Abstract
To investigate whether pigmentation genes involved in the melanogenic pathway (melanogenesis) contributed to melanoma predisposition, we compared pigmentary genetics with quantitative skin pigmentation measurements, the number of atypical nevi, the total nevus count, and the familial atypical multiple mole and melanoma (FAMMM) syndrome. We typed 32 pigmentary SNP markers and sequenced MC1R in 246 healthy individuals and 116 individuals attending periodic control for malignant melanoma development, 50 of which were diagnosed with FAMMM. It was observed that individuals with any two grouped MC1R variants (missense, NM_002386:c. 456C > A (p.TYR152*), or NM_002386:c.83_84insA (p.Asn29Glnfs*14) had significantly (p<0.001) lighter skin pigmentation of the upper-inner arm than those with none or one MC1R variant. We did not observe any significant association of the MC1R variants with constitutive pigmentation measured on the buttock area. We hypothesize that the effect of MC1R variants on arm pigmentation is primarily reflecting the inability to tan when subjected to UVR. A gender specific effect on skin pigmentation was also observed, and it was found that the skin pigmentation of females on average were darker than that of males (p<0.01). We conclude that MC1R variants are associated with quantitative skin colour in a lightly pigmented Danish population. We did not observe any association between any pigmentary marker and the FAMMM syndrome. We suggest that the genetics of FAMMM is not related to the genetics of the pigmentary pathway.
Introduction
Human skin colour is greatly influenced by environmental factors, including ultraviolet radiation (UVR). UVR is high in equatorial regions of the world, where darker skinned populations are found, and lower in regions distant to equator, where lighter skinned populations are found [1]. Excessive UVR of the skin induces DNA damage and is considered one of the main risk factors of developing various skin cancers. Dark skin, with high levels of the pigment eumelanin, protects against UVR, because eumelanin block the effects of UVR. On the other hand, vitamin D synthesis requires UVR and human skin colour diversity reflects the balance of letting enough UVR through the skin to produce vitamin D and preventing DNA-damage caused by UVR [2].
Melanin is produced in melanosomes, which are lysosome-related organelles present in melanocytes. Two types of melanin are produced, pheomelanin (yellow/red) and eumelanin (brown/black). Pheomelanin levels are rather constant among people with different skin colours whereas eumelanin levels vary [3]. Eumelanin is more efficient than pheomelanin in blocking UVR and in scavenging the reactive oxygen species produced from UVR of the skin (reviewed in [4]). Pheomelanin confers a carcinogenic risk in mice independently of UVR [5]. Melanocytes are found in the basal layer of the skin together with keratinocytes and the two cell types form the epidermal melanocyte unit [6]. Keratinocyte cells function as the primary barrier against environmental damage, including UVR. DNA damage in the keratinocyte induces expression of tumour protein p53 (P53), which induces melanin synthesis by increasing the level of α-melanocyte stimulating hormone (αMSH) that binds to the melanocyte-membrane-bound melanocortin 1 receptor (MC1R) and increases the cAMP level in the melanocytes. This results in increased transcription of several pigmentation genes via microphtalmia associated transcription factor (MITF) (reviewed in [7]).
Many of the key genes involved in the melanogenic pathway are associated with skin colour and skin cancer risk including agouti signalling protein (ASIP), oculocutaneous albinism 2 (OCA2), solute carrier family 45 member 2 (SLC45A2), tyrosinase (TYR), tyrosinase-related protein 1 (TYRP1), and melanocortin 1 receptor MC1R [8–17]. Variants in the MC1R have been studied extensively in relation to the risk of developing various skin cancers [18–21], and certain MC1R variants are associated with increased survival of melanoma patients [22]. Mutations in MC1R cause the red hair colour phenotype that is characterized by red hair, fair skin, and inability to tan [23–27]. Mutations in MC1R may result in either diminished α-MSH binding or decreased cAMP signalling resulting in decreased production of eumelanin.
UVR exposure is a major health risk factor, especially in light skinned populations [28,29]. Lightly pigmented skin and a large number of atypical nevi is associated with increased risk of developing malignant melanoma [11,30–33]. Atypical nevi are more prevalent in melanoma patients from light than from dark skinned populations [34]. The grade of cytological atypia of the nevi is an important risk factor for the development of malignant melanoma [35,36]. The familial atypical multiple-mole and melanoma (FAMMM) syndrome is defined by (1) occurrence of melanoma in one or more first or second degree relatives, (2) large numbers of moles (often greater than 50) some of which are atypical and often variable in size, and (3) moles that demonstrate certain distinct histologic features [37,38]. Genetic mutations have been identified in melanoma prone families including mutations in cyclin kinase 2A (CDKN2A) and cyclin kinase 4 (CDK4) genes, both of which encode proteins involved in the retinoblastoma pathway [39,40]. Genetic variants of CDKN2A are associated with the number and distribution of atypical nevi [41].
MC1R variants can modify the penetrance of CDKN2A mutations, as carriers of mutations in both genes are at increased risk of developing malignant melanoma compared to individuals carrying a mutation in only one gene [31,39,42–47].
Many nevi, including common nevi, harbour the V600E mutation in the v-raf murine sarcoma viral oncogene homolog, B1 (BRAF) [48]. V600E was observed in 60% of all melanomas [49]. BRAF can regulate melanoma proliferation through MITF [50] that is a transcription factor and regulator of pigmentation.
Even though high melanoma risk alleles clearly exist in e.g. CDKN2A, it is speculated if the melanoma risk might be attributable to combinations of low to moderate risk alleles in e.g. MC1R, TYR, and ASIP [51].
In this study, we investigated the associations between SNPs in pigmentary genes and (1) quantitative skin colour measurements in 246 healthy, light skinned Danish individuals, (2) total nevi, (3) total atypical nevi count, and (4) the FAMMM syndrome in 116 Danish individuals at increased risk of developing malignant melanoma.
Results
Associations between quantitative skin colour and genetic markers
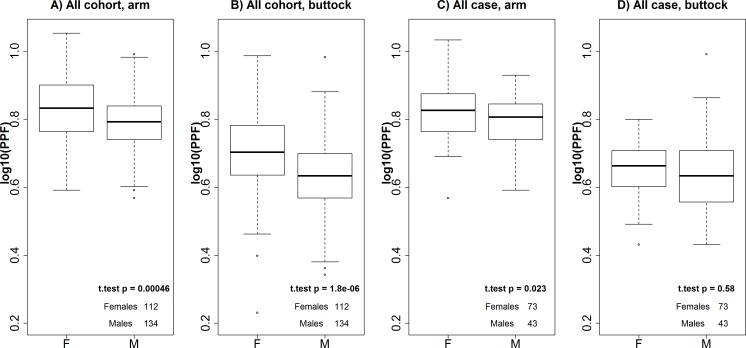
The Pearson correlation coefficient between logarithmic transformed arm and buttock PPF measurements was 0.51 (r2 = 0.26, data not shown). Buttock PPF measurements were statistically significantly lower than those of arm measurements for both genders in the cohort group (p<2.3x10-16). The skin was signifcantly darker in females than in males among the cohorts at both measurement sites (p<0.001, Figs 1A and 2B). Furthermore, female cases had statistically significantly higher arm PPF measurements than male cases (Fig 1C). However, we did not detect any statistically significant difference between the buttock PPF measurements of males and females in the case group (Fig 1D). Female cases had significantly lighter skin on the buttock than female cohorts (p = 0.0004). However, this tendency was not observed in males (p = 0.7), and there was no significant difference in the pigmentation levels on the arms between cases and cohorts in either sex (p = 0.8 and p = 0.9 in females and males, respectively).
Fig 1. Comparison between skin color measurements in females and males.
Arm (A) and buttock (B) measurements in females and males of the cohort. Arm (C) and buttock (D) measurements of female and male cases. P-values were calculated using Welch’s two sample t-test.
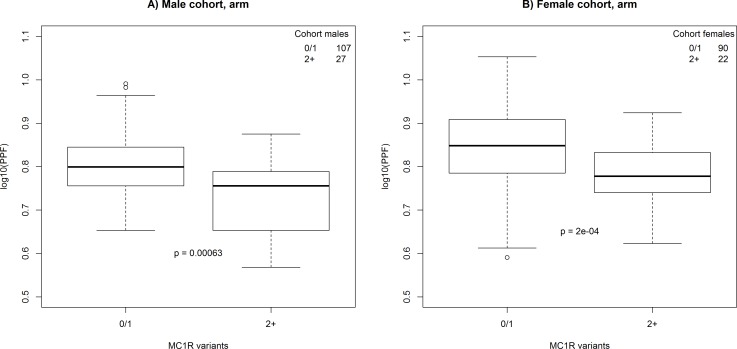
Fig 2. Comparison between skin colour measurements of the arm in male and female cohort individuals with ≤1 or ≥2 MC1R variants.
P-values were calculated using Welch’s two sample t-test.
Influence of genetic variations, gender, age, and month of measurement on PPF
Individuals from the cohort group with two or more MC1R variants (missense, p.TYR152*, or p.Asn29Glnfs*14, S2 Table) had statistically significantly lower PPF measurements of the arm (Fig 2) than those having one or no MC1R variant. We did not observe this relationship for buttock measurements (data not shown). We performed multi linear regression model analysis of the cohorts (n = 246) with log10(PPF) as response and gender, age, month of measurement, 22 SNPs with sufficient observations, and MC1R variants as explanatory variables (Pane 1 S3 Table and Pane 2 S3 Table). For arm measurements, we only observed grouped MC1R variants to deviate statistically significantly (p<0.01). The MC1R association was not observed when investigating a similar multi linear regression model with buttock PPF as response. Among the tested variables, only gender was statistically significantly associated (p<0.01) with the buttock PPF.
Associations with nevi and FAMMM
We evaluated the logistic regression model including age, gender, 22 SNPs, and MC1R for association with nevi count (Pane 3 S3 Table). A statistically significant association with total nevi count prior to correction for multiple testing was observed for the G and AG genotypes of rs6475555 on chromosome 9 (non-corrected p = 0.004 (corrected 0.18) and p = 0.005 (corrected 0.24), respectively). None of the parameters were statistically significantly associated with the total nevi count after correcting for multiple testing.
When evaluating a logistic regression model with atypical nevi count as response (Pane 4 S3 Table), we observed statistically significant association with age (p<0.05). Healthy cohorts had a mean age of 45.1, years and cases with atypical nevi had a mean age of 38.4 years, which may explain the statistically significant results of the model. The G and GA genotypes of rs1015362 in ASIP were statistically significantly associated with the atypical nevi count prior to correction for multiple testing (non-corrected p = 0.008 (0.36 corrected) and p = 0.003 (0.13 corrected), respectively).
When performing the logistic regression model including FAMMM (Pane 5 S3 Table), we observed a statistically significant association with age. This was most likely caused by the fact that patients with FAMMM had a mean age of 36.9 years, whereas the healthy cohorts had a mean age of 45.1 years. We did not observe any statistically significant association between the 22 SNPs or MC1R with the FAMMM syndrome, allthough that we prior to Bonferoni correction observed statistically significant associations with the vitamin D receptor (VDR) SNP rs1544410 G and the agouti signalling protein (ASIP) SNP rs1015362 G (non-corrected p = 0.007 (0.33 corrected) and p = 0.009 (0.43 corrected), respectively).
Discussion
The measurements of pigmentation levels demonstrated that the buttock area was statistically significantly less pigmented than the upper-inner arm area. This implies that the buttock is a better site for measuring constitutive pigmentation levels than the upper-inner arm, and that the upper-inner arm measurements often reflect the tanning response of the individuals. We observed a statistically significant difference of the logarithmic transformed PPF values between males and females of the cohort group on both arm and buttock. This observation supports previous observations in eye colour studies indicating that there may exist a yet unknown biological, sex-determined mechanism controlling pigmentation levels [52,53]. On the other hand, the darker skin pigmentation levels observed in females could also be an environmental effect due to e.g. different sun or clothing habits (Fig 1A and 1B). The individuals in the case group are expected to avoid extensive UV-exposure. In this group, females and males did not have statistically significantly different PPF values of the buttock (Fig 1D), and the effect of gender on the arm pigmentation was less pronounced (Fig 1C). This seemed to indicate that the observed gender effect on pigmentation wass caused by an environmental effect.
When performing multi linear regression analysis among the cohorts, we observed that individuals with two or more MC1R variants had statistically significantly lower PPF values of the arm than those with none or one MC1R variants. MC1R variants were not statistically significantly associated with PPF measurements on the buttock. This shows, in concordance with established genetic theories concerning MC1R (reviewed in [54]), that MC1R variants are involved in human pigmentation. Since MC1R variants are not statistically significantly associated with pigmentation measurements of the buttock, we hypothesize that the effect of MC1R variants on arm pigmentation is primarily reflecting an inability to tan when subjected to UVR. Two of the genotypes of rs6475555 on chromosome 9 were, prior to correction for multiple testing, statistically significantly associated with the total nevi count. This SNP was previously found to be associated with the total nevus count in a study comprised of twins from the UK and twins of European ancestry in Australia [55]. Falchi and colleagues hypothesized that the gene methythoiadenosine phosphorylase (MTAP) or the adjacent CDKN2A gene are involved in the nevus formation [55]. A SNP in MTAP was reported to be associated with an increased number of nevi [33] indicating the importance of the MTAP region in nevus formation.
Two of the genotypes of ASIP rs1015362 were, prior to correction for multiple testing, found to be statistically significantly associated with the atypical nevi count. ASIP was described as being associated with skin colour and melanoma risk [55].
Neither the tested SNPs nor MC1R was statistically significantly associated with FAMMM after correction for multiple comparisons. This most likely reflects that FAMMM is caused by defects in non-pigmentary systems, e.g. the retinoblastoma or P53 pathways [39,40]. It is possible that the atypical nevi observed in these individuals are by-products of signalling through MITF rather than being caused by a specific function of the pigmentary system.
Material and Methods
Samples and DNA purification
Blood samples from 362 unrelated Danish individuals were collected. A sample of 246 healthy individuals (cohorts) was collected at the Section of Forensic Genetics, Department of Forensic Medicine, Faculty of Health and Medical Sciences, University of Copenhagen, and at the Blood Bank, Glostrup Hospital. Blood samples from 116 individuals (cases) were collected at the Department of Dermatology, Bispebjerg Hospital, Copenhagen, Denmark. The case group contained individuals attending regular inspection at the Department of Dermatology at Bispebjerg Hospital due to an increased risk of developing malignant melanoma. Not all case group individuals had a definite diagnosis. All individuals in the case group were evaluated by a trained dermatologist. Nevi count (> 50 or<50) and atypical nevi count (> 5 or < 5) were evaluated by the Dermatologist. Atypical nevi are large nevi (>5mm in diameter) with irregular borders and colour variations. Cases were heterogeneous and presented multiple indications; 71 cases had more than 5 atypical nevi and 59 cases had more than 50 nevi. Fifty cases were diagnosed with FAMMM using the following criteria; 1) 100 or more melanocytic nevi, 2) one or more melanocytic nevi greater than or equal to 8mm in its largest diameter, and 3) one or more clinically atypical melanocytic nevi (clinically defined entities, >5 mm in diameter, with irregular pigmentation and an irregular or diffuse edge), according to the description defined in [56].
The individuals in the cohort group were asked to fill in a questionnaire and answer if they had more than five large nevi (>5mm) on their skin. Individuals in the cohort group that reported more than five large nevi were excluded from the study of atypical nevi and FAMMM, as these individuals may represent undiagnosed case individuals. Individuals in the cohort group that reported less than five large nevi were classified as belonging to the non-FAMMM and the low nevi count category groups. This left 159 cohort individuals (87 provided no nevi information) and 116 case group individuals for analysis of nevi and FAMMM and a total of 246 cohort individuals and 116 cases for analysis of quantitative skin colour (Table 1).
Table 1. Descriptive statistics of the participants.
F: females. M: males. Sd; Standard deviation.
| Gender | Status | No of individuals | Age mean (years) | Age sd (years) | log(PPF),arm, mean | log(PPF), arm, sd | log(PPF), buttock, mean | log(PPF), buttock, sd |
|---|---|---|---|---|---|---|---|---|
| F | Cohorts | 112 | 41 | 11.5 | 0.83 | 0.091 | 0.70 | 0.11 |
| F | Case | 73 | 38 | 8.4 | 0.82 | 0.081 | 0.65 | 0.08 |
| M | Cohorts | 134 | 43 | 11.1 | 0.79 | 0.084 | 0.63 | 0.12 |
| M | Case | 43 | 41 | 9.0 | 0.79 | 0.077 | 0.64 | 0.11 |
DNA was purified from 200μL whole blood using the DNA Blood Mini Kit (Qiagen) as recommended by the manufacturer. DNA was eluted in 50μL AE Buffer (Qiagen). Blood samples were collected between March 2010 and January 2012. Samples were grouped by the month of collection, regardless of year, in order to analyse the possible seasonal variation in quantitative skin reflectance measurements.
The study was approved by the Ethical Committee of the Capital region of Denmark, H-4-2009-125 and M-20090237. All participants provided written informed consent. All individuals included in the study reported Scandinavian parental origin. Individuals did not report regular use of tanning beds.
Skin reflectance measurements
The quantitative skin reflectance measurements were performed using a UV-Optimize Scientific 555 (Chromo Light APS, Espergærde, Denmark). The instrument is a non-invasive spectrophotometer that calculates the reflectance of skin redness and pigmentation from the measured area in percentage [57]. The instrument was calibrated with a white standard (ISO 2469).
The reflectance of a wavelength at 555nm measures the haemoglobin content and is scaled as 0%-100% skin redness. Null percent skin redness corresponds to skin without blood, and 100 percent corresponds to highly vascular skin lesions. The reflectance of 660 nm indicates the melanin content termed skin pigmentation when the skin redness is eliminated. A null percentage skin pigmentation measurement corresponds to the reflection of no pigmentation (pale), and a 100 percentage skin pigmentation measurement corresponds to the reflection of black skin [57]. The pigment protection factor (PPF) is a value for the protection against UVR provided by skin pigmentation and the top layer of epidermis (stratum corneum). The PPF can be used in parallel with the Fitzpatrick skin types [58]. Measurements were performed on the upper, inner arm and on the buttock in triplicates for each participant. The medians of each PPF triplicates were used for statistical analyses. All measured skin areas were free from nevi, freckles, tattoos, and hair.
SNP typing
All 362 samples were typed for 32 SNPs, which were chosen based on their associations to various skin malignancies and skin colours (S1 Table), using the iPLEX® Gold kit (Sequenom) (S1 Table). The PCR contained 2μL DNA, 0.5μL 10x Buffer, 0.8μL 25mM MgCl2, 0.1μL 25mM dNTP mix, 1.3μL 0.5μM primer mix (DNA Technology), 0.2μL 5U/μl HotStarTaq, and 1.1μL H2O. The PCR was performed in a GeneAmp® PCR system 9700 thermal cycler (Applied Biosystems) with the following conditions: denaturation at 94°C for 2 min followed by 45 cycles of 94°C for 20 s, 62°C for 30 s, 72°C for 1 min, followed by 72°C for 3 min. The PCR products were treated with Shrimp Alkaline Phosphatase (SAP) (Sequenom) in a GeneAmp® PCR system 9700 thermal cycler at 37°C for 40 min and 85°C for 5 min. The SBE reaction contained 8μl SAP treated PCR products and 2μl iPLEX® mix (Sequenom). The iPLEX® mix contained 0.2μL 10x iPLEX® buffer, 0.2μL iPLEX®-Termination mix, 0.94μL primer mix (DNA Technology), 0.04μL iPLEX® enzyme, and 0.62μL H2O.
The SBE reaction was performed in a GeneAmp® PCR system 9700 thermal cycler with the following conditions: denaturation at 94°C for 30 s followed by 40 cycles of 94°C for 5 s, 52°C for 5 s and 80°C 5 s, 52°C for 5 s and 80°C for 5 s, 52°C for 5 s and 80°C for 5 s, 52°C for 5 s and 80°C for 5 s, 52°C for 5 s, 80°C 5 s, and 72°C for 3 min.
A total of 40μL of molecular grade water and ion exchange resin (Sequenom) was added to each sample. Samples were rotated for approximately 4 h and kept in the refrigerator for up to 4 days before spotting. The samples were spotted in duplicates using the RS1000 Nanospotter (Sequenom) and visualized on the MassARRAY® analyzer 4 system (Sequenom) using the autorun settings.
Samples were analysed with Typer Analyzer 4 (Sequenom) and were autoclustered using a signal to noise ratio of 7. Clusterplots were visually inspected, and outliers were further investigated.
All samples were run in duplicates. Genotypes were compared between spots and duplicate typings as described in [53] and implemented with the statistic software R (R core team, version 2.11.0, URL http://www.R-project.org). The SNPs rs26722, rs28777, rs1426654, rs1800407, rs7495174, rs8059973, rs12203592, rs12913032, rs16891982 and rs36118030 all had minor allele frequencies (MAF)<0.01 and ≤4 observations, and were removed from the analysis.
Sequencing of MC1R
The MC1R gene of all 362 individuals was sequenced. A region encompassing the MC1R exon and the promoter region was amplified in a single PCR amplicon of 1,963bp. Prior to library preparation, we employed a multiplexing approach using endonuclease digestion of the PCR product as previously described [59]. Libraries were prepared for sequencing with the TruSeq® DNA Sample Preparation kit (Illumina) and paired-end sequenced (2x250 cycles) on the Illumina Miseq platform using the Illumina MiSeq Reagent Nano kit v2 (500 cycles).
Illumina adaptors were trimmed using Flexbar [60]. The FASTQ files were aligned to the MC1R reference sequence assembly Feb.2009 GRCh37/hg19 (UCSC Genome Browser) with the Burrows-Wheeler Aligner (BWA) MEM algorithm [61] to generate BAM files. Variant calling was carried out using GATK ver. 2.6.5 [62]. Variants were accepted if they had a minimum coverage of 50. Variants, where two different alleles were observed, were accepted as heterozygote variant calls if the frequency of the minor variant allele was >0.15. Variants were analysed using Alamut Batch (Interactive Biosoftware, France). MC1R variants were grouped according to potential significance. All missense, non-sense, and frameshift mutations (S2 Table) were assigned to the MC1R variant group and analysed together (Table 2). Due to a low number of participants and a number of unclassified variants detected, the R/r system of MC1R mutation classification [63] was disregarded.
Table 2. Number of MC1R variants.
| Group/MC1R variants | No of individuals | 0 | 1 | 2 | 3 |
|---|---|---|---|---|---|
| Cohort | 246 | 89 | 108 | 49 | 0 |
| Cases | 116 | 36 | 65 | 13 | 2 |
Statistical analyses
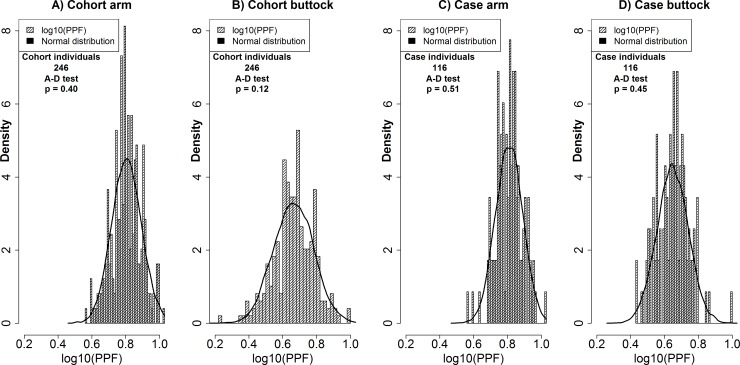
All statistic calculations were performed using R (R core team, version 3.1.1, URL http://www.R-project.org). The PPF data was log transformed as this gave the best approximation to the normal distribution. All PPF data groups passed the Anderson-Darling test for normality (Fig 3). Correlations between logaritmically transformed PPF measurements of arm and buttock areas were investigated using the Pearson correlation coefficient. Multi linear models were performed using the lm command of R. Models were performed with log10(PPF) as response and gender, age, month of measurement, 22 SNPs with sufficient observations (MAF>0.01), and grouped MC1R variants as explanatory variables (25 variables, 53 comparisons). Logistic regression models were performed using the glm command of R. Models were constructed for age, gender, 22 SNPs and as MC1R to test for association with grouped nevi count or FAMMM (24 variables, 48 comparisons). Welch’s two sample t-test was used due to differences in sample sizes. P-values were corrected for multiple comparisons using Bonferroni correction.
Fig 3.
Distributions of arm (A) and buttock (B) measurements in the cohort. Distribution of arm (C) and buttock (D) measurements in the cases. The black line represents a simulated normal distribution with 10,000 observations with the same mean and standard deviation as that of the underlying data.
Supporting Information
(DOCX)
(DOCX)
(XLSX)
Acknowledgments
We thank scientific director Thomas F. Hansen, PhD, Institute of Biological Psychiatry, MHC Sct. Hans, Copenhagen Mental Health Services; Department of Clinical Medicine, University of Copenhagen. for initiating contacts and collecting samples.
Data Availability
Data may compromise the privacy of study participants and may not be shared publicly. Data are available upon request to the authors.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Relethford JH. Hemispheric difference in human skin color. Am J Phys Anthropol 1997;104:449–57. [DOI] [PubMed] [Google Scholar]
- 2.Yuen AWC, Jablonski NG. Vitamin D: In the evolution of human skin colour. Med Hypotheses 2010;74:39–44. 10.1016/j.mehy.2009.08.007 [DOI] [PubMed] [Google Scholar]
- 3.Wakamatsu K, Kavanagh R, Kadekaro AL, Terzieva S, Sturm RA, Leachman S, et al. Diversity of pigmentation in cultured human melanocytes is due to differences in the type as well as quantity of melanin. Pigment Cell Res Spons Eur Soc Pigment Cell Res Int Pigment Cell Soc 2006;19:154–62. 10.1111/j.1600-0749.2006.00293.x [DOI] [PubMed] [Google Scholar]
- 4.Brenner M, Hearing VJ. The protective role of melanin against UV damage in human skin. Photochem Photobiol 2008;84:539–49. 10.1111/j.1751-1097.2007.00226.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mitra D, Luo X, Morgan A, Wang J, Hoang MP, Lo J, et al. An ultraviolet-radiation-independent pathway to melanoma carcinogenesis in the red hair/fair skin background. Nature 2012;491:449–53. 10.1038/nature11624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ando H, Niki Y, Ito M, Akiyama K, Matsui MS, Yarosh DB, et al. Melanosomes are transferred from melanocytes to keratinocytes through the processes of packaging, release, uptake, and dispersion. J Invest Dermatol 2012;132:1222–9. 10.1038/jid.2011.413 [DOI] [PubMed] [Google Scholar]
- 7.Rouzaud F, Kadekaro AL, Abdel-Malek ZA, Hearing VJ. MC1R and the response of melanocytes to ultraviolet radiation. Mutat Res 2005;571:133–52. 10.1016/j.mrfmmm.2004.09.014 [DOI] [PubMed] [Google Scholar]
- 8.Bishop DT, Demenais F, Iles MM, Harland M, Taylor JC, Corda E, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet 2009;41:920–5. 10.1038/ng.411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brudnik U, Branicki W, Wojas-Pelc A, Kanas P. The contribution of melanocortin 1 receptor gene polymorphisms and the agouti signalling protein gene 8818A>G polymorphism to cutaneous melanoma and basal cell carcinoma in a Polish population. Exp Dermatol 2009;18:167–74. 10.1111/j.1600-0625.2008.00760.x [DOI] [PubMed] [Google Scholar]
- 10.Duffy DL, Zhao ZZ, Sturm RA, Hayward NK, Martin NG, Montgomery GW. Multiple pigmentation gene polymorphisms account for a substantial proportion of risk of cutaneous malignant melanoma. J Invest Dermatol 2010;130:520–8. 10.1038/jid.2009.258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez LP, Milne RL, Pita G, Floristan U, Sendagorta E, Feito M, et al. Pigmentation-related genes and their implication in malignant melanoma susceptibility. Exp Dermatol 2009;18:634–42. 10.1111/j.1600-0625.2009.00846.x [DOI] [PubMed] [Google Scholar]
- 12.Gudbjartsson DF, Sulem P, Stacey SN, Goldstein AM, Rafnar T, Sigurgeirsson B, et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat Genet 2008;40:886–91. 10.1038/ng.161 [DOI] [PubMed] [Google Scholar]
- 13.Guedj M, Bourillon A, Combadières C, Rodero M, Dieudé P, Descamps V, et al. Variants of the MATP/SLC45A2 gene are protective for melanoma in the French population. Hum Mutat 2008;29:1154–60. 10.1002/humu.20823 [DOI] [PubMed] [Google Scholar]
- 14.Han J, Qureshi AA, Nan H, Zhang J, Song Y, Guo Q, et al. A germline variant in the interferon regulatory factor 4 gene as a novel skin cancer risk locus. Cancer Res 2011;71:1533–9. 10.1158/0008-5472.CAN-10-1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jannot A- S, Meziani R, Bertrand G, Gérard B, Descamps V, Archimbaud A, et al. Allele variations in the OCA2 gene (pink-eyed-dilution locus) are associated with genetic susceptibility to melanoma. Eur J Hum Genet EJHG 2005;13:913–20. 10.1038/sj.ejhg.5201415 [DOI] [PubMed] [Google Scholar]
- 16.Kanetsky PA, Swoyer J, Panossian S, Holmes R, Guerry D, Rebbeck TR. A polymorphism in the agouti signaling protein gene is associated with human pigmentation. Am J Hum Genet 2002;70:770–5. 10.1086/339076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nan H, Kraft P, Hunter DJ, Han J. Genetic variants in pigmentation genes, pigmentary phenotypes, and risk of skin cancer in Caucasians. Int J Cancer J Int Cancer 2009;125:909–17. 10.1002/ijc.24327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Box NF, Duffy DL, Irving RE, Russell A, Chen W, Griffyths LR, et al. Melanocortin-1 receptor genotype is a risk factor for basal and squamous cell carcinoma. J Invest Dermatol 2001;116:224–9. 10.1046/j.1523-1747.2001.01224.x [DOI] [PubMed] [Google Scholar]
- 19.Kennedy C, ter Huurne J, Berkhout M, Gruis N, Bastiaens M, Bergman W, et al. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J Invest Dermatol 2001;117:294–300. 10.1046/j.0022-202x.2001.01421.x [DOI] [PubMed] [Google Scholar]
- 20.Pérez Oliva AB, Fernéndez LP, Detorre C, Herráiz C, Martínez-Escribano JA, Benítez J, et al. Identification and functional analysis of novel variants of the human melanocortin 1 receptor found in melanoma patients. Hum Mutat 2009;30:811–22. 10.1002/humu.20971 [DOI] [PubMed] [Google Scholar]
- 21.Raimondi S, Sera F, Gandini S, Iodice S, Caini S, Maisonneuve P, et al. MC1R variants, melanoma and red hair color phenotype: a meta-analysis. Int J Cancer J Int Cancer 2008;122:2753–60. 10.1002/ijc.23396 [DOI] [PubMed] [Google Scholar]
- 22.Davies JR, Randerson-Moor J, Kukalizch K, Harland M, Kumar R, Madhusudan S, et al. Inherited variants in the MC1R gene and survival from cutaneous melanoma: a BioGenoMEL study. Pigment Cell Melanoma Res 2012;25:384–94. 10.1111/j.1755-148X.2012.00982.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beaumont KA, Shekar SN, Cook AL, Duffy DL, Sturm RA. Red hair is the null phenotype of MC1R. Hum Mutat 2008;29:E88–94. 10.1002/humu.20788 [DOI] [PubMed] [Google Scholar]
- 24.Duffy DL, Box NF, Chen W, Palmer JS, Montgomery GW, James MR, et al. Interactive effects of MC1R and OCA2 on melanoma risk phenotypes. Hum Mol Genet 2004;13:447–61. 10.1093/hmg/ddh043 [DOI] [PubMed] [Google Scholar]
- 25.Mengel-Jørgensen N. EH J.. M BC. Genetic screening of 15 SNPs in the MC1R gene in relation to hair colour in Danes. Int Congr Ser 2006;1288:55–7. 10.1016/j.ics.2005.11.037 [DOI] [Google Scholar]
- 26.Sulem P, Gudbjartsson DF, Stacey SN, Helgason A, Rafnar T, Magnusson KP, et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat Genet 2007;39:1443–52. 10.1038/ng.2007.13 [DOI] [PubMed] [Google Scholar]
- 27.Valverde P, Healy E, Jackson I, Rees JL, Thody AJ. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat Genet 1995;11:328–30. 10.1038/ng1195-328 [DOI] [PubMed] [Google Scholar]
- 28.Lucas RM, McMichael AJ, Armstrong BK, Smith WT. Estimating the global disease burden due to ultraviolet radiation exposure. Int J Epidemiol 2008;37:654–67. 10.1093/ije/dyn017 [DOI] [PubMed] [Google Scholar]
- 29.Zhang M, Qureshi AA, Geller AC, Frazier L, Hunter DJ, Han J. Use of tanning beds and incidence of skin cancer. J Clin Oncol Off J Am Soc Clin Oncol 2012;30:1588–93. 10.1200/JCO.2011.39.3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang Y, Newton-Bishop JA, Bishop DT, Armstrong BK, Bataille V, Bergman W, et al. A pooled analysis of melanocytic nevus phenotype and the risk of cutaneous melanoma at different latitudes. Int J Cancer J Int Cancer 2009;124:420–8. 10.1002/ijc.23869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demenais F, Mohamdi H, Chaudru V, Goldstein AM, Newton Bishop JA, Bishop DT, et al. Association of MC1R variants and host phenotypes with melanoma risk in CDKN2A mutation carriers: a GenoMEL study. J Natl Cancer Inst 2010;102:1568–83. 10.1093/jnci/djq363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elder DE, Goldman LI, Goldman SC, Greene MH, Clark WH. Dysplastic nevus syndrome: a phenotypic association of sporadic cutaneous melanoma. Cancer 1980;46:1787–94. [DOI] [PubMed] [Google Scholar]
- 33.Newton-Bishop JA, Chang Y-M, Iles MM, Taylor JC, Bakker B, Chan M, et al. Melanocytic nevi, nevus genes, and melanoma risk in a large case-control study in the United Kingdom. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol 2010;19:2043–54. 10.1158/1055-9965.EPI-10-0233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Friedman RJ, Farber MJ, Warycha MA, Papathasis N, Miller MK, Heilman ER. The “dysplastic” nevus. Clin Dermatol 2009;27:103–15. 10.1016/j.clindermatol.2008.09.008 [DOI] [PubMed] [Google Scholar]
- 35.Ahmed I, Piepkorn MW, Rabkin MS, Meyer LJ, Feldkamp M, Goldgar DE, et al. Histopathologic characteristics of dysplastic nevi. Limited association of conventional histologic criteria with melanoma risk group. J Am Acad Dermatol 1990;22:727–33. [PubMed] [Google Scholar]
- 36.Arumi-Uria M, McNutt NS, Finnerty B. Grading of atypia in nevi: correlation with melanoma risk. Mod Pathol Off J U S Can Acad Pathol Inc 2003;16:764–71. 10.1097/01.MP.0000082394.91761.E5 [DOI] [PubMed] [Google Scholar]
- 37.NIH Consens Statement Online. Diagnosis and Treatment of Early Melanoma. n.d.;1992. January 27–29:1–26. [Google Scholar]
- 38.Eckerle Mize D, Bishop M, Resse E, Sluzevich J. Familial Atypical Multiple Mole Melanoma Syndrome In: Riegert-Johnson DL, Boardman LA, Hefferon T, Roberts M, editors. Cancer Syndr., Bethesda (MD): National Center for Biotechnology Information (US); 2009. [PubMed] [Google Scholar]
- 39.Goldstein AM, Chaudru V, Ghiorzo P, Badenas C, Malvehy J, Pastorino L, et al. Cutaneous phenotype and MC1R variants as modifying factors for the development of melanoma in CDKN2A G101W mutation carriers from 4 countries. Int J Cancer J Int Cancer 2007;121:825–31. 10.1002/ijc.22712 [DOI] [PubMed] [Google Scholar]
- 40.Soufir N, Avril MF, Chompret A, Demenais F, Bombled J, Spatz A, et al. Prevalence of p16 and CDK4 germline mutations in 48 melanoma-prone families in France. The French Familial Melanoma Study Group. Hum Mol Genet 1998;7:209–16. [DOI] [PubMed] [Google Scholar]
- 41.Bishop JA, Wachsmuth RC, Harland M, Bataille V, Pinney E, MacK P, et al. Genotype/phenotype and penetrance studies in melanoma families with germline CDKN2A mutations. J Invest Dermatol 2000;114:28–33. 10.1046/j.1523-1747.2000.00823.x [DOI] [PubMed] [Google Scholar]
- 42.Chaudru V, Laud K, Avril M-F, Minière A, Chompret A, Bressac-de Paillerets B, et al. Melanocortin-1 receptor (MC1R) gene variants and dysplastic nevi modify penetrance of CDKN2A mutations in French melanoma-prone pedigrees. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol 2005;14:2384–90. 10.1158/1055-9965.EPI-04-0777 [DOI] [PubMed] [Google Scholar]
- 43.Debniak T, Scott R, Masojc B, Serrano-Fernández P, Huzarski T, Byrski T, et al. MC1R common variants, CDKN2A and their association with melanoma and breast cancer risk. Int J Cancer J Int Cancer 2006;119:2597–602. 10.1002/ijc.22210 [DOI] [PubMed] [Google Scholar]
- 44.Ghiorzo P, Pastorino L, Pizzichetta MA, Bono R, Queirolo P, Talamini R, et al. CDKN2A and MC1R analysis in amelanotic and pigmented melanoma. Melanoma Res 2009;19:142–5. 10.1097/CMR.0b013e32832a1e18 [DOI] [PubMed] [Google Scholar]
- 45.Landi MT, Kanetsky PA, Tsang S, Gold B, Munroe D, Rebbeck T, et al. MC1R, ASIP, and DNA repair in sporadic and familial melanoma in a Mediterranean population. J Natl Cancer Inst 2005;97:998–1007. 10.1093/jnci/dji176 [DOI] [PubMed] [Google Scholar]
- 46.Pastorino L, Bonelli L, Ghiorzo P, Queirolo P, Battistuzzi L, Balleari E, et al. CDKN2A mutations and MC1R variants in Italian patients with single or multiple primary melanoma. Pigment Cell Melanoma Res 2008;21:700–9. 10.1111/j.1755-148X.2008.00512.x [DOI] [PubMed] [Google Scholar]
- 47.van der Velden PA, Sandkuijl LA, Bergman W, Pavel S, van Mourik L, Frants RR, et al. Melanocortin-1 receptor variant R151C modifies melanoma risk in Dutch families with melanoma. Am J Hum Genet 2001;69:774–9. 10.1086/323411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu J, Rosenbaum E, Begum S, Westra WH. Distribution of BRAF T1799A(V600E) mutations across various types of benign nevi: implications for melanocytic tumorigenesis. Am J Dermatopathol 2007;29:534–7. 10.1097/DAD.0b013e3181584950 [DOI] [PubMed] [Google Scholar]
- 49.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–54. 10.1038/nature00766 [DOI] [PubMed] [Google Scholar]
- 50.Wellbrock C, Rana S, Paterson H, Pickersgill H, Brummelkamp T, Marais R. Oncogenic BRAF regulates melanoma proliferation through the lineage specific factor MITF. PloS One 2008;3:e2734 10.1371/journal.pone.0002734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes Dev 2012;26:1131–55. 10.1101/gad.191999.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Martinez-Cadenas C, Peña-Chilet M, Ibarrola-Villava M, Ribas G. Gender is a major factor explaining discrepancies in eye colour prediction based on HERC2/OCA2 genotype and the IrisPlex model. Forensic Sci Int Genet 2013;7:453–60. 10.1016/j.fsigen.2013.03.007 [DOI] [PubMed] [Google Scholar]
- 53.Pietroni C, Andersen JD, Johansen P, Andersen MM, Harder S, Paulsen R, et al. The effect of gender on eye colour variation in European populations and an evaluation of the IrisPlex prediction model. Forensic Sci Int Genet 2014;11:1–6. 10.1016/j.fsigen.2014.02.002 [DOI] [PubMed] [Google Scholar]
- 54.Dessinioti C, Antoniou C, Katsambas A, Stratigos AJ. Melanocortin 1 receptor variants: functional role and pigmentary associations. Photochem Photobiol 2011;87:978–87. 10.1111/j.1751-1097.2011.00970.x [DOI] [PubMed] [Google Scholar]
- 55.Falchi M, Bataille V, Hayward NK, Duffy DL, Bishop JAN, Pastinen T, et al. Genome-wide association study identifies variants at 9p21 and 22q13 associated with development of cutaneous nevi. Nat Genet 2009;41:915–9. 10.1038/ng.410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Albert LS, Rhodes AR, Sober AJ. Dysplastic melanocytic nevi and cutaneous melanoma: markers of increased melanoma risk for affected persons and blood relatives. J Am Acad Dermatol 1990;22:69–75. [DOI] [PubMed] [Google Scholar]
- 57.Kongshoj B, Thorleifsson A, Wulf HC. Pheomelanin and eumelanin in human skin determined by high-performance liquid chromatography and its relation to in vivo reflectance measurements. Photodermatol Photoimmunol Photomed 2006;22:141–7. 10.1111/j.1600-0781.2006.00215.x [DOI] [PubMed] [Google Scholar]
- 58.Wulf HC, Philipsen PA, Ravnbak MH. Minimal erythema dose and minimal melanogenesis dose relate better to objectively measured skin type than to Fitzpatricks skin type. Photodermatol Photoimmunol Photomed 2010;26:280–4. 10.1111/j.1600-0781.2010.00544.x [DOI] [PubMed] [Google Scholar]
- 59.Andersen JD, Pereira V, Pietroni C, Mikkelsen M, Johansen P, Børsting C, et al. Next-generation sequencing of multiple individuals per barcoded library by deconvolution of sequenced amplicons using endonuclease fragment analysis. BioTechniques 2014;57:91–4. 10.2144/000114200 [DOI] [PubMed] [Google Scholar]
- 60.Dodt M, Roehr JT, Ahmed R, Dieterich C. FLEXBAR-Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms. Biology 2012;1:895–905. 10.3390/biology1030895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinforma Oxf Engl 2009;25:1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duffy DL, Box NF, Chen W, Palmer JS, Montgomery GW, James MR, et al. Interactive effects of MC1R and OCA2 on melanoma risk phenotypes. Hum Mol Genet 2004;13:447–61. 10.1093/hmg/ddh043 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(XLSX)
Data Availability Statement
Data may compromise the privacy of study participants and may not be shared publicly. Data are available upon request to the authors.