Abstract
Most small genera containing yeast species in the Pucciniomycotina (Basidiomycota, Fungi) are monophyletic, whereas larger genera including Bensingtonia, Rhodosporidium, Rhodotorula, Sporidiobolus and Sporobolomyces are polyphyletic. With the implementation of the “One Fungus = One Name” nomenclatural principle these polyphyletic genera were revised. Nine genera, namely Bannoa, Cystobasidiopsis, Colacogloea, Kondoa, Erythrobasidium, Rhodotorula, Sporobolomyces, Sakaguchia and Sterigmatomyces, were emended to include anamorphic and teleomorphic species based on the results obtained by a multi-gene phylogenetic analysis, phylogenetic network analyses, branch length-based methods, as well as morphological, physiological and biochemical comparisons. A new class Spiculogloeomycetes is proposed to accommodate the order Spiculogloeales. The new families Buckleyzymaceae with Buckleyzyma gen. nov., Chrysozymaceae with Chrysozyma gen. nov., Microsporomycetaceae with Microsporomyces gen. nov., Ruineniaceae with Ruinenia gen. nov., Symmetrosporaceae with Symmetrospora gen. nov., Colacogloeaceae and Sakaguchiaceae are proposed. The new genera Bannozyma, Buckleyzyma, Fellozyma, Hamamotoa, Hasegawazyma, Jianyunia, Rhodosporidiobolus, Oberwinklerozyma, Phenoliferia, Pseudobensingtonia, Pseudohyphozyma, Sampaiozyma, Slooffia, Spencerozyma, Trigonosporomyces, Udeniozyma, Vonarxula, Yamadamyces and Yunzhangia are proposed to accommodate species segregated from the genera Bensingtonia, Rhodosporidium, Rhodotorula, Sporidiobolus and Sporobolomyces. Ballistosporomyces is emended and reintroduced to include three Sporobolomyces species of the sasicola clade. A total of 111 new combinations are proposed in this study.
Key words: Fungi, GMYC approach, Molecular phylogeny, Phylogenetic rank boundary optimisation, Pucciniomycotina, Taxonomy, Yeasts
Taxonomic novelties: New class: Spiculogloeomycetes Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout
New families: Buckleyzymaceae Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Chrysozymaceae Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Colacogloeaceae Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Microsporomycetaceae Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Ruineniaceae Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Sakaguchiaceae Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Symmetrosporaceae Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout
New genera: Bannozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Buckleyzyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Chrysozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Fellozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Hamamotoa Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Hasegawazyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Jianyunia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Microsporomyces Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Oberwinklerozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Phyllozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Phenoliferia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Pseudobensingtonia F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout; Pseudohyphozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Rhodosporidiobolus Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Ruinenia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Sampaiozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Slooffia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Spencerozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Symmetrospora Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Trigonosporomyces Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Udeniozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Vonarxula Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Yamadamyces Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Yunzhangia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout
New combinations: Ballistosporomycessasicola (Nakase & M. Suzuki) F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout; B. taupoensis (Hamam. & Nakase) F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout; Bannoabischofiae (Hamam., Thanh & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; B. syzygii (Hamam., Thanh & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; B. ogasawarensis (Hamam., Thanh & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Bannozymaarctica (Vishniac & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; B. yamatoana (Nakase, M. Suzuki & M. Itoh) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Buckleyzymaarmeniaca (R.G. Shivas & Rodr. Mir.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; B. aurantiaca (Saito) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; B. kluyveri-nielii (van der Walt) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; B. phyllomatis (van der Walt & Y. Yamada) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; B. salicina (B.N. Johri & Bandoni) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Chrysozymafushanensis (Nakase, F.L. Lee & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; C. griseoflava (Nakase & M. Suzuki) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Colacogloeacycloclastica (Thanh, M.S. Smit, Moleleki & Fell) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; C. diffluens (Ruinen) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; C. eucalyptica (C.H. Pohl, M.S. Smit & Albertyn) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; C. falcata (Nakase, M. Itoh & M. Suzuki) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; C. foliorum (Ruinen) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; C. philyla (van der Walt, Klift & D.B. Scott) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; C. retinophila (Thanh, M.S. Smit, Moleleki & Fell) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; C. terpenoidalis (Thanh, M.S. Smit, Moleleki & Fell) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Cystobasidiopsislactophilus (Nakase, M. Itoh, M. Suzuki & Bandoni) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; C. lophatheri (Nakase, Tsuzuki, F.L. Lee, Jindam. & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Cystobasidiumportillonense (F. Laich, I. Vaca & R. Chávez) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Erythrobasidiumelongatum (R.G. Shivas & Rodr. Mir.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; E. yunnanense (F.Y. Bai, M. Takash., Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Fellozymainositophila (Nakase & M. Suzuki) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Hamamotoalignophila (Dill, C. Ramírez & González) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; H. singularis (Phaff & do Carmo-Sousa) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Hasegawazymalactosa (Hasegawa) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Jianyuniasakaguchii (Sugita, M. Takash., Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Kondoachangbaiensis (F.Y. Bai & Q.M. Wang) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout; K. miscanthi (Nakase & M. Suzuki) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout; K. phyllada (van der Walt & Y. Yamada) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout; K. sorbi (F.Y. Bai & Q.M. Wang) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout; K. subrosea (Nakase & M. Suzuki) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout; K. thailandica (Fungsin, Hamam. & Nakase ) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout; K. yuccicola (Nakase & M. Suzuki) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout; Microsporomycesbloemfonteinensis (Pohl, M.S. Smit & Albertyn) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; M. magnisporus (Nakase, Tsuzuki, F.L. Lee, Sugita, Jindam. & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; M. pini (Pohl, M.S. Smit & Albertyn) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; M. orientis (Pohl, M.S. Smit & Albertyn) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Oberwinklerozymasilvestris (Golubev & Scorzetti) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; O. straminea (Golubev & Scorzetti) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; O. yarrowii (Á. Fonseca & van Uden) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Phenoliferiapsychrophenolica (Margesin & J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; P. psychrophila (Margesin & J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; P. glacialis (Margesin & J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; P. himalayensis (Shivaji, Bhadra & Rao) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Phyllozymacoprosmicola (Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; P. corallina (N. Furuya & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; P. dimennae (Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; P. linderae (Nakase, M. Takash. & Hamam.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; P. novozealandica (Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; P. producta (N. Furuya & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; P. subbrunnea (Nakase & M. Suzuki) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Pseudobensingtoniaingoldii (Nakase & Itoh.) F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout; P. musae (M. Takash., S.O. Suh & Nakase) F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout; Pseudohyphozymabogoriensis (Deinema) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; P. buffonii (C. Ramírez) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; P. pustula (Buhagiar) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Rhodotorulaalborubescens (Derx) Q.M. Wang, F.Y. Bai, Groenew. & Boekhout; R. babjevae (Golubev) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. diobovata (S.Y. Newell & I.L. Hunter) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. kratochvilovae (Hamam., Sugiy. & Komag.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. paludigena (Fell & Tallman) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. sphaerocarpa (S.Y. Newell & Fell) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. toruloides (I. Banno) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Rhodosporidiobolus fluvialis (Fell, Kurtzman, Tallman & J.D. Buck) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. azoricus (J.P. Samp. & Gadanho) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. microsporus (Higham ex Fell, Blatt & Statzell) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. nylandii (M. Takash. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. ruineniae (Holzschu, Tredick & Phaff) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. lusitaniae (Á. Fonseca & J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. colostri (T. Castelli) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. odoratus (J.P. Samp., Á. Fonseca & Valério) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. poonsookiae (M. Takash. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Ruineniaclavata (F.Y. Bai & Q.M. Wang) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. diospyroris (Nakase, Tsuzuki, F.L. Lee, Jindam. & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. dracophylli (Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. pyrrosiae (Nakase, Tsuzuki, F.L. Lee, Jindam. & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; R. rubra (Nakase, Oakada & Sugiy.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Sakaguchiacladiensis (Fell, Statzell & Scorzetti) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; S. lamellibrachiae (Nagah., Hamam., Nakase & Horikoshi) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; S. meli (Libkind, van Broock & J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; S. oryzae (F.Y. Bai & Y.M. Cai) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Sampaiozymaingeniosa (Di Menna) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; S. vanillica (J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Slooffia cresolica (Middelhoven & Spaaij) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; S. pilati (F.H. Jacob, Faure-Raynaud & Berton) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; S. tsugae (Phaff & do Carmo-Sousa) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Spencerozymacrocea (Shifrine & Phaff) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Sporobolomyceslongiusculus (Libkind, van Broock & J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; S. johnsonii (Nyland) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Sterigmatomyceshyphaenes (Har. & Pat.) F.Y. Bai, Q.M. Wang, Groenewald & Boekhout; S. pulcherrima (J.E. Wright) F.Y. Bai, Q.M. Wang, Groenewald & Boekhout; S. novozelandica (W.B. Kendr. & X.D. Gong) F.Y. Bai, Q.M. Wang, Groenewald & Boekhout; Symmetrosporacoprosmae (Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; S. foliicola (R.G. Shivas & Rodr. Mir.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; S. gracilis (Derx) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; S. vermiculata (M. Takash. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; S. marina (Phaff, Mrak & Williams) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; S. symmetrica (F.Y. Bai & Q.M. Wang) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Trigonosporomyceshylophilus (van der Walt, van der Klift & D.B. Scott) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Ustilentylomagraminis (Rodr. Mir. & Diem) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Udeniozymaferulica (J.P. Samp. & van Uden) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Vonarxulajavanica (Ruinen) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Yamadamycesrosulatus (Golubev & Scorzetti) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Yunzhangiaauriculariae (Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout; Y. sonckii (Hopsu-Havu, Tunnela & Yarrow) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout
Introduction
The subphylum Pucciniomycotina (Phylum Basidiomycota, Kingdom Fungi) presently includes eight classes, and four of these, namely the Agaricostilbomycetes, Cystobasidiomycetes, Microbotryomycetes and Mixiomycetes, contain taxa with a dominant yeast stage (Aime et al., 2006, Aime et al., 2014, Bauer et al., 2006, Hibbett et al., 2007, Boekhout et al., 2011, Wang et al., 2015a). So far 28 genera with yeast states have been proposed within Pucciniomycotina (Boekhout et al., 2011, Turchetti et al., 2011, Toome et al., 2013). Most of these genera are monophyletic, whereas five genera, namely Bensingtonia, Rhodosporidium, Rhodotorula, Sporidiobolus and Sporobolomyces are polyphyletic (Fell et al., 2000, Scorzetti et al., 2002, Boekhout et al., 2011, Wang et al., 2015a, Wang et al., 2015b). Bensingtonia, Rhodotorula and Sporobolomyces species are placed in various classes of Pucciniomycotina (Fell et al., 2000, Scorzetti et al., 2002, Boekhout et al., 2011, Hamamoto et al., 2011, Sampaio, 2011, Yurkov et al., 2015). Species of the teleomorphic genera Rhodosporidium and Sporidiobolus together with some asexual Rhodotorula and Sporobolomyces species are nested within Sporidiobolales (Fell et al., 2000, Scorzetti et al., 2002, Boekhout et al., 2011, Sampaio, 2011). With the implementation of the “One Fungus = One Name” nomenclatural principle (Hawksworth, 2011, Taylor, 2011, McNeill et al., 2012) these five polyphyletic genera need to be revised.
Several studies using molecular analyses of ribosomal DNA sequences have provided a detailed grouping of species in clades among the four classes that contain yeast and yeast-like species (Hamamoto and Nakase, 2000, Nakase, 2000, Fell et al., 2000, Scorzetti et al., 2002, Boekhout et al., 2011), but many species remained unassigned (Boekhout et al. 2011). Thus the boundaries of the clades and genera have to be reassessed by analysing a robust molecular dataset. In another study we analysed seven gene fragments, namely SSU (18S) rRNA, LSU (26S/28S) rRNA D1/D2 domains, the ITS region (including the 5.8S rRNA), RPB1 (the largest subunit of DNA polymerase II), RPB2 (the second largest subunit of DNA polymerase II), TEF1 (translation elongation factor 1-α) and CYTB (cytochrome b) that placed most pucciniomycetous yeast species into 51 well-supported clades (Wang et al. 2015a). These data are used here to address the taxonomic affiliations of those fungi. We propose 26 of the 51 recognised clades as new taxa at the genus, family and class levels based on a phylogenetic and taxonomic analysis of the combined seven genes-based and the enlarged LSU rRNA gene datasets. The assessment of taxonomic ranks followed the branch length-based methods as described in Liu et al. (2015).
Materials and methods
Strains and molecular phylogenetic analyses
Multi-gene data of the yeast strains used were taken from a previous study (Wang et al. 2015a). As described previously (Wang et al. 2015a) multi-gene phylogenetic trees constructed from Maximum likelihood (ML), Maximum parsimony (MP) and Bayesian inference (BI) analyses of a dataset comprising nucleotide sequences of the ITS region (including the 5.8S rRNA), the D1/D2 domains of the LSU rRNA, the SSU rRNA, and the RPB1, RPB2, TEF1 and CYTB genes is used here to address the taxonomy of the pucciniomycetous yeasts. Fifty-one clades that may be equal to the generic rank, including 16 single-species lineages, were recognised among the pucciniomycetous yeasts used in the previous study with strong statistical support values in all trees drawn using different phylogenetic algorithms (Wang et al. 2015a). In order to detect the reliability of those 51 clades, a phylogenetic network approach was employed to infer the relationships between those pucciniomycetous yeasts. The seven genes-based phylogenetic network was constructed in SplitsTree4 (Huson & Bryant 2006) using the ConsensusNetwork algorithm with default parameter settings. The seven single-gene ML trees used in the phylogenetic network analysis were constructed using RAxML-HPC 7.2.8 (Stamatakis 2006) using the parameter settings described previously (Wang et al. 2015a).
The supplementary LSU rRNA gene (D1/D2 domains) sequence dataset containing data from newly published pucciniomycetous yeast species and a few additional filamentous teleomorphic taxa was constructed and subjected to constrained maximum likelihood (ML) and maximum parsimony (MP) analyses based on the topology of a seven genes-based dataset taken from Wang et al. (2015a). The LSU sequences were aligned with MAFFT version 7 and the G-INS-i option (Standley K 2013). Constrained phylogenetic analyses were only inforced for species previously analysed using seven DNA loci. Only bipartitions that received at least 85% bootstrap support during fast bootstrapping of the seven genes-based dataset (Wang et al. 2015a) conducted with Pthreads-parallelised RAxML version 8.1.24 (Stamatakis 2014) were used as a backbone constraint for LSU phylogenetic inference. Fast bootstrapping in conjunction with the autoMRE bootstopping criterion (Pattengale et al. 2009) and subsequent search for the best tree (Stamatakis et al. 2008) were conducted using the GTRCAT model approximation. MP bootstrapping with 1 000 replicates was conducted with TNT version 1.1/June 2015 (Goloboff et al. 2008). The alignments and trees were deposited in TreeBASE (No. 18537).
Quantitative assessment of taxonomic ranks
The modified Generalized Mixed Yule Coalescent (GMYC) method (Humphreys & Barraclough 2014) was applied iteratively to identify higher evolutionary significant units (higher ESUs) above the species levels in the pucciniomycetous yeasts as done before for tremellomycetous fungi (Liu et al. 2015). Firstly, the overall GMYC analysis was carried out for the simulation at class level, and secondly nested analyses were run for the higher ESUs identification at family level for each clade. Outgroup samples were excluded from the dataset using the drop.tip command in ape (Paradis 2006). A chronogram was calculated from the ML-based tree using the penalised likelihood method (Sanderson 2002) as implemented in the chronopl command in ape (Paradis 2006). The chronogram was then analysed using a modified GMYC package in SPLITS in R (version 2.10, www.cran.r-project.org) using the single threshold method. In the case of clades with a small number of samples the modified GMYC approach would not result in significant differences simply because of sampling size: these were marked as ‘NA’ (not analysed).
Phylogenetic rank boundary optimisation (PRBO), a phylogenetic variant of clustering optimisation (Göker et al., 2009, Göker et al., 2010, Stielow et al., 2011), was conducted based on taxonomy-based reference information as described in Liu et al. (2015). A reduced classification including twelve putatively reliable genera of pucciniomycetous yeasts was chosen as reference taxonomy. The resulting optimal upper boundaries for the divergence of each taxonomic rank were then applied back to the entire dataset. These boundaries for each taxonomic rank were compared with boundaries estimated from the entire classification (Table 1). For each newly proposed or already established taxon, maximum subtree height (MaSH) of its corresponding clade, absolute deviation and significant deviation (Sigdev) from the threshold optimal for the reliable taxa were calculated. One hundred bootstrap replicates were applied to obtain the 95% confidence intervals for the boundaries to detect the significances of the divergences from the optimal range for each taxonomic rank.
Table 1.
PRBO results showing the divergences, if any, of the proposed genera from the optimal range of divergences for their rank as inferred from selected reference data.
| Taxa | Rank | MaSH | Deviation | Sigdev |
|---|---|---|---|---|
| Agaricostilbomycetes | Class | 0.71223 | 0.07318 | 0 |
| Agaricostilbum clade | Genus | 0.22827 | 0 | 0 |
| ingoldii clade | Genus | 0.14859 | 0 | 0 |
| Chionosphaera* | Genus | 0.29924 | 0 | 0 |
| Kurtzmanomyces* | Genus | 0.33350 | 0.02844 | 0 |
| lactophilus clade | Genus | 0.25804 | 0 | 0 |
| sasicola clade | Genus | 0.15364 | 0 | 0 |
| Kondoa clade* | Genus | 0.28874 | 0 | 0 |
| Bensingtonia* | Genus | 0.24787 | 0 | 0 |
| ruber clade | Genus | 0.29828 | 0 | 0 |
| subbrunneus clade | Genus | 0.39731 | 0.09225 | 0 |
| Cystobasidiomycetes | Class | 0.51968 | 0 | 0 |
| Erythrobasidium clade* | Genus | 0.16907 | 0 | 0 |
| Bannoa clade* | Genus | 0.15301 | 0 | 0 |
| aurantiaca clade | Genus | 0.16265 | 0 | 0 |
| marina clade | Genus | 0.23897 | 0 | 0 |
| Sakaguchia clade* | Genus | 0.29017 | 0 | 0 |
| magnisporus clade | Genus | 0.33920 | 0.03414 | 0 |
| Cystobasidium (minuta) clade* | Genus | 0.18036 | 0 | 0 |
| Microbotryomycetes | Class | 0.72324 | 0.08419 | 0 |
| Sporidiobolus clade | Genus | 0.34525 | 0.04020 | 0 |
| Rhodosporidium clade | Genus | 0.40759 | 0.10253 | 0 |
| mixed Rhodosporidium/Sporidiobolus clade | Genus | 0.16275 | 0 | 0 |
| Kriegeria* | Genus | 0.21892 | 0 | 0 |
| glacialis clade | Genus | 0.16560 | 0 | 0 |
| buffonii clade | Genus | 0.10602 | 0 | 0 |
| yarrowii clade | Genus | 0.09543 | 0 | 0 |
| tsugae clade | Genus | 0.23831 | 0 | 0 |
| singularis clade | Genus | 0.05035 | 0 | 0 |
| yamatoana clade | Genus | 0.06763 | 0 | 0 |
| griseoflavus clade | Genus | 0.16370 | 0 | 0 |
| Curvibasidium clade* | Genus | 0.04099 | 0 | 0 |
| Colacogloea clade* | Genus | 0.23771 | 0 | 0 |
| sonckii clade | Genus | 0.06955 | 0 | 0 |
| vanillica clade | Genus | 0.07445 | 0 | 0 |
| Leucosporidium clade* | Genus | 0.15399 | 0 | 0 |
| Microbotryum clade | Genus | 0.14270 | 0 | 0 |
Note: MaSH: Maximum Subtree Height; Deviation: deviation from the point estimate for the upper (positive value) or lower (negative value) threshold of the rank of the taxon; Sigdev: significant deviation, i.e. a deviation even outside the upper or lower 95 % confidence band of the upper or lower threshold, respectively. Zero indicates taxa with the appropriate divergence, negative values indicate taxa that are too small, positive values taxa that are too large. An asterisk (*) indicates the well-established taxa that were used as a reference classification for PRBO.
The seven genes-based ML tree used in Wang et al. (2015a) was employed as the basis for the PRBO and iterative modified GMYC analyses. The taxa within Ustilaginomycotina were used as outgroup and taxa within Pucciniomycetes were used as ingroup in the above two analyses. Note that none of the two methods was followed strictly in the current study. Where possible, wider circumscriptions of taxa were chosen to lower the number of taxonomic changes suggested; moreover, where possible, clades with distinct phenotypic or ecological features were proposed as new taxa (Liu et al. 2015). Additionally, already established taxa were kept unless they appeared evidently non-monophyletic.
Results and discussion
Taxonomic units addressed by the iterative modified GMYC and PRBO analyses
The pucciniomycetous yeast species belong to four recognised classes, namely Agaricostilbomycetes, Cystobasidiomycetes, Microbotryomycetes and Mixiomycetes (Bauer et al., 2006, Hibbett et al., 2007, Boekhout et al., 2011). The overall modified GMYC analysis supported the Agaricostilbomycetes without Spiculogloeales as a class in agreement with the indication that Spiculogloeales may represent a new class within Pucciniomycotina based on the seven genes-based phylogenetic analyses (Wang et al. 2015a). Thus Spiculogloeomycetes is proposed as a new class to accommodate the order Spiculogloeales.
The nested analyses of the GMYC approach identified five families in the class Agaricostilbomycetes (Table 2), including the recognised families Agaricostilbaceae, Chionosphaeraceae and Kondoaceae, a new Agaricostilbales family 1 (ruber clade) and a new Agaricostilbales family 2 (Bensingtonia sakaguchii lineage). Because only B. sakaguchii occurs in the new Agaricostilbales family 2, this family is not proposed in this study. Consequently, B. sakaguchii is placed into a new genus (see Taxonomy), which is presently treated as ‘incertae sedis’ in the Agaricostilbales.
Table 2.
Analyses with the modified GMYC approach showing the supported classification of the pucciniomycetous yeast at family and class levels.
| Class/Order | family | Genus | GMYC |
|---|---|---|---|
| Agaricostilbomycetes | supported | ||
| Agaricostilbales | |||
| Kondoaceae | supported | ||
| Kondoa | |||
| Bensingtonia | |||
| Agaricostilbaceae | supported | ||
| Sterigmatomyces (Agaricostilbum clade) | |||
| Pseudobensingtonia (ingoldii clade) | |||
| Chionosphaeraceae | supported | ||
| Chionosphaera | |||
| Kurtzmanomyces | |||
| Mycogloea nipponica | |||
| Ballistosporomyces (sasicola clade) | |||
| Cystobasidiopsis (lactophilus clade) | |||
| Ruineniaceae | new family | ||
| Ruinenia (ruber clade) | |||
| incertae sedis in the Agaricostilbales | Jianyunia (Bensingtonia sakaguchii) | new family | |
| Spiculogloeomycetes | not supported | ||
| Spiculogloeales | |||
| Spiculogloeaceae | supported | ||
| Phyllozyma (subbrunneus clade) | |||
| Cystobasidiomycetes | not supported | ||
| Cystobasidiales | |||
| Cystobasidiaceae | not supported | ||
| Occultifur | |||
| Cystobasidium (minuta clade) | |||
| Erythrobasidiales | |||
| Erythrobasidiaceae | supported | ||
| Erythrobasidium | |||
| Bannoa | |||
| incertae sedis in the Erythrobasidiales | Hasegawazyma (Rhodotorula lactosa) | new family | |
| Cyrenella | new family | ||
| Naohideales | |||
| Naohideaceae | supported | ||
| Naohidea | |||
| incertae sedis in the Cystobasidiomycetes | |||
| Buckleyzymaceae | not supported | ||
| Buckleyzyma (aurantiaca clade) | |||
| Symmetrosporaceae | not supported | ||
| Symmetrospora (marina clade) | |||
| Sakaguchiaceae | new family | ||
| Sakaguchia | |||
| Microsporomycetaceae | new family | ||
| Microsporomyces (magnisporus clade) | |||
| Microbotryomycetes | not supported | ||
| Sporidiobolales | |||
| Sporidiobolaceae | not supported | ||
| Rhodotorula(Rhodosporidium clade) | new family | ||
| Rhodosporidiobolus (mixed Rhodosporidium/Sporidiobolus clade) | new family | ||
| Sporobolomyces (Sporidiobolus clade) | new family | ||
| Kriegeriales | |||
| Kriegeriaceae | not supported | ||
| Kriegeria | |||
| Meredithblackwellia | |||
| Phenoliferia (glacialis clade) | |||
| Yamadamyces (Rhodotorula rosulata) | |||
| Camptobasidiaceae | not supported | ||
| Glaciozyma | |||
| Leucosporidiales | |||
| Leucosporidiaceae | supported | ||
| Leucosporidium | |||
| Microbotryales | |||
| Microbotryaceae | supported | ||
| Microbotryum | |||
| Ustilentylomataceae | supported | ||
| Ustilentyloma | |||
| Heterogastridiales | |||
| Heterogastridiaceae | supported | ||
| Heterogastridium | |||
| incertae sedis in the Microbotryomycetes | |||
| Chrysozymaceae | new family | ||
| Chrysozyma (griseoflavus clade) | |||
| Bannozyma (yamatoana clade) | |||
| Hamamotoa (singularis clade) | |||
| Fellozyma (Sporobolomyces inositophilus) | |||
| Colacogloeaceae | new family | ||
| Colacogloea | |||
| Genera incertae sedis in the Microbotryomycetes | |||
| Pseudohyphozyma (buffonii clade) | new family | ||
| Slooffia (tsugae clade) | |||
| Oberwinklerozyma (yarrowii clade) | |||
| Sampaiozyma (vanillica clade) | new family | ||
| Yunzhangia (sonckii clade) | new family | ||
| Curvibasidium | new family | ||
| Pseudoleucosporidium (Leucosporidium fasciculatum) | |||
| Udeniozyma (Rhodotorula ferulica) | new family | ||
| Reniforma | new family | ||
| Trigonosporomyces (Rhodotorula hylophila) | new family | ||
| Vonarxula (Rhodotorula javanica) | new family | ||
| Spencerozyma (Rhodotorula crocea) | |||
| Mixiomycetes | not supported | ||
| Mixiales | |||
| Mixiaceae | NA | ||
| Mixia |
Note: NA means “not analysed”. In the overall GMYC analysis, Naohideales is a separate class from Cystobasidiomycetes; Spiculoglomycetes and Mixiomycetes were identified as one class; Heterogastridium, Rhodotorula hylophila, and Reniforma form a separate class from Microbotryomycetes; Rhodotorula javanica and Rhodotorula crocea form another separate class from Microbotryomycetes.
Nine clades were indentified at the family level within Cystobasidiomycetes by the nested analyses of GMYC (Table 2). The presently accepted families Cystobasidiaceae, Erythrobasidiaceae and Naohideaceae belong to Cystobasidiales, Erythrobasidiales and Naohideales, respectively. The genus Occultifur is separated from Cystobasidiaceae as a family in the GMYC analyses, but we presently prefer to keep this genus in the Cystobasidiaceae due to the low number of taxa in this genus. The Cyrenella and Rhodotorula lactosa lineages were suggested as two new families in the Erythrobasidiales by the GMYC nested analyses, however, these two lineages represent single species each, and, therefore, we temporarily placed them as ‘incertae sedis’ in the Erythrobasidiales. The aurantiaca and marina clades were grouped into one family in the GMYC nested analyses, which is not supported by the phylogenetic analysis of seven genes that showed the two clades as a paraphyletic group (Wang et al. 2015a). The magnisporus and Sakaguchia clades were identified as families in agreement with the phylogenetic analysis of seven genes (Wang et al. 2015a).
Within Microbotryomycetes two families, namely Leucosporidiaceae and Microbotryaceae, were supported by the nested analyses of GMYC approach (Table 2). The family Sporidiobolaceae in the Sporidiobolales was divided into three families represented by the Sporidiobolus clade, the Rhodosporidium clade and the mixed Rhodosporidium/Sporidiobolus clade, respectively. We preserve the current taxonomic status of Sporidiobolaceae because the phenotype of these three clades is similar and it forms a strongly supported lineage in the phylogenetic analysis of seven genes (Wang et al. 2015a). The Kriegeriaceae and Camptobasidiaceae in the Kriegeriales were grouped into a single family in the nested GMYC analyses. However, the Camptobasidiaceae, including Glaciozyma antarctica, clustered together with the Kriegeriaceae lacking support value in the ML analysis and they did not occur in the same cluster in the MP and BI analyses (Wang et al. 2015a, Fig. 1 of this study). Consequently the two families are maintained in this study.
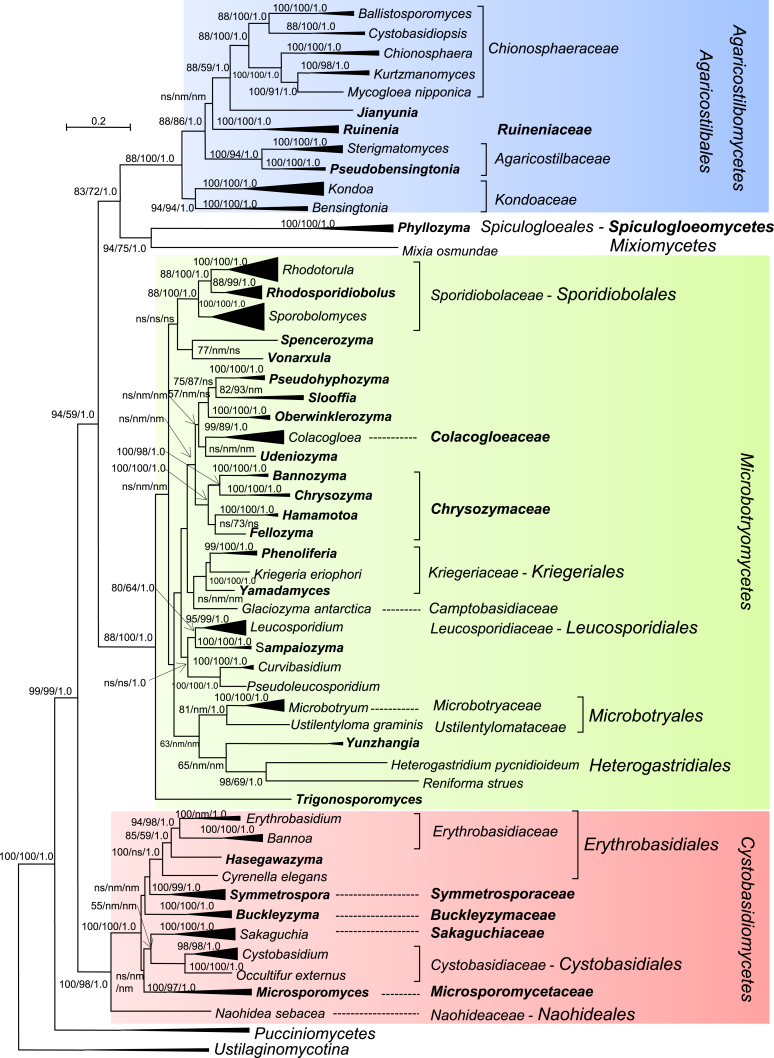
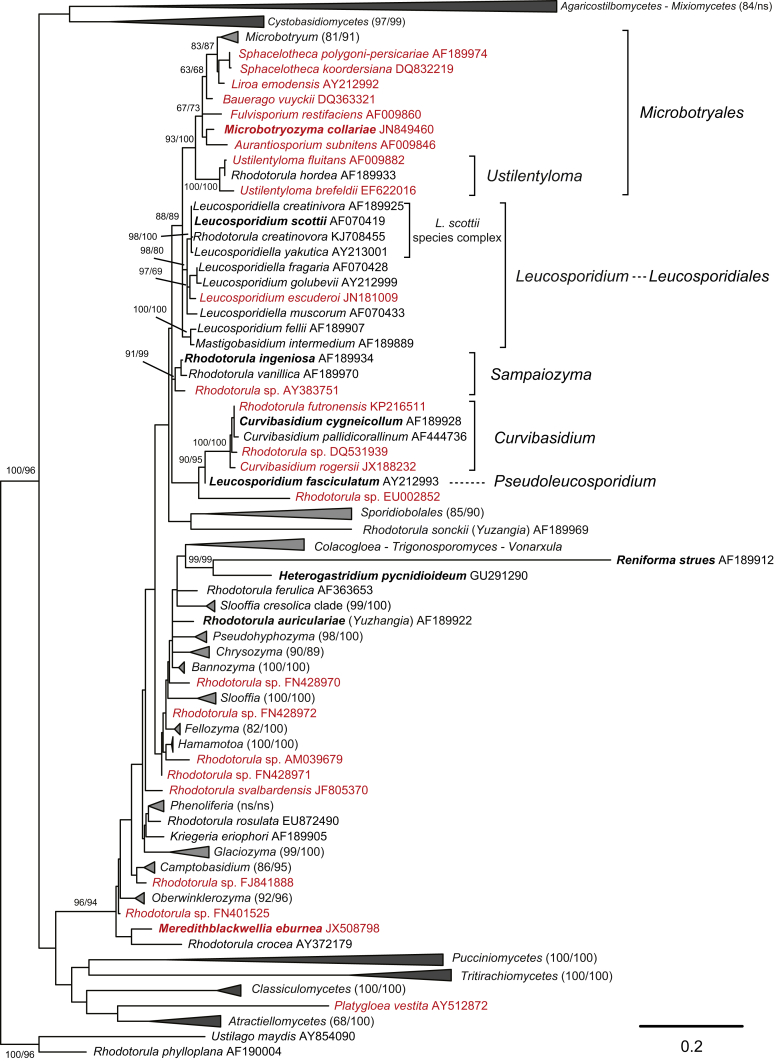
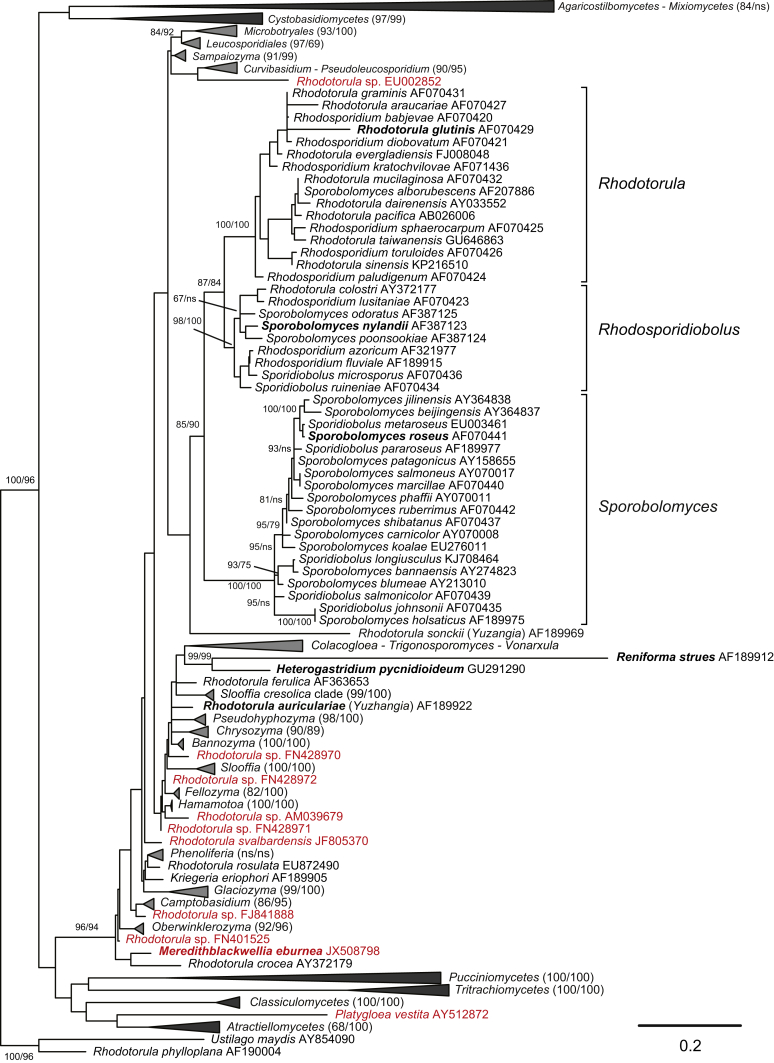
Fig. 1.
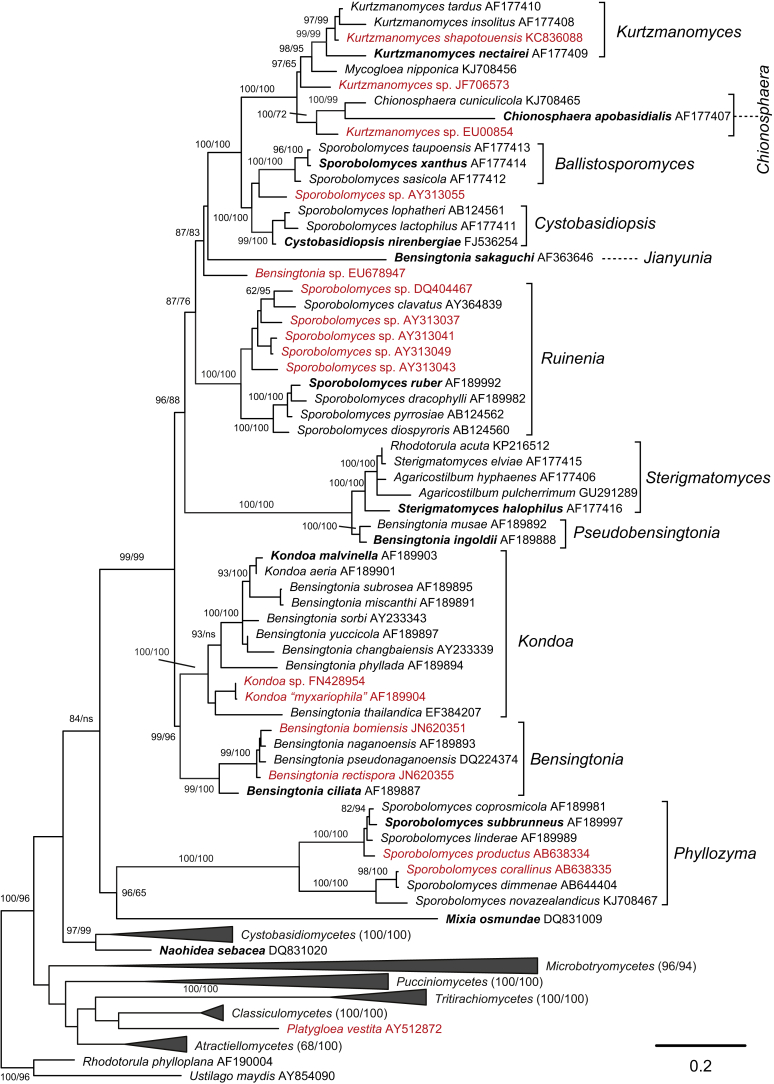
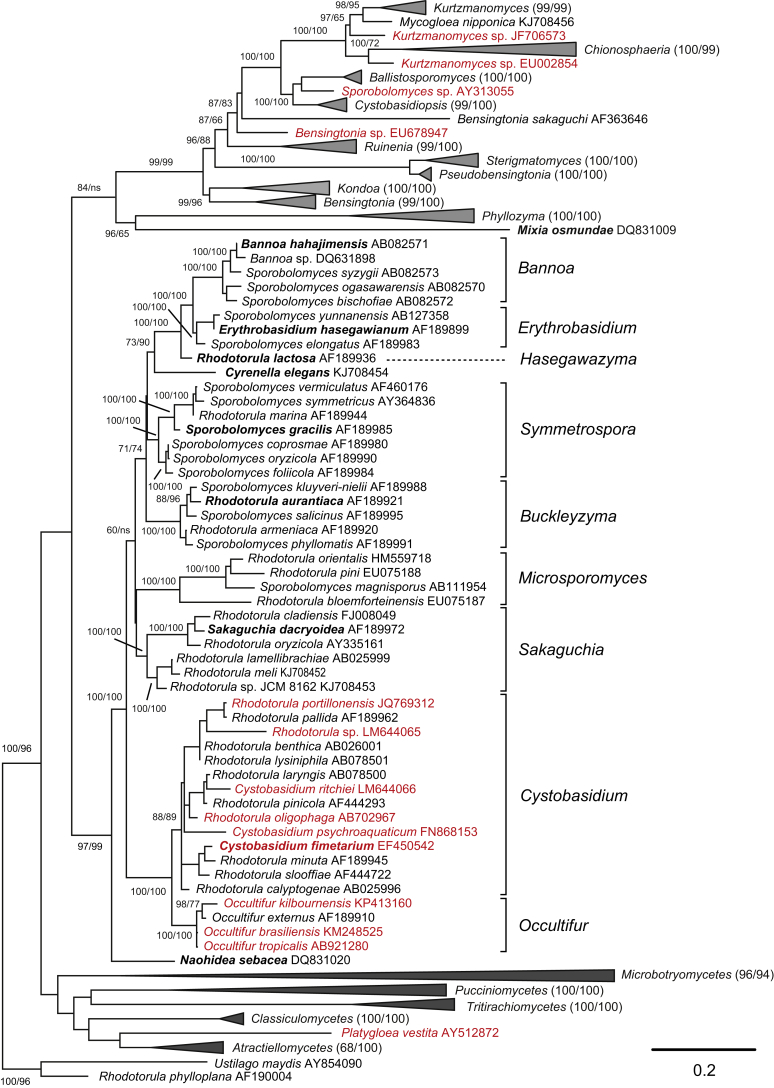
Phylogenetic tree inferred using the combined sequences of SSU rRNA, LSU rRNA D1/D2 domains, ITS regions (including 5.8S rRNA), RPB1, RPB 2, TEF1 and CYTB, depicting the phylogenetic positions of existing yeast taxa and new genera (in bold) within Pucciniomycotina. The tree backbone was constructed using maximum likelihood analysis. Bootstrap percentages of maximum likelihood and maximum parsimony analyses over 50 % from 1 000 bootstrap replicates and posterior probabilities of Bayesian inference above 0.95 are shown respectively from left to right on the deep and major branches. Bar = 0.2 substitutions per nucleotide position. Note: ns, not supported (BP < 50 % or PP < 0.9); nm, not monophyletic. The new taxa are in bold.
Ten clades and seven single-species lineages in Microbotryomycetes could not be assigned to presently recognised families and orders (Wang et al. 2015a). These clades are strongly divergent from each other and seem to have a sister relationship to the known families and orders within Microbotryomycetes (Fig. 1). The species Rhodotorula hylophila, R. javanica, R. crocea and Reniforma strues were not included in the nested GMYC analyses because they occurred outside the Microbotryomycetes in the overall GMYC analysis. In the nested GMYC analyses, the griseoflavus, yamatoana, singularis clades and Sporobolomyces inositophilus were identified as one family that was supported by the phylogenetic analysis of seven genes with strong support values (Wang et al. 2015a); the buffonii, tsugae and yarrowii clades were assigned to one family, but this was weakly supported by the seven genes ML analysis (57 % BP), lacking support in the BI analysis and were not supported by the MP analysis (Fig. 1), and consequently, they are not treated as a single family in this study; the other clades and the single-species lineages were identified as separate families in the nested GMYC analyses (Table 2).
Among 51 pucciniomycetous yeast clades suggested as genera in the previous multi-gene phylogenetic study (Wang et al. 2015a), only few were found to deviate from the optimal range of divergences as determined in the PRBO analysis, and were found to significantly deviate (Table 1), which supports the preliminary taxonomic conclusions from the multi-gene phylogenetic analysis. Twenty-six of them represent currently described genera. The others are proposed as new genera (Fig. 1) based on the phylogenetic analyses, PRBO analysis and phenotypic comparisons (Table 1, Table 3 and Fig. 2, Fig. 3) presented in this study.
Table 3.
Selected physiological and biochemical characteristics of different clades within the Pucciniomycotina.
| Genus or species | Sucrose | Raffinose | Lactose | Trehalose | Maltose | Melezitose | Methyl-α-d-glucoside | Soluble Starch | l-Arabinose | d-Arabinose | Glycerol | myo-Inositol | dl-glucoside | Nitrate | Nitrite | CoQ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Agaricostilbomycetes | ||||||||||||||||
| Agaricostilbales | ||||||||||||||||
| Kondoaceae | ||||||||||||||||
| Kondoa | v | v | v | + | v | v | v | v | v | v | + | − | v | v | v | 9 |
| Bensingtonia | v | v | v | v | v | v | − | v | v | v | + | − | v | + | v | 9 |
| Agaricostilbaceae | ||||||||||||||||
| Sterigmatomyces (Agaricostilbum clade) | v | v | v | + | − | v | v | − | v | + | + | − | v | v | v | 9 |
| Pseudobensingtonia (ingoldii clade) | v | v | v | + | − | v | − | v | v | + | + | − | v | v | v | 9 |
| Chionosphaeraceae | ||||||||||||||||
| Ballistosporomyces (sasicola clade) | + | v | v | + | + | v | v | + | v | − | − | − | − | v | v | 10 |
| Cystobasidiopsis (lactophilus clade) | + | v | v | + | + | v | v | + | v | + | + | − | + | v | v | 10 |
| Kurtzmanomyces | v | v | v | v | v | v | − | − | v | v | v | v | v | + | + | 10 |
| Chionosphaera | − | − | − | v | v | − | − | v | v | v | v | v | v | − | − | 10 |
| Jianyunia (Bensingtonia sakaguchii) | − | − | + | + | + | + | v | + | − | − | − | − | − | − | − | 9 |
| Ruinenia (ruber clade) | + | + | v | v | v | + | v | v | v | − | v | − | − | − | − | 10 |
| Spiculogloeales | ||||||||||||||||
| Phyllozyma (subbrunneus clade) | v | v | v | v | − | − | − | − | − | v | v | − | v | + | v | 10 |
| Cystobasidiomycetes | ||||||||||||||||
| Cystobasidiales | ||||||||||||||||
| Cystobasidium (minuta clade) | v | v | v | v | v | v | v | v | v | v | + | v | v | − | − | 9,10 |
| Occultifur externus | + | − | + | + | + | + | − | − | + | + | + | − | + | − | − | n |
| Erythrobasidiales | ||||||||||||||||
| Erythrobasidium | + | − | − | + | + | + | − | v | + | + | + | − | v | v | v | 10(H2) |
| Bannoa | + | + | v | + | + | + | v | + | v | v | + | v | v | − | − | 10 (H2) |
| Hasegawazyma (Rhodotorula lactosa) | + | + | + | + | + | + | − | + | + | + | + | − | + | + | + | 9 |
| Cyrenella elegans | + | + | − | + | + | + | − | + | + | − | + | − | − | + | + | n |
| Microsporomyces (magnisporus clade) | v | v | v | + | v | + | v | v | v | v | v | v | v | v | v | 10 |
| Buckleyzyma (aurantiaca clade) | v | v | − | v | v | v | − | − | + | + | + | − | v | v | v | 10 |
| Symmetrospora (marina clade) | v | v | v | v | v | v | v | v | v | v | + | − | v | v | v | 10 |
| Sakaguchia | v | v | − | + | v | v | − | v | v | v | + | − | v | − | − | 10 |
| Naohideales | ||||||||||||||||
| Naohidea sebacea | − | − | − | − | − | − | − | − | − | − | + | − | + | − | − | n |
| Mixiomycetes | ||||||||||||||||
| Mixia osmundae | − | − | − | − | − | − | − | + | − | v | − | − | − | − | − | 10 |
| Microbotryomycetes | ||||||||||||||||
| Sporidiobolales | ||||||||||||||||
| Rhodotorula (Rhodosporidium clade) | + | v | − | + | v | v | v | − | v | v | + | − | v | v | v | 9,10 |
| Rhodosporidiobolus (mixed Rhodosporidium/Sporidiobolus clade) | v | v | − | + | v | v | v | v | v | v | + | − | v | + | + | 9,10 |
| Sporobolomyces (Sporidiobolus clade) | + | v | − | v | v | v | v | v | v | v | v | − | v | v | v | 10 |
| Leucosporidiales | ||||||||||||||||
| Leucosporidium | v | v | − | v | v | v | v | − | v | v | + | − | v | v | v | 9,10 |
| Kriegeriales | ||||||||||||||||
| Kriegeriaceae | ||||||||||||||||
| Phenoliferia (glacialis clade) | + | + | − | − | − | + | − | n | − | − | − | − | − | + | − | n |
| Meredithblackwellia eburnea | + | − | − | + | + | + | + | − | w | + | + | − | + | − | − | n |
| Yamadamyces (Rhodotorula rosulata) | + | − | − | w | + | + | − | w | − | − | w | + | d | + | + | n |
| Kriegeria eriophori | + | − | − | + | + | + | + | − | + | v | + | − | + | + | + | n |
| Camptobasidiaceae | ||||||||||||||||
| Glaciozyma antarctica | v | − | − | − | v | − | − | v | − | − | v | − | − | + | + | 10 |
| Sampaiozyma (vanillica clade) | + | + | + | + | + | + | v | + | − | v | + | − | + | + | + | 10 |
| Curvibasidium | v | v | v | v | v | v | − | − | + | v | v | v | v | − | − | 9 |
| Chrysozyma (griseoflavus clade) | v | − | − | + | + | + | v | v | − | v | v | − | − | v | v | 10 |
| Bannozyma (yamatoana clade) | + | − | − | + | v | + | − | v | − | − | + | − | − | − | − | 9 |
| Hamamotoa (singularis clade) | − | − | + | + | − | − | − | − | v | v | + | − | + | − | − | n |
| Fellozyma (Sporobolomyces inositophilus) | + | − | − | + | + | + | − | − | − | − | + | + | − | + | + | 10 |
| Colacogloea | v | v | − | + | v | v | v | − | − | v | v | − | v | v | v | 10 |
| Udeniozyma (Rhodotorula ferulica) | + | − | + | + | + | + | + | − | − | + | + | − | v | + | + | 10 |
| Pseudohyphozyma (buffonii clade) | − | − | − | v | v | v | − | v | v | + | + | − | v | v | v | 10 |
| Slooffia (tsugae clade) | + | − | v | + | + | + | v | − | − | v | + | − | + | + | + | 10 |
| Oberwinklerozyma (yarrowii clade) | + | + | − | + | + | + | v | v | − | v | + | + | + | + | + | 9 |
| Microbotryum | + | − | + | + | + | + | + | + | − | − | + | − | + | + | + | n |
| Ustilentyloma graminis (Rhodotorula hordea) | + | − | + | + | + | + | + | + | − | − | + | − | + | + | + | n |
| Yunzhangia (sonckii clade) | v | − | − | + | v | v | − | − | − | − | + | − | v | v | v | n |
| Trigonosporomyces (Rhodotorula hylophila) | − | − | − | − | − | − | − | − | − | − | + | − | + | − | − | n |
| Vonarxula (Rhodotorula javanica) | − | + | − | + | + | − | − | + | + | + | + | − | − | + | + | 9 |
| Spencerozyma (Rhodotorula crocea) | + | − | − | + | + | + | − | − | v | v | + | − | − | + | + | 10 |
| Reniforma strues | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 7 |
Note: V: variable; +: positive; −: negative; w: weak; d: delay; n: not tested.
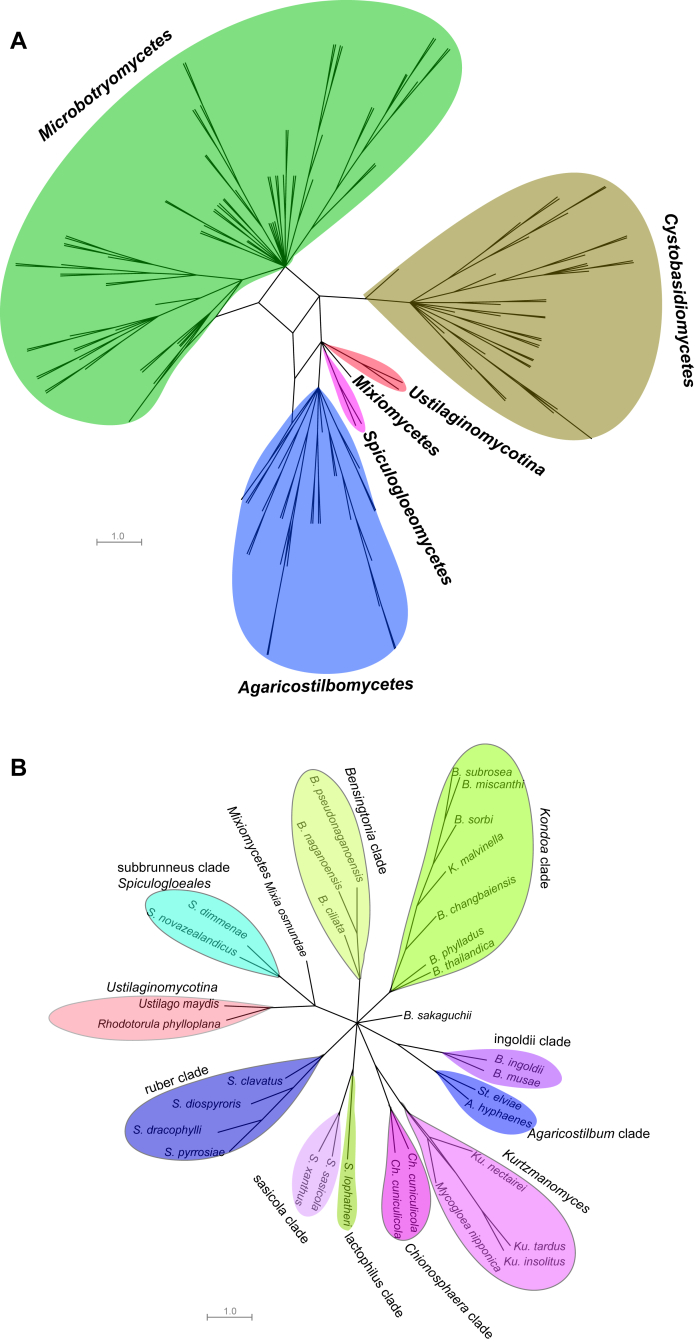
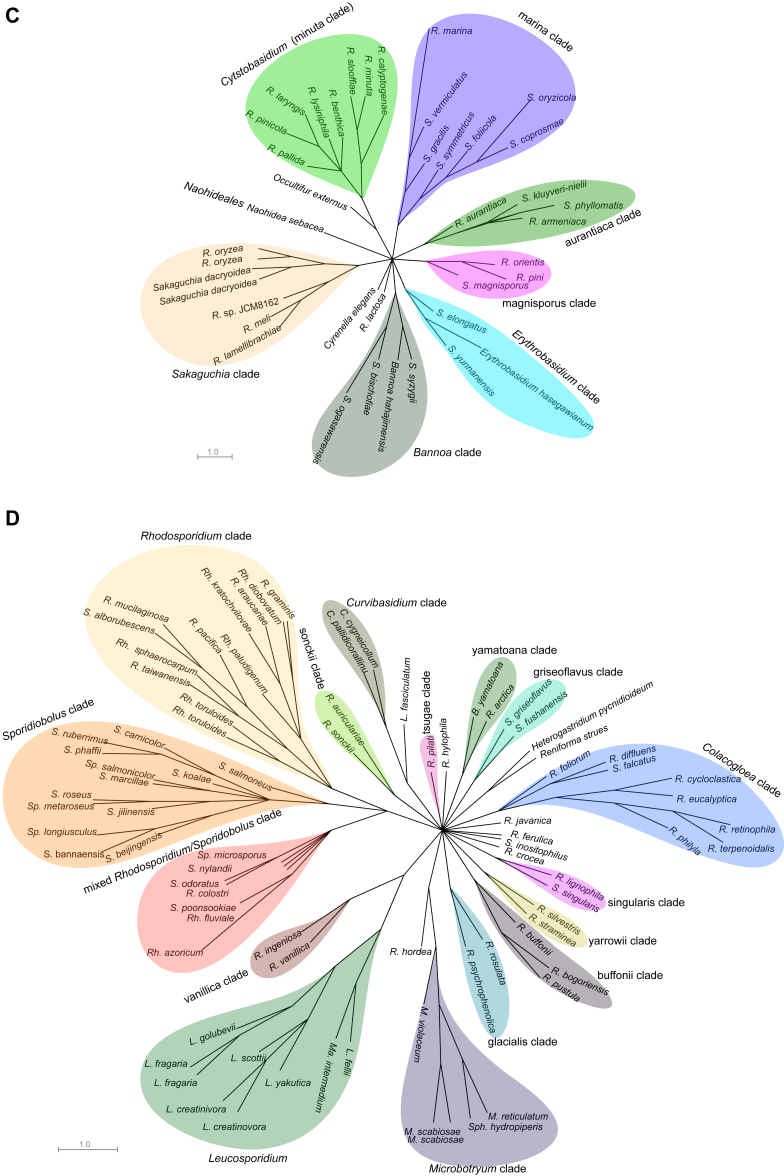
Fig. 2.
Phylogenetic network of the yeast species within Pucciniomycotina. Single gene ML trees inferred with RAxML were investigated in Splitstree 4.13.1 using the ConsensusNetwork algorithm under default settings. A): the phylogenetic network of the Agaricostilbomycetes, Mixiomycetes, Microbotryomycetes, Cystobasidiomycetes and Spiculogloeomycetes; B): the phylogenetic network of Agaricostilbomycetes, Mixiomycetes and Spiculogloeomycetes; C): the phylogenetic network of Cystobasidiomycetes; D): the phylogenetic network of Microbotryomycetes. Abbreviations: A: Agaricostilbum; B: Bensingtonia; C: Curvibasidium; Ch: Chionosphaera; K: Kondoa; Ku: Kurtzmanomyces; L: Leucosporidium; Le: Leucosporidiella; M: Microbotryum; Ma: Mastigobasidium; R: Rhodotorula; Rh: Rhodosporidium; S: Sporobolomyces; Sa: Sakaguchia; Sp: Sporidiobolus; Sph: Sphacelotheca; Ster: Sterigmatomyces.
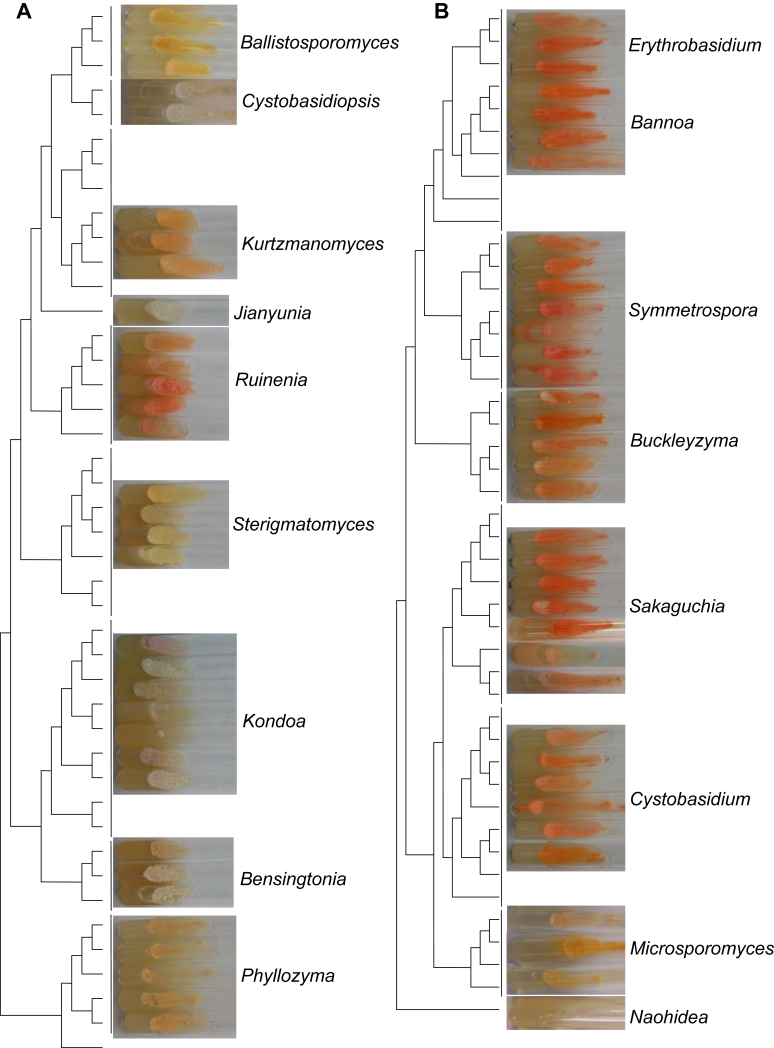
Fig. 3.
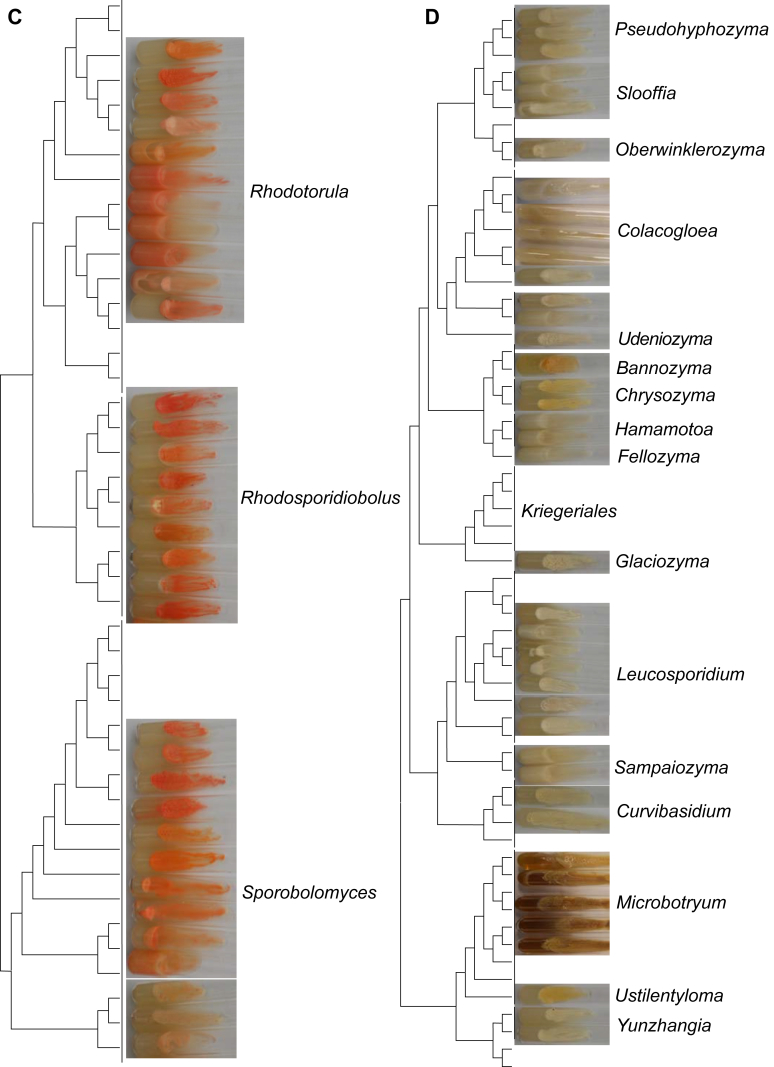
Comparison of the colony characteristics in different clades within the Pucciniomycotina. All strains were cultured on slants with potato dextrose agar (PDA) medium for one month at 17 °C. The tree was inferred using maximum likelihood analysis from the combined seven genes (Wang et al. 2015a). A): The colony characteristics in different clades of the classes Agaricostilbomycetes and Spiculogloeomycetes; B): The colony characteristics in different clades of the class Cystobasidiomycetes; C): The colony characteristics in different clades of the order Sporidiobolales of Microbotryomycetes; D): The colony characteristics in different clades of the class Microbotryomycetes, except those belonging to order Sporidiobolales.
Phylogenetic analyses
Only 156 species from 184 ones used in the seven genes-based tree (Wang et al. 2015a) were selected to construct the phylogenetic network, because some protein genes were not available for all species. As a result all clades recognised in the seven genes-based tree could be recognised in the network approach. The network result showed that the five classes containing yeast species, viz. Agaricostilbomycetes, Cystobasidiomycetes, Microbotryomycetes, Mixiomycetes and Spiculogloeomycetes, remain separated (Fig. 2A). The 51 clades in the seven genes-based tree are also separated without any reticulation (Fig. 2B–D). This result confirmed the reliability of the combined phylogenetic analysis of the seven genes.
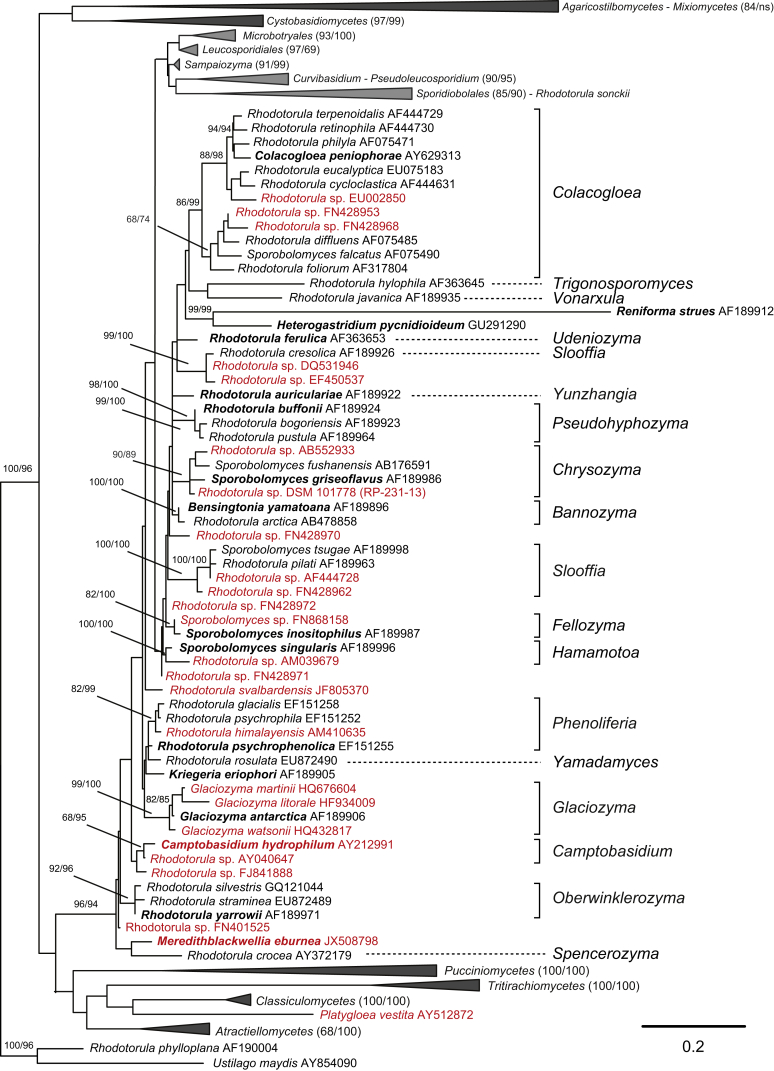
For a better understanding of the phylogenetic relationships between Agaricostilbomycetes, Cystobasidiomycetes, Microbotryomycetes, Mixiomycetes and Spiculogloeomycetes, and to include recently described species, an enlarged LSU rRNA gene dataset was analysed. Thereby, the LSU dataset analysed by Wang et al. (2015a) was enlarged from 184 to 242 sequences containing both sexual (e.g. Camptobasidium, Cystobasidium, Glaciozyma, Kondoa and Ustilentyloma) and asexual (e.g. Rhodotorula, Sporobolomyces and Occultifur) genera and species (Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8). The constrained ML analysis of the enlarged LSU dataset was used to place species known from LSU rRNA gene sequences in the phylogenetic clades previously recognised in the analysis based on the seven DNA-loci. Results from the LSU analysis were not used to challenge the results of the multi-gene study (Wang et al. 2015a), but to investigate the stability of the taxonomy in undersampled clades.
Fig. 4.
Phylogenetic relationships of yeasts and related taxa from the Agaricostilbomycetes and Mixiomycetes lineages obtained by Maximum-Likelihood analysis of the LSU (D1/D2 domains) rRNA gene. Tree topology was constrained according to the topology of the seven genes-based tree (Wang et al. 2015a) with nodes showing bootstrap values >85 % inforced to be monophyletic. The type species of each genus is in bold. Taxa not included in the phylogenetic analysis of the seven genes (Wang et al. 2015a) are indicated in red. Note: ns, not supported (BP < 50 %).
Fig. 5.
Phylogenetic relationships of yeasts and related taxa from the Cystobasidiomycetes lineage obtained by Maximum-Likelihood analysis of the LSU (D1/D2 domains) rRNA gene. Tree topology was constrained according to the topology of the seven genes-based tree (Wang et al. 2015a) with nodes showing bootstrap values >85 % inforced to be monophyletic. The type species of each genus is in bold. Taxa not included in the phylogenetic analysis of the seven genes (Wang et al. 2015a) are indicated in red. Note: ns, not supported (BP < 50 %).
Fig. 6.
Phylogenetic relationships of yeasts and related taxa from the Microbotryomycetes (‘incertae sedis’ lineages) obtained by Maximum-Likelihood analysis of the LSU (D1/D2 domains) rRNA gene. Tree topology was constrained according to the topology of the seven genes-based tree (Wang et al. 2015a) with nodes showing bootstrap values >85 % inforced to be monophyletic. The type species of each genus is in bold. Taxa not included in the phylogenetic analysis of the seven genes (Wang et al. 2015a) are indicated in red. Note: ns, not supported (BP < 50 %).
Fig. 7.
Phylogenetic relationships of yeasts and related taxa from the Microbotryomycetes (Microbotryales, Leucosporidiales and related ‘incertae sedis’ lineages) obtained by Maximum-Likelihood analysis of the LSU (D1/D2 domains) rRNA gene. Tree topology was constrained according to the topology of the seven genes-based tree (Wang et al. 2015a) with nodes showing bootstrap values >85 % inforced to be monophyletic. The type species of each genus is in bold. Taxa not included in the phylogenetic analysis of the seven genes (Wang et al. 2015a) are indicated in red. Note: ns, not supported (BP < 50 %).
Fig. 8.
Phylogenetic relationships of yeasts and related taxa from the Microbotryomycetes (Sporidiobolales and selected ‘incertae sedis’ lineages) obtained by Maximum-Likelihood analysis of the LSU (D1/D2 domains) rRNA gene. Tree topology was constrained according to the topology of the seven genes-based tree (Wang et al. 2015a) with nodes showing bootstrap values >85 % inforced to be monophyletic. The type species of each genus is in bold. Taxa not included in the phylogenetic analysis of the seven genes (Wang et al. 2015a) are indicated in red. Note: ns, not supported (BP < 50 %).
The enlarged analysis of the LSU rRNA gene dataset suggests that the number of single-species lineages in Microbotryomycetes is likely to increase in the future, since many sequences representing potentially new species could not be assigned to any of the clades recognised (Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8). In agreement with previous studies (Scorzetti et al., 2002, Weiß et al., 2004, Boekhout et al., 2011, Wang et al., 2015a), our results showed that LSU alone is not sufficient to resolve many clades in Microbotryomycetes (Fig. 6).
The addition of the supplemental species or sequences representing potential new species resulted in the enlargement of several clades, especially single species lineages recognised in the phylogenetic analysis of seven genes (Wang et al. 2015a), e.g. Fellozyma, Glaciozyma, Occultifur and Rhodotorula hordea (Fig. 5, Fig. 6, Fig. 7). In addition, a few new clades and single-species lineages were identified such as Camptobasidium hydrophilum, Meredithblackwellia eburnea and Rhodotorula svalbardensis (Fig. 6). Most supplemental sequences (34 out of 58) were located in Microbotryomycetes (Fig. 6, Fig. 7). The following type species were added to the dataset, namely Cystobasidium fimetarium, Camptobasidium hydrophilum, Meredithblackwellia eburnea and Microbotryozyma collariae (Fig. 5, Fig. 6, Fig. 7). Newly added sequences substantially expanded the following clades recognised in Wang et al. (2015a), viz. Curvibasidium (Fig. 7), Glaciozyma (Fig. 6), Ruinenia (Fig. 4) and Slooffia (Fig. 6).
Taxonomy
Class Agaricostilbomycetes R. Bauer et al., Mycol. Progr. 5: 45. 2006.
Type order: Agaricostilbales Oberw. & R. Bauer
This class contains the order Agaricostilbales. Our previous multi-gene sequence analyses indicated that nine well support clades, namely Agaricostilbum, Bensingtonia, Chionosphaera, Kondoa, Kurtzmanomyces, ingoldii, lactophilus, ruber, sasicola, and two species Bensingtonia sakaguchii and Mycogloea nipponica, occurred in the Agaricostilbales (Wang et al. 2015a). These clades which are delimited at the generic rank are supported by the PRBO analysis (Table 1), the phylogenetic network analysis (Fig. 2B), and the analysis of the enlarged LSU rRNA gene dataset (Fig. 4). The genera Chionosphaera and Kurtzmanomyces are well-established genera. The genera Sterigmatomyces, Cystobasidiopsis and Kondoa are emended to include both teleomorphic and anamorphic species in the Agaricostilbum, lactophilus and Kondoa clades, respectively. Ballistosporomyces is emended and reintroduced to include species in the sasicola clade. Jianyunia gen. nov., Pseudobensingtonia gen. nov. and Ruinenia gen. nov. are proposed to accommodate the species in the Bensingtonia sakaguchii, ingoldii and ruber clades, respectively. Ruineniaceae fam. nov. is proposed to accommodate the genus Ruinenia based on results from the phylogenetic analysis of seven genes (Fig. 1) and GMYC analyses (Table 2).
Order Agaricostilbales Oberw. & R. Bauer, Sydowia 41: 240. 1989.
Type family: Agaricostilbaceae Oberw. & R. Bauer.
This order was proposed to accommodate the family Agaricostilbaceae (Oberwinkler & Bauer 1989). The Agaricostilbaceae, Chionosphaeraceae and Kondoaceae were accepted in this order by Bauer et al. (2006). Here we propose Ruineniaceae and Jianyunia as ‘incertae sedis’ in the Agaricostilbales.
Family Agaricostilbaceae Oberw. & R. Bauer, Sydowia 41: 240. 1989.
Type genus: Agaricostilbum J.E. Wright.
This family is characterised by septal pores without microbodies, aseptate basidiospores produced in a yeast-like manner and lack of tremelloid haustorial cells (Oberwinkler and Bauer, 1989, Bauer et al., 2006).
Genera accepted: Sterigmatomyces Fell, Pseudobensingtonia F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout.
Sterigmatomyces Fell, Antonie van Leeuwenhoek 32: 101. 1966. emend. F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout.
= Agaricostilbum J.E. Wright, Mycologia 62: 679. 1970.
Type species: Sterigmatomyces halophilus Fell.
This genus is emended to include species of Agaricostilbum and Sterigmatomyces, which occurred as a well supported Agaricostilbum clade within Agaricostilbaceae (Fig. 1, Fig. 4). The name Sterigmatomyces was published before Agaricostilbum (Fell, 1966, Fell, 2011, Wright, 1970, Bandoni and Boekhout, 2011), so the merged genus is named Sterigmatomyces.
Sexual reproduction observed in some species. Basidia occur predominantly in synnemata-like basidiomata. Hyphae, basidia and basidiospores relatively thick-walled. Basidiospores often attached to a budding locus (Bandoni & Boekhout 2011). Colonies cream and butyrous. Budding cells present or not, some of them produce one or more stalk-like conidiophores with blastoconidia separating at a septum in the mid-region of the stalk on the parent cell. Ballistoconidia not produced. Major CoQ system Q-9.
Species accepted:
-
1)
Sterigmatomyces elviae Sonck & Yarrow, Antonie van Leeuwenhoek 35: 172. 1969.
-
2)
Sterigmatomyces halophilus Fell, Antonie van Leeuwenhoek 32: 101. 1966.
-
3)Sterigmatomyces hyphaenes (Har. & Pat.) F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout, comb. nov. MycoBank MB813385.
- Basionym: Pilacre hyphaenes Har. & Pat., Bull. Mus. Hist. Nat. 17: 370. 1911.
- ≡ Agaricostilbum hyphaenes (Har. & Pat.) Oberw. & Bandoni.
-
4)Sterigmatomyces pulcherrimus (J.E. Wright) F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout, comb. nov. MycoBank MB813386.
- Basionym: Isaria pulcherrima Berk. & Broome, J. Linn. Soc. Bot. 14: 96. 1873.
- ≡ Agaricostilbum pulcherrimum (Berk. & Broome) B.L. Brady, B. Sutton & Samson.
- = Agaricostilbum palmicola J.E. Wright.
-
5)Sterigmatomyces novozelandicus (W.B. Kendr. & X.D. Gong) F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout, comb. nov. MycoBank MB813388.
- Basionym: Agaricostilbum novozelandicum (as nova-zelandica) W.B. Kendr. & X.D. Gong, Mycotaxon 54: 21. 1995.
Pseudobensingtonia F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout, gen. nov. MycoBank MB813078.
Etymology: The genus is named because of a similar morphology as present in the genus Bensingtonia.
This genus is proposed to accommodate the ingoldii clade containing two species that previously belonged to the genus Bensingtonia (Wang et al. 2015a). Member of the Agaricostilbaceae. The genus is mainly circumscribed by the phylogenetic analysis of seven genes, in which it formed a sister lineage to the genus Sterigmatomyces within Agaricostilbaceae (Fig. 1, Fig. 4). This genus is phylogenetically distinct from the Bensingtonia clade that contains the type species of Bensingtonia, B. ciliata, that belongs to the family Kondoaceae (Wang et al. 2015a, Fig. 4 of this study).
Sexual reproduction not known. Colonies greyish-yellow or dark yellow and butyrous. Budding cells present. Pseudohyphae present or not. Ballistoconidia present, ellipsoidal or kidney-shaped. Major CoQ system Q-9.
Type species: Pseudobensingtonia ingoldii (Nakase & Itoh.) F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout.
Note: The species of Pseudobensingtonia do not form conidiogenous stalks, which are present in the anamorphic species of Sterigmatomyces (Nakase et al., 1989, Nakase et al., 2011, Takashima et al., 1995).
Species accepted:
-
1)Pseudobensingtonia ingoldii (Nakase & Itoh.) F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout, comb. nov. MycoBank MB813079.
- Basionym: Bensingtonia ingoldii Nakase & Itoh., J. Gen. Appl. Microbiol. 35: 53. 1989.
-
2)Pseudobensingtonia musae (M. Takash., S.O. Suh & Nakase) F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout, comb. nov. MycoBank MB813080.
- Basionym: Bensingtonia musae M. Takash. et al., J. Gen. Appl. Microbiol. 41: 143. 1995.
Family Chionosphaeraceae Oberw. & Bandoni, Can. J. Bot. 60: 1732. 1982.
Type genus: Chionosphaera D.E. Cox.
This family is characterised by teleomorphic members with gasteroid basidia with simultaneous basidiospore production per basidium (Oberwinkler and Bandoni, 1982, Bauer et al., 2006).
Genera accepted: Ballistosporomyces Nakase et al. emend. F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout, Chionosphaera D.E. Cox, Cystobasidiopsis R. Bauer et al. emend. F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout, Kurtzmanomyces Y. Yamada et al., Stilbum Tode.
Notes: Mycogloea nipponica was placed in this family based on a multi-gene analyses (Wang et al. 2015a) and an analysis of the enlarged LSU rRNA gene dataset (Fig. 4). The species of the genus Stilbum are not listed here because living cultures of Stilbum are not available at present.
Ballistosporomyces Nakase et al., J. Gen. Appl. Microbiol. 35: 291. 1989. emend. F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout.
Type species: Ballistosporomyces xanthus Nakase et al.
This genus is emended and reintroduced to include species of the sasicola clade (Wang et al. 2015a), which occurred as a well-supported clade related to the genus Cystobasidiopsis within Chionosphaeraceae (Fig. 1, Fig. 4). The genus Ballistosporomyces was erected by Nakase et al. (1989) and included Ba. xanthus (= Sporobolomyces xanthus), the type of Ballistosporomyces, and Ba. ruber (= Sporobolomyces ruber). This genus was treated as a synonym of Sporobolomyces (Boekhout 1991). Our analyses showed that Ba. xanthus (S. xanthus) is located in the sasicola clade, whereas Ba. ruber (S. ruber) occurs in the ruber clade that is phylogenetically distinct from the family Chionosphaeraceae (Fig. 1). Thus, here we emend and reintroduce Ballistosporomyces as a genus to include the species of the sasicola clade.
Sexual reproduction unknown. Colonies orange to pale yellowish-brown and butyrous. Budding cells present. Hyphae and pseudohyphae not formed. Ballistoconidia present, allantoid. Major CoQ system Q-10.
Note: Sporobolomyces ruber (Ba. ruber), which is located in the ruber clade (Fig. 1), is proposed as a new combination in Ruinenia (Fig. 4).
Species accepted:
-
1)Ballistosporomyces sasicola (Nakase & M. Suzuki) F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout, comb. nov. MycoBank MB813081.
- Basionym: Sporobolomyces sasicola Nakase & M. Suzuki, J. Gen. Appl. Microbiol. 33: 171. 1987.
-
2)Ballistosporomyces taupoensis (Hamam. & Nakase) F.Y. Bai, Q.M. Wang, M. Groenew. & Boekhout, comb. nov. MycoBank MB813082.
- Basionym: Sporobolomyces taupoensis Hamam. & Nakase, Antonie van Leeuwenhoek 67: 163. 1995.
-
3)
Ballistosporomyces xanthus Nakase et al., J. Gen. Appl. Microbiol. 35: 292. 1989.
Chionosphaera D.E. Cox, Mycologia 68: 503. 1976.
Type species: Chionosphaera apobasidialis D.E. Cox.
Species accepted:
-
1)
Chionosphaera apobasidialis D.E. Cox, Mycologia 68: 503. 1976.
-
2)
Chionosphaera coppinsii P. Roberts, Mycotaxon 63: 195. 1997.
-
3)
Chionosphaera cuniculicola R. Kirschner et al., Mycol. Res. 105: 1404. 2001.
-
4)
Chionosphaera erythrinae (Hansf.) R. Kirschner, Fungal Science Taipei 23: 50. 2008.
-
5)
Chionosphaera lichenicola Alstrup et al., Graphis Scripta 5: 97. 1993.
-
6)
Chionosphaera phylaciicola (Seifert & Bandoni) R. Kirschner & Oberw., Mycol. Res. 105: 1406. 2001.
Note: Living cultures have been obtained only from Ch. apobasidialis and Ch. cuniculicola, which have an asexual yeast stage.
Cystobasidiopsis R. Bauer et al., Mycol. Res. 113: 962. 2009. emend. Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Type species: Cystobasidiopsis nirenbergiae R. Bauer et al.
This genus is emended to include species of Cystobasidiopsis and anamorphic species of the lactophilus clade (Wang et al. 2015a), which occurred as a well supported clade related to the genus Ballistosporomyces within Chionosphaeraceae (Fig. 1, Fig. 4).
Sexual reproduction observed in some species. Teleomorphic taxa produce probasidia with stipitate, transversely septate basidia. Basidiospores sessile. Colonies cream white and butyrous. Budding cells present or not. Ballistoconidia present or not, ellipsoidal, amygdaliform or falcate. Major CoQ system Q-10.
Species accepted:
-
1)Cystobasidiopsis lactophilus (Nakase, M. Itoh, M. Suzuki & Bandoni) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813083.
- Basionym: Sporobolomyces lactophilus Nakase et al., Trans. Mycol. Soc. 31: 161. 1990.
-
2)Cystobasidiopsis lophatheri (Nakase, Tsuzuki, F.L. Lee, Jindam. & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813084.
- Basionym: Sporobolomyces lophatheri Nakase et al., J. Gen. Appl. Microbiol. 51: 282. 2005.
-
3)
Cystobasidiopsis nirenbergiae R. Bauer et al., Mycol. Res. 113: 962. 2009.
Note: The emended genera Cystobasidiopsis and Ballistosporomyces can be distinguished by colony morphology and some physiological characteristics (Table 3, Fig. 3A). The species of Cystobasidiopsis form white colonies and assimilate d-arabinose, glycerol and dl-lactate, whereas the species of Ballistosporomyces form yellow-brown colonies and do not assimilate these three carbon sources.
Kurtzmanomyces Y. Yamada et al., J. Gen. Appl. Microbiol. 34: 505. 1988.
Type species: Kurtzmanomyces nectairei (Rodr. Mir.) Y. Yamada et al.
Species accepted:
-
1)
Kurtzmanomyces insolitus J.P. Samp. & Fell, Syst. Appl. Microbiol. 22: 62. 1999.
-
2)
Kurtzmanomyces nectairei (Rodr. Mir.) Y. Yamada et al., J. Gen. Appl. Microbiol. 34: 505. 1988.
-
3)
Kurtzmanomyces shapotouensis T. Zhang & L.Y. Yu, Int. J. Syst. Evol. Microbiol. 63: 3894. 2013.
-
4)
Kurtzmanomyces tardus Gim.-Jurado & van Uden, Antonie van Leeuwenhoek 58: 130. 1990.
Note: Kurtzmanomyces shapotouensis was not included in our previous phylogenetic study (Wang et al. 2015a); the sequence analysis of the LSU rRNA D1/D2 domains and ITS (including 5.8S rRNA) region indicated that it belongs to the genus Kurtzmanomyces (Zhang et al. 2013, Fig. 4 of this study).
Family Kondoaceae R. Bauer et al., Mycol. Progr. 5: 45. 2006.
Type genus: Kondoa Y. Yamada et al.
This family was proposed to accommodate the genus Kondoa that has ballistosporic phragmobasidia, as well as members of the genus Bensingtonia (Bauer et al. 2006).
Genera accepted: Bensingtonia Ingold, Kondoa Y. Yamada et al. emend. Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout.
Bensingtonia Ingold, Trans. Br. mycol. Soc. 86: 325. 1986.
Type species: Bensingtonia ciliata Ingold
Species accepted:
-
1)
Bensingtonia bomiensis F.Y. Bai & Q.M. Wang, Int. J. Syst. Evol. Microbiol. 62: 2043. 2012.
-
2)
Bensingtonia ciliata Ingold, Trans. Br. mycol. Soc. 86: 325. 1986.
-
3)
Bensingtonia naganoensis (Nakase & M. Suzuki) Nakase & Boekhout, J. Gen. Appl. Microbiol. 34: 435. 1988.
-
4)
Bensingtonia pseudonaganoensis F.Y. Bai & Q.M. Wang, Antonie van Leeuwenhoek 89: 262. 2006.
-
5)
Bensingtonia rectispora F.Y. Bai & Q.M. Wang, Int. J. Syst. Evol. Microbiol. 62: 2042. 2012.
Note: B. rectispora and B. bomiensis were not included in our previous phylogenetic study (Wang et al. 2015a), but the ITS and the D1/D2 domains of LSU rRNA sequences analysis demonstrated that they belong to Bensingtonia (Wang et al. 2012, Fig. 4 of this study).
Kondoa Y. Yamada et al., J. Gen. Appl. Microbiol. 35: 383. 1989. emend. Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout.
Type species: Kondoa malvinella (Fell & Hunter) Y. Yamada et al.
This genus is emended to include species of the genera Kondoa and Bensingtonia hitherto classified in the Kondoa clade (Wang et al. 2015a), which occurred as a well supported clade that is phylogenetically distinct from the genus Bensingtonia within Kondoaceae (Fig. 1, Fig. 4). Thus all Bensingtonia species included in the Kondoa clade will be transferred into the genus Kondoa based on the “One Fungal = One Name” principle (Hawksworth, 2011, Taylor, 2011, McNeill et al., 2012).
Sexual reproduction observed in some species. Transversely septate basidia arise directly on the hyphae. Sexual structures not known on agar media. Teliospores are not formed. Colonies cream to pinkish-cream and butyrous. Budding cells present. Pseudohyphae or true hyphae present or not. Septal pores in true hyphae ‘simple’ and uniperforate. Major CoQ system Q-9.
Species accepted:
-
1)
Kondoa aeria Á. Fonseca, J.P. Samp. & Fell, Antonie van Leeuwenhoek 77: 295.
-
2)Kondoa changbaiensis (F.Y. Bai & Q.M. Wang) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout, comb. nov. MycoBank MB813085.
- Basionym: Bensingtonia changbaiensis F.Y. Bai & Q.M. Wang, Int. J. Syst. Evol. Microbiol. 53: 2086. 2003.
-
3)
Kondoa malvinella (Fell & Hunter) Y. Yamada et al., J. Gen. Appl. Microbiol. 35: 384. 1989.
-
4)Kondoa miscanthi (Nakase & M. Suzuki) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout, comb. nov. MycoBank MB813086.
- Basionym: Sporobolomyces miscanthi Nakase & M. Suzuki, J. Gen. Appl. Microbiol. 33: 183. 1987.
- ≡ Bensingtonia miscanthi (Nakase & M. Suzuki) Nakase & Boekhout.
-
5)Kondoa phyllada (van der Walt & Y. Yamada) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout, comb. nov. MycoBank MB813087.
- Basionym: Sporobolomyces phylladus van der Walt & Y. Yamada, Antonie van Leeuwenhoek 55: 190. 1989.
- ≡ Bensingtonia phyllada (van der Walt & Y. Yamada) van der Walt et al. ex Boekhout.
- ≡ Bensingtonia phylladus (van der Walt & Y. Yamada) van der Walt et al., Nom. inval.
-
6)Kondoa sorbi (F.Y. Bai & Q.M. Wang) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout, comb. nov. MycoBank MB813088.
- Basionym: Bensingtonia sorbi F.Y. Bai & Q.M. Wang, Int. J. Syst. Evol. Microbiol. 53: 2087. 2003.
-
7)Kondoa subrosea (Nakase & M. Suzuki) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout, comb. nov. MycoBank MB813089.
- Basionym: Sporobolomyces subroseus Nakase & M. Suzuki, J. Gen. Appl. Microbiol. 33: 186. 1987.
- ≡ Bensingtonia subrosea (Nakase & M. Suzuki) Nakase & Boekhout.
-
8)Kondoa thailandica (Fungsin, Hamam. & Nakase) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout, comb. nov. MycoBank MB813090.
- Basionym: Bensingtonia thailandica Fungsin et al., Int. J. Syst. Evol. Microbiol. 51: 1209. 2001.
-
9)Kondoa yuccicola (Nakase & M. Suzuki) Q.M. Wang, M. Groenew., F.Y. Bai & Boekhout, comb. nov. MycoBank MB813091.
- Basionym: Sporobolomyces yuccicola Nakase & M. Suzuki, Antonie van Leeuwenhoek 54: 48. 1988.
- ≡ Bensingtonia yuccicola (Nakase & M. Suzuki) Nakase & Boekhout.
Note: Two sequences representing the not yet described species Kondoa myxariophila (Scorzetti et al., 2002, Fonseca, 2011) were obtained from public databases (Fig. 4).
Family Ruineniaceae Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, fam. nov. MycoBank MB813092.
Member of Agaricostilbales (Agaricostilbomycetes). The diagnosis of the family Ruineniaceae is based on the description of the genus Ruinenia. The nomenclature of the family is based on the genus Ruinenia.
Type genus: Ruinenia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Genus accepted: Ruinenia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Ruinenia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813093.
Etymology: The genus is named in honour of J. Ruinen for her contributions to the biology of yeasts from the phyllosphere.
This genus agrees with the ruber clade (Wang et al. 2015a). Member of Ruineniaceae (Agaricostilbales, Agaricostilbomycetes). The genus is mainly circumscribed by the phylogenetic analysis of seven genes, in which it occurred as a well supported clade distinct from the other genera within Agaricostilbales (Fig. 1, Fig. 4).
Sexual reproduction not known. Colonies orange-red or salmon-pink, and butyrous. Budding cells present. Hyphae and pseudohyphae present or not. Ballistoconidia present, ellipsoidal, reniform to falcate. Major CoQ system Q-10.
Type species: Ruinenia rubra (Nakase, Oakada & Sugiy.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Ruinenia clavata (F.Y. Bai & Q.M. Wang) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813094.
- Basionym: Sporobolomyces clavatus F.Y. Bai & Q.M. Wang, FEMS Yeast Res. 4: 583. 2004.
-
2)Ruinenia diospyroris (Nakase, Tsuzuki, F.L. Lee, Jindam. & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813095.
- Basionym: Sporobolomyces diospyroris (as diospyri) Nakase et al., J. Gen. Appl. Microbiol. 51: 280. 2005.
-
3)Ruinenia dracophylli (Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813096.
- Basionym: Sporobolomyces dracophylli (as dracophyllus) Hamam. & Nkase, Antonie van Leeuwenhoek 67: 168. 1995.
-
4)Ruinenia pyrrosiae (Nakase, Tsuzuki, F.L. Lee, Jindam. & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813097.
- Basionym: Sporobolomyces pyrrosiae Nakase et al., J. Gen. Appl. Microbiol. 51: 284. 2005.
-
5)Ruinenia rubra (Nakase, G. Oakada & Sugiy.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813098.
- Basionym: Ballistosporomyces ruber Nakase et al., J. Gen. Appl. Microbiol. 35: 295. 1989.
- ≡ Sporobolomyces ruber (Nakase et al.) Boekhout.
Note: The species of Ruinenia (i.e. ruber clade) form salmon-orange to red colonies, which are a unique feature in the Agaricostilbomycetes (Fig. 3A). Additionally, five sequences representing potential new species of this genus were obtained from public databases (Fig. 4).
Taxa incertae sedis in the Agaricostilbales
Jianyunia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813099.
Etymology: The genus is named in honour of Jian-Yun Zhuang, former professor at the Institute of Microbiology, Chinese Academy of Sciences, for his contributions to the taxonomic study of Pucciniales in China.
This genus agrees with the Bensingtonia sakaguchii lineage (Wang et al. 2015a). Member of Agaricostilbales (Agaricostilbomycetes). The genus is mainly circumscribed by the phylogenetic analysis of seven genes, in which it occurred as a single-species lineage distinct from the other genera within Agaricostilbales (Fig. 1, Fig. 4).
Sexual reproduction not known. Colonies ivory and butyrous. Budding cells present. Pseudohyphae present. Ballistoconidia present, kidney-shaped. Major CoQ system Q-9.
Type species: Jianyunia sakaguchii (Sugita, M. Takash., Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Jianyunia sakaguchii (Sugita, M. Takash., Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813100.
- Basionym: Bensingtonia sakaguchii Sugita et al., J. Gen. Appl. Microbiol. 43: 232. 1997.
Class Spiculogloeomycetes Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, class. nov. MycoBank MB813101.
Type order: Spiculogloeales R. Bauer et al.
Member of Pucciniomycotina. The class is mainly circumscribed by the phylogenetic analysis of seven genes, in which it formed a deep well supported lineage with affinity to Mixiomycetes within Pucciniomycotina (Fig. 1). The diagnosis of the class Spiculogloeomycetes is based on the description of the order Spiculogloeales (Bauer et al. 2006). The nomenclature of this class is based on the Spiculogloeales.
This class contains species of Spiculogloea and some species of Mycogloea and Sporobolomyces (Aime et al., 2006, Aime et al., 2014, Bauer et al., 2006, Wang et al., 2015a). Phyllozyma gen. nov. is proposed to accommodate the Sporobolomyces species in the subbrunneus clade based on the phylogenetic analysis of seven genes (Fig. 1), PRBO (Table 1), phylogenetic network analysis (Fig. 2B) and the analysis of the enlarged LSU rRNA gene dataset (Fig. 4).
Order Spiculogloeales R. Bauer et al., Mycol. Prog. 5: 41. 2006.
Type family: Spiculogloeaceae Denchev.
This order is characterised by teleomorphic members that may form tremelloid haustorial cells (nanometer-fusion mycoparasitism) and includes species of the sexual genera Spiculogloea and Mycogloea, as well as asexual species previously classified in the genus Sporobolomyces (Bauer et al. 2006).
Spiculogloeaceae Denchev, Mycol. Balcanica 6: 87. 2009.
Type genus: Spiculogloea P. Roberts.
The name Spiculogloeaceae was validated by Denchev (2009) to include the taxa of Spiculogloeales (Bauer et al. 2006).
Genera accepted: Spiculogloea P. Roberts, Phyllozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Mycogloea L.S. Olive (pro parte).
Note: The species of the genus Spiculogloea and Mycogloea are not listed here because cultures of Spiculogloea and Mycogloea, except for Mycogloea nipponica that is located in the Chionosphaeraceae, are presently not available. Moreover, nucleotide sequence data for type species of these genera are not available from public databases.
Phyllozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813102.
Etymology: The genus is named based on the habitat as all species from this clade were isolated from the leaves of plants.
This genus agrees with the subbrunneus clade (Wang et al. 2015a). Member of the Spiculogloeaceae (Spiculogloeales, Spiculogloeomycetes). The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a well supported clade (Fig. 1, Fig. 4). Species in the subbrunneus clade are associated with plant leaves (Hamamoto et al. 2011) and are ecologically different from the teleomorphic species Spiculogloea spp. and Mycogloea spp., which are mycoparasites with tremelloid haustorial cells (Roberts, 1996, Bauer, 2004, Weiß et al., 2004).
Sexual reproduction not known. Colonies pale yellowish-brown, reddish-orange and butyrous. Budding cells present. Hypha and pseudohyphae present or not. Ballistoconidia present, ellipsoidal, fusiform or sickle-shaped. Major CoQ system Q-10.
Type species: Phyllozyma subbrunnea (Nakase & M. Suzuki) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout
Species accepted:
-
1)Phyllozyma coprosmicola (Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813103.
- Basionym: Sporobolomyces coprosmicola Hamam. & Nakase, Antonie van Leeuwenhoek 67: 162. 1995.
-
2)Phyllozyma corallina (N. Furuya & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813321.
- Basionym: Sporobolomyces corallinus N. Furuya & M. Takash., Mycoscience 53: 261. 2012.
-
3)Phyllozyma dimennae (Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813104.
- Basionym: Sporobolomyces dimennae Hamam. & Nakase, Antonie van Leeuwenhoek 67: 159. 1995.
-
4)Phyllozyma linderae (Nakase, M. Takash. & Hamam.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813105.
- Basionym: Sporobolomyces linderae Nakase et al., J. Gen. Appl. Microbiol. 40: 98. 1994.
-
5)Phyllozyma novozealandica (Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813106.
- Basionym: Sporobolomyces novazealandicus Hamam. & Nakase, Antonie van Leeuwenhoek 67: 156. 1995.
-
6)Phyllozyma producta (N. Furuya & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813322.
- Basionym: Sporobolomyces productus N. Furuya & M. Takash., Mycoscience 53: 261. 2012.
-
7)Phyllozyma subbrunnea (Nakase & M. Suzuki) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813107.
- Basionym: Sporobolomyces subbrunneus Nakase & M. Suzuki, J. Gen. Appl. Microbiol. 31: 468. 1985.
Note: S. productus and S. corallinus were not included in our previous phylogenetic study (Wang et al. 2015a). These two species were placed in the subbrunneus lineage closely related to S. subbrunneus and S. dimnenae based on the sequence analysis of the D1/D2 domains of LSU rRNA (Furuya et al. 2012, Fig. 4 of the present study), and hence, they are recombined in the genus Phyllozyma.
Class Cystobasidiomycetes R. Bauer et al., Mycol. Progr. 5: 46. 2006.
Type order: Cystobasidiales R. Bauer et al.
Three orders, namely Cystobasidiales, Erythrobasidiales and Naohideales, as well as the aurantiaca, magnisporus, marina and Sakaguchia clades occur within Cystobasidiomycetes (Wang et al. 2015a). As the relative positioning of these five groups could not be resolved with certainty (Fig. 1) we suggest to treat the four clades as families but not to assign them to any order. Instead, we place them as ‘incertae sedis’ within Cystobasidiomycetes.
Buckleyzymaceae fam. nov. with Buckleyzyma gen. nov, Microsporomycetaceae fam. nov. with Microsporomyces gen. nov., Symmetrosporaceae fam. nov. with Symmetrospora gen. nov., and Sakaguchiaceae fam. nov. are proposed to accommodate the taxa in the aurantiaca, magnisporus, marina and Sakaguchia clades, respectively (Fig. 1, Fig. 5).
Order Cystobasidiales R. Bauer et al., Mycol. Progr. 5: 46. 2006.
Type family: Cystobasidiaceae Gäum.
This order was proposed to accommodate the family Cystobasidiaceae that is characterised by nanometer-fusion mycoparasitism with tremelloid haustorial cells and septal pores with cystosomes (Bauer et al. 2006).
Family Cystobasidiaceae Gäum., Vergl. Morph. Pilze (Jena): 411. 1926.
Type genus: Cystobasidium Lagerh. emend. A.M. Yurkov et al.
The family comprises the genera Occultifur and Cystobasidium, as well as species previously classified in the genus Rhodotorula in Bauer et al. (2006).
Genera accepted: Cystobasidium Lagerh. emend. A.M. Yurkov et al., Occultifur Oberw.
Cystobasidium (Lagerh.) emend. A.M. Yurkov et al., Antonie van Leeuwenhoek 107: 179. 2015.
= Jola subgen. Cystobasidium Lagerh., Bihang till Kungliga svenska Vetenskaps-Akademiens Handlingar 24: 15. 1898.
Type species: Cystobasidium fimetarium (Schumach.) P. Roberts
Species accepted:
-
1)
Cystobasidium benthicum (Nagah. et al.) A.M. Yurkov et al., Antonie van Leeuwenhoek 107: 180. 2015.
-
2)
Cystobasidium calyptogenae (Nagah. et al.) A.M. Yurkov et al., Antonie van Leeuwenhoek 107: 181. 2015.
-
3)
Cystobasidium fimetarium (Schumach.) P. Roberts, Mycologist 13: 171. 1999.
-
4)
Cystobasidium hypogymniicola Diederich & Ahti, Biblthca Lichenol. 61: 21. 1996.
-
5)
Cystobasidium laryngis (Reiersöl) A.M. Yurkov et al., Antonie van Leeuwenhoek 107: 181. 2015.
-
6)
Cystobasidium lysinophilum (Nagah. et al.) A.M. Yurkov et al., Antonie van Leeuwenhoek 107: 181. 2015.
-
7)
Cystobasidium minuta (Saito) A.M. Yurkov et al., Antonie van Leeuwenhoek 107: 180. 2015.
-
8)
Cystobasidium oligophagum (Satoh & Makimura) A.M. Yurkov et al., Antonie van Leeuwenhoek 107: 181. 2015.
-
9)
Cystobasidium pallidum (Lodder) A.M. Yurkov et al., Antonie van Leeuwenhoek 107: 181. 2015.
-
10)
Cystobasidium pinicola (F.Y. Bai et al.) A.M. Yurkov et al., Antonie van Leeuwenhoek 107: 181. 2015.
-
11)Cystobasidium portillonense (F. Laich, I. Vaca & R. Chávez) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout comb. nov. MycoBank MB813108.
- Basionym: Rhodotorula portillonensis F. Laich et al., Int. J. Syst. Evol. Microbiol. 63: 3889. 2013.
-
12)
Cystobasidium proliferans L.S. Olive, Mycologia 44: 564. 1952.
-
13)
Cystobasidium psychroaquaticum A.M. Yurkov et al., Antonie van Leeuwenhoek 107: 181. 2015.
-
14)
Cystobasidium ritchiei A.M. Yurkov et al., Antonie van Leeuwenhoek 107: 182. 2015.
-
15)
Cystobasidium sebaceum G.W. Martin, Mycologia 31: 507. 1939.
-
16)
Cystobasidium slooffiae (E.K. Novák & Vörös-Felkai) A.M. Yurkov et al., Antonie van Leeuwenhoek 107: 180. 2015.
-
17)
Cystobasidium usneicola Diederich & Alstrup, Biblthca Lichenol. 61: 25. 1996.
Note: R. portillonensis that is phylogenetically located in the Cystobasidium clade was recently proposed by Laich et al. (2013). This species was placed in this genus in the analysis of the enlarged LSU rRNA gene dataset (Fig. 5) and we transfer R. portillonensis to the genus Cystobasidium.
Occultifur Oberw., Rep. Tottori Mycol. Inst. 28: 119. 1990.
Type species: Occultifur internus (L.S. Olive) Oberw.
Species accepted:
-
1)
Occultifur brasiliensis Gomes et al., Antonie van Leeuwenhoek 107: 608. 2015.
-
2)
Occultifur corticiorum P. Roberts, Mycotaxon 63: 202. 1997.
-
3)
Occultifur externus J.P. Samp. et al., Mycologia 91: 1095. 1999.
-
4)
Occultifur internus (L.S. Olive) Oberw., Rep. Tottori Mycol. Inst. 28: 120. 1990.
-
5)
Occultifur kilbournensis Kurtzman & Robnett, Antonie Van Leeuwenhoek 107: 1325. 2015.
-
6)
Occultifur tropicalis Khunnamwong et al., Int. J. Syst. Evol. Microbiol. 65: 1580. 2015.
Notes: Three species of the genus Occultifur, O. corticiorum, O. externus and the generic type O. internus, were accepted by Sampaio & Oberwinkler (2011), but only O. externus was included in our previous phylogenetic study (Wang et al. 2015a) as neither living cultures nor molecular data are available for the other two species. Three new asexual members of the genus Occultifur (Fig. 5), namely O. brasiliensis, O. kilbournensis and O. tropicalis, were recently described by Gomes et al., 2015, Kurtzman and Robnett, 2015 and Khunnamwong et al. (2015), respectively.
Order Erythrobasidiales R. Bauer et al., Mycol. Progr. 5: 46. 2006.
Type family: Erythrobasidiaceae Denchev.
This order accommodates the genera Erythrobasidium and Bannoa that have non-tremelloid haustorial cells, septal pores without cystosomes, and coenzyme CoQ10 (H2), as well as some Rhodotorula and Sporobolomyces species (Bauer et al. 2006). The genus Cyrenella was not placed in this order by Aime et al., 2006, Bauer et al., 2006 and Hibbett et al. (2007). This genus, however, formed a well supported clade with the genera Erythrobasidium and Bannoa, and Rhodotorula lactosa, which is placed in a new genus Hasegawazyma (Fig. 1). Thus, this order includes the family Erythrobasidiaceae and the genera Cyrenella and Hasegawazyma, which are treated as ‘incertae sedis’ in the Erythrobasidiales.
Family Erythrobasidiaceae Denchev, Mycol. Balcanica 6: 87. 2009.
Type genus: Erythrobasidium Hamam. et al.
The name Erythrobasidiaceae was validated by Denchev (2009) to include the taxa of Erythrobasidiales (Bauer et al. 2006).
Genera accepted: Bannoa Hamam., Erythrobasidium Hamam. et al.
Bannoa Hamam., Int. J. Syst. Evol. Microbiol. 52: 1027. 2002. emend. Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Type species: Bannoa hahajimensis Hamam. et al.
This genus is emended to include species of Bannoa and related Sporobolomyces species that hitherto were classified in the Bannoa clade (Wang et al. 2015a), which occurred as a well supported clade related to Erythrobasidium within Erythrobasidiales (Fig. 1, Fig. 5).
Sexual reproduction observed in some species. Clamp connections present. Teliospores not formed. Unicellular basidia arise laterally on a clamp connection, or terminally at the hyphae. Cells of the basidia germinate with hyphae, from which yeast cells originate. Colonies orange to salmon-red. Budding cells present. Pseudohyphae absent. Ballistoconidia present or not, ovoid and ellipsoidal. Major CoQ system Q-10(H2).
Species accepted:
-
1)Bannoa bischofiae (Hamam., Thanh & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813109.
- Basionym: Sporobolomyces bischofiae Hamam. et al., Int. J. Syst. Evol. Microbiol. 52: 1029. 2002.
-
2)
Bannoa hahajimensis Hamam. et al., Int. J. Syst. Evol. Microbiol. 52: 1028. 2002.
-
3)Bannoa syzygii (Hamam., Thanh & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813110.
- Basionym: Sporobolomyces syzygii Hamam. et al., Int. J. Syst. Evol. Microbiol. 52: 1031. 2002.
-
4)Bannoa ogasawarensis (Hamam., Thanh & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813111.
- Basionym: Sporobolomyces ogasawarensis Hamam. et al., Int. J. Syst. Evol. Microbiol. 52: 1030. 2002.
Note: Our analyses suggest that Bannoa sp. MP3490 (AFTOL-ID 1921) represents a potentially new species of this genus (Wang et al. 2015a, Fig. 5 of this study).
Erythrobasidium Hamam., Sugiy. & Komag., J. Gen. Appl. Microbiol. 34: 285. 1988. emend. Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout
Type species: Erythrobasidium hasegawianum Hamam. et al.
This genus is emended to include species of Erythrobasidium and related Sporobolomyces species that hitherto were classified in the Erythrobasidium clade (Wang et al. 2015a), which occurred as a well supported clade closely related to Bannoa within Erythrobasidiales (Fig. 1, Fig. 5).
Sexual reproduction observed in some species. Hyphae form from single cells without mating. Clamp connections present or absent. Septal pores ‘simple’. Teliospores not formed. Unicellular basidia (holobasidia) arise by the formation of lateral protrusions on the hyphae. Sessile basidiospores produced terminally on the holobasidia and not forcibly discharged. Colonies orange-red. Budding cells present. Pseudohyphae absent. Ballistoconidia present or not, ellipsoidal or ovoid. Major CoQ system Q-10(H2).
Species accepted:
-
1)Erythrobasidium elongatum (R.G. Shivas & Rodr. Mir.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813112.
- Basionym: Sporobolomyces elongatus R.G. Shivas & Rodr. Mir., Antonie van Leeuwenhoek 49: 160. 1983.
-
2)
Erythrobasidium hasegawianum Hamam. et al., J. Gen. Appl. Microbiol. 37: 131. 1991.
-
3)Erythrobasidium yunnanense (F.Y. Bai, M. Takash., Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813113.
- Basionym: Sporobolomyces yunnanensis F.Y. Bai et al., Int. J. Syst. Evol. Microbiol. 51: 234. 2001.
Taxa incertae sedis in the Erythrobasidiales
Cyrenella Goch., Mycotaxon 13: 268. 1981.
Type species: Cyrenella elegans Goch.
Species accepted:
-
1)
Cyrenella elegans Goch., Mycotaxon 13: 268. 1981.
Hasegawazyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813114.
Etymology: The genus is named in honour of T. Hasegawa who firstly described the species Rhodotorula lactosa.
This genus corresponds to the Rhodotorula lactosa lineage (Wang et al. 2015a, Fig. 5 of this study). Member of Erythrobasidiales (Cystobasidiomycetes). The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a single-species lineage distinct from the other genera within Cystobasidiomycetes (Fig. 1, Fig. 5). This genus differs from Bannoa, Cyrenella and Erythrobasidium by having ubiquinone Q-9.
Sexual reproduction not known. Colonies pink-coloured and butyrous. Budding cells present. Pseudohyphae and true hyphae not observed. Ballistoconidia absent. Major CoQ system Q-9.
Type species: Hasegawazyma lactosa (Hasegawa) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout
Species accepted:
-
1)Hasegawazyma lactosa (Hasegawa) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813115.
- Basionym: Rhodotorula lactosa Hasegawa, J. Gen. Appl. Microbiol. 5: 31. 1959.
Order Naohideales R. Bauer et al., Mycol. Progr. 5: 46. 2006.
Type family: Naohideaceae Denchev
This order was proposed to accommodate the genus Naohidea, which is characterised by nanometer-fusion mycoparasitism with intracellular haustoria (tremelloid) and septal pores without cystosomes (Bauer et al. 2006).
Family Naohideaceae Denchev, Mycol. Balcanica 6: 87. 2009.
Type genus: Naohidea Oberw.
The name Naohideaceae was validated by Denchev (2009) to include the taxa of Naohideales (Bauer et al. 2006).
Genus accepted: Naohidea Oberw.
Naohidea Oberw., Rep. Tottori Mycol. Inst. 28: 114. 1990.
Type species: Naohidea sebacea (Berk. & Broome) Oberw.
The genus Naohidea, representing the order Naohideales, produces cream-coloured cultures that are different from all other taxa in the Cystobasidiomycetes, which form pink to orange-red pigmented colonies (Sampaio & Chen 2011, Fig. 3B of this study).
Species accepted:
-
1)
Naohidea sebacea (Berk. & Broome) Oberw., Rep. Tottori Mycol. Inst. 28: 114. 1990.
Taxa incertae sedis in the Cystobasidiomycetes
Family Symmetrosporaceae Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, fam. nov. MycoBank MB813116.
Member of the Cystobasidiomycetes. The diagnosis of the family Symmetrosporaceae is based on the description of the genus Symmetrospora. The nomenclature of the family is based on the genus Symmetrospora.
Type genus: Symmetrospora Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Genus accepted: Symmetrospora Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Symmetrospora Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813117.
Etymology: The genus is named by the (almost) symmetrical ballistoconidia that are formed by ballistoconidia-forming species of this clade.
This genus agrees with the marina clade (Wang et al. 2015a). Member of the Symmetrosporaceae (Cystobasidiomycetes). The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a sister clade to Erythrobasidiales within Cystobasidiomycetes (Fig. 1, Fig. 5).
Sexual reproduction not known. Colonies orange-red and butyrous. Budding cells present. Hypha and pseudohyphae not observed. Ballistoconidia present or not, symmetrical or nearly symmetrical, ellipsoidal or ovoidal. Major CoQ system Q-10.
Type species: Symmetrospora gracilis (Derx) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout
Species accepted:
-
1)Symmetrospora coprosmae (Hamam. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813118.
- Basionym: Sporobolomyces coprosmae Hamam. & Nakase, Antonie van Leeuwenhoek 67: 166. 1995.
-
2)Symmetrospora foliicola (R.G. Shivas & Rodr. Mir.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813119.
- Basionym: Sporobolomyces foliicola R.G. Shivas & Rodr. Mir., Antonie van Leeuwenhoek 49: 162. 1983.
-
3)Symmetrospora gracilis (Derx) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813120.
- Basionym: Sporobolomyces gracilis Derx, Annls mycol. 28: 18. 1930.
-
4)Symmetrospora vermiculata (M. Takash. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813121.
- Basionym: Sporobolomyces vermiculatus M. Takash. & Nakase, Mycoscience 41: 367. 2000.
-
5)Symmetrospora marina (Phaff, Mrak & Williams) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813123.
- Basionym: Rhodotorula marina Phaff et al., Mycologia 44: 436. 1952.
-
6)Symmetrospora symmetrica (F.Y. Bai & Q.M. Wang) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813125.
- Basionym: Sporobolomyces symmetricus F.Y. Bai & Q.M. Wang, FEMS Yeast Res. 4: 584. 2004.
Family Buckleyzymaceae Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, fam. nov. MB813126.
Member of the Cystobasidiomycetes. The diagnosis of the family Buckleyzymaceae is based on the description of the genus Buckleyzyma. The nomenclature of the family is based on the genus Buckleyzyma.
Type genus: Buckleyzyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Genus accepted: Buckleyzyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Buckleyzyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813127.
Etymology: The genus is named in honour of Helen R. Buckley for her contributions to yeast taxonomy.
This genus agrees with the aurantiaca clade (Wang et al. 2015a). Member of Buckleyzymaceae (Cystobasidiomycetes). The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a sister clade to Erythrobasidiales and Symmetrospora within Cystobasidiomycetes (Fig. 1, Fig. 5).
Sexual reproduction not known. Colonies brownish-orange or orange and butyrous. Budding cells present. Hyphae and pseudohyphae present or not. Ballistoconidia present or not, ellipsoidal allantoid to amygdaliform. Major CoQ system Q-10.
Type species: Buckleyzyma aurantiaca (Saito) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout
Species accepted:
-
1)Buckleyzyma armeniaca (R.G. Shivas & Rodr. Mir.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813133.
- Basionym: Rhodotorula armeniaca R.G. Shivas & Rodr. Mir., Antonie van Leeuwenhoek 49: 163. 1983.
-
2)Buckleyzyma aurantiaca (Saito) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813128.
- Basionym: Torula aurantiaca Saito, Jap. J. Bot. 1: 45. 1922.
- ≡ Rhodotorula aurantiaca (Saito) Lodder
-
3)Buckleyzyma kluyveri-nielii (van der Walt) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813130.
- Basionym: Sporobolomyces kluyveri-nielii van der Walt, Antonie van Leeuwenhoek 52: 432. 1986.
-
4)Buckleyzyma phyllomatis (van der Walt & Y. Yamada) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813131.
- Basionym: Sporobolomyces phyllomatis van der Walt & Y. Yamada, Antonie van Leeuwenhoek 54: 202. 1988.
-
5)Buckleyzyma salicina (B.N. Johri & Bandoni) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813132.
- Basionym: Bullera salicina B.N. Johri & Bandoni, Taxonomy of Fungi (Proc. int. Symp. Madras 1973) Part 2 (Madras) 2: 544. 1984.
- ≡ Sporobolomyces salicinus (B.N. Johri & Bandoni) Nakase & M. Itoh
Family Microsporomycetaceae Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, fam. nov. MycoBank MB813134.
Member of Cystobasidiomycetes. The diagnosis of the family Microsporomycetaceae is based on the description of the genus Microsporomyces. The nomenclature of the family is based on the genus Microsporomyces.
Type genus: Microsporomyces Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Genus accepted: Microsporomyces Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Microsporomyces Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813135.
Etymology: The genus name relates to the type species, Sporobolomyces magnisporus that produces small (micro-)ballistoconidia.
This genus agrees with the magnisporus clade (Wang et al. 2015a). Member of the Microsporomycetaceae (Cystobasidiomycetes). The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a sister clade to Cystobasidiales and Sakaguchia within the Cystobasidiomycetes (Fig. 1, Fig. 5).
Sexual reproduction not known. Colonies orange or salmon-coloured and butyrous. Budding cells present. Pseudohyphae present or not. Ballistoconidia present or not, ellipsoidal, allantoid to amygdaliform. Major CoQ system Q-10.
Type species: Microsporomyces magnisporus (Nakase, Tsuzuki, F.L. Lee, Sugita, Jindam. & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Microsporomyces bloemfonteinensis (Pohl, M.S. Smit & Albertyn) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB814789.
- Basionym: Rhodotorula bloemfonteinensis Pohl et al., Int. J. Syst. Evol. Microbiol. 61: 2324. 2011.
-
2)Microsporomyces magnisporus (Nakase, Tsuzuki, F.L. Lee, Sugita, Jindam. & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813137.
- Basionym: Sporobolomyces magnisporus Nakase et al., J. Gen. Appl. Microbiol. 49: 341. 2003.
-
3)Microsporomyces pini (Pohl, M.S. Smit & Albertyn) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813138.
- Basionym: Rhodotorula pini Pohl et al., Int. J. Syst. Evol. Microbiol. 61: 2323. 2011.
-
4)Microsporomyces orientalis (Pohl, M.S. Smit & Albertyn) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813327.
- Basionym: Rhodotorula orientalis (as orientis) Pohl et al., Int. J. Syst. Evol. Microbiol. 61: 2325. 2011.
Family Sakaguchiaceae Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, fam. nov. MycoBank MB813142.
Member of the Cystobasidiomycetes. The diagnosis of the family Sakaguchiaceae is based on the description of the genus Sakaguchia. The nomenclature of the family is based on the genus Sakaguchia.
Type genus: Sakaguchia Y. Yamada et al.
Genus accepted: Sakaguchia Y. Yamada et al. emend. Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Sakaguchia Y. Yamada et al., Biosc. Biotechn. Biochem. 58: 102. 1994. emend. Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Type species: Sakaguchia dacryoidea (Fell et al.) Y. Yamada et al.
This genus is emended to include Sakaguchia dacryoidea and related Rhodotorula species that hitherto were classified in the Sakaguchia clade (Wang et al. 2015a), which occurred as a well supported clade within the Cystobasidiomycetes (Fig. 1, Fig. 5).
Sexual reproduction in some species. Clamp connections present. Teliospores laterally or terminally on the hyphae. Teliospores germinate with two- to four-celled metabasidium with lateral and terminal basidiospores. Colonies red or orange-red and butyrous. Budding cells present. Pseudohyphae or true hyphae present or not. Ballistoconidia not produced. Major CoQ system Q-10.
Species accepted:
-
1)Sakaguchia cladiensis (Fell, Statzell & Scorzetti) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813144.
- Basionym: Rhodotorula cladiensis Fell et al., Antonie van Leeuwenhoek 99: 546. 2011.
-
2)
Sakaguchia dacryoidea (Fell et al.) Y. Yamada et al., Biosc. Biotechn. Biochem. 58: 102. 1994.
-
3)Sakaguchia lamellibrachiae (Nagah., Hamam., Nakase & Horikoshi) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813147.
- Basionym: Rhodotorula lamellibrachiae (as lamellibrachii) Nagah. et al., Antonie van Leeuwenhoek 80: 320. 2001.
-
4)Sakaguchia meli (Libkind, van Broock & J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813148.
- Basionym: Rhodotorula meli Libkind et al., Int. J. Syst. Evol. Microbiol. 60: 2253. 2010.
-
5)Sakaguchia oryzae (F.Y. Bai & Y.M. Cai) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813150.
- Basionym: Rhodotorula oryzae F.Y. Bai & Y.M. Cai, Antonie van Leeuwenhoek 86: 296. 2004.
Note: Our analyses suggest that Rhodotorula sp. JCM 8162 represents a potential new species of this genus, as revealed by the multi-gene analyses (Wang et al. 2015a) and the analysis of the enlarged LSU rRNA gene dataset (Fig. 5).
Class Microbotryomycetes R. Bauer et al., Mycol. Progr. 5: 47. 2006.
Type order: Microbotryales R. Bauer & Oberw.
This class presently contains four orders that contain yeast species, namely Kriegeriales, Leucosporidiales, Microbotryales and Sporidiobolales (Aime et al., 2006, Aime et al., 2014, Bauer et al., 2006, Hibbett et al., 2007, Toome et al., 2013). Ten clades and seven single-species lineages do not belong to the above listed orders (Wang et al. 2015a), and they may be assigned into eleven families based on modified GMYC analysis (Table 2) in conjunction with the phylogenetic analysis of seven genes (Fig. 1). However, most of these suggested families are monotypic or poorly sampled (Table 2). Thus, we propose Chrysozymaceae fam. nov. to accommodate the griseoflavus, yamatoana, singularis and Sporobolomyces inositophilus clades, and Colacogloeaceae fam. nov. to cover the Colacogloea clade because these two proposed families have more taxa than the other clades and have strong support values (Fig. 1). The other six clades and six single-species lineages are proposed to be included in twelve genera that at the higher rank are considered as ‘incertae sedis’ within the Microbotryomycetes at present. The species Rhodotorula svalbardensis (Singh et al. 2014) was not included in the seven genes-based phylogenetic analysis (Wang et al. 2015a) and has no proper placement in the constrained LSU analysis (Fig. 6). The phylogenetic placement of this species needs to be addressed by a robust molecular analysis. Thus, we leave it as Rhodotorula svalbardensis pro tem. in this study. The term “pro tem.” (pro tempore) was proposed in Wang et al. (2015b) and indicates a temporary taxonomic placement.
Order Kriegeriales Toome & Aime, Mycologia 105: 489. 2013.
Type family: Kriegeriaceae Toome & Aime.
This order was proposed to contain the families Kriegeriaceae and Camptobasidiaceae (Toome et al. 2013). The teleomorphic members of this order are characterised by the presence of ‘simple’ septal pores and subgloboid spindle pole bodies (Toome et al. 2013).
Family Kriegeriaceae Toome & Aime, Mycologia 105: 489. 2013.
Type genus: Kriegeria Bres.
This family was proposed to accommodate the sexual plant parasite Kriegeria, the asexual genus Meredithblackwellia, and six related Rhodotorula species (Toome et al. 2013). The species R. pilati was placed in the Kriegeriaceae by Toome et al. (2013), but this affiliation was not supported in the muligene study by Wang et al. (2015a). This species was located in the tsugae clade (Wang et al. 2015a). The other five Rhodotorula species in the Kriegeriaceae are reclassified into two new genera Phenoliferia and Yamadamyces.
Genera accepted: Kriegeria Bres., Meredithblackwellia Toome & Aime, Phenoliferia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Yamadamyces Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout
Kriegeria Bres., Revue mycol. Toulouse 13: 14. 1891.
= Zymoxenogloea D.J. McLaughlin & Doublés, Mycologia 84: 671. 1992.
Type species: Kriegeria eriophori Bres.
Species accepted:
-
1)
Kriegeria eriophori Bres., Revue mycol. Toulouse 13: 14. 1891.
Meredithblackwellia Toome & Aime, Mycologia 105: 490. 2013.
Type species: Meredithblackwellia eburnea Toome & Aime.
Species accepted:
-
1)
Meredithblackwellia eburnea Toome & Aime, Mycologia 105: 491. 2013.
Phenoliferia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813152.
Etymology: The genus is named because the species in this clade can assimilate phenol.
This genus agrees with the glacialis clade (Wang et al. 2015a). Member of the Kriegeriaceae (Kriegeriales, Microbotryomycetes). The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a well supported clade related to the genus Kriegeria within Kriegeriaceae (Fig. 1, Fig. 6).
Sexual reproduction not known. Colonies creamy-white and butyrous. Budding cells present. Pseudomycelium or true hyphae not observed. Ballistoconidia not produced. Major CoQ system unknown.
Type species: Phenoliferia psychrophenolica (Margesin & J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout
Species accepted:
-
1)Phenoliferia psychrophenolica (Margesin & J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813153.
- Basionym: Rhodotorula psychrophenolica Margesin & J.P. Samp., Int. J. Syst. Evol. Microbiol. 57: 2183. 2007.
-
2)Phenoliferia psychrophila (Margesin & J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813154.
- Basionym: Rhodotorula psychrophila Margesin & J.P. Samp., Int. J. Syst. Evol. Microbiol. 57: 2181. 2007.
-
3)Phenoliferia glacialis (Margesin & J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813155.
- Basionym: Rhodotorula glacialis Margesin & J.P. Samp., Int. J. Syst. Evol. Microbiol. 57: 2183. 2007.
-
4)Phenoliferia himalayensis (Shivaji, Bhadra & Rao) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813156.
- Basionym: Rhodotorula himalayensis Shivaji et al., Extremophiles 12: 380. 2008.
Notes: Species of Phenoliferia (i.e. the glacialis clade) have similar assimilation patterns of carbon and nitrogen compounds (Margesin et al. 2007). They share the ability to grow on raffinose as the sole carbon source but not on maltose. In contrast, closely related taxa, such as Kriegeria eriophori and Rhodotorula rosulata, have different assimilation properties and are able to utilise maltose, but not raffinose (Table 3). R. himalayensis was not included in our previous phylogenetic study (Wang et al. 2015a). This species was found to be closely related to R. psychrophila and R. glacialis based on the sequence analyses of LSU rRNA D1/D2 domains and ITS (Shivaji et al., 2008, Turchetti et al., 2011, Toome et al., 2013, Singh et al., 2014, Fig. 6 of this study), and, hence, it is recombined in the genus Phenoliferia.
Yamadamyces Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813326.
Etymology: The genus is named in honour of Y. Yamada because of his contributions to the taxonomy of yeasts.
This genus corresponds to the Rhodotorula rosulata lineage (Wang et al. 2015a). Member of the Kriegeriaceae (Kriegeriales, Microbotryomycetes). The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a single-species lineage within the Kriegeriaceae (Fig. 1, Fig. 6).
Sexual reproduction not known. Colonies greyish-cream and butyrous. Budding cells present. Pseudomycelium present. Ballistoconidia not produced. Major CoQ system unknown.
Type species: Yamadamyces rosulatus (Golubev & Scorzetti) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Yamadamyces rosulatus (Golubev & Scorzetti) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813578.
- Basionym: Rhodotorula rosulata Golubev & Scorzetti, Int. J. Syst. Evol. Microbiol. 60: 2503. 2010.
Notes: The species R. rosulata was found to differ from K. eriophori in the utilisation of three carbon sources, namely methyl-α-d-glucoside, l-arabinose and myo-inositol (Table 3). The micromorphology of the species resembles that of Me. eburnea and K. eriophori with budding cells remaining connected to each other at the base and forming clusters (or rosettes) of cells.
Family Camptobasidiaceae R.T. Moore, Mycotaxon 59: 8. 1996.
Type genus: Camptobasidium Marvanová & Suberkr.
The family Camptobasidiaceae was described to accommodate a teleomorphic aquatic fungus Camptobasidium (Marvanová & Suberkropp 1990) that was placed in the Atractiellales (Atractiellomycetes) by Moore (1996). Aime et al. (2006) indicated that Camptobasidium belongs to Microbotryomycetes rather than Atractiellomycetes based on rRNA sequence analysis.
Genera accepted: Camptobasidium Marvanová & Suberkr., Glaciozyma Turchetti et al.
Note: Our previous study (Wang et al. 2015a) did not support the family Camptobasidiaceae as belonging to the Kriegeriales, and, therefore, we consider the family as ‘incertae sedis’ in Microbotryomycetes.
Camptobasidium Marvanová & Suberkr., Mycologia 82: 209. 1990.
= Crucella Marvanová & Suberkr., Mycologia 82: 212. 1990.
Type species: Camptobasidium hydrophilum Marvanová & Suberkr.
Species accepted:
-
1)
Camptobasidium hydrophilum Marvanová & Suberkr., Mycologia 82: 209. 1990.
Notes: Camptobasidium hydrophilum is a slow-growing psychrophilic fungus without a yeast stage but with a tetraradiate anamorph, Crucella subtilis. In co-culturing experiments with aquatic hyphomycetes Camptobasidium hydrophilum behaves like a contact biotrophic mycoparasite and its hyphae coil around host hyphae or conidia. The penetration of the host by hyphae has, however, not been reported (Marvanová & Suberkropp 1990).
Glaciozyma Turchetti et al., Extremophiles 15: 579. 2011.
Type species: Glaciozyma antarctica (Fell et al.) Turchetti et al.
Species accepted:
-
1)
Glaciozyma antarctica (Fell et al.) Turchetti et al., Extremophiles 15: 579. 2011.
-
2)
Glaciozyma litoralis A.V. Kachalkin, Antonie van Leeuwenhoek 105: 1080. 2014.
-
3)
Glaciozyma martinii Turchetti et al., Extremophiles 15: 579. 2011.
-
4)
Glaciozyma watsonii Turchetti et al., Extremophiles 15: 582. 2011.
Note: Only one species of the genus Glaciozyma, namely G. antarctica, was employed in our previous study (Wang et al. 2015a) because G. litoralis, G. martini and G. watsonii were only recently published (Turchetti et al., 2011, Kachalkin, 2014). These species were included in the analysis of the enlarged LSU rRNA gene dataset (Fig. 6), and the genus Glaciozyma received high support in ML and MP analyses.
Order Leucosporidiales J.P. Samp. et al., Mycol. Prog. 2: 61. 2003.
Type family: Leucosporidiaceae J.P. Samp. et al.
This order was proposed to accommodate the asexual or sexual, non-phytoparasitic members of the Microbotryomycetidae with white to cream-coloured colonies (Sampaio et al. 2003). The order includes Mastigobasidium and Leucosporidium fellii, and the family Leucosporidiaceae in Sampaio et al. (2003). Members of this order have colacosomes (lenticular bodies) and represent potential mycoparasites (Sampaio et al. 2003).
Family Leucosporidiaceae J.P. Samp. et al., Mycol. Prog. 2: 63. 2003.
Type genus: Leucosporidium Fell et al. emend. V. de Garcia et al.
This family was proposed to accommodate the genera Leucosporidiella and Leucosporidium excluding the species L. fellii and L. fasciculatum, and Mastigobasidium (Sampaio et al. 2003). The genera Mastigobasidium and Leucosporidiella were recently proposed as a synonym of Leucosporidium (de García et al. 2015). Thus, the family Leucosporidiaceae presently includes only the genus Leucosporidium (Fig. 7).
Genus accepted: Leucosporidium Fell et al. emend. V. de Garcia et al.
Leucosporidium Fell et al., Antonie van Leeuwenhoek 35: 438. 1969. emend. V. de Garcia et al., FEMS Yeast Res. 15: 9. 2015.
= Mastigobasidium Golubev, Int. J. Syst. Bacter. 49: 49. 1999
= Leucosporidiella Sampaio, Mycol. Progr. 2: 63. 2003
Type species: Leucosporidium scottii Fell et al.
Species accepted:
-
1)
Leucosporidium creatinivorum (Golubev) V. de Garcia et al., FEMS Yeast Res. 15: 9. 2015.
-
2)
Leucosporidium drummii A.M. Yurkov et al., Int. J. Syst. Evol. Microbiol. 62: 730. 2012.
-
3)
Leucosporidium escuderoi Vaca et al., Antonie van Leeuwenhoek 105: 599. 2013.
-
4)
Leucosporidium fellii Gim.-Jurado & Uden, Antonie van Leeuwenhoek 55: 134. 1989.
-
5)
Leucosporidium golubevii Gadanho et al., Mycol. Progr. 2: 57. 2003.
-
6)
Leucosporidium scottii Fell et al., Antonie van Leeuwenhoek 35: 440. 1969.
-
7)
Leucosporidium yakuticum (Golubev) V. de Garcia et al., FEMS Yeast Res. 15: 9. 2015.
-
8)
Leucosporidium muscorum (Di Menna) V. de Garcia et al., FEMS Yeast Res. 15: 9. 2015.
-
9)
Leucosporidium intermedium (Golubev) V. de Garcia et al., FEMS Yeast Res. 15: 9. 2015.
-
10)
Leucosporidium fragarium (J.A. Barnett & Buhagiar) V. de Garcia et al., FEMS Yeast Res. 15: 9. 2015.
Order Microbotryales R. Bauer & Oberw., Can. J. Bot. 75: 1309. 1997.
Type family: Microbotryaceae R.T. Moore.
This order was proposed to accommodate phytoparasitic taxa lacking colacosomes (lenticular bodies) and teliospores (Bauer et al., 1997, Bauer et al., 2006). The order includes the families Microbotryaceae and Ustilentylomataceae. No yeast species are included in the family Microbotryaceae, but yeast stages of taxa in this family occur (Fig. 3D).
Family Ustilentylomataceae R. Bauer & Oberw., Can. J. Bot. 75: 1311. 1997.
Type genus: Ustilentyloma Savile.
This family was proposed to include taxa with ‘simple’ septal pores in the Microbotryales (Bauer et al. 1997).
Genera accepted: Aurantiosporium M. Piepenbr. et al., Fulvisporium Vánky, Ustilentyloma Savile, Microbotryozyma S.O. Suh et al.
Note: The species of the genera Aurantiosporium and Fulvisporium are not listed here because no yeast phase has been observed in these two genera. Representatives of these genera were included in the analysis of the enlarged LSU rRNA gene dataset (Fig. 7).
Microbotryozyma S.O. Suh et al., Antonie van Leeuwenhoek 102: 102. 2012.
Type species: Microbotryozyma collariae S.O. Suh et al.
Species accepted:
-
1)
Microbotryozyma collariae S.O. Suh et al., Antonie van Leeuwenhoek 102: 103. 2012.
Note: Mi. collariae is a yeast species within Ustilentylomataceae, which was recently described to accommodate two strains isolated from the intestine of a plant bug (Collaria oleosa, Miridae, Heteroptera, Insecta) by Suh et al. (2012). This anamorphic species is distantly related to any teleomorphic parasitic taxa in this family (Fig. 7).
Ustilentyloma Savile, Can. J. Bot. 42: 708. 1964. emend. Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Type species: Ustilentyloma pleuropogonis Savile.
This genus was originally described for teleomorphic fungi occurring on Poaceae (Savile and Parmelee, 1964, Vánky, 2002). Here it is emended to include free-living yeast species with unknown sexual states.
Species accepted:
-
1)
Ustilentyloma pleuropogonis Savile, Can. J. Bot. 42: 708. 1964.
-
2)Ustilentyloma brefeldii (Krieg.) Vánky, Mycotaxon 41: 491. 1991.
- Basionym: Entyloma brefeldii Krieg., Hedwigia 35(Beibl.): 145. 1896.
-
3)
Ustilentyloma fluitans (Liro) Vánky, Microbiologia Bucuresti 1: 328. 1970.
-
4)
Ustilentyloma oreochloae (Durrieu) Vánky, Mycotaxon 78: 319. 2001.
-
5)Ustilentyloma graminis (Rodr. Mir. & H.G. Diem) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813157.
- Basionym: Candida graminis Rodr. Mir. & H.G. Diem, Can. J. Bot. 52: 279. 1974.
- = Rhodotorula hordea Rodr. Mir. & Weijman.
Notes: The close relatives of R. hordea are plant parasite species of Ustilentyloma based on the sequence analysis of the D1/D2 domains of the LSU rRNA (Sampaio 2011, Fig. 7 of this study) and the ITS (including 5.8S rRNA) region. Nucleotide sequences of R. hordea differ from Ustilentyloma fluitans (GenBank number: AF009882 and KC994459) and Ustilentyloma brefeldii (GenBank number: EF622016) by 1 and 15 positions in the D1/D2 domains of the LSU rRNA, respectively. However, R. hordea differs from U. fluitans by 78 mismatches and gaps (12 %) in the ITS region (GenBank number: AY212990 and KC994460), and by the assimilation of d-galactose, l-sorbose, d-ribose, lactose, erythritol, ribitol and protocatechuic acid (Sampaio 2011). The genus Ustilentyloma established by Savile & Parmelee (1964) contains U. brefeldii, U. fluitans, U. oreochloae and U. pleuropogonis. All species of this genus are parasites infecting leaves of Poaceae. R. hordea isolated from the leaves of six-row barley (Hordeum vulgare subsp. hexastichium, Poaceae) may have a similar ecology as a phyllosphere inhabitant as the species of Ustilentyloma, although it is not known to be a parasite on barley. One possibility may be that R. hordea represents a yeast stage of a dimorphic phytoparasite that is distinct from U. fluitans. Sequences of U. pleuropogonis, the type species of Ustilentyloma, are not available at present, but the above mentioned sequence analysis of U. brefeldii and U. fluitans indicated that this genus may be monophyletic (Fig. 7). The morphological characters of this genus also support its monophyletic nature (Vánky 2002). Based on the above discussion we prefer to propose R. hordea as a new combination of Ustilentyloma. The name Rhodotorula hordea was chosen to replace Candida graminis when it was transferred to Rhodotorula by Weijman et al. (1998) because the specific epithet ‘graminis’ was already used for the species Rhodotorula graminis. Here we reuse this epithet for Ustilentyloma graminis.
Order Sporidiobolales J.P. Samp. et al., Mycol. Progr. 2: 66. 2003.
Type family: Sporidiobolaceae (R.T. Moore) J.P. Samp. et al.
This order was proposed to include the family Sporidiobolaceae (Sampaio et al. 2003).
Family Sporidiobolaceae R.T. Moore, Bot. Mar. 23: 361. 1980. emend. J.P. Samp. et al., Mycol. Progr. 2: 66. 2003.
Type genus: Sporidiobolus Nyland.
This family was emended to include Sporidiobolus and Rhodosporidium that have colacosomes (lenticular bodies), ‘simple’ septal pores and teliospores, and some species that are hitherto classified in Sporobolomyces and Rhodotorula (Sampaio et al. 2003). All known species in this family have pink-coloured cultures (Sampaio et al. 2003, Fig. 3C of this study).
Genera accepted: Rhodotorula F.C. Harrison emend. Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Rhodosporidiobolus Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Sporobolomyces Kluyver & C.B. Niel emend. Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Rhodotorula F.C. Harrison, Proc. & Trans. Roy. Soc. Canada ser. 3 21: 349. 1927. emend. Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
= Rhodosporidium I. Banno, J. Gen. Appl. Microbiol. 13: 192. 1967.
Type species: Rhodotorula glutinis (Fresen.) F.C. Harrison.
This genus is emended to include Rhodotorula species and their sexual counterpart Rhodosporidium in the Rhodosporidium clade (Wang et al. 2015a), which is a well supported clade within Sporidiobolaceae (Sporidiobolales). The Rhodosporidium clade is composed of Rhodotorula glutinis, the type species of Rhodotorula, and Rhodosporidium toruloides, the type species of Rhodosporidium (Fig. 1, Fig. 8). The name Rhodotorula is older than Rhodosporidium, and has taxonomic priority over the latter.
Sexual reproduction observed in some species. Clamp connections present. Teliospores may be formed and produce transversely septate basidia. The basidiospores ovoid, passively released and germinate by budding. Colonies red and butyrous. Budding cells present. Pseudohyphae or true hyphae present or not. Ballistoconidia formed or not, ellipsoidal. Major CoQ systems Q-9 or Q-10.
Species accepted:
-
1)Rhodotorula alborubescens (Derx) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813323.
- Basionym: Sporobolomyces alborubescens Derx, Annls mycol. 28: 15. 1930.
-
2)
Rhodotorula araucariae Grinb. & Yarrow, Antonie van Leeuwenhoek 36: 455. 1970.
-
3)Rhodotorula babjevae (Golubev) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813324.
- Basionym: Rhodosporidium babjevae Golubev, Syst. Appl. Microbiol. 16: 445. 1993.
-
4)
Rhodotorula dairenensis (T. Haseg. & I. Banno) Fell, J.P. Samp. & Gadanho, FEMS Yeast Res. 2: 56. 2002.
-
5)Rhodotorula diobovata (S.Y. Newell & I.L. Hunter) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813325.
- Basionym: Rhodosporidium diobovatum S.Y. Newell & I.L. Hunter, J. Bact. 104: 504. 1970.
- = Rhodotorula glutinis (Fresenius) Harrison var. lusitanica Marcilla.
-
6)
Rhodotorula evergladensis Fell, Statzell & Scorzetti, Antonie van Leeuwenhoek 99: 547. 2011.
-
7)
Rhodotorula glutinis (Fresen.) F.C. Harrison, Proc. & Trans. Roy. Soc. Canada ser. 3 21: 349. 1928.
-
8)
Rhodotorula graminis Di Menna, J. Gen. Microbiol. 18: 270. 1958.
-
9)Rhodotorula kratochvilovae (Hamam., Sugiy. & Komag.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813353.
- Basionym: Rhodosporidium kratochvilovae Hamam. et al., J. Gen. Appl. Microbiol. 34: 122. 1988.
-
10)
Rhodotorula mucilaginosa (A. Jörg.) F.C. Harrison, Proc. & Trans. Roy. Soc. Canada ser. 3 21: 349. 1928.
-
11)
Rhodotorula pacifica Nagah. & Hamam., Int. J. Syst. Evol. Microbiol. 56: 297. 2006.
-
12)Rhodotorula paludigena (Fell & Tallman) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813354.
- Basionym: Rhodosporidium paludigenum Fell & Tallman, Int. J. Syst. Bacteriol. 30: 658. 1980.
-
13)Rhodotorula sphaerocarpa (S.Y. Newell & Fell) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813356.
- Basionym: Rhodosporidium sphaerocarpum S.Y. Newell & Fell, Mycologia 62: 276. 1970.
-
14)
Rhodotorula taiwanensis F.L. Lee & C.H. Huang, Antonie van Leeuwenhoek 99: 300. 2011.
-
15)Rhodotorula toruloides (I. Banno) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813358.
- Basionym: Rhodosporidium toruloides I. Banno, J. Gen. Appl. Microbiol. 13: 193. 1967.
Rhodosporidiobolus Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813359.
Etymology: The genus name refers to the fact that the species were hitherto classified in the genera Rhodosporidium or Sporidiobolus.
This genus agrees with the mixed Rhodosporidium/Sporidiobolus clade (Wang et al. 2015a) and includes asexual states classified in the genera Rhodotorula and Sporobolomyces, and their sexual counterparts Rhodosporidium and Sporidiobolus (Fig. 8). Member of the Sporidiobolaceae (Sporidiobolales). The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a well supported clade within Sporidiobolales (Fig. 1, Fig. 8). Because the type species of the genera Rhodotorula, Rhodosporidium, Sporobolomyces and Sporidiobolus are located in other clades, species in this clade will be transferred into a new genus.
Sexual reproduction observed in some species. Clamp connections present. Teliospores may be formed and produce transversely septate basidia. Colonies pink to red and butyrous. Budding cells present. Pseudohyphae or true hyphae present or not. Ballistoconidia formed or not, ellipsoidal, allantoid or amygdaliform. Major CoQ systems Q-9 or Q-10.
Type species: Rhodosporidiobolus nylandii (M. Takash. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Rhodosporidiobolus fluvialis (Fell, Kurtzman, Tallman & J.D. Buck) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813360.
- Basionym: Rhodosporidium fluviale Fell et al., Mycologia 80: 562. 1988.
-
2)Rhodosporidiobolus azoricus (J.P. Samp. & Gadanho) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813364.
- Basionym: Rhodosporidium azoricum J.P. Samp. & Gadanho, Can. J. Microbiol. 47: 214. 2001.
-
3)Rhodosporidiobolus microsporus (Higham ex Fell, Blatt & Statzell) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813366.
- Basionym: Sporidiobolus microsporus Higham ex Fell et al., Antonie van Leeuwenhoek 74: 268. 1998.
-
4)Rhodosporidiobolus nylandii (M. Takash. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813369.
- Basionym: Sporobolomyces nylandii M. Takash. & Nakase, Mycoscience 41: 364. 2000.
-
5)Rhodosporidiobolus ruineniae (Holzschu, Tredick & Phaff) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813371.
- Basionym: Sporidiobolus ruineniae Holzschu et al., Curr. Microbiol. 5: 75. 1981.
-
6)Rhodosporidiobolus lusitaniae (Á. Fonseca & J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813374.
- Basionym: Rhodosporidium lusitaniae Á. Fonseca & J.P. Samp., Syst. Appl. Microbiol. 15: 48. 1992.
-
7)Rhodosporidiobolus colostri (T. Castelli) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813375.
- Basionym: Mycotorula colostri T. Castelli, Giorn. Biol. App. alla Indust. Chim. ad Alm. 2: 1. 1932.
-
8)Rhodosporidiobolus odoratus (J.P. Samp., Á. Fonseca & Valério) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813377.
- Basionym: Sporobolomyces odoratus J.P. Samp. et al., FEMS Yeast Res. 2: 15. 2002.
-
9)Rhodosporidiobolus poonsookiae (M. Takash. & Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813379.
- Basionym: Sporobolomyces poonsookiae M. Takash. & Nakase, Mycoscience 41: 365. 2000.
Sporobolomyces Kluyver & C.B. Niel, Centbl. Bakt. ParasitKde Abt. II 63: 19. 1924. emend. Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
= Sporidiobolus Nyland, Mycologia 41: 686. 1949.
Type species: Sporobolomyces roseus Kluyver & C.B. Niel
This genus is emended to include Sporobolomyces species and their sexual counterparts Sporidiobolus, which belong to the Sporobolomyces clade (Wang et al. 2015a) that occurred as a well supported clade within the Sporidiobolaceae (Sporidiobolales). The Sporidiobolus clade contains the type species of Sporobolomyces, S. salmonicolor, and the type species of Sporidiobolus, Sp. johnsonii (Fig. 1, Fig. 8). From this perspective the name Sporobolomyces has taxonomic priority over Sporidiobolus, as the former was published in 1924 and the latter in 1949. Thus we propose to keep the genus name Sporobolomyces for this clade.
Sexual reproduction observed in some species. Clamp connections present. Teliospores are formed and germinate to produce transversely septate basidia. Colonies salmon-pink, red and butyrous. Budding cells present. Pseudohyphae or true hyphae present or not. Ballistoconidia formed, ellipsoidal, allantoid or amygdaliform. Major CoQ system Q-10.
Species accepted:
-
1)
Sporobolomyces bannaensis F.Y. Bai & J.H. Zhao, Int. J. Syst. Evol. Microbiol. 53: 2092. 2003.
-
2)
Sporobolomyces beijingensis F.Y. Bai & Q.M. Wang, FEMS Yeast Res. 4: 582. 2004.
-
3)
Sporobolomyces blumeae M. Takash. & Nakase, Mycoscience 41: 366. 2000.
-
4)
Sporobolomyces carnicolor Yamasaki & H. Fujii ex F.Y. Bai & Boekhout, Int. J. Syst. Evol. Microbiol. 52: 2313. 2002.
-
5)Sporobolomyces longiusculus (Libkind, van Broock & J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813380.
- Basionym: Sporidiobolus longiusculus Libkind et al., Int. J. Syst. Evol. Microbiol. 55: 505. 2005.
-
6)
Sporobolomyces japonicus Iizuka & Goto, J. Gen. Appl. Microbiol. 11: 333. 1965.
-
7)
Sporobolomyces jilinensis F.Y. Bai & Q.M. Wang, FEMS Yeast Res. 4: 584. 2004.
-
8)Sporobolomyces johnsonii (Nyland) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813382.
- Basionym: Sporidiobolus johnsonii Nyland, Mycologia 41: 687. 1949.
-
9)
Sporobolomyces koalae Satoh & Makimura, Int. J. Syst. Evol. Microbiol. 58: 2985. 2008.
-
10)
Sporobolomyces patagonicus Libkind et al., Int. J. Syst. Evol. Microbiol. 55: 506. 2005.
-
11)
Sporobolomyces phaffii F.Y. Bai et al., Int. J. Syst. Evol. Microbiol. 52: 2313. 2002.
-
12)
Sporobolomyces roseus Kluyver & C.B. Niel, Centbl. Bakt. ParasitKde Abt. II 63: 19. 1924.
-
13)
Sporobolomyces ruberrimus Yamasaki & H. Fujii ex Fell et al., FEMS Yeast Res. 1: 267. 2002
-
14)
Sporobolomyces salmonicolor (B. Fisch. & Brebeck) Kluyver & C.B. Niel, Centbl. Bakt. ParasitKde Abt. II 63: 19. 1924.
-
15)
Sporobolomyces salmoneus Derx, Annls mycol. 28: 17. 1930.
Taxa incertae sedis in the Microbotryomycetes
Family Colacogloeaceae Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, fam. nov. MycoBank MB813158.
Member of the Microbotryomycetes. The diagnosis of the family Colacogloeaceae is based on the description of the genus Colacogloea. The nomenclature of the family is based on the genus Colacogloea.
Type genus: Colacogloea Oberw. & Bandoni.
Genus accepted: Colacogloea Oberw. & Bandoni.
Colacogloea Oberwinkler & Bandoni, Can. J. Bot. 68: 2532. 1990. emend. Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Type species: Colacogloea peniophorae (Bourdot & Galzin) Oberwinkler & Bandoni.
This genus is emended to include species of Colacogloea and related Rhodotorula species in the Colacogloea clade (Wang et al. 2015a), which occurred as a well supported clade within Microbotryomycetes (Fig. 1, Fig. 6).
Sexual reproduction observed in some species. Teleomorphic taxa mycoparasitic, and the sexual state develops only in the host. Basidiocarps minute, pulvinate to effuse, and mucoid-gelatinous. Basidia auricularioid (i.e., transversely septate). Hyphae thin-walled, hyaline, with clamp connections, and grow intrahymenially in host fructifications. Septal pores ‘simple’. Colacosomes (or lenticular body) occur at the interface between the parasite and the host (Sampaio et al. 2011). Colonies cream, mucoid or butyrous. Budding cells present. Ballistoconidia not formed. Major CoQ system Q-10.
Species accepted:
-
1)Colacogloea cycloclastica (Thanh, M.S. Smit, Moleleki & Fell) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813159.
- Basionym: Rhodotorula cycloclastica Thanh et al., FEMS Yeast Res. 4: 858. 2004.
-
2)Colacogloea diffluens (Ruinen) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813160.
- Basionym: Candida diffluens Ruinen, Antonie van Leeuwenhoek 29: 437. 1963.
- ≡ Rhodotorula diffluens (Ruinen) von Arx & Weijman
- ≡ Vanrija diffluens (Ruinen) R.T. Moore
-
3)Colacogloea eucalyptica (C.H. Pohl, M.S. Smit & Albertyn) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813161.
- Basionym: Rhodotorula eucalyptica C.H. Pohl et al., Int. J. Syst. Evol. Microbiol. 61: 2326. 2011.
-
4)Colacogloea falcata (Nakase, M. Itoh & M. Suzuki) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813162.
- Basionym: Sporobolomyces falcatus Nakase et al., Trans. Mycol. Soc. Japan 28: 296. 1987.
-
5)Colacogloea foliorum (Ruinen) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813163.
- Basionym: Candida foliarum Ruinen, Antonie van Leeuwenhoek 29: 436. 1963.
- ≡ Rhodotorula foliorum (Ruinen) Rodr. Mir. & Weijman
-
6)
Colacogloea peniophorae (Bourdot & Galzin) Oberwinkler & Bandoni, Can. J. Bot. 68: 2532. 1990.
-
7)Colacogloea philyla (van der Walt, Klift & D.B. Scott) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813164.
- Basionym: Torulopsis philyla van der Walt et al., Antonie van Leeuwenhoek 37: 464. 1971.
- ≡ Rhodotorula philyla (van der Walt et al.) Rodr. Mir. & Weijman
-
8)Colacogloea retinophila (Thanh, M.S. Smit, Moleleki & Fell) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813166.
- Basionym: Rhodotorula retinophila Thanh et al., FEMS Yeast Res. 4: 859. 2004.
-
9)Colacogloea terpenoidalis (Thanh, M.S. Smit, Moleleki & Fell) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813169.
- Basionym: Rhodotorula terpenoidalis Thanh et al., FEMS Yeast Res. 4: 860. 2004.
Note: Additional sequences representing three potential new species of this genus were found in public databases (Fig. 6).
Family Chrysozymaceae Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, fam. nov. MycoBank MB813171.
Member of the Microbotryomycetes. The diagnosis of the family Chrysozymaceae is based on the description of the genus Chrysozyma. The nomenclature of the family is based on the genus Chrysozyma.
Type genus: Chrysozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Genera accepted: Bannozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Chrysozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Hamamotoa Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, Fellozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Bannozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813173.
Etymology: The genus is named in honour of I. Banno for his contributions to yeast taxonomy.
This genus corresponds to the yamatoana clade (Wang et al. 2015a). Member of Chrysozymaceae (Microbotryomycetes). The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a well supported clade within Microbotryomycetes (Fig. 1, Fig. 6).
Sexual reproduction not known. Colonies pale yellow to greyish-yellow, butyrous. Budding cells present. Pseudohyphae and septate hyphae present or not. Ballistoconidia present or not, kidney-shaped, allantoid or elongate. Major CoQ system Q-9.
Type species: Bannozyma yamatoana (Nakase, M. Suzuki & M. Itoh) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Bannozyma arctica (Vishniac & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813176.
- Basionym: Rhodotorula arctica Vishniac & M. Takash., Int. J. Syst. Evol. Microbiol. 60: 1215. 2010.
-
2)Bannozyma yamatoana (Nakase, M. Suzuki & M. Itoh) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813179.
- Basionym: Sporobolomyces yamatoanus Nakase et al., J. Gen. Appl. Microbiol. 33: 446. 1987.
- ≡ Bensingtonia yamatoana (Nakase et al.) Nakase & Boekhout.
Chrysozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813180.
Etymology: The genus is named because the type species produces yellowish colonies.
This genus agrees with the griseoflavus clade (Wang et al. 2015a). Member of Chrysozymaceae (Microbotryomycetes). The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a well supported clade closely related to the yamatoana clade within Microbotryomycetes (Fig. 1, Fig. 6).
Sexual reproduction not known. Colonies greyish-white to yellowish-cream, butyrous. Budding cells present. Hyphae and pseudohyphae not observed. Ballistoconidia present, ellipsoidal, allantoid or lunate. Major CoQ system Q-10.
Type species: Chrysozyma griseoflava (Nakase & M. Suzuki) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Chrysozyma fushanensis (Nakase, F.L. Lee & M. Takash.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813181.
- Basionym: Sporobolomyces fushanensis Nakase et al., J. Gen. Appl. Microbiol. 51: 43. 2005.
-
2)Chrysozyma griseoflava (Nakase & M. Suzuki) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813183.
- Basionym: Sporobolomyces griseoflavus Nakase & M. Suzuki, J. Gen. Appl. Microbiol. 33: 168. 1987.
Note: Species of Chrysozyma and Bannozyma can be distinguished from each other by the presence of Q-10 and Q-9, respectively.
Fellozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813184.
Etymology: The genus is named in honour of Jack.W. Fell for his contributions to yeast taxonomy.
This genus agrees with the Sporobolomyces inositophilus lineage (Wang et al. 2015a). Member of the Chrysozymaceae (Microbotryomycetes). The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a single-species lineage within the Microbotryomycetes (Fig. 1, Fig. 6). S. inositophilus is related to the singularis clade in the MP analysis of the combined seven genes-based dataset. This, however, was not supported in the ML and Bayesian analyses (Wang et al. 2015a).
Sexual reproduction not known. Colonies greyish-cream, butyrous. Budding cells present. Hyphae and pseudohyphae not observed. Ballistoconidia present, amygdaliform to falcate. Major CoQ system Q-10.
Type species: Fellozyma inositophila (Nakase & M. Suzuki) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout
Species accepted:
-
1)Fellozyma inositophila (Nakase & M. Suzuki) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813185.
- Basionym: Sporobolomyces inositophilus Nakase & M. Suzuki, Antonie van Leeuwenhoek 53: 246. 1987.
Note: Additional sequence representing a potential new species of this genus was found in public databases (Fig. 6).
Hamamotoa Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813186.
Etymology: The genus is named in honour of Dr. Makiko Hamamoto for her contributions to yeast taxonomy.
This genus agrees with the singularis clade (Wang et al. 2015a). Member of Chrysozymaceae (Microbotryomycetes). The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a well supported clade closely related to the griseoflavus and yamatoana clades within Microbotryomycetes (Fig. 1, Fig. 6).
Sexual reproduction not known. Colonies cream-coloured to pale yellowish-brown, mucoid. Budding cells present. Hyphae and pseudohyphae not present. Ballistoconidia present or not, ellipsoidal or kidney-shaped. Major CoQ system Q-10.
Type species: Hamamotoa singularis (Phaff & do Carmo-Sousa) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Hamamotoa lignophila (Dill, C. Ramírez & González) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813187.
- Basionym: Candida lignophila Dill et al., Antonie van Leeuwenhoek 50: 220. 1984.
- ≡ Rhodotorula lignophila (Dill et al.) Roeijmans et al.
-
2)Hamamotoa singularis (Phaff & do Carmo-Sousa) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813188.
- Basionym: Sporobolomyces singularis Phaff & do Carmo-Sousa, Antonie van Leeuwenhoek 28: 205. 1962.
- ≡ Bullera singularis (Phaff & do Carmo-Sousa) Rodr. Mir.
Note: Species of Hamamotoa (i.e. the singularis clade) assimilate lactose and dl-lactate, but not melezitose and form highly mucoid colonies (Fig. 3D). The species of Chrysozyma (i.e. the griseoflavus clade) and Bannozyma (i.e. the yamatoana clade) are not able to grow on the former two carbon sources and have colonies with a butyrous texture (Fig. 3D).
Pseudohyphozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813189.
Etymology: The name of the genus refers to the presence of pseudohyphae in all known species of this clade.
This genus agrees with the buffonii clade (Wang et al. 2015a). Member of the Microbotryomycetes. The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a well supported clade within the Microbotryomycetes (Fig. 1, Fig. 6).
Sexual reproduction not known. Colonies cream-coloured and butyrous. Budding cells present. Pseudohyphae of branched chains of ovoid to cylindrical cells. Ballistoconidia not produced. Major CoQ system Q-10.
Type species: Pseudohyphozyma buffonii (C. Ramírez) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Pseudohyphozyma bogoriensis (Deinema) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813190.
- Basionym: Candida bogoriensis Deinema, J. Econ. Biol. 61: 40. 1961.
- ≡ Rhodotorula bogoriensis (Deinema) von Arx & Weijman.
- ≡ Candida bogoriensis Deinema var. lipolytica Ruinen.
- ≡ Vanrija bogoriensis (Deinema) Moore.
-
2)Pseudohyphozyma buffonii (C. Ramírez) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813191.
- Basionym: Torulopsis buffonii C. Ramírez, Microbiol. 10: 236. 1957.
- ≡ Rhodotorula buffonii (C. Ramírez) Roeijmans.
-
3)Pseudohyphozyma pustula (Buhagiar) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813192.
- Basionym: Torulopsis pustula Buhagiar, J. Gen. Microbiol. 86: 3. 1975.
- ≡ Rhodotorula pustula (Buhagiar) Rodr. Mir. & Weijman.
Note: Species of the genus Pseudohyphozyma (i.e. the buffonii clade) have a butyrous colony texture on slants with potato dextrose agar (PDA) and differ from members of the genus Slooffia (the tsugae clade), which are usually mucoid on PDA (Fig. 3D).
Pseudoleucosporidium V. de Garcia et al., FEMS Yeast Res. 15: 11. 2015.
Type species: Pseudoleucosporidium fasciculatum (Babeva & Lisichk.) V. de Garcia et al.
Species accepted:
-
1)
Pseudoleucosporidium fasciculatum (Babeva & Lisichk.) V. de Garcia et al., FEMS Yeast Res. 15: 13. 2015.
Note: Our analyses suggest a close relationship between the genera Pseudoleucosporidium and Curvibasidium, as revealed by the multi-gene analyses (Wang et al. 2015a) and the analysis of the enlarged LSU rRNA gene dataset (Fig. 7).
Oberwinklerozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813193.
Etymology: The genus is named in honour of Franz. Oberwinkler for his contributions to the taxonomy of Basidiomycota.
This genus agrees with the yarrowii clade (Wang et al. 2015a). Member of the Microbotryomycetes. The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a well supported clade within the Microbotryomycetes (Fig. 1, Fig. 6). The yarrowii, buffonii and tsugae clades clustered together with low support in the ML analysis, but were not supported by the MP and BI analyses (Wang et al. 2015a).
Sexual reproduction not known. Colonies cream-coloured. Budding cells present. Pseudohyphae present. Ballistoconidia not produced. Major CoQ system Q-9.
Type species: Oberwinklerozyma yarrowii (Á. Fonseca & van Uden) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Oberwinklerozyma silvestris (Golubev & Scorzetti) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813194.
- Basionym: Rhodotorula silvestris Golubev & Scorzetti, Int. J. Syst. Evol. Microbiol. 60: 2501. 2010.
-
2)Oberwinklerozyma straminea (Golubev & Scorzetti) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813195.
- Basionym: Rhodotorula straminea Golubev & Scorzetti, Int. J. Syst. Evol. Microbiol. 60: 2501. 2010.
-
3)Oberwinklerozyma yarrowii (Á. Fonseca & van Uden) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813196.
- Basionym: Cryptococcus yarrowii Á. Fonseca & van Uden, Antonie van Leeuwenhoek 59: 177. 1991.
- ≡ Rhodotorula yarrowii (Á. Fonseca & van Uden) Boekhout et al.
Note: Species of the genus Oberwinklerozyma (i.e. the yarrowii clade) have major coenzyme Q system Q-9 and are able to assimilate raffinose and myo-inositol, whereas members of the genera Pseudohyphozyma (i.e. the buffonii clade) and Slooffia (i.e. the tsugae clade) have coenzyme Q-10 and do not use these two carbon sources (Table 3).
Sampaiozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813197.
Etymology: The genus is named in honour of J.P. Sampaio for his contributions to yeast taxonomy.
This genus agrees with the vanillica clade (Wang et al. 2015a). Member of the Microbotryomycetes. The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a well supported clade within Microbotryomycetes (Fig. 1, Fig. 7).
Sexual reproduction not known. Colonies cream or yellowish. Budding cells present. Pseudohyphae and true hyphae absent. Ballistoconidia not produced. Major CoQ system Q-10.
Type species: Sampaiozyma ingeniosa (Di Menna) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Sampaiozyma ingeniosa (Di Menna) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813198.
- Basionym: Torulopsis ingeniosa Di Menna, J. Gen. Microbiol. 19: 581. 1958.
- ≡ Rhodotorula ingeniosa (Di Menna) von Arx & Weijman.
- ≡ Candida ingeniosa (Di Menna) Meyer & Yarrow.
- ≡ Vanrija ingeniosa (Di Menna) Moore.
-
2)Sampaiozyma vanillica (J.P. Samp.) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813199.
- Basionym: Rhodotorula vanillica J.P. Samp., Syst. Appl. Microbiol. 17: 616. 1994.
Spencerozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813200.
Etymology: The genus is named in honour of I. Spencer-Martins for her contributions to yeast taxonomy and physiology.
This genus agrees with the Rhodotorula crocea lineage (Wang et al. 2015a). Member of the Microbotryomycetes. The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a single-species lineage within the Microbotryomycetes (Fig. 1, Fig. 6).
Sexual reproduction not known. Colonies yellowish-cream, butyrous. Budding cells present. Pseudohyphae and true hyphae absent. Ballistoconidia not produced. Major CoQ system Q-10.
Type species: Spencerozyma crocea (Shifrine & Phaff) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Spencerozyma crocea (Shifrine & Phaff) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813201.
- Basionym: Rhodotorula crocea Shifrine & Phaff, Mycologia 48: 50. 1956.
Slooffia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813202.
Etymology: The genus is named in honour of W.C. Slooff for her contributions to yeast taxonomy.
This genus corresponds to the tsugae clade (Wang et al. 2015a). Member of the Microbotryomycetes. The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a well supported clade within Microbotryomycetes (Fig. 1, Fig. 6).
Sexual reproduction not known. Colonies cream-coloured and mucoid. Budding cells present. Hyphae and pseudohyphae not formed. Ballistoconidia present or not, ellipsoidal. Major CoQ system Q-10
Type species: Slooffia tsugae (Phaff & do Carmo-Sousa) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout
Species accepted:
-
1)Slooffia cresolica (Middelhoven & Spaaij) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813203.
- Basionym: Rhodotorula cresolica Middelhoven & Spaaij, Int. J. Syst. Bacteriol. 47: 324. 1997.
-
2)Slooffia pilati (F.H. Jacob, Faure-Raynaud & Berton) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813204.
- Basionym: Torulopsis pilati F.H. Jacob et al., Mycopathologia 69: 83. 1979.
- ≡ Rhodotorula pilati (F.H. Jacob et al.) Barnett et al.
-
3)Slooffia tsugae (Phaff & do Carmo-Sousa) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813206.
- Basionym: Bullera tsugae Phaff & do Carmo-Sousa, Antonie van Leeuwenhoek 28: 205. 1962.
- ≡ Sporobolomyces tsugae (Phaff & do Carmo-Sousa) Nakase & M. Itoh.
Notes: Species of Slooffia can be distinguished from those of Pseudohyphozyma (the buffonii clade) by their colony texture (see above, Fig. 3D). Additional sequences representing two potential new species of this genus were found in public databases (Fig. 6).
Trigonosporomyces Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813207.
Etymology: The genus is named based on the morphology of the type species that forms triangular cells on pseudohyphae.
This genus agrees with the Rhodotorula hylophila (Wang et al. 2015a). Member of the Microbotryomycetes. The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a single-species lineage within the Microbotryomycetes (Fig. 1, Fig. 6).
Sexual reproduction not known. Colonies cream-coloured. Budding cells present. Pseudohyphae of long, slender cells, often triangular. Ballistoconidia not produced. Major CoQ system unknown.
Type species: Trigonosporomyces hylophilus (van der Walt, van der Klift & D.B. Scott) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Trigonosporomyces hylophilus (van der Walt, van der Klift & D.B. Scott) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813208.
- Basionym: Candida hylophila van der Walt et al., Antonie van Leeuwenhoek 37: 449. 1971.
- ≡ Rhodotorula hylophila (van der Walt et al.) Rodr. Mir. & Weijman.
Yunzhangia Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813209.
Etymology: The genus is named in honour of Yun-Zhang Wang, former professor at the Institute of Microbiology, Chinese Academy of Sciences, for his contributions to the taxonomic study of Pucciniales in China.
This genus agrees with the sonckii clade (Wang et al. 2015a). Member of the Microbotryomycetes. The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a well supported clade diverged from the other clades within the Microbotryomycetes (Fig. 1, Fig. 6).
Sexual reproduction not known. Colonies cream-coloured, mucoid or butyrous. Budding cells present. Pseudohyphae and true hyphae not observed. Ballistoconidia not produced. Major CoQ system unknown.
Type species: Yunzhangia auriculariae (Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Yunzhangia auriculariae (Nakase) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813210.
- Basionym: Torulopsis auriculariae Nakase, J. Gen. Appl. Microbiol. 17: 413. 1971.
- ≡ Rhodotorula auriculariae (Nakase) Rodr. Mir. & Weijman.
- ≡ Candida auriculariae (Nakase) Meyer & Yarrow.
-
2)Yunzhangia sonckii (Hopsu-Havu, Tunnela & Yarrow) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813212.
- Basionym: Candida sonckii Hopsu-Havu et al., Antonie van Leeuwenhoek 44: 436. 1978.
- ≡ Rhodotorula sonckii (Hopsu-Havu et al.) Rodr. Mir. & Weijman.
Udeniozyma Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813213.
Etymology: The genus is named in honour of N.J. van Uden for his contributions to the study and taxonomy of basidiomycetous yeasts.
This genus agrees with the Rhodotorula ferulica lineage (Wang et al. 2015a). Member of the Microbotryomycetes. The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as single-species lineage within the Microbotryomycetes (Fig. 1, Fig. 6). R. ferulica seems to have affiliation to Colacogloea, but did not receive high bootstrap values and Bayesian posterior probability.
Sexual reproduction not known. Colonies cream-coloured, mucoid. Budding cells present. Hyphae and pseudohyphae present or not. Ballistoconidia not produced. Major CoQ system Q-10.
Type species: Udeniozyma ferulica (J.P. Samp. & van Uden) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Udeniozyma ferulica (J.P. Samp. & van Uden) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813214.
- Basionym: Rhodotorula ferulica J.P. Samp. & van Uden, Syst. Appl. Microbiol. 14: 146. 1991.
Vonarxula Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813216.
Etymology: The genus is named in honour of J.A. von Arx for his contributions to fungal taxonomy.
This genus agrees with the Rhodotorula javanica lineage (Wang et al. 2015a). Member of the Microbotryomycetes. The genus is mainly circumscribed by the phylogenetic analysis of seven genes and the analysis of the enlarged LSU rRNA gene dataset, in which it occurred as a single-species lineage within the Microbotryomycetes (Fig. 1, Fig. 6).
Sexual reproduction not known. Colonies cream-coloured. Budding cells present. Pseudohyphae of branched chains of fusiform cells. Ballistoconidia not produced. Major CoQ system Q-9.
Type species: Vonarxula javanica (Ruinen) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout.
Species accepted:
-
1)Vonarxula javanica (Ruinen) Q.M. Wang, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813218.
- Basionym: Candida javanica Ruinen, Antonie van Leeuwenhoek 29: 436. 1963.
- ≡ Rhodotorula javanica (Ruinen) von Arx & Weijman.
Mixiomycetes R. Bauer et al., Mycol. Progr. 5: 47. 2006.
Type order: Mixiales R. Bauer et al.
Mixiales R. Bauer et al., Mycol. Progr. 5: 47. 2006.
Type family: Mixiaceae C.L. Kramer.
This order is characterised by multinucleate hyphae and multiple spores produced simultaneously on sporogenous cells (Bauer et al. 2006).
Mixiaceae C.L. Kramer, Stud. Mycol. 30: 159. 1987.
Type genus: Mixia C.L. Kramer.
This family was proposed to accommodate the Taphrina-like genus Mixia (Kramer 1987), which is an intracellular parasite of ferns belonging to the genus Osmunda.
Genus accepted: Mixia C.L. Kramer
Mixia C.L. Kramer, Mycologia 50: 924. 1958.
Type species: Mixia osmundae (Nishida) C.L. Kramer.
Species accepted:
-
1)
Mixia osmundae (Nishida) C.L. Kramer, Mycologia 50: 924. 1958.
Suggestion for new species descriptions
In the future descriptions of new species in the genera Rhodotorula, Sporobolomyces and Bensingtonia should be restricted to the clades containing the respective type species (Fig. 1, Fig. 4, Fig. 8). In the case of unclassified Microbotryomycetes or 'incertae sedis’, none of the aforementioned generic names should be used to describe new species, and new genera have to be introduced following a robust phylogenetic analysis utilising several independent DNA loci or whole-genome comparisons (e.g. Wang et al. 2015a). Our results show that using the LSU rRNA gene alone is not sufficient to resolve the high-level phylogenetic relationships in Microbotryomycetes.
Acknowledgements
We thank Masako Takashima for providing some strains and Walter Gams for his advice on nomenclatural matters; we also thank José Paulo Sampaio and Diego Libkind for their critical comments for this manuscript and Nathalie van der Wiele and Vincent Robert for their help with the registration of taxa in MycoBank. This study was supported by grants No. 30970013 and No. 31010103902 from the National Natural Science Foundation of China (NSFC), No. 2012078 from the Youth Innovation Promotion Association of the Chinese Academy of Sciences and No. 10CDP019 from the Royal Netherlands Academy of Arts and Sciences (KNAW). AY acknowledges a grant from the Fundação para a Ciência e a Tecnologia, Portugal (grant number PTDC/BIA-BIC/4585/2012). TB is supported by grant NPRP 5-298-3-086 of Qatar Foundation. The authors are solely responsible for the content of this work.
Footnotes
Peer review under responsibility of CBS-KNAW Fungal Biodiversity Centre.
Contributor Information
T. Boekhout, Email: t.boekhout@cbs.knaw.nl.
F.-Y. Bai, Email: baify@im.ac.cn.
References
- Aime M.C., Matheny P.B., Henk D.A. An overview of the higher level classification of Pucciniomycotina based on combined analyses of nuclear large and small subunit rRNA sequences. Mycologia. 2006;98:896–905. doi: 10.3852/mycologia.98.6.896. [DOI] [PubMed] [Google Scholar]
- Aime M.C., Toome M., McLaughlin D.J. Pucciniomycotina. In: McLaughlin D.J., Spatafora J.W., editors. 2nd edn. Springer-Verlag; Berlin: 2014. pp. 271–294. (The mycota, Vol. VII, Part A: Systematics and evolution). [Google Scholar]
- Bandoni R.J., Boekhout T. Agaricostilbum Wright (1970) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts, a taxonomic study. 5th edn. Elsevier; Amsterdam: 2011. pp. 1375–1378. [Google Scholar]
- Bauer R. Basidiomycetous interfungal cellular interactions – a synopsis. In: Agerer R., Piepenbring M., Blanz P., editors. Frontiers in basidiomycote mycology. IHW-Verlag; Eching: 2004. pp. 325–337. [Google Scholar]
- Bauer R., Begerow D., Sampaio J.P. The simple-septate basidiomycetes: a synopsis. Mycological Progress. 2006;5:41–66. [Google Scholar]
- Bauer R., Oberwinkler F., Vánky K. Ultrastructural markers and systematics in smut fungi and allied taxa. Canadian Journal of Botany. 1997;75:1273–1314. [Google Scholar]
- Boekhout T. A revision of ballistoconidia-forming yeasts and fungi. Studies in Mycology. 1991;33:1–194. [Google Scholar]
- Boekhout T., Fonseca Á., Sampaio J.P. Discussion of teleomorphic and anamorphic basidiomycetous yeasts. In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts, a taxonomic study. 5th edn. Elsevier; Amsterdam: 2011. pp. 1339–1372. [Google Scholar]
- de García V., Coelho M.A., Maia T.M. Sex in the cold: taxonomic reorganization of psychrotolerant yeasts in the order Leucosporidiales. FEMS Yeast Research. 2015;15 doi: 10.1093/femsyr/fov019. pii: fov019. [DOI] [PubMed] [Google Scholar]
- Denchev C.M. Validation of three names of families in the Pucciniomycotina. Mycologia Balcanica. 2009;6:87–88. [Google Scholar]
- Fell J.W. Sterigmatomyces, a new fungal genus from marine areas. Antonie van Leeuwenhoek. 1966;32:99–104. doi: 10.1007/BF02097449. [DOI] [PubMed] [Google Scholar]
- Fell J.W. Sterigmatomyces Fell (1966) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts, a taxonomic study. 5th edn. Elsevier; Amsterdam: 2011. pp. 1991–1994. [Google Scholar]
- Fell J.W., Boekhout T., Fonseca Á. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rRNA D1/D2 domain sequence analysis. International Journal of Systematic and Evolutionary Microbiology. 2000;50:1351–1371. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- Fonseca Á. Kondoa Y. Yamada, Nakagawa & Banno emend. Á. Fonseca, Sampaio & Fell (2000) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts, a taxonomic study. 5th edn. Elsevier; Amsterdam: 2011. pp. 1471–1475. [Google Scholar]
- Furuya N., Takashima M., Shiotani H. Reclassification of citrus pseudo greasy spot causal yeasts, and a proposal of two new species, Sporobolomyces productus sp. nov. and S. corallinus sp. nov. Mycoscience. 2012;53:261–269. [Google Scholar]
- Göker M., García-Blázquez G., Voglmayr H. Molecular taxonomy of phytopathogenic fungi: a case study in Peronospora. PLoS One. 2009;4:e6319. doi: 10.1371/journal.pone.0006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göker M., Grimm G.W., Auch A.F. A clustering optimisation strategy for molecular taxonomy applied to planktonic foraminifera SSU rRNA. Evolutionary Bioinformatics. 2010;6:97–112. doi: 10.4137/ebo.s5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloboff P.A., Farris J.S., Nixon K.C. TNT, a free program for phylogenetic analysis. Cladistics. 2008;24:774–786. [Google Scholar]
- Gomes F.C., Safar S.V., Marques A.R. The diversity and extracellular enzymatic activities of yeasts isolated from water tanks of Vriesea minarum, an endangered bromeliad species in Brazil, and the description of Occultifur brasiliensis f.a., sp. nov. Antonie van Leeuwenhoek. 2015;107:597–611. doi: 10.1007/s10482-014-0356-4. [DOI] [PubMed] [Google Scholar]
- Hamamoto M., Boekhout T., Nakase T. Sporobolomyces Kluyver & van Niel (1924) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts, a taxonomic study. 5th edn. Elsevier; Amsterdam: 2011. pp. 1929–1990. [Google Scholar]
- Hamamoto M., Nakase T. Phylogenetic analysis of the ballistoconidium-forming yeast genus Sporobolomyces based on 18S rRNA sequences. International Journal of Systematic and Evolutionary Microbiology. 2000;50:1373–1380. doi: 10.1099/00207713-50-3-1373. [DOI] [PubMed] [Google Scholar]
- Hawksworth D.L. A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication and regulation of fungal names. MycoKeys. 2011;1:7–20. doi: 10.5598/imafungus.2011.02.02.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbett D.S., Binder M., Bischoff J.F. A higher-level phylogenetic classification of the Fungi. Mycological Research. 2007;111:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Humphreys A.M., Barraclough T.G. The evolutionary reality of higher taxa in mammals. Proceedings of the Royal Society B: Biological Sciences. 2014;281:20132750. doi: 10.1098/rspb.2013.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Kachalkin A.V. Yeasts of the White Sea intertidal zone and description of Glaciozyma litorale sp. nov. Antonie van Leeuwenhoek. 2014;105:1073–1083. doi: 10.1007/s10482-014-0165-9. [DOI] [PubMed] [Google Scholar]
- Khunnamwong P., Surussawadee J., Jindamorakot S. Occultifur tropicalis f.a., sp. nov., a novel cystobasidiomycetous yeast species isolated from tropical regions. International Journal of Systematic and Evolutionary Microbiology. 2015;65:1578–1582. doi: 10.1099/ijs.0.000140. [DOI] [PubMed] [Google Scholar]
- Kramer C.L. The Taphrinales. Studies in Mycology. 1987;30:151–166. [Google Scholar]
- Kurtzman C.P., Robnett C.J. Occultifur kilbournensis f.a. sp. nov., a new member of the Cystobasidiales associated with maize (Zea mays) cultivation. Antonie van Leeuwenhoek. 2015;107:1323–1329. doi: 10.1007/s10482-015-0427-1. [DOI] [PubMed] [Google Scholar]
- Laich F., Vaca I., Chávez R. Rhodotorula portillonensis sp. nov., a basidiomycetous yeast isolated from Antarctic shallow-water marine sediment. International Journal of Systematic and Evolutionary Microbiology. 2013;63:3884–3891. doi: 10.1099/ijs.0.052753-0. [DOI] [PubMed] [Google Scholar]
- Liu X.Z., Wang Q.M., Groenewald M. Towards an integrated phylogenetic classification of tremellomycetous yeasts. Studies in Mycology. 2015;81:85–147. doi: 10.1016/j.simyco.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margesin R., Fonteyne P.A., Schinner F. Rhodotorula psychrophila sp. nov., Rhodotorula psychrophenolica sp. nov. and Rhodotorula glacialis sp. nov., novel psychrophilic basidiomycetous yeast species isolated from alpine environments. International Journal of Systematic and Evolutionary Microbiology. 2007;57:2179–2184. doi: 10.1099/ijs.0.65111-0. [DOI] [PubMed] [Google Scholar]
- Marvanová L., Suberkropp K. Camptobasidium hydrophilum and its anamorph, Crucella subtilis: a new Heterobasidiomycete from streams. Mycologia. 1990;82:208–217. [Google Scholar]
- McNeill J., Barrie F.R., Buck W.R. A.R.G. Gantner Verlag KG; Ruggell, Liechtenstein: 2012. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code). Regnum Vegetabile 154.http://www.iapt-taxon.org/nomen/main.php Available at: [Google Scholar]
- Moore R.T. An inventory of the phylum Ustomycota. Mycotaxon. 1996;59:1–31. [Google Scholar]
- Nakase T. Expanding world of ballistosporous yeasts: distribution in the phyllosphere, systematics and phylogeny. The Journal of General and Applied Microbiology. 2000;46:189–216. doi: 10.2323/jgam.46.189. [DOI] [PubMed] [Google Scholar]
- Nakase T., Bai F.Y., Boekhout T. Bensingtonia Ingold emend. Nakase & Boekhout (1986) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts, a taxonomic study. 5th edn. Elsevier; Amsterdam: 2011. pp. 1607–1622. [Google Scholar]
- Nakase T., Itoh M., Sugiyama J. Bensingtonia ingoldii sp. nov., a ballistospore-forming yeast isolated from Knightia excelsa collected in New Zealand. The Journal of General and Applied Microbiology. 1989;35:53–58. [Google Scholar]
- Oberwinkler F., Bandoni R.J. A taxonomic survey of the gasteroid, auricularioid heterobasidiomycetes. Canadian Journal of Botany. 1982;60:1726–1750. [Google Scholar]
- Oberwinkler F., Bauer R. The systematics of gasteroid, auricularioid heterobasidiomycetes. Sydowia. 1989;41:224–256. [Google Scholar]
- Paradis E. Analysis of phylogenetics and evolution with R. In: Gentleman R., Hornik K., Parmigiani G., editors. Springer Science; New York: 2006. [Google Scholar]
- Pattengale N.D., Alipour M., Bininda-Emonds O.R.P. How many bootstrap replicates are necessary? Lecture Notes in Computer Science. 2009;5541:184–200. [Google Scholar]
- Roberts P. Heterobasidiomycetes from Majorca & Cabrera (Balearic Islands) Mycotaxon. 1996;60:111–123. [Google Scholar]
- Sampaio J.P. Rhodotorula Harrison (1928) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts, a taxonomic study. 5th edn. Elsevier; Amsterdam: 2011. pp. 1873–1927. [Google Scholar]
- Sampaio J.P., Chen C.J. Naohidea Oberwinkler (1990) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts, a taxonomic study. 5th edn. Elsevier; Amsterdam: 2011. pp. 1511–1513. [Google Scholar]
- Sampaio J.P., Gadanho M., Bauer R. Taxonomic studies in the Microbotryomycetidae: Leucosporidium golubevii sp. nov., Leucosporidiella gen. nov. and the new orders Leucosporidiales and Sporidiobolales. Mycological Progress. 2003;2:53–68. [Google Scholar]
- Sampaio J.P., Kirschner R., Oberwinkler F. Colacogloea Oberwinkler & Bandoni (1990) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts, a taxonomic study. 5th edn. Elsevier; Amsterdam: 2011. pp. 1403–1408. [Google Scholar]
- Sampaio J.P., Oberwinkler F. Cystobasidium (Lagerheim) Neuhoff (1924) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts, a taxonomic study. 5th edn. Elsevier; Amsterdam: 2011. pp. 1419–1422. [Google Scholar]
- Sanderson M.J. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Molecular Biology and Evolution. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Savile D.B.O., Parmelee J.A. Parasitic fungi of the Queen Elisabeth Islands. Canadian Journal of Botany. 1964;42:699–722. [Google Scholar]
- Scorzetti G., Fell J.W., Fonseca Á. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rRNA regions. FEMS Yeast Research. 2002;2:495–517. doi: 10.1111/j.1567-1364.2002.tb00117.x. [DOI] [PubMed] [Google Scholar]
- Shivaji S., Bhadra B., Rao R.S. Rhodotorula himalayensis sp. nov., a novel psychrophilic yeast isolated from Roopkund Lake of the Himalayan mountain ranges, India. Extremophiles. 2008;12:375–381. doi: 10.1007/s00792-008-0144-z. [DOI] [PubMed] [Google Scholar]
- Singh P., Singh S.M., Tsuji M. Rhodotorula svalbardensis sp. nov., a novel yeast species isolated from cryoconite holes of Ny-Ålesund, Arctic. Cryobiology. 2014;68:122–128. doi: 10.1016/j.cryobiol.2014.01.006. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Systematic Biology. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Standley K. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stielow B., Bratek Z., Orczán A.K.I. Species delimitation in taxonomically difficult fungi: the case of Hymenogaster. PLoS One. 2011;6:e15614. doi: 10.1371/journal.pone.0015614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh S.-O., Maslov D.A., Molestina R.E. Microbotryozyma collariae gen. nov., sp. nov., a basidiomycetous yeast isolated from a plant bug Collaria oleosa (Miridae) Antonie van Leeuwenhoek. 2012;102:99–104. doi: 10.1007/s10482-012-9717-z. [DOI] [PubMed] [Google Scholar]
- Takashima M., Suh S.-O., Nakase T. Bensingtonia musae sp. nov. isolated from a dead leaf of Musa paradisiaca and its phylogenetic relationship among basidiomycetous yeasts. The Journal of General and Applied Microbiology. 1995;41:143–151. [Google Scholar]
- Taylor J.W. One Fungus = One Name: DNA and fungal nomenclature twenty years after PCR. IMA Fungus. 2011;2:113–120. doi: 10.5598/imafungus.2011.02.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toome M., Roberson R.W., Aime M.C. Meredithblackwellia eburnea gen. et sp. nov., Kriegeriaceae fam. nov. and Kriegeriales ord. nov. – toward resolving higher-level classification in Microbotryomycetes. Mycologia. 2013;105:486–495. doi: 10.3852/12-251. [DOI] [PubMed] [Google Scholar]
- Turchetti B., Thomas Hall S.R., Connell L.B. Psychrophilic yeasts from Antarctica and European glaciers: description of Glaciozyma gen. nov., Glaciozyma martinii sp. nov. and Glaciozyma watsonii sp. nov. Extremophiles. 2011;15:573–586. doi: 10.1007/s00792-011-0388-x. [DOI] [PubMed] [Google Scholar]
- Vánky K. 2nd edn. APS Press; St. Paul, MN: 2002. Illustrated genera of smut fungi. [Google Scholar]
- Wang Q.M., Boekhout T., Bai F.Y. Bensingtonia rectispora sp. nov. and Bensingtonia bomiensis sp. nov., novel ballistoconidium-forming yeast species from plant leaves collected in Tibet. International Journal of Systematic and Evolutionary Microbiology. 2012;62:2039–2044. doi: 10.1099/ijs.0.038117-0. [DOI] [PubMed] [Google Scholar]
- Wang Q.M., Groenewald M., Takashima M. Phylogeny of yeasts and related taxa within Pucciniomycotina determined from multigene sequence analyses. Studies in Mycology. 2015;81:27–53. doi: 10.1016/j.simyco.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.M., Begerow D., Groenewald M. Multigene phylogeny and taxonomic revision of yeasts and related fungi in the Ustilaginomycotina. Studies in Mycology. 2015;81:54–80. doi: 10.1016/j.simyco.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiß M., Bauer R., Begerow D. Spotlights on heterobasidiomycetes. In: Agerer R., Piepenbring M., Blanz P., editors. Frontiers in basidiomycote mycology. IHW-Verlag; Eching: 2004. pp. 7–48. [Google Scholar]
- Weijman A.C.M., Rodrigues de Miranda L., van der Walt J.P. Redefinition of Candida Berkhout and the consequent emendation of Cryptococcus Kützing and Rhodotorula Harrison. Antonie van Leeuwenhoek. 1998;54:545–553. doi: 10.1007/BF00588390. [DOI] [PubMed] [Google Scholar]
- Wright J.E. Agaricostilbum, a new genus of Deuteromycetes on palm spathes from Argentina. Mycologia. 1970;62:679–682. [Google Scholar]
- Yurkov A.M., Kachalkin A.V., Daniel H.M. Two yeast species Cystobasidium psychroaquaticum f.a. sp. nov. and Cystobasidium rietchieii f.a. sp. nov. isolated from natural environments, and the transfer of Rhodotorula minuta clade members to the genus Cystobasidium. Antonie van Leeuwenhoek. 2015;107:173–185. doi: 10.1007/s10482-014-0315-0. [DOI] [PubMed] [Google Scholar]
- Zhang T., Jia R.L., Zhang Y.Q. Kurtzmanomyces shapotouensis sp. nov., an anamorphic, basidiomycetous yeast isolated from desert soil crusts. International Journal of Systematic and Evolutionary Microbiology. 2013;63:3892–3895. doi: 10.1099/ijs.0.053058-0. [DOI] [PubMed] [Google Scholar]