Abstract
Purpose
MYCN gene amplification (MNA) is a hallmark of aggressive neuroblastoma. We sought to determine the univariate and multivariate predictors of tumor MNA.
Patients and Methods
Data from the International Neuroblastoma Risk Group (INRG) were analyzed from the subset of 7,102 patients with known MYCN status. We used chi-squared testing and logistic regression to identify univariate and multivariate predictors of MYCN status. Recursive partitioning was used to identify groups of patients with maximal difference in rates of MNA.
Results
All clinical [age >18 months; high ferritin; high lactate dehydrogenase (LDH); INSS stage 4; adrenal site] and pathology/biology [DNA index ≤1; high MKI, undifferentiated/poorly differentiated grade; unfavorable histology by International Neuroblastoma Pathology Classification (INPC); segmental chromosomal aberrations (SCA)] features were significantly associated with MNA. LDH (OR=8.4; p<0.001) and chromosomal 1p LOH (OR 19.8; p<0.001) respectively were the clinical and biologic variables most strongly associated with MYCN-Amp. In logistic regression, all variables except chromosome 17q aberration and pooled SCAs were independently predictive of MNA. Recursive partitioning identified subgroups with disparate rates of MNA, including subgroups with 85.7% MNA [patients with high LDH who had poorly differentiated adrenal tumors with chromosome 1p deletion] and 0.6% MNA [localized tumors with hyperdiploidy, low MKI, and lacking chromosome 1p aberration].
Conclusion
MNA is strongly associated with other clinical and biologic variables in neuroblastoma. Recursive partitioning identifies subgroups of neuroblastoma patients with highly disparate rates of MNA. These findings can be used to inform investigations of molecular mechanisms of MNA.
Keywords: neuroblastoma, neuroblastoma/genetics, gene amplification, oncogene proteins/genetics, decision trees
Introduction
Neuroblastoma (NB) is a childhood tumor with marked clinical and biological heterogeneity.1,2,3 Stage, age4, histology3,5, lactate dehydrogenase (LDH) and ferritin levels3, MYCN status6,7, ploidy8,9, segmental chromosomal aberrations (SCA)3, and primary site10 have all been shown to be independently prognostic. In particular, MYCN amplification (MNA) is associated with rapid disease progression in patients of all ages or stages2,6,11 and is one of the strongest independent adverse prognostic factors.3
MYCN status is also associated with other adverse prognostic factors. For example, MNA is associated with unfavorable histology5, mitotic karyorrhectic index (MKI)12, and diploid/tetraploid tumors.13 MNA is associated with the presence of a range of SCAs, especially 1p deletion1,14,15, though 11q aberration shows a strong inverse relationship.16,17,18 The adverse prognostic effect of MNA can supersede otherwise favorable tumor genetics (e.g., near-triploid DNA content).2 MNA is found in a higher proportion of adrenal primary tumors10 and its incidence may also differ based upon the involvement of specific metastatic sites.19,20 Tumor MNA also varies by patient age, though this has been a complex nonlinear relationship in prior reports.21,22,23 To our knowledge, a comprehensive and definitive analysis of clinical and pathology/biology predictors of MNA in a large cohort of patients has not been reported. Such an analysis has the potential to inform to what extent variables associated with MNA interact, or are independent predictors, and therefore whether or not these associations are stronger in specific patient subgroups.
In the current study, we used the largest available international neuroblastoma dataset to define comprehensively the association of MNA with clinical and pathology/biology factors. We identified variables independently associated with MNA and patient subgroups with significantly different rates of MNA. These findings will provide a valuable resource for the field, will facilitate care of patients with neuroblastoma, and may lead to greater biological insight into the propensity for MNA. In particular, the use of recursive partitioning has the potential to reveal differences in the relative impact of predictors of MNA depending upon specific contexts defined by other variables.
Patients and Methods
Patients
The International Neuroblastoma Risk Group (INRG) database includes 8,800 individual patients <21 years of age with pathologically confirmed neuroblastoma who were diagnosed between January 1, 1990 and December 31, 2002.3 All patients were consented and enrolled on a neuroblastoma study within INRG member countries (France, Germany, Japan, Italy, Spain, United Kingdom) or within a cooperative group (Children’s Oncology Group, SIOPEN LNESG1 study). Only patients with known MYCN status were included.
Outcome Variable
MYCN status was the primary dichotomous outcome variable for this analysis. Within each INRG member group or country, MNA was determined by either interphase fluorescence in situ hydridization (I-FISH)24, polymerase chain reaction (PCR)25, array-based comparative genomic hybridization (aCGH)2, or multiplex ligation-dependent probe amplification (MPLA)2. For I-FISH, MNA has been defined as a more than 4-fold increase in the MYCN signal number compared to the reference probe on chromosome 2q.2 Data on number of copies of MYCN were not available for this analysis.
Predictor Variables
The INRG collects data on the following predictor variables used in this analysis: age; International Neuroblastoma Staging System (INSS) stage26,27; ploidy; presence of SCAs (1p, 11q, or 17q); ferritin level; LDH; primary site; sites of metastasis; histology (favorable vs. unfavorable by International Neuroblastoma Pathology Classification [INPC] system5); tumor diagnostic category (neuroblastoma vs ganglioneuroblastoma [intermixed, maturing, well-differentiated, or nodular]); grade (differentiating vs. poorly or undifferentiated); and MKI. Each continuous predictor variable was transformed into a binary variable for the analysis. Ferritin was categorized as high (≥92 ng/mL) versus low (<92 ng/mL) and LDH was categorized as high (≥587 U/L) versus low (<587 U/L), based on the mean values of ferritin and LDH in the overall INRG cohort, as previously reported3. Tumor primary site was categorized as thoracic vs. non-thoracic and adrenal vs. non-adrenal primary site. These site categories were chosen based on our previous work demonstrating the greatest effects of these sites on rates of MNA and because they are the most common primary sites10. We created a binary variable to reflect the presence of any SCA, defined as having 1p, 11q, and/or 17q aberrations.
Statistical Analyses
We used chi-squared testing to perform univariate analyses comparing the frequency of MNA as a function of each categorical predictor variable. We used a t-test to compare the distribution of ages as a continuous variable between patients with and without MNA.
We evaluated appropriate variables for inclusion in multivariate testing (logistic regression and recursive partitioning). Most potential predictor variables had missing data for >10% of patients; predictor variables ranged from 0% missing values (age) to 95.2% (17q chromosomal aberration). Within each predictor variable, we performed an assessment to determine if MYCN status was missing at random. The proportion of patients with MNA was calculated for the favorable category (decreased rates of MNA expected), the unfavorable category (increased rates of MNA expected), and the missing category. If MYCN status was missing at random, we expected the proportion of MNA in patients with missing data for the variable to fall between the proportions seen in the favorable and unfavorable groups. Tumor diagnosis category violated this pattern and was excluded from multivariate models.
INPC histology classification system incorporates age as well as grade and MKI. To avoid confounding of INPC histology and age in multivariate models, only the underlying components (grade, age, and MKI) were included.
We performed multivariate logistic regression to select variables independently associated with MNA. Excluding 17q chromosomal aberration, only 188/7102 patients had known data for all other predictors. To utilize the full sample size of the dataset without selection bias and allow inclusion of patients for whom some predictor variables had missing values, we created a series of “dummy” variables that fully describe each predictor variable as unfavorable (yes/no), favorable (yes/no), and missing (yes/no). The “dummy” variables for unfavorable and missing categories were included in the model, leaving the favorable category as the reference. This approach prevents bias that might occur if only the subset of patients with complete data was utilized in the model.28,29 We used a backward selection approach with p<0.05 to enter the model and p<0.05 to remain in the final model. Three multivariate logistic regression models were built: Model A - testing all covariates; Model B – testing clinical variables only; and, Model C – testing pathology/biology variables only. Model A was repeated including a binary variable for vital status (alive vs. dead), to see which baseline variables of interest remain associated with MNA after adjustment for vital status.
We used the classification and regression tree (CART) analysis method to identify age cut points associated with lower and higher rates of MNA.30 In this analysis, age as a continuous variable was the only predictor variable and MNA was the outcome variable. The first three (most highly predictive) age cut points from CART were eligible for selection in future models.
For multivariate recursive partitioning models, we used the univariate odds ratios for MNA to prioritize variables for selection. Each predictor variable was tested within the overall cohort and the statistically significant predictor with the largest odds ratio was selected manually to form the split. Each split created two nodes; odds ratios were recalculated and the process was repeated within each node. This process proceeded iteratively until either or both of the following pre-specified conditions were satisfied: 1) no remaining variables with a statistically significant (p<0.05) odds ratio; and/or 2) further split yielded a subgroup with fewer than 15 total patients.
We utilized CART in R (http://www.R-project.org/) for recursive partitioning using age as the only predictor variable. STATA version 12 (StataCorp, College Station, TX) was used for all other analyses.
Results
Patient Characteristics
The characteristics of the 7,102 patients with known MYCN status are shown in Table 1 and are similar to the full INRG cohort reported previously.3 MNA was reported in 1,155 patients in our cohort (16.3%).
Table 1.
Clinical and pathology/biology features of 7,102 patients with neuroblastoma according to MYCN amplification status.
| Clinical features | Sample size (N)1 | All (%) |
MYCN
Non-amplified (%)2 |
MYCN
Amplified (%)2 |
P-value3 | Odds Ratio4 |
|---|---|---|---|---|---|---|
| Mean age at diagnosis, months |
7102 | 25.1 | 24.3 | 28.1 | <0.001 | N/A |
| Age at diagnosis | 7102 | <0.001 | 3.0 | |||
| ≥ 18 months | 42.8 | 75.3 | 24.7 | |||
| < 18 months | 57.2 | 90.1 | 9.9 | |||
| INSS Stage | 7003 | <0.001 | 5.1 | |||
| 4 | 36.9 | 69.6 | 30.4 | |||
| Not Stage 4 | 63.1 | 92.1 | 7.9 | |||
| INSS Stage | 7003 | <0.001 | N/A | |||
| 1 | 23.9 | 97.1 | 2.9 | |||
| 2 | 14.1 | 96.0 | 4.0 | |||
| 3 | 17.6 | 82.4 | 17.6 | |||
| 4S | 7.5 | 91.1 | 8.9 | |||
| 4 | 36.9 | 69.6 | 30.4 | |||
| Primary tumor site | 6815 | <0.001 | 2.5 | |||
| Adrenal5 | 45.9 | 77.1 | 22.9 | |||
| Non-Adrenal | 54.1 | 89.2 | 10.8 | |||
| Primary tumor site | 6815 | <0.001 | 7.1 | |||
| Non-Thoracic | 84.7 | 81.2 | 18.8 | |||
| Thoracic5 | 15.3 | 96.9 | 3.1 | |||
| Lactate dehydrogenase (U/L) | 4354 | <0.001 | 8.4 | |||
| ≥ 587 | 48.6 | 70.9 | 29.1 | |||
| < 587 | 51.4 | 95.3 | 4.7 | |||
| Serum ferritin (ng/mL) | 3602 | <0.001 | 4.2 | |||
| ≥ 92 | 48.8 | 74.0 | 26.0 | |||
| < 92 | 51.2 | 92.2 | 7.8 | |||
| Pathology/Biology features | ||||||
| International Neuroblastoma Pathology Classification |
3866 | <0.001 | 14.8 | |||
| Unfavorable | 35.0 | 66.7 | 33.3 | |||
| Favorable | 65.0 | 96.7 | 3.3 | |||
| Ploidy | 3516 | <0.001 | 3.9 | |||
| ≤ 1 (diploid, hypodiploid) | 29.2 | 71.2 | 28.8 | |||
| > 1 (hyperdiploid) | 70.8 | 90.5 | 9.5 | |||
| Tumor diagnosis7 | 3349 | <0.001 | 10.1 | |||
| Neuroblastoma
or ganglioneuroblastoma, nodular |
95.3 | 83.6 | 16.4 | |||
| Ganglioneuroblastoma
(except nodular) or ganglioneuroma (maturing subtype) |
4.7 | 98.1 | 1.9 | |||
| Tumor diagnosis (N=3349) 7 | 3349 | <0.001 | 9.3 | |||
| Neuroblastoma | 89.1 | 82.7 | 17.3 | |||
| Ganglioneuroblastoma
or ganglioneuroma (maturing subtype) |
10.9 | 97.8 | 2.2 | |||
| Grade of differentiation | 2871 | <0.001 | 4.6 | |||
| Undifferentiated/Poorly differentiated |
83.8 | 82.1 | 17.9 | |||
| Differentiating | 16.2 | 95.5 | 4.5 | |||
| MKI | 2738 | <0.001 | 16.8 | |||
| High | 11.8 | 39.6 | 60.4 | |||
| Low/Intermediate | 88.2 | 91.7 | 8.3 | |||
| LOH at 1p | 2026 | <0.001 | 19.8 | |||
| Present | 22.2 | 46.3 | 53.7 | |||
| Absent | 77.8 | 94.5 | 5.5 | |||
| Pooled segmental chromosomal aberration 6 |
2060 | <0.001 | 10.4 | |||
| Present | 31.7 | 61.5 | 38.5 | |||
| Absent | 68.3 | 94.3 | 5.7 | |||
| Aberration at 11q | 1033 | 0.001 | 0.4 | |||
| Present | 20.9 | 91.7 | 8.3 | |||
| Absent | 79.1 | 82.1 | 17.9 | |||
| Gain of 17q | 340 | 0.01 | 2.1 | |||
| Present | 47.7 | 73.5 | 26.5 | |||
| Absent | 52.3 | 84.8 | 15.2 | |||
N represents total with known data for the given variable.
Row percentages sum to 100%.
P-value refers to a student’s T-test (for continuous age variable) or a chi-squared test for all other variables.
Odds ratio for univariate logistic regression with MYCN amplification as dependent variable.
Primary tumor site [adrenal or thoracic] versus all other primary tumor sites.
LOH at 1p, gain of 17q, and/or aberration at 11q.
International Neuroblastoma Pathology Classification tumor diagnosis category (includes neuroblastoma, ganglioneuroblastoma intermixed, ganglioneuroma maturing subtype or ganglioneuroblastoma well differentiated, ganglioneuroblastoma nodular, or unknown).
Incidence of MNA Varies with Age at Diagnosis
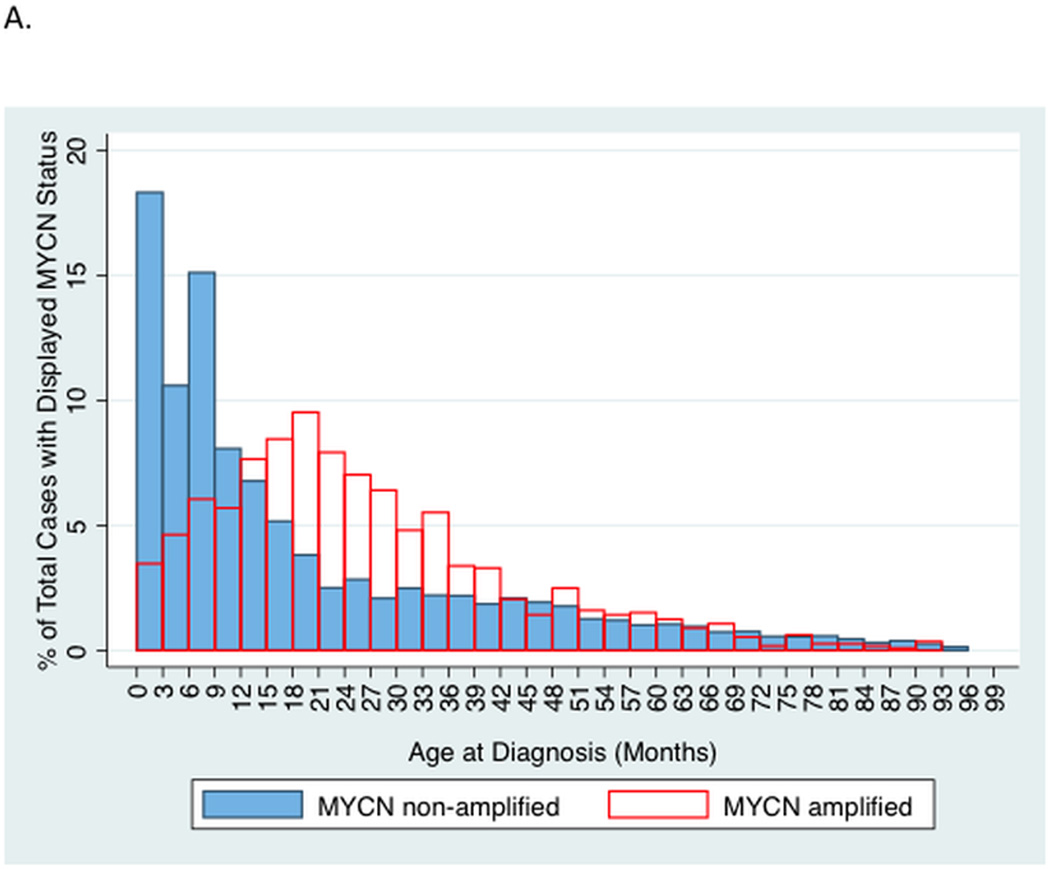
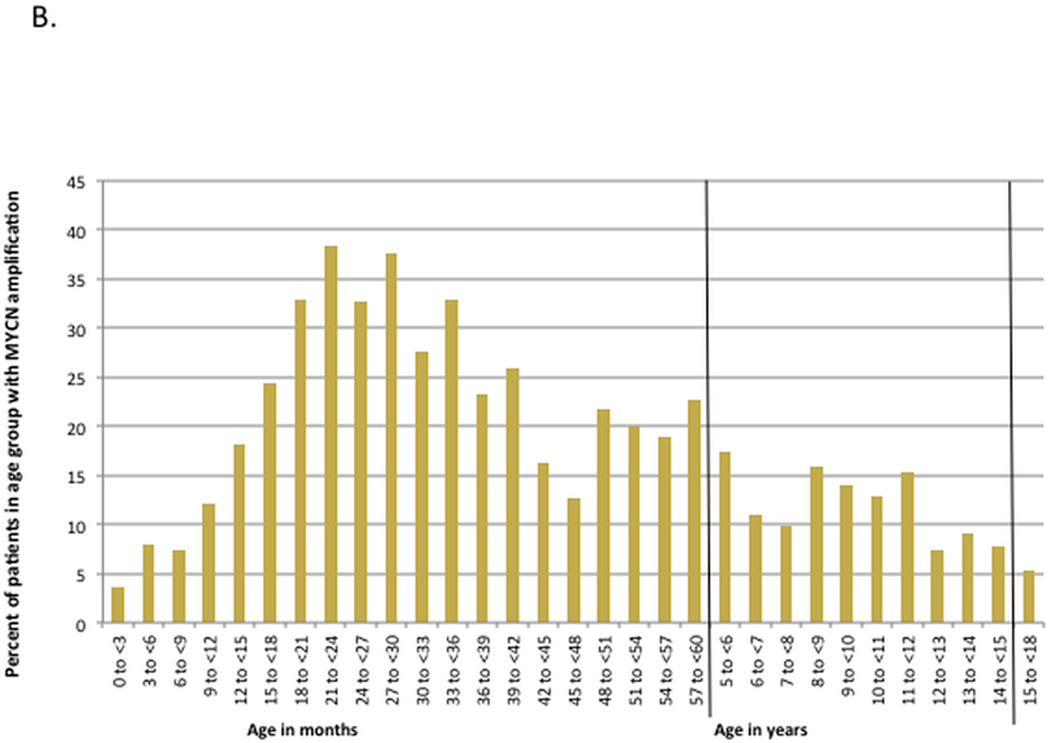
Patients with MYCN amplified tumors were older compared to patients with MYCN non-amplified tumors (mean age 28.1 versus 24.3 months, p<0.001). A higher percentage of patients ≥18 months had MNA compared to patients <18 months (24.7% versus 9.9%); in addition, 65% of patients with MNA were ≥18 months of age. To further characterize the relationship of MNA with age, we analyzed the proportion of total cases with and without MNA as a function of age (Figure 1A). Patients 18–20 months old contributed the highest proportion of cases with MNA (9.3% of all cases with MNA). In contrast, patients 0–2.9 months old contributed the highest proportion of cases without MNA (17.6% of all cases without MNA). We then plotted the percent of patients in a given age interval with MNA (Figure 1B). The 3-month interval with the highest incidence of MNA was in patients 21–23.9 months old (38.4%) and the lowest incidence was in patients 0–2.9 months old (3.6%).
Figure 1.
A. Two-way histogram comparing the proportion of total cases with MYCN amplification to those without, by age (sum of bars for each histogram = 100%).
B. Histogram showing percent of patients within a given age group that have MYCN-amplified tumors.
We used CART to identify optimal age cut points associated with the most disparate rates of MNA (Supplemental Figure 1). The top three age cutoffs identified by recursive partitioning were 367 days (12 months), 110 days (3.5 months), and 1282 days (42 months). Patients <110 days had an incidence of MNA of 3.6%, whereas patients 516–1086 days (17–35.5 months) had an incidence of MNA of 33.5%.
Additional Clinical and Pathology/Biology Features Are Associated with MNA
All clinical and pathology/biology variables were significantly associated with MYCN status in univariate analyses (Table 1). The clinical factor most strongly associated with MNA was high LDH [odds ratio (OR) 8.4; p<0.001], though multiple other variables had OR>4. The pathology/biology variable most strongly associated with MNA was LOH at 1p [OR 19.8; p<0.001], though multiple other variables had OR>10.
We created three separate logistic regression models (Table 2). When building a model that tested all clinical and pathology/biology variables, we demonstrated that all variables except gain of 17q and pooled SCAs were independently associated with MNA (Model A). After the inclusion of vital status, the results were similar to Model A except age was no longer independently associated with MNA (data not shown). Model B tested only clinical variables, and demonstrated that all clinical variables were independently associated with MNA. Model C tested only pathology/biology variables, and demonstrated that all pathology/biology variables except gain of 17q and pooled SCAs were independent predictors of MNA. In Model C, the point estimate for the OR for 1p LOH was similar to the univariate OR (17.8 from Table 2 model C vs. 19.8 from Table 1).
Table 2.
Results of multivariate logistic regression models predicting MYCN amplification in 7,102 patients with neuroblastoma and known MYCN status.
| Predictor variable (Unfavorable category shown / Favorable category is reference) |
Number of patients
with unfavorable variable / Number of patients with available data |
Odds Ratio for
MYCN Amplification in Presence of Unfavorable Predictor Variable (95% CI) |
P-value |
|---|---|---|---|
| Model A (N=7,102) - All Variables1 | |||
| Age ≥ 18 Months | 3041 / 7102 | 1.5 (1.3–1.8) |
<0.001 |
| Ferritin ≥ 92 (ng/mL) | 1757 / 3602 | 1.6 (1.3–2.1) |
<0.001 |
| LDH ≥ 587 (U/L) | 2114 / 4354 | 5.0 (4.0–6.4) |
<0.001 |
| INSS Stage 4 | 2585 / 7003 | 2.2 (1.9–2.6) |
<0.001 |
| Adrenal Primary Site | 3129 / 6815 | 1.5 (1.3–1.8) |
<0.001 |
| Non-thoracic Primary Site | 5771 / 6815 | 3.5 (2.4–5.1) |
<0.001 |
| Diploid or Hypodiploid | 1025 / 3516 | 2.6 (2.1–3.2) |
<0.001 |
| High MKI | 323 / 2738 | 9.9 (7.2–13.4) |
<0.001 |
| Poorly Differentiated
or Undifferentiated |
2407 / 2871 | 1.9 (1.1–3.1) |
0.014 |
| Chromosome 1p LOH Present |
449 / 2026 | 11.4 (8.2–15.9) |
<0.001 |
| Chromosome 11q Aberration Present |
216 / 1033 | 0.1 (0.06–0.2) |
<0.001 |
| Model B (N=7,102) - Clinical Variables Only1 | |||
| Age ≥ 18 Months | 3041 / 7102 | 1.7 (1.5–2.0) |
<0.001 |
| Ferritin ≥ 92 (ng/mL) | 1757 / 3602 | 1.7 (1.4–2.2) |
<0.001 |
| LDH ≥ 587 (U/L) | 2114 / 4354 | 5.5 (4.4–6.9) |
<0.001 |
| INSS Stage 4 | 2585 / 7003 | 2.7 (2.3–3.1) |
<0.001 |
| Adrenal Primary Site | 3129 / 6815 | 1.6 (1.4–1.9) |
<0.001 |
| Non-thoracic Primary Site | 5771 / 6815 | 3.8 (2.6–5.6) |
<0.001 |
| Model C (N=7,102) - Pathology/Biology Variables Only1 | |||
| Diploid or Hypodiploid | 1025 / 3516 | 3.3 (2.7–4.1) |
<0.001 |
| High MKI | 323 / 2738 | 13.7 (10.2–18.4) |
<0.001 |
| Poorly Differentiated
or Undifferentiated |
2407 / 2871 | 2.6 (1.6–4.3) |
<0.001 |
| Chromosome 1p LOH Present |
449 / 2026 | 17.8 (13.1–24.3) |
<0.001 |
| Chromosome 11q Aberration Present |
216 / 1033 | 0.2 (0.1–0.4) |
<0.001 |
Variables tested in the model and found non-significant were: A: Gain of 17q & pooled segmental chromosomal aberration; B: None; C: Gain of 17q & pooled segmental chromosomal aberration.
Recursive Partitioning Identifies Patient Subgroups with Disparate Rates of MNA
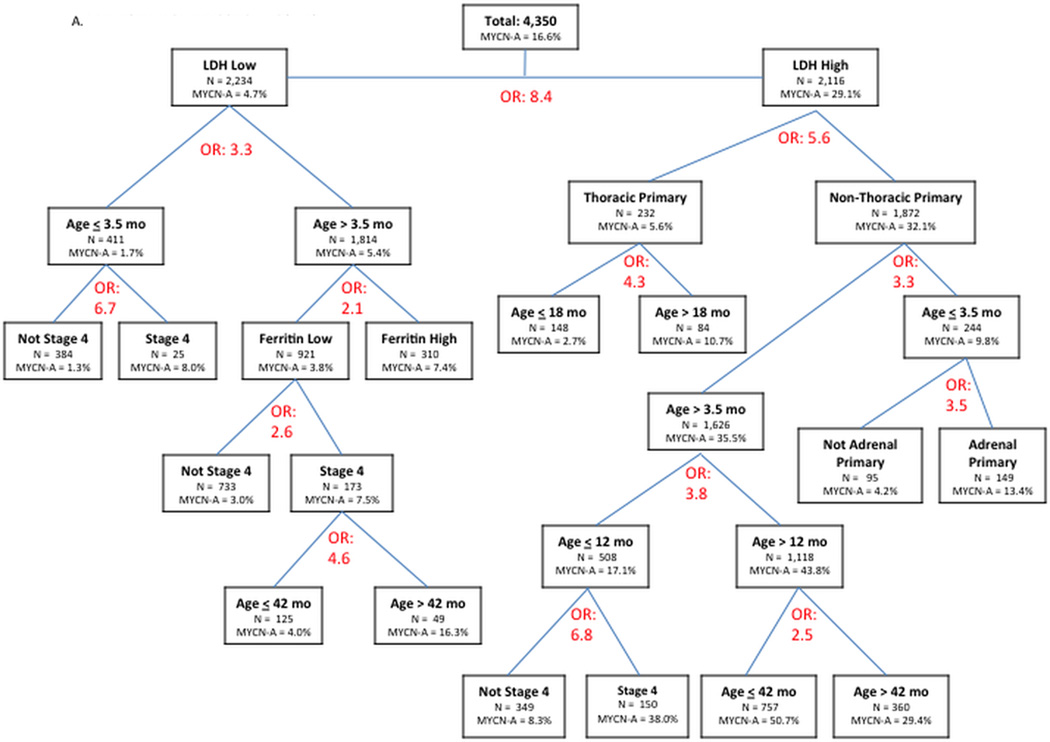
In multivariate recursive partitioning using only clinical variables (Figure 2A), we identified two extreme patient subgroups. The first subgroup with low LDH level, age <3.5 months, and non-stage 4 were MYCN amplified in 1.3%. In contrast, patients with high LDH, non-thoracic sites, and age >12 months but <42 months were MYCN amplified in 50.7%. The tree continued to split beyond these groups, but we made a post hoc decision to truncate the model after 5 splits to increase the practicality of the model (full tree with all splits shown in Supplemental Figure 2).
Figure 2.
A. Manual recursive partitioning using logistic regression to identify factors associated with MYCN amplification, testing only clinical variables (tree truncated at 5 levels of variables; see Supplemental Figure 2 for full tree). Odds Ratio (OR) listed refers to odds ratio for MYCN amplification for the variable that resulted in each node (e.g. OR 8.4 indicates patients with high LDH had 8.4 fold higher odds of MYCN amplification compared to patients with low LDH).
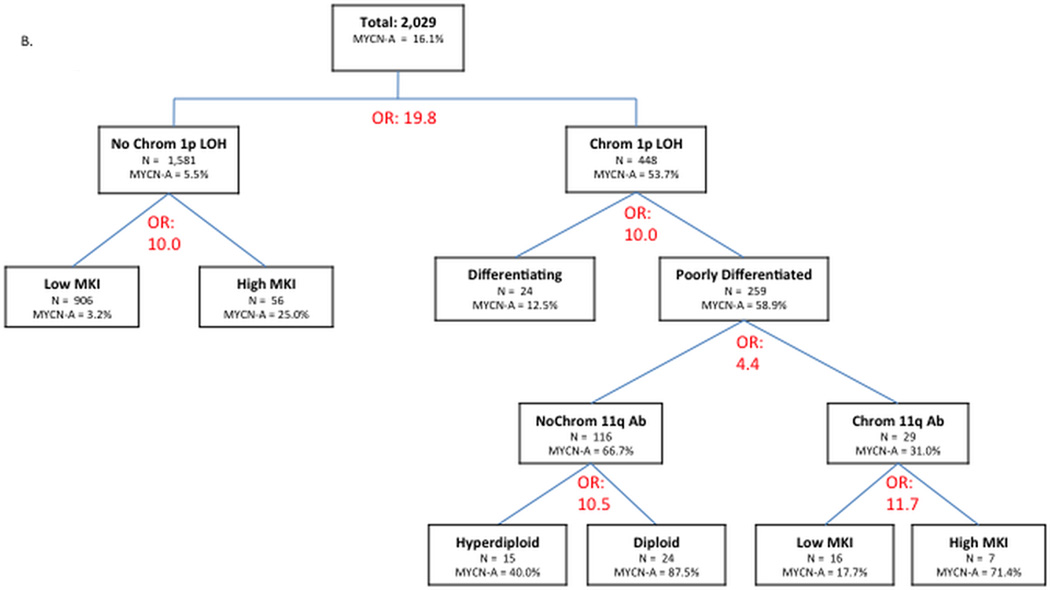
B. Manual recursive partitioning using logistic regression to identify factors associated with MYCN amplification, testing only pathology/biology variables. Variables tested and not included in the final tree were 17q gain and pooled segmental chromosomal aberrations.
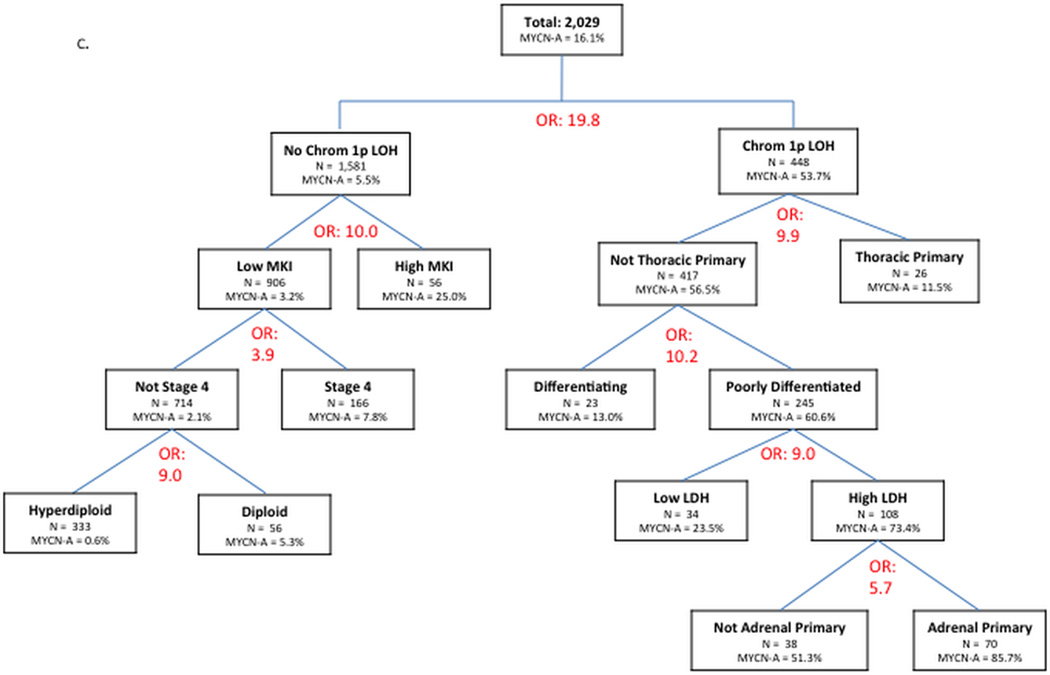
C. Manual recursive partitioning using logistic regression to identify factors associated with MYCN amplification, testing all potential predictor variables. Variables tested and not included in the final tree were: age; ferritin; 17q gain; pooled segmental chromosomal aberrations; and 11q aberration.
Our second tree utilized only pathology/biology variables (Figure 2B) and two extreme subgroups were identified. Tumors without 1p LOH and low MKI had a rate of MNA of 3.2%. In contrast, tumors with 1p LOH that were poorly differentiated, lacked 11q aberration and were diploid were MYCN amplified in 87.5%.
In our final recursive partitioning tree, all clinical and pathology/biology variables were available for selection (Figure 2C). This tree showed that non-stage 4 patients with low MKI, hyperdiploid tumors lacking 1p LOH were MYCN amplified in 0.6%. In contrast, patients with high LDH and adrenal primary tumors that were poorly differentiated and had 1p LOH were MYCN amplified in 85.7%. In this final tree with all variables available for selection, age was not chosen as a significant variable.
Metastatic Sites Correlate with MYCN Status
We repeated univariate analyses among the 2,176 patients with stage 4 disease and at least one known specific metastatic site to assess whether metastatic sites are associated with MNA (Table 3). Lung metastasis was the site most strongly associated with MNA (OR 3.0; p<0.001). Bone marrow and bone metastases were also significantly associated with MNA (OR 1.4 and 1.3, respectively; p<0.01), while skin metastases had lower likelihood of MNA (OR 0.4; p=0.01).
Table 3.
Univariate analysis of metastatic sites as predictors of MYCN status in patients with stage 4 neuroblastoma.1
| Metastatic site1 | Sample size (N)2 |
Percent with metastatic site (%) |
MYCN-Non
amplified (%)3 |
MYCN-amplified (%)3 |
P-value4 |
Odds Ratio5 |
|---|---|---|---|---|---|---|
| Bone marrow | 2171 | 0.002 | 1.4 | |||
| Yes | 72.1 | 68.1 | 31.9 | |||
| No | 27.9 | 74.9 | 25.1 | |||
| Bone | 2164 | 0.006 | 1.3 | |||
| Yes | 58.8 | 67.9 | 32.2 | |||
| No | 41.2 | 73.3 | 26.7 | |||
| Distant lymph node | 2165 | 0.76 | 1.0 | |||
| Yes | 33.9 | 70.4 | 29.6 | |||
| No | 66.1 | 69.8 | 30.2 | |||
| Liver | 2172 | 0.74 | 1.0 | |||
| Yes | 18.1 | 69.4 | 30.6 | |||
| No | 81.9 | 70.2 | 29.8 | |||
| Skin | 1958 | 0.01 | 0.4 | |||
| Yes | 3.3 | 84.6 | 15.4 | |||
| No | 96.7 | 70.1 | 29.9 | |||
| Lung | 1923 | <0.001 | 3.0 | |||
| Yes | 4.1 | 46.2 | 53.9 | |||
| No | 95.9 | 71.7 | 28.3 | |||
| Central Nervous System | 1923 | 0.6 | 1.2 | |||
| Yes | 2.6 | 67.4 | 32.7 | |||
| No | 97.4 | 70.8 | 29.2 | |||
| Other | 2128 | <0.001 | 1.5 | |||
| Yes | 27.8 | 64.2 | 35.8 | |||
| No | 72.2 | 72.5 | 27.5 | |||
At least one known metastatic site specified in the INRG database.
N represents total with known data for the given variable.
Row percentages sum to 100%.
p-value refers to a chi-squared test.
Odds ratio for univariate logistic regression with MYCN amplification as dependent variable.
In this same group, a multivariate logistic regression model, testing all of the potential clinical and pathology/biology variables previously evaluated, demonstrated that LOH at 1p, high LDH, absence of 11q aberration, high MKI, non-thoracic site, lung metastases, diploid/hypodiploid tumors, absence of skin metastases, and adrenal site were independently significantly associated with MNA (Table 4).
Table 4.
Multivariate predictors of MYCN status in 2,585 patients with stage 4 neuroblastoma.1
| Predictor variable2 (Unfavorable category shown / Favorable category is reference) |
Number of patients with unfavorable variable / Number of patients with available data |
Odds Ratio
(95% CI) |
P-value |
|---|---|---|---|
| Chromosome 1p LOH Present |
263 / 650 | 8.6 (5.4–14.0) |
<0.001 |
| LDH > 587 (U/L) | 1028 / 1504 | 6.8 (4.7–9.8) |
<0.001 |
| Chromosome 11q Aberration Absent |
151 / 379 | 6.1 (2.7–13.9) |
<0.001 |
| High MKI | 216 / 869 | 4.8 (3.1–7.5) |
<0.001 |
| Non-thoracic Primary Site |
2300 / 2508 | 3.1 (1.8–5.6) |
<0.001 |
| Lung Metastasis | 78 / 2270 | 2.4 (1.4–4.1) |
0.001 |
| No Skin Metastasis |
2240 / 2305 | 2.1 (1.0–4.4) |
0.05 |
| Diploid or Hypodiploid |
489 / 1166 | 1.8 (1.3–2.5) |
<0.001 |
| Adrenal Primary Site |
1450 / 2508 | 1.6 (1.2–2.0) |
0.001 |
At least one known metastatic site specified in the INRG database.
Variables tested in the model and found non-significant were: age; ferritin level; grade of differentiation; aberration at 11q; gain of 17q; pooled segmental chromosomal aberrations; and metastasis to bone, bone marrow, distant lymph node, liver, and central nervous system.
Discussion
In this comprehensive analysis of predictors of MYCN amplified status, we demonstrate in a comprehensive and definitive manner the complex interaction between MNA and other features of this disease. The presence of each prognostic factor was statistically associated with MYCN status, with most remaining significant on multivariate testing. This novel finding demonstrating the independent association between MNA and almost all variables was unexpected. The result of our multivariable logistic regression model adjusting for vital status demonstrates that our findings cannot be fully explained by an association between MNA and prognosis. Pathology/biology variables demonstrated the strongest associations with MNA. Tumors with 1p LOH were almost 20 times as likely to have MNA compared to tumors without 1p LOH, with a similar association even after controlling for other biological predictors. This is consistent with prior observations1,14,15, and in context with our other findings, highlights the critical association between MNA and tumor genetic features compared to clinical features. For example, of the clinical variables, elevated LDH was the strongest predictor with a univariate odds ratio of 8.4 compared to 19.8 for 1p LOH.
Our novel recursive partitioning approach revealed dramatic differences in the rates of MNA between identified subgroups of patients, including two trees that yielded >80% absolute differences in rates of MNA between subgroups. In addition to highlighting the importance of SCA in prediction of MNA, this approach also enables us to illustrate the relative importance of some variables over others in predicting MNA as well as the importance of context of other predictor variables in the impact of a specific variable. For example, in the absence of LOH at 1p, the presence of high MKI is associated with a maximum incidence of MNA of 25% whereas, in the presence of LOH at 1p (along with other features), the presence of high MKI is associated with a maximum incidence of MNA of 71.4%. Moreover, among poorly differentiated tumors with LOH at 1p, aberration at chromosome 11q is associated with MNA, though the difference in maximum incidence of MNA based upon presence or absence of this aberration is not large (maximum possible incidence for those without 11q aberration of 87.5% compared to 71.4% for those with 11q aberration).
Along these same lines, it is noteworthy that age was a key predictor of differential rates of MNA in models relying solely on clinical variables, but age was not selected as a predictive variable in the recursive partitioning model that included all clinical and biologic/pathologic variables as potential predictors. This finding, along with the results of our multivariate logistic regression models, suggest that other variables associated with age account in part for some of the association between MYCN status and age, which fits with the concept that age behaves as a surrogate for the effects of other clinical and biological variables. Graphical representations of the relationship of age with MNA show a complex relationship. London and colleagues showed that any age cutoff between 15–20 months appropriately separates neuroblastoma patients into high and low risk groups for treatment decisions, with 18 months selected for future risk stratification.23 We complement these findings by showing that MNA peaks in incidence at approximately this same age. Our results demonstrate the nonlinear relationship between age and MNA and should motivate additional studies into the developmental and molecular pathways involved in this association.
Our findings extend previous observations demonstrating positive associations between MNA and 1p LOH and gain of 17q, as well as a negative association between MNA and 11q aberration.2,14–16,31,32 Our multivariate logistic regression model focused solely on pathology/biology variables showed that the 1p and 11q associations with MYCN status were independent of each other, while the 17q association was not. This finding may be due to the fact that 17q and 1p aberrations are themselves correlated,2 or more likely reflect small sample size with available 17q data. The composite variable that included 1p, 11q, and/or 17q segmental aberrations was also not independently associated with MYCN status, likely due to opposing effects of 1p and 11q aberrations on the incidence of MNA. The overall pattern observed in this and other studies33 is noteworthy as it indicates that specific loci rather than non-specific SCAs are associated with the presence or absence of MNA, though the underlying biologic mechanisms for these associations are yet unknown.
Our results address the association between MNA and sites of primary and metastatic disease. We previously reported the association of primary site with MYCN status.10 We have extended that finding to demonstrate that adrenal tumors are more likely and thoracic tumors are less likely to be MYCN amplified even after controlling for other predictors of MYCN status. Likewise, we have previously demonstrated that patients with lung metastases are more likely to have MYCN amplified tumors.20 We now demonstrate that this association is independent of other predictors of MYCN status, though the underlying mechanisms for this association remain obscure. Unlike previous analyses, we did not find an association between MNA and central nervous system metastases, though this may be due to the rarity of this metastatic site at diagnosis.34 We also demonstrate that skin metastases are predictive of being MYCN non-amplified. This site of disease is more common in stage 4S disease and also in young infants, groups which are less likely to have MYCN amplified tumors.35
Although we utilized the largest available patient database, with 7,102 patients with known MYCN status, our work has certain limitations. Our overall rate of MNA of 16% is lower than widely cited estimates of 20–25%.1 It is possible that the comprehensive nature of the INRG database provides a more accurate estimate of the rate of MNA across neuroblastoma. We note that our estimate is identical to that reported by a large analysis from the COG16, though acknowledge that data from the COG are included within the INRG database and may bias the estimate towards those previously reported by the COG. It is also possible that the contribution of low-risk patients identified from national neuroblastoma screening efforts and/or higher rates of MYCN testing in localized patients enriched the population with biologically favorable tumors thereby reducing the proportion with MNA. Although our primary outcome, MYCN status, was not determined by identical techniques in each INRG member country, all those used are standard validated methods2,24,25. We chose to dichotomize LDH and ferritin based upon the median of the initial INRG cohort to maintain consistency with prior INRG analyses, though acknowledge that other cutpoints could be explored in future analysis for their association with MYCN status. The multivariate logistic regression analyses were limited by missing data. We attempted to circumvent this issue by utilizing dummy variables to allow the full data set to be used, thereby reducing the risk of selection bias. This approach could not be used as part of our recursive partitioning models and therefore certain variables with limited data, such as gain of 17q, may have been less likely to be selected, thus potentially decreasing the generalizability of our findings. Although the differences in patient subgroups revealed by recursive partitioning are innovative and can both inform insights into the biological and pathological factors that lead to MYCN amplification and lead to further research in the field, some of the resulting subgroups are small and therefore one must use caution in interpreting these subgroups for clinical decisions.
Despite these limitations, the current study is the largest analysis of predictors of MNA. Our work highlights the importance of obtaining adequate tissue for detailed molecular testing as part of the diagnostic process. While we identified groups of patients with very low probabilities of MNA, no subgroups that completely lack MNA could be identified. Moreover, other pathology/biology variables, particularly SCAs, were key to identifying groups of patients with maximally different rates of MNA and improved our stratification beyond one restricted to the use of only clinical variables. Our findings should stimulate additional laboratory studies into the mechanisms of MNA as a function of age, as well as the genetics or epigenetics that may be orchestrating the complex interaction between the described molecular findings. Finally, additional studies exploring the ways that molecular changes associated with MNA interact with clinical features associated with MNA will be critical.
Supplementary Material
Supplemental Figure 1. Recursive partitioning using CART to identify age subgroups associated with MYCN amplification by testing age as a continuous variable.
Supplemental Figure 2. Manual recursive partitioning using logistic regression to identify factors associated with MYCN amplification, testing only clinical variables (full tree without truncation).
Acknowledgments
Support: The INRG database is supported in part by the William Guy Forbeck Research Foundation, the Little Heroes Cancer Research Fund, Children’s Neuroblastoma Cancer Foundation, Neuroblastoma Children’s Cancer Foundation, and the Super Jake Foundation. Data included in the INRG database were provided by Children’s Oncology Group (COG), Pediatric Oncology Group (POG), Children’s Cancer Study Group (CCSG), German Gesellschaft für Pädiatrische Onkologie und Hämatologie (GPOH), European Neuroblastoma Study Group (ENSG), International Society of Paediatric Oncology Europe Neuroblastoma Group (SIOPEN), Japanese Advanced Neuroblastoma Study Group (JANB), Japanese Infantile Neuroblastoma Co-operative Study Group (JINCS), Spanish Neuroblastoma Group and the Italian Neuroblastoma Group. Supported also in part by the Alex’s Lemonade Stand Foundation (KTV, KKM, SGD), Frank A. Campini Foundation (KKM and SGD), Edward Conner Fund (KKM), Dougherty Foundation (KKM and SGD), NIH grant CA039771 and the Audrey Evans Endowed Chair (GMB), and the Mildred V. Strouss Chair (KKM). The contents are solely the responsibility of the authors and do not necessarily represent the official views of the funding sources listed above.
Footnotes
Disclaimers/Disclosures: The authors have no conflicts of interest and/or acknowledgements to disclose.
References
- 1.Brodeur GM, Bagatell R. Mechanisms of neuroblastoma regression. Nat Rev Clin Oncol. 2014;11(12):704–713. doi: 10.1038/nrclinonc.2014.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambros PF, Ambros IM, Brodeur GM, et al. International consensus for neuroblastoma molecular diagnostics: report from the International Neuroblastoma Risk Group (INRG) Biology Committee. BMJ Cancer. 2009;100:1471–1482. doi: 10.1038/sj.bjc.6605014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohn SL, Pearson AD, London WB, et al. The International Neuroblastoma Risk Group (INRG) classification system: an INRG Task Force report. J Clin Oncol. 2009;27:289–297. doi: 10.1200/JCO.2008.16.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlsen NL, Christensen IJ, Schroeder H, et al. Prognostic factors in neuroblastomas treated in Denmark from 1943 to 1980. A statistical estimate of prognosis based 253 cases. Cancer. 1986;58:2726–2735. doi: 10.1002/1097-0142(19861215)58:12<2726::aid-cncr2820581229>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 5.Shimada H, Ambros IM, Dehner LP, et al. The international neuroblastoma pathology classification (the Shimada system) Cancer. 1999;86:364–372. [PubMed] [Google Scholar]
- 6.Brodeur GM, Seeger RC, Schwab M, et al. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 7.George RE, Variend S, Cullinane C, et al. Relationship between histopathological features, MYCN amplification, and prognosis: A UKCCSG study. Med Pediatr Oncol. 2001;36:169–176. doi: 10.1002/1096-911X(20010101)36:1<169::AID-MPO1041>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 8.Oppedal BR, Storm-Mathison I, Lie SO, et al. Prognostic factors in neuroblastoma: Clinical, histopathologic, and immunohistochemical features and DNA ploidy in relation to prognosis. Cancer. 1988;62(4):772–780. doi: 10.1002/1097-0142(19880815)62:4<772::aid-cncr2820620422>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 9.Look AT, Hayes FA, Shuster JJ, et al. Clinical relevance of tumor cell ploidy and N-myc gene amplification in childhood neuroblastomas: a Pediatric Oncology Group study. J Clin Oncol. 1991;9:581–591. doi: 10.1200/JCO.1991.9.4.581. [DOI] [PubMed] [Google Scholar]
- 10.Vo K, Matthay KK, Neuhaus J, et al. Clinical, biological, and prognostic differences based upon primary tumor site in neuroblastoma: a report from the International Neuroblastoma Risk Group (INRG) project. J Clin Oncol. 2014;32:3169–3176. doi: 10.1200/JCO.2014.56.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeger RC, Brodeur GM, Sather H, et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 12.Teshiba R, Kawano S, Wang L, et al. Age-dependent prognostic effect by mitosis-karyorrhexis index in neuroblastoma: A report from the children’s oncology group. Pediatr Dev Pathol. 2014;17(6):441–449. doi: 10.2350/14-06-1505-OA.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bowman LC, Castleberry RP, Cantor A, et al. Genetic staging of unresectable or metastatic neuroblastoma in infants: a Pediatric Oncology Group study. J Natl Cancer Inst. 1997;89(5):373–380. doi: 10.1093/jnci/89.5.373. [DOI] [PubMed] [Google Scholar]
- 14.Fong CT, Dracopoli NC, White PS, et al. Loss of heterozygosity for the short arm of chromosome 1 in human neuroblastomas: correlation with N-myc amplification. Proc Natl Acad Sci U S A. 1989;86(10):3753–3757. doi: 10.1073/pnas.86.10.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maris JM, Guo C, Blake D, et al. Comprehensive analysis of chromosome 1p deletions in neuroblastoma. Med Pediatr Oncol. 2001;36(1):32–36. doi: 10.1002/1096-911X(20010101)36:1<32::AID-MPO1009>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 16.Attiyeh EF, London WB, Mosse YP, et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N Engl J Med. 2005;353(21):2243–2253. doi: 10.1056/NEJMoa052399. [DOI] [PubMed] [Google Scholar]
- 17.Plantaz D, Vandesompele J, Van Roy N, et al. Comparative genomic hybridization (CGH) analysis of stage 4 neuroblastoma reveals high frequency of 11q deletion in tumors lacking MYCN amplification. Int J Cancer. 2001;91(5):680–686. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1114>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 18.Spitz R, Hero B, Simon T, et al. Loss in chromosome 11q identifies tumors with increased risk for metastatic relapses in localized and 4S neuroblastoma. Clin Cancer Res. 2006;12:3368–3373. doi: 10.1158/1078-0432.CCR-05-2495. [DOI] [PubMed] [Google Scholar]
- 19.Morgenstern DA, London WB, Stephens D, et al. Metastatic neuroblastoma confined to distant lymph nodes (stage 4N) predicts outcome in patients with stage 4 disease: a study from the International Neuroblastoma Risk Group database. J Clin Oncol. 2014;32(12):1228–1235. doi: 10.1200/JCO.2013.53.6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DuBois SG, London WB, Zhang Y, et al. Lung metastases in neuroblastoma at initial diagnosis: a report from the international neuroblastoma risk group (INRG) project. Pediatr Blood Cancer. 2008;51:589–592. doi: 10.1002/pbc.21684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mosse YP, Deyell RJ, Berthold F, et al. Neuroblastoma in older children, adolescents and young adults: a report from the International Neuroblastoma Risk Group project. Pediatr Blood Cancer. 2014;61:627–635. doi: 10.1002/pbc.24777. [DOI] [PubMed] [Google Scholar]
- 22.Cetinkaya C, Martinsson T, Sandgren J, et al. Age dependence of tumor genetics in unfavorable neuroblastoma: arrayCGH profiles of 34 consecutive cases, using a Swedish 25-year neuroblastoma cohort for validation. BMC Cancer. 2013;13:231–242. doi: 10.1186/1471-2407-13-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.London WB, Castleberry RP, Matthay KK, et al. Evidence for an age cutoff greater than 365 days for neuroblastoma risk stratification in the Children’s Oncology Group. J Clin Oncol. 2005;23:6459–6465. doi: 10.1200/JCO.2005.05.571. [DOI] [PubMed] [Google Scholar]
- 24.Shapiro DN, Valentine MB, Rowe ST, et al. Detection of N-myc gene amplification by fluorescence in situ hybridization: diagnostic utility for neuroblastoma. Am J Pathol. 1993;142(5):1339–1346. [PMC free article] [PubMed] [Google Scholar]
- 25.Crabbe DC, Peters J, Seeger RC. Rapid detection of MYCN gene amplification in neuroblastoma using the polymerase chain reaction. Diagn Mol Pathol. 1992;1(4):229–234. [PubMed] [Google Scholar]
- 26.Brodeur GM, Seeger RC, Barrett A, et al. International criteria for diagnosis, staging and response to treatment in patients with neuroblastoma. J Clin Oncol. 1988;6:1874–1881. doi: 10.1200/JCO.1988.6.12.1874. [DOI] [PubMed] [Google Scholar]
- 27.Brodeur GM, Pritchard J, Berthold F, et al. Revisions of the international criteria for neuroblastoma diagnosis, staging and response to treatment. J Clin Oncol. 1993;11:1466–1477. doi: 10.1200/JCO.1993.11.8.1466. [DOI] [PubMed] [Google Scholar]
- 28.Machin D, Cheung Y-B, Parmar MKB. Survival Analysis: A Practical Approach. Chichester: Wiley; 2006. [Google Scholar]
- 29.Moroz V, Machin D, Faldum A, et al. Changes over three decades in outcome and the prognostic influence of age-at-diagnosis in young patients with neuroblastoma: A report from the International Neuroblastoma Risk Group Project. Eur J Cancer. 2011;47(4):561–571. doi: 10.1016/j.ejca.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 30.Cook EF, Goldman L. Empiric comparison of multivariate analytic techniques: advantages and disadvantages of recursive partitioning techniques. Journal of Chronic Dis. 1984;37(9):721–731. doi: 10.1016/0021-9681(84)90041-9. [DOI] [PubMed] [Google Scholar]
- 31.Maris JM, Weiss MJ, Guo C, et al. Loss of heterozygosity as chromosome 1p36 predicts for disease progression but no decreased overall survival probability in neuroblastoma patients: a Children’s Cancer Group study. J Clin Oncol. 2000;18(9):1888–1899. doi: 10.1200/JCO.2000.18.9.1888. [DOI] [PubMed] [Google Scholar]
- 32.Guo C, White PS, Weiss MJ, et al. Allelic deletion at 11q23 is common in MYCN single copy neuroblastomas. Oncogene. 1999;18(35):4948–4957. doi: 10.1038/sj.onc.1202887. [DOI] [PubMed] [Google Scholar]
- 33.Mosse YP, Diskin SJ, Wasserman N, et al. Neuroblastomas have distinct genomic DNA profiles that predict clinical phenotype and regional gene expression. Genes Chromosomes Cancer. 2007;46(10):936–949. doi: 10.1002/gcc.20477. [DOI] [PubMed] [Google Scholar]
- 34.Matthay KM, Brisse H, Couanet D, et al. Central nervous system metastases in neuroblastoma: radiologic, clinical, and biologic features in 23 patients. Cancer. 2003;1(98):155–165. doi: 10.1002/cncr.11448. [DOI] [PubMed] [Google Scholar]
- 35.Taggart DR, London WB, Schmidt ML, et al. Prognostic value of the stage 4S metastatic pattern and tumor biology in patients with metastatic neuroblastoma diagnosed between birth and 18 months of age. J Clin Oncol. 2011;29(33):4358–4364. doi: 10.1200/JCO.2011.35.9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Recursive partitioning using CART to identify age subgroups associated with MYCN amplification by testing age as a continuous variable.
Supplemental Figure 2. Manual recursive partitioning using logistic regression to identify factors associated with MYCN amplification, testing only clinical variables (full tree without truncation).