Abstract
Metastatic tumor cells colonize the pre-metastatic niche, which is a complex microenvironment consisting partially of extracellular matrix (ECM) proteins. We sought to identify and validate novel contributors to tumor cell colonization using ECM coated poly(ε-caprolactone) (PCL) scaffolds as mimics of the pre-metastatic niche. Utilizing orthotopic breast cancer mouse models, fibronectin and collagen IV-coated scaffolds implanted in the subcutaneous space captured colonizing tumor cells, showing a greater than 2-fold increase in tumor cell accumulation at the implant site compared to uncoated scaffolds. As a strategy to identify additional ECM colonization contributors, decellularized matrix (DCM) from lungs and livers containing metastatic tumors were characterized. In vitro, metastatic cell adhesion was increased on DCM coatings from diseased organs relative to healthy DCM. Furthermore, in vivo implantations of diseased DCM-coated scaffolds had increased tumor cell colonization relative to healthy DCM coatings. Mass-spectrometry proteomics was performed on healthy and diseased DCM to identify candidates associated with colonization. Myeloperoxidase was identified as abundantly present in diseased organs and validated as a contributor to colonization using myeloperoxidase-coated scaffold implants. This work identified novel ECM proteins associated with colonization using decellularization and proteomics techniques and validated candidates using a scaffold to mimic the pre-metastatic niche.
Keywords: Metastasis, Colonization, Extracellular matrix, Organ decellularization, Pre-metastatic niche
1. INTRODUCTION
The likelihood of patient survival significantly decreases with the spread of metastatic tumor cells from the primary tumor to distant organs. The process of spreading to distant organs is not random but predetermined, as indicated by Paget’s “seed and soil” hypothesis [1]. In breast cancer, metastatic tumor cells tend to colonize the liver, lung, brain, and bone [2]. This targeting of specific organs implies that metastatic cells have a proclivity for specific microenvironments. These microenvironments – termed pre-metastatic niches – are formed to prime the target organ for tumor cell colonization and provide a initial site for tumor cell growth [3,4]. A myriad of factors promotes tumor cell colonization at pre-metastatic niche sites, including interactions with tumor-associated neutrophils [5], chemoattractants [6], immune cell secreted factors due to hypoxic stress at the site [7], integrin interactions with the surrounding matrix [8], and the mechanical topography and stiffness of the niche microenvironment [9]. As the local organ microenvironment is remodeled to become permissive for tumor cell arrival, tumor cells undergo genetic and epigenetic alterations at the niche to allow for organ-specific metastasis [10–12]. Since the environment is permissive to metastatic cell colonization, identifying the specific components of the niche may provide strategic targets to block the spread of metastatic cells.
Aberrant accumulations of extracellular matrix (ECM) proteins in organs contribute to the establishment of a pre-metastatic niche, which are critical for organ-specific metastasis. The adhesion of circulating tumor cells to the remodeled ECM at the pre-metastatic niche can transition tumor cells to a proliferative state [13]. Changes in the ECM composition of the niche during metastatic progression include the excess production of fibronectin [4] and collagen IV [14]. ECM remodeling is mediated in part by immune cells that localize to target organs, and ultimately promote the recruitment of metastatic cells to the site [15]. Additionally, tumor supportive fibroblasts present at the niche are also known to deposit matrix proteins that prepare the site for tumor cell arrival [4]. Each component of the pre-metastatic niche contributes to the generation of a complex, permissive ECM microenvironment for tumor cell colonization. While fibronectin and collagen IV have been identified in the niche previously, additional proteins are expected to contribute to the inherent complexity of this microenvironment.
In this report, we characterized the matrix composition of target organs containing metastatic tumors and validated ECM proteins that contribute to colonization of metastatic cells using biomaterial mimics of the pre-metastatic niche. Using orthotopic mouse models of breast cancer, we isolated and decellularized lungs and livers from diseased mice to generate functional decellularized matrix (DCM) protein coatings that promote tumor cell adhesion in vitro. ECM and DCM coatings were also coated onto porous poly(ε-caprolactone) (PCL) scaffolds that were implanted in tumor-bearing mice to evaluate the role of ECM and organ DCM in metastatic cell colonization to a synthetic site in vivo. Using proteomics, we characterized the DCM from diseased lungs and livers relative to healthy controls to identify potential candidates of metastatic cell colonization. One candidate, myeloperoxidase, was identified and validated as a mediator of metastatic cell colonization. The synthetic scaffold environment and DCM coatings can be readily applied to identify proteins that mediate metastatic cell colonization, which may ultimately provide targets for inhibiting pre-metastatic niche formation.
2. MATERIALS AND METHODS
2.1. PCL scaffold fabrication and imaging
Poly(ε-caprolactone) (PCL, Durect Corporation) was dissolved to a 6% weight/volume solution in dichloromethane overnight. Using a single emulsion approach, the PCL solution was homogenized in a 10% solution of polyvinyl alcohol (PVA) and mixed with water for 3 hours to yield PCL microspheres. Particles were centrifuged at 2000g and rinsed with dH2O to wash away residual PVA. After rinsing, particles were flash frozen in liquid nitrogen and lyophilized overnight to yield dry PCL particles.
For scaffold fabrication, PCL particles and NaCl crystals (250–425 µm crystal size) were mixed at a 1:30 PCL/NaCl mass ratio. The particle mixture was pressed using a steel die at 1500 psi. Scaffolds were heat-treated at 55°C for 10 minutes and gas-foamed in a CO2 chamber at a constant pressure of 800 psi for 24 hours. Scaffolds were leached in dH2O to remove NaCl crystals and treated in a 0.5 M NaOH solution for 3 hours. Scaffolds were washed with dH2O until a neutral pH was reached and sterilized with 70% EtOH for 1 minute. Scaffolds were dried and stored at room temperature in a dessicator until use.
For protein coatings, 30 µL of 1 mg/mL solutions of collagen IV (Sigma), fibronectin (Sigma), decellularized matrix, or recombinant myeloperoxidase (R&D Labs) were slowly added to scaffolds and incubated overnight for protein adsorption. For imaging, protein-coated scaffolds were allowed to dry completely, mounted and sputter-coated with 10 nm gold particles, and imaged using a scanning electron microscope (NOVA 600 NanoLab, FEI company). Images were taken using a voltage of 20 kV and a current of 48 pA.
2.2. Cell culture
Human LM2-4 cells derived from MDA-MB-231 cells [16] and mouse 4T1 cells were used for all experiments. LM2-4 cells were routinely cultured on tissue culture polystyrene flasks in RPMI 1640 media supplemented with 10% fetal bovine serum (FBS), 1% penicillinstreptomycin solution, and 0.02% gentamicin (Life Technologies). 4T1 cells were cultured in RPMI 1640 media supplemented with 10% FBS and 1% penicillin-streptomycin solution (Life Technologies). Cells used for mouse tumor inoculations were cultured in penicillin-streptomycin free media. Media was exchanged every other day. Once ~80% confluent, cells were harvested with 0.05% Trypsin/EDTA (Life Technologies) solution and counted using a Trypan blue stain (Sigma Aldrich) and a Cell Countess automated hemocytometer (Life Technologies). Cells were cultured in a humidified 5% CO2 incubator at 37°C.
2.3. Tumor inoculations and organ harvest
Animal studies were performed in accordance with institutional guidelines and protocols were approved by the Northwestern University Institutional Animal Care and Use Committee (IACUC). Tumor inoculation was performed by injecting 2 × 106 LM2-4 or 4T1 cells in a volume of 50 µL PBS (Life Technologies) into the number four right L PBS (Life Technologies) into the number four right mammary fat pads of female NOD/SCID-IL2Rã −/− (NSG) or Balb/C mice (The Jackson Laboratory), respectively. Diseased lung and livers were harvested from mice 28 days post-inoculation for NSG mice and 21 days post inoculation for Balb/C mice. Healthy lungs and livers were harvested from mice without tumor inoculation. Organs were stored at −80°C until use.
2.4. Scaffold implantation and flow cytometry
Female NSG or Balb/C mice were inoculated with LM2-4 or 4T1 cells respectively as described above. One week post-inoculation, two protein-coated scaffolds were implanted subcutaneously in the left and right dorsal regions. One week post-scaffold implant, scaffolds were harvested from euthanized mice and minced with microscissors in a 0.38 mg/mL solution of Liberase TL in HBSS. Samples were incubated at 37 °C for 20 minutes and neutralized with 0.125 M EDTA (Life Technologies). The digested sample was passed through a 70 µm filter (BD Biosciences) and rinsed with FACS buffer. For leukocyte population analysis, scaffold and spleen cells were blocked with anti-CD16/32 (1:50, BioLegend) and stained for viability using fixable violet dead cell stain kit (Life Technologies). Cells were then stained with Alexa Fluor® 700-conjugated anti-CD45 (30-F11, 1:125; Biolegend), Pacific Blue-conjugated anti-Gr-1 (RB6-8C5, 1:70; Biolegend), FITC-conjugated anti-Ly-6C (HK1.4, 1:100; Biolegend), PE-Cyanine7-conjugated anti-F4/80 (BM8, 1:70; Biolegend), APC-conjugated anti-CD11c (N418, 1:85; eBioscience), v500-conjugated anti-CD11b (M1/70, 1:100; BD Biosciences), Pacific Blue-conjugated anti-CD19 (6D5, 1:100; Biolegend), v500-conjugated anti-CD4 (RM4-5, 1:100; BD Biosciences), and FITC-conjugated anti-CD8a (53–6.7, 1:25; Biolegend). For tumor cell quantification, PE+ events were counted and checked for auto-fluorescence against the APC-Cy7 channel. For Ki67 analysis, cells were permeabilized with 70% EtOH, washed with cell staining buffer (BioLegend), and stained with FITC-conjugated anti-Ki67 antibody (16A8, 1:50; Biolegend). All samples were re-suspended in FACS buffer and analyzed using a BD LSR Fortessa flow cytometer (Becton Dickinson Immunocytometry Systems, BDIS).
2.5. Organ decellularization
Healthy and diseased organs were slightly thawed and cut into ~1 mm thick slices using a razor. Slices were decellularized using a sequential 30 minute treatments of 1%, 2%, and 3% Triton X-100 solutions, followed by an overnight treatment of 0.1% sodium dodecyl sulfate (SDS) on a rocker with vigorous shaking at 4°C. Treatments were repeated the following day until tissues were completely decellularized. Samples were rinsed liberally with PBS and flash frozen in liquid nitrogen. The tissue was lyophilized overnight to yield decellularized matrix (DCM) and stored at 4°C until use.
2.6. Histology and immunofluorescence staining
Decellularized and control organ samples were fixed in 10% formalin for 24 hours and embedded in paraffin wax for sectioning. Tissue samples were sliced into 4 µm thick sections, heat-fixed and xylene-treated prior to staining. For cellular staining, sections were stained for 10 minutes with Gill III Hematoxylin and Eosin Y (Leica). For immunofluorescence staining, sections were treated with boiling 0.01 M sodium citrate solution supplemented with 0.05% Tween-20 and brought to a pH of 6.0 for antigen retrieval. Sections were blocked with a 10% normal goat serum solution and stained with a 1:200 dilution of myeloperoxidase antibody (Abcam) and 1:500 dilution of Alexa Fluor 488 secondary antibody (Abcam). Tissues were co-stained with a 1:100 dilution of collagen IV or fibronectin antibody (Santa Cruz Biotechnology) and a 1:200 dilution of Alexa Fluor 555 secondary antibody (Santa Cruz Biotechnology). Stained sections were preserved with Vectashield DAPI mounting medium (Vector Labs) and imaged using a Zeiss upright fluorescence microscope. Fluorescence exposure was adjusted as needed to avoid over-saturated images of tissue samples.
2.7. Decellularized matrix coatings
Lyophilized DCM was minced to a fine powder using a razor blade. A 1 mg/mL solution of DCM was prepared using a 10 mg/mL solution of pepsin from porcine mucosa (Sigma) dissolved in 0.1 M of hydrochloric acid under constant gentle stirring for 72 hours. Aliquots of DCM solutions were stored at −20°C until use. DNA concentration in decellularized samples was tested using a PicoGreen dsDNA Assay Kit (Life Technologies) according to the manufacturer’s instructions. DCM solutions were diluted in 0.25% acetic acid and allowed to adsorb to 96-well cell culture plates for 24 hours. Adsorbed myeloperoxidase on tissue culture plastic was visualized using immunofluorescence with manufacturer recommended primary antibody dilutions (1:200, Abcam).
2.8. Adhesion assays
Tumor cell adhesion was quantified using two-dimensional tissue culture treated polystyrene 96-well plates coated with various DCM solutions from both NSG and Balb/C organs. Prepared DCM coated plates were blocked with a 10 mg/mL solution of bovine serum albumin in supplement-free RPMI media for 1 hour at 37°C. After washing with PBS, 40,000 cells (4T1 or LM2-4) were added to each well and allowed to adhere for 45 minutes at 37°C. Unattached cells were washed away with PBS and attached cells were fixed and stained in a 0.5% wt/vol crystal violet solution using a 60% EtOH/40% PBS solvent. Plates were washed with dH2O and imaged using a Zeiss brightfield microscope. After imaging, the crystal violet inside of attached cells was solubilized using a 10% solution of acetic acid, and absorbance readings at 595 nm were quantified using a Cytation3 plate reader (Bio-Tek).
2.9. Proteomics of DCM
Lyophilized DCM powder was deglycosylated using a protein deglycosylation enzyme mix (New England Biolabs) according to the manufacturer’s instructions. Deglycosylated proteins were separated using a 4–20% gradient SDS-page gel (BioRad) and stained using Imperial Protein Stain (Pierce).
For each sample, gel lanes were cut from top to bottom into 18–20 pieces. Protein digestion was performed with sequencing grade trypsin (Promega, Madison WI) and extracted peptides from each condition were pooled together. The pooled samples were loaded directly onto a 15 cm long, 75 µM reversed phase capillary column (ProteoPep™ II C18, 300 Å, 5 µm size, New Objective, Woburn MA) and separated with a 70 minute gradient from 5% acetonitrile to 100% acetonitrile on a Proxeon Easy n-LC II (Thermo Scientific, San Jose, CA). The peptides were directly eluted into an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific, San Jose, CA) with electrospray ionization at a 350 nl/minute flow rate. The mass spectrometer was operated in data dependent mode, and for each MS1 precursor ion scan, the ten most intense ions were selected from fragmentation by CID (collision induced dissociation). The other parameters for mass spectrometry analysis were: (i) resolution of MS1 was set at 60,000, (ii) normalized collision energy 35%, (iii) activation time 10 ms, (iv) isolation width 1.5, and (v) +4 and higher charge states were rejected. Proteomic experiments were performed in duplicate.
The data were processed using Proteome Discoverer (version 1.4, Thermo Scientific, San Jose, CA) and searched using embedded SEQUEST HT search engine. The data were searched against reference proteome of homo sapiens (uniprot.org, Download date: June 2014). The other parameters were as follows: (i) enzyme specificity: trypsin; (ii) fixed modification: cysteine carbamidomethylation; (iii) variable modification: methionine oxidation and N-terminal acetylation; (iv) precursor mass tolerance was ±10 ppm; and (v) fragment ion mass tolerance was ±0.8 Da. All the spectra were searched against target/decoy databases and results were used to estimate the q values in Percolator algorithm as embedded in Proteome discoverer 1.4. The peptide identification was considered valid at q value < 0.1 and were grouped for protein inference to satisfy the rule of parsimony. In the final protein list, protein identification was considered only valid if supported by minimum of one unique peptide. The log2 fold change for each identified protein was calculated using the average peptide spectral matches from diseased DCM samples relative to healthy DCM samples.
2.10. Statistical significance
Data are shown as mean ± standard error (SEM) unless otherwise noted. Significance was claimed with p-values less than 0.05, determined using unpaired Student’s t-tests for single comparisons or one-way ANOVA with post-hoc testing for multiple comparisons. Statistical analysis was performed using GraphPad Prism.
3. RESULTS
3.1. Extracellular matrix coated PCL scaffolds increase tumor cell colonization
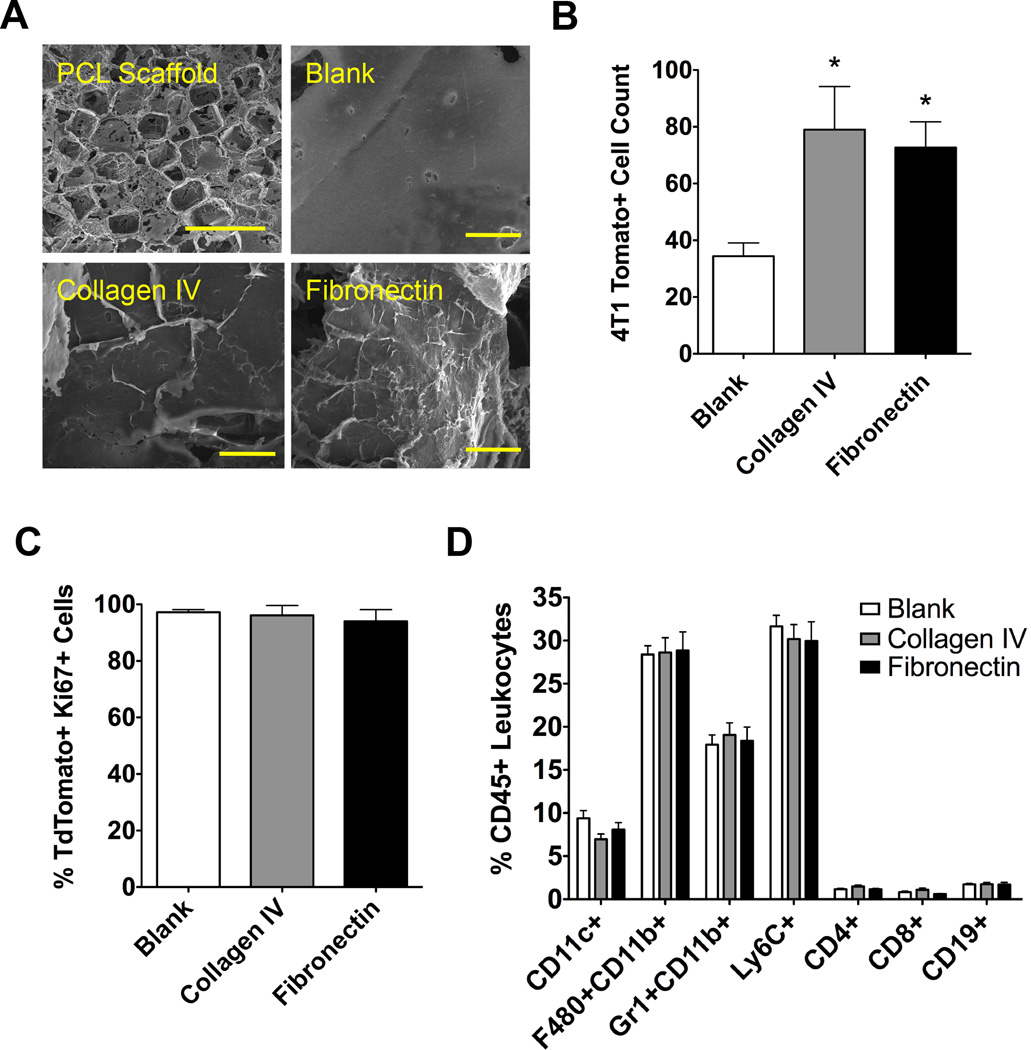
ECM proteins present in the pre-metastatic niche were utilized to enhance tumor cell colonization to PCL scaffolds implanted subcutaneously, which is a location to which metastatic cells do not normally colonize in our breast cancer models [17]. The ability of ECM to promote colonization was investigated by preparing PCL scaffolds coated with fibronectin and collagen IV, two ECM proteins previously shown to accumulate at the niche. Scanning electron microscopy images reveal the micro-porous structure of the PCL scaffold. Following overnight adsorption, collagen IV and fibronectin, fibers could be visualized on the struts of the porous scaffold (Fig. 1A). These scaffolds were then implanted subcutaneously in the dorsal region of Balb/C mice previously inoculated with 4T1 metastatic mouse tumor cells. ECM coatings of collagen IV and fibronectin resulted in a greater than 2-fold increase in the number of tumor cells recruited to the scaffold, with 79.0 ± 15.2 recruited cells to a collagen IV coated scaffold and 72.7 ± 9.0 cells recruited to a fibronectin coated scaffold, compared to 34.4 ± 4.7 cells recruited to a blank uncoated scaffold (Fig. 1B). Next, the proliferative capacity of the tumor cells was quantified using a Ki67 antibody stain and quantified via flow cytometry. No differences were observed in the proliferative capacity of the tdTomato+ tumor cells recruited to the scaffolds, with the percentage of Ki67+ cells at blank, collagen IV, and fibronectin scaffolds equaling 97.2 ± 1.0%, 96.1 ± 3.5%, and 94.0 ± 4.2% respectively (Fig. 1C). Additionally, no observed differences were observed in innate and adaptive immune cell populations at the scaffolds (Fig. 1D).
Figure 1. ECM-coated scaffolds for metastatic tumor cell colonization.
(A) SEM images of a porous PCL scaffold (top left scale bar = 1 mm) with zoomed in details of an uncoated blank scaffold and coated collagen IV and fibronectin scaffolds (scale bars = 50 µm). (B) Flow cytometric analysis of 4T1 tumor cell recruitment to coated PCL scaffolds implanted subcutaneously. Asterisk indicates statistical significance compared to blank control, P < 0.05 (n = 10). (C) Percent of Ki67-positive tumor cells present at implanted scaffolds analyzed using flow cytometry (n = 10). (D) Flow cytometry quantification of immune cell populations at ECM-coated scaffolds (n = 10).
3.2. Lung and liver DCM coatings enhance tumor cell adhesion in vitro
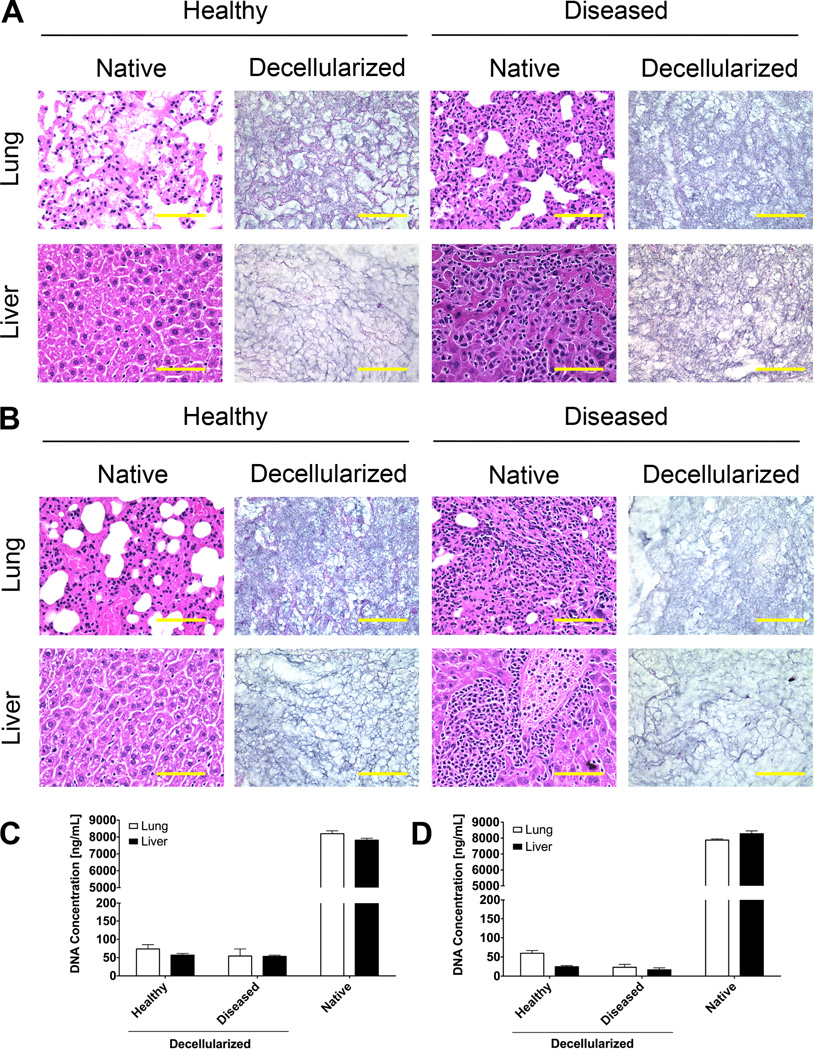
The decellularization of lungs and livers from healthy and diseased NSG and Balb/C mice yielded matrix representative of the organ ECM. Chemical treatments removed visible traces of cellular material, leaving behind the intact ECM for both healthy and diseased tissues (Fig. 2A, 2B). Upon digesting the DCM into solution, measurements of DNA concentration indicated low concentrations of DNA material for decellularized samples [18]. The decellularized lungs and livers contained less than 1% of the DNA present in native healthy tissue controls (Fig. 2C, 2D). These studies confirmed the DCM solutions contained minor detectable amounts of cellular material.
Figure 2. Decellularization of lungs and livers of healthy and diseased mice.
H&E sections of healthy and diseased lungs and livers from (A) NSG mice inoculated with LM2-4 cells and (B) BalbC mice inoculated with 4T1 cells (scale bar = 100 µm). DNA concentrations of decellularized matrix and native tissue digests of (C) NSG and (D) BalbC lungs and livers (n = 4).
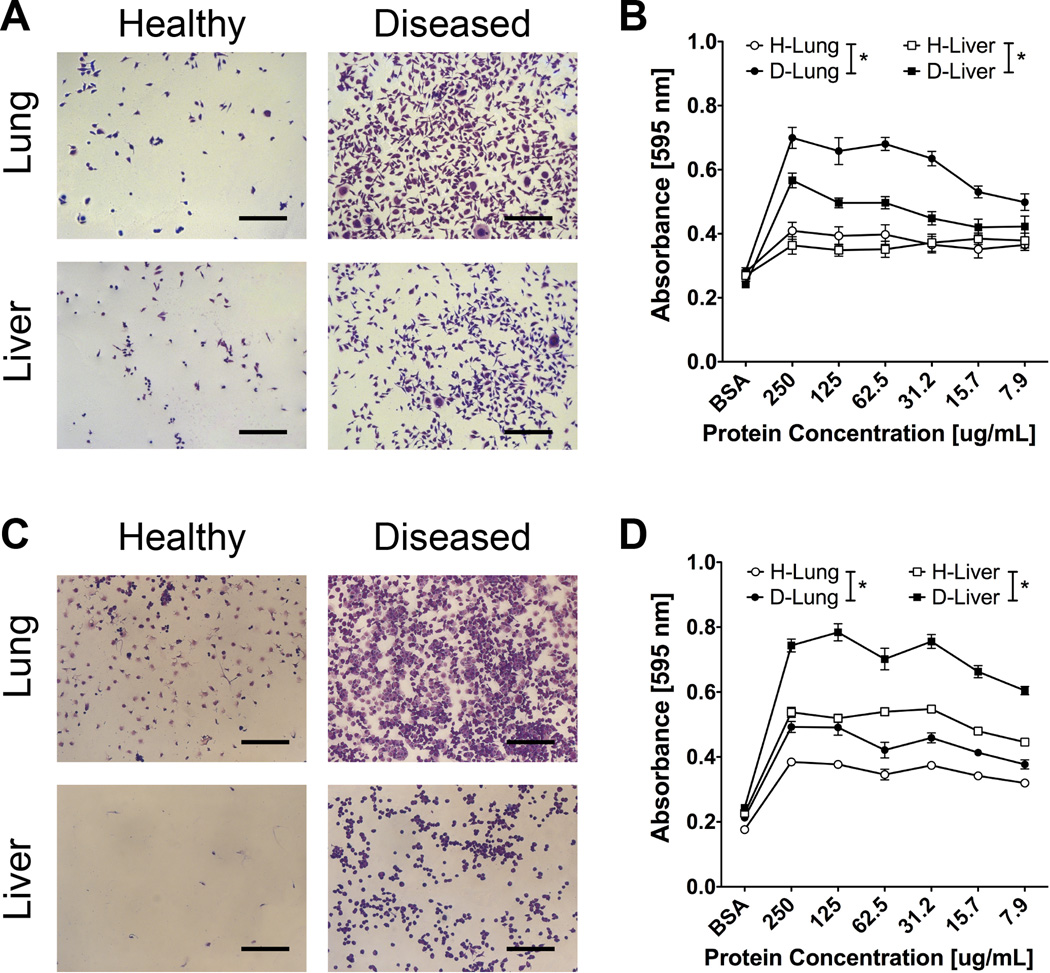
The DCM coatings were subsequently utilized for LM2-4 and 4T1 tumor cell adhesion studies. A greater number of LM2-4 tumor cells adhered to D-Lung and D-Liver DCM coatings compared to H-Lung and H-Liver coatings (Fig. 3A), which was consistent across multiple dilutions of DCM (Fig. 3B). Similarly, increased 4T1 tumor cell adhesion was observed on D-Lung and D-Liver matrix from Balb/C mice compared to H-Lung and H-Liver matrix (Fig. 3C), with studies again performed across a range of DCM concentrations (Fig. 3D). These results indicate diseased lung and liver decellularized matrix coatings enhance tumor cell adhesion for both LM2-4 and 4T1 metastasis models.
Figure 3. In vitro adhesion of LM2-4 and 4T1 cells on DCM coatings.
(A) Representative images of LM2-4 cells adhered to tissue culture plastic coated with lung and liver DCM from healthy and diseased NSG mice (scale bars = 300 µm, protein concentration of coating solution = 250 µg/mL). (B) Crystal violet absorbance values of LM2-4 cells adhered on DCM matrix coatings (n = 12), asterisk indicates P < 0.05 comparison at each protein concentration value (excluding BSA control). (C) Representative images of 4T1 cells adhered to tissue culture plastic coated with lung and liver DCM from healthy and diseased BalbC mice (scale bars = 300 µm, protein concentration of coating solution = 250 µg). (D) Crystal violet absorbance values of 4T1 cells adhered on DCM matrix coatings (n = 12), asterisk indicates P < 0.05 comparison at each protein concentration value (excluding BSA control).
3.3. Decellularized matrix coated PCL scaffolds enhance tumor cell colonization in vivo
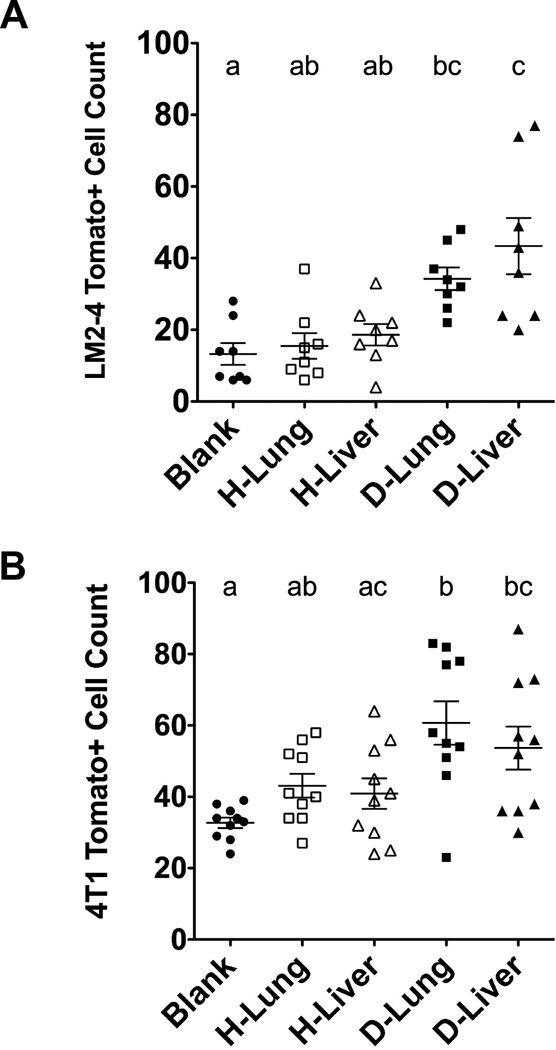
The DCM was subsequently used to coat PCL scaffolds, which were then implanted in mouse models of breast cancer to test the effects of DCM coatings on colonizing tumor cells in vivo. Seven days post-implantation, significantly more tumor cells colonized PCL scaffolds coated with DCM from D-Lung and D-Liver relative to coatings of H-Lung and H-Liver in both NSG and Balb/C mouse models inoculated with LM2-4 and 4T1 cells, respectively. For NSG mice, 13.3 ± 8.6 LM-2 cells were identified in uncoated PCL scaffolds compared to 15.5 ± 10.1 cells to H-Lung, 18.6 ± 8.5 cells to H-Liver, 34.2 ± 8.9 cells to D-Lung, and 43.4 ± 7.8 cells to D-Liver coated scaffolds (Fig. 4A). Similarly in Balb/C mice, 4T1 cell colonization to uncoated scaffolds was 32.7 ± 4.6 cells compared to 43.1 ± 3.3 cells to H-Lung, 40.9 ± 4.3 cells to H-Liver, 60.7 ± 6.1 cells to D-Lung, and 53.7 ± 6.0 cells to D-Liver (Fig. 4B). These results indicate that proteins present in the DCM of diseased organs enhance tumor cell colonization to the PCL scaffold.
Figure 4. In vivo tumor cell colonization to PCL scaffolds coated with various DCM solutions implanted subcutaneously.
(A) LM2-4 tumor cell colonization in NSG mice (n = 8) and (B) 4T1 tumor cell colonization in BalbC mice (n = 10) show increased tumor cell recruitment on diseased lung and liver DCM coatings compared to blank scaffolds. Groups with different letters indicate statistical significance (P < 0.05).
3.4. Proteomics analysis of DCM samples identifies myeloperoxidase as a mediator of tumor cell colonization
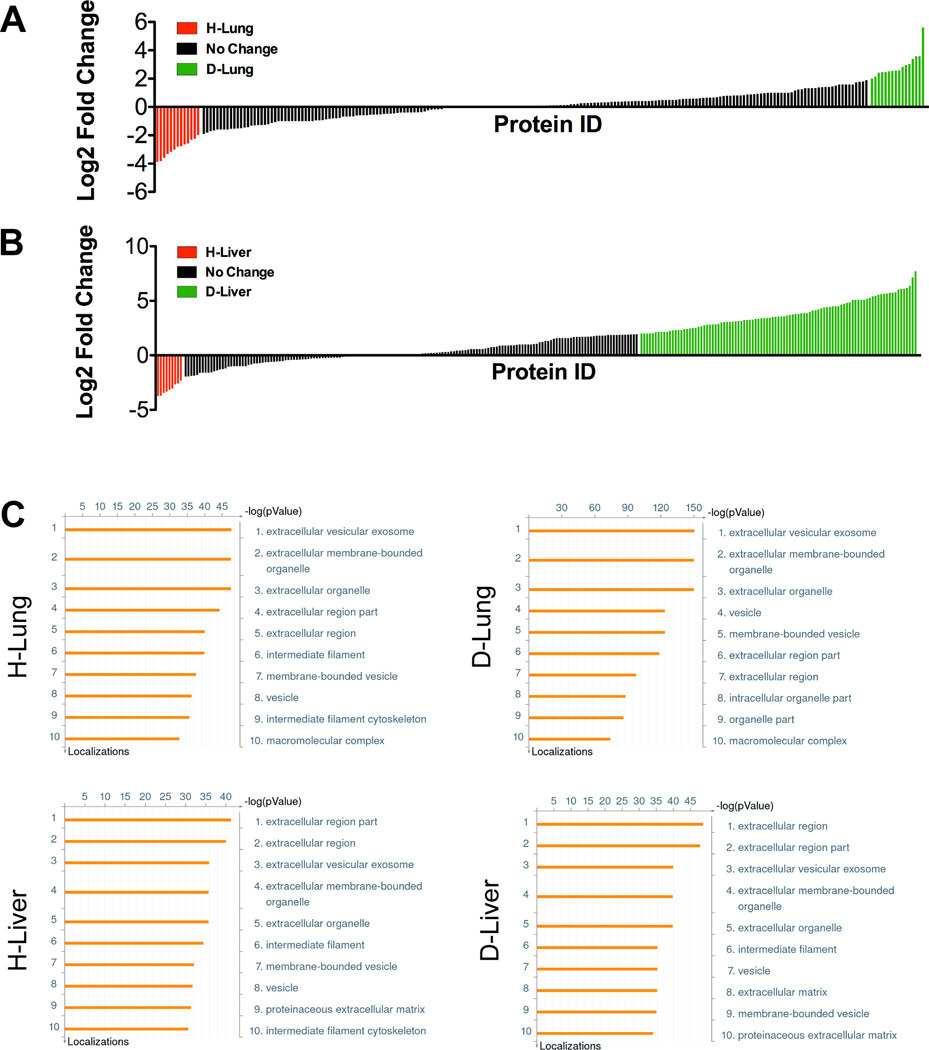
A proteomics approach to analyze the protein composition within DCM samples was implemented to identify factors associated with increasing tumor cell colonization on diseased DCM coatings. For NSG H-Lung and D-Lung, a total of 569 proteins were identified, with 207 proteins uniquely identified in D-Lung, 136 proteins uniquely identified in H-Lung, and 226 proteins identified in both samples. For the proteins that were identified in both H-Lung and D-Lung, 16 proteins in D-Lung had a log2 fold difference greater than 2 when compared to H-Lung, and 13 proteins in H-Lung had a log2 fold increase greater than 2 when compared to D-Lung (Fig. 5A). For NSG H-Liver and D-Liver, a total of 1020 proteins were identified, with 95 proteins identified uniquely in H-Liver, 672 proteins uniquely identified in D-Liver, and 253 proteins identified in both samples. For the proteins that were identified in both H-Liver and D-Liver, 96 proteins in D-Liver had a two-fold log2 fold difference when compared to H-Liver, and 9 proteins in H-Liver had a two-fold log2 fold increase when compared to D-Liver (Fig. 5B). Ontological analysis using MetaCore software indicated most of the proteins in H-Lung, D-Lung, H-Liver, and D-Liver as belonging to extracellular matrix locations (Fig. 5C).
Figure 5. Proteomic characterization of DCM coatings.
Graph showing log2 fold changes of healthy and diseased (A) lungs and (B) livers peptide spectral matches, where each tick on the x-axis represents an identified protein in the matrix. Log2 fold change values shaded in red indicate proteins 2-fold more abundant in healthy DCM, and log2 fold change values shaded in green indicate proteins 2-fold more abundant in diseased DCM. (C) Gene ontology analysis of lung and liver DCM coatings using MetaCore software show protein localizations.
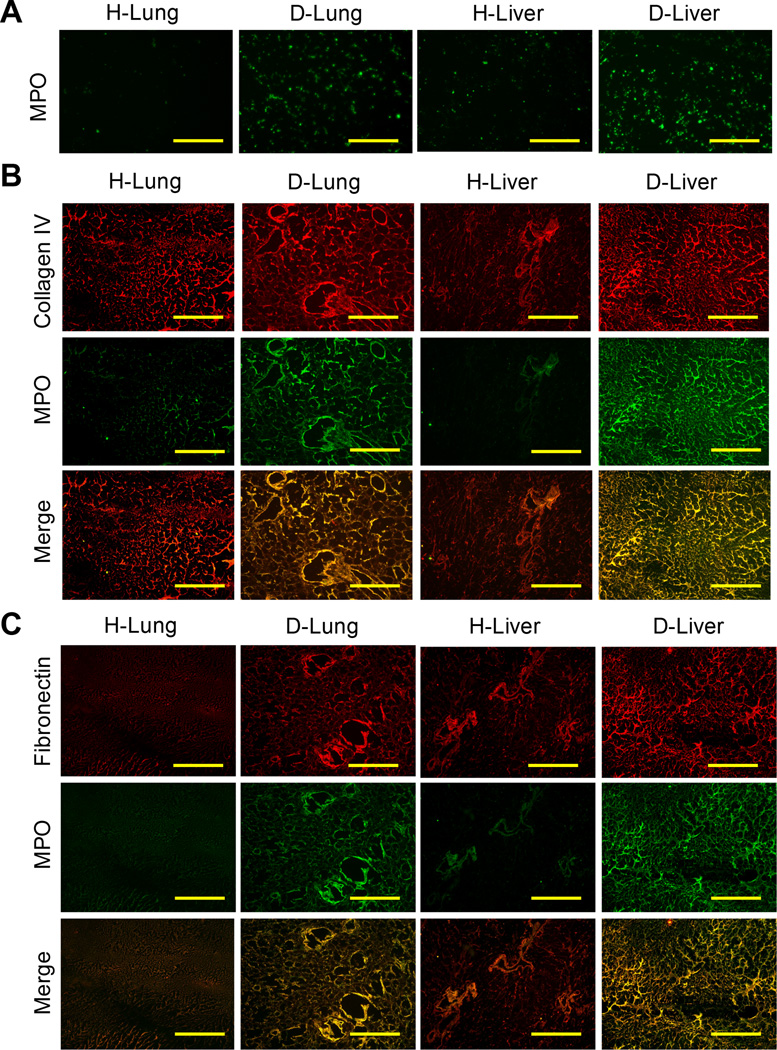
In both D-Lung and D-Liver protein catalogs, myeloperoxidase (MPO) was identified as most abundant compared to H-Lung and H-Liver samples. Myeloperoxidase abundance had a 2.96-fold increase in D-Lung relative to H-Lung and a 6.39-fold increase in D-Liver relative to H-Liver (Supplemental Table 1). Excessive expression of MPO is involved in the progression of many inflammatory-based diseases, including breast cancer [19]. However, the role of MPO in mediating metastatic spread remains unclear, and warranted further investigation in our study. Using DCM coatings of H-Lung, H-Liver, D-Lung, and D-Liver from NSG mice, MPO was confirmed to be more abundant on D-Lung and D-Liver coatings using immunofluorescence stains (Fig. 6A). Additionally, immunofluorescence stains revealed MPO was more abundantly present in diseased decellularized organs relative to healthy. Furthermore, the presence of MPO was shown to localize in regions rich with collagen IV and fibronectin (Fig. 6B, 6C).
Figure 6. Presence of myeloperoxidase in decellularized lung and liver tissues.
(A) Confirmation of increased abundance of myeloperoxidase in diseased lung and liver DCM compared to healthy DCM using immunofluorescence stains of coatings on tissue culture plastic (scale bars = 100 µm). Co-immunofluorescence stains for (B) collagen IV or (C) fibronectin with myeloperoxidase, showing increased abundance of myeloperoxidase in diseased tissues (scale bars = 100 µm).
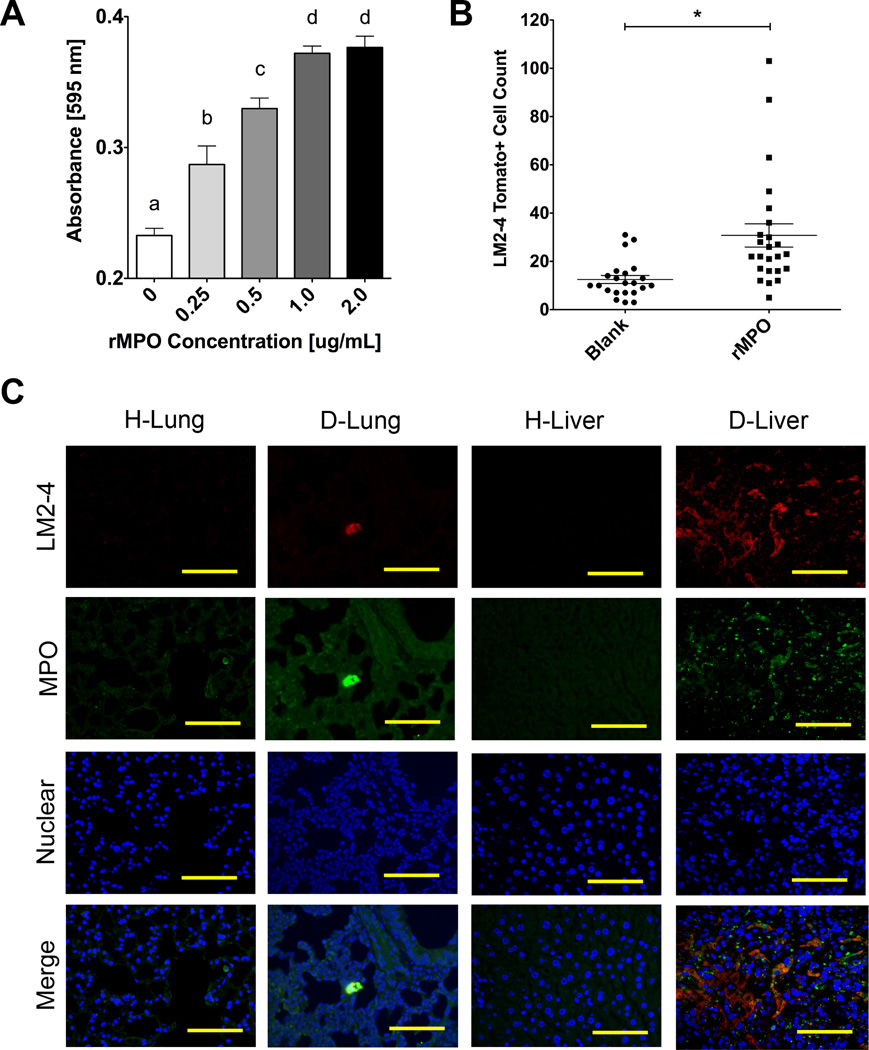
An adhesion assay was utilized to determine the role of myeloperoxidase on the adhesion of LM-2 cells. After coating tissue-cultured polystyrene with recombinant MPO (rMPO), LM2-4 cells showed concentration-dependent adhesion on rMPO-coated polystyrene (Fig. 7A). The role of MPO on tumor cell colonization was also investigated in vivo using MPO-coated PCL scaffolds implanted in NSG mice. Flow cytometry results showed a ~2.5-fold increase in tumor cell colonization to 30.7 ± 4.8 LM2-4 cells on rMPO-coated PCL scaffolds compared to 12.5 ± 1.7 LM2-4 cells on uncoated controls (Fig. 7B). Lung and liver sections were subsequently stained for MPO to determine co-localization with tumor cell clusters. Immunofluorescence (IF) stains revealed the presence of MPO localized around tdTomato+ tumor cell clusters in both D-Lung and D-Liver tissue sections (Fig. 7C). The increased abundance of MPO in diseased tissues compared to healthy tissues was visually confirmed, further validating the proteomic identification of MPO as a more abundant protein in diseased tissues.
Figure 7. Validation of myeloperoxidase as a contributor to metastatic cell colonization.
(A) Adhesion assay absorbance values showing concentration dependent adhesion of LM2-4 cells on tissue culture plastic treated with myeloperoxidase solutions at indicated protein concentrations (n = 4). Groups with different letters indicate statistical significance (P < 0.05). (B) In vivo colonization of LM2-4 cells on MPO-coated PCL scaffolds 7 days post subcutaneous implantation (n = 24). Asterisk indicates significance at P < 0.002. (D) Immunofluorescence stains showing MPO localization near tumor cell clusters in D-Lung and D-Liver sections compared to H-Lung and H-Liver controls (scale bars = 100 µm).
4. DISCUSSION
In this report, we utilized an implantable polymer scaffold as a platform to identify ECM-associated proteins as mediators of tumor cell colonization. Few reports have demonstrated metastatic tumor cell recruitment in animal models to implanted niches that mimic characteristics of the pre-metastatic niche. Biomaterials have been used as scaffolds to model inflammation-mediated metastasis [20,21], to identify stromal-derived factors associated with metastasis [22], and to mimic the bone marrow microenvironment as a metastatic niche [23]. Herein, we coated polymer scaffolds with ECM proteins typically present in the pre-metastatic niche to validate that the scaffolds can recapitulate the role of ECM on tumor cell colonization. ECM proteins are critical regulators of tumor cell colonization and assist in providing an optimal environment for tumor cell adhesion and growth in the pre-metastatic niche [24]. Using collagen IV and fibronectin coatings, we observed increases in the amount of tumor cells able to colonize to a PCL scaffold within seven days of implantation. Our results suggest the presence of ECM proteins mediate increased colonization of tumor cells at the ECM-coated scaffolds independent of proliferative capacity or immune cell mediated effects on tumor cell homing to the scaffold.
Decellularized organs were employed as a source to obtain and ultimately identify novel ECM proteins associated with metastatic colonization. Organ decellularization is a common method used in the tissue engineering field to obtain organ-specific ECM proteins [18]. Organ decellularization has been employed to study modifications to ECM associated with disease progression [25]. In previous metastasis models, decellularized lung tissue has been used to model metastatic tumor nodule formation ex vivo [26,27]. In this study, we implanted decellularized ECM coated scaffolds into NSG mice inoculated with a human breast cancer line that is known to metastasize to the lung and liver [16]. The organ microenvironment of lungs and livers was reproducibly modified following tumor cell inoculation and was employed to generate an abundant source of disease-modified ECM for coating purposes. Organ ECM has been used to coat tissue culture well plates to promote stem cell differentiation [28] and primary cell culture [29]. In our study, decellularized coatings were used to model organ-specific tumor cell adhesion and colonization. Our functional decellularized coatings demonstrated that diseased lungs and livers contained proteins that promote tumor cell adhesion in vitro and colonization in vivo. Our results broadly suggest that the decellularization of organs from diseased hosts can retain functional proteins that are associated with and likely contribute to disease progression.
Using mass-spectrometry proteomics, we have identified dozens of proteins more abundant in diseased organs than healthy organs, offering a list of ECM candidates that may contribute to organ-specific metastatic cell colonization. Numerous other studies have characterized ECM-associated proteins in decellularized matrix samples using mass-spectrometry proteomics [25,30,31]. By comparing the log2 fold change of quantified peptide spectral matches, we have identified multiple proteins present in diseased lung and liver samples (Supplementary Table 1). In these lists, ECM-associated proteins such as tenascin [32], MMP9 [33], and vitronectin [34] have been associated previously with colonization and metastasis. Interestingly, MPO was the only protein identified in both our D-Lung and D-Liver top 10 lists. MPO is known to enzymatically generate reactive oxygen species to destroy pathogens engulfed by phagocytes, and plays a role in mediating the progression of various inflammatory diseases [35]. MPO is expressed in Gr1+CD11b+ myeloid derived suppressor cells (MDSCs), and excessive expression of MPO has been linked to acute and chronic inflammatory diseases [35,36]. MDSCs are known to accumulate in pre-metastatic niches [33], suggesting increased levels of MPO at the niche may result from the accumulation of MDSCs and other immune cell types, such as neutrophils and monocytes found at target organs during metastatic progression [37]. Furthermore, MPO was recently identified by our laboratory as a candidate mediator of metastatic cell homing to the pre-metastatic niche after an analysis of the immune cell secretome in a MDA-MB-231 model of breast cancer [21]. MPO is also reported to electrostatically bind to ECM proteins including collagen IV and fibronectin, two ECM proteins known to be present in abundant quantities at pre-metastatic niche sites [38]. This prior knowledge motivated our validation studies to visualize the retention and localization of MPO in fibronectin- and collagen IV-rich sites in our decellularized organ samples, as well as test the role of MPO in colonization using our scaffold technology. To further demonstrate the versatility of PCL scaffolds, we implanted MPO-coated scaffolds in our NSG model and demonstrated increased metastatic cell colonization. Given these observations, our scaffolds may provide a novel strategy for validating proteins associated with metastatic colonization and disease progression. Through the effective use of proteomics, MPO was identified and validated as a novel mediator of tumor cell colonization using our scaffold technology. Further, our proteomics and decellularization approach may be utilized to identify additional contributors to metastatic disease progression, and may be extended to discover tissue-specific proteins that contribute to the progression of other diseases.
Our work also suggests that ECM-coated scaffolds may be used to collect colonizing tumor cells more effectively for downstream clinical applications. Circulating tumor cell capture at ECM-coated scaffolds is enhanced relative to blank scaffolds in vivo, suggesting that ECM coatings allow for a more permissive environment for tumor cell colonization. A previous report by Azarin et al. demonstrates that implanted polymer scaffolds can be used for the early detection of circulating tumor cells in orthotopic breast cancer mouse models, prior to their arrival at target organs [17]. The potential for early detection may be improved due to the increased accumulation of tumor cells to our defined niche that could facilitate the application of imaging tools to identify cancer cells within the implant. Furthermore, our biomaterials-based technique for capturing colonizing tumor cells may also facilitate current efforts in determining biomarkers associated with colonization, given the increased abundance of colonizing metastatic cells on the scaffold available for analysis [39]. The genomic analysis of colonizing tumor cells at the scaffold relative to primary tumor cells may provide insights on tumor cell heterogeneity during disease progression to determine more effective treatments against metastasis in a future clinical setting. Taken together, our ECM-coated scaffolds may improve the likelihood of collecting colonizing tumor cells in vivo, which may facilitate early detection and personalized therapies for metastatic cancer patients.
5. CONCLUSIONS
This report demonstrates the use of implantable biomaterial scaffolds, decellularized organ matrix, and proteomics to identify and validate novel ECM contributors to metastatic cell colonization. Our work demonstrates that PCL scaffolds may be used to mimic components of the pre-metastatic niche and thereby provide a platform to investigate mediators of tumor cell colonization. We confirmed the functionality of ECM protein coatings in enhancing the amount of tumor cells recruited to the scaffold implant site. Knowing ECM coatings impact cell colonization to the scaffold, we explored the role of organ ECM on adhesion and colonization using DCM coatings from diseased lungs and livers. The complexity of the diseased organ DCM was further characterized with proteomics to yield dozens of protein candidates associated with colonization. To validate our approach, myeloperoxidase was shown to localize to target tissues and mediate tumor cell colonization through in vitro adhesion assays and in vivo scaffold implantation studies. Collectively, our scaffold technology represents a novel juxtaposition of proteomics, metastasis models, and tissue engineering approaches to uncover target proteins associated with tumor cell colonization.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
The pre-metastatic niche consists partially of ECM proteins that promote metastatic cell colonization to a target organ. We present a biomaterials-based approach to mimic this niche and identify ECM mediators of colonization. Using murine breast cancer models, we implant microporous PCL scaffolds to recruit colonizing tumor cells in vivo. As a strategy to modulate colonization, we coated scaffolds with various ECM proteins, including decellularized lung and liver matrix from tumor-bearing mice. After characterizing the organ matrices using proteomics, myeloperoxidase was identified as an ECM protein contributing to colonization and validated using our scaffold. Our scaffold provides a platform to identify novel contributors to colonization, and allows for the capture of colonizing tumor cells for a variety of downstream clinical applications.
Acknowledgments
We thank Dr. Jason Wertheim at Northwestern University for insightful conversations and advice on decellularization techniques. We also thank Matt Bury for providing antibodies for ECM immunofluorescence stains and Dr. Ji Yi for his assistance with SEM imaging, both at Northwestern University. The Tumor Biology Core at Northwestern University provided LM2-4 cells for the study. The National Institutes of Health (R01CA173745) and the Northwestern H Foundation Cancer Research Award supported this research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the H Foundation. B.A.A. and G.G.B. are recipients of National Science Foundation Graduate Research Fellowships. The Northwestern Flow Cytometry Facility and a Cancer Center Support Grant (NCI CA060553) supported flow cytometry work. The Northwestern University Proteomics Core Facility supported secretomics work and analysis. The Simpson Querrey Institute Equipment Core provided access and support to the Cytation3 and microscopy equipment.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Fidler IJ. The pathogenesis of cancer metastasis: the “seed and soil” hypothesis revisited. Nat. Rev. Cancer. 2003;3:1–6. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 2.Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ-specific colonization. Nat. Rev. Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 3.Psaila B, Lyden D. The metastatic niche: adapting the foreign soil. Nat. Rev. Cancer. 2009;9:285–293. doi: 10.1038/nrc2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, et al. VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature. 2005;438:820–827. doi: 10.1038/nature04186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wculek SK, Malanchi I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature. 2015;528:413–417. doi: 10.1038/nature16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiratsuka S, Watanabe A, Aburatani H, Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat. Cell Biol. 2006;8:1369–1375. doi: 10.1038/ncb1507. [DOI] [PubMed] [Google Scholar]
- 7.Cox TR, Rumney RMH, Schoof EM, Perryman L, Høye AM, Agrawal A, et al. The hypoxic cancer secretome induces pre-metastatic bone lesions through lysyl oxidase. Nature. 2015;522:106–110. doi: 10.1038/nature14492. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Thibaudeau L, Taubenberger AV, Theodoropoulos C, Holzapfer BM, Ramuz O, Straub M, et al. New mechanistic insights of integrin β1 in breast cancer bone colonization. Oncotarget. 2014:1–13. doi: 10.18632/oncotarget.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balachander GM, Balaji SA, Rangarajan A, Chatterjee K. Enhanced metastatic potential in a 3D tissue scaffold toward a comprehensive in vitro model for breast cancer metastasis. Appl. Mater. Interfaces. 2015 doi: 10.1021/acsami.5b09064. [DOI] [PubMed] [Google Scholar]
- 10.Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu W, Giri DD, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 12.Bos PD, Zhang XH-F, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barkan D, Green JE, Chambers AF. Extracellular matrix: a gatekeeper in the transition from dormancy to metastatic growth. Eur. J. Cancer. 2010;46:1181–1188. doi: 10.1016/j.ejca.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erler JT, Bennewith KL, Cox TR, Lang G, Bird D, Koong A, et al. Hypoxia-induced lysyl oxidase Is a critical mediator of bone marrow cell recruitment to form the premetastatic niche. Cancer Cell. 2009;15:35–44. doi: 10.1016/j.ccr.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeNardo DG, Johansson M, Coussens LM. Immune cells as mediators of solid tumor metastasis. Cancer Metastasis Rev. 2008;27:11–18. doi: 10.1007/s10555-007-9100-0. [DOI] [PubMed] [Google Scholar]
- 16.Munoz R, Man S, Shaked Y, Lee CR, Wong J, Francia G, et al. Highly efficacious nontoxic preclinical treatment for advanced metastatic breast cancer using combination oral UFT-cyclophosphamide metronomic chemotherapy. Cancer Res. 2006;66:3386–3391. doi: 10.1158/0008-5472.CAN-05-4411. [DOI] [PubMed] [Google Scholar]
- 17.Azarin SM, Yi J, Gower RM, Aguado BA, Sullivan ME, Goodman AG, et al. In vivo capture and label-free detection of early metastatic cells. Nat. Commun. 2015;6:8094. doi: 10.1038/ncomms9094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32:3233–3243. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mika D, Guruvayoorappan C. Myeloperoxidase: the yin and yang in tumour progression. J. Exp. Ther. Oncol. 2011;9:93–100. [PubMed] [Google Scholar]
- 20.Ko CY, Wu L, Nair AM, Tsai YT, Lin VK, Tang L. The use of chemokine-releasing tissue engineering scaffolds in a model of inflammatory response-mediated melanoma cancer metastasis. Biomaterials. 2012;33:876–885. doi: 10.1016/j.biomaterials.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguado BA, Wu JJ, Azarin SM, Nanavati D, Rao SS, Bushnell GG, et al. Secretome identification of immune cell factors mediating metastatic cell homing. Sci. Rep. 2015;5:17566. doi: 10.1038/srep17566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bersani F, Lee J, Yu M, Morris R, Desai R, Ramaswamy S, et al. Bioengineered implantable scaffolds as a tool to study stromal-derived factors in metastatic cancer models. Cancer Res. 2014:7229–7239. doi: 10.1158/0008-5472.CAN-14-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seib FP, Berry JE, Shiozawa Y, Taichman RS, Kaplan DL. Tissue engineering a surrogate niche for metastatic cancer cells. Biomaterials. 2015;51:313–319. doi: 10.1016/j.biomaterials.2015.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barkan D, Kleinman H, Simmons JL, Asmussen H, Kamaraju AK, Hoenorhoff MJ, et al. Inhibition of metastatic outgrowth from single dormant tumor cells by targeting the cytoskeleton. Cancer Res. 2008;68:6241–6250. doi: 10.1158/0008-5472.CAN-07-6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner DE, Bonenfant NR, Parsons CS, Sokocevic D, Brooks EM, Borg ZD, et al. Comparative decellularization and recellularization of normal versus emphysematous human lungs. Biomaterials. 2014;35:3281–3297. doi: 10.1016/j.biomaterials.2013.12.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra DK, Thrall MJ, Baird BN, Ott HC, Blackmon SH, Kurie JM, et al. Human lung cancer cells grown on acellular rat lung matrix create perfusable tumor nodules. Ann. Thorac. Surg. 2012;93:1075–1081. doi: 10.1016/j.athoracsur.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong G, Flynn T, Chen J, Trinkle CA, Xu R. Development of an ex vivo breast cancer lung colonization model utilizing decellularized lung matrix. Integr. Biol. 2015:1518–1525. doi: 10.1039/c5ib00157a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DeQuach JA, Mezzano V, Miglani A, Lange S, Keller GM, Sheikh F, et al. Simple and high yielding method for preparing tissue specific extracellular matrix coatings for cell culture. PLoS One. 2010;5:e13039. doi: 10.1371/journal.pone.0013039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee JS, Shin J, Park H-M, Kim Y-G, Kim B-G, Oh J-W, et al. Liver extracellular matrix providing dual functions of two-dimensional substrate coating and three-dimensional injectable hydrogel platform for liver tissue engineering. Biomacromolecules. 2014;15:206–218. doi: 10.1021/bm4015039. [DOI] [PubMed] [Google Scholar]
- 30.Didangelos A, Yin X, Mandal K, Baumert M, Jahangiri M, Mayr M. Proteomics characterization of extracellular space components in the human aorta. Mol. Cell. Proteomics. 2010;9:2048–2062. doi: 10.1074/mcp.M110.001693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lü WD, Zhang L, Wu CL, Liu ZG, Lei GY, Liu J, et al. Development of an acellular tumor extracellular matrix as a three-dimensional scaffold for tumor engineering. PLoS One. 2014;9:1–13. doi: 10.1371/journal.pone.0103672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oskarsson T. Extracellular matrix components in breast cancer progression and metastasis. The Breast. 2013;22(Suppl 2):S66–S72. doi: 10.1016/j.breast.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 33.Ye XZ, Yu SC, Bian XW. Contribution of myeloid-derived suppressor cells to tumorinduced immune suppression, angiogenesis, invasion and metastasis. J. Genet, nomics. 2010;37:423–430. doi: 10.1016/S1673-8527(09)60061-8. [DOI] [PubMed] [Google Scholar]
- 34.Carter RZ, Micocci KC, Natoli A, Redvers RP, Paquet-Fifield S, Martin ACBM, et al. Tumour but not stromal expression of β3 integrin is essential, and is required early, for spontaneous dissemination of bone-metastatic breast cancer. J. Pathol. 2015;235:760–772. doi: 10.1002/path.4490. [DOI] [PubMed] [Google Scholar]
- 35.van der Veen BS, de Winther MP, Heeringa P. Myeloperoxidase: molecular mechanisms of action and their relevance to human health and disease. Antioxid Redox Signal. 2009;11:2899–2937. doi: 10.1089/ars.2009.2538. [DOI] [PubMed] [Google Scholar]
- 36.Youn J-I, Collazo M, Shalova IN, Biswas SK, Gabrilovich DI. Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. J. Leukoc. Biol. 2012;91:167–181. doi: 10.1189/jlb.0311177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith HA, Kang Y. The metastasis-promoting roles of tumor-associated immune cells. J. Mol. Med. 2013;91:411–429. doi: 10.1007/s00109-013-1021-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubala L, Kolárová H, Vítecek J, Kremserová S, Klinke A, Lau D, et al. The potentiation of myeloperoxidase activity by the glycosaminoglycan-dependent binding of myeloperoxidase to proteins of the extracellular matrix. Biochim Biophys Acta. 2013;1830:4524–4536. doi: 10.1016/j.bbagen.2013.05.024. [DOI] [PubMed] [Google Scholar]
- 39.Krebs MG, Metcalf RL, Carter L, Brady G, Blackhall FH, Dive C. Molecular analysis of circulating tumour cells—biology and biomarkers. Nat. Rev. Clin. Oncol. 2014;11:129–144. doi: 10.1038/nrclinonc.2013.253. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.