Abstract
Roquin-1 (Rc3h1) is an E3 ubiquitin ligase originally discovered in a mutational screen for genetic factors contributory to systemic lupus erythematosus-like symptoms in mice. A single base pair mutation in the Rc3h1 gene resulted in the manifestation of autoantibody production and sustained immunological inflammation characterized by excessive T follicular helper cell activation and formation of germinal centers. Subsequent studies have uncovered a multifactorial process by which Roquin-1 contributes to the maintenance of immune homeostasis. Through its interactions with partner proteins, Roquin-1 targets mRNAs for decay with inducible costimulator being a primary target. In this review, we discuss newly discovered functions of Roquin-1 in the immune system and inflammation, and in disease manifestation, and discuss avenues of further research. A model is presented for the role of Roquin in health and disease.
Keywords: autoimmunity, costimulation, T follicular helper, inflammation, T cell
Roquin-1 as an Atypical E3 Ubiquitin Ligase
Structural analysis of Roquin-1 reveals the presence of a RING (Really Interesting New Gene) finger domain, a CCCH-type (C3H1) zinc finger domain, and a ROQ domain (1, 2). The RING domain is a hallmark of the RING Finger E3 ubiquitin ligase family while the C3H1 and ROQ domains are emblematic of RNA binding proteins. E3 ubiquitin ligases play a critical role in ubiquitination (3). Ubiquitination, or ubiquitylation, is a post-translational modification to regulate protein expression. It involves the concerted efforts of three distinct classes of enzymatic proteins to attach the ubiquitin molecule to a target protein (substrate). Via an ATP-dependent process, the E1 ubiquitin-activating enzyme initiates the ubiquitination cascade by binding free ubiquitin molecules via a thioester bond with its catalytic cysteine. The E2 ubiquitin conjugating enzyme transfers the activated ubiquitin from the E1 active site cysteine to the E2 active site cysteine. The E3 ubiquitin ligase binds both the E2 enzyme and the target substrate to catalyze the transfer of the activated ubiquitin to the target protein. The ubiquitin cascade may be repeated to poly-ubiquinate the target or end with a single ubiquitin molecule (mono-ubiquitination). Dependent upon the number of ubiquitins, distinct signals and physiological outcomes are generated. While poly-ubiquitination generally results in proteosomal-mediated decay of the target protein, both mono- and poly-ubiquitination can modulate other biological processes, including direct receptor signaling and trafficking from the cell surface, and can affect protein-protein interactions and alter enzymatic activity.
The vast majority of the over 600 identified E3 ligases are of the RING finger family. The Homologous to E6-AP Carboxyl Terminus (HECT) E3 ubiquitin ligase and U-box E3 ligase comprise the other E3 ligase families (3, 4). Whereas RING finger E3 ligases directly transfer the ubiquitin molecule from the E2 enzyme to the target substrate, the HECT E3 ligases act as intermediaries temporarily transferring the ubiquitin to the E3 active site before conjugating the ubiquitin to the target substrate. BRCA1, FANC, and Mdm2 are three well known E3 ligases.(3, 4) Mutations and altered expression of those proteins are associated breast cancer, Fanconi anemia, and colorectal cancer, respectively. The tumor suppressor p53 is a target of Mdm2; amplification of Mdm2 leads to excessive turnover of p53 resulting in cancer (3, 4).
Despite the presence of the RING finger domain, to date there have been no reports indicating that either the human or mouse Roquin-1 functions as an E3 ligase, nor has a function in the ubiquitination pathway been described yet. In contrast, the Roquin-2 paralog and the RLE-1 (regulation of longevity by E3) C. elegans homolog of Roquin-1 are the exceptions as both have been accorded roles and activities in the ubiquitination cascade. Studies in which rle-1 was deleted in C. elegans resulted in an extended life span, reduced offspring, and increased resistance to stressors including UV radiation, heat, and certain pathogens. Protein levels but not the transcript levels of the DAF-16 transcription factor were elevated in mutants (5). Subsequent studies in wild type C. elegans revealed that RLE-1 colocalized with DAF-16 while in vitro studies demonstrated that DAF-16 coimmunoprecipitated with RLE-1 (5). Further, those in vitro studies revealed poly-ubiquitination of DAF-16 with elevated RLE-1 expression (5).
In a small interfering RNA (siRNA) screen for regulators of the reactive oxygen species (ROS) response and the apoptosis signal-regulating kinase 1 (ASK1 or MAP3K5), Roquin-2 was identified as a candidate (6). In response to H2O2 treatment, Roquin-2 coimmunoprecipitated with ASK1 in HeLa-S3 cells (6). Roquin-2-specific siRNAs treatment reduced the Roquin-2-ASK1 interaction, prolonged the half-life of ASK1 protein, and reduced the ubiquitination of ASK1 (5). Interestingly, overexpression of Roquin-2 had the opposite effects, specifically, promoting the ubiquitination and turnover of ASK1 (5).
In combination, these studies clearly illustrate that both RLE-1 and Roquin-2 have E3 ligase activity. Whether or not the mouse and human Roquin-1 demonstrates a similar activity remains to be seen. And, it raises the question as to why this function has not been conserved evolutionarily.
Roquin-1 RNA targets
There is ample evidence that Roquin-1 functions as an RNA binding protein to regulate gene expression post-transcriptionally. In the seminal manuscripts, Roquin-1 was shown to limit the expression of inducible costimulator (ICOS) (1, 2). Subsequent analyses confirmed that Roquin-1 specifically recognized and bound to the ICOS 3’UTR to regulate its expression. Although this was initially thought to involve miR-101 and the RNA-induced silencing complex (RISC) as ancillary control factors, later experiments with Dicer-deficient mouse embryonic fibroblasts ruled out the miRNA involvement (7).
Recent studies have focused on identifying additional RNA targets of Roquin-1 regulation as well as the mechanism by which Roquin-1 recognizes its targets. Using Icos as the standard, a conserved cis-regulatory element (CRE) was identified in the 3′UTR as the recognition element for Roquin-1. This RNA secondary structure, alternatively identified as a constitutive decay element (CDE) or a stem-loop structure (SL), is AU-enriched with the consensus sequence being 5′-NNNNNUUCYRYGAANNNNN-3′ (8–10). Although RNA immunoprecipitations (IP), including HITS-CLIP (High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation) and PAR-CLIP (Photoactivatable-ribonucleoside-enhanced crosslinking and immunoprecipitation) have identified approximately 3,800 target mRNAs for Roquin-1, only A20, TNFA, OX40, NFKBID, and NFKBIZ have subsequently been validated as Roquin-1 targets (Table 1) (1, 10–12).
Table 1.
Targets of Roquin-1
Deletion studies identified the ROQ domain as both the structure critical for RNA binding and localization to stress granules (13). Crystallography of the ROQ domain in contact with the TNFα CDE revealed three sub-domains within the ROQ domain (8). Two of these subdomains were confirmed to be RNA binding sites for stem loop RNA and dsRNA, respectively, as determined by mutagenesis analysis (8). The ROQ domain is the site of the point mutation in sanroque mice (1, 2). Crystallography analysis revealed the presence of a novel RNA interaction domain, the HEPN (higher eukaryotes and prokaryotes nucleotide-binding) domain, and further defined the structural consequences of the M199R mutation in which a previously buried hydrophobic residue (F234) is turned out and exposed (14).
Interestingly, Regnase-1 (Reg1), a ribonuclease with a C3H zinc finger domain, has recently been proposed to work in conjunction with Roquin-1 and Roquin-2 to regulate the inflammatory response. Similar to Roquin-1, Reg1 recognizes a common SL structure in the 3′UTR of mRNAs to promote RNA turn over (15, 16). Whereas Roquin-1 localizes to stress granules and processing (P) bodies, Reg1 localizes to ribosomes and the endoplasmic reticulum, thereby providing a spatiotemporally distinct manner of operation for Reg1 (15, 16). However, RNA-IP sequencing (RIP-seq) analysis identified seven target mRNAs that overlapped Roquin-1 and Reg1, including ICOS, OX40, and TNFA (16). The consensus recognition sequence for Reg1, 5′-UUGGAAAGYRYCUUUCCAA-3′, is similar to that for Roquin-1, which may account for the overlap in mRNA targets. This again illustrates how Roquin-1 interacts with multiple players to adjust the expression of genes in an inflammatory response.
Roquin-1 and microRNAs
MicroRNAs (miRNAs) are small non-coding RNAs that post-transcriptionally regulate gene expression (17). Given their regulatory roles, it is not surprising that studies aimed at understanding the relationship between Roquin-1 and miRNAs have been conducted. A recent study examined the effects of the sanroque mutation on miRNA expression. miR-146a-5p (24.9 fold) and miR-21 (13.9 fold) showed the greatest elevation in expression among 15 miRNAs that had > 2-fold increases in expression in Roquinsan/san T cells compared to Roquin+/+ T cells (14). Interestingly, no miRNAs were found to have decreased expression. RNA stability assays demonstrated that Roquin-1 controlled miR-146a stability. miR-146a had a longer half-life in Roquinsan/san T cells in comparison to Roquin+/+ T cells (14). Further, Roquin-1 bound both miR-146a and the RISC subunit Ago2 (argonaute RISC catalytic component 2) (14). The binding to Ago2 suggests that this may be a common mechanism used to regulate other miRNAs.
Elevated expression of both miR-146a-5p and miR-21 has been associated with inflammatory disease (18–27). miR-146a-5p has been shown to be highly expressed in T follicular helper (Tfh) cells and regulate ICOS expression (28). Another miRNA of note that had elevated expression in these studies was miR-223 (3.4 fold) (14). miR-223 is a validated regulator of Roquin-1 expression and has been demonstrated to have elevated expression in patients with inflammatory bowel disease and the interleukin-10 knockout (IL-10−/−) mouse model of chronic intestinal inflammation (24, 25). Those studies indicate that, in addition to target mRNAs, Roquin-1 regulates miRNA stability, including several miRNAs that regulate its own expression.
Regulation of Roquin proteins
A recent study investigating the paracaspase MALT1 in immune function has shed some light on the regulatory control of the Roquin-1 protein. It began with the initial observation that when CD4+ T cells were stimulated with phorbol myristate acetate (PMA) and ionomycin (IO), Roquin-1 and Roquin-2 protein levels decreased over the three hour time course.(29) Previous studies established Reg1 as a substrate for MALT1-mediated proteolytic cleavage to regulate gene expression (30, 31). Based on those studies, Jeltsch et al. investigated the ability of MALT1 to cleave the Roquin proteins. When PMA/IO stimulation of CD4+ T cells was repeated in the presence of pharmacological inhibitors of MALT1 activity (z-VRPR-fmk, mepazine, and thioridazine), cleavage of the Roquin proteins was reduced (29). A subsequent series of genetic and molecular studies confirmed that MALT1 was able to cleave Roquin-1 and Roquin-2 as well (29). Those studies further reinforce the importance of Roquin-1 in Th17 cell differentiation and function, and IL-17 expression (25, 29), thus adding an additional post-transcriptional method for regulating Roquin-1 expression.
Roquin Involvement in Inflammation and Autoimmunity
The Roquin-1 gene mutation was identified in C57BL/6 mice treated with N-ethyl-N-nitrosourea, a mutagen that induces single base pair alterations in spermatogonial cells at a rate of ~1 per 0.5 megabases (32). Animals were bred and screened for systemic lupus erythematosus (SLE) based on the presence of antinuclear antibodies (ANAs) as a predictive biomarker. The mutation was linked to the Rc3h1 (Roquin-1) gene, a gene of previous unknown function. Animals homozygous for the mutation, referred to as sanroque or Roquinsan/san mice, were found to have extensive immunological alterations that included antibodies to double-stranded DNA, IgG immune complexes, anemia, glomerulonephritis and necrotizing hepatitis, splenomegaly and lymphadenopathy, and autoimmune thrombocytopenia and plasmacytosis. CD44hi, CD62lo CD4+ and CD8+ T cells were present, and animals had high level ICOS expression (2) in both male and female animals that was evident as early as 6–8 weeks of age. Sanroque mice have dysregulated IL-5 and IFN-γ synthesis (2).
It is notable that despite the extreme immunological perturbations in sanroque mice, they show minimal if any changes in the Th1 or Th2 responses to foreign antigen compared to wildtype animals. T cell tolerance is normal and deletion of intrathymic CD4+ T cell and self-reactive T cells is unaffected. Activation-induced cell death and the proportional numbers of CD4+ CD25+ Tregs remain unaltered in sanroque mice (2). Those changes are T cells autonomous as seen from studies of mixed bone marrow chimeras (2), and in radiation chimaeras using bone marrow from Roquin-deficient mice (33).
A prominent immunological feature of the sanroque phenotype is lymph node germinal center disorganization resulting in large numbers of follicular helper T (Tfh) cells. Additionally, sanroque mice have elevated levels of several molecules involved in immune activation or regulation. These include ICOS, Regulated upon Activation, Normal T cell Expressed (RANTES, CCL5), CXCR5, and the programmed cell death 1 (PCD1) molecules, among others (2, 34, 35). Mesenteric lymph node (MLN) T cells in sanroque mice have increased ICOS and OX40 expression on CD44+ T cells (36), and CD44+ CD62L− cells have increased expression of KLRG1 (36), a population of short-lived effector cells (37).
Both CD28 and ICOS can function as costimulatory signals during T cell activation. CD28 is constitutively expressed on most T cells, whereas ICOS is upregulated during activation on effector and memory T cells (38), and is present on peripheral Tregs (39, 40). The regulation of ICOS by the action of Roquin-1 occurs by binding to ICOS but not CD28 mRNA, a system that may provide a failsafe for both immune protection and autoimmunity. Sanroque x CD28−/− mice retain high levels of ICOS, as might be expected, despite the lack of CD28, and have germinal center formation, Tfh cells, and peripheral Tregs that are mostly lacking in CD28−/− mice (34), thus demonstrating targeted action of Roquin-1 on ICOS.
The finding of small intestinal inflammation in sanroque mice (36) is particularly interesting given that, whereas there are numerous models of inflammatory bowel disease (IBD) for the large intestine inflammation (41–44), few models have been identified for small intestinal IBD. One such model is the SAMP1/YitFc mouse, in which most animals develop ileitis by 20–30 weeks of age. In sanroque mice, the T cell profile in the mucosal epithelium and lamina propria differs from that of the MLNs in that ICOS+ cells are present in the epithelium but not the lamina propria, and because OX40 is not appreciably expressed on T cells in either mucosal site whereas it is present on MLN T cells (36). Moreover, the extensive chemokine dysregulation consisting of elevated levels of CCL1, CCL24, CCL25, CCL20, CXCL1, and IL-13 would favor chronic inflammation in the small intestine of sanroque mice (36).
Roquin and Immune-Based Disorders
Two divergent sets of findings have been reported with regard to the role of Roquin-1 in angioimmunoblastic T cell lymphoma (AITL). Mice Roquinsan/+ heterozygous mice were reported to develop AITL-like disease at about 4 months of age in ~50 percent of the animals (45). Animals had lymph node tumors (enlarged lymph nodes) with characteristic histopathologic and cellular features of AITL in humans (45). In a study of twelve human AITL lymph node biopsies, although expression of CXCL13, ICOS, and PD1 was noted as determined by histocytochemistry, Roquin-1 and miR101 gene expression remained unaltered compared to tissues from control patients (46). Thus, whereas Roquinsan/+ heterozygous mice display some of the phenotypic and pathologic features of AITL, this may reflect a concomitant rather than a direct effect of partial Roquin-1 impairment.
Although much information has been gleaned from studies using Roquin-1 mutant sanroque mice, findings using T cell lines or transgenic (Tg) mice with Roquin-1 overexpression have been equally illuminating. Stimulation of EL-4 via CD3/CD28 in cells that overexpressed Roquin-1 resulted in lower ICOS expression as expected, but elevated CD28 expression with increased IL-2 and TNFα synthesis (47). Elevated levels of IL-2 are consistent with the role of CD28 as a primary IL-2 signal through Grb2 (48, 49). Inhibition of AKT and JNK signaling suppressed CD3/CD28 increases caused by Roquin-1 overexpression (47). Consistent with those findings, Tg mice that over-expressed Roquin-1 had suppressed ICOS and elevated CD28 T cell expression, and had more IL-2 and TNFα synthesis following CD3/CD28 stimulation (47).
In a model of collagen-induced arthritis (CIA), Roquin-1 Tg mice mounted stronger CIA responses and had suppressed levels of ICOS, elevated CD28, and an imbalance in the Th1/Th2 that favored Th1 cells (50). By day 45 post-CIA induction, levels of IFN-γ, TNFα, IL-6, and IL-17 were elevated in Tg mice (50). Tfh cell numbers and germinal center B cells were unaffected in Roquin-1 Tg mice (50). Selective overexpression of Roquin-1 in T cells using a T cell-specific promoter also led to more severe T cell-mediated hepatitis, increased numbers of Th17 cells, and production of proinflammatory cytokines (51). Taken together, those findings are at odds with the notion that the primary function of Roquin-1 is to limit and modulate the immune response. Rather, Roquin-1 may have a dual function as both an activator and suppressor of immunity, as discussed below.
Sanroque Mice vs. Roquin-deficient Mice
One of the most enigmatic findings involving Roquin comes from studies in which the Roquin-1 gene has been disrupted leading to complete Roquin-1 protein ablation. Roquin-1 knockout mice are born with a caudal spine defect and impaired lung development, resulting in poor perinatal survival (52), thus implying a role for Roquin-1 in embryonic development that extends beyond the immune system. Rc3h1−/− mice had higher levels of ICOS expression on CD4+ and CD8+ T cells and an increase in overall numbers of CD8+ T cells bearing an effector-like phenotype (52). Numbers of eosinophils and cells of the monocyte/macrophage lineages also were elevated. Notably, splenic germinal centers were otherwise unaffected in Rc3h1−/− mice (52).
In mice in which Roquin-1 ablation targeted the entire hematopoietic system or B cells, immunological changes were similar to what was seen in T cell-targeted mice. Animals did not develop autoantibodies and lacked autoimmunity in the kidney, liver, and lung (52). Those finding indicate that Roquin-1 ablation in and of itself is not sufficient to cause a breach in self-tolerance or promote autoimmunity. Rc3h2−/− mice displayed the same perinatal lethality as Rc3h1−/− mice (53). ICOS expression was elevated in Rc3h2−/− mice (11). Roquin-2 has been shown to regulate inflammatory signals mediated by ASK-1 (6).
Several interpretations can be drawn from those studies. Because the Roquin-1 protein in sanroque mice is present but mutated, consisting of a M119R substitution, whereas the protein is fully absent in Rc3h1−/− mice, it may be inferred that the mutated version of the protein is more detrimental in the context of immunobiological changes than a complete lack of the protein (52). The basis for that remains unclear; however, it has been demonstrated that the mutated form of Roquin-1 binds more than three-fold tighter to Icos mRNA than WT Roquin-1 (13), thus perhaps causing long-lasting disturbances within the immune system. Additional studies will be needed to address this.
The possibility that the Roquin-1 paralogue, Rc3h2, might compensate for the lack of autoimmunity in Rc3h1−/− has been addressed in mice carrying deletions in both Rc3h1 and Rc3h2. Ablation of Rc3h1 and Rc3h2 resulted in an immunophysiologic phenotype that mimicked that of sanroque mice and, unlike individual knockout animals, double knockouts had increased numbers of Tfh cells (11, 53).
At odds with those findings are studies in which Rc3h1−/− mice were generated by insertion of a gene-trap into intron 1 (Rc3h1gt/gt mice), which resulted in a phenotype more typical of that seen in sanroque animals (33, 36). Although Rc3h1gt/gt mice had caudal spine deformity and poor post-birth survival (36) similar to that of Rc3h1−/− and Rc3h2−/− mice (11, 52), Rc3h1gt/gt mice developed extensive inflammation in the small intestine, though not in the colon, and had a destructive inflammatory response in the kidney, lungs, liver, and spleen (33) that was reminiscent of sanroque mice. That phenotype was similarly retained in chimeric mice made from bone marrow of Rc3h1gt/gt mice injected into irradiated syngeneic animals (33), confirming that hematopoietic and not systemic disruption of Roquin-1 expression was the primary factor responsible for the development of immunopathology. The basis for the differences in the immunological findings of Rc3h1−/−, Rc3h2−/−, and Rc3h1gt/gt mice is unclear; however, it could reflect variations in the methods used to render mice Roquin deficient. Yet, this seems unlikely given that in all three animal systems, mice were devoid of Roquin gene and protein expression.
Regulation of Roquin-1 Gene Expression
The factors that regulate Roquin-1 gene expression are only now beginning to come to light. Colonic tissues of Il10−/− mice, an animal model extensively used in studies of human IBD, have decreased Roquin-1 gene expression and elevated ICOS and IL-17 gene expression (25). Those patterns were reversed, however, following in vitro treatment of colonic intraepithelial lymphocytes (cIELs) with exogenous IL-10, suggesting that IL-10 functions in some manner to regulate Roquin-1 expression either directly or indirectly. This is further supported by findings that Th17 stimulation of MLN cells lowered Roquin-1 gene expression, and because IL-10 was able to counteract that effect, resulting in increased Roquin-1 and Socs3 gene expression (54), the latter being susceptible to attenuation by IL-10 (55).
Several transcription factors (TFs) have been identified that act on the Roquin-1 promoter. These include STAT1, STAT3, GATA2, and c-Rel, which have positive regulatory effects, and IKZF2, which has a negative regulatory effect on Roquin-1. Of interest, treatment of EL4 T cells with IL-10 resulted in increased gene expression of all five TFs, and inhibition of TF expression by siRNA suppressed Roquin-1 gene expression. In that context, it was recently reported that a defect in IKZF2 is a factor in the development of autoimmunity in mice caused by a failure of CD8+ Tregs (56). The extent to which IKZF2 similarly influences Treg function in sanroque mice remains to be determined.
Mechanistic Model of Roquin in Health and Disease
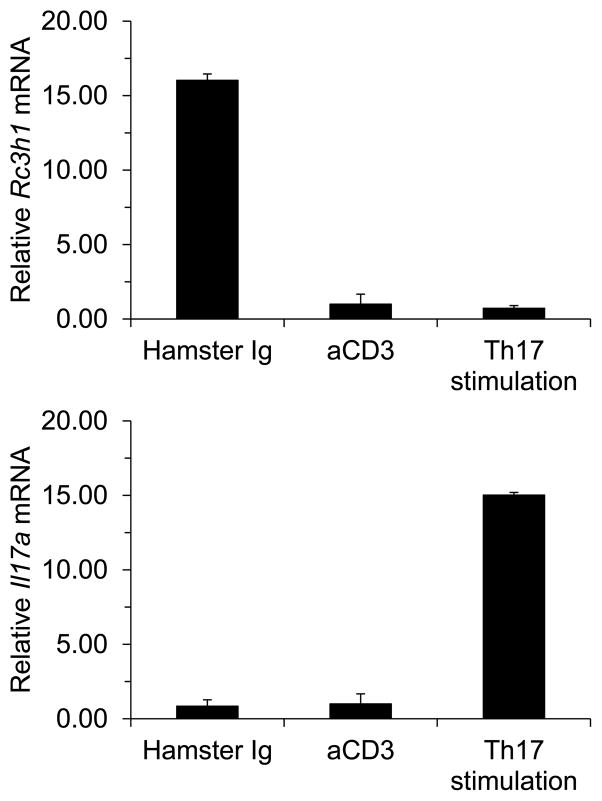
In drawing upon the information presented here, we offer a biological model of Roquin that incorporates both positive and negative regulatory elements during the generation of a homeostatic immune response, as well as during chronic inflammation and autoimmunity. Using MLN T cells, we observed that stimulation through the TCR/CD3 complex or following exposure to a Th17-inducing cocktail, Rc3h1 expression was suppressed compared to control Ig stimulation (Figure 1). In extending those findings, lower levels of Roquin-1 during a primary immune response would predictably lead to elevated ICOS and OX40 levels (1, 2, 36), thus promoting strong T cell activation. The benefit to the host would be a robust immune response in dealing with infection and foreign antigen challenge. A potent Roquin-1 response at this stage, however, could have negative consequences by holding ICOS expression down, as seen in studies in Roquin-1 Tg mice, although that response could be partially compensated for by elevated IL-2, IFN-γ, TNFα, IL-6, and IL-17 synthesis (47, 50). Interestingly, a unique population of CD4+ CD25+ FoxP3+ ICOS+ regulatory T cells has been identified (40), demonstrating that immunological significance of ICOS regulation by Roquin-1 may be more complex than previously appreciated.
Figure 1.
(Top panel) Mesenteric lymph node T cells were cultured in the presence of control Ig, anti-CD3 antibody, or a cocktail for stimulation of Th17 cells. T cell stimulation by either pan-T cell activation or Th-17 cell activation resulted in decreased Rc3h1 gene expression, suggesting that during the normal process of T cell immune activation, Rc3h1 expression is suppressed. (Bottom panel) Confirmation that Th17 stimulation specifically induces Th17 gene expression.
The natural immune response to antigen under homeostatic conditions proceeds systematically through expansion and contraction phases. The patterns just described would largely define the role for Roquin-1 during the initial expansion phase. While beneficial overall, failure to shut down the response would have significant deleterious consequences by preventing movement into the contraction phase, potentially leading to chronic inflammation or autoimmunity. Thus, upregulation of Roquin-1 expression would serve to suppress the expression of ICOS, RANTES, CXCR5, and CCL5 on peripheral T cells, thereby limiting effector cell activity.
Additional studies will be needed to understand how the cells of the immune system lose and gain Roquin expression. Clearly, Roquin-1 gene regulation through TFs such as STAT1, STAT3, GATA2, c-Rel, and IKZF2, as discussed above, along with production of immune-modulating cytokines, would likely come into play. IL-10, in particular, may be a key regulatory factor in this pathway by independently curtailing the expression of TNFα, IFN-γ, and IL-17 (57). Secondarily, however, IL-10 may directly modulate Roquin-1 gene expression by increasing the expression of Roquin-1 TFs (54). The net effect of IL-10-mediated Roquin-1 modulation would be to further limit ICOS and OX40 expression and influence Tfh cell regulation. The latter is supported by studies demonstrating that T cells deficient in IL-10 signaling are more likely to have increases in Tfh cells (58). A model of how Roquin-1 expression contributes to the immune response is shown in Figure 2.
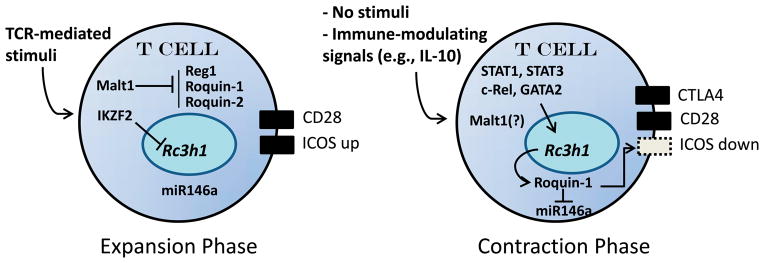
Figure 2.
A model of a biphasic role for Roquin during the expansion and contraction phases of the immune response. Upon encounter with foreign antigen, Rc3h1 gene expression is suppressed by the IKZF2 (and possibly other) transcription factor, down-regulating Roquin-1 protein expression. Concomitantly, Malt1 blocks Reg1, Roquin-1, and Roquin-2 proteins intracellularly, leading to increased surface expression of ICOS and a robust immune response. Upon removal of immune stimuli, and possibly due to immune-modulating cytokines such as IL-10, the Rc3h1 gene is transcriptionally upregulated by STAT1, STAT3, c-Rel, and GATA2. Malt1 activity also may be suppressed. Rc3h1 gene expression and Roquin-1 block miR146a and suppress surface ICOS expression. Contributing to this process is CTLA4, which competes for binding to the CD28 ligands, CD80 and CD86. Failure to re-activate Rc3h1 gene expression would result in elevated ICOS expression, however, potentially leading to autoimmunity and chronic inflammation.
In summary, Roquin poses to be an interesting molecular switch used in the regulation of the immune response. Although in some areas differences exist in findings regarding the role of Roquin in immunity, these will likely be resolved as work continues into this exciting aspect of immunobiology.
Acknowledgments
This work was supported by NIH grant DK035566 and a grant from the Crohn’s and Colitis Foundation of America.
Footnotes
The authors declare no conflict of interest.
References
- 1.Yu D, Tan AH, Hu X, Athanasopoulos V, Simpson N, Silva DG, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450(7167):299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- 2.Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435(7041):452–8. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- 3.Metzger MB, Pruneda JN, Klevit RE, Weissman AM. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochimica et biophysica acta. 2014;1843(1):47–60. doi: 10.1016/j.bbamcr.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernassola F, Karin M, Ciechanover A, Melino G. The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer cell. 2008;14(1):10–21. doi: 10.1016/j.ccr.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Li W, Gao B, Lee SM, Bennett K, Fang D. RLE-1, an E3 ubiquitin ligase, regulates C. elegans aging by catalyzing DAF-16 polyubiquitination. Developmental cell. 2007;12(2):235–46. doi: 10.1016/j.devcel.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Maruyama T, Araki T, Kawarazaki Y, Naguro I, Heynen S, Aza-Blanc P, et al. Roquin-2 promotes ubiquitin-mediated degradation of ASK1 to regulate stress responses. Science signaling. 2014;7(309):ra8. doi: 10.1126/scisignal.2004822. [DOI] [PubMed] [Google Scholar]
- 7.Glasmacher E, Hoefig KP, Vogel KU, Rath N, Du L, Wolf C, et al. Roquin binds inducible costimulator mRNA and effectors of mRNA decay to induce microRNA-independent post-transcriptional repression. Nature immunology. 2010;11(8):725–33. doi: 10.1038/ni.1902. [DOI] [PubMed] [Google Scholar]
- 8.Tan D, Zhou M, Kiledjian M, Tong L. The ROQ domain of Roquin recognizes mRNA constitutive-decay element and double-stranded RNA. Nature structural & molecular biology. 2014;21(8):679–85. doi: 10.1038/nsmb.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schlundt A, Heinz GA, Janowski R, Geerlof A, Stehle R, Heissmeyer V, et al. Structural basis for RNA recognition in roquin-mediated post-transcriptional gene regulation. Nature structural & molecular biology. 2014;21(8):671–8. doi: 10.1038/nsmb.2855. [DOI] [PubMed] [Google Scholar]
- 10.Leppek K, Schott J, Reitter S, Poetz F, Hammond MC, Stoecklin G. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell. 2013;153(4):869–81. doi: 10.1016/j.cell.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 11.Vogel KU, Edelmann SL, Jeltsch KM, Bertossi A, Heger K, Heinz GA, et al. Roquin paralogs 1 and 2 redundantly repress the Icos and Ox40 costimulator mRNAs and control follicular helper T cell differentiation. Immunity. 2013;38(4):655–68. doi: 10.1016/j.immuni.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Murakawa Y, Hinz M, Mothes J, Schuetz A, Uhl M, Wyler E, et al. RC3H1 post-transcriptionally regulates A20 mRNA and modulates the activity of the IKK/NF-kappaB pathway. Nature communications. 2015;6:7367. doi: 10.1038/ncomms8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Athanasopoulos V, Barker A, Yu D, Tan AH, Srivastava M, Contreras N, et al. The ROQUIN family of proteins localizes to stress granules via the ROQ domain and binds target mRNAs. The FEBS journal. 2010;277(9):2109–27. doi: 10.1111/j.1742-4658.2010.07628.x. [DOI] [PubMed] [Google Scholar]
- 14.Srivastava M, Duan G, Kershaw NJ, Athanasopoulos V, Yeo JH, Ose T, et al. Roquin binds microRNA-146a and Argonaute2 to regulate microRNA homeostasis. Nature communications. 2015;6:6253. doi: 10.1038/ncomms7253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mino T, Takeuchi O. Regnase-1 and Roquin regulate inflammatory mRNAs. Oncotarget. 2015;6(20):17869–70. doi: 10.18632/oncotarget.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mino T, Murakawa Y, Fukao A, Vandenbon A, Wessels HH, Ori D, et al. Regnase-1 and Roquin Regulate a Common Element in Inflammatory mRNAs by Spatiotemporally Distinct Mechanisms. Cell. 2015;161(5):1058–73. doi: 10.1016/j.cell.2015.04.029. [DOI] [PubMed] [Google Scholar]
- 17.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 18.Nakasa T, Miyaki S, Okubo A, Hashimoto M, Nishida K, Ochi M, et al. Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis and rheumatism. 2008;58(5):1284–92. doi: 10.1002/art.23429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coskun M, Bjerrum JT, Seidelin JB, Nielsen OH. MicroRNAs in inflammatory bowel disease--pathogenesis, diagnostics and therapeutics. World journal of gastroenterology: WJG. 2012;18(34):4629–34. doi: 10.3748/wjg.v18.i34.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu F, Guo NJ, Tian H, Marohn M, Gearhart S, Bayless TM, et al. Peripheral blood microRNAs distinguish active ulcerative colitis and Crohn’s disease. Inflamm Bowel Dis. 2011;17(1):241–50. doi: 10.1002/ibd.21450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu F, Zhang S, Dassopoulos T, Harris ML, Bayless TM, Meltzer SJ, et al. Identification of microRNAs associated with ileal and colonic Crohn’s disease. Inflamm Bowel Dis. 2010;16(10):1729–38. doi: 10.1002/ibd.21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, et al. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135(5):1624–35 e24. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 23.Folini M, Gandellini P, Longoni N, Profumo V, Callari M, Pennati M, et al. miR-21: an oncomir on strike in prostate cancer. Molecular cancer. 2010;9:12. doi: 10.1186/1476-4598-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaefer JS, Attumi T, Opekun AR, Abraham B, Hou J, Shelby H, et al. MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC immunology. 2015;16:5. doi: 10.1186/s12865-015-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaefer JS, Montufar-Solis D, Vigneswaran N, Klein JR. Selective upregulation of microRNA expression in peripheral blood leukocytes in IL-10−/− mice precedes expression in the colon. Journal of immunology. 2011;187(11):5834–41. doi: 10.4049/jimmunol.1100922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pauley KM, Stewart CM, Gauna AE, Dupre LC, Kuklani R, Chan AL, et al. Altered miR-146a expression in Sjogren’s syndrome and its functional role in innate immunity. European journal of immunology. 2011;41(7):2029–39. doi: 10.1002/eji.201040757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10(4):R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pratama A, Srivastava M, Williams NJ, Papa I, Lee SK, Dinh XT, et al. MicroRNA-146a regulates ICOS-ICOSL signalling to limit accumulation of T follicular helper cells and germinal centres. Nature communications. 2015;6:6436. doi: 10.1038/ncomms7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeltsch KM, Hu D, Brenner S, Zoller J, Heinz GA, Nagel D, et al. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nature immunology. 2014;15(11):1079–89. doi: 10.1038/ni.3008. [DOI] [PubMed] [Google Scholar]
- 30.Akira S. Regnase-1, a ribonuclease involved in the regulation of immune responses. Cold Spring Harbor symposia on quantitative biology. 2013;78:51–60. doi: 10.1101/sqb.2013.78.019877. [DOI] [PubMed] [Google Scholar]
- 31.Uehata T, Iwasaki H, Vandenbon A, Matsushita K, Hernandez-Cuellar E, Kuniyoshi K, et al. Malt1-induced cleavage of regnase-1 in CD4(+) helper T cells regulates immune activation. Cell. 2013;153(5):1036–49. doi: 10.1016/j.cell.2013.04.034. [DOI] [PubMed] [Google Scholar]
- 32.Vinuesa CG, Goodnow CC. Illuminating autoimmune regulators through controlled variation of the mouse genome sequence. Immunity. 2004;20(6):669–79. doi: 10.1016/j.immuni.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 33.Montufar-Solis D, Vigneswaran N, Nakra N, Schaefer JS, Klein JR. Hematopoietic not systemic impairment of Roquin expression accounts for intestinal inflammation in Roquin-deficient mice. Scientific reports. 2014;4:4920. doi: 10.1038/srep04920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Linterman MA, Rigby RJ, Wong R, Silva D, Withers D, Anderson G, et al. Roquin differentiates the specialized functions of duplicated T cell costimulatory receptor genes CD28 and ICOS. Immunity. 2009;30(2):228–41. doi: 10.1016/j.immuni.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 35.Hondowicz BD, Batheja AO, Metzgar MH, Caton AJ, Erikson J. ICOS expression by effector T cells influences the ability of regulatory T cells to inhibit anti-chromatin B cell responses in recipient mice. Journal of autoimmunity. 2010;34(4):460–8. doi: 10.1016/j.jaut.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaefer JS, Montufar-Solis D, Nakra N, Vigneswaran N, Klein JR. Small intestine inflammation in Roquin-mutant and Roquin-deficient mice. PloS one. 2013;8(2):e56436. doi: 10.1371/journal.pone.0056436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, et al. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27(2):281–95. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beier KC, Hutloff A, Dittrich AM, Heuck C, Rauch A, Buchner K, et al. Induction, binding specificity and function of human ICOS. Eur J Immunol. 2000;30(12):3707–17. doi: 10.1002/1521-4141(200012)30:12<3707::AID-IMMU3707>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 39.Salomon B, Lenschow DJ, Rhee L, Ashourian N, Singh B, Sharpe A, et al. B7/CD28 costimulation is essential for the homeostasis of the CD4+CD25+ immunoregulatory T cells that control autoimmune diabetes. Immunity. 2000;12(4):431–40. doi: 10.1016/s1074-7613(00)80195-8. [DOI] [PubMed] [Google Scholar]
- 40.Vocanson M, Rozieres A, Hennino A, Poyet G, Gaillard V, Renaudineau S, et al. Inducible costimulator (ICOS) is a marker for highly suppressive antigen-specific T cells sharing features of TH17/TH1 and regulatory T cells. The Journal of allergy and clinical immunology. 2010;126(2):280–9. 9 e1–7. doi: 10.1016/j.jaci.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 41.Sadlack B, Merz H, Schorle H, Schimpl A, Feller AC, Horak I. Ulcerative colitis-like disease in mice with a disrupted interleukin-2 gene. Cell. 1993;75(2):253–61. doi: 10.1016/0092-8674(93)80067-o. [DOI] [PubMed] [Google Scholar]
- 42.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75(2):263–74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 43.Claesson MH, Rudolphi A, Kofoed S, Poulsen SS, Reimann J. CD4+ T lymphocytes injected into severe combined immunodeficient (SCID) mice lead to an inflammatory and lethal bowel disease. Clinical and experimental immunology. 1996;104(3):491–500. doi: 10.1046/j.1365-2249.1996.48757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int Immunol. 1993;5(11):1461–71. doi: 10.1093/intimm/5.11.1461. [DOI] [PubMed] [Google Scholar]
- 45.Ellyard JI, Chia T, Rodriguez-Pinilla SM, Martin JL, Hu X, Navarro-Gonzalez M, et al. Heterozygosity for Roquinsan leads to angioimmunoblastic T-cell lymphoma-like tumors in mice. Blood. 2012;120(4):812–21. doi: 10.1182/blood-2011-07-365130. [DOI] [PubMed] [Google Scholar]
- 46.Auguste T, Travert M, Tarte K, Ame-Thomas P, Artchounin C, Martin-Garcia N, et al. ROQUIN/RC3H1 alterations are not found in angioimmunoblastic T-cell lymphoma. PloS one. 2013;8(6):e64536. doi: 10.1371/journal.pone.0064536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim HJ, Ji YR, Kim MO, Yu DH, Shin MJ, Yuh HS, et al. The role of Roquin overexpression in the modulation of signaling during in vitro and ex vivo T-cell activation. Biochem Biophys Res Commun. 2012;417(1):280–6. doi: 10.1016/j.bbrc.2011.11.101. [DOI] [PubMed] [Google Scholar]
- 48.Harada Y, Ohgai D, Watanabe R, Okano K, Koiwai O, Tanabe K, et al. A single amino acid alteration in cytoplasmic domain determines IL-2 promoter activation by ligation of CD28 but not inducible costimulator (ICOS) J Exp Med. 2003;197(2):257–62. doi: 10.1084/jem.20021305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe R, Harada Y, Takeda K, Takahashi J, Ohnuki K, Ogawa S, et al. Grb2 and Gads exhibit different interactions with CD28 and play distinct roles in CD28-mediated costimulation. J Immunol. 2006;177(2):1085–91. doi: 10.4049/jimmunol.177.2.1085. [DOI] [PubMed] [Google Scholar]
- 50.Ji YR, Kim HJ, Yu DH, Bae KB, Park SJ, Yi JK, et al. Enforced expression of roquin protein in T cells exacerbates the incidence and severity of experimental arthritis. J Biol Chem. 2012;287(50):42269–77. doi: 10.1074/jbc.M112.374835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ji YR, Kim HJ, Yu DH, Bae KB, Park SJ, Park SJ, et al. Over-expression of Roquin aggravates T cell mediated hepatitis in transgenic mice using T cell specific promoter. Biochem Biophys Res Commun. 2014;452(3):822–7. doi: 10.1016/j.bbrc.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 52.Bertossi A, Aichinger M, Sansonetti P, Lech M, Neff F, Pal M, et al. Loss of Roquin induces early death and immune deregulation but not autoimmunity. J Exp Med. 2011;208(9):1749–56. doi: 10.1084/jem.20110578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pratama A, Ramiscal RR, Silva DG, Das SK, Athanasopoulos V, Fitch J, et al. Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity. 2013;38(4):669–80. doi: 10.1016/j.immuni.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 54.Schaefer JS, Montufar-Solis D, Klein JR. A role for IL-10 in the transcriptional regulation of Roquin-1. Gene. 2014;549(1):134–40. doi: 10.1016/j.gene.2014.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen X, Hong F, Nguyen VA, Gao B. IL-10 attenuates IFN-alpha-activated STAT1 in the liver: involvement of SOCS2 and SOCS3. FEBS letters. 2000;480(2–3):132–6. doi: 10.1016/s0014-5793(00)01905-0. [DOI] [PubMed] [Google Scholar]
- 56.Kim HJ, Barnitz RA, Kreslavsky T, Brown FD, Moffett H, Lemieux ME, et al. Stable inhibitory activity of regulatory T cells requires the transcription factor Helios. Science. 2015;350(6258):334–9. doi: 10.1126/science.aad0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schaefer JS, Montufar-Solis D, Vigneswaran N, Klein JR. ICOS promotes IL-17 synthesis in colonic intraepithelial lymphocytes in IL-10−/− mice. J Leukoc Biol. 2010;87(2):301–8. doi: 10.1189/jlb.0409238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai G, Nie X, Zhang W, Wu B, Lin J, Wang H, et al. A regulatory role for IL-10 receptor signaling in development and B cell help of T follicular helper cells in mice. J Immunol. 2012;189(3):1294–302. doi: 10.4049/jimmunol.1102948. [DOI] [PubMed] [Google Scholar]