Abstract
The Tremellomycetes (Basidiomycota) contains a large number of unicellular and dimorphic fungi with stable free-living unicellular states in their life cycles. These fungi have been conventionally classified as basidiomycetous yeasts based on physiological and biochemical characteristics. Many currently recognised genera of these yeasts are mainly defined based on phenotypical characters and are highly polyphyletic. Here we reconstructed the phylogeny of the majority of described anamorphic and teleomorphic tremellomycetous yeasts using Bayesian inference, maximum likelihood, and neighbour-joining analyses based on the sequences of seven genes, including three rRNA genes, namely the small subunit of the ribosomal DNA (rDNA), D1/D2 domains of the large subunit rDNA, and the internal transcribed spacer regions (ITS 1 and 2) of rDNA including 5.8S rDNA; and four protein-coding genes, namely the two subunits of the RNA polymerase II (RPB1 and RPB2), the translation elongation factor 1-α (TEF1) and the mitochondrial gene cytochrome b (CYTB). With the consideration of morphological, physiological and chemotaxonomic characters and the congruence of phylogenies inferred from analyses using different algorithms based on different data sets consisting of the combined seven genes, the three rRNA genes, and the individual protein-coding genes, five major lineages corresponding to the orders Cystofilobasidiales, Filobasidiales, Holtermanniales, Tremellales, and Trichosporonales were resolved. A total of 45 strongly supported monophyletic clades with multiple species and 23 single species clades were recognised. This phylogenetic framework will be the basis for the proposal of an updated taxonomic system of tremellomycetous yeasts that will be compatible with the current taxonomic system of filamentous basidiomycetes accommodating the ‘one fungus, one name’ principle.
Key words: Fungi, Basidiomycota, Tremellomycetes, Yeasts, Multigene phylogeny
Introduction
Unicellular basidiomycetes and dimorphic basidiomycetes with a stable free-living unicellular state during their life cycles are recognised as basidiomycetous yeasts (Boekhout et al. 2011). They occur in all three subphyla of the Basidiomycota, namely Agaricomycotina, Pucciniomycotina and Ustilaginomycotina, which are presently recognised on the basis of molecular phylogenetic analyses (Fell et al., 2000, Scorzetti et al., 2002, James et al., 2006, Hibbett et al., 2007, Wuczkowski et al., 2011). Yeast taxa in the Agaricomycotina occur only in the basal Tremellomycetes lineage (Hibbett, 2006, Boekhout et al., 2011, Weiß et al., 2014). Phenotypic and molecular analyses revealed a close affiliation of basidiomycetous yeasts with various groups of filamentous basidiomycetes (Millanes et al. 2011). However, yeasts and filamentous fungi have conventionally been studied by different scientific communities and classified using different criteria, resulting in the developments of hitherto independent taxonomic systems of the two groups of fungi. Recent molecular phylogenetic studies have shown the incompatibility between the taxonomic system of basidiomycetous yeasts and that of filamentous basidiomycetes. Furthermore many currently recognised genera of tremellomycetous yeasts, which are mainly defined based on phenotypic characters, are strikingly polyphyletic. For example, species of the genus Cryptococcus are located in all currently recognised orders of Tremellomycetes and occur intermingled with species of other genera, such as Bullera (Fell et al., 2000, Boekhout et al., 2011, Fonseca et al., 2011, Millanes et al., 2011, Weiß et al., 2014). The polyphyletic nature of the traditionally defined teleomorphic genus Tremella, which is usually dimorphic, is also remarkable. Several monophyletic clades have been recognised among Tremella species which occur interspersed with other teleomorphic and anamorphic genera (Boekhout et al., 2011, Millanes et al., 2011, Weiß et al., 2014). These problems existing in the current taxonomic systems of both yeasts and filamentous fungi in the Tremellomycetes remain to be resolved.
The high-level classification of Basidiomycota has been updated with results from the Assembling the Fungal Tree of Life (AFTOL) project that used a multigene sequence analysis approach (Lutzoni et al., 2004, James et al., 2006, Hibbett et al., 2007). However, only a limited number of basidiomycetous yeast taxa were included in that project, making it impossible to propose a corresponding revision of the taxonomic system of basidiomycetous yeasts. Consequently, the artificial classification system of these organisms largely remained in the latest edition of ‘The Yeasts, a Taxonomic Study’ [hereafter referred to as ‘The Yeasts’] (Kurtzman et al. 2011), due to the lack of reliable multigene phylogenetic studies of these yeasts.
The requirement for revising the taxonomic system of tremellomycetous yeasts has been emphasised by recent progress in biodiversity studies of yeasts and by the change of fungal nomenclature adopting the ‘one fungus = one name’ concept (Hawksworth 2011). Molecular taxonomic studies have resulted in the availability of a comprehensive sequence database of the large subunit of the ribosomal RNA gene (LSU rDNA) D1/D2 domains and the ITS (including 5.8S) regions of rDNA for almost all known basidiomycetous yeast species (Fell et al., 2000, Scorzetti et al., 2002, Wang and Bai, 2008, Schoch et al., 2012). The rDNA sequence databases have become a molecular platform for rapid identification of yeasts, resulting in continued discovery of new taxa in recent years (Boekhout, 2005, Wang and Bai, 2008, Wuczkowski et al., 2011). The addition of these new species has contributed to the increase of the polyphyletic nature of many basidiomycetous yeast genera. For example, the distribution of Bullera species has expanded from Tremellales and Filobasidiales (Boekhout & Nakase 1998) to Trichosporonales (Nakase et al., 2002, Fungsin et al., 2006). With each new species being taxonomically misplaced, the chaos of the taxonomic system increases. Therefore, an updated taxonomic system is imperative for the correct placement of the vast amount of hidden yeast diversity.
Similar to filamentous fungi, many yeast species have separate teleomorphic and anamorphic names. As regulated by the new International Code of Nomenclature for algae, fungi and plants (Melbourne Code) (McNeill et al. 2012), after January 1 2013, only one name is legitimate regardless of whether or not a sexual state exists. An updated taxonomic system, especially the redefinition of genera based on a robust multigene phylogeny, will be required for the name choices and to minimise the possibility of name changes in the future. The purpose of this study is to confidently resolve the phylogenetic relationships among tremellomycetous yeasts and dimorphic fungi based on multiple gene sequence analyses, resulting in a framework that allows us to update the taxonomic system of yeasts and related taxa in the Tremellomycetes.
Materials and methods
Taxon sampling
A total of 294 tremellomycetous yeast strains were included in this study, which covered the type strains of 286 currently recognised species and varieties, the type strains of six synonyms, and two additional strains with mating types opposite to those of the type strains (Table 1). From the 240 tremellomycetous yeast species and varieties included in the latest edition of ‘The Yeasts’ (Kurtzman et al. 2011), 234 were included in this study. In addition, 52 tremellomycetous yeast species which were published too late for inclusion in the book were also employed in this study. The taxa sampled covered 16 teleomorphic and 19 anamorphic genera. The type strains of two pucciniomycetous and one ustilaginomycetous yeast species were employed as outgroup (Table 1).
Table 1.
List of tremellomycetous yeasts and dimorphic taxa employed. The sequences with GenBank numbers in bold are determined in this study.
The asterisks indicate teleomorphic taxa; CBS database: sequences are available from the CBS database at http://www.cbs.knaw.nl/Collections/Biolomics.aspx?Table=CBS strain database.
DNA extraction, PCR, and sequencing
Genomic DNA was extracted from yeast cells actively growing on YPD medium using the method described in Bolano et al. (2001) with minor modifications. The UltraClean® Microbial DNA Isolation Kit (MO BIO, CA) was used when high quality DNA templates were required for PCR amplification of some protein genes. A set of six genes was selected and sequenced based on previous studies of the Assembling the Fungal Tree of Life (AFTOL-1) project (James et al., 2006, Hibbett et al., 2007). These genes included three rRNA genes, namely the small subunit (SSU or 18S) of the ribosomal DNA (rDNA), D1/D2 domains of the large subunit (LSU or 26S) rDNA, and the internal transcribed spacer regions (ITS 1 and 2) of the rDNA, including the 5.8S rDNA; and three nuclear protein-coding genes, namely the two subunits of RNA polymerase II (RPB1 and RPB2) and translation elongation factor 1-α (TEF1). In addition, the mitochondrial gene cytochrome b (CYTB) was also included. The primers used for PCR amplification and sequencing of these genes are listed in Table 2. Because of the degenerate nature of the primers used for PCR amplification of the protein-coding genes, sometimes faint or multiple PCR bands were generated from PCR amplification or direct sequencing of amplicons failed. In these cases, amplicons were cloned using the pGEM®-T Easy Vector Systems (Promega Corporation, Madison) following the protocol of the kit. Positive colonies with an insert of expected size were chosen for sequencing.
Table 2.
PCR and sequencing primers used in this study.
| Primer name | Nucleotide sequence (5′–3′) | Reference |
|---|---|---|
| ITS and D1/D2 | ||
| V9 | TGC GTT GAT TAC GTC CCT GC→ | Boekhout et al. 2003 |
| RLR3R | ←GGT CCG TGT TTC AAG AC | Boekhout et al. 2003 |
| ITS4 | ←TCC TCC GCT TAT TGA TAT GC | White et al. 1990 |
| NL1 | GCA TAT CAA TAA GCG GAG GAA AAG→ | O'Donnell 1993 |
| SSU | ||
| NS1 | GTA GTC ATA TGC TTG TCT→ | White et al. 1990 |
| NS24 | ←AAA CCT TGT TAC GAC TTT TA | Gargas & Taylor 1992 |
| Oligo3 | ←GTA CAC ACC GCC CGT C | Hendriks et al. 1989 |
| Oligo10 | ←TGG YRA ATG CTT TCG C | Hendriks et al. 1989 |
| Oligo13 | ←ATA ACA GGT CTG TGA TGC CC | Hendriks et al. 1989 |
| Oligo14 | ATA ACA GGT CTG TGA TGC CC→ | Hendriks et al. 1989 |
| RPB1 | ||
| RPB1-Af | GAR TGY CCD GGD CAY TTY GG→ | Stiller & Hall 1997 |
| RPB 1-Cr | ←CCN GCD ATN TCR TTR TCC ATR TA | Matheny et al. 2002 |
| RPB2 | ||
| f RPB2-5F | GAY GAY MGW GAT CAY TTY GG→ | Liu et al. 1999 |
| RPB2-6F | TGG GGK WTG GTY TGY CCT GC→ | Liu et al. 1999 |
| RPB2-6R | ←GCA GGR CAR ACC AWM CCC CA | Liu et al. 1999 |
| RPB2-7R | ←CCC ATW GCY TGC TTM CCC AT | Liu et al. 1999 |
| bRPB2-7.1R | ←CCC ATR GCY TGY TTM CCC ATD GC | Matheny 2005 |
| TEF1 | ||
| EF1-983F | GCY CCY GGH CAY CGT GAY TTY AT→ | Rehner & Buckley 2005 |
| EF1-2218R | ←ATG ACA CCR ACR GCR ACR GTY TG | Rehner & Buckley 2005 |
| EF1-2212R | ←CCR ACR GCR ACR GTY YGT CTC AT | Rehner & Buckley 2005 |
| 1577F | CAR GAY GTB TAC AAG ATY GGT GG→ | Rehner & Buckley 2005 |
| 1567R | ←ACH GTR CCR ATA CCA CCR ATC TT | Rehner & Buckley 2005 |
| CYTB | ||
| E1M4 | TGR GGW GCW ACW GTT ATT ACT A→ | Biswas et al. 2003 |
| E2M4 | ←GGW ATA GMW SKT AAW AYA GCA TA | Biswas et al. 2003 |
Molecular phylogenetic analyses
Sequences were inspected and assembled using the SeqMan program in the Lasergene 7 software package (DNASTAR Inc., Madison) and were then aligned with Clustal X 1.83 (Thompson et al. 1997). Spliceosomal intron regions were inferred from the insertions with canonical splice sites (GT-AG, GC-AG, AT-AC) (Babenko et al. 2004) in the nucleotide sequence alignments between our data and reference cDNA sequences from GenBank. Exon sequences of the protein-encoding genes RPB1, RPB2, TEF1 and CYTB were manually aligned using MEGA 5 (Tamura et al. 2011). Positions deemed ambiguous to align were excluded manually. Thereafter, multiple sequence alignments for ITS, D1/D2, SSU, RPB1, RPB2, TEF1, and CYTB were concatenated as a combined file.
Maximum likelihood (ML), neighbour-joining (NJ), and Bayesian analyses were conducted for separate and combined nucleotide data sets using RAxML v8.1.X (Stamatakis 2014), MEGA 5.0 (Tamura et al. 2011) and MrBayes 3.2.1 (Ronquist et al. 2012), respectively. ML analysis was implemented with the novel fast bootstrap algorithm with 100 replicates and a subsequent search for the best maximum-likelihood tree in conjunction with the GTRGAMMAI model approximation (Stamatakis 2014). NJ analysis was performed on the evolutionary distance data calculated from Kimura's two-parameter model (Kimura 1980). Bootstrap analyses (Felsenstein 1985) were performed from 1 000 random re-samplings in both ML and NJ analyses. A bootstrap proportion (BP) support above 70 % obtained from the ML and NJ analyses was considered as significant (Hillis & Bull 1993).
Bayesian analysis was implemented using heterogeneous models to the data set with seven unlinked partitions, one for each gene. The best-fit evolution model of each gene fragment in the data set was determined using the Bayesian Information Criterion (BIC) in jModeltest (Posada 2008). The ITS, D1D2, and SSU rDNA gene sequences were fitted to TPM3uf+G, TIM3+G, and TIM2+T+G models, respectively. The protein-coding genes RPB1 and CYTB both used the GTR+I+G model; whereas RPB2 and TEF1 used the TPM3uf+I+G and TPM1uf+G models, respectively. Six to fifty million generations were run with four Markov chains (three heated and one cold), sampling every 500 generations. The average standard deviation of split frequencies, below 0.01, was examined to identify the convergence of the two independent runs. Clades with posterior probabilities (PP) above 0.95 were considered as significantly supported (Larget & Simon 1999).
Results
Sequences generated and data sets constructed for phylogenetic analyses
A total of 1 147 new sequences were produced in this study, including 21 ITS, 123 SSU, 269 RPB1, 270 RPB2, 249 TEF1, and 215 CYTB sequences. In addition, a total of 777 previously published sequences of these genes from the type strains of tremellomycetous yeast taxa were retrieved from GenBank (Table 1). Different data sets consisting of the three rRNA genes (rDNA), the individual protein-coding genes, and the combined seven genes were constructed from the 1 924 sequences employed in this study (Table 3). In addition, a data set of 5.8S and LSU rDNA D1/D2 domain sequences was constructed to include more Tremella species whose sequences were determined from herbarium specimens (Millanes et al. 2011).
Table 3.
Nucleotide sequence data sets constructed for phylogenetic analyses.
| Data set | No. of strains | No. of taxa | Length of alignment | Parsimony informative characters (%) |
|---|---|---|---|---|
| rDNA1 | 297 | 285 | 3 208 | 1 447 (45) |
| RPB1 | 271 | 262 | 758 | 615 (81) |
| RPB2 | 273 | 263 | 1 133 | 872 (77) |
| TEF1 | 249 | 238 | 909 | 498 (55) |
| CYTB | 246 | 238 | 388 | 279 (71) |
| Seven-gene | 281 | 269 | 6 298 | 3 623 (57) |
The rDNA data set includes 296 ITS, 297 LSU D1/D2, and 292 SSU rDNA sequences.
These data sets were subjected to phylogenetic analyses using Bayesian, ML and NJ algorithms, respectively. The topologies of the trees obtained were compared visually to inspect the phylogenetic concordance among the taxa analysed, based on which monophyletic lineages and clades were recognised and defined (Table 4). As expected, among the trees drawn from different data sets analysed, the seven-gene trees exhibited the clearest resolution and strongest supports; and among the algorithms employed, the Bayesian analysis usually showed the most robust phylogeny (Table 4). Thus, the Bayesian tree constructed from the seven-gene data set was used as the primary basis for lineage and clade recognition and definition, and as the starting point for the subsequent comparison and discussion. The phylogenetic trees inferred from the rDNA data set containing all the taxa employed in this study were used as references to judge the phylogenetic positions of a minority of taxa which were absent in the seven-gene tree because of failure in sequencing of the protein coding genes.
Table 4.
Monophyletic clades resolved in tremellomycetous yeasts and dimorphic taxa based on different data sets using different algorithms.
| Lineage/Clade |
RPB1 |
RPB2 |
TEF1 |
CYTB |
rDNA |
Seven-gene |
|---|---|---|---|---|---|---|
| PP/BP1/BP2 | PP/BP1/BP2 | PP/BP1/BP2 | PP/BP1/BP2 | PP/BP1/BP2 | PP/BP1/BP2 | |
| Cystofilobasidiales | nm/nm/nm | 1.0/100/99 | .90/64/70 | nm/nm/nm | 1.0/87/100 | 1.0/100/100 |
| Cystofilobasidium | 1.0/100/100 | 1.0/100/99 | nm/nm/nm | nm/nm/nm | 1.0/100/100 | 1.0/100/100 |
| Guehomyces | S | S | S | S | 1.0/100/nm | S |
| huempii | 1.0/100/100 | 1.0/100/100 | 1.0/100/100 | 1.0/100/99 | 1.0/100/100 | 1.0/100/100 |
| Itersonilia | S | 1.0/100/100 | S | S | 1.0/100/100 | 1.0/100/100 |
| Mrakia | 1.0/90/83 | 1.0/96/89 | nm/nm/ns | nm/nm/nm | 1.0/100/100 | 1.0/100/100 |
| Udeniomyces | 1.0/100/87 | 1.0/100/99 | S | – | 1.0/100/100 | 1.0/100/100 |
| Phaffia | S | 1.0/100/100 | 1.0/100/100 | 1.0/99/99 | 1.0/100/100 | 1.0/100/100 |
| Filobasidiales | 1.0/100/98 | 1.0/100/100 | nm/nm/ns | nm/nm/nm | 1.0/83/98 | 1.0/100/100 |
| aerius | 1.0/100/100 | 1.0/100/100 | ns/nm/nm | nm/nm/nm | nm/nm/nm | 1.0/100/85 |
| albidus | 1.0/100/100 | 1.0/100/100 | 1.0/90/99 | nm/nm/nm | 1.0/94/99 | 1.0/100/100 |
| cylindricus | 1.0/100/100 | 1.0/100/100 | 1.0/100/100 | ns/55/ns | 1.0/100/100 | 1.0/100/100 |
| Filobasidium | 1.0/99/100 | 1.0/100/100 | 1.0/97/92 | nm/nm/nm | 1.0/100/100 | 1.0/100/100 |
| gastricus | 1.0/100/99 | 1.0/100/98 | nm/nm/nm | nm/nm/nm | 1.0/99/91 | 1.0/100/100 |
| Holtermanniales | 1.0/100/99 | 1.0/100/100 | 1.0/100/100 | .99/66/ns | 1.0/100/100 | 1.0/100/56 |
| Holtermanniella | 1.0/62/78 | 1.0/100/100 | 1.0/100/100 | nm/nm/nm | 1.0/99/100 | 1.0/100/80 |
| Tremellales/Trichosporonales | 1.0/100/99 | 1.0/100/97 | ns/ns/68 | nm/nm/nm | .99/99/nm | 1.0/100/55 |
| Tremellales | nm/nm/nm | .95/72/nm | nm/nm/nm | nm/nm/nm | nm/nm/nm | ns/nm/nm |
| amylolyticus | 1.0/83/84 | 1.0/100/100 | 1.0/97/88 | 1.0/99/99 | 1.0/100/100 | 1.0/100/100 |
| aurantia | 1.0/62//99 | 1.0/100/100 | ns/ns/63 | nm/nm/nm | nm/nm/nm | 1.0/100/100 |
| aureus | 1.0/100/100 | 1.0/100/100 | .99/85/98 | nm/nm/nm | 1.0/100/100 | 1.0/100/100 |
| Auriculibuller | ns/59/nm | 1.0/100/100 | ns/ns/89 | 1.0/94/64 | 1.0/100/nm | 1.0/100/100 |
| Bandoniozyma | 1.0/100/100 | 1.0/100/100 | 1.0/96/95 | 1.0/100/99 | 1.0/100/100 | 1.0/100/100 |
| Bulleribasidium | .94/57/nm | 1.0/67/56 | nm/nm/nm | nm/nm/nm | 1.0/100/100 | 1.0/100/100 |
| Bulleromyces | 1.0/100/100 | S | 1.0/94/98 | 1.0/99/99 | 1.0/100/100 | 1.0/100/100 |
| Cryptococcus | 1.0/98/80 | 1.0/100/99 | nm/nm/nm | 1.0/85/92 | 1.0/100/100 | 1.0/100/100 |
| Derxomyces | .93/ns/nm | ns/ns/nm | nm/nm/nm | ns/ns/57 | 1.0/100/100 | 1.0/100/100 |
| dimennae | 1.0/96/93 | 1.0/99/100/ | nm/nm/nm | nm/nm/nm | 1.0/99/83 | 1.0/100/100 |
| Dioszegia | 1.0/96/89 | 1.0/93/99 | nm/nm/nm | 1.0/98/99 | 1.0/100/100 | 1.0/100/100 |
| Fellomyces | nm/nm/nm | nm/nm/nm | 1.0/69/63 | S | 1.0/87/98 | 1.0/64/nm |
| Fibulobasidium | 1.0/100/100 | 1.0/100/100 | 1.0/99/100 | 1.0/100/99 | 1.0/100/100 | 1.0/100/100 |
| flavus | nm/nm/nm | nm/nm/nm | S/S/S | 1.0/96/99 | .99/83/98 | .97/72/82 |
| foliacea | 1.0/100/100 | 1.0/100/99 | nm/nm/nm | nm/nm/nm | 1.0/100/100 | 1.0/100/100 |
| hannae | 1.0/100/100 | 1.0/100/100 | 1.0/100/100 | S | 1.0/100/100 | 1.0/100/100 |
| Hannaella | nm/nm/ns | 1.0/99/95 | nm/nm/nm | 1.0/72/97 | 1.0/100/100 | 1.0/100/100 |
| Kockovaella | 1.0/62/74 | nm/nm/nm | 1.0/100/99 | nm/nm/nm | 1.0/100/100 | 1.0/96/96 |
| Kwoniella | 1.0/75/92 | 1.0/100/99 | nm/nm/nm | nm/nm/nm | 1.0/100/100 | 1.0/100/100 |
| laurentii | 1.0/100/99 | 1.0/100/69 | .97/ns/nm | nm/nm/nm | 1.0/100/92 | 1.0/100/100 |
| melastomae | 1.0/96/98 | S | – | 1.0/96/98 | 1.0/99/100 | 1.0/100/100 |
| moriformis | S | S | S | S | 1.0/100/95 | S |
| Papiliotrema | 1.0/100/100 | 1.0/100/100 | 1.0/95/97 | 1.0/99/99 | 1.0/100/nm | 1.0/100/100 |
| pseudoalba | .98/89/96 | 1.0/100/98 | 1.0/75/59 | .97/100/99 | .96/89/nm | 1.0/100/100 |
| Tremella | 1.0/100/99 | 1.0/100/100 | nm/nm/nm | nm/nm/nm | 1.0/100/99 | 1.0/100/55 |
| Trichosporonales | 1.0/95/65 | 0.92/ns/nm | nm/nm/ns | nm/nm/nm | nm/nm/nm | 1.0/100/100 |
| gracile/brassicae | .90/61/nm | 1.0/97/91 | nm/nm/nm | ns/ns/nm | 1.0/98/100 | 1.0/100/100 |
| cutaneum | .96/ns/76 | ns/ns/61 | nm/60/86 | nm/nm/nm | 1.0/100/100 | 1.0/100/98 |
| formosensis | 1.0/100/99 | 1.0/100/95 | 1.0/92/66 | 1.0/100/99 | 1.0/100/100 | 1.0/100/100 |
| Vanrija | 1.0/100/100 | 1.0/100/100 | 1.0/100/100 | 1.0/98/99 | 1.0/97/77 | 1.0/100/100 |
| haglerorum | nm/ns/nm | nm/ns/nm | nm/nm/ns | nm/nm/nm | 1.0/91/100 | 1.0/78/91 |
| porosum | ns/80/100 | 1.0/100/100 | 1.0/51/86 | nm/nm/nm | 1.0/100/100 | 1.0/100/100 |
| Trichosporon | 1.0/100/99 | 1.0/100/100 | 1.0/100/99 | nm/nm/nm | 1.0/100/100 | 1.0/100/100 |
Note. PP, Bayesian posterior probability; BP1 and BP2, bootstrap values from the maximum likelihood and neighbour-joining analyses, respectively; nm: not monophyletic; ns, not supported (PP < 0.9 or BP < 50 %); S: single species clade. Data sets that produce both significant PP (≥0.95) and BP (≥70 %) values have dark grey shaded cells; and data sets that produce either a significant PP or BP support value have light grey shaded cells.
Major lineages recognised among tremellomycetous yeasts
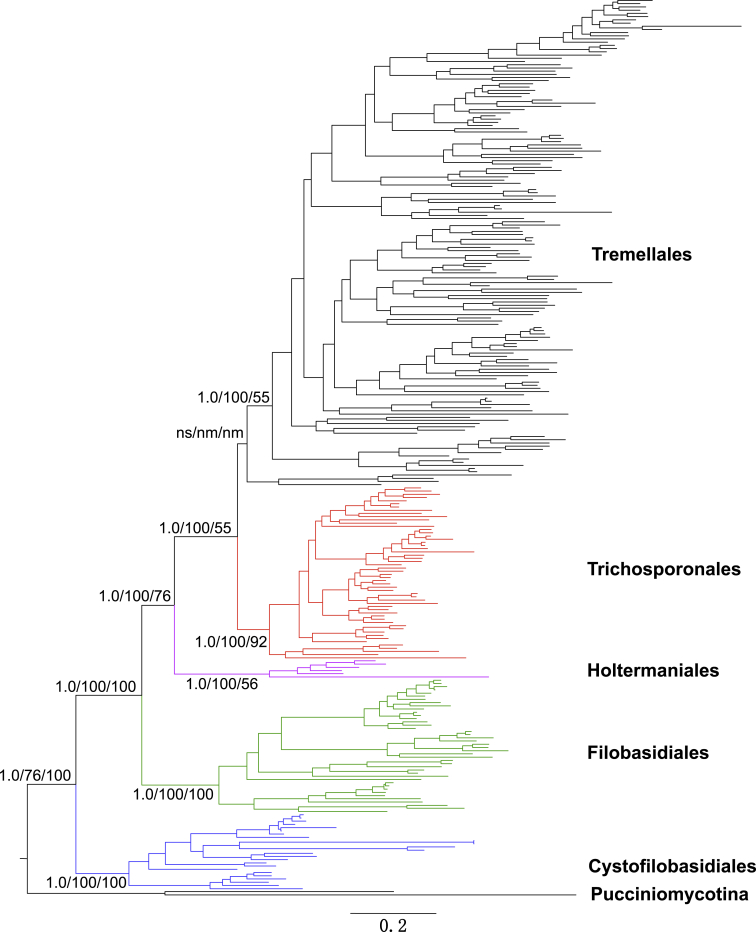
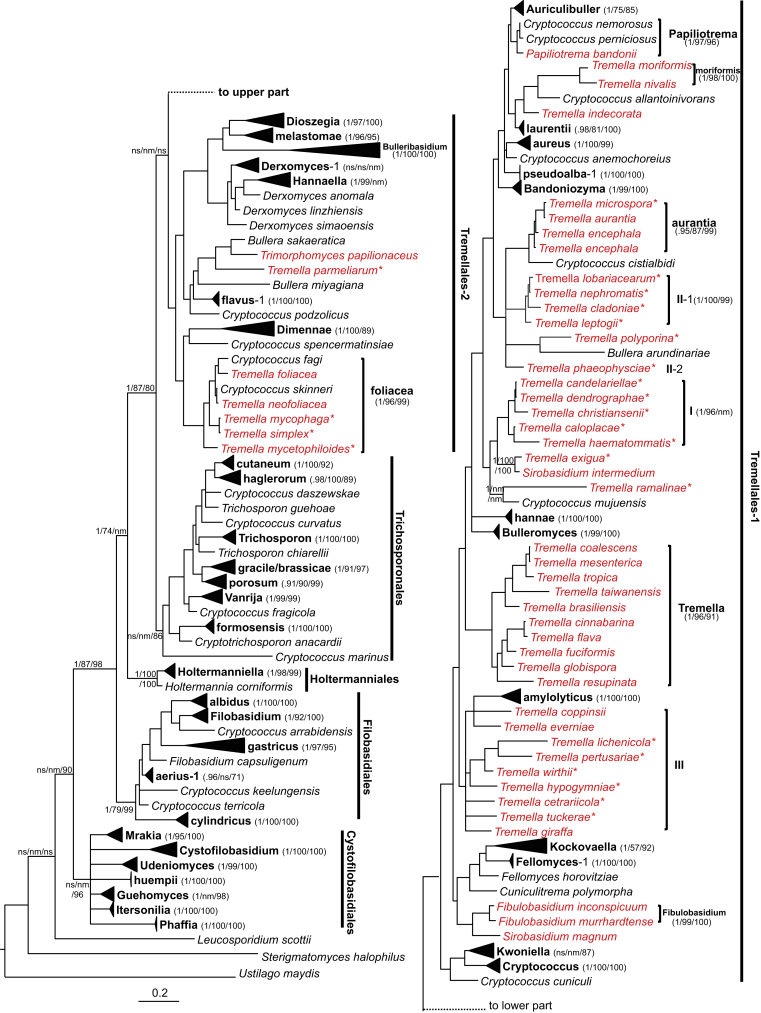
In the Bayesian tree constructed from the seven-gene data set, five lineages corresponding to the orders Tremellales, Trichosporonales, Filobasidiales and Cystofilobasidiales recognised by Boekhout et al. (2011) and the order Holtermanniales proposed by Wuczkowski et al. (2011) were resolved (Table 4, Fig. 1). The posterior probability for each of the Trichosporonales, Holtermanniales, Filobasidiales and Cystofilobasidiales lineages was 1.0. The support for the Tremellales was weak (PP = 0.51) when the basal foliacea clade of the lineage formed by Cryptococcus fagi, C. skinneri, C. spencermartinsiae, and Tremella foliacea was included. However, when this clade was not included, the Tremellales taxa formed a well-supported lineage with a PP value of 1.0 (Fig. 1).
Fig. 1.
An outline of the phylogeny of tremellomycetous yeasts and dimorphic taxa inferred from a seven-gene data set including sequences of three rDNA genes, RPB1, RPB2, TEF1 and CYTB. The tree backbone is constructed using Bayesian analysis. Branch lengths are scaled in terms of expected numbers of nucleotide substitutions per site. The Bayesian posterior probabilities (PP) and bootstrap percentages (BP) of maximum likelihood and neighbour-joining analyses from 1 000 replicates are shown respectively from left to right on the deep and major branches resolved. Note: ns, not supported (PP < 0.9 or BP < 50 %); nm, not monophyletic.
The five lineages were also clearly recognised in the ML and NJ trees constructed from the seven-gene data set (Table 4, Fig. 1), though the statistic support values varied. The Cystofilobasidiales, Filobasidiales, and Trichosporonales lineages received 100 % bootstrap supports, while the Holtermanniales received a moderate bootstrap support (56 %) in the NJ tree, but a strong support (100 % BP) in the ML tree. In the ML tree, the foliacea clade was located basal to the Trichosporonales lineage. In the NJ tree, the foliacea clade was located basal to the Trichosporonales and Tremellales lineages, but the bootstrap support for this topology was weak (Fig. 1).
Cystofilobasidiales
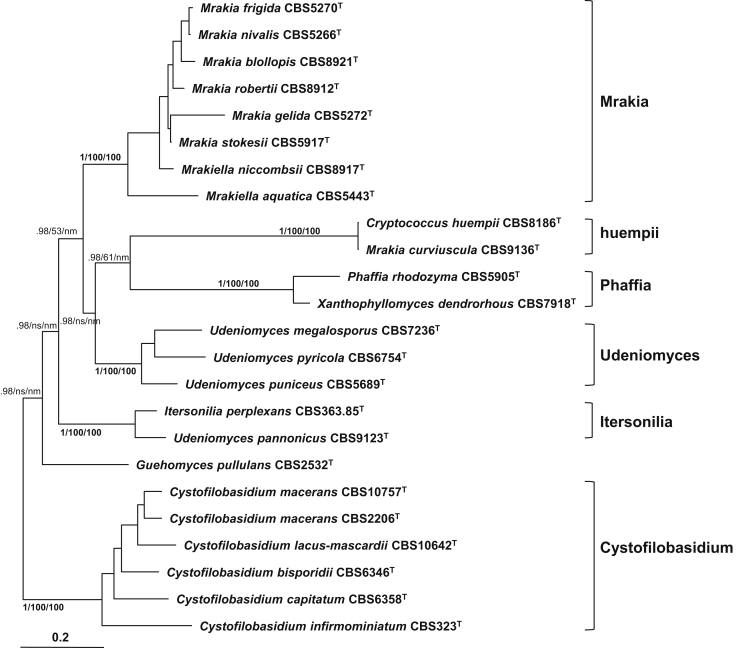
The species of the Cystofilobasidiales clustered into seven well-supported clades in the Bayesian tree drawn from the seven-gene data set (Table 4, Fig. 2), being in agreement with Boekhout et al. (2011). Each of the clades was strongly supported with a posterior probability of 1.0. The Mrakia clade contained all the Mrakia and Mrakiella species, except Mrakia curviuscula that formed a separated clade together with Cryptococcus huempii. Three of the four currently recognised Udeniomyces species formed the Udeniomyces clade, while the other species of the genus, U. pannonicus, formed a clade together with Itersonilia perplexans. All six recognised Cystofilobasidium species clustered together in a single clade. Phaffia rhodozyma CBS 5905T and its proposed teleomorph, Xanthophyllomyces dendrorhous CBS 7918T, formed a well separated clade, but the type strains of the two taxa differ clearly in protein gene sequences, suggesting that they may represent different species. Previous studies showed that Guehomyces pullulans CBS 2532T and Tausonia pamirica CBS 8428T clustered together (Boekhout et al., 2011, Fell and Guého-Kellermann, 2011, Sampaio, 2011b). Unfortunately, due to the unsuccessful amplification and sequencing of the protein genes of T. pamirica CBS 8428T, this species was not included in the seven-gene data set. However, in the tree drawn from the rDNA data set, CBS 2532T and CBS 8428T formed a well-supported clade (Fig. 3). The seven clades were also all recognised and well-supported with bootstrap value of 100 % in the trees drawn from the ML and NJ analyses (Table 4).
Fig. 2.
The phylogenetic relationships among species of the Cystofilobasidiales inferred from a seven-gene data set including sequences of three rDNA genes, RPB1, RPB2, TEF1 and CYTB. The tree backbone is constructed using Bayesian analysis. The Bayesian posterior probabilities (PP) and bootstrap percentages (BP) of maximum likelihood and neighbour-joining analyses from 1 000 replicates are shown respectively from left to right on the deep and major branches and clades resolved. Note: nm, not monophyletic; ns, not supported (PP < 0.9 or BP < 50 %).
Fig. 3.
Phylogeny of tremellomycetous yeasts and dimorphic taxa based on the rDNA data set containing ITS, D1/D2, and SSU rDNA sequences. The tree backbone is constructed using Bayesian analysis. The Bayesian posterior probabilities (PP) and bootstrap percentages (BP) of maximum likelihood and neighbour-joining analyses from 1 000 replicates are shown respectively from left to right on the deep and major branches and in brackets following the clades resolved. Notes: nm, not monophyletic; ns, not supported (PP < 0.9 or BP < 50 %).
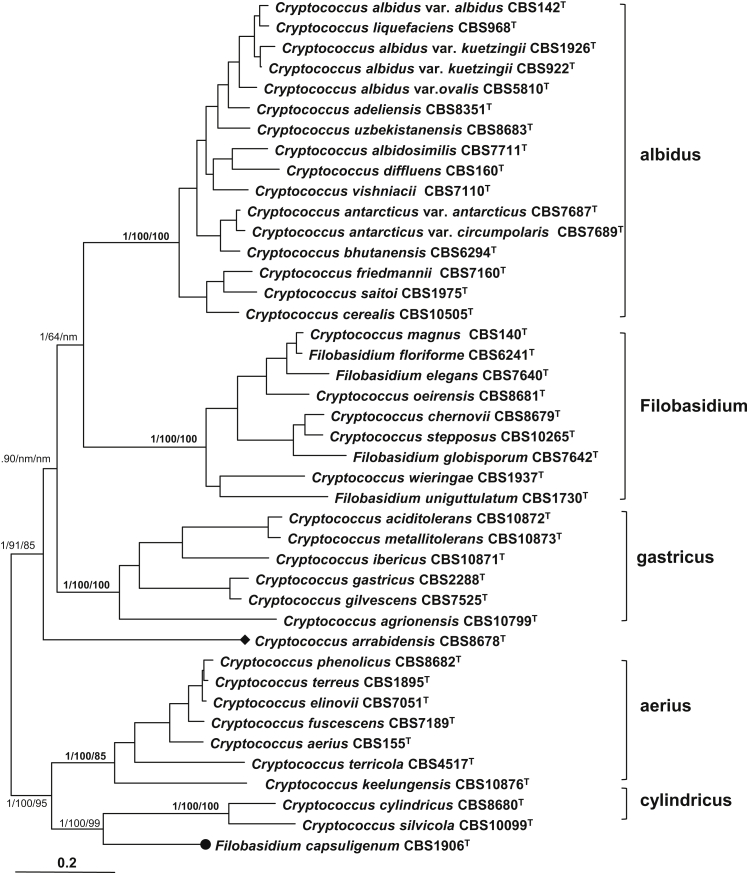
Filobasidiales
Bayesian analysis on the seven-gene data set recognised five strongly supported clades within the Filobasidiales, namely aerius, albidus, cylindricus, Filobasidium, and gastricus, being largely in agreement with Boekhout et al. (2011) (Fig. 4). The albidus clade containing 17 Cryptococcus species and varieties was clearly separated from the rest of the Filobasidiales. The Filobasidium clade contained four teleomorphic Filobasidium species including the type species of the genus, F. floriforme, and five Cryptococcus species. The affinity of F. uniguttulatum to this clade was not supported in Boekhout et al. (2011) and Weiß et al. (2014), but this study clearly showed that this species belongs to the Filobasidium clade with 1.0 posterior probability and 100 % bootstrap supports. This species was located in a basal branch of the Filobasidium clade together with C. wieringae (Fig. 4).
Fig. 4.
The phylogenetic relationships among species of the Filobasidiales inferred from a seven-gene data set including sequences of three rDNA genes, RPB1, RPB2, TEF1 and CYTB sequences. The tree backbone is constructed using Bayesian analysis. The Bayesian posterior probabilities (PP) and bootstrap percentages (BP) of maximum likelihood and neighbour-joining analyses from 1 000 replicates are shown respectively from left to right on the deep and major branches and clades resolved. The branches ending with a filled cycle and a diamond represent single-species clades with a stable and unstable position, respectively. Note: nm, not monophyletic.
The gastricus clade contained six Cryptococcus species, including three species isolated from acid rock drainage (ARD) from a pyrite mine in Portugal. The three Cryptococcus species were recognised as the ARD ecoclade (Gadanho & Sampaio 2009). This ecoclade was supported by Bayesian and ML analyses based on the seven-gene data set, but not by NJ analysis. Therefore, we included this ecoclade in the gastricus clade.
The aerius and cylindricus clades contained seven and two Cryptococcus species in the seven gene tree, respectively (Fig. 4). Analyses based on the rDNA data set showed that Bullera taiwanensis, whose protein gene sequences were not successfully determined, also clustered in the cylindricus clade with strong statistical supports (PP = 1.0) (Table 4, Fig. 3). The two clades together with Filobasidium capsuligenum, which represent a separate monotypic clade, formed a well-supported lineage (PP = 1.0; BP > 95 %). Cryptococcus arrabidensis was not included in any of the clades recognised in the Filobasidiales and remained as a separate branch in the trees constructed using different algorithms (Fig. 4).
Holtermaniales
Five anamorphic Holtermanniella species proposed by Wuczkowski et al. (2011) and the teleomorphic species Holtermannia corniformis were included in this small lineage (Fig. 5). This lineage was well separated from other groups of tremellomycetous yeasts and strongly supported in the seven-gene Bayesian and ML trees, though it was weakly supported in the NJ tree. Holtermannia corniformis was located as a basal branch in this lineage and its affinity with the Holtermanniella species was weakly supported by NJ analysis (Fig. 5), implying that this teleomorphic species may represent a distinct clade.
Fig. 5.
The phylogenetic relationships among species of the Trichosporonales and Holtermaniales inferred from a seven-gene data set including sequences of three rDNA gene, RPB1, RPB2, TEF1 and CYTB sequences. The tree backbone is constructed using Bayesian analysis. The Bayesian posterior probabilities (PP) and bootstrap percentages (BP) of maximum likelihood and neighbour-joining analyses from 1 000 replicates are shown respectively from left to right on the deep and major branches resolved. The branches ending with filled cycles and diamonds represent single-species clades with a stable and unstable position, respectively. Note: nm, not monophyletic; ns, not supported (PP < 0.9 or BP < 50 %).
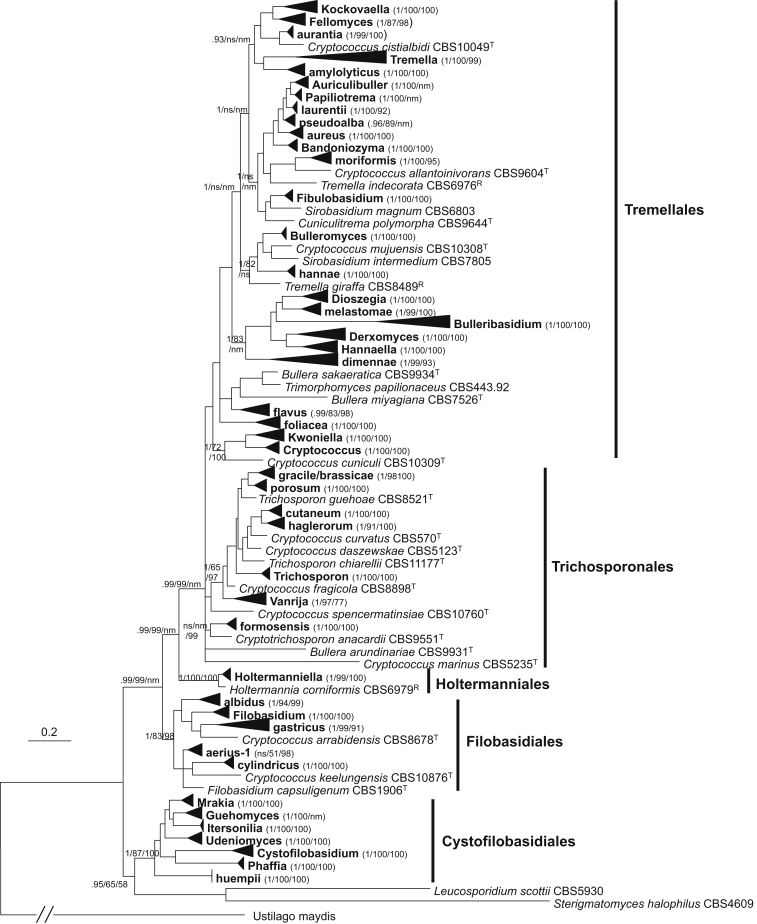
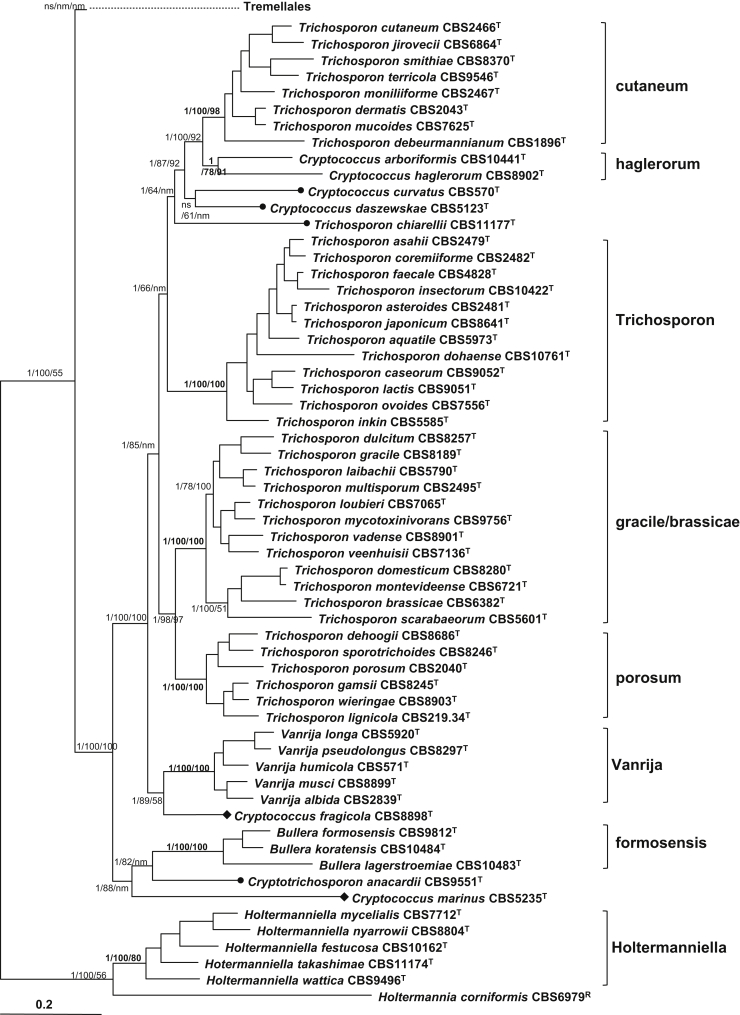
Trichosporonales
Seven well-supported clades with multiple species and seven single species clades were recognised in this order (Table 4, Fig. 5). The Trichosporon species were separated into four clades, namely cutaneum, gracile, porosum, and Trichosporon (also referred to as ovoides), supporting the classification of Middelhoven et al. (2004). The brassicae clade recognised in Sugita et al., 2004, Boekhout et al., 2011 and Sugita (2011) was also resolved in the Bayesian and ML trees based on the seven-gene data set, however, its separation from the gracile clade was only weakly supported in the seven-gene NJ tree. Furthermore, the distinction of the two clades was not supported in the Bayesian tree drawn from the rDNA data set. Therefore, we combined these two clades into a single clade. Among the Trichosporon species employed in this study, T. chiarellii could not be assigned to any clade. Trichosporon guehoae, whose protein gene sequences were not successfully amplified, was also located in a single branch in the tree drawn from the rDNA data set (Fig. 3).
In addition to the Trichosporon species, seven Cryptococcus, three Bullera and five Vanrija species and the monotypic genus Cryptotrichosporon (Okoli et al. 2007) were included in the Trichosporonales lineage (Table 1, Fig. 5). The genus Vanrija which was recently reinstalled by Weiß et al. (2014) for the five Cryptococcus species in the humicola clade recognised before (Boekhout et al., 2011, Fonseca et al., 2011) was confirmed to be a monophyletic group (Fig. 5). Two Cryptococcus species, C. arboriformis and C. haglerorum, formed the haglerorum clade which were resolved and well-supported in the seven-gene and the rDNA trees (Table 3, Fig. 3, Fig. 5). Other four Cryptococcus species, C. curvatus, C. daszewskae, C. fragicola, and C. marinus, occurred in single species branches. The three Bullera species formed a basal formosensis clade with strong statistical support (Table 4, Fig. 5). The thermotolerant species Cryptococcus tepidarius was located in this clade with a close relationship to B. lagerstroemiae based on rDNA sequence analysis (Fig. 3), being in agreement with Takashima et al. (2009). The protein gene sequences of C. tepidarius were not successfully determined. A close phylogenetic relationship of the formosensis clade with Cryptococcus marinus and Cryptotrichosporon anacardii was shown in the seven-gene Bayesian and ML trees, but the latter two species were located in separate clusters in the NJ tree (Fig. 5). In the trees drawn from the rDNA and single protein gene data sets, these two species did not cluster together, suggesting they represent different clades.
The affinity of Cryptococcus marinus within the Trichosporonales was strongly supported in the seven gene tree. It was located in a basal cluster of the order together with the formosensis clade and Cryptotrichosporon anacardii with strong support values from the Bayesian and ML analyses, but its phylogenetic position was not resolved by the NJ analysis (Fig. 5).
Tremellales
The majority of the taxa employed in this study belong to this lineage. Most of the clades recognised in Boekhout et al. (2011) were confirmed here with improved resolution and stronger support values. While most of the species can be assigned into clear clades, some remained undetermined and the boundaries of some clades need to be examined further.
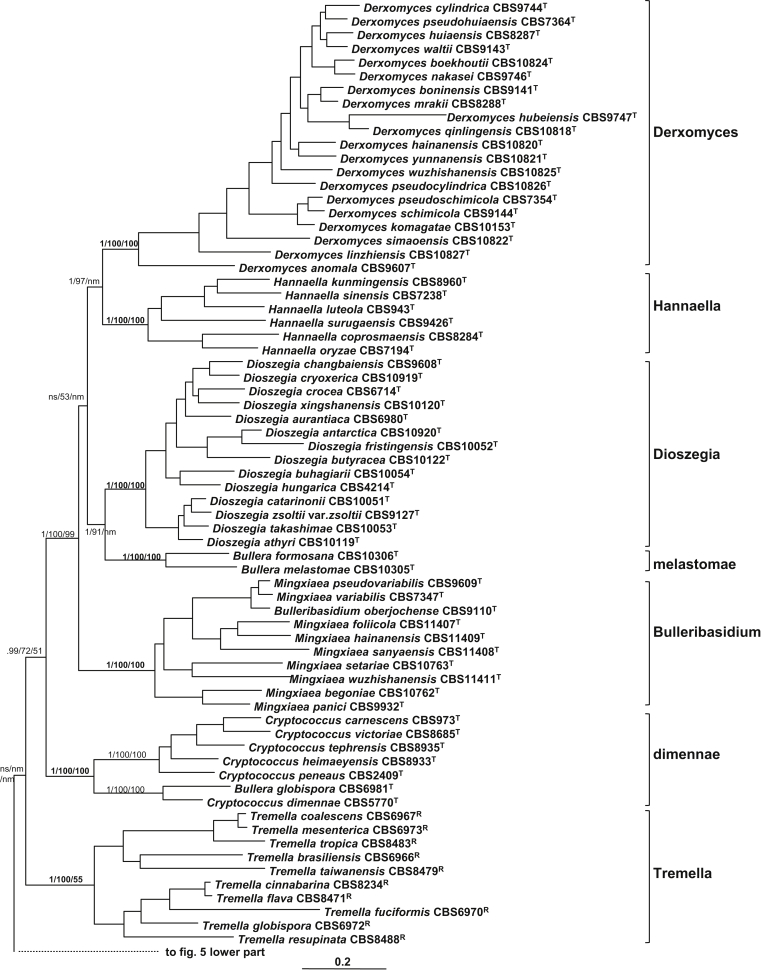
Twenty five well-supported clades were recognised among the 160 strains included in the Bayesian tree drawn from the seven-gene data set (Table 1, Table 4, Fig. 6). Five recently proposed or redefined genera based on molecular phylogenetic analyses were confirmed as monophyletic groups, including Bandoniozyma (Valente et al. 2012), Bulleribasidium/Mingxiaea (Sampaio et al., 2002, Wang et al., 2011), Derxomyces, Dioszegia, and Hannaella (Takashima et al., 2001, Wang and Bai, 2008). Each of these clades received a posterior probability value of 1.0 in the Bayesian tree and bootstrap values of 100 % in the ML and NJ trees drawn from the seven-gene data set, respectively (Table 4, Fig. 6). These clades were also clearly resolved in the analyses using the rDNA and single protein gene data sets (Table 4).
Fig. 6.
The phylogenetic relationships among species of the Tremellales inferred from a seven-gene data set including sequences of three rDNA genes, RPB1, RPB2, TEF1 and CYTB sequences. The tree backbone is constructed using Bayesian analysis. The Bayesian posterior probabilities (PP) and bootstrap percentages (BP) of maximum likelihood and neighbour-joining analyses from 1 000 replicates are shown respectively from left to right on the deep and major branches resolved. The branches ending with filled cycles and diamonds represent single-species clades with a stable and unstable position, respectively. Note: nm, not monophyletic; ns, not supported (PP < 0.9 or BP < 50 %).
In addition to the monotypic teleomorphic genus Cuniculitrema, the Cuniculitremaceae designated by Kirschner et al. (2001) contained Fellomyces and Kockovaella species. The species of the latter two anamorphic genera clustered into a well-supported cluster. However, two subclades represented by the type species of the two genera, F. polyborus and K. thailandica, respectively, could be recognised in the seven-gene Bayesian and ML trees (Fig. 6). The two subclades were also resolved in the NJ tree, with F. horovitziae being located as a basal branch to the two subclades. In the Bayesian and ML trees, this species was basal to the Fellomyces subclade with a PP and BP value of 1.0 and 64 %, respectively (Fig. 6).
The phylogenetic relationships among the species tentatively assigned to the Bulleromyces/Papiliotrema/Auriculibuller group by Boekhout et al. (2011) were resolved in this study (Fig. 6). The teleomorphic species Bulleromyces albus and three anamorphic Bullera species occurred in a distinct group with two clades being recognised, namely the Bulleromyces clade containing the anamorphic species Bullera unica, and the hannae clade formed by B. hannae and B. penniseticola. However, in the trees drawn from the rDNA data set, the close relationship of the two clades was not resolved (Fig. 3).
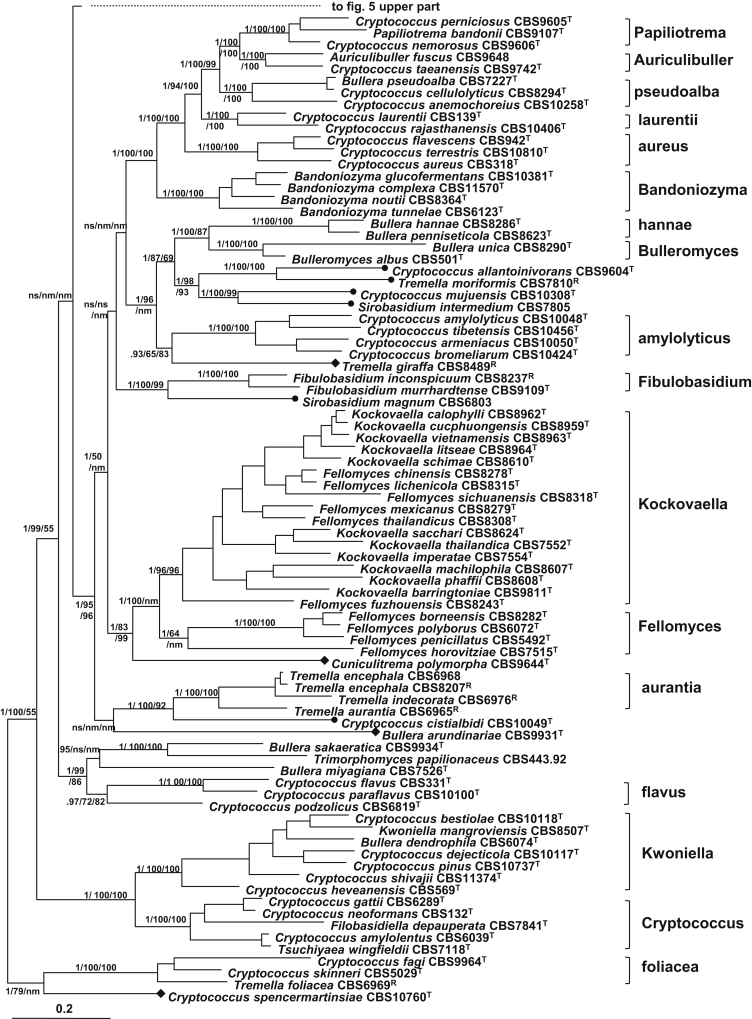
The monotypic teleomorphic genera Papiliotrema and Auriculibuller formed a well-supported group with one Bullera and 10 Cryptococcus species. This group showed a close relationship to the Bandoniozyma clade with strong support (Fig. 6). Five clades were recognised in this group (Table 4, Fig. 6). The Papiliotrema clade contained two other Cryptococcus species, namely C. nemorosus and C. perniciosus; C. taeanensis showed a close affinity to the Auriculibuller clade. The pseudoalba clade contained a Bullera species and two Cryptococcus species, C. anemochoreius and C. cellulolyticus. The laurentii and the aureus clades contained two and three Cryptococcus species, respectively.
Four recently described Cryptococcus species with orange coloured colonies (Inácio et al., 2005, Wang et al., 2007, Landell et al., 2009) clustered together in a well-supported amylolyticus clade. Two Bullera species described from Taiwan (Nakase et al. 2004), which were assigned to the Dioszegia clade in Boekhout et al. (2011), formed a distinct melastomae clade closely related with the Dioszegia clade. Other clearly supported clades consisting of species with only or mainly yeast forms were the Cryptococcus, dimennae, and Kwoniella clades. The dimennae clade, which was also resolved by Boekhout et al. (2011) but was referred to as the victoriae clade by Fonseca et al. (2011), consisted of six Cryptococcus species and one Bullera species (B. globispora). In addition to the teleomorphic species Kwoniella mangroviensis, five Cryptococcus and one Bullera species were included in the Kwoniella clade. The opportunistically pathogenic species in the Cryptococcus neoformans complex and their teleomorphs were included in the Cryptococcus clade together with Filobasidiella depauperata, C. amylolentus and Tsuchiyaea wingfieldii.
The Tremella species employed in the present study separated into different clades. Ten of them, including the type species of the genus, T. mesenterica, clustered in the Tremella clade. No species with mainly yeast forms in their life cycle were located in this clade. Three Tremella species formed the aurantia clade. Tremella moriformis was located in a group containing two Cryptococcus species (C. allantoinivorans and C. mujuensis) and Sirobasidium intermedium, a teleomorphic species. This group, which was tentatively included in the Bulleromyces/Papiliotrema/Auriculibuller group in Boekhout et al. (2011), was also resolved as a separate group in the ML and NJ trees with 93–98 % bootstrap supports (Fig. 6). However, C. mujuensis and S. intermedium were separated from the other species of this group in the tree drawn from the rDNA data set (Fig. 3, Fig. 7). With the consideration that the four species in this group exhibit quite different morphological characters from each other, they were regarded as representing four separate single species clades. Tremella nivalis and T. moriformis formed the moriformis clade with 1.0 PP and with over 95 % bootstrap support in the tree drawn from the rDNA sequence data set (Fig. 3). Another Sirobasidium species employed in this study, S. magnum, was located in a branch basal to the Fibulobasidium clade (Fig. 3, Fig. 6). Tremella foliacea and two Cryptococcus species (C. fagi and C. skinneri) clustered in the foliacea clade, which was located at the basal position of the Tremellales lineage in the seven-gene Bayesian tree (Fig. 5). The rDNA tree showed that T. neofoliacea was also located in this clade (Fig. 3).
Fig. 7.
Phylogeny of tremellomycetous yeasts and dimorphic taxa based on 5.8S and LSU D1/D2 rDNA sequences from strains employed in this study and 26 more Tremella species employed in Millanes et al. (2011). The tree backbone is constructed using Bayesian analysis. The Bayesian posterior probabilities (PP) and bootstrap percentages (BP) of maximum likelihood and neighbour-joining analyses from 1 000 replicates are shown respectively from left to right on the deep and major branches and in brackets following the clades resolved. The species names in red represent fruiting-body forming taxa and those with a star superscript indicate that the sequences are from herbarium specimens of lichen-inhabiting species. Note: nm, not monophyletic; ns, not supported (PP < 0.9 or BP < 50 %).
Another group containing both yeast and filamentous taxa is the Trimorphomyces group. Two Bullera species and three Cryptococcus species were located in this group together with Trimorphomyces papilionaceus, a basidiocarp-forming species with a yeast state (Fig. 6). T. papilionaceus was regarded as representing a distinct clade because of its unique sexual reproductive structures (Bandoni and Boekhout, 2011, Boekhout et al., 2011). The three Cryptococcus species, C. flavus, C. paraflavus and C. podzolicus, were assigned to the flavus clade since they clustered together in the seven-gene and rDNA Bayesian trees with 0.97–0.99 PP supports (Fig. 3, Fig. 6). The two Bullera species in this group, B. sakaeratica and B. miyagiana, was separated by T. papilionaceus in the seven-gene and the rDNA trees (Fig. 3, Fig. 6). Therefore, they were regarded as representing two different single species clades.
The following species in the Tremellales lineage, Bullera arundinariae, Cryptococcus cistialbidi, Cryptococcus spencermatinsiae, Cuniculitrema polymorpha, and Tremella giraffe, could not be assigned to any recognised clade or group, because of their unstable or unresolved phylogenetic positions, or their unique phenotypic characters. Bullera arundinariae and C. cistialbidi were located as basal branches to the aurantia clade formed by four Tremella species in the seven-gene Bayesian tree (Fig. 6). While the close relationship of C. cistialbidi to the aurantia clade was consistent in different trees, B. arundinariae was located in different positions in the seven-gene NJ tree and the trees resulting from the rDNA data set (Fig. 3, Fig. 6). Cryptococcus spencermartinsiae was located in a branch basal to the foliacea clade with strong statistical support in the Bayesian and ML trees drawn from the seven-gene data set, but the species was located at a different position in the seven-gene NJ tree and the trees drawn from the rDNA data set (Fig. 3, Fig. 6). The teleomorphic species Cuniculitrema polymorpha (anamorph: Sterigmatosporidium polymorphum) was located in a branch basal to the Fellomyces/Kockovaella group. Tremella giraffa was located as a basal branch to the amylolyticus clade in the seven-gene tree with weak to moderate support values (Fig. 6), but its position was not resolved in the rDNA tree (Fig. 3).
In order to investigate further the relationships of yeasts with filamentous taxa in the Tremellomyetes, we retrieved the 5.8S and LSU rDNA sequences of 26 lichen-inhabiting Tremella species employed in Millanes et al. (2011) that were absent in the current data set. These sequences were determined from herbarium specimens (Millanes et al. 2011). The Bayesian tree obtained from the combined 5.8S and LSU D1/D2 rDNA sequence data set showed a largely identical topology with that obtained from the seven-gene data set and the five major lineages were also clearly resolved (Fig. 7). The majority of the additional 26 Tremella species were located in clades I, II and III as defined by Millanes et al. (2011) which mainly contained lichen-inhibiting Tremella species; one in the aurantia clade containing Tremella taxa only; three in the foliacea clade containing both Tremella and Cryptococcus species; and one in the Trimorphomyces group (Fig. 7).
Discussion
In this study, we inferred the phylogeny of basidiomycetous yeasts and related dimorphic and filamentous basidiomycetes in the Tremellomycetes based on analyses of seven gene sequences using different phylogenetic algorithms. The majority of the yeast taxa and dimorphic basidiomycetes that have free-living unicellular states in their life cycles in the Agaricomycotina were employed. Five major lineages corresponding to the five orders currently recognised in the Tremellomycetes (Boekhout et al., 2011, Millanes et al., 2011, Weiß et al., 2014) were resolved. A total of 45 strongly supported monophyletic clades with multiple species and 23 single species clades were recognised. This phylogenetic framework will be the basis for an improved modern taxonomy unifying both yeast-like and filamentous species in the Tremellomycetes as well as anamorphs and teleomorphs occurring in this class. The result is also helpful for a better understanding of the evolution of characters and different life styles by integrating the phylogeny with biochemical, morphological and reproductive characteristics of unicellular, dimorphic and filamentous basidiomycetes in the Tremellomycetes.
Congruence of phylogenies inferred from analyses using different algorithms and data sets
Almost all currently recognised teleomorphic and anamorphic yeast species and dimorphic taxa in the Agaricomycotina were obtained from culture collections and revived for DNA isolation and PCR amplification in this study. Despite our best effort to obtain a complete sequence data set for all the genes and strains employed, the sequence of some genes, especially the nuclear protein-coding genes and the mitochondrial gene CYTB, could not be determined for a small percentage of strains because of failure in the PCR amplification or sequencing reactions. Specifically, 8.8 %, 8.1 %, 16.2 % and 17.2 % of the total 297 strains employed failed in the sequence determination of the RPB1, RPB2, TEF1 and CYTB genes, respectively. This problem is known from all groups of fungi (Schoch et al. 2012). A previous study has shown that an inferred phylogeny is not sensitive to 25 % or even 50 % missing data for a sufficiently large alignments (e.g., ∼30 000 positions and 36 species) (Philippe et al. 2004). Though the length of the seven-gene alignment in this study is only about 6 300 positions, the amount of missing data is also much less. Thus, we assume that the relative minor amount of missing data in our study will not significantly influence the reliability of the resulting phylogeny.
The phylogenies of the taxa compared in this study were inferred from analyses using different data sets and algorithms. The topologies of the trees constructed using different algorithms performed on different data sets were largely congruent as examined visually, which make the delimitation of major lineages and clades more clear and confident. In addition to the Clustal X, we also used the MAFFT program (Katoh & Standley 2013) to align the sequences and the alignments generated were subjected to ML analysis. The topologies of the trees obtained from the Clustal X and the MAFFT alignments were almost the same (data not shown). This further supports the notion that our inferred trees are reliable and not greatly influenced by the missing data as discussed above.
Bayesian analysis is usually believed to be more reliable compared to parsimony and neighbour-joining methods, especially for an extensive sampling with a high divergence occurring among the sequences (Alfaro et al., 2003, Holder and Lewis, 2003, James et al., 2006). As expected, the Bayesian analysis of the seven-gene data set showed the most robust phylogeny among the analyses performed (Table 4). However, analyses aiming at comparing Bayesian and ML supports have revealed that PP and BP values show significant correlation, but the strength of this correlation is highly variable and sometimes very low. ML BP values are generally lower than PP values, and thus, ML BP might be less prone to strongly supporting a wrong phylogenetic hypothesis (Douady et al. 2003). Therefore, the boundaries of the lineages and clades recognised in this study were determined based not only on Bayesian analysis, but also on ML and NJ analyses, aiming to recognize reliable monophyletic groups.
Conflicts between phylogenies obtained from rDNA and protein-coding gene sequences have been observed in different studies on basidiomycetes (Matheny et al., 2002, Froslev et al., 2005, Matheny, 2005, Matheny et al., 2006, Matheny et al., 2007). However, in this study, the topologies of the trees and the clades resolved from the data sets of RPB1 and RPB2 were similar to those obtained from the rDNA data set (Table 4), except for the position of the Trichosporonales which was nested into the Tremellales in the RPB1-based phylogeny. Furthermore, RPB1 and RPB2 had an equivalent resolution power in the Cystofilobasidiales and Filobasidiales lineages. The Holtermanniales lineage was supported strongly (100 % BP) by the ML algorithm in the RPB2-based phylogeny but only received moderate support (62 % BP) in the RPB1-based phylogeny. The RPB1 and RPB2-based phylogenies constructed from Bayesian analysis supported the same number of clades in the Tremellales, while the RPB1-based phylogeny constructed from ML or NJ analyses resolved one more clade if compared to the RPB2-based phylogeny. The RPB1 and RPB2-based phylogenies drawn from Bayesian and ML analyses also resolved the same number of clades in the Trichosporonales. The TEF1 and CYTB sequences showed less parsimony-informative characters for the inference of phylogenetic relationship in the tremellomycetous yeasts compared to the RPB1 and RPB2 sequences. The TEF1 and CYTB data sets generated the lowest resolution across the Bayesian, ML and NJ trees, in which only 19 and 16 strongly supported clades were resolved with high BP and PP values, respectively (Table 4). The TEF1 and CYTB data sets were unable to resolve higher level taxonomic relationships, such as the five orders in the Tremellomycetes, and they did not show strong support to some clades, such as the Cystofilobasidium, Derxomyces, foliacea, Hannaella, Kwoniella, and Trichosporon clades, which were strongly supported by the analyses based on the other data sets. Our results suggest that RPB1 and RPB2 are more useful to infer reliable phylogeny of tremellomycetous yeasts than the TEF1 and CYTB genes. A previous study of basidiomycetes phylogeny also showed that the major clades at higher and lower taxonomic levels were more clearly resolved based on RPB2 than on TEF1 sequence data (Matheny et al. 2007).
More robust topologies and higher resolution were achieved in this study than those obtained in previous studies based on the LSU rDNA D1/D2 domains or ITS-5.8S sequences (Fell et al., 2000, Scorzetti et al., 2002, Boekhout et al., 2011). The consensus is that the major groups recognised in the previous studies were confirmed in the present study. Fell et al. (2000) studied 171 hymenomycetous yeast strains representing 116 species. They recognised four major lineages including the Cystofilobasidiales, Filobasidiales, Tremellales and Trichosporonales. However, the clades within each lineage were largely unresolved. In addition to the four major lineages, Scorzetti et al. (2002) recognised clades within each lineage. Most of the clades recognised in the Cystofilobasidiales, Filobasidiales and Trichosporonales were in agreement to those recognised in this study. However, the fine phylogenetic relationships among the taxa in the Tremellales remained largely unresolved in the previous studies. Boekhout et al. (2011) employed more strains and designated a fifth lineage containing the Holtermanniella clade and a teleomorphic species Holtermannia corniformis that was described as a separate order (Wuczkowski et al. 2011). Our study confirmed this fifth lineage as a separate order Holtermanniales with 1.0 PP and 100 % ML BP supports. The phylogenetic position of Cryptococcus marinus has been debated. It was considered to belong to the Tremellales according to a phylogenetic analysis of SSU rDNA sequences (Takashima & Nakase 1999). The phylogenetic position in the LSU rDNA D1/D2 tree suggested that this species may represent a separate order within the Tremellomycetes (Scorzetti et al., 2002, Fonseca et al., 2011, Weiß et al., 2014). However, the affinity of this species with the Trichosporonales lineage was strongly supported in this study (Fig. 5).
The major lineages and clades recognised in this study are similar to those recognised in Millanes et al. (2011) and Weiß et al. (2014), which sampled more teleomorphic and filamentous taxa in the Tremellomycetes. In their molecular phylogenetic study on the jelly fungi based on nuclear SSU, 5.8S and LSU rDNA sequences, Millanes et al. (2011) employed three more teleomorphic genera Biatoropsis, Syzygospora and Tetragoniomyces, but limited yeast taxa. In addition to the teleomorphic genera employed in Millanes et al., 2011, Weiß et al., 2014 listed seven other teleomorphic genera that were not employed in our study, including Carcinomyces, Rhynchogastrema, Phyllogloea, Phragmoxenidium, Sigmogloea, Sirotrema, and Xenolachne in the Tremellomycetes. However, the latter five genera were not included in their phylogenetic analysis based on LSU D1/D2 sequences, because no DNA data were available from these genera. In the trees presented in Millanes et al. (2011) and Weiß et al. (2014), the species of the teleomorphic and filamentous genera that were not included in this study were located in separated clades from those formed by yeast taxa.
Correlation between morphology, physiology and molecular phylogeny
Because of the morphological simplicity, it is not easy to find morphological characters that distinguish the five major lineages of tremellomycetous yeasts recognised by molecular phylogenetic analyses. Teleomorphic taxa belonging to the Tremellales usually form tremella-type basidia, e.g., phragmobasidia with longitudinal primary septa; whereas those of the Cystofilobasidiales and Filobasidiales are usually characterised by forming holobasidia (Wells and Bandoni, 2001, Boekhout et al., 2011). However, some species with holobasidia or transversely septate basidia, like Auriculibuller fuscus (Sampaio et al. 2004), Papiliotrema bandonii (Sampaio et al. 2002), Tremella fuciformis, T. hypogymniae (Millanes et al. 2011) and Bulleribasidium oberjochense (Sampaio et al. 2002) are also present in Tremellales. These observations show that different types of basidial septation can coexist in the same lineage. The sexual stage of the Trichosporonales species has not yet been observed. The majority of the species in this order are characterised by forming abundant true hyphae that disarticulate into arthroconidia. However, the filamentous species Tetragoniomyces uliginosus which was tentatively assigned to the Trichosporonales in Millanes et al. (2011) and Weiß et al. (2014) forms basidia in pustulate basidiocarps (Oberwinkler & Bandoni 1981).
The species in the genera Fellomyces and Kockovaella share a special morphological character of forming conidia on stalks (Nakase et al. 1991). These species were located together in a cluster with strong PP and ML BP supports (Fig. 6). The affinity of F. horovitziae to the Fellomyces clade was weakly supported in ML analysis and not supported in NJ analysis. We tentatively assign F. horovitziae to the Fellomyces clade with the consideration of minimising name changes in the subsequent taxonomic treatment.
The ability to form ballistoconidia has since long been shown to be an unreliable phylogenetic marker (Nakase et al. 1993). This observation is confirmed by the intermixture of species of the ballistoconidia-forming genera Bullera and Kockovaella with those of non ballistoconidia-forming genera Cryptococcus and Fellomyces. However, the morphology of ballistoconidia seems to be phylogenetically relevant. Ballistoconidia formed by species in the Cystofilobasidiales and Trichosporonales are usually bilaterally symmetrical, whereas those formed by species in the Tremellales and Filobasidiales are usually rotationally symmetrical (Boekhout et al. 2011).
Within the Tremellales, some clades may be distinguished by colony morphology. For example, the anamorphic genera Derxomyces, Hannaella and Dioszegia are closely related, but are distinguishable by forming whitish to yellowish colonies with a butyrous texture, whitish colonies with a highly mucoid texture, and orange-coloured colonies with a butyrous texture, respectively (Wang & Bai 2008). The two Bullera species in the melastomae clade were assigned to the Dioszegia clade by Boekhout et al. (2011). However, they are morphologically different by forming yellowish to brownish colonies compared to the orange-coloured colonies of Dioszegia species (Takashima et al., 2001, Wang and Bai, 2008).
The physiological and biochemical differences among the major lineages are also quite elusive, though some trends have been observed (Sampaio and Fonseca, 1995, Sampaio, 2004). The majority of the Cystofilobasidiales and Filobasidiales species can utilise nitrate; whereas the Tremellales and Trichosporonales taxa are usually nitrate negative. The coenzyme Q (CoQ) system has been used as an important taxonomic criterion at the genus level in yeasts (Yamada & Kondo 1973). The major CoQ systems of the tremellomycetous yeasts are CoQ-8, CoQ-9 and CoQ-10 (Fell, 2011, Fell and Guého-Kellermann, 2011, Sampaio, 2011a, Sampaio, 2011b). The taxa with CoQ-8 are concentrated in the Cystofilobasidiales. The species within a strongly supported clade usually possess the same major CoQ type, which may be helpful to recognize and define homogenous clades. The species with the ability to ferment sugars, a rare trait among basidiomycetous yeasts, are concentrated in a few clades in the Cystofilobasidiales (Mrakia and Phaffia/Xanthophyllomyces) and Tremellales (Bandoniozyma). One species in the Filobasidiales, Filobasidium capsuligenum, can also ferment glucose and maltose, while the other known Filobasidium species can not ferment glucose (Kwon-Chung 2011). F. capsuligenum was separated from the Filobasidium clade and located in a branch closely related with the cylindricus clade containing two Cryptococcus species with strong PP and BP supports (Fig. 4). Ultrastructurally, F. capsuligenum is also special by having cone-shaped vesicular parenthesomes (Moore & Kreger-van Rij 1972). Thus, we recognised this species as representing a distinct clade. Consequently, the cylindricus clade and the closely related aerius clade were recognised as separate clades.
Serological characteristics of Trichosporon species correspond to some extent with their phylogenetic clustering. Species in the cutaneum, Trichosporon and brassicae clades have serotypes I, II and III, respectively, while species in the gracile and porosum clades have serotype I-III, which is a serotype that reacts to both antisera I and III (Ikeda et al., 1996, Sugita and Nakase, 1998, Sugita et al., 2004, Sugita, 2011). However, the phylogenetic separation between the brassicae and gracile clades, which have different serotypes (III and I-III, respectively) was not supported in this study. The gracile and brassicae clades were recognised as separate clades based on D1/D2 rDNA sequence analyses and serological characteristics (Sugita et al., 2004, Boekhout et al., 2011, Sugita, 2011). However, both clades lacked bootstrap supports in the NJ trees drawn from D1/D2 sequences (Boekhout et al., 2011, Sugita, 2011). In this study, the monophyly of the gracile clades was not resolved and supported in the Bayesian tree drawn from the rDNA data set. Therefore, we combined the gracile and brassicae clades.
Life strategy evolution in Tremellomycetes
The multiple gene phylogeny of tremellomycetous yeasts is helpful for a better understanding on the evolution of different life styles and strategies. The tremellomycetous fungi present a high diversity of lifestyles, with many species being dimorphic, including both unicellular and filamentous growth forms (Bandoni, 1995, Sampaio, 2004, Boekhout et al., 2011). They are also nutritionally heterogeneous, comprising saprotrophs, animal parasites, and fungal-inhabiting (including lichen-inhabiting) species (Millanes et al., 2011, Weiß et al., 2014). A previous study on phylogeny and character evolution in tremellomycetous fungi based on three rDNA markers (nSSU, 5.8S and nLSU) showed that, in a broad sense, a specific life style or strategy is usually homoplastic; however, taxa with the same life strategy, for example, fungal- or lichen-inhabiting, usually form distinct clades (e.g., clades I, II and III in Millanes et al. (2011)). The results of this study also show that taxa with different life styles (e.g., dominated by unicellular and filamentous growth stages, respectively) usually form different clades, though clades with species having the same life styles may not be closely related.
This observation is also shown by the analysis based on an integrated 5.8S and LS D1/D2 sequence data set containing additional Tremella species as employed in Millanes et al. (2011). Though fruiting-body forming species were intermingled with yeast species throughout the Tremellales (Fig. 7), the former usually clustered into different groups from the latter. A few fruiting-body forming species, e.g., Papiliotrema bandonii, Tremella parmeliarum, T. polyporina, T. ramalinae, T. foliacea, and Trimorphomyces papilionaceus, were located in the same clusters together with some yeast taxa, but they usually formed distinct branches or clades. These results suggest that tremellomycetous fungi with the same life styles or nutritional strategies may be the result of convergent evolution as a result of early adaptation to different ecological niches or habitats.
Taxonomic consequences
As with many other groups of fungi, the taxonomic system of basidiomycetous yeasts needs to be updated to reflect the evolutionary relationships of the taxa concerned and to accommodate the requirements of the new nomenclatural code (McNeill et al. 2012). Based on the results of this study, we will propose an updated taxonomic system for tremellomycetous yeasts which will have the best approximation of the molecular phylogeny and that will be compatible with the current taxonomic system of filamentous basidiomycetes. A considerable number of genera need to be redefined to include only the species in the monophyletic clades that contain the type species of those genera, and, secondly, many new genera need to be proposed to accommodate monophyletic clades that do not include any generic type species. The names of many species will be changed due to the proposal of new genera and adaptation of the ‘one fungus = one name’ principle at this stage. We believe that this updated taxonomic system based on a reliable phylogeny and extensive phenotypical comparisons will be relatively stable and minimise the necessity of future name changes.
Acknowledgments
We thank Wendy Epping, Diana Vos and Hanslin Stasia from the CBS collection for retrieving the vast number of strains used during this study from the CBS collection. This study was supported by grants No. 31010103902, No. 30700001 and No. 30970013 from the National Natural Science Foundation of China (NSFC), KSCX2-YW-Z-0936 from the Knowledge Innovation Program of the Chinese Academy of Sciences and grant No. 10CDP019 from the Royal Netherlands Academy of Arts and Sciences (KNAW). TB is supported by grant NPRP 5-298-3-086 of Qatar Foundation. The authors are solely responsible for the content of this manuscript.
Footnotes
Peer review under responsibility of CBS-KNAW Fungal Biodiversity Centre.
Contributor Information
F.-Y. Bai, Email: baify@im.ac.cn.
T. Boekhout, Email: t.boekhout@cbs.knaw.nl.
References
- Alfaro M.E., Zoller S., Lutzoni F. Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Molecular Biology and Evolution. 2003;20:255–266. doi: 10.1093/molbev/msg028. [DOI] [PubMed] [Google Scholar]
- Babenko V.N., Rogozin L.B., Mekhedov S.L. Prevalence of intron gain over intron loss in the evolution of paralogous gene families. Nucleic Acids Research. 2004;32:3724–3733. doi: 10.1093/nar/gkh686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandoni R.J. Dimorphic heterobasidiomycetes, taxonomy and parasitism. Studies in Mycology. 1995;38:13–27. [Google Scholar]
- Bandoni R.J., Boekhout T. Trimorphomyces Bandoni & Oberwinkler. In: Kurtzman C.P., Fell J.W., editors. The yeasts: a taxonomic study. Elsevier; London: 2011. pp. 1591–1594. [Google Scholar]
- Biswas S.K., Wang L., Yokoyama K. Molecular analysis of Cryptococcus neoformans mitochondrial cytochrome b gene sequences. Journal of Clinical Microbiology. 2003;41:5572–5576. doi: 10.1128/JCM.41.12.5572-5576.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boekhout T. Gut feeling for yeasts. Nature. 2005;434:449–450. doi: 10.1038/434449a. [DOI] [PubMed] [Google Scholar]
- Boekhout T., Fonseca A., Sampaio J.P. Discussion of teleomorphic and anamorphic basidiomycetous yeasts. In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; London: 2011. pp. 1339–1372. [Google Scholar]
- Boekhout T., Nakase T. Bullera Derx. In: Kurtzman C.P., Fell J.W., editors. The yeasts: a taxonomic study. Elsevier; London: 1998. pp. 1623–1659. [Google Scholar]
- Boekhout T., Theelen B., Houbraken J. Novel anamorphic mite-associated fungi belonging to the Ustilaginomycetes: Meira geulakonigii gen. nov., sp. nov., Meira argovae sp. nov. and Acaromyces ingoldii gen. nov., sp. nov. International Journal of Systematic and Evolutionary Microbiology. 2003;53:1655–1664. doi: 10.1099/ijs.0.02434-0. [DOI] [PubMed] [Google Scholar]
- Bolano A., Stinchi S., Preziosi R. Rapid methods to extract DNA and RNA from Cryptococcus neoformans. FEMS Yeast Research. 2001;1:221–224. doi: 10.1111/j.1567-1364.2001.tb00037.x. [DOI] [PubMed] [Google Scholar]
- Douady C.J., Delsuc F., Boucher Y. Comparison of Bayesian and maximum likelihood bootstrap measures of phylogenetic reliability. Molecular Biology and Evolution. 2003;20:248–254. doi: 10.1093/molbev/msg042. [DOI] [PubMed] [Google Scholar]
- Fell J.W. Mrakia Y. Yamada & Komagata (1987) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; London: 2011. pp. 1503–1510. [Google Scholar]
- Fell J.W., Boekhout T., Fonseca A. Biodiversity and systematic of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 sequence analysis. International Journal of Systematic and Evolutionary Microbiology. 2000;50:1351–1371. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- Fell J.W., Guého-Kellermann E. Guehomyces Fell & Scorzetti (2004) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; London: 2011. pp. 1773–1775. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Fonseca Á., Boekhout T., Fell J.W. Cryptococcus Vuillemin (1901) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; London: 2011. pp. 1661–1737. [Google Scholar]
- Froslev T.G., Matheny P.B., Hibbett D.S. Lower level relationships in the mushroom genus Cortinarius (Basidiomycota, Agaricales): a comparison of RPB1, RPB2, and ITS phylogenies. Molecular Phylogenetics and Evolution. 2005;37:602–618. doi: 10.1016/j.ympev.2005.06.016. [DOI] [PubMed] [Google Scholar]
- Fungsin B., Takashima M., Sugita T. Bullera koratensis sp. nov. and Bullera lagerstroemiae sp. nov., two new ballistoconidium-forming yeast species in the Trichosporonales-clade isolated from plant leaves in Thailand. The Journal of General and Applied Microbiology. 2006;52:73–81. doi: 10.2323/jgam.52.73. [DOI] [PubMed] [Google Scholar]
- Gadanho M., Sampaio J.P. Cryptococcus ibericus sp. nov., Cryptococcus aciditolerans sp. nov. and Cryptococcus metallitolerans sp.nov., a new ecoclade of anamorphic basidiomycetous yeast species from an extreme environment associated with acid rock drainage in Sao Domingos pyrite mine, Portugal. International Journal of Systematic and Evolutionary Microbiology. 2009;59:2375–2379. doi: 10.1099/ijs.0.008920-0. [DOI] [PubMed] [Google Scholar]
- Gargas A., Taylor J.W. Polymerase chain reaction (PCR) primers for amplifying and sequencing nuclear 18S rDNA from lichenized fungi. Mycologia. 1992;84:589–592. [Google Scholar]
- Hawksworth D.L. A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. MycoKeys. 2011;1:7–20. doi: 10.5598/imafungus.2011.02.02.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks L., Goris A., Neefs J.M. The nucleotide sequence of the small ribosomal subunit RNA of the yeast Candida albicans and the evolutionary position of the fungi amongst the Eukaryotes. Systematic and Applied Microbiology. 1989;12:223–229. [Google Scholar]
- Hibbett D.A., Binder M., Bischoff J.F. A high-level phylogenetic classification of the fungi. Mycological Research. 2007;3:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Hibbett D.S. A phylogenetic overview of the Agaricomycotina. Mycologia. 2006;98:917–925. doi: 10.3852/mycologia.98.6.917. [DOI] [PubMed] [Google Scholar]
- Hillis D.M., Bull J.J. An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analyses. Systematic Biology. 1993;42:182–192. [Google Scholar]
- Holder M., Lewis P.O. Phylogeny estimation: traditional and Bayesian approaches. Nature Reviews. 2003;4:275–284. doi: 10.1038/nrg1044. [DOI] [PubMed] [Google Scholar]
- Ikeda R., Yokota M., Shinoda T. Serological characterization of Trichosporon cutaneumand related species. Microbiology and Immunology. 1996;40:813–819. doi: 10.1111/j.1348-0421.1996.tb01146.x. [DOI] [PubMed] [Google Scholar]
- Inácio J., Portugal L., Spencer-Martins I. Phylloplane yeasts from Portugal: seven novel anamorphic species in the Tremellales lineage of the Hymenomycetes (Basidiomycota) producing orange-coloured colonies. FEMS Yeast Research. 2005;5:1167–1183. doi: 10.1016/j.femsyr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- James T.Y., Kauff F., Schoch C.L. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kirschner R., Sampaio J.P., Gadanho M. Cuniculitrema polymorpha (Tremellales, gen. nov. and sp. nov.), a heterobasidiomycete vectored by bark beetles, which is the teleomorph of Sterigmatosporidium polymorphum. Antonie Van Leeuwenhoek. 2001;80:149–161. doi: 10.1023/a:1012275204498. [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P., Fell J.W., Boekhout T. Elsevier; London: 2011. The yeasts: a taxonomic study. [Google Scholar]
- Kwon-Chung K.J. Filobasidium Olive (1968) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; London: 2011. pp. 1457–1465. [Google Scholar]
- Landell F.M., Inácio J., Fonseca Á Cryptococcus bromeliarum sp. nov., an orange-coloured basidiomycetous yeast isolated from bromeliads in Brazil. International Journal of Systematic and Evolutionary Microbiology. 2009;59:910–913. doi: 10.1099/ijs.0.005652-0. [DOI] [PubMed] [Google Scholar]
- Larget B., Simon D.L. Markov chain Monte Carlo algorithms for the Bayesian analysis of phylogenetic trees. Molecular Biology and Evolution. 1999;16:750–759. [Google Scholar]
- Liu Y.J., Whelen S., Hall B.D. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Lutzoni F., Kauff F., Cox C.J. Assembling the fungal tree of life: progress, classification, and evolution of subcellular traits. American Journal of Botany. 2004;91:1446–1480. doi: 10.3732/ajb.91.10.1446. [DOI] [PubMed] [Google Scholar]
- Matheny P.B. Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales) Molecular Phylogenetics and Evolution. 2005;35:1–20. doi: 10.1016/j.ympev.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Matheny P.B., Gossmann J.A., Zalar P. Resolving the phylogenetic position of the Wallemiomycetes: an enigmatic major lineage of Basidiomycota. Canadian Journal of Botany. 2006;84:1794–1805. [Google Scholar]
- Matheny P.B., Liu Y.J., Ammirati J.F. Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales) American Journal of Botany. 2002;89:688–698. doi: 10.3732/ajb.89.4.688. [DOI] [PubMed] [Google Scholar]
- Matheny P.B., Wang Z., Binder M. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi) Molecular Phylogenetics and Evolution. 2007;43:430–451. doi: 10.1016/j.ympev.2006.08.024. [DOI] [PubMed] [Google Scholar]
- McNeill J., Barrie F.R., Buck W.R. A.R.G. Gantner Verlag KG; Ruggell, Liechtenstein: 2012. International Code of Nomenclature for Algae, Fungi, and Plants (Melbourne Code). Regnum Vegetabile 154.http://www.iapt-taxon.org/nomen/main.php Available from: [Google Scholar]
- Middelhoven W.J., Scorzetti G., Fell J.W. Systematics of the anamorphic basidiomycetous yeast genus Trichosporon Behrend with the description of five novel species: Trichosporon vadense, T. smithiae, T. dehoogii, T. scarabaeorum and T. gamsii. International Journal of Systematic and Evolutionary Microbiology. 2004;54:975–986. doi: 10.1099/ijs.0.02859-0. [DOI] [PubMed] [Google Scholar]
- Millanes A.M., Diederich P., Ekman S. Phylogeny and character evolution in the jelly fungi (Tremellomycetes, Basidiomycota, Fungi) Molecular Phylogenetics and Evolution. 2011;61:12–28. doi: 10.1016/j.ympev.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Moore R.T., Kreger-van Rij N.J.W. Ultrastructure of Filobasidium Olive. Canadian Journal of Microbiology. 1972;18:1949–1951. doi: 10.1139/m72-301. [DOI] [PubMed] [Google Scholar]
- Nakase T., Itoh M., Takematsu A. Kockovaella, a new ballistospore-forming anamorph yeast genus. Journal of General and Applied Microbiology. 1991;37:175–197. [Google Scholar]
- Nakase T., Jan-ngam H., Tsuzuki S. Two new ballistoconidium-forming yeast species, Bullera melastomae and Bullera formosana, found in Taiwan. Systematic and Applied Microbiology. 2004;27:558–564. doi: 10.1078/0723202041748118. [DOI] [PubMed] [Google Scholar]
- Nakase T., Takematsu A., Yamada Y. Molecular approaches to the taxonomy of ballistosporous yeasts based on the analysis of the partial nucleotide sequences of 18S ribosomal ribonucleic acids. The Journal of General and Applied Microbiology. 1993;39:107–134. [Google Scholar]
- Nakase T., Tsuzuki S., Takashima M. Bullera taiwanensis sp. nov. and Bullera formosensis sp. nov., two new ballistoconidium-forming yeast species isolated from plant leaves in Taiwan. The Journal of General and Applied Microbiology. 2002;48:345–355. doi: 10.2323/jgam.48.345. [DOI] [PubMed] [Google Scholar]
- Oberwinkler F., Bandoni R.J. Tetragoniomyces gen. nov. and Tetragoniomycetaceae fam. nov. (Tremellales) Canadian Journal of Botany. 1981;59:1034–1040. [Google Scholar]
- O'Donnell K. Fusarium and its near relatives. In: Reynolds D.R., Taylor J.W., editors. The fungal holomorph: mitotic, meiotic and pleomorphic speciation in fungal systematics. CAB International; Wallingford, UK: 1993. pp. 225–233. [Google Scholar]
- Okoli I., Oyeka C.A., Kwon-Chung K.J. Cryptotrichosporon anacardii gen. nov., sp. nov., a new trichosporonoid capsulate basidiomycetous yeast from Nigeria that is able to form melanin on niger seed agar. FEMS Yeast Research. 2007;7:339–350. doi: 10.1111/j.1567-1364.2006.00164.x. [DOI] [PubMed] [Google Scholar]
- Philippe H., Snell E.A., Bapteste E., Lopez P., Holland P.W., Casane D. Phylogenomics of Eukaryotes: impact of missing data on large alignments. Molecular Biology and Evolution. 2004;21:1740–1752. doi: 10.1093/molbev/msh182. [DOI] [PubMed] [Google Scholar]
- Posada D. JModelTest: phylogenetic model averaging. Molecular Biology and Evolution. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Rehner S.A., Buckley E. A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia. 2005;97:84–98. doi: 10.3852/mycologia.97.1.84. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio J.P. Diversity, phylogeny and classification of basidiomycetous yeasts. In: Agerer R., Piepenbring M., Blanz P., editors. Frontiers in basidiomycote mycology. IHW-Verlag; Eching, Germany: 2004. pp. 49–80. [Google Scholar]
- Sampaio J.P. Cystofilobasidium Oberwinkler & Bandoni (1983) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; London: 2011. pp. 1423–1431. [Google Scholar]
- Sampaio J.P. Tausonia Bab'eva (1998) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; London: 2011. pp. 1999–2001. [Google Scholar]
- Sampaio J.P., Fonseca A. Physiological aspects in the systematics of heterobasidiomycetous yeasts. Studies in Mycology. 1995;38:29–46. [Google Scholar]
- Sampaio J.P., Inacio J., Fonseca A. Auriculibuller fuscus gen. nov., sp. nov. and Bullera japonica sp. nov., novel taxa in the Tremellales. International Journal of Systematic and Evolution Microbiology. 2004;54:987–993. doi: 10.1099/ijs.0.02970-0. [DOI] [PubMed] [Google Scholar]
- Sampaio J.P., Weiβ M., Gadanho M. New taxa in the Tremellales: Bulleribasidium oberjochense gen. et sp. nov., Papiliotrema bandonii gen. et sp. nov. and Fibulobasidium murrhardtense sp. nov. Mycologia. 2002;94:873–887. [PubMed] [Google Scholar]
- Schoch C.L., Seifert K.A., Huhndorf S. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:6241–6246. doi: 10.1073/pnas.1117018109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorzetti G., Fell J.W., Fonseca A. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Research. 2002;2:495–517. doi: 10.1111/j.1567-1364.2002.tb00117.x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller J.W., Hall B.D. The origin of red algae: implications for plastid evolution. Proceedings of the National Academy of Sciences. 1997;94:4520–4525. doi: 10.1073/pnas.94.9.4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita T. Trichosporon Behrend (1890) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; London: 2011. pp. 2015–2061. [Google Scholar]
- Sugita T., Ikeda R., Nishikawa A. Analysis of Trichosporon isolates obtained from the houses of patients with summer-type hypersensitivity pneumonitis. Journal of Clinical Microbiology. 2004;42:5467–5471. doi: 10.1128/JCM.42.12.5467-5471.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita T., Nakase T. Molecular phylogenetic study of the basidiomycetous anamorphic yeast genus Trichosporon and related taxa based on small subunit ribosomal DNA sequences. Mycoscience. 1998;39:7–13. [Google Scholar]
- Takashima M., Deak T., Nakase T. Emendation of Dioszegia with redescription of Dioszegia hungarica and two new combinations, Dioszegia aurantiaca and Dioszegia crocea. The Journal of General and Applied Microbiology. 2001;47:75–84. doi: 10.2323/jgam.47.75. [DOI] [PubMed] [Google Scholar]
- Takashima M., Nakase T. Molecular phylogeny of the genus Cryptococcus and related species based on the sequences of SSU rDNA and internal transcribed spacer regions. Microbiology and Culture Collections. 1999;15:35–47. [Google Scholar]
- Takashima M., Sugita T., Toriumi Y. Cryptococcus tepidarius sp. nov., a thermotolerant yeast species isolated from a stream from a hotspring area in Japan. International Journal of Systematic and Evolutionary Microbiology. 2009;59:181–185. doi: 10.1099/ijs.0.004515-0. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente P., Boekhout T., Landell M.F. Bandoniozyma gen. nov., a genus of fermentative and non-fermentative Tremellacetous yeast species. PLOS One. 2012;7:1–11. doi: 10.1371/journal.pone.0046060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.M., Bai F.Y. Molecular phylogeny of basidiomycetous yeasts in the Cryptococcus luteolus lineage (Tremellales) based on nuclear rDNA and mitochonodrial cytochrome b gene sequence analyses, proposal of Derxomyces gen. nov. and Hannaella gen. nov., and description of eight novel Derxomyces species. FEMS Yeast Research. 2008;8:799–814. doi: 10.1111/j.1567-1364.2008.00403.x. [DOI] [PubMed] [Google Scholar]
- Wang Q.M., Bai F.Y., Fungsin B. Proposal of Mingxiaea gen. nov. for the anamorphic basidiomycetous yeast species in the Bulleribasidium clade (Tremellales) based on molecular phylogenetic analysis, with six new combinations and four novel species. International Journal of Systematic and Evolutionary Microbiology. 2011;61:210–219. doi: 10.1099/ijs.0.019299-0. [DOI] [PubMed] [Google Scholar]
- Wang Q.M., Wang S.A., Jia J.H. Cryptococcus tibetensis sp. nov., a novel basidiomycetous anamorphic yeast species isolated from plant leaves. The Journal of General and Applied Microbiology. 2007;53:281–285. doi: 10.2323/jgam.53.281. [DOI] [PubMed] [Google Scholar]
- Weiß M., Bauer R., Sampaio J.P. Tremellomycetes and related groups. In: McLaughlin D.J., Spatafora J.W., editors. The mycota, Vol. VII, Part A: Systematics and evolution. 2nd edn. Springer-Verlag; Berlin: 2014. pp. 331–355. [Google Scholar]
- Wells K., Bandoni R.J. Heterobasidiomycetes. In: McLaughlin D.J., McLaughlin E.G., Lemke P.A., editors. The mycota VII, Systematics and evolution, Part B. Springer-Verlag; Berlin: 2001. pp. 85–120. [Google Scholar]
- White T.J., Bruns T., Lee S.B. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR protocols, a guide to methods and applications. Academic; San Diego, USA: 1990. pp. 315–322. [Google Scholar]
- Wuczkowski M., Passoth V., Turchetti B. Description of Holtermanniella takashimae sp. nov., Holtermanniella gen. nov. and proposal of the order Holtermanniales to accommodate Tremellomycetous yeasts of the Holtermanniales clade. International Journal of Systematic and Evolutionary Microbiology. 2011;61:680–689. doi: 10.1099/ijs.0.019737-0. [DOI] [PubMed] [Google Scholar]
- Yamada Y., Kondo K. Coenzyme Q system in the classification of the yeast genera Rhodotorula and Cryptococcus and the yeast like genera Sporobolomyces and Rhodosporidium. The Journal of General and Applied Microbiology. 1973;19:59–77. [Google Scholar]