those systems, and there are thus many good possibilities for rapid improvement in precision.
In particular, to address variability in fluorescent measurement, we recommend pursuit of three methods for increasing precision, listed here in order of ease of adoption, are:
Dissemination, training, and standardization around improved protocols for calibration of assays. There are well-established protocols for calibration of flow cytometers to absolute units [12, 23–26], but these have not yet been widely adopted outside of the medical diagnostic community. Other calibration protocols exist for measurement of population fluorescence (e.g., [13, 27, 28]), but these also have not been effectively disseminated through the community.
Removing “craft” from protocol execution. Much of the training that scientists receive in wet-lab protocols has an aspect of “apprenticeship” about it: important “craft” information is transmitted that is not formally written down in any location, and this makes it more difficult for protocols to be correctly replicated. A number of systems have been developed which make attempt to regularize the manner in which humans execute protocols (e.g., [14, 29]). These approaches still have lab work being done by humans, but apply techniques such as detailed checklists and computer monitoring of protocol execution to decrease the skill required and the variability of results produced by humans executing protocols.
Increased automation of protocols. Variability in protocol execution can be reduced yet further by entirely or almost-entirely automating the execution of protocols. The two main approaches currently being developed in these areas are robotics (e.g., [30–33]) and microfluidics (e.g., [15, 34]). In the long run, this is where most of the field will likely go, just as automation has produced gains in productivity and precision in most other fields. At present, adopting protocol automation is impractical for most laboratories, as doing so generally requires a large investment of time and resources. Ongoing miniaturization and models for outsourcing to “cloud labs,” however, may soon make this technology much more widely accessibly.
Effective use of any of these approaches also requires deepening our understanding of both the systems we study and the means applied for studying them. We need to know which aspects of protocols are most sensitive, and which are susceptible to optimization and modification to fit circumstances, we need better controls and procedures for eliminating outliers, and we also need to extend and apply all of these principles beyond constitutive fluorescence in E. coli to a much wider range of organisms and engineered systems. Toward this end, we are making the dataset collected in this study available to the scientific community for deeper, multifactorial analysis, and intend to organize further interlaboratory studies putting our hypotheses to the test and extending their range of study.
Supporting Information
(PDF)
(PDF)
(PDF)
Document provided is a snapshot of the page as of October 4th, 2015, which includes information presented on participation and the preliminary results of the study.
(PDF)
Document provided is a snapshot of the page as of October 4th, 2015.
(PDF)
(CSV)
(CSV)
Some teams returned their forms (or portions thereof) manually for a number of reasons, most frequently due to Internet censorship in their home countries; entries from these teams are not included in the attached form. Only technical entries for the form are included, and team names have been replaced by numbers corresponding to the numbers in the summary data table.
(CSV)
Some teams returned their forms (or portions thereof) manually for a number of reasons, most frequently due to Internet censorship in their home countries; entries from these teams are not included in the attached form. Only technical entries for the form are included, and team names have been replaced by numbers corresponding to the numbers in the summary data table.
(CSV)
- Medium14/Weak14
- Medium15/Positive15
- Medium15/Weak15
- Positive15/Weak15
(PDF)
- Medium14/Weak14
- Medium15/Positive15
- Medium15/Weak15
- Positive15/Weak15
(PDF)
Acknowledgments
The authors wish to thank Sarah Munro and Marc Salit of NIST for help in designing this study.
Consortium authors include all persons self-identified by contributing teams as deserving co-authorship credit. Contributors are listed alphabetically within team, and teams alphabetically and by year. Note that some persons may be credited as contributing in both years. Team names are given as identified in iGEM records: full details of each team’s institution and additional members may be found online in the iGEM Foundation archives at:
http://year.igem.org/Team:name
e.g.: full information on the 2015 ETH_Zurich team may be found at:
http://2015.igem.org/Team:ETH_Zurich
2014 iGEM Interlab Study Contributors
Aachen: Philipp Demling, Rene Hanke, Michael Osthege, Anna Schechtel, Suresh Sudarsan, Arne Zimmermann
Aalto-Helsinki: Bartosz Gabryelczyk, Martina Ikonen, Minnamari Salmela
ATOMS Turkiye: Muradıye Acar, Muhammed Fatih Aktas, Furkan Bestepe, Furkan Sacit Ceylan, Sadık Cigdem, Mikail Dohan, Mustafa Elitok, Mehmet Gunduz, Esra Gunduz, Omer Faruk Hatipoglu, Turan Kaya, Orhan Sayin, Safa Tapan, Osman Faruk Tereci, Abdullah Uçar, Mustafa Yilmaz
Austin_Texas: Jeffrey Barrick, Alex Gutierrez, Dennis Mishler, Jordan Monk, Kate Mortensen, Nathan Shin, Ella Watkins
BIT: Yintong Chen, Yuji Jin, Yuanjie Shi, Haoqian Myelin Zhang
Brasil-SP: Bruno Ono, Ieda Maria Martinez Paino, Lais Ribovski, Ivan Silva, Danilo Keiji Zampronio
Braunschweig: Nils Birkholz, Rudiger Frederik Busche, Oliver Konzock, Steffen Lippold, Carsten Ludwig, Melanie Philippi, Lukas Platz, Christian Sigismund, Susanne Weber, Maren Wehrs, Niels Werchau, Anna Wronska, Zen-Zen Yen
BostonU: Yash Agarwal, Evan Appleton, Douglas Densmore, Ariela Esmurria, Kathleen Lewis, Alan Pacheco
Carnegie_Mellon: Marcel Bruchez, Danielle Peters, Cheryl Telmer, Lena Wang
Colombia: Silvia Canas-Duarte, Daniel Giraldo-Perez, Camilo Gomez-Garzon, Jorge Madrid-Wolff, Nathaly Marin-Medina, Valentina Mazzanti, Laura Rodriguez-Forero, Eitan Scher
CU-Boulder: Robin Dowell, Samantha O’Hara, Cloe Simone Pogoda, Kendra Shattuck
DTU-Denmark: Ali Altintas, Anne Pihl Bali, Rasmus Bech, Anne Egholm, Anne Sofie Laerke Hansen, Kristian Jensen, Kristian Barreth Karlsen, Caroline Mosbech
Evry: Sophia Belkhelfa, Noemie Berenger, Romain Bodinier, Cecile Jacry, Laura Matabishi-Bibi, Pierre Parutto, Julie Zaworski
Groningen: Andries de Vries, Freek de Wijs, Rick Elbert, Lisa Hielkema, Chandhuru Jagadeesan, Bayu Jayawardhana, Oscar Kuipers, Anna Lauxen, Thomas Meijer, Sandra Mous, Renske van Raaphorst, Aakanksha Saraf, Otto Schepers, Oscar Smits, Jan-Willem Veening, Ruud Detert Oude Weme, Lianne Wieske
Imperial: Catherine Ainsworth, Xenia Spencer-Milnes
ITESM-CEM: Alejandro GómezÁvila, Eddie Cano Gamez, Ana Laura Torres Huerta, Carlos Alejandro Meza Ramirez
LMU-Munich: Philipp Popp, Jara Radeck, Anna Sommer
LZU-China: Xiangkai Li, Qi Wu, Hongxia Zhao, Ruixue Zhao
METU_Turkey: Irem Bastuzel, Yasemin Ceyhan, Mayda Gursel, Burak Kizil, Ilkem Kumru, Yasemin Kuvvet, Helin Tercan, Seniz Yuksel
NU_Kazakhstan: Luiza Niyazmetova
Oxford: Timothy Ang, Lucas Black, Ciaran Kelly, George Wadhams
Paris_Bettencourt: Clovis Basier, Urszula Czerwinska
Sumbawagen: Cindy Suci Ananda, Muhammad Al Azhar, Adelia Elviantari, Maya Fitriana, Arief Budi Witarto, Yulianti
SUSTC-Shenzhen: Jia Fangxing, Qingfeng Hou, Wan Pei, Chen Rifei, Wang Rong, Huang Wei, Zhang Yushan
SYSU-China: He Jianguo, Dengwen Lai, Pai Li, Jianheng Liu, Chunyang Ni, Qianbin Zhang
Tec-Monterrey: Cinthya Cadenas, Eduardo J. Zardain Canabal, Claudia Nallely Alonso Cantu, Mercedes Alejandra Vazquez Cantu, Eduardo Cepeda Canedo, Cesar Miguel Valdez Cordova, Jose Alberto de la Paz Espinosa, Carlos Enrique Alavez Garcia, Ana Laura Navarro Heredia, Adriana Hernandez, Sebastian Valdivieso Jauregui, Eduardo Ramirez Montiel, Eduardo Serna Morales, Yamile Minerva Castellanos Morales, Omar Alonso Cantu Pena, Eduardo A. Ramirez-Rodríguez, Elizabeth Vallejo Trejo
UANL_Mty-Mexico: Jesus Gilberto Rodriguez Ceja, Jesus Eduardo Martinez Hernandez, Mario Alberto Pena Hernandez, Enrique Amaya Perez, Rebeca Paola Torres Ramirez, J. Claudio Moreno Rocha, Alber Sanchez, Claudia Melissa Guerra Vazquez
uOttawa: Martin Hanzel, Sarah Mohand-Said, Shihab Sawar, Dylan Siriwardena, Alex Tzahristos
Uppsala: Nils Anlind, Martin Friberg, Erik Gullberg, Stephanie Herman
Utah_State: Dallin Christensen, Sara Gertsch, Cody Maxfield, Charles Miller, Ryan Putman
Valencia_UPV: Christine Bauerl, Lucia T Estelles Lopez, Estefania Huet-Trujillo, Marta Vazquez Vilar
Wageningen_UR: Marlène Sophie Birk, Nico Claassens, Walter de Koster, Rik van Rosmalen, Wen Ying Wu
Warwick: Sian Davies, Dan Goss, William Rostain, Chelsey Tye, Waqar Yousaf
WPI-Worcester: Natalie Farny, Chloe LaJeunesse, Alex Turland
XMU-China: An Chen, Jielin Chen, Yahong Chen, Zehua Chen, Baishan Fang, Xiaotong Fu, Xifeng Guo, Yue Jiang, Yiying Lei, Jianqiao Li, Zhe Li, Chang Liu, Weibing Liu, Yang Liu, Yizhu Lv, Qingyu Ruan, Yue Su, Chun Tang, Yushen Wang, Fan Wu, Xiaoshan Yan, Ruihua Zhang, Tangduo Zhang
Yale: Farren Isaacs, Ariel Leyva-Hernandez, Natalie Ma, Stephanie Mao, Yamini Naidu
2015 iGEM Interlab Study Contributors
Aalto-Helsinki: Tuukka Miinalainen
Aix-Marseille: Marion Aruanno, Daniel Calendini, Yoann Chabert, Gael Chambonnier, Myriam Choukour, Ella de Gaulejac, Camille Houy, Axel Levier, Loreen Logger, Sebastien Nin, Valerie Prima, James N. Sturgis
Amoy: Beibei Fang
ATOMS-Turkiye: Sadik Cigdem, Turan Kaya, Abdullah Ucar
Austin_UTexas: Alejandro Gutierrez, Dennis Mishler, Revanth Poondla, Sanjana Reddy, Tyler Rocha, Natalie Schulte, Devin Wehle
Bielefeld-CeBiTec: Marta Eva Jackowski
Birkbeck: Sean Ross Craig, Ariana Mirzarafie-Ahi, Elliott Parris, Luba Prout, Barbara Steijl, Rachel Wellman,
BIT: Zhao Fan, Zhang Jing, Yang Wei, Yang Yuanzhan, Wen Zhaosen
BostonU: Evan Appletion, Jeffrey Chen, Abha Patil, Shaheer Priracha, Kate Ryan, Nick Salvador, John Viola
Brasil-USP: Camila Maria S. Boralli, Camila Barbosa Bramorski, Juliana Cancino-Bernardi, Ana Laura de Lima, Paula Maria Pincela Lins, Cristiane Casonato Melo, Deborah Cezar Mendonca, Thiago Mosqueiro, Lais Ribovski, Everton Silva, Graziele Vasconcelos
Carnegie_Mellon: Ruchi Asthana, Donna Lee, Cheryl Telmer, Michelle Yu
CityU_HK: Peter Choi, Effie Lau, Kenneth Lau, Oscar Ying
Cork_Ireland: Brandon Malone, Paul Young
CSU_Fort_Collins: Aidan Ceney, Dakota Hawthorne, Sharon Lian, Sam Mellentine, Dylan Miller, Barbara Castro Moreira, Christie Peebles, Olivia Smith, Kevin Walsh, Allison Zimont
CU_Boulder: Michael Brasino, Michael Donovan, Hannah Young
Czech_Republic: Jan Bejvl, Daniel Georgiev, Hynek Kasl, Katerina Pechotova, Vaclav Pelisek, Anna Sosnova, Pavel Zach
Duke: Anthony Ciesla, Benjamin Hoover
Edinburgh: Elliott Chapman, Jon Marles-Wright, Vicky Moynihan, Liusaidh Owen, Brooke Rothschild-Mancinelli
EPF_Lausanne: Emilie Cuillery, Joseph Heng, Vincent Jacquot, Paola Malsot, Rocco Meli, Cyril Pulver, Ari Sarfatis, Loic Steiner, Victor Steininger, Nina van Tiel, Gregoire Thouvenin, Axel Uran
ETH_Zurich: Lisa Baumgartner, Anna Fomitcheva, Daniel Gerngross, Verena Jagger, Michael Meier, Anja Michel
Exeter University: Jasmine Bird, Bradley Brown, Todd Burlington, Daniel Herring, Joseph Slack, Georgina Westwood, Emilia Wojcik
Freiburg: Julian Bender, Julia Donauer, Ramona Emig, Rabea Jesser, Julika Neumann, Lara Stuhn
Gifu: Takema Hasegawa, Tomoya Kozakai, Haruka Maruyama
Glasgow: Sean Colloms, Charlotte Flynn, Vilija Lomeikaite, James Provan
HUST-China: Kang Ning, Shuyan Tang, Guozhao Wu, Yunjun Yang, Zhi Zeng, Yi Zhan
HZAU-China: Pan Chu, Jun Li, Keji Yan
IISER_Pune: Chaitanya A. Athale, Swapnil Bodkhe, Manasi Gangan, Harsh Gakhare, Yash Jawale, Snehal Kadam, Prachiti Moghe, Gayatri Mundhe, Neha Khetan, Ira Phadke, Prashant Uniyal, Siddhesh Zadey
KU Leuven: Ines Cottignie, Eline Deprez, Astrid Deryckere, Jasper Janssens, Frederik Jonnaert, Katarzyna Malczewska, Thomas Pak, Johan Robben, Ovia Margaret Thirukkumaran, Vincent Van Deuren, Laurens Vandebroek, Laura Van Hese, Laetitia Van Wonterghem, Leen Verschooten, Moritz Wolter
Leicester: Joss Auty, Richard Badge, Liam Crawford, Raymond Dalgleish, Amy Evans, Cameron Grundy, Charlie Kruczko, Payal Karia
Lethbridge: Graeme Glaister, Rhys Hakstol, Seme Mate, Karin Otero, Dustin Smith, Jeff Tingley, Hans-Joachim Wieden
LZU-China: Xiangkai Li, Haotian Wang, Qi Wu, Ningning Yao, Ruixue Zhao
Marburg: Matthias Franz, Anna Knoerlein, Nicolas Koutsoubelis, Anne C. Loechner, Max Mundt, Alexandra Richter, Oliver Schauer
METU_Turkey: Ilkem Kumru
MIT: Marjorie Buss, Sivateja Tangirala, Brian Teague
Nankai: Tianyi Huang, Xinhao Song, Yibing Wei, Zhaoran Zhang
NEAU-China: Longzhi Cao, Cheng Li, Kang Yang
NJAU_China: Zhiqin Chen, Yuxing Fang, Libo Sun, Weiyi Wang, Yang Yang
Northeastern_Boston: David Adams, Joshua Colls, Ariela Esmurria, Joshua Timmons, David Urick
NTNU_Trondheim: Julia Anna Adrian, Madina Akan, Youssef Chahibi, Rahmi Lale, Typhaine Le Doujet, Marit Vaagen Roee
NU_Kazakhstan: Altynay Abdirakhmanova, Askarbek Orakov, Azhar Zhailauova
OUC-China: Jinyang Liang, Yu Ma, Qikai Qin, Yetian Su
Oxford: Ju Yeon Han, Raphaella Hull, Wei Chung Kong, Li Chieh Lu, Duke Quinton
Paris-Saclay: Pauline Aubert, Johan Bourdarias, Olivier Bugaud, Coralie Demon-Chaine, Isabelle Hatin, Ibtissam Kaid-Slimane, Seong Koo Kang, Audrey Moatti, Cheikh Fall Ndiaye
Pasteur_Paris: Mathilde Ananos, Alexander Arkhipenko, Valentin Bailly, Jules Caput, Javier Castillo, Alma Chapet-Batlle, Floriane Cherrier, Claudia Demarta-Gatsi, Deshmukh Gopaul, Muriel Gugger, Caroline Lambert, Lucas Krauss, Amelie Vandendaele
SCUT: Li Xiaojing, Lin Xiaomei, Luo Xunxun
SDU-Denmark: Anders Chr. Hansen, Tina Kronborg, Jens S. Pettersen
Stanford-Brown: Charles Calvet, Tyler Dae Devlin, Kosuke Fujishima, Danny Greenberg, Tina Ju, Ryan Kent, Daniel Kunin, Erica Lieberman, Griffin McCutcheon, Thai Nguyen, Lynn Rothschild, Joseph D. Shih, Jack Takahashi, Kirsten Thompson, Forrest Tran, Daniel Xiang
Stockholm: Felix Richter
SYSU-Software: Yang Xiaoran, Hu Xiangyue
SZMS_15_Shenzhen: Changyuan Deng, Shuyu Hua, Yumeng Li, Xinyu Meng, Boxiang Wang, Yingqi Wang, Xuan Wang, Zixuan Xu, Jieyu Yan, Ming Yan, Yineng Zhou
TecCEM: Edgar Alberto Alcalá Orozco, José Alberto Cristerna Bermúdez, Daniela Flores Gómez, José Ernesto Hernández Castañeda, Diana Clarisse Montaño Navarro, Juana Yessica PérezÁvila, María Fernanda Salazar Figueroa, María Fernanda Sánchez Arroyo, Oliva Angélica Sánchez Montesinos, Ana Laura Torres Huerta
TecCEM_HS:Ángel Farid Rojas Cruz, Daniela Flores Gómez, Carlos Ramos Gutiérrez, Alonso Pérez Lona, Carlos Alejandro Meza Ramírez, Fernanda Sotomayor Olivares, Jorge Sebastián Rodríguez Iniesta, Juan Carlos Rueda Silva, Oliva Angélica Sánchez Montesinos, Ana Laura Torres Huerta
Tokyo_Tech: Shotaro Ayukawa, Takahiro Kashiwagi, Daisuke Kiga, Misa Minegishi, Riku Shinohara, Hiraku Tokuma, Yuta Yamazaki, Shuhei Yasunishi, Erinn Sim Zixuan
TrinityCollegeDublin: Remsha Afzal, Matthew Carrigan, Barry Moran, Marlena Mucha, Arnas Petrauskas
TU_Delft: Stefan Marsden, Michelle Post, Anne Rodenburg, Hector Sanguesa, Marit van der Does, Erwin van Rijn, Max van’t Hof
TU_Eindhoven: Yeshi de Bruin, Hans de Ferrante, Elles Elschot, Laura Jacobs, Jan-Willem Muller, Sjoerd Nooijens, Femke Vaassen, Cas van der Putten, Esther van Leeuwen, Laura van Smeden, Kwan Kwan Zhu
Tuebingen: Kevin Sabath, Katharina Sporbeck, Nicolai von Kügelgen, Lisa Wellinger
UCL: Stefanie Braun, Jack Ho, Yash Mishra, Mariola Sebastian, Lucas von Chamier
UCLA: Fasih M. Ahsan, Megan A. Satyadi
UC_San_Diego: Vivienne Gunadhi, Phillip Kyriakakis, Jenny Lee, Walter Thavarajah
UMaryland: Kimia Abtahi, Robert Hand, Chun Mun Loke, Adam Wahab, Iowis Zhu
UNITN-Trento: Cristina Del Bianco, Fabio Chizzolini, Elisa Godino, Roberta Lentini, Sheref S. Mansy, Noel Yeh Martin, Claudio Oss Pegorar
Utah_State: Alexander Cook, Sara Gertsch, Timothy Kerns, Charles Miller, Chad Nielsen, Michael Paskett, Alexander Torgesen
Vanderbilt: Stephen Lee, Ophir Ospovat, Sikandar Raza, Daniel Shaykevich, Jarrod Shilts
Vilnius-Lithuania: Barbora Bajorinaite, Mykolas Bendorius, Ieva Rauluseviciute, Ieva Savickyte, Sarunas Tumas
William_and_Mary: William Buchser, Elli Cryan, Caroline Golino, Andrew Halleran, Taylor Jacobs, Michael LeFew, Joe Maniaci, John Marken, Margaret Saha, Panya Vij
WPI-Worcester: Kayla DeSanty, Natalie Farny, Julie Mazza
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Raytheon BBN Technologies, Synthace, and Agilent provided support in the form of salaries for authors JB, MG, and JH, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific roles of these authors are articulated in the author contributions section.
References
- 1. Ro DK, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440(10.1038):940–943. 10.1038/nature04640 [DOI] [PubMed] [Google Scholar]
- 2. Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333(6047):1307–1311. 10.1126/science.1205527 [DOI] [PubMed] [Google Scholar]
- 3. Dunlop MJ, Keasling JD, Mukhopadhyay A. A model for improving microbial biofuel production using a synthetic feedback loop. Systems and synthetic biology. 2010;4(2):95–104. 10.1007/s11693-010-9052-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nature Reviews Genetics. 2010;11(5):367–379. 10.1038/nrg2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Endy D. Foundations for engineering biology. Nature. 2005;438(7067):449–453. 10.1038/nature04342 [DOI] [PubMed] [Google Scholar]
- 6. Kwok R. Five hard truths for synthetic biology. Nature. 2010;463(7279):288–90. 10.1038/463288a [DOI] [PubMed] [Google Scholar]
- 7. Prinz F, Schlange T, Asadullah K. Believe it or not: how much can we rely on published data on potential drug targets? Nature reviews Drug discovery. 2011;10(9):712–712. 10.1038/nrd3439-c1 [DOI] [PubMed] [Google Scholar]
- 8. Begley CG, Ellis LM. Drug development: Raise standards for preclinical cancer research. Nature. 2012;483(7391):531–533. 10.1038/483531a [DOI] [PubMed] [Google Scholar]
- 9. Mobley A, Linder SK, Braeuer R, Ellis LM, Zwelling L. A survey on data reproducibility in cancer research provides insights into our limited ability to translate findings from the laboratory to the clinic. PLOS ONE. 2013. 10.1371/journal.pone.0063221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lazebnik Y. Can a biologist fix a radio? x2014;Or, what I learned while studying apoptosis. Cancer cell. 2002;2(3):179–182. 10.1016/S1535-6108(02)00133-2 [DOI] [PubMed] [Google Scholar]
- 11. Kelly JR, Rubin AJ, Davis JH, Ajo-Franklin CM, Cumbers J, Czar MJ, et al. Measuring the activity of BioBrick promoters using an in vivo reference standard. Journal of Biological Engineering. 2009;3(4). 10.1186/1754-1611-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beal J, Weiss R, Yaman F, Davidsohn N, Adler A. A Method for Fast, High-Precision Characterization of Synthetic Biology Devices. MIT; 2012. MIT-CSAIL-TR-2012-008. [Google Scholar]
- 13. Rosenfeld N, Perkins TJ, Alon U, Elowitz MB, Swain PS. A fluctuation method to quantify in vivo fluorescence data. Biophysical journal. 2006;91(2):759–766. 10.1529/biophysj.105.073098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klavins E. The Aquarium Project; Retrieved Oct. 2, 2014. http://klavinslab.org/aquarium.html.
- 15. Huang H, Densmore D. Integration of microfluidics into the synthetic biology design flow. Lab on a Chip. 2014;14(18):3459–3474. 10.1039/C4LC00509K [DOI] [PubMed] [Google Scholar]
- 16.Anderson JC. Anderson promoter collection; Retrieved Oct. 30, 2015. http://parts.igem.org/Promoters/Catalog/Anderson.
- 17. Shetty RP, Endy D, Knight TF Jr. Engineering BioBrick vectors from BioBrick parts. Journal of biological engineering. 2008;2(1):1–12. 10.1186/1754-1611-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Friedman N, Cai L, Xie XS. Linking stochastic dynamics to population distribution: an analytical framework of gene expression. Physical review letters. 2006;97(16):168302 10.1103/PhysRevLett.97.168302 [DOI] [PubMed] [Google Scholar]
- 19. Stanton BC, Nielsen AA, Tamsir A, Clancy K, Peterson T, Voigt C. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nature Chemical Biology. 2014;10(2):99–105. 10.1038/nchembio.1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bonnet J, Yin P, Ortiz ME, Subsoontorn P, Endy D. Amplifying genetic logic gates. Science. 2013;340(6132):599–603. 10.1126/science.1232758 [DOI] [PubMed] [Google Scholar]
- 21. Davidsohn N, Beal J, Kiani S, Adler A, Yaman F, Li Y, et al. Accurate predictions of genetic circuit behavior from part characterization and modular composition. ACS Synthetic Biology. 2014;. [DOI] [PubMed] [Google Scholar]
- 22. Iverson SV, Haddock TL, Beal J, Densmore DM. CIDAR MoClo: Improved MoClo Assembly Standard and New E. coli Part Library Enable Rapid Combinatorial Design for Synthetic and Traditional Biology. ACS synthetic biology. 2015;. [DOI] [PubMed] [Google Scholar]
- 23. Beal J. Bridging the Gap: A Roadmap to Breaking the Biological Design Barrier. Frontiers in Bioengineering and Biotechnology. 2014;2(87). 10.3389/fbioe.2014.00087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang L, Gaigalas AK, Marti G, Abbasi F, Hoffman RA. Toward quantitative fluorescence measurements with multicolor flow cytometry. Cytometry Part A. 2008;73(4):279–288. 10.1002/cyto.a.20507 [DOI] [PubMed] [Google Scholar]
- 25. Hoffman RA, Wang L, Bigos M, Nolan JP. NIST/ISAC standardization study: Variability in assignment of intensity values to fluorescence standard beads and in cross calibration of standard beads to hard dyed beads. Cytometry Part A. 2012;81(9):785–796. 10.1002/cyto.a.22086 [DOI] [PubMed] [Google Scholar]
- 26.SpheroTech. Measuring Molecules of Equivalent Fluorescein (MEFL), PE (MEPE) and RPE-CY5 (MEPCY) using Sphero Rainbow Calibration Particles. SpheroTech; 2001. SpheroTechnical Notes: STN-9, Rev C 071398.
- 27. Model MA, Burkhardt JK. A standard for calibration and shading correction of a fluorescence microscope. Cytometry. 2001;44(4):309–316. [DOI] [PubMed] [Google Scholar]
- 28. Harris DL, Mutz M. Debunking the myth: validation of fluorescein for testing the precision of nanoliter dispensing. Journal of the Association for Laboratory Automation. 2006;11(4):233–239. 10.1016/j.jala.2006.05.006 [DOI] [Google Scholar]
- 29. Hu G, Chen L, Okerlund J, Shaer O. Exploring the Use of Google Glass in Wet Laboratories In: Proceedings of the 33rd Annual ACM Conference on Human Factors in Computing Systems. ACM; 2015. p. 2103–2108. [Google Scholar]
- 30. Chen B, Cahoon D, Canton B, Che A. Software for Engineering Biology in a Multi-Purpose Foundry In: International Workshop on Bio-Design Automation; 2015. [Google Scholar]
- 31. Linshiz G, Stawski N, Poust S, Bi C, Keasling JD, Hillson NJ. PaR-PaR laboratory automation platform. ACS synthetic biology. 2012;2(5):216–222. 10.1021/sb300075t [DOI] [PubMed] [Google Scholar]
- 32.Synthace. Antha Programming language; Retrieved 2015. https://www.antha-lang.org.
- 33. Vasilev V, Liu C, Haddock T, Bhatia S, Adler A, Yaman F, et al. A Software Stack for Specification and Robotic Execution of Protocols for Synthetic Biological Engineering 3rd International Workshop on Bio-Design Automation. 2011;. [Google Scholar]
- 34. Gulati S, Rouilly V, Niu X, Chappell J, Kitney RI, Edel JB, et al. Opportunities for microfluidic technologies in synthetic biology. Journal of The Royal Society Interface. 2009; p. rsif20090083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
Document provided is a snapshot of the page as of October 4th, 2015, which includes information presented on participation and the preliminary results of the study.
(PDF)
Document provided is a snapshot of the page as of October 4th, 2015.
(PDF)
(CSV)
(CSV)
Some teams returned their forms (or portions thereof) manually for a number of reasons, most frequently due to Internet censorship in their home countries; entries from these teams are not included in the attached form. Only technical entries for the form are included, and team names have been replaced by numbers corresponding to the numbers in the summary data table.
(CSV)
Some teams returned their forms (or portions thereof) manually for a number of reasons, most frequently due to Internet censorship in their home countries; entries from these teams are not included in the attached form. Only technical entries for the form are included, and team names have been replaced by numbers corresponding to the numbers in the summary data table.
(CSV)
- Medium14/Weak14
- Medium15/Positive15
- Medium15/Weak15
- Positive15/Weak15
(PDF)
- Medium14/Weak14
- Medium15/Positive15
- Medium15/Weak15
- Positive15/Weak15
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.