Abstract
Families and genera assigned to Tremellomycetes have been mainly circumscribed by morphology and for the yeasts also by biochemical and physiological characteristics. This phenotype-based classification is largely in conflict with molecular phylogenetic analyses. Here a phylogenetic classification framework for the Tremellomycetes is proposed based on the results of phylogenetic analyses from a seven-genes dataset covering the majority of tremellomycetous yeasts and closely related filamentous taxa. Circumscriptions of the taxonomic units at the order, family and genus levels recognised were quantitatively assessed using the phylogenetic rank boundary optimisation (PRBO) and modified general mixed Yule coalescent (GMYC) tests. In addition, a comprehensive phylogenetic analysis on an expanded LSU rRNA (D1/D2 domains) gene sequence dataset covering as many as available teleomorphic and filamentous taxa within Tremellomycetes was performed to investigate the relationships between yeasts and filamentous taxa and to examine the stability of undersampled clades. Based on the results inferred from molecular data and morphological and physiochemical features, we propose an updated classification for the Tremellomycetes. We accept five orders, 17 families and 54 genera, including seven new families and 18 new genera. In addition, seven families and 17 genera are emended and one new species name and 185 new combinations are proposed. We propose to use the term pro tempore or pro tem. in abbreviation to indicate the species names that are temporarily maintained.
Key words: Jelly fungi, Morphology, Multigene phylogeny, Ranks, Taxonomy, Tremellomycetes, Yeasts
Taxonomic novelties: New families: Bulleraceae X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Bulleribasidiaceae X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Mrakiaceae X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Naemateliaceae X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Phaeotremellaceae A.M. Yurkov & Boekhout; Piskurozymaceae X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Trimorphomycetaceae X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout
New genera: Bandonia A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Carlosrosaea A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cutaneotrichosporon X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Dimennazyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Effuseotrichosporon A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Fonsecazyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Gelidatrema A.M. Yurkov, X.Z. Liu, F.Y. Bai; M. Groenew. & Boekhout; Genolevuria X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Goffeauzyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Haglerozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Krasilnikovozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Nielozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Piskurozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Pseudotremella X.Z. Liu, F.Y. Bai, A.M. Yurkov, M. Groenew. & Boekhout; Saitozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Sugitazyma A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Solicoccozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Vishniacozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout
New species: Kockovaellaprillingeri (Prillinger, G. Kraep. & Lopandic) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout
New combinations: Apiotrichumbrassicae (Nakase) A.M. Yurkov & Boekhout; A. cacaoliposimilis (J.L. Zhou, S.O. Suh & Gujjari) Kachalkin, A.M. Yurkov & Boekhout; A. dehoogii (Middelhoven, Scorzetti & Fell) A.M. Yurkov & Boekhout; A. domesticum (Sugita, A. Nishikawa & Shinoda) A.M. Yurkov & Boekhout; A. dulcitum (Berkhout) A.M. Yurkov & Boekhout; A. gamsii (Middelhoven, Scorzetti, Sigler & Fell) A.M. Yurkov & Boekhout; A. gracile (Weigmann & A. Wolff) A.M. Yurkov & Boekhout; A. laibachii (Windisch) A.M. Yurkov & Boekhout; A. lignicola (Diddens) A.M. Yurkov & Boekhout; A. loubieri (Morenz) A.M. Yurkov & Boekhout; A. montevideense (L.A. Queiroz) A.M. Yurkov & Boekhout; A. mycotoxinivorans (O. Molnár, Schatzm. & Prillinger) A.M. Yurkov & Boekhout; A. scarabaeorum (Middelhoven, Scorzetti & Fell) A.M. Yurkov & Boekhout; A. siamense (Nakase, Jindam., Sugita & H. Kawas.) Kachalkin, A.M. Yurkov & Boekhout; A. sporotrichoides (van Oorschot) A.M. Yurkov & Boekhout; A. vadense (Middelhoven, Scorzetti & Fell) A.M. Yurkov & Boekhout; A. veenhuisii (Middelhoven, Scorzetti & Fell) A.M. Yurkov & Boekhout; A. wieringae (Middelhoven) A.M. Yurkov & Boekhout; A. xylopini (S.O. Suh, Lee, Gujjari & Zhou) Kachalkin, A.M. Yurkov & Boekhout; Bandoniamarina (van Uden & Zobell) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Bulleribasidiumbegoniae (Nakase, Tsuzuki, F.L. Lee & M. Takash.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Bu. foliicola (Q.M. Wang, F.Y. Bai, Boekhout & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Bu. hainanense (Q.M. Wang, F.Y. Bai, Boekhout & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Bu. panici (Fungsin, M. Takash. & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Bu. pseudovariabile (F.Y. Bai, M. Takash. & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Bu. sanyaense (Q.M. Wang, F.Y. Bai, Boekhout & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Bu. setariae (Nakase, Tsuzuki, F.L. Lee & M. Takash.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Bu. siamense (Fungsin, M. Takash. & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Bu. variabile (Nakase & M. Suzuki) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Bu. wuzhishanense (Q.M. Wang, F.Y. Bai, Boekhout & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Carcinomycesarundinariae (Fungsin, M. Takash. & Nakase) A.M. Yurkov; Carc. polyporina (D.A. Reid) A.M. Yurkov; Carlosrosaeavrieseae (Landell, Brandão, Safar, Gomes, Félix, Santos, Pagani, Ramos, Broetto, Mott, Valente & Rosa) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cryptococcusdepauperatus (Petch) Boekhout, Liu, Bai & M. Groenew.; Cr. luteus (Roberts) Boekhout, Liu, Bai & M. Groenew.; Cutaneotrichosporonarboriformis (Sugita, M. Takash., Sano, Nishim., Kinebuchi, S. Yamag. & Osanai) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cu. curvatus (Diddens & Lodder) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cu. cutaneum (de Beurmann, Gougerot & Vaucher) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cu. cyanovorans (Motaung, Albertyn, J.L.F. Kock et Pohl) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cu. daszewskae (Takash., Sugita, Shinoda & Nakase) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cu. debeurmannianum (Sugita, Takash., Nakase & Shinoda) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cu. dermatis (Sugita, Takash., Nakase, Ichikawa, Ikeda & Shinoda) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cu. guehoae (Middelhoven, Scorzettii & Fell) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cu. haglerorum (Middelhoven, Á. Fonseca, S.C. Carreiro, Pagnocca & O.C. Bueno) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cu. jirovecii (Frágner) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cu. moniliiforme (Weigmann & A. Wolff) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cu. mucoides (E. Guého & M.T. Smith) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cu. oleaginosus (J.J. Zhou, S.O. Suh & Gujjari) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cu. smithiae (Middelhoven, Scorzetti, Sugita & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Cu. terricola (Sugita, M. Takash. & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Dimennazyma cistialbidi (Á. Fonseca, J. Inácio & Spenc.-Mart.) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Effuseotrichosporon vanderwaltii (Motaung, Albertyn, Kock, C.F. Lee, S.O. Suh, M. Blackwell & C.H. Pohl) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Filobasidium chernovii (Á. Fonseca, Scorzetti & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Fil. magnum (Lodder & Kreger-van Rij) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Fil. oeirense (Á. Fonseca, Scorzetti & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Fil. stepposum (Golubev & J.P. Samp.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Fil. wieringae (Á. Fonseca, Scorzetti & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Fonsecazyma betulae (K. Sylvester, Q.M. Wang, C. T. Hittinger) A.M. Yurkov, A.V. Kachalkin & Boekhout; Fon. mujuensis (K.S. Shin & Y.H. Park) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Fon. tronadorensis (V. De Garcia, Zalar, Brizzio, Gunde-Cim. & van Brook) A.M. Yurkov; Gelidatrema spencermartinsiae (Garcia, Brizzio, Boekhout, Theelen, Libkind & van Broock) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Genolevuria amylolytica (Á. Fonseca, J. Inácio & Spenc.-Mart.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Gen. armeniaca (Á. Fonseca & J. Inácio) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Gen. bromeliarum (Landell & P. Valente) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Gen. tibetensis (F.Y. Bai & Q.M. Wang) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Goffeauzyma aciditolerans (Gadanho & J.P. Samp.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Gof. agrionensis (Russo, Libkind, Samp. & van Broock) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Gof. gastrica (Reiersöl & di Menna) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Gof. gilvescens (Chernov & Babeva) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Gof. iberica (Gadanho & J.P. Samp.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Gof. metallitolerans (Gadanho & J.P. Samp.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Haglerozymachiarellii (Pagnocca, Legaspe, Rodrigues & Ruivo) A. M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Heterocephalacriaarrabidensis (Á. Fonseca, Scorzetti & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Het. bachmannii (Diederich & M.S. Christ.) Millanes & Wedin; Het. physciacearum (Diederich) Millanes & Wedin; Itersoniliapannonica (Niwata, Takash., Tornai-Lehoczki, T. Deák & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Kockovaellachinensis (Prillinger, G. Kraep. & Lopandic) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Ko. distylii (Hamam., Kuroy. & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Ko. fuzhouensis (J.Z. Yue) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Ko. lichenicola (Prillinger, G. Kraep. & Lopandic) X.Z. Liu, F.Y. Bai; M. Groenew. & Boekhout; Ko. mexicana (Lopandic, O. Molnár & Prillinger) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Ko. ogasawarensis (Hamam., Kuroy. & Nakase) X.Z. Liu, F.Y. Bai, Groenew. & Boekhout; Ko. sichuanensis (Prillinger, G. Kraep. & Lopandic) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Krasilnikovozymahuempii (C. Ramírez & A. E. González) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Kr. tahquamenonensis (Wang, Hulfachor, Sylvester and Hittinger) A.M. Yurkov; Kwoniellabestiolae (Thanh, Hai & Lachance) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Kw. dejecticola (Thanh, Hai & Lachance) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Kw. dendrophila (Van der Walt & D.B. Scott) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Kw. pini (Golubev & Pfeiffer) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Kw. shivajii (S.R. Ravella, S.A. James, C.J. Bond, I.N. Roberts, K. Cross, Retter & P.J. Hobbs) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Mrakiaaquatica (E.B.G. Jones & Slooff) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; M. cryoconiti (Margesin & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; M. niccombsii (Thomas-Hall) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Naemateliaaurantialba (Bandoni & M. Zang) Millanes & Wedin; Naem. microspora (Lloyd) Millanes & Wedin; Naganishiaadeliensis (Scorzetti, I. Petrescu, Yarrow & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Nag. albida (Saito) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Nag. albidosimilis (Vishniac & Kurtzman) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Nag. antarctica (Vishniac & Kurtzman) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Nag. bhutanensis (Goto & Sugiy.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Nag. cerealis (Passoth, A.-C. Andersson, Olstorpe, Theelen, Boekhout & Schnürer) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Nag. diffluens (Zach) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Nag. friedmannii (Vishniac) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Nag. liquefaciens (Saito & M. Ota) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Nag. onofrii (Turchetti, Selbmann & Zucconi) A.M. Yurkov; Nag. randhawae (Z.U. Khan, S.O. Suh. Ahmad, F. Hagen, Fell, Kowshik, Chandy & Boekhout) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Nag. uzbekistanensis (Á. Fonseca, Scorzetti & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Nag. vaughanmartiniae (Turchetti, Blanchette & Arenz) A.M. Yurkov; Nag. vishniacii (Vishniac & Hempfling) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Nielozymaformosana (Nakase, Tsuzuki, F.L. Lee & M. Takash.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Niel. melastomae (Nakase, Tsuzuki, F.L. Lee & M. Takash.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Papiliotremaanemochoreius (C.H. Pohl, Kock, P.W.J. van Wyk & Albertyn) F.Y. Bai, M. Groenew. & Boekhout; Pap. aspenensis (K. Ferreira-Paim, T.B. Ferreira, L. Andrade-Silva, D.J. Mora, D.J. Springer, J. Heitman, F.M. Fonseca, D. Matos, M.S.C. Melhem & M.L. Silva-Vergara) X.Z. Liu, F.Y. Bai, A.M. Yurkov & Boekhout; Pap. aurea (Saito) M. Takash., Sugita, Shinoda & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Pap. baii (A.M. Yurkov, M.A. Guerreiro & Á. Fonseca) A.M. Yurkov; Pap. flavescens (Saito) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Pap. fonsecae (V. de García, Zalar, Braizzio, Gunde-Cim. & van Brollck) A.M. Yurkov; Pap. frias (V. de García, Zalar, Braizzio, Gunde-Cim. & van Brollck) A.M. Yurkov; Pap. fuscus (J.P. Samp., J. Inácio, Fonseca & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Pap. hoabinhensis (D.T. Luong, M. Takash., Ty. Dung & Nakase) A.M. Yurkov; Pap. japonica (J.P. Samp., Fonseca & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Pap. laurentii (Kuff.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Pap. mangalensis (Fell, Statzell & Scorzett) A.M. Yurkov; Pap. nemorosus (Golubev, Gadanho, J.P. Samp. & N.W. Golubev) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Pap. perniciosus (Golubev, Gadanho, J.P. Samp. & N.W. Golubev) X.Z. Liu, F.Y. Bai; M. Groenew. & Boekhout; Pap. pseudoalba (Nakase & M. Suzuki) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Pap. rajasthanensis (Saluja & G.S. Prasad) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Pap. ruineniae (A.M. Yurkov, M.A. Guerreiro & Á. Fonseca) A.M. Yurkov; Pap. taeanensis (K.S. Shin & Y.H. Park) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Pap. terrestris (Crestani, Landell, Faganello, Vainstein, Vishniac & P. Valente) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Pap. wisconsinensis (Crestani, Landell, Faganello, Vainstein, Vishniac & P. Valente) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Phaeotremellafagi (Middelhoven & Scorzetti) A.M. Yurkov & Boekhout; Ph. mycetophiloides (Kobayasi) Millanes & Wedin; P. mycophaga (G.W. Martin) Millanes & Wedin; Ph. neofoliacea (Chee J. Chen) Millanes & Wedin; Ph. simplex (H.S. Jacks. & G.W. Martin) Millanes & Wedin; Ph. skinneri (Phaff & Carmo Souza) A.M. Yurkov & Boekhout; Pseudotremellaallantoinivorans (Middelhoven) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Ps. lacticolor (Satoh & Makimura) A.M. Yurkov; Ps. moriformis (Berk.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Ps. nivalis (Chee J. Chen) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Piskurozymacapsuligena (Fell, Statzell, I.L. Hunter & Phaff) A.M. Yurkov; Pis. cylindrica (Á. Fonseca, Scorzetti & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Pis. fildesensis (T. Zhang & L.-Y. Yu) A.M. Yurkov; Pis. filicatus (Golubev & J.P. Samp.) Kachalkin; Pis. silvicola (Golubev & J.P. Samp.) X.Z. Liu, F.Y. Bai, Groenew. & Boekhout; Pis. sorana (Hauerslev) A.M. Yurkov; Pis. taiwanensis (Nakase, Tsuzuki & M. Takash.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Rhynchogastremaaquatica (Brandao, Valente, Pimenta & Rosa) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov; R. complexa (Landell, Pagnocca, Sette, Passarini, Garcia, Ribeiro, Lee, Brandao, Rosa & Valente) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov; R. fermentans (Lee) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov; R. glucofermentans (S.O. Suh & Blackwell) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov; R. nanyangensis (F.L. Hui & Q.H. Niu) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov; R. noutii (Boekhout, Fell, Scorzett & Theelen) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov; R. tunnelae (Boekhout, Fell, Scorzetti & Theelen) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov; R. visegradensis (Peter & Dlauchy) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov; Saitozymaflava (Saito) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Sait. ninhbinhensis (Luong, Takash., Dung & Nakase) A.M. Yurkov; Sait. paraflava (Golubev & J.P. Samp.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Sait. podzolica (Babeva & Reshetova) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Solicoccozymaaeria (Saito) A.M. Yurkov; Sol. fuscescens (Golubev) A.M. Yurkov; Sol. keelungensis (C.F. Chang & S.M. Liu) A.M. Yurkov; Sol. phenolicus (Á. Fonseca, Scorzetti & Fell) A.M. Yurkov; Sol. terreus (Di Menna) A.M. Yurkov; Sol. terricola (T.A. Pedersen) A.M. Yurkov; Sugitazymamiyagiana (Nakase, Itoh, Takem. & Bandoni) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Tausoniapullulans (Lindner) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Tremellayokohamensis (Alshahni, Satoh & Makimura) A.M. Yurkov; Trimorphomycessakaeraticus (Fungsin, M. Takash. & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov; Vanrijafragicola (M. Takash., Sugita, Shinoda & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Van. meifongana (C.F. Lee) Kachalkin, A.M. Yurkov & Boekhout; Van. nantouana (C.F. Lee) Kachalkin, A.M. Yurkov & Boekhout; Van. thermophila (Vogelmann, Chaves & Hertel) Kachalkin, A.M. Yurkov & Boekhout; Vishniacozymacarnescens (Verona & Luchetti) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Vis. dimennae (Fell & Phaff) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Vis. foliicola (Q.M. Wang & F.Y. Bai) A.M. Yurkov; Vis. globispora (B.N. Johri & Bandoni) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Vis. heimaeyensis (Vishniac) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Vis. nebularis (Vishniac) A.M. Yurkov; Vis. peneaus (Phaff, Mrak & O.B. Williams) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Vis. psychrotolerans (V. de García, Zalar, Brizzio, Gunde-Cim. & van Broock) A.M. Yurkov; Vis. taibaiensis (Q.M. Wang & F.Y. Bai) A.M. Yurkov; Vis. tephrensis (Vishniac) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout; Vis. victoriae (M.J. Montes, Belloch, Galiana, M.D. García, C. Andrés, S. Ferrer, Torr.-Rodr. & J. Guinea) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout
Introduction
Tremellomycetes is a class of Agaricomycotina (Hibbett et al., 2007, Boekhout et al., 2011a) and encompasses yeasts, dimorphic taxa and species that form hyphae and/or complex fruiting bodies. Five orders, namely Cystofilobasidiales, Filobasidiales, Holtermanniales, Tremellales and Trichosporonales, are currently recognised in Tremellomycetes based on phenotypic and phylogenetic properties (Fell et al., 2000, Scorzetti et al., 2002, Boekhout et al., 2011a, Wuczkowski et al., 2011, Weiss et al., 2014, Liu et al., 2015). However, the separation of Trichosporonales from Tremellales remains a matter of debate (Hibbett et al., 2007, Millanes et al., 2011).
Basidiomycetous yeasts, like ascomycetous yeasts, were conventionally classified based on morphological features, including sexual and asexual reproductive structures and morphology; chemotaxonomic criteria, including cell-wall composition data and ubiquinone types; and physiological properties characterised by standardised tests on the assimilation and fermentation of carbon and nitrogen compounds, production of starch-like compounds, and other growth tests (Lodder and Kreger-van Rij, 1952, Van der Walt, 1970, Van der Walt and Yarrow, 1984, Boekhout et al., 1993, Boekhout et al., 2011a, Kurtzman et al., 2011b, Prillinger et al., 1993, McLaughlin et al., 1995, Sampaio and Fonseca, 1995, Nguyen et al., 1998, Yarrow, 1998, Takashima et al., 2000, Bauer et al., 2006, Celio et al., 2006). Basidiocarp-forming Tremellomycetes were mainly classified using morphological features, such as form and consistency of the fruiting bodies, and details of hyphae, basidia, basidiospores etc. The classification based on these phenotypical features, however, was in many cases not consistent with the results obtained from molecular phylogenetic analyses. This non-concordance is one of the most prominent problems in the classification of this group of fungi, as illustrated by the polyphyletic nature of many currently recognised genera. Bullera and Cryptococcus are just two examples of genera that are highly polyphyletic with species belonging to more than one order of Tremellomycetes (Boekhout et al., 2011b, Fonseca et al., 2011). Further discovery of new species belonging to these genera not only worsened the problem of systematics of Tremellomycetes, but also impeded the communication of researchers from different fields. For instance, a large number of environmental sequences have been obtained by microbial ecologists and were identified to species belonging to these polyphyletic genera. The NCBI GenBank database (http://www.ncbi.nlm.nih.gov/) is the most widely used tool to identify these molecular reads, but the taxon names presented in the results from GenBank searches are often confusing because they refer to such non-monophyletic groups or genera.
Molecular phylogenetic analyses of multiple genes have become an effective approach to reconstruct fungal phylogenies. As the result of the Assembling the Fungal Tree of Life (AFTOL) project, James et al. (2006) used a six-gene dataset to resolve a kingdom-wide fungal phylogeny and Hibbett et al. (2007) proposed a comprehensive phylogenetic re-classification of the fungi down to the ordinal level. Yeast species formed only a small part of the nearly 200 fungal species involved in the AFTOL project. The majority of teleomorphic ascomycetous yeast taxa have independently been reclassified based on results obtained from multigene phylogenetic studies (Kurtzman, 2003, Kurtzman and Robnett, 2003, Kurtzman and Robnett, 2007, Kurtzman et al., 2007, Kurtzman et al., 2008), but most basidiomycetous yeast taxa remain to be studied. In the fifth edition of ‘The Yeasts, a Taxonomic Study’ (Kurtzman et al. 2011a) many of the basidiomycetous yeast genera included are still highly polyphyletic.
The recent changes in the dual nomenclature for pleomorphic fungi prompted us to modify the classification of Tremellomycetes to fulfil the “One Fungus = One Name” principle (McNeill et al. 2012). As is the case in other groups of fungi, many species of this class have separate teleomorphic and anamorphic names. A further change in the new nomenclature is that electronic publication of new taxa and their names is permitted from 1 January 2012 in the absence of a printed hard copy (Knapp et al. 2011). So far, approximately 190 new fungal species have been e-published in Index Fungorum since then (http://www.indexfungorum.org/). The e-publication rule will accelerate the description of new fungal species, and this further urges us to revise the taxonomy of the Tremellomycetes in order to provide a stable taxonomic framework that reflects our knowledge of the phylogenetic diversity and relationships of these fungi.
Several studies across the eukaryotic tree of life have shown that there is poor equivalence of taxonomic ranks across the groups studied (Johns and Avise, 1998, Avise and Johns, 1999, Castresana, 2001, Lumbsch, 2002, Avise and Liu, 2011, Talavera et al., 2013). Clades of the same rank often show high variance with regard to genetic distances and temporal aspects. Despite the arbitrary nature of taxonomic ranks above the species level, they have importance in an evolutionary framework for cataloguing and communicating about biological diversity and, hence, taxonomic decisions should be made on a consistent basis (Holt & Jonsson 2014). Different methods have been suggested to level off ranks within taxonomic groups, including a temporal approach (Hennig, 1966, Farris, 1976, Avise and Johns, 1999, Holt and Jonsson, 2014), but they have not commonly been applied in botany, mycology and zoology (Vences et al. 2013). In bacteriology, a quantitative interpretation of ranks is much more common, particularly by means of traditional (Tindall et al. 2010) or digital (Meier-Kolthoff et al. 2013) DNA:DNA hybridisation thresholds for species and 16S rRNA gene thresholds for higher ranks (Yarza et al. 2014). However, thresholds for pairwise (dis-)similarities are not a phylogenetic criterion (Wiley & Lieberman 2011) and can yield inconsistencies (Meier-Kolthoff et al. 2014). Moreover, the method used by Yarza et al. (2014) to estimate similarity thresholds does not guarantee maximum agreement with the existing assignment to ranks, which could be obtained using clustering optimisation (Göker et al., 2009, Göker et al., 2010, Stielow et al., 2011). Another question regarding Linnaean classification is whether higher taxa are real; this has usually been denied in the literature (Wiley and Lieberman, 2011, Vences et al., 2013), but recent studies found statistical tests to identify evolutionary significant units above the species level (Humphreys and Barraclough, 2014, Barraclough and Humphreys, 2015).
Recently, we reconstructed the phylogeny of tremellomycetous yeasts and related dimorphic and filamentous Tremellomycetes by analysing sequences from seven genes (Liu et al. 2015), resulting in a relatively robust framework that allows us to update the taxonomic system of the Tremellomycetes. Here, we employed two quantitative methods, namely a phylogenetic variant of clustering optimisation (Göker et al., 2009, Göker et al., 2010, Stielow et al., 2011) and the iterative application of a modified general mixed Yule coalescent (GMYC) (Humphreys & Barraclough 2014), to test and circumscribe the taxonomic units at the order, family and genus levels recognised from this multigene phylogeny. Results of either method were not followed strictly, however, but were modified where necessary to decrease the number of new taxa to be introduced. Moreover, a comprehensively sampled LSU rRNA gene phylogeny, including taxa that were not studied in the seven-gene phylogeny, was constructed for Tremellomycetes integrating both yeasts and filamentous fungi, incorporating information from the seven-gene phylogeny by using backbone constraints. An updated taxonomic system for Tremellomycetes is consequently proposed based on the integrated phylogenetic evidence combined with morphological and physiological criteria.
Materials and methods
Organisms
A total of 294 tremellomycetous yeast strains, including the type strains of 286 currently recognised species and varieties as listed in Table 1 of Liu et al. (2015), were employed in this study. In addition, 47 tremellomycetous yeast species which were published too late to be included in the study of Liu et al. (2015) and 47 more fruiting body forming species from the genera Tremella, Syzygospora, Rhynchogastrema, Tetragoniomyces and Trimorphomyces were employed in this study (Table 1). Additionally, 23 novel but undescribed species retrieved from the public dataset were also included. Five Cryptococcus species recently described by Hagen et al. (2015) were listed in Table 1 of accepted species names but not included in the phylogenetic analyses. Large ribosomal subunit (LSU) rRNA gene sequences were additionally sampled specifically from related and filamentous taxa in Tremellomycetes, for which no culture material or other nucleotide data are available. All together, a total of 435 taxa were compared in this study.
Table 1.
List of accepted tremellomycetous yeast and dimorphic taxa.
| Taxon | Basionym or important synonym | Strain | LSU D1D2 |
|---|---|---|---|
| Cystofilobasidiales | |||
| Cystofilobasidiaceae emend. | |||
| Cystofilobasidium | |||
| C. bisporidii | CBS 6346T | EU085532 | |
| C. capitatumT | Rhodosporidium capitatum | CBS 6358T | AF075465 |
| C. ferigula | CBS 7202T | CBS database | |
| C. infirmominiatum | CBS 323T | AF075505 | |
| C. lacus-mascardii | CBS 10642T | AY158642 | |
| C. macerans | Cryptococcus macerans | CBS 10757T | EU082225 |
| Mrakiaceae fam. nov. | |||
| Itersonilia emend. | |||
| I. pannonicus comb. nov. | Udeniomyces pannonicus | CBS 9123T | AB077382 |
| I. perplexansT | CBS 363.85T | AJ235274 | |
| Krasilnikovozyma gen. nov. | |||
| K. huempiiT comb. nov. |
Candida huempii C. huempii Mrakia curviuscula |
CBS 8186T | AF189844 |
| K. tahquamenonensis comb. nov. | C. tahquamenonensis | CBS 13897T | KM408125 |
| Mrakia emend. | |||
| M. aquatica comb. nov. | Mrakiella aquatica | CBS 5443T | AF075470 |
| M. blollopis | CBS 8921T | AY038814 | |
| M. cryoconiti comb. nov. | M. cryoconiti | CBS 10834T | GQ911524 |
| M. frigidaT | Leucosporidium frigidum | CBS 5270T | AF075463 |
| M. gelida | L. gelidum | CBS 5272T | AF189831 |
| M. niccombsii comb. nov. | M. niccombsii | CBS 8917T | AY029345 |
| M. psychrophila | CBS 10828T | EU224266 | |
| M. robertii | CBS 8912T | AY038811 | |
| Phaffia emend. | Xanthophyllomyces | ||
| P. rhodozymaT | X. dendrorhous | CBS 5905T | AF189871 |
| Tausonia emend. | |||
| T. pamiricaT | CBS 8428T | EF118825 | |
| T. pullulans comb. nov. | Guehomyces pullulans | CBS 2532T | EF551318 |
| Udeniomyces | |||
| U. kanasensis | CBS 12488T | JQ002681 | |
| U. megalosporus | CBS 7236T | AF075510 | |
| U. puniceus | CBS 5689T | AF075519 | |
| U. pyricolaT | CBS 6754T | AF075507 | |
| Filobasidiales | |||
| Filobasidiaceae emend. | |||
| Filobasidium emend. | |||
| F. chernovii comb. nov. | C. chernovii | CBS 8679T | AF181530 |
| F. elegans | CBS 7640EXT | AF181548 | |
| F. floriformeT | CBS 6241EXT | AF075498 | |
| F. globisporum | CBS 7642EXT | AF075495 | |
| F. magnum comb. nov. | C. magnus | CBS 140T | AF181851 |
| F. oeirensis comb. nov. | C. oeirensis | CBS 8681T | AF181519 |
| F. stepposus comb. nov. | C. stepposus | CBS 10265T | DQ222456 |
| F. uniguttulatum | CBS 1730T | AF075468 | |
| F. wieringae comb. nov. | C. wieringae | CBS 1937T | AF181541 |
| Goffeauzyma gen. nov. | |||
| G. aciditolerans comb. nov. | Cryptococcus aciditolerans | CBS 10872T | AY731790 |
| G. agrionensis comb. nov. | C. agrionensis | CBS 10799T | EU627786 |
| G. gastricusT comb. nov. | C. gastricus | CBS 2288T | AF137600 |
| G. gilvescens comb. nov. | C. gilvescens | CBS 7525T | AF181547 |
| G. ibericus comb. nov. | C. ibericus | CBS 10871T | AY731791 |
| G. metallitolerans comb. nov. | C. metallitolerans | CBS 10873T | AY731789 |
| Heterocephalacria emend. | |||
| H. arrabidensis comb. nov. | C. arrabidensis | CBS 8678T | AF181535 |
| H. bachmannii comb. nov. | Syzygospora bachmannii | AM72 | JN043613 |
| H. physciacearum comb. nov. | S. physciacearum | AM17 | JN043614 |
| H. solida | S. solida | ||
| Naganishia emend. | |||
| N. adeliensis comb. nov. | C. adeliensis | CBS 8351T | AF137603 |
| N. albida comb. nov. | C. albidus | CBS 142T | AF075474 |
| N. albidosimilis comb. nov. | C. albidosimilis | CBS 7711T | AF137601 |
| N. antarctica comb. nov. | C. antarcticus | CBS 7687T | AF075488 |
| N. bhutanensis comb. nov. | C. bhutanensis | CBS 6294T | AF137599 |
| N. cerealis comb. nov. | C. cerealis | CBS 10505T | FJ473376 |
| N. diffluens comb. nov. | C. diffluens | CBS 160T | AF075502 |
| N. friedmannii comb. nov. | C. friedmannii | CBS 7160T | AF075478 |
| N. globosaT | C. saitoi | CBS 5106T | AF181539 |
| N. liquefaciens comb. nov. | C. liquefaciens | CBS 968T | AF181515 |
| N. onofrii comb. nov. | C. onofrii | DBVPG 5303T | KC433831 |
| N. randhawae comb. nov. | C. randhawai | CBS 10160T | AJ876599 |
| N. uzbekistanensis comb. nov. | C. uzbekistanensis | CBS 8683T | AF181508 |
| N. vaughanmartiniae comb. nov. | C. vaughanmartiniae | DBVPG4736T | KF861779 |
| N. vishniacii comb. nov. | C. vishniacii | CBS 7110T | AF075473 |
| Syzygospora | |||
| S. albaT | AM147 | JN043616 | |
| S. pallida | FO31621 | AJ406403 | |
| Piskurozymaceae fam. nov. | |||
| Piskurozyma gen. nov. | |||
| P. capsuligenum comb. nov. | Filobasidium capsuligenum | CBS 1906T | AF363642 |
| P. cylindricusT comb. nov. | C. cylindricusT | CBS 8680T | AF181534 |
| P. fildesensis comb. nov. | C. fildesensis | CBS12705 | KC894161 |
| P. filicatus comb. nov. | C. filicatus | CBS 10874T | EU433983 |
| P. silvicola comb. nov. | C. silvicola | CBS 10099T | AY898955 |
| P. sorana comb. nov. | S. sorana | UBC_F16310 | EU541305 |
| P. taiwanensis comb. nov. | Bullera taiwanensis | CBS 9813T | AB079065 |
| Solicoccozyma gen. nov. | |||
| S. aeriusT comb. nov. | C. aerius | CBS 155T | AF075486 |
| S. fuscescens comb. nov. | C. fuscescens | CBS 7189T | AF075472 |
| S. keelungensis comb. nov. | C. keelungensis | CBS 10876T | EF621562 |
| S. phenolicus comb. nov. | C. phenolicus | CBS 8682T | AF181523 |
| S. terreus comb. nov. | C. terreus | CBS 1895T | AF075479 |
| S. terricola comb. nov. | C. terricola | CBS 4517T | AF181520 |
| Holtermanniales | |||
| Holtermannia | |||
| H. corniformis | CBS 6979R | AF189843 | |
| Holtermanniella | |||
| H. festucosa | Cryptococcus festucosus | CBS 10162T | AY462119 |
| H. mycelialis | C. mycelialis | CBS 7712T | AJ311450 |
| H. nyarrowii | C. nyarrowii | CBS 8804T | AY006480 |
| H. takashimaeT | CBS 11174T | FM242574 | |
| H. wattica | C. watticus | CBS 9496T | AY138478 |
| Tremellales | |||
| Bulleraceae fam. nov. | |||
| Bullera emend. | Bulleromyces | ||
| B. albaT | B. albus | CBS 501T | AF075500 |
| B. hannae | CBS 8286T | AF363661 | |
| B. penniseticola | CBS 8623T | AF363649 | |
| B. unica | CBS 8290T | AF075524 | |
| Fonsecazyma gen. nov. | |||
| F. betulae comb. nov. | Kwoniella betulae | CBS 13896T | KM408130 |
| F. mujuensisT comb. nov. | C. mujuensis | CBS 10308T | DQ333884 |
| F. tronadorensis comb. nov. | C. tronadorensis | CBS 12691T | GU560003 |
| Genolevuria gen. nov. | |||
| G. amylolyticusT comb. nov. | C. amylolyticus | CBS 10048T | AY562134 |
| G. armeniacus comb. nov. | C. armeniacus | CBS 10050T | AY562140 |
| G. bromeliarum comb. nov. | C. bromeliarum | CBS 10424T | DQ784566 |
| G. tibetensis comb. nov. | C. tibetensis | CBS 10456T | EF363143 |
| Pseudotremella gen. nov. | |||
| P. allantoinivorans comb. nov. | C. allantoinivorans | CBS 9604T | AY315662 |
| P. lacticolor comb. nov. | C. lacticolor | CBS 10915T | AB375775 |
| P. moriformisT comb. nov. | Tremella moriformis | CBS 7810R | AF075493 |
| P. nivalis comb. nov. | T. nivalis | CBS 8487R | AF042232 |
| Tremella Clade I (Millanes et al. 2011) | |||
| Cryptococcus cuniculi pro tem | CBS 10309T | DQ333885 | |
| Sirobasidium brefeldianum | AM71 | JN043578 | |
| S. intermedium | CBS 7805 | AF075492 | |
| Tremella caloplacae | AM31 | JN043573 | |
| T. candelariellae | AM34 | JN043575 | |
| T. christiansenii | AM36 | JN043577 | |
| T. dendrographae | AM39 | JN043576 | |
| T. exigua | RB6623-15 | AF042248 | |
| Tremella Clade III (Millanes et al. 2011) | |||
| Biatoropsis usnearum | Sweden, Hagner s.n. (S-F92134) | JN043592 | |
| Tremella cetrariicola | Finland, Suija s.n. (S-F102413) | JN043596 | |
| T. coppinsii | UK, Diederich 15628 (S-F102414) | JN043601 | |
| T. everniae | USA, 2005, Kneiper s.n. (S) | JN043599 | |
| T. giraffa | CBS 8489R | AF042271 | |
| T. huuskonenii | Canada, B.C., Goward 11-50 (UBC), L321S/L321 | KR857095 | |
| T. hypogymniae | Sweden, Wedin 6892 (UPS) | JN043590 | |
| T. lichenicola | Germany, John & Diederich s.n. (UPS-256878) | JN043611 | |
| T. pertusariae | France, Diederich 16331 (S-F102502) | JN043600 | |
| T. tuckerae | Mexico, Tucker 37335 (SBBG) | JN043588 | |
| T. wirthii | The Netherlands, 1996, Herk s.n. (herb. Diederich) | JN043598 | |
| Single-species lineages | |||
| Tremella ‘indecorata’ I pro tem | AM5 | JN043610 | |
| Tremella haematommatis pro tem | AM41 | JN043617 | |
| Tremella ramalinae pro tem | Spain, Etayo s.n. (UPS 158799) | JN043619 | |
| Bulleribasidiaceae fam. nov. | |||
| Bulleribasidium emend. | Mingxiaea | ||
| B. begoniae comb. nov. | Bullera begoniae | CBS 10762T | AB119462 |
| B. foliicola comb. nov. | M. foliicola | CBS 11407T | GQ438834 |
| B. hainanense comb. nov. | M. hainanensis | CBS 11409T | GQ438828 |
| B. oberjochenseT | CBS 9110T | AF416646 | |
| B. panici comb. nov. | B. panici | CBS 9932T | AY188387 |
| B. pseudovariabile comb. nov. | B. pseudovariabilis | CBS 9609T | AF544247 |
| B. sanyaense comb. nov. | M. sanyaensis | CBS 11408T | GQ438831 |
| B. setariae comb. nov. | B. setariae | CBS 10763T | AB119463 |
| B. siamense comb. nov. | B. siamensis | CBS 9933T | AY188388 |
| B. variabile comb. nov. | B. variabilis | CBS 7347T | AF189855 |
| B. wuzhishanense comb. nov. | M. wuzhishanensis | CBS 11411T | GQ438830 |
| Derxomyces | |||
| D. amylogenes | CBS 12233T | HQ890372 | |
| D. anomala | B. anomala | CBS 9607T | EF682504 |
| D. bambusicola | CBS 12234T | HQ890376 | |
| D. boekhoutii | CBS 10824T | EU517057 | |
| D. boninensis | B. boninensis | CBS 9141T | AY487568 |
| D. corylopsis | CBS 12259T | HQ890374 | |
| D. cylindrica | B. cylindrica | CBS 9744T | AY487563 |
| D. hainanensis | CBS 10820T | EU517056 | |
| D. hubeiensis | B. hubeiensis | CBS 9747T | AY487566 |
| D. huiaensis | B. huiaensis | CBS 8287T | AB118870 |
| D. komagatae | B. komagatae | CBS 10153T | AF544249 |
| D. linzhiensis | CBS 10827T | EU517058 | |
| D. mrakiiT | B. mrakii | CBS 8288T | AB118871 |
| D. nakasei | B. nakasei | CBS 9746T | AY487564 |
| D. pseudocylindrica | CBS 10826T | EU517059 | |
| D. pseudohuiaensis | B. pseudohuiaensis | CBS 7364T | AF544250 |
| D. pseudoschimicola | B. pseudoschimicola | CBS 7354T | AF416647 |
| D. qinlingensis | CBS 10818T | EU517060 | |
| D. schimicola | B. schimicola | CBS 9144T | AY487570 |
| D. simaoensis | CBS 10822T | EU517062 | |
| D. waltii | B. waltii | CBS 9143T | AY487569 |
| D. wuzhishanensis | CBS 10825T | EU517063 | |
| D. yunnanensis | CBS 10821T | EU517064 | |
| Dioszegia | |||
| D. antarctica | CBS 10920T | FJ640575 | |
| D. athyri | CBS 10119T | EU070931 | |
| D. aurantiaca | Bullera aurantiaca | CBS 6980T | AB104689 |
| D. buhagiarii | CBS 10054T | AY562151 | |
| D. butyracea | CBS 10122T | EU070929 | |
| D. catarinonii | CBS 10051T | AY562142 | |
| D. changbaiensis | CBS 9608T | AY242819 | |
| D. crocea | B. crocea | CBS 6714T | AF075508 |
| D. cryoxerica | CBS 10919T | FJ640562 | |
| D. fristingensis | CBS 10052T | AY562146 | |
| D. hungaricaT | Cryptococcus hungaricus | CBS 4214T | AF075503 |
| D. rishiriensis | CBS 11844T | AB545810 | |
| D. statzelliae | CBS 8925T | AY029341 | |
| D. takashimae | CBS 10053T | AY562149 | |
| D. xingshanensis | CBS 10120T | EU070928 | |
| D. zsoltii | CBS 9127T | AF544245 | |
| Hannaella | |||
| H. coprosmaensis | B. coprosmae | CBS 8284T | AF363660 |
| H. kunmingensis | B. kunmingensis | CBS 8960T | AB109558 |
| H. luteola | C. luteolus | CBS 943T | AF075482 |
| H. oryzae | B. oryzae | CBS 7194T | AF075511 |
| H. pagnoccae | CBS 11142T | FJ828959 | |
| H. phetchabunensis | CBS 13386T | AB922849 | |
| H. phyllophila | CBS 13921T | AB934929 | |
| H. siamensis | CBS 13533T | AB922844 | |
| H. sinensisT | B. sinensis, B. derxii | CBS 7238T | AF189884 |
| H. surugaensis | C. surugaensis | CBS 9426T | AB100440 |
| Nielozyma gen. nov. | |||
| N. formosana comb. nov. | B. formosana | CBS 10306T | AB119465 |
| N. melastomaeT comb. nov. | B. melastomae | CBS 10305T | AB119464 |
| Vishniacozyma gen. nov. | |||
| V. carnescensT comb. nov. |
Torulopsis carnescens C. carnescens |
CBS 973T | AB035054 |
| V. dimennae comb. nov. | C. dimennae | CBS 5770T | AF075489 |
| V. foliicola comb. nov. | C. foliicola | CBS 9920T | AY557599 |
| V. globispora comb. nov. | B. globispora | CBS 6981T | AF075509 |
| V. heimaeyensis comb. nov. | C. heimaeyensis | CBS 8933T | DQ000317 |
| V. nebularis comb. nov. | Trimorphomyces nebularis | CBS12283 | EU266921 |
| V. peneaus comb. nov. |
Rhodotorula peneaus C. peneaus |
CBS 2409T | AB035051 |
| V. psychrotolerans comb. nov. | C. psychrotolerans | CBS 12690T | JN193445 |
| V. taibaiensis comb. nov. | C. taibaiensis | CBS 9919T | AY557601 |
| V. tephrensis comb. nov. | C. tephrensis | CBS 8935T | DQ000318 |
| V. victoriae comb. nov. | C. victoriae | CBS 8685T | AF363647 |
| Carcinomycetaceae emend. | |||
| Carcinomyces emend. | |||
| C. arundinariae comb. nov. | B. arundinariae | CBS 9931T | AF547661 |
| C. effibulatusT | Syzygospora effibulata | AM6 | JN043605 |
| C. polyporinus comb. nov. | Tremella polyporina | AM20 | JN043607 |
| Tremella Clade II (Millanes et al. 2011) | |||
| Tremella cladoniae | AM125 | JN043583 | |
| T. leptogii | AM81 | JN043582 | |
| T. lobariacearum | AM80 | JN043579 | |
| T. nephromatis | AM133 | JN043581 | |
| T. phaeophysciae | AM98 | JN043585 | |
| Cryptococcaceae emend. | |||
| Cryptococcus emend. | |||
| C. amylolentus |
Candida amylolenta Filobasidiella amylolenta Tsuchiyaea wingfieldii Sterigmatomyces wingfieldii |
CBS 6039T | AF105391 |
| C. bacillisporus | F. bacillispora | CBS 6955T | JN939485 |
| C. decagattii | CBS 11687T | ||
| C. deneoformans |
F. neoformans F. neoformans var. neoformans |
CBS 6900T | |
| C. depauperatus comb. nov. | F. depauperata | CBS 7841T | FJ534911 |
| C. deuterogattii | CBS 10514T | FJ534907 | |
| C. gattii | CBS 6289T | AF075526 | |
| C. luteus comb. nov. | F. lutea | ||
| C. neoformansT | Cryptococcus neoformans var. grubii | CBS 8710T | FJ534909 |
| C. tetragattii | CBS 10101T | ||
| Kwoniella emend. | |||
| K. bestiolae comb. nov. | C. bestiolae | CBS 10118T | FJ534903 |
| K. botswanensis | CBS 12716T | HF545769 | |
| K. dejecticola comb. nov. | C. dejecticola | CBS 10117T | AY917102 |
| K. dendrophila comb. nov. | Bullera dendrophila | CBS 6074T | AF189870 |
| K. europaea | CBS 12714T | AY167602 | |
| K. heveanensis | C. heveanensis | CBS 569T | AF075467 |
| K. mangroviensisT | CBS 8507T | AF444742 | |
| K. newhampshirensis | CBS 13917T | KM408127 | |
| K. pinus comb. nov. | C. pinus | CBS 10737T | EF672245 |
| K. shandongensis | CBS 12478T | JN160602 | |
| K. shivajii comb. nov. | C. shivajii | CBS 11374T | FM212446 |
| Cuniculitremaceae | |||
| Fellomyces | |||
| F. borneensis | CBS 8282T | AF189877 | |
| F. horovitziae | CBS 7515T | AF189856 | |
| F. penicillatus | CBS 5492T | AF177405 | |
| F. polyborusT | CBS 6072T | AF189859 | |
| Kockovaella emend. | |||
| K. barringtoniae | CBS 9811T | AB292854 | |
| K. calophylli | CBS 8962T | AB292852 | |
| K. chinensis comb. nov. | Fellomyces chinensis | CBS 8278T | AF189878 |
| K. cucphuongensis | CBS 8959T | AB292853 | |
| K. distylii comb. nov. | F. distylii | CBS 8545T | AF363652 |
| K. fuzhouensis comb. nov. | F. fuzhouensis | CBS 8243T | AF363659 |
| K. imperatae | CBS 7554T | AF189862 | |
| K. lichenicola comb. nov. | F. lichenicola | CBS 8315T | AF363643 |
| K. litseae | CBS 8964T | AB292850 | |
| K. machilophila | CBS 8607T | AF363654 | |
| K. mexicanus comb. nov. | F. mexicanus | CBS 8279T | AJ627906 |
| K. ogasawarensis comb. nov. | Fellomyces ogasawarensis | CBS 8544T | AF363651 |
| K. phaffii | CBS 8608T | AF363655 | |
| K. prillingeri sp. nov. | F. thailandicus | CBS 8308T | AF363644 |
| K. sacchari | CBS 8624T | AF363650 | |
| K. schimae | CBS 8610T | AF363656 | |
| K. sichuanensis comb. nov. | F. sichuanensis | CBS 8318T | AF189879 |
| K. thailandicaT | CBS 7552T | AF075516 | |
| K. vietnamensis | CBS 8963T | AB292851 | |
| Sterigmatosporidium | Cuniculitrema | ||
| S. polymorphumT | C. polymorpha | CBS 9644T | AY032662 |
| Naemateliaceae fam. nov. | |||
| Dimennazyma gen. nov. | |||
| D. cistialbidiT comb. nov. | Cryptococcus cistialbidi | CBS 10049T | AY562135 |
| Naematelia | |||
| N. aurantia | Tremella aurantia | CBS 6965R | AF189842 |
| N. aurantialba comb. nov. | T. aurantialba | strain9102 | EF010939 |
| N. encephalaT | T. encephala | CBS 8207R | AF042220 |
| N. microspora comb. nov. | T. microspora | BPI702328 | AF042253 |
| Single-species lineage | |||
| Tremella ‘indecorata’ II pro tem. | CBS 6976R | AF042250 | |
| Phaeotremellaceae fam. nov. | |||
| Gelidatrema gen. nov. | |||
| G. spencermartinsiaeT comb. nov. | C. spencermartinsiae | CBS 10760T | DQ513279 |
| Phaeotremella emend. | |||
| P. fagi comb. nov. | C. fagi | CBS 9964T | DQ054535 |
| P. mycetophiloides comb. nov. | T. mycetophiloides | AM23 | JN043608 |
| P. mycophaga comb. nov. | T. mycophaga | RB6539-4 | AF042249 |
| P. neofoliacea comb. nov. | T. neofoliacea | CBS 8475R | AF042236 |
| P. pseudofoliaceaT | T. foliacea | CBS 6969R | AF189868 |
| P. simplex comb. nov. | T. simplex | FO31782 | AF042246 |
| P. skinneri comb. nov. | C. skinneri | CBS 5029T | AF189835 |
| Rhynchogastremaceae emend. | |||
| Papiliotrema emend. | |||
| P. anemochoreius comb. nov. | C. anemochoreius | CBS 10258T | DQ384929 |
| P. aspenensis comb. nov. | C. aspenensis | CBS 13867T | KC485500 |
| P. aureus comb. nov. |
Torula aurea C. aureus |
CBS 318T | AB035041 |
| P. baii comb. nov. | C. baii | PYCC 6523T | LK023766 |
| P. bandoniiT | CBS 9107T | AF416642 | |
| P. flavescens comb. nov. |
T. flavescens C. flavescens |
CBS 942T | AB035042 |
| P. fonsecae comb. nov. | C. fonsecae | CBS 12692T | JN193447 |
| P. frias comb. nov. | C. frias | CBS 12693T | LK023834 |
| P. fuscus comb. nov. | Auriculibuller fuscus | CBS 9648 | AF444763 |
| P. hoabinhensis comb. nov. | Bullera hoabinhensis | JCM 10835T | AB193347 |
| P. japonica comb. nov. | B. japonica | CBS 2013T | AF444760 |
| P. laurentii comb. nov. | C. laurentii | CBS 139T | AF075469 |
| P. mangaliensis comb. nov. | C. mangaliensis | CBS 10870T | FJ008046 |
| P. nemorosus comb. nov. | C. nemorosus | CBS 9606T | AF472625 |
| P. perniciosus comb. nov. | Cryptococcus perniciosus | CBS 9605T | AF472624 |
| P. pseudoalba comb. nov. | Bullera pseudoalba | CBS 7227T | AF075504 |
| P. rajasthanensis comb. nov. | C. rajasthanensis | CBS 10406T | AM262324 |
| P. ruineniae comb. nov. | C. ruineniae | PYCC 6170T | LK023764 |
| P. siamense | CBS 13330T | AB909023 | |
| P. taeanensis comb. nov. | C. taeanensis | CBS 9742T | AY422719 |
| P. terrestris comb. nov. | C. terrestris | CBS 10810T | EF370393 |
| P. wisconsinensis comb. nov. | C. wisconsinensis | CBS 13895T | KM408131 |
| Rhynchogastrema emend. | |||
| R. aquatica comb. nov. | Bandoniozyma aquatica | CBS 12527T | JN979992 |
| R. complexa comb. nov. | B. complexa | CBS 11570T | GU321090 |
| R. coronatumT | BBA 65155T | KJ170152 | |
| R. fermentans comb. nov. | B. fermentans | CBS 12399T | HM461720 |
| R. glucofermentans comb. nov. | B. glucofermentans | CBS 10381T | AY520334 |
| R. nanyangensis comb. nov. | C. nanyangensis | CBS 12474T | JN564592 |
| R. noutii comb. nov. | B. noutii | CBS 8364T | AF444700 |
| R. tunnelae comb. nov. | B. tunnelae | CBS 6123T | AF444687 |
| R. visegradensis comb. nov. | B. visegradensis | CBS 12505T | GU195658 |
| Sirobasidiaceae | |||
| Fibulobasidium | |||
| F. inconspicuum | CBS 8237R | AF363641 | |
| F. murrhardtense | CBS 9109T | AF416648 | |
| F. sirobasidioides | RJB12787 | AF416644 | |
| Single-species lineages | |||
| Sirobasidium japonicum pro tem. | MY111_05 | LC016573 | |
| Sirobasidium magnum pro tem. | CBS 6803 | AF075475 | |
| Tremellaceae | |||
| Tremella | |||
| T. brasiliensis | CBS 6966R | AF189864 | |
| T. cinnabarina | CBS 8234R | AF189866 | |
| T. coalescens | CBS 6967R | AF189865 | |
| T. flava | CBS 8471R | AF042221 | |
| T. fuciformis | CBS 6970R | AF075476 | |
| T. globispora | CBS 6972R | AF189869 | |
| T. laurisilvae | TFC Mic.24580 | ||
| T. mesentericaT | CBS 6973R | AF075518 | |
| T. resupinata | CBS 8488R | AF042239 | |
| T. taiwanensis | CBS 8479R | AF042230 | |
| T. tropica | CBS 8483R | AF042251 | |
| T. yokohamensis comb. nov. | C. yokohamensis | JCM 16989T | HM222927 |
| Trimorphomycetaceae fam. nov. | |||
| Carlosrosaea gen. nov. | |||
| C. vrieseaeT comb. nov. | B. vrieseae | CBS 13870T | JX280388 |
| Saitozyma gen. nov. | |||
| S. flavaT comb. nov. | C. flavus | CBS 331T | AF075497 |
| S. ninhbinhensis comb. nov. | B. ninhbinhensis | JCM 10836T | AB261011 |
| S. paraflava comb. nov. | C. paraflavus | CBS 10100T | AY395799 |
| S. podzolica comb. nov. | C. podzolicus | CBS 6819T | AF075481 |
| Sugitazyma gen. nov. | |||
| S. miyagianaT comb. nov. | Bullera miyagiana | CBS 7526T | AF189858 |
| Trimorphomyces emend. | |||
| T. papilionaceusT | CBS 443.92 | AF075491 | |
| T. sakaeratica comb. nov. | B. sakaeratica | CBS 9934T | AY211546 |
| Incertae sedis | |||
| Tremella diploschistina pro tem. | AM199T | JN790588 | |
| Tremella parmeliarum pro tem. | Spain, Diederich 16574 (S-F102497) | JN043618 | |
| Trichosporonales | |||
| Tetragoniomycetaceae emend. | |||
| Bandonia gen. nov. | |||
| B. marinaT comb. nov. | Cryptococcus marinus | CBS 5235T | AF189846 |
| Cryptotrichosporon | |||
| C. anacardiiT | CBS 9551T | AY550002 | |
| C. tibetense | CBS 10455T | KP020115 | |
| Takashimella | |||
| T. formosensisT | B. formosensis | CBS 9812T | AY787858 |
| T. koratensis | B. koratensis | CBS 10484T | AY313006 |
| T. lagerstroemiae | B. lagerstroemiae | CBS 10483T | AY313010 |
| T. tepidarius | C. tepidarius | CBS 9427T | AB094046 |
| Tetragoniomyces | |||
| T. uliginosusT | Tremella uliginosa | AM186 | JN043621 |
| Trichosporonaceae emend. | |||
| Apiotrichum emend. | |||
| A. brassicae comb. nov. | Trichosporon brassicae | CBS 6382T | AF075521 |
| A. cacaoliposimilis comb. nov. | T. cacaoliposimilis | ATCC 20505T | HM755978 |
| A. dehoogii comb. nov. | T. dehoogii | CBS 8686T | AF444718 |
| A. domesticum comb. nov. | T. domesticum | CBS 8280T | AF075512 |
| A. dulcitum comb. nov. |
Oospora dulcita T. dulcitum |
CBS 8257T | AF075517 |
| A. gamsii comb. nov. | T. gamsii | CBS 8245T | AF444708 |
| A. gracile comb. nov. |
Oidium gracile T. gracile |
CBS 8189T | AF105399 |
| A. laibachii comb. nov. |
Endomyces laibachii T. laibachii T. multisporum |
CBS 5790T | AF075514 |
| A. lignicola comb. nov. |
Hyalodendron lignicola T. lignicola |
CBS 219.34T | AY370685 |
| A. loubieri comb. nov. |
Geotrichum loubieri T. loubieri |
CBS 7065T | AF075522 |
| A. montevideense comb. nov. |
Endomycopsis montevideensis T. montevideense |
CBS 6721T | AF105397 |
| A. mycotoxinivorans comb. nov. | T. mycotoxinivorans | CBS 9756T | AJ601388 |
| A. porosumT | T. porosum | CBS 2040T | AF189833 |
| A. scarabaeorum comb. nov. | T. scarabaeorum | CBS 5601T | AF444710 |
| A. siamense comb. nov. | T. siamense | JCM 12478T | AB164370 |
| A. sporotrichoides comb. nov. | T. sporotrichoides | CBS 8246T | AF189885 |
| A. vadense comb. nov. | T. vadense | CBS 8901T | AY093426 |
| A. veenhuisii comb. nov. | T. veenhuisii | CBS 7136T | AF105400 |
| A. wieringae comb. nov. | T. wieringae | CBS 8903T | AY315666 |
| A. xylopini comb. nov. | Trichosporon xylopini | CBS 11841T | HQ005757 |
| Cutaneotrichosporon gen. nov. | |||
| C. arboriformis comb. nov. | Cryptococcus arboriformis | CBS 10441T | AB260936 |
| C. curvatus comb. nov. | C. curvatus | CBS 570T | AF189834 |
| C. cutaneumT comb. nov. |
Oidium cutaneum T. cutaneum |
CBS 2466T | AF075483 |
| C. cyanovorans comb. nov. | C. cyanovorans | CBS 11948T | JF680899 |
| C. daszewskae comb. nov. | C. daszewskae | CBS 5123T | AB126588 |
| C. debeurmannianum comb. nov. | T. debeurmannianum | CBS 1896T | AY143554 |
| C. dermatis comb. nov. | T. dermatis | CBS 2043T | AY143555 |
| C. guehoae comb. nov. | T. guehoae | CBS 8521T | AF105401 |
| C. hagleorum comb. nov. | C. haglerorum | CBS 8902T | AF407276 |
| C. jirovecii comb. nov. | T. jirovecii | CBS 6864T | AF105398 |
| C. moniliiforme comb. nov. |
O. moniliiforme T. moniliiforme |
CBS 2467T | AF105392 |
| C. mucoides comb. nov. | T. mucoides | CBS 7625T | AF075515 |
| C. oleaginosus comb. nov. | T. oleaginosus | ATCC 20509T | HM802135 |
| C. smithiae comb. nov. | T. smithiae | CBS 8370T | AF444706 |
| C. terricola comb. nov. | T. terricola | CBS 9546T | AB086382 |
| Effuseotrichosporon gen. nov. | |||
| E. vanderwaltii comb. nov. | T. vanderwaltii | CBS 12124T | JF680903 |
| Haglerozyma gen. nov. | |||
| H. chiarelliiT comb. nov. | T. chiarellii | CBS 11177T | EU030272 |
| Trichosporon | |||
| T. aquatile | CBS 5973T | AF075520 | |
| T. asahii | CBS 2479T | AF105393 | |
| T. asteroides | CBS 2481T | AF075513 | |
| T. caseorum | CBS 9052T | AJ319757 | |
| T. coremiiforme | CBS 2482T | AF139983 | |
| T. dohaense | CBS 10761T | FJ228471 | |
| T. faecale | CBS 4828T | AF105395 | |
| T. inkin | CBS 5585T | AF105396 | |
| T. insectorum | CBS 10422T | AY520383 | |
| T. japonicum | CBS 8641T | AF308657 | |
| T. lactis | CBS 9051T | AJ319756 | |
| T. ovoidesT | CBS 7556T | AF075523 | |
| Vanrija | |||
| V. albida | Sporobolomyces albidus | CBS 2839T | AB126584 |
| V. fragicola comb. nov. | C. fragicola | CBS 8898T | AB126585 |
| V. humicolaT |
Torula humicola Candida humicola |
CBS 571T | AF189836 |
| V. longa | C. longus | CBS 5920T | AB126589 |
| V. meifongana comb. nov. | Asterotremella meifongana | CBS 11424T | EU289356 |
| V. musci | C. musci | CBS 8899T | AB126586 |
| V. nantouana comb. nov. | Asterotremella nantouana | CBS 10890T | EF653952 |
| V. pseudolongus | C. pseudolongus | CBS 8297T | AB126587 |
| V. thermophila comb. nov. | C. thermophilus | CBS 10687T | AM746982 |
Phylogenetic analysis
The phylogenetic analysis used for the taxonomic backbone in this study was presented in Liu et al. (2015). The phylogeny was inferred from a seven-gene dataset comprising nucleotide sequences of the internal transcribed spacer region (ITS) rRNA gene, the D1/D2 domains of the large subunit (LSU or 26S) rRNA gene, the small subunit (SSU or 18S) rRNA gene, two subunits of RNA polymerase II (RPB1 and RPB2), translation elongation factor 1-α (TEF1) and cytochrome b (CYTB), using Bayesian inference, maximum likelihood (ML) and neighbour-joining (NJ) analyses (Liu et al. 2015). The supplementary LSU rRNA gene (D1/D2 domains) sequence dataset containing newly published tremellomycetous yeast species and additional filamentous teleomorphic taxa was constructed and subjected to constrained maximum likelihood (ML) and maximum parsimony (MP) analyses based on the topology of a seven-genes dataset taken from Liu et al. (2015). The LSU sequences were aligned with MAFFT version 7 and the G-INS-i option. Constrained phylogenetic analyses were only inforced for species previously analysed using seven DNA loci. Only bipartitions that received at least 85 % bootstrap support during fast bootstrapping of the seven-genes dataset (Liu et al. 2015) conducted with Pthreads-parallelised RAxML version 8.1.24 (Stamatakis 2014) were used as a backbone constraint for LSU phylogenetic inference. Fast bootstrapping in conjunction with the autoMRE bootstopping criterion (Pattengale et al. 2009) and subsequent search for the best tree (Stamatakis et al. 2008) were conducted using the GTRCAT model approximation. MP bootstrapping with 1 000 replicates was conducted with TNT version 1.1/June 2015 (Goloboff et al. 2008).
Quantitative assessment of taxonomic ranks
Two methods were used for the assessment of taxonomic ranks on the basis of the maximum likelihood tree obtained from the concatenated sequences of the seven genes (Liu et al. 2015). Firstly, we used a phylogeny-based variant of clustering optimisation called phylogenetic rank boundary optimisation (PRBO). The goal of clustering optimisation (Göker et al., 2009, Göker et al., 2010, Stielow et al., 2011) is to detect distance thresholds (and clustering parameters) that yield non-hierarchical clusterings that are in maximal agreement with a given reference clustering (such as a classification into taxa of a single rank). The limitations of the approach are that it is rather a clustering method than a phylogenetic approach, even though the resulting clusters are often monophyletic, and that it estimates the boundaries for each taxonomic rank separately. PRBO instead is based on rooted phylogenies. It measures the divergence of each clade as maximum subtree height (MaSH). Taxonomic ranks are made quantitatively comparable by assigning an upper MaSH boundary to each rank, which also serves as the lower MaSH boundary of the next higher rank. Using an existing classification as template, these MaSH ranges for each rank are chosen so as to minimise the number of taxa whose MaSH is outside the MaSH range of their respective rank. Clades to which no taxon is assigned can also enter the optimisation, because their MaSH value must be higher than the upper boundary for the rank of any taxon they contain but lower than the upper boundary for the rank of the taxon assigned to the closest parent clade. This allows for estimating the boundaries for all ranks from an entire tree in a single run. Confidence intervals for the boundaries can be obtained by bootstrapping the set of clades that enters the boundary estimation. Afterwards taxa that did not enter the calculation can be judged as too large or too small, as long as their MaSH values are on the same scale.
The implementation used is the one of the forthcoming MaSH package for the R statistical software environment (R core team 2015). The boundary optimisation is done using the rpart package (Therneau et al. 2015) with the MaSH values of each clade as dependent and the rank as independent variable. One hundred bootstrap replicates were applied to obtain 95 % confidence intervals. A taxon was regarded as too large or too small in a strict sense when its MaSH was outside the MaSH range of its rank, and as deviating in even a relaxed sense (i.e. significantly deviating) when its MaSH was not even located within the outer confidence bounds. Ten genera (Table 2) of tremellomycetous yeasts and their parent taxa were chosen as references. These genera are considered as being well delimited phenotypically and phylogenetically because they were resolved as strongly supported monophyletic clades with stable positions in the trees derived from different datasets using different algorithms (Liu et al. 2015) and the species in each genus exhibit similar morphological or physiological properties. The resulting optimal MaSH thresholds and their 95 % confidence intervals were then applied to the entire dataset.
Table 2.
PRBO results showing the divergences, if any, of the proposed taxa (except for the families) from the optimal range of divergences for their rank as inferred from the selected reference taxa.
| Taxa | Rank | MaSH | Deviation | Sigdev |
|---|---|---|---|---|
| Cystofilobasidiales | Order | 0.7211 | −0.1226 | 0 |
| Cystofilobasidium* | Genus | 0.2518 | 0 | 0 |
| Krasilnikovozyma (huempii clade) | Genus | 0.0003 | 0 | 0 |
| Itersonilia | Genus | 0.0672 | 0 | 0 |
| Mrakia* | Genus | 0.2071 | 0 | 0 |
| Phaffia | Genus | 0.1028 | 0 | 0 |
| Tausonia | Genus | – | – | – |
| Udeniomyces* | Genus | 0.1326 | 0 | 0 |
| Filobasidiales | Order | 0.5866 | −0.2570 | −0.1242 |
| Filobasidium | Genus | 0.2465 | 0 | 0 |
| Naganishia (albidus clade) | Genus | 0.1835 | 0 | 0 |
| Goffeauzyma (gastricus clade) | Genus | 0.3281 | 0 | 0 |
| Solicoccozyma (aerius clade) | Genus | 0.2537 | 0 | 0 |
| Piskurozyma (cylindricus clade) | Genus | 0.1235 | 0 | 0 |
| Cryptococcus arrabidensis | Genus | – | – | – |
| Filobasidium capsuligenum | Genus | – | – | – |
| Holtermanniales | Order | 0.4607 | 0 | 0 |
| Holtermanniella* | Genus | 0.1832 | 0 | 0 |
| Holtermannia | Genus | – | – | – |
| Tremellales | Order | 0.9475 | 0 | 0 |
| Naematelia (aurantia clade and C. cistialbidi) | Genus | 0.3098 | 0 | 0 |
| Rhynchogastrema | Genus | 0.1336 | 0 | 0 |
| Bulleribasidium* | Genus | 0.3380 | 0 | 0 |
| Bullera (Bullera and hannae clades) | Genus | 0.4009 | 0.0597 | 0.0476 |
| Cryptococcus | Genus | 0.2325 | 0 | 0 |
| Derxomyces* | Genus | 0.6263 | 0.2851 | 0.2730 |
| Dioszegia* | Genus | 0.3296 | 0 | 0 |
| Genolevuria (amylolyticus clade) | Genus | 0.1985 | 0 | 0 |
| Fellomyces | Genus | 0.3096 | 0 | 0 |
| Fibulobasidium* | Genus | 0.0971 | 0 | 0 |
| Phaeotremella (foliacea clade) | Genus | 0.2697 | 0 | 0 |
| Hannaella* | Genus | 0.2876 | 0 | 0 |
| Kockovaella | Genus | 0.3573 | 0.0161 | 0.0040 |
| Kwoniella | Genus | 0.3335 | 0 | 0 |
| Papiliotrema (aureus, Auriculibuller, Papiliotrema, pseudoalba, laurentii clades) | Genus | 0.3345 | 0 | 0 |
| Pseudotremella (moriformis clade and C. allantoinivorans) | Genus | 0.2068 | 0 | 0 |
| C. mujuensis and S. intermedium | Genus | 0.2123 | 0 | 0 |
| Saitozyma (flavus clade) | Genus | 0.3187 | 0 | 0 |
| Nielozyma (melastomae clade) | Genus | 0.1478 | 0 | 0 |
| Tremella | Genus | 0.4809 | 0.1397 | 0.1276 |
| Vishniacozyma (dimennae clade) | Genus | 0.3674 | 0.0262 | 0.0141 |
| Trichosporonales | Order | 0.4386 | −0.4050 | −0.2722 |
| Cutaneotrichosporon (cutaneum clade and haglerorum clade) | Genus | 0.2107 | 0 | 0 |
| Takashimella (formosensis clade) | Genus | 0.1490 | 0 | 0 |
| Apiotrichum (gracile/brassicae and porosum clade) | Genus | 0.2377 | 0 | 0 |
| Trichosporon* | Genus | 0.2250 | 0 | 0 |
| Vanrija (humicola clade) | Genus | 0.1955 | 0 | 0 |
| Cryptotrichosporon | Genus | – | – | – |
Note. MaSH: Maximum Subtree Height; Deviation: deviation from the point estimate for the upper (positive value) or lower (negative value) threshold of the rank of the taxon; Sigdev: significant deviation, i.e. a deviation even outside the upper or lower 95 % confidence band of the upper or lower threshold, respectively. Zero indicates taxa with the appropriate divergence, negative values indicate taxa that are too small, positive values taxa that are too large. An asterisk (*) indicates the well-established taxa that were used as a reference classification for PRBO. A dash (–) indicates taxa that are monotypic in the investigated sampling, for which MaSH is not calculated. Numbers in bold indicate significant deviation from the range defined by the optimal boundaries calculated for the rank.
In addition, we used a modified GMYC (generalised mixed Yule coalescent) method designed to identify evolutionary significant units (Humphreys & Barraclough 2014). The GMYC method was originally developed for species delimitation (Pons et al., 2006, Monaghan et al., 2009) and subsequently extended for identifying higher taxonomic units, also called evolutionary significant units (Humphreys & Barraclough 2014). The method allows, without prior expectations, locating independently evolving lineages as a transition from intra- to intertaxa relationships on a phylogenetic tree. It aims at detecting shifts in branching rates between relationships within and among evolutionary significant units. The most recent common ancestral node at the transition point is interpreted as distinguishing taxa at a specific level. Within a likelihood framework it uses chronograms to compare two models: a) a null model under which the whole sample derives from a single evolutionary significant unit and b) an alternative GMYC model. The latter combines equations that separately describe branching patterns within and among evolutionary units. A likelihood ratio test (LRT) is used to evaluate whether the null model can be significantly rejected. If the GMYC model fits the data significantly better than the null model, the threshold T allows estimating the number of evolutionary significant units (to be interpreted as higher taxa) present in the dataset. Outgroup samples were excluded from the dataset using the drop.tip command in ape (Paradis 2006). A chronogram was calculated from the ML tree using the penalised likelihood method (Sanderson 2002) as implemented in the chronopl function in ape (Paradis 2006). The chronogram was then analysed using a modified GMYC package in SPLITS in R (R core team 2015) version 2.10, using the single-threshold method. Since the method identifies order units, we subsequently performed nested analyses for each major order to identify evolutionary significant units. That is, the modified GMYC model was applied iteratively to subtrees identified as evolutionary significant units in the respective last step.
The major differences in perspective between the two approaches are as follows. The modified GMYC method presupposes an ultrametric tree but is independent of an existing classification. When using the single-threshold approach, the modified GMYC method is capable of identifying taxa of a similar divergence, measured as evolutionary age. If applied iteratively, however, there is no guarantee even under the single-threshold model that the thresholds of all taxa obtained in the second, third etc. iteration are at a comparable level of divergence, because the thresholds are independently estimated for each tree obtained by splitting the tree of the last iteration. The iteratively applied modified GMYC method is ideal for obtaining taxa of several ranks that correspond to evolutionarily significant units in the sense of Humphreys & Barraclough (2014). It does not guarantee to obtain taxa of the same rank that are quantitatively comparable and cannot minimise the deviations from an existing classification.
In contrast, PRBO is independent of a molecular clock (can be applied to either ultrametric or non-ultrametric trees) but as used here it assumes an existing classification for at least some of the organisms under study. Boundaries for missing ranks could be derived by interpolation but this was not applied in this study. It is capable of obtaining taxa of the same rank that are quantitatively comparable, measured as evolutionary age or just as maximum subtree height, because the boundaries of all ranks are inferred from the entire dataset at once. It is also able to minimise the deviations from an existing classification, but only as the secondary criterion. Moreover, PRBO often allows the taxonomist, within the limits set by the estimated boundaries, several choices for delineating taxa. It does not guarantee, however, to find taxa that are “real” or evolutionary significant units.
It must be emphasised, however, that none of the two methods was followed strictly in the current study, even where they agreed on taxon boundaries. As far as possible, broader circumscriptions of taxa were chosen to lower the number of suggested names, particularly if these taxa were newly introduced, had strong branch support, or showed diagnostic phenotypical features. These principles will be illustrated by examples given below.
Results and discussion
Taxonomic units assessed by PRBO and a modified GMYC approach
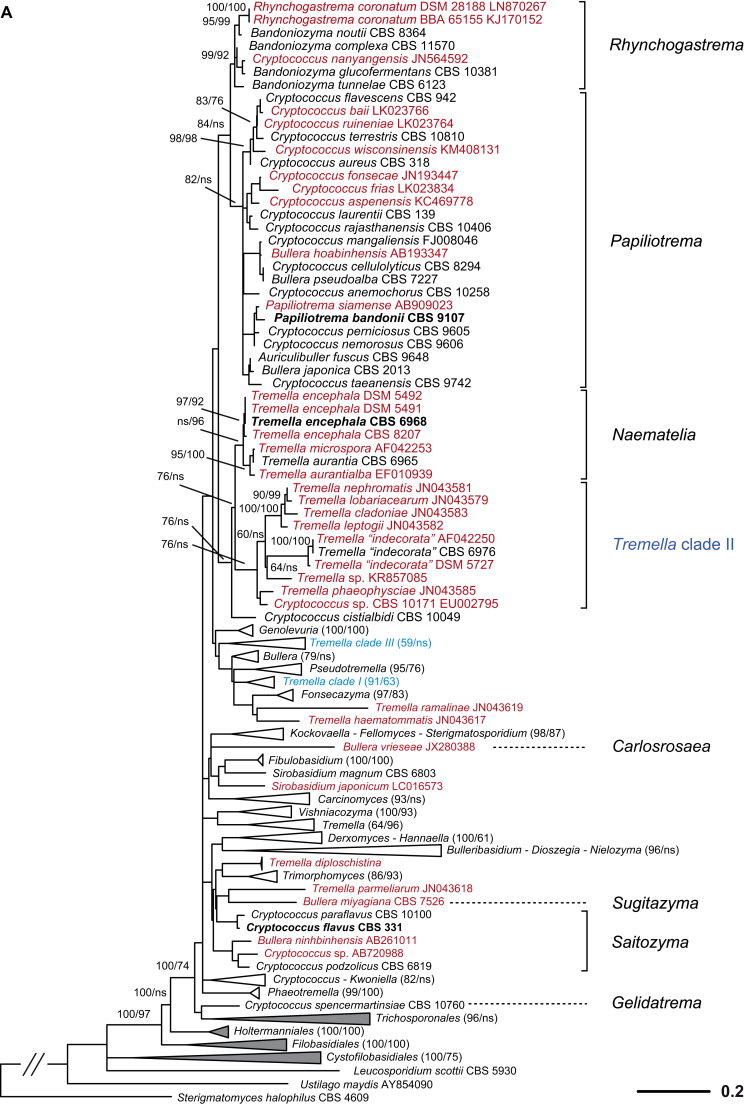
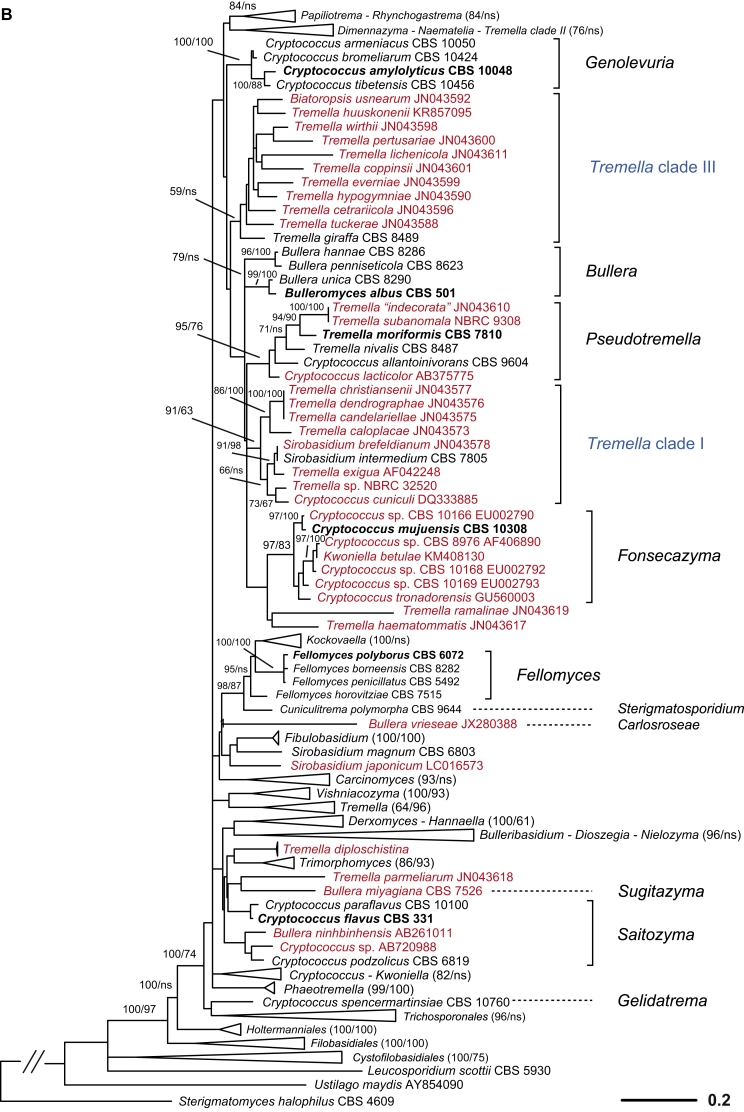
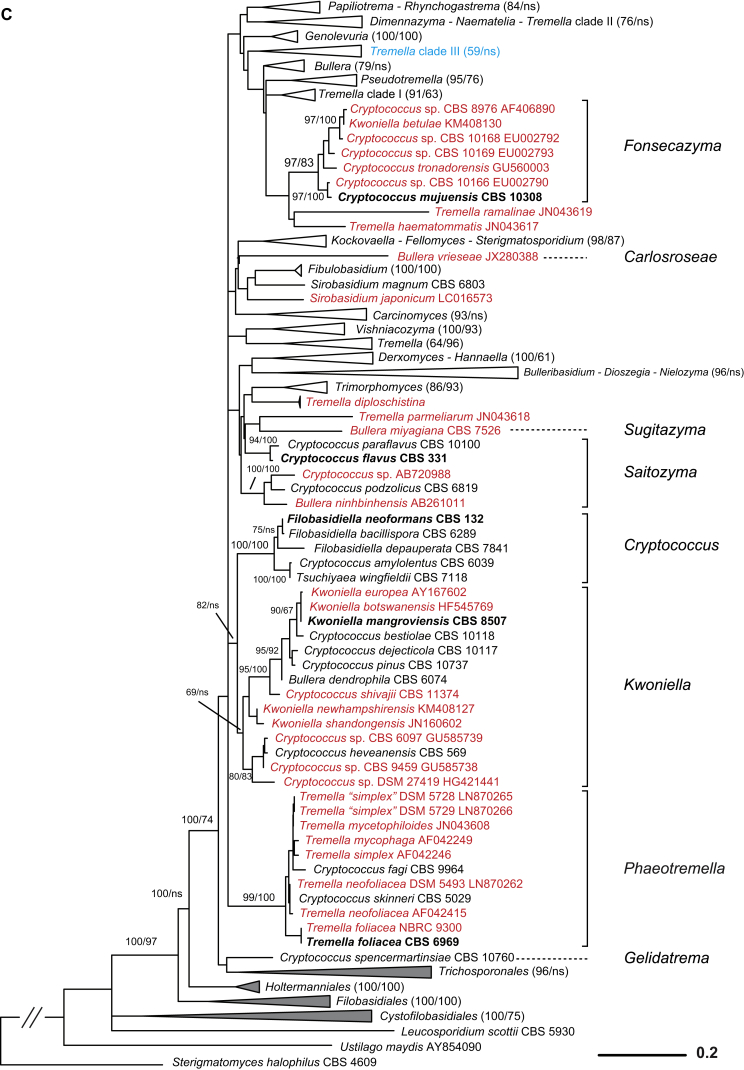
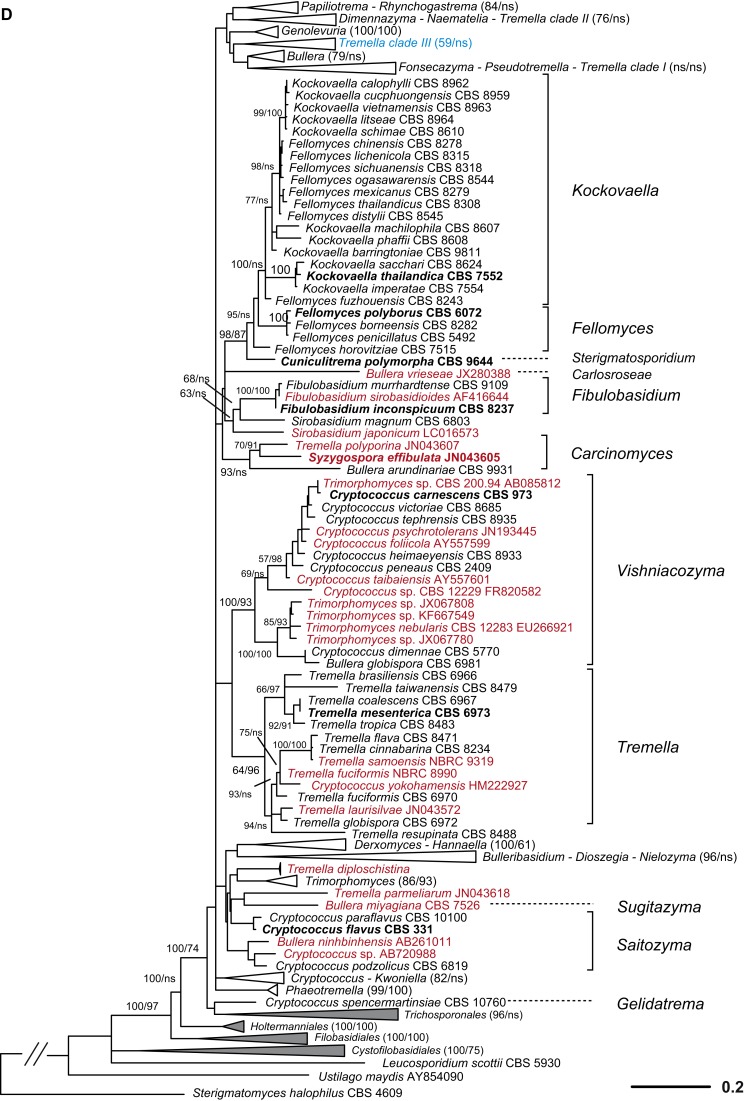
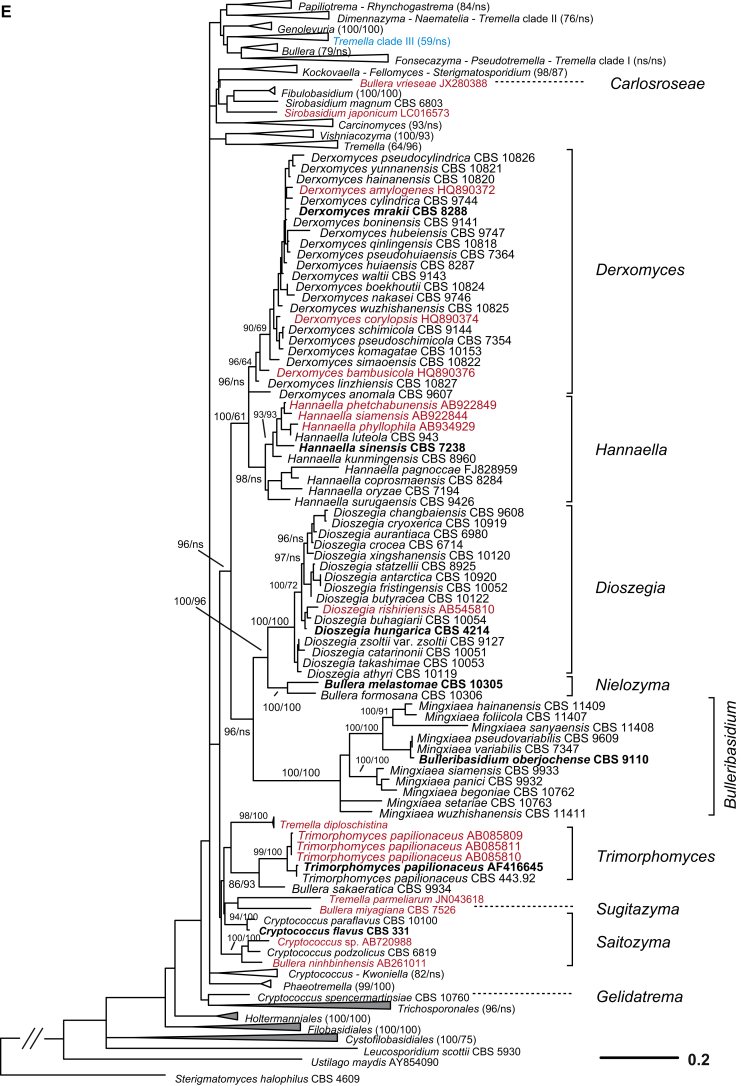
A taxonomic framework was firstly proposed based on the seven-genes phylogeny (Liu et al. 2015) as shown in Fig. 1, which was the basis for further taxonomic unit assessment and expanded LSU dataset analysis. Based on the PRBO analysis including 286 taxa represented by 294 strains of tremellomycetous yeasts, strong delimitation evidence was found for the orders Cystofilobasidiales, Holtermanniales and Tremellales, which showed no significant deviation from the range defined by the optimal boundaries calculated for their rank (Table 2). The demarcation of orders Filobasidiales and Trichosporonales showed a small negative, significant deviation (Sigdev = −0.1242 and −0.2722, respectively), i.e. these orders were judged too small, most likely because of the overlap between the divergences of taxa of distinct ranks. The recognition of Filobasidiales as a distinct monophyletic lineage in Tremellomycetes is consistent in many studies (Fell et al., 2000, Scorzetti et al., 2002, Weiss et al., 2004, Hibbett et al., 2007, Boekhout et al., 2011a, Millanes et al., 2011, Liu et al., 2015). Therefore, and for reasons of conservatism we keep it as a single order. The recognition of Trichosporonales remains a matter of debate. Some authors placed this order within Tremellales (Weiss et al., 2004, Hibbett et al., 2007, Millanes et al., 2011), while others accepted it as a sister group to Tremellales (Scorzetti et al., 2002, Boekhout et al., 2011a, Weiss et al., 2014). The problem was probably caused by the unstable phylogenetic position of the foliacea clade. It appeared as an early branching clade of the Tremellales in the Bayesian tree but posterior probability (PP) support for this positioning was lacking. In the ML and NJ tree, the foliacea clade was branching before the Trichosporonales and the Tremellales lineages with strong to moderate bootstrap support values (data not shown). When the foliacea clade was considered as an independent lineage as implied by the results shown above, the monophyly of both Trichosporonales and Tremellales was resolved by Bayesian and ML analyses of a seven-genes dataset with 1.0 posterior probability and 100 % bootstrap support (Liu et al. 2015). In the LSU tree the node delimiting Tremellales was not originally constrained and received poor to moderate support in ML and MP analyses, respectively, while other nodes in Tremellales received lower support (Fig. 2). This implies that undersampling may substantially influence the delimitation of clades, and, therefore, we presently consider Trichosporonales as a distinct order based on its phylogenetic position and phenotypic characters.
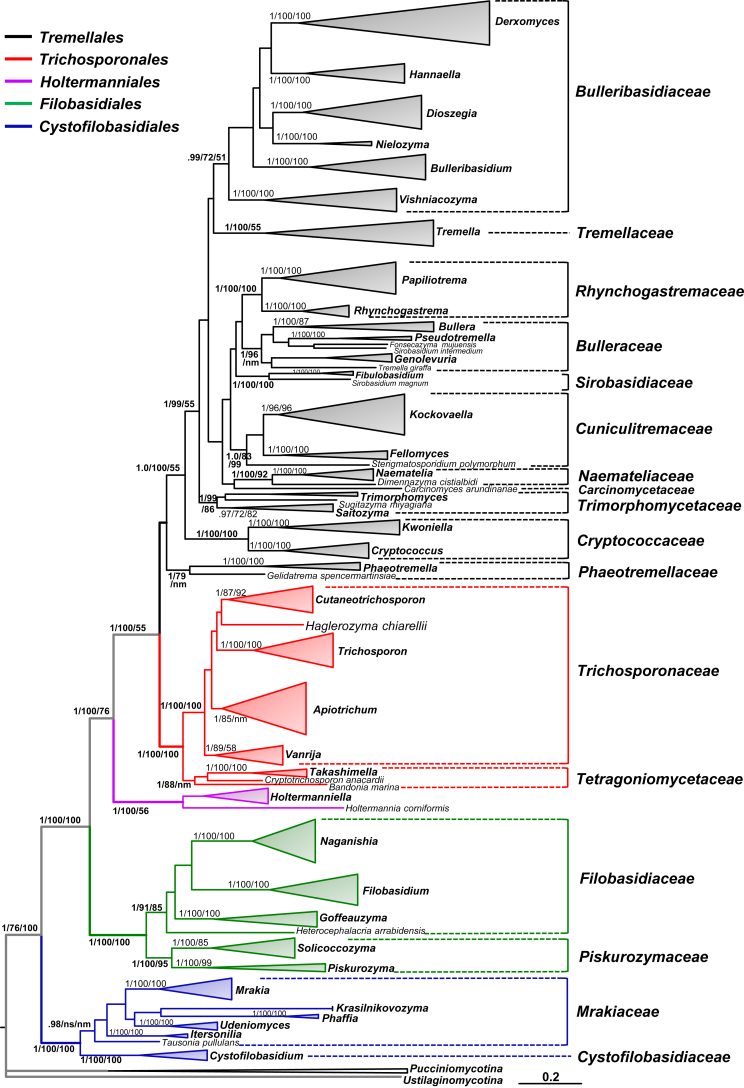
Fig. 1.
A taxonomic framework of genera and higher levels in Tremellomycetes based on the seven-genes phylogeny (Liu et al. 2015). The tree backbone is constructed using Bayesian analysis and branch lengths are scaled in terms of expected numbers of nucleotide substitutions per site. The Bayesian posterior probabilities (PP) and bootstrap percentages (BP) of maximum likelihood and neighbour-joining analyses from 1 000 replicates are shown respectively from left to right on the deep and major branches resolved. Note: ns, not supported (PP < 0.9 or BP < 50 %); nm, not monophyletic.
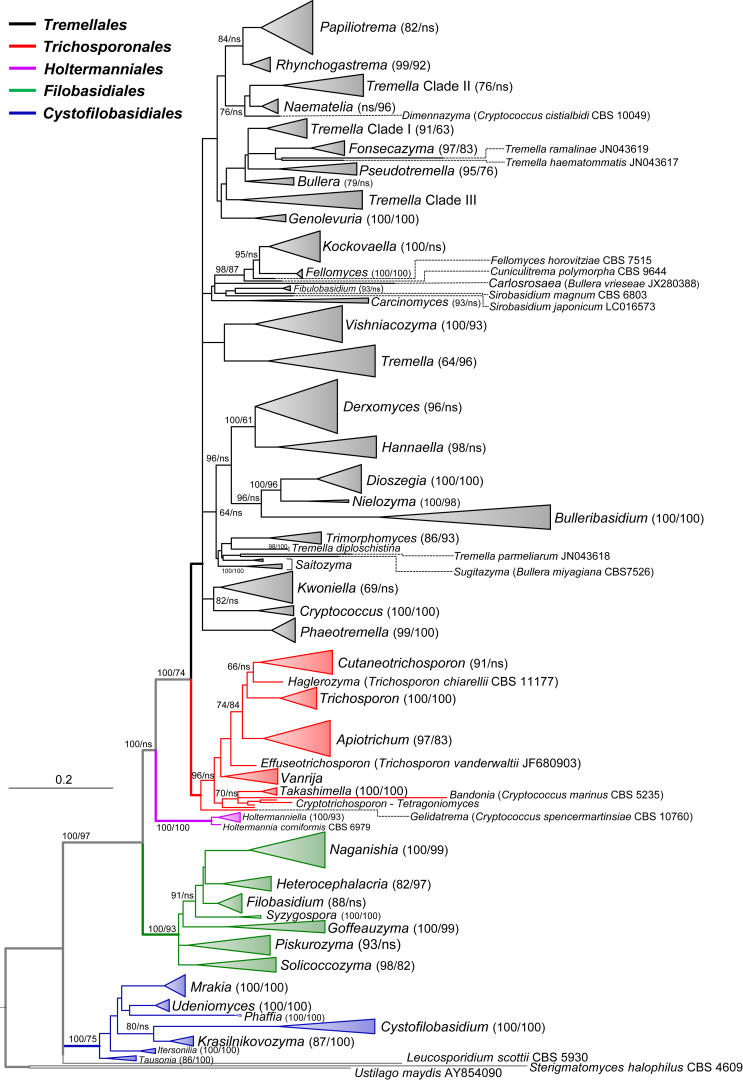
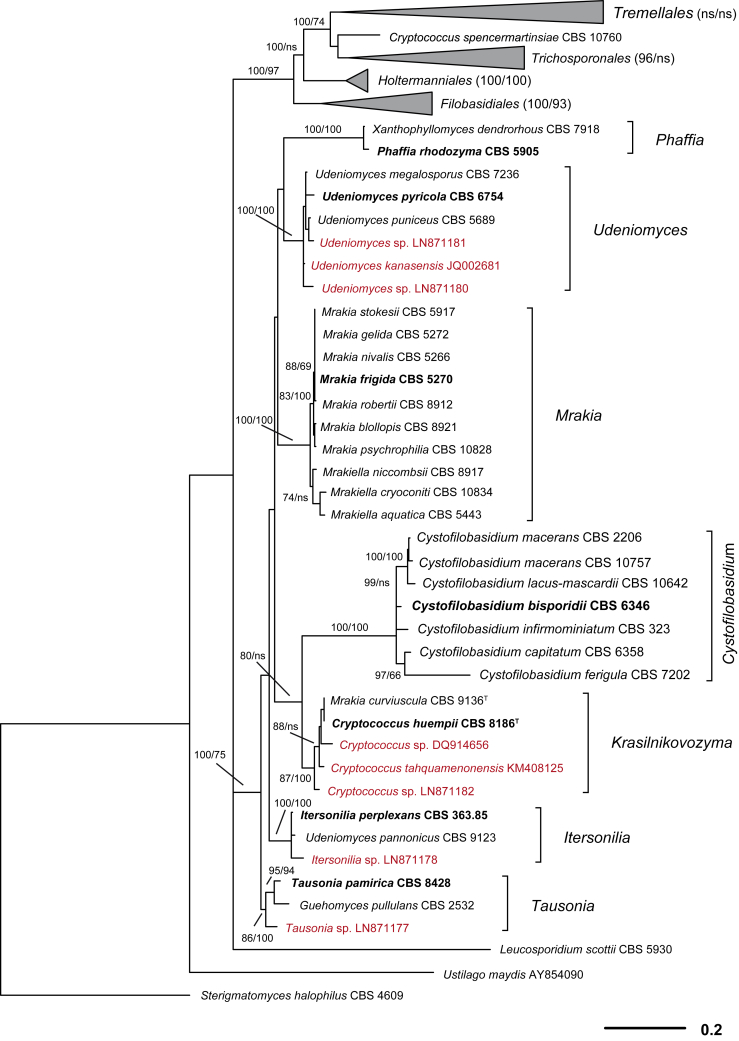
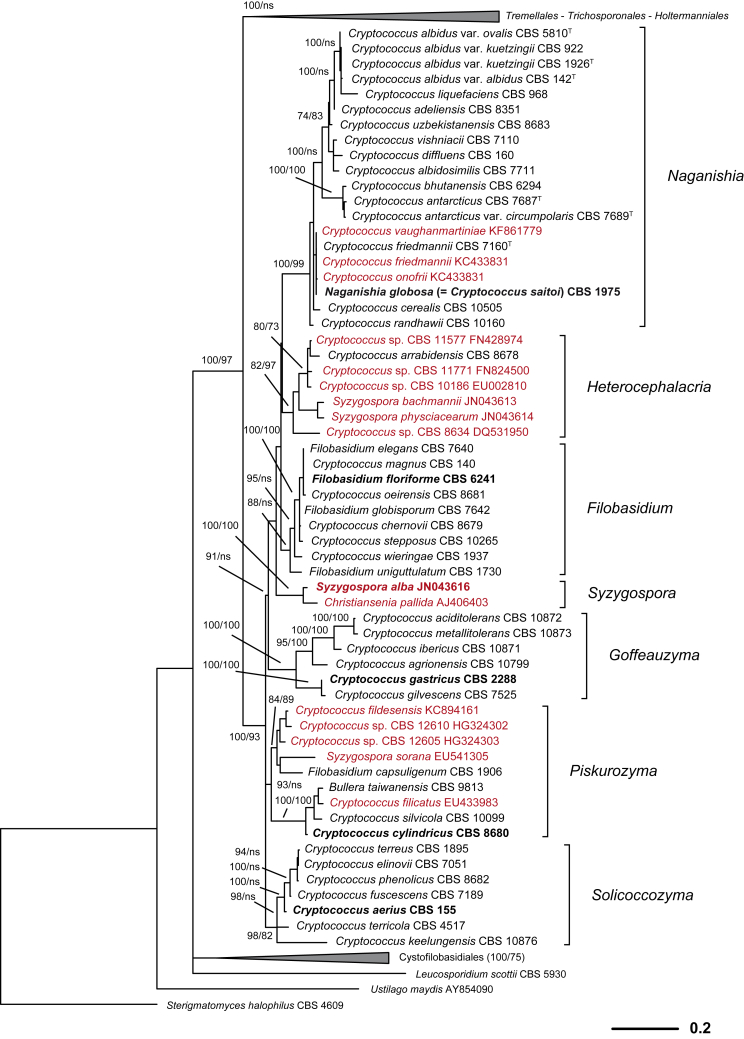
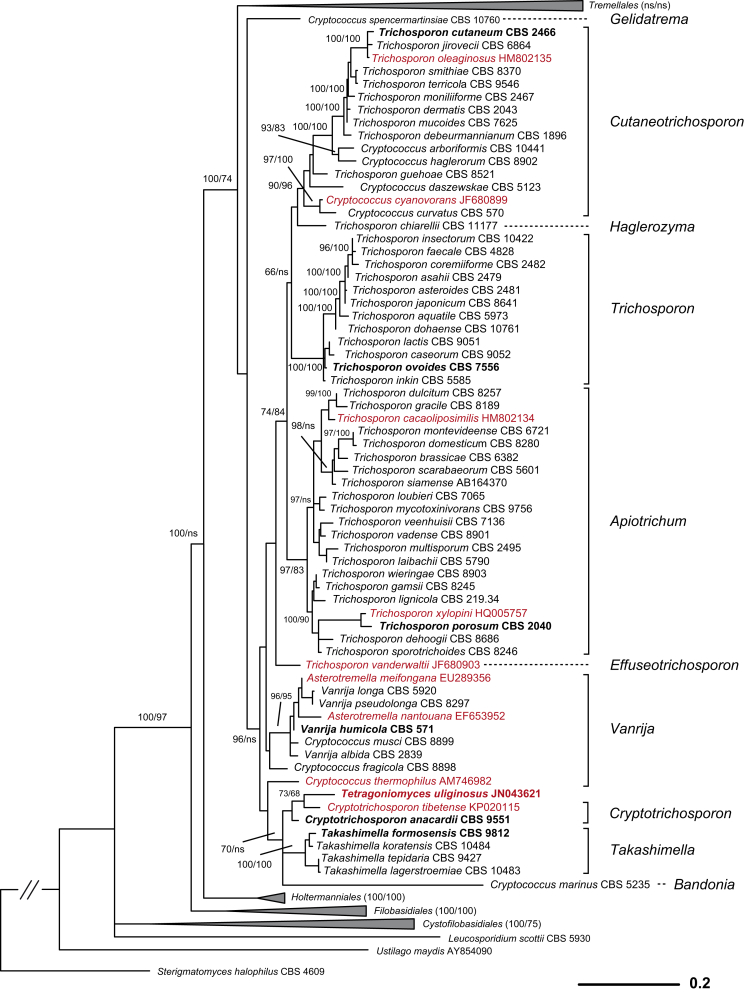
Fig. 2.
Phylogenetic relationships of yeasts and related taxa in Tremellomycetes (major lineages) obtained by maximum-likelihood analysis of LSU (D1/D2 domains) rRNA gene. Tree topology was backbone-constrained with the well-supported (>85 %) bipartitions of the topology of the seven-genes tree (Liu et al. 2015). Bootstrap percentages (BP) of maximum likelihood and neighbour-joining analyses from 1 000 replicates are shown respectively from left to right on the deep and major branches resolved and in brackets following recognised clades. Note: ns, not supported (BP < 50 %).
Ten genera or clades of tremellomycetous yeasts which were consistently resolved as monophyletic groups with strong statistical support in the trees constructed using different datasets and algorithms (Liu et al. 2015) were used as references in the PRBO test (Table 2). Six of these genera, Bulleribasidium emend., Derxomyces, Dioszegia, Fibulobasidium, Hannaella and Holtermanniella, have been accepted as well-established genera based on their distinct morphological characters and phylogenetic positions (Bandoni, 1979, Wang and Bai, 2008, Wang et al., 2011, Wuczkowski et al., 2011). Of the 44 monotypic genera or clades identified in this study based on the multigene phylogeny (Liu et al. 2015), the majority (34 genera) was in agreement with the PRBO results (Table 2). Five genera showed significant deviation from the optimal range calculated for the genus rank, namely Bullera emend., Derxomyces, Kockovaella emend., Tremella sensu stricto and Vishniacozyma gen. nov. (i.e. the dimennae clade). The emended genus Bullera contains four species that were located in two clades (i.e. Bulleromyces and hannae clades) in Liu et al. (2015). The GC contents of the species in the Bulleromyces and hannae clades are 53.3–55.4 % and 42.7.6–44.5 %, respectively. These data support the significant deviation of the four species assigned to a single genus. We nevertheless propose to keep them in the emended genus Bullera at present to accommodate their phenotypic similarity and to avoid suggesting a new genus with only two species. The significant deviation of the genus Derxomyces resulted from the inclusion of the early branching species D. anomola, which showed a remarkable divergence from the other species of the genus in the tree inferred from the seven genes (Liu et al. 2015). We prefer to maintain the current taxonomic status of this species to avoid creating a single-species clade or a monotypic genus without specific phenotypic characters. The significant deviation of the genus Kockovaella emend. was caused by the inclusion of Fellomyces fuzhouensis in this genus. This species branches first within the Kockovaella clade with strong support (Liu et al. 2015) and is morphologically similar to the other species of the genus by forming conidia on stalks (Lopandic et al. 2011). Thus, at present we prefer to assign this species to the emended genus Kockovaella. The new genus Vishniacozyma is proposed for the strongly supported dimennae clade (Table 1, Fig. 1) (i.e. victoriae clade in Fonseca et al. 2011) including one Bullera and six Cryptococcus species in the seven-genes tree (Fig. 1). Two strongly supported sub-clades containing two and five species, respectively, were recognised in this clade (Liu et al. 2015). The combination of these two sub-clades into a single genus is due to the consideration that they are phenotypically indistinguishable. The Tremella sensu stricto clade (i.e. the mesenterica clade in Boekhout et al. 2011a) exclusively contains Tremella species, including the type species of the genus, T. mesenterica, together with other representatives of the mesenterica and fuciformis subclades as distinguished by Chen (1998) and Liu et al. (2015). Several ecological and morphological features can be used to distinguish both subclades (Chen 1998): 1) basidiocarps are frequently associated with fungi of Russulales (Peniophora) in the mesenterica subclade, and with fungi of Xylariaceae (Hypoxilon or Xylaria) in the fuciformis subclade; 2) hymenial and subhymenial structures are loose in the mesenterica subclade in contrast to firm with numerous anastomoses present in the fuciformis subclade; 3) hyphidia are present in the mesenterica subclade but lacking in the fuciformis subclade; and 4) haustorial hyphae are not branched in the mesenterica subclade in contrast to those of the species in the fuciformis subclade. The genus Tremella containing approximately 90 species (Kirk et al. 2008) is highly polyphyletic and remains to be recircumscribed (Millanes et al., 2011, Weiss et al., 2014, Liu et al., 2015). We propose to keep the species in the Tremella sensu stricto clade in this genus at present. We also caution against the use of the generic name Tremella for newly described species outside that clade.
Phylogenetic demarcation of families in the Fungi is usually difficult due to the lack of molecular data from many taxa (Hibbett et al. 2007). This is also the case for Tremellomycetes at present because the rRNA gene sequences of many filamentous members of this class are not yet available for an integrated phylogenetic analysis (e.g. Millanes et al., 2011, Lindgren et al., 2015). The boundaries of the existing families within Tremellomycetes have not been subjected to robust phylogenetic analyses based on sufficient taxon sampling, thus family boundaries could only be derived by PRBO via interpolation, which was not applied here because the number of established genera is also small. The modified GMYC approach (Humphreys & Barraclough 2014), which does not need the reference information, was used to assign families, and the resulting taxa were then assessed using PRBO. The LRT test resulted in the recognition of two families in Cystofilobasidiales, five in Filobasidiales, 15 in Tremellales and two in Trichosporonales (Table 3). Some of the families identified in Filobasidiales and Tremellales using the GMYC approach lacked phylogenetic support. Therefore, we adjusted the family boundaries from a conservative point of view and a combined consideration on phylogenetic and phenotypic criteria, resulting in the proposal of two families in Filobasidiales and ten in Tremellales (Table 3).
Table 3.
The family identification using single thresholds model in mixed Yule coalescent method (GMYC) and its comparison with the taxonomic units delimited according to phylogeny and morphological characters.
| Order | Genus (or single-species clade) | GMYC ST | PP/BP1/BP2 | Accepted | PP/BP1/BP2 |
|---|---|---|---|---|---|
| Cystofilobasidiales | Cystofilobasidium | new family 1 | 1.0/100/100 | Cystofilobasidiaceae | 1.0/100/100 |
| Tausonia | new family 2 | 0.98/ns/nm | Mrakiaceae | 0.98/ns/nm | |
| Krasilnikovozyma | |||||
| Itersonilia | |||||
| Mrakia | |||||
| Udeniomyces | |||||
| Xanthophyllomyces | |||||
| Filobasidiales | Piskurozyma | new family 3 | 1.0/100/99 | Piskurozymaceae | 1.0/100/95 |
| Filobasidium capsuligenum | |||||
| Solicoccozyma | new family 4 | 1.0/100/85 | |||
| Goffeauzyma | new family 5 | 1.0/100/100 | Filobasidiaceae | 1.0/91/85 | |
| Filobasidium | new family 6 | 1.0/100/100 | |||
| Fonsecazyma | new family 7 | 1.0/100/100 | |||
| Cryptococcus arrabidensis | Single-species clade | / | |||
| Tremellales | Phaeotremella | new family 8 | 1.0/79/nm | Phaeotremellaceae | 1.0/79/nm |
| C. spencermartinsiae | |||||
| Cryptococcus | new family 9 | 1.0/100/100 | Cryptococcaceae | 1.0/100/100 | |
| Kwoniella | new family 10 | 1.0/100/100 | |||
| Naematelia | new family 11 | ns/nm/nm | Naemateliaceae | ns/nm/nm | |
| B. arundinariae | |||||
| Rhynchogastrema | new family 12 | 1.0/100/100 | Rhynchogastremaceae | 1.0/100/100 | |
| Papiliotrema | new family 13 | 1.0/100/100 | |||
| Bullera | new family 14 | nm/nm/nm | Bulleraceae | 1.0/96/nm | |
| Genolevuria | |||||
| Pseudotremella | |||||
| Cryptococcus mujuensis, Sirobasidium intermedium, Tremella giraffa | |||||
| Fibulobasidium | Sirobasidiaceae | 1.0/100/99 | |||
| Sirobasidium magnum | |||||
| Fellomyces | new family 15 | 1.0/83/99 | Cuniculitremaceae | 1.0/83/99 | |
| Kockovaella | |||||
| Cuniculitrema polymorpha | |||||
| Tremella | new family 17 | 1.0/100/55 | Tremellaceae | 1.0/100/55 | |
| Saitozyma | new family 18 | 1.0/99/86 | Trimorphomycaceae | 1.0/99/86 | |
| Bullera miyagiana, B. sakaeratica, Trimorphomyces papilionaceus | |||||
| Vishniacozyma | new family 16 | 1.0/100/100 | Bulleribasidiaceae | .99/72/51 | |
| Derxomyces | new family 19 | 1.0/100/100 | |||
| Hannaella | new family 20 | 1.0/100/100 | |||
| Bulleribasidium | new family 21 | 1.0/100/100 | |||
| Dioszegia | new family 22 | 1.0/91/nm | |||
| Nielozyma | |||||
| Trichosporonales | Takashimella | new family 23 | 1.0/88/nm | Tetragoniomycetaceae | 1.0/88/nm |
| Cryptotrichosporon anacardii, Cryptococcus marinus | |||||
| Cutaneotrichosporon | new family 24 | 1.0/100/100 | Trichosporaceae | 1.0/100/100 | |
| Apiotrichum | |||||
| Haglerozyma | |||||
| Trichosporon | |||||
| Vanrija |
Note. GMYC ST: the family delimitation using GMYC for single thresholds (ST) model across seven-gene ML tree; PP, Bayesian posterior probability; BP1 and BP2, bootstrap values from the maximum likelihood and neighbour-joining analyses, respectively; nm: not monophyletic; ns, not supported (PP < 0.9 or BP < 50 %).
The two families recognised in Cystofilobasidiales by the GMYC test are accepted here. Family 1 is equivalent to Cystofilobasidiaceae (Fell et al. 1999) and Mrakiaceae fam. nov. is proposed to accommodate family 2. They are supported phenotypically by the morphology and size of basidia and phylogenetically by the seven-genes Bayesian analysis. The former usually produces long and slender (up to 65 μm) holobasidia, while the teleomorphic taxa of the latter usually form short and tubular (8–12 μm) holo- or phragmometabasidia. The two families recognised in Trichosporonales by the GMYC test are also accepted and are assigned to Trichosporonaceae (family 23) and Tetragoniomycetaceae (family 24). The majority of the taxa in the former can produce arthroconidia while the latter can not. Each of them also received strong phylogenetic support (Table 3).
The five clades resolved in the Filobasidiales obtained from the seven-genes phylogeny (Liu et al. 2015) were recognised as representing a family each (families 3–7) by the GMYC test (Table 3). However, discriminative phenotypic criteria among these clades were difficult to find from the morphological, biochemical and physiological characters that have been investigated (Kurtzman et al. 2011b). From a practical point of view, we suggest to combine families 3 and 4 (i.e. the aerius and cylindricus clades in Fonseca et al. 2011 and Liu et al. 2015) into a single new family Piskurozymaceae fam. nov. and combine families 5–7 together with the single-species lineage Cryptococcus arrabidensis into Filobasidiaceae.
The family recognition in the order Tremellales was more complicated and five (families 8, 11, 15, 17 and 18) of the 15 families recognised by the GMYC test are accepted as well delimited families with phenotypic and phylogenetic support (Table 3). The genera Cryptococcus sensu stricto (i.e. the Filobasidiella clade in Fonseca et al. 2011) and Kwoniella were identified as representing two separate families (families 9 and 10) by the GMYC test, but we suggest to combine them as a single family Cryptococcaceae in view of their close phylogenetic relationship with strong support (Liu et al. 2015) and both contain human or arthropod-associated species (Findley et al. 2009). We propose to combine families 12 and 13 (Table 3) identified by the GMYC test into a single family Auriculibulleraceae, which is phylogenetically strongly supported (Liu et al. 2015). Family 14 determined by the GMYC analysis was a paraphyletic group in the seven-genes tree (Liu et al. 2015), thus we divided it into two separate families, namely Bulleraceae and Sirobasidiaceae. The dimennae, Derxomyces, Hannaella, Bulleribasidium and Dioszegia (together with the melostomae clade) clades were recognised as representing a family each (families 16 and 19–22) by the GMYC test (Table 3). These clades consist of species previously belonging to the genera Bullera or Cryptococcus (Wang and Bai, 2008, Fonseca et al., 2011, Wang et al., 2011), suggesting their phenotypic similarity. Therefore, we propose to combine these five clades into a single family Bulleribasidiaceae, which showed good support values in the seven-genes analyses, both in Bayesian and ML trees (Fig. 1).
Taxonomic impact of the expanded LSU rDNA sequence dataset analysis
The phylogenetic analysis using the seven-genes sequence dataset contained limited teleomorphic and filamentous species of Tremellomycetes. For a better understanding of the phylogenetic relationships between anamorphic (mostly well represented in the analysis in Liu et al. 2015) and teleomorphic species in Tremellomycetes, more teleomorphic species were added to the expanded LSU rDNA dataset. In public databases these taxa are often known from a limited number of rRNA gene sequences derived from herbarium specimens. Furthermore, several recently published yeast species were also added to the LSU dataset. The constrained ML analysis of the expanded LSU dataset was used to place species known from LSU sequences in the phylogenetic clades previously recognised in the analysis of the seven genes (Fig. 1). Results from the LSU analysis were not used to challenge the results of multi-gene studies (Findley et al., 2009, Millanes et al., 2011, Liu et al., 2015), but to enlarge undersampled clades in Liu et al. (2015) and to investigate the stability of these clades.
In agreement with the few available multigene studies (Findley et al., 2009, Millanes et al., 2011, Liu et al., 2015), our results showed that LSU alone is not sufficient to resolve many clades in Tremellomycetes and the topology of the best tree from the unconstrained ML analysis (data not shown) differed from the seven-genes tree in Liu et al. (2015). Enforcing topological constraints and enlarging step-wise the dataset from 396 (data not shown) to 435 taxa, resulted in a slightly altered topology and, on average, acceptable bootstrap values (Fig. 2). Sequences containing ambiguities and missing data (e.g. from herbarium specimens) and species with unclear phylogenetic placement resulted in decreased average bootstrap values from 79.04 % in the 396-taxa-dataset (data not shown) to 69.05 % in the 435-taxa-dataset, respectively. In particular, support values of the aurantia clade comprised by Tremella aurantia, T. indecorata and T. encephala and strongly supported in Liu et al. (2015) decreased substantially, while support values for the clade containing Cryptococcus mujuensis increased. The constrained ML tree obtained from the expanded LSU rRNA gene sequence dataset did not change the main taxonomic conclusions (Fig. 2). Among the clades supported in Liu et al. (2015) poor or no support was observed for the clade (Saitozyma) comprised by C. flavus, C. paraflavus and C. podzolicus (Fig. 1, Fig. 2).
The addition of the supplemental species or strains resulted in the enlargement of several clades, especially single-species lineages recognised in the seven-genes tree, e.g. Bullera arundinariae, Cryptococcus arrabidensis, C. mujuensis, Filobasidium capsuligenum, and Tremella giraffa (Fig. 2). In addition, a few new clades were identified (Fig. 2, Table 1). The addition of the recently described species Cryptococcus tahquamenonensis (Sylvester et al. 2015) to the monotypic huempii clade recognised in the seven-genes dataset in the Cystofilobasidiales supports the proposal of a new genus (Krasilnikovozyma gen. nov.) for this clade (Fig. 3). Thirteen of the supplemental sequences corresponding to nine species were located in the Filobasidales (Fig. 4). The single-species clade represented by Cryptococcus arrabidensis in the seven-genes tree was expanded to contain eight species including two Syzygospora and five undescribed Cryptococcus species with high (82 % in ML and 97 % in MP, respectively) bootstrap support (Fig. 4), confirming that this clade represents a distinct genus, for which the older name Heterocephalacria is reinstalled (Fig. 4, see Taxonomy section). The cylindricus clade containing two Cryptococcus species recognised in the seven-genes tree (Liu et al. 2015) was expanded to include Syzygospora sorana, Filobasidium capsuligenum and three additional Cryptococcus species. The expanded clade received 93 % and 92 % bootstrap support in the ML and MP trees, respectively (Fig. 4). The teleomorphic species Christiansenia pallida and Syzygospora alba, the type species of the genus Syzygospora, formed a distinct new clade with 100 % bootstrap support (Fig. 4). This result supports the transfer of C. pallida to the genus Syzygospora as proposed previously by Ginns (1986) and shown by Millanes et al. (2011). The remaining four clades recognised in the Filobasidales in Liu et al. (2015) remain unchanged (Fig. 1, Fig. 2, Fig. 4).
Fig. 3.
Phylogenetic relationships of yeasts and related taxa from the order Cystofilobasidiales in Tremellomycetes obtained by maximum-likelihood analysis of LSU (D1/D2 domains) rRNA gene. Tree topology was backbone-constrained with the well-supported (>85 %) bipartitions of the topology of the seven-genes tree (Liu et al. 2015). Bootstrap percentages (BP) of maximum likelihood and neighbour-joining analyses from 1 000 replicates are shown respectively from left to right on the deep and major branches resolved and in brackets following recognised clades. The type species of accepted genera are in bold and the taxa not included in the seven-genes dataset (Liu et al. 2015) are in red. Note: ns, not supported (BP < 50 %).
Fig. 4.
Phylogenetic relationships of yeasts and related taxa from the order Filobasidiales in Tremellomycetes obtained by maximum-likelihood analysis of LSU (D1/D2 domains) rRNA gene. Tree topology was backbone-constrained with the well-supported (>85 %) bipartitions of the topology of the seven-genes tree (Liu et al. 2015). Bootstrap percentages (BP) of maximum likelihood and neighbour-joining analyses from 1 000 replicates are shown respectively from left to right on the deep and major branches resolved and in brackets following recognised clades. The type species of accepted genera are in bold and the taxa not included in the seven-genes dataset (Liu et al. 2015) are in red. Note: ns, not supported (BP < 50 %).
In the Tremellales, the majority of the newly added Tremella species were located in the Tremella-containing clades recognised previously, including clades I, II and III, which were distinguished by Millanes et al. (2011) for lichenicolous taxa. These three clades exclusively or mainly contained Tremella species (Fig. 5A, B). The strongly supported aurantia clade in the seven-genes tree, comprising T. aurantia, T. indecorata and T. encephala, was extended with two species, namely T. aurantialba and T. microspora (Fig. 5A). The phylogenetic position of T. indecorata in the LSU tree was controversial because the support for this clade was decreasing with increasing dataset size and because sequences representing this taxon appeared to be separated in the tree. Nucleotide sequences of T. indecorata derived from strains CBS 6976 (used in Liu et al. 2015) and DSM 5727 grouped together, while a sequence derived from a herbarium specimen (used in Millanes et al. 2011) was placed close to T. moriformis (Fig. 5B). The latter sequence was also identical to the sequence of T. subanomala strain NBRC 9308 (collected by R.J. Bandoni), which is available in the online catalogue of the NBRC collection (NITE, Japan). Thus additional sampling is needed to assess the phylogenetic position of T. indecorata. We recognise the resolving power of multi-gene trees and despite lack of support in the LSU phylogeny, the aurantia clade will be treated as a separate genus, for which the name Naematelia is available (Fig. 5A). The foliacea clade formed by one Tremella and two Cryptococcus species recognised previously (Liu et al. 2015) was expanded to include four more Tremella species represented by eight strains with high (99 % in ML and 100 % in MP) bootstrap support (Fig. 5C). This clade will also receive generic rank and the available name Phaeotremella will be assigned. The single-species clade Tremella moriformis recognised in Liu et al. (2015) was expanded to include three Tremella and two Cryptococcus species with moderate bootstrap support (95 % and 76 % in ML and MP analyses, respectively) and the new genus Pseudotremella is proposed to accommodate these fungi (Fig. 5B). Bullera arundinariae, which was placed in an isolated position in the seven-genes tree (Liu et al. 2015), joined a well-supported clade together with Tremella polyporina and Syzygospora effibulata (syn. Carcinomyces effibulatus) in the LSU analysis, for which the genus Carcinomyces is resurrected (Fig. 5D). Tremella exigua together with Sirobasidium brefeldianum and Sirobasidium intermedium formed a strongly supported new clade with a close relationship to clade I in Millanes et al. (2011). The remaining five Tremella species (T. diploschistina, T. haematommatis, T. ramalinae, T. parmeliarum, and T. phaeophysciae) newly added to the LSU tree were located in independent branches in the Tremellales. The Tremella sensu stricto clade resolved in Liu et al. (2015) (i.e. the mesenterica clade in Boekhout et al. 2011a) containing the type species of the genus, T. mesenterica, was also revealed in the LSU tree and received moderate bootstrap support (64 % and 96 % in ML and MP analyses, respectively) (Fig. 5D).
Fig. 5.
Phylogenetic relationships of yeasts and related taxa from the order Tremellales in Tremellomycetes obtained by maximum-likelihood analysis of LSU (D1/D2 domains) rRNA gene. Tree topology was backbone-constrained with the well-supported (>85 %) bipartitions of the topology of the seven-genes tree (Liu et al. 2015). Bootstrap percentages (BP) of maximum likelihood and neighbour-joining analyses from 1 000 replicates are shown respectively from left to right on the deep and major branches resolved and in brackets following recognised clades. The type species of accepted genera are in bold and the taxa not included in the seven-genes dataset (Liu et al. 2015) are in red. Note: ns, not supported (BP < 50 %).
The Bandoniozyma clade recognised in the seven-genes tree was expanded to include the sexual species Rhynchogastrema coronatum and a recently described Cryptococcus species with strong bootstrap support (99 % and 92 % in ML and MP analyses, respectively) (Fig. 5A). Hence, the Bandoniozyma species will be renamed as Rhynchogastrema.
Our analysis also cautions against the description of new species in the genus Kwoniella solely based on LSU sequence data. The support for this rapidly growing clade was low in all ML analyses and varied from 70 % to 64 % (data not shown). From the recently described asexual species placed in the genus based on the similarity to known Kwoniella species, K. newhampshirensis and K. shandongensis were grouped close to K. heveanensis (Fig. 5C). However, two other recently described anamorphic species, Cryptococcus tronadorensis, which was assigned to the Kwoniella clade by de Garcia et al. (2012) and K. betulae described by Sylvester et al. (2015) formed a separate clade together with Cryptococcus mujuensis with high bootstrap support (97 % in ML and 93 % in MP analyses) (Fig. 5C). These taxa were in isolated positions in Liu et al. (2015). The affinity of this clade with the Kwoniella clade was not supported in this study, therefore a new genus Fonsecazyma is proposed to accommodate these species (Fig. 5C).
The five strains of Trimorphomyces papilionaceus, which is the type species of the genus, clustered together and were found to be closely related to Bullera sakaeratica with bootstrap support values of 86 % and 93 % in the ML and MP analyses, respectively (Fig. 5E). The close relationship of T. papilionaceus and B. sakaeratica was also resolved in the seven-genes tree (Liu et al. 2015), suggesting that B. sakaeratica has to be transferred to the genus Trimorphomyces. The other additional Trimorphomyces species employed here were placed in another well supported clade (100 % and 93 % in ML and MP analyses, respectively) with some Cryptococcus and Bullera species (Fig. 5D). This clade is also referred to as victoriae clade (Fonseca et al. 2011) or dimennae clade (Liu et al. 2015) and is not closely related to T. papilionaceus. The result suggests that the H-shaped conidia, as a morphological character of the genus Trimorphomyces, can be found in different phylogenetic clades. Thus different yeast-like taxa in Tremellomycetes may have a Trimorphomyces-like morph in their life cycles. In addition to the two Trimorphomyces strains, the dimennae clade originally holding six Cryptococcus and one Bullera species as recognised from the seven-genes tree (Liu et al. 2015) was expanded to contain four more Cryptococcus species in the tree constructed from the LSU dataset (Fig. 5D). The new Trimorphomyces strains were located in two sub-groups in the dimennae clade, indicating that this clade may have a trimorphomyces-like sexual state and might be mycoparasitic.
Ten of the supplemental sequences were located in the Trichosporonales (Fig. 6). Two Asterotremella species and one recently described species Cryptococcus thermophilus (Vogelmann et al. 2012) clustered in the Vanrija clade. The teleomorphic species, Tetragoniomyces uliginosus, was located in this order with a phylogenetic position being in agreement with that obtained by Millanes et al. (2011) based on the combined rRNA analysis. The present and previous results suggest that Tetragoniomyces holds an early-branching position in Trichosporonales together with the genera Cryptotrichosporon (Okoli et al. 2007) and Takashimella (Wang & Wang 2015). Three of the four newly added Trichosporon species were scattered in different clades formed by other species of the genus; the other species, T. wanderwaltii, formed a single species lineage (Fig. 6).
Fig. 6.
Phylogenetic relationships of yeasts and related taxa from the orders Trichosporonales in Tremellomycetes obtained by maximum-likelihood analysis of LSU (D1/D2 domains) rRNA gene. Tree topology was backbone-constrained with the well-supported (>85 %) bipartitions of the topology of the seven-genes tree (Liu et al. 2015). Bootstrap percentages (BP) of maximum likelihood and neighbour-joining analyses from 1 000 replicates are shown respectively from left to right on the deep and major branches resolved and in brackets following recognised clades. The type species of accepted genera are in bold and the taxa not included in the seven-genes dataset (Liu et al. 2015) are in red. Note: ns, not supported (BP < 50 %).
Taxonomy
Based on 1) the multigene phylogenetic framework presented by in Liu et al. (2015), 2) the quantitative assessment of taxonomic units using the PRBO approach as well as the iterative modified GMYC tests, 3) a further phylogenetic analysis on an expanded LSU rRNA (D1/D2 domains) gene sequence dataset containing as many as available teleomorphic and filamentous taxa within Tremellomycetes, and finally 4) phenotypical criteria, we propose to update the taxonomic system for tremellomycetous yeasts and related filamentous taxa (Table 1). The phylogenetic classification includes five orders, 17 families and 54 genera. Among the genera accepted here, 18 are newly described and 18 are emended (Table 1, Fig. 1, Fig. 2). Seven of the families are newly proposed and seven are emended from existing ones. Among the accepted species, one is newly proposed and 185 are new combinations. The new and emended families and genera and new combinations are described and discussed within their respective orders. We propose to use the term pro tempore or pro tem. in abbreviation to indicate the species names that are temporarily maintained.
Order Cystofilobasidiales Fell, Roeijmans & Boekhout, Int. J. Syst. Bacteriol. 49: 911. 1999.
The order Cystofilobasidiales was proposed for teleomorphic yeasts with teliospores, holobasidia and septa with dolipores, but without parenthesomes (Fell et al. 1999). This order was accepted as family Cystofilobasidiaceae in Filobasidiales by Wells & Bandoni (2001). Our phylogenetic analysis demonstrated the rationality of its taxonomic rank as an order (Liu et al. 2015). Accordingly a new family Mrakiaceae is described and family Cystofilobasidiaceae is emended to accommodate the two major lineages recognised in the order. A new genus Krasilnikovozyma is described and four genera are emended.
Mrakiaceae X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, fam. nov. MycoBank MB812173.
Etymology: In reference to the name of the type genus Mrakia.
This new family is proposed for the monophyletic group formed by the Mrakia, huempii, Phaffia, Udeniomyces, Itersonilia and Tausonia clades recognised in the trees constructed from the Bayesian and ML analyses of the seven-gene dataset (Liu et al. 2015) as shown in Fig. 1 of this study.
Type genus: Mrakia Y. Yamada & Komag. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout
Genera accepted: Mrakia emend., Krasilnikovozyma gen. nov., Phaffia emend., Udeniomyces, Itersonilia emend. and Tausonia emend.
Mrakia Y. Yamada & Komag., J. Gen. Appl. Microbiol. 33: 456. 1987. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
= Mrakiella Margesin & Fell, Int. J. Syst. Evol. Microbiol. 58: 2980. 2008.
This genus is emended to accommodate the Mrakia clade recognised in Liu et al. (2015) which includes five Mrakia and two Mrakiella species in the seven-genes tree.
Basidiocarps absent. Pseudohyphae occasionally produced, branched. True hyphae occasionally produced. Clamp connections present. Sexual reproduction observed in some species. Teliospores terminal or intercalary produced, developing directly from a single cell; teliospores germination by formation of short (8–12 μm), tubular holometabasidia or phragmobasidia. Budding cells present. Chlamydospores and ballistoconidia may be present. Glucose and sucrose may be fermented. Nitrate and nitrite usually utilised. Major CoQ systems CoQ-8, CoQ-9 or CoQ-10.
Type species: Mrakia frigida (Fell, Statzell, I.L. Hunter & Phaff) Y. Yamada & Komag., J. Gen. Appl. Microbiol. 33: 457. 1987.
Notes: Mrakia curviuscula is excluded from the emended genus Mrakia because this species forms a distinct clade together with the anamorph species Cryptococcus huempii (Liu et al. 2015). The genus Mrakiella was proposed for anamorphic species closely related to Mrakia (Margesin & Fell 2008). The three Mrakiella species are transferred here to the genus Mrakia. The affinity of Mrakiella cryoconiti, which was not included in the seven-genes phylogeny, to Mrakia is shown in the trees obtained from the LSU rRNA gene sequences in this study (Fig. 3) as well as in Boekhout et al. (2011a) and Weiss et al. (2014). The emended genus Mrakia currently contains eight species (Table 1).
New combinations for Mrakia
Mrakia aquatica (E.B.G. Jones & Slooff) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812175.
Basionym: Candida aquatica E.B.G. Jones & Slooff, Antonie van Leeuwenhoek 32: 223. 1966.
≡ Mrakiella aquatica (E.B.G. Jones & Sloof) Margesin & Fell, Int. J. Syst. Evol. Microbiol. 58: 2981. 2008.
Mrakia cryoconiti (Margesin & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812176.
Basionym: Mrakiella cryoconiti Margesin & Fell, Int. J. Syst. Evol. Microbiol. 58: 2981. 2008.
Mrakia niccombsii (Thomas-Hall) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812177.
Basionym: Mrakiella niccombsii Thomas-Hall, Extremophiles 14: 56. 2010.
Krasilnikovozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB812178.
Etymology: The genus is named in honour of the Russian microbiologist N. A. Krasil'nikov for his contribution to the ecology and systematics of yeasts.
This genus is proposed for the huempii clade containing Mrakia curviuscula, its anamorph Cryptococcus huempii (Liu et al. 2015) and the recently described Cryptococcus tahquamenonensis (Sylvester et al. 2015).
Basidiocarps absent. Pseudohyphae and true hyphae occasionally produced. Sexual reproduction observed in some species. Teliospores with pseudoclamps terminal produced; teliospores germination by formation of holometabasidia or hyphal structures. Budding cells present. Chlamydospores may be present. Fermentation absent. d-glucose, d-galactose, cellobiose, lactose, d-xylose, and citric acid are usually utilised. Glycerol and low-weight aromatic compounds are not utilised. Nitrate and nitrite usually utilised. Starch-like compounds can be produced. Major CoQ system CoQ-8.
Type species: Krasilnikovozyma huempii (C. Ramírez & A. E. González) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout
Notes: Mrakia curviuscula was considered to be the teleomorph of C. huempii based on similar physiological and biochemical characters (Bab'eva et al. 2002) and identical D1/D2 LSU rRNA gene sequences (Boekhout et al., 2011a, Weiss et al., 2014). Liu et al. (2015) showed that they also exhibited similar protein-coding gene sequences, confirming their conspecificity. Cryptococcus tahquamenonensis was recently described as a close relative of Mrakia curviuscula and Cryptococcus huempii (Sylvester et al. 2015). This relationship was confirmed in this study by the ML analysis of the D1/D2 sequence dataset (Fig. 3). Thus, C. tahquamenonensis is transferred here to Krasilnikovozyma gen. nov. Phenotypically, the species in Krasilnikovozyma can grow at 25 °C and thus differ from the species of the genus Mrakia emend., which are psychrophilic with a maximum growth temperature below 20 °C (Fell 2011). Krasilnikovozyma currently contains two species (Table 1) and two additional sequences representing potential new species were obtained from public databases (Fig. 3).
New combinations for Krasilnikovozyma
Krasilnikovozyma huempii (C. Ramírez & A. E. González) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812179.
Basionym: Candida huempii C. Ramírez & A. E. González, Mycopathologia 88: 167. 1984.
= Mrakia curviuscula Bab'eva, Lisichk., Reshetova & Danilev., Microbiology, Moscow 71: 450. 2002.
Krasilnikovozyma tahquamenonensis (Q.M. Wang, A.B. Hulfachor, K. Sylvester & C.T. Hittinger) A.M. Yurkov, comb. nov. MycoBank MB813656.
Basionym: Cryptococcus tahquamenonensis Wang et al., FEMS Yeast Res. 15: http://dx.doi.org/10.1093/femsyr/fov002. 2015.
Phaffia M.W. Mill., Yoney. & Soneda, Int. J. Syst. Bacteriol. 26: 287. 1976. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
= Xanthophyllomyces Golubev, Yeast 11: 105. 1995.
The genus Phaffia is emended to include the teleomorphic species Xanthophyllomyces dendrorhous (Golubev 1995) based on molecular phylogenetic analyses (Fell et al., 2000, Scorzetti et al., 2002, Boekhout et al., 2011a, David-Palma et al., 2014, Weiss et al., 2014, Liu et al., 2015).
Basidiocarps absent. Cultures often pigmented and pink to orange due to carotenoid pigments. Holobasidia slender cylindrical with apical basidiospores. True hyphae not produced. Rudimentary pseudohyphae occasionally produced. Budding cells present. Ballistoconidia not produced. Fermentation present. Nitrate and nitrite not utilised. Major CoQ system CoQ-10.
Type species: Phaffia rhodozyma M.W. Miller, Yoneyama & Soneda, Int. J. Syst. Bacteriol. 26: 287. 1976.
Notes: The teleomorphic genus Xanthophyllomyces (Golubev 1995) was described later than the anamorphic genus Phaffia (Miller et al. 1976), thus the name Phaffia has priority according to the code of nomenclature (McNeill et al. 2012). Phaffia rhodozyma was regarded as the anamorph of X. dendrorhous because the type strains of the two species had similar D1/D2 sequences (Fell et al., 2007, Fell et al., 2011). However, ITS and IGS sequence analyses generated by Fell & Blatt (1999) and protein-coding gene sequence comparisons performed in Liu et al. (2015) suggest that they may represent different species. A recent analysis performed by David-Palma et al. (2014) showed a rather complex population structure within the species P. rhodozyma. These authors also observed that the type strain of P. rhodozyma contains genetic material from two different populations. Thus, DNA sequencing attempts may result in two different PCR-products for marker genes, which show a substantial nucleotide difference (David-Palma et al., 2014, Liu et al., 2015). It has been suggested to consider sexual and asexual strains in a single species with four genetic lineages, and we adopt this proposal in the present study. The genus currently contains one species (Table 1) and the discovery of two more potential novel species was recently reported by David-Palma et al. (2014).
Itersonilia Derx, Bull. bot. Gdns Buitenz. 17: 471. 1948. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
This genus is emended to include the Itersonilia clade recognised from the rRNA and multigene sequence analyses (Niwata et al., 2002, Boekhout et al., 2011a, Weiss et al., 2014, Liu et al., 2015) and to incorporate a species previously assigned to the genus Udeniomyces.
Basidiocarps absent. Dikaryotic hyphae occasionally produced, regularly branched, inflated cells may be present. Monokaryotic hyphae may be present. Clamp connections present. Septa with dolipores lacking parenthesomes. Pseudohyphae without clamp connections occasionally present. Sexual reproduction not observed. Budding (monokaryotic) cells may be present. Ballistoconidia present. Chlamydospores occasionally produced. Fermentation absent. Low-weight aromatic compounds not utilised. Nitrate and nitrite utilised. Starch-like compounds may be produced. The major CoQ system CoQ-9 or CoQ-10.
Type species: Itersonilia perplexans Derx., Bull. Bot. Gdns Buitenz. 17: 471. 1948.
Notes: The species Udeniomyces pannonicus was separated from the Udeniomyces clade and located together with Itersonilia perplexans based on molecular data (Niwata et al., 2002, Boekhout et al., 2011a, Weiss et al., 2014, Liu et al., 2015). I. perplexans and U. pannonicus are phenotypically similar by forming grayish to yellowish colonies, whereas the other Udeniomyces species form pinkish-white to orange-white colonies. The emended genus Itersonilia currently contains two species (Table 1) and an additional sequence representing a potential new species was obtained from public databases (Fig. 3).
New combination for Itersonilia
Itersonilia pannonica (Niwata, Tornai-Leh., T. Deák & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812189.
Basionym: Udeniomyces pannonicus Niwata et al., Int. J. Syst. Evol. Microbiol. 52: 1890. 2002.
Tausonia Babeva, Mikrobiologiya 67: 231. 1998. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
= Guehomyces Fell & Scorzetti, Int. J. Syst. Evol. Microbiol. 54: 997. 2004.
This genus is emended to include the monotypic genus Guehomyces based on rRNA sequence analyses (Boekhout et al., 2011a, Fell and Guého-Kellermann, 2011, Sampaio, 2011b, Liu et al., 2015).
Basidiocarps absent. Cultures may be pigmented and pinkish. Hyphae disarticulate into arthroconidia. Chlamydospores occasionally produced. Sexual reproduction not observed. Budding cells present. Fermentation absent. Nitrate and nitrite utilised. Major CoQ system CoQ-9.
Type species: Tausonia pamirica Bab'eva, Mikrobiologiya 67: 232. 1998.
Notes: Guehomyces pullulans (formerly Trichosporon pullulans) and T. pamirica were located together in a well-supported clade named Guehomyces based on rRNA sequence analyses (Boekhout et al., 2011a, Fell and Guého-Kellermann, 2011, Sampaio, 2011b, Liu et al., 2015). Tausonia (Bab'eva 1998) has nomenclatural priority over Guehomyces (Fell & Scorzetti 2004). The two species are phenotypically similar in producing arthroconidia. The genus currently contains two species (Table 1) and an additional sequence representing a potential new species was obtained from public databases (Fig. 3).
New combination for Tausonia
Tausonia pullulans (Lindner) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812190.
Basionym: Oidium pullulans Lindner, Mikrosk. Betriebsk. Gährung. 3te Aufl.: 286. 1901.
≡ Trichosporon pullulans (Lindner) Diddens & Lodder, Die anaskosporogenen Hefen, II Hälfte: 410. 1942. MycoBank MB291595.
≡ Guehomyces pullulans (Lindner) Fell & Scorzetti, Int. J. Syst. Evol. Microbiol. 54: 997. 2004.
Cystofilobasidiaceae K. Wells & Bandoni, The Mycota 7(B): 113. 2001. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
This family is emended to include the Cystofilobasidium clade recognised from the seven-gene phylogeny (Liu et al. 2015).
Basidiocarps absent. Cultures often pink to orange due to carotenoid pigments. True hyphae produced. Pseudohyphae occasionally produced. Clamp connections may be present. Sexual reproduction observed in some species. Teliospores terminal or intercalary produced; teliospores germination by holobasidia. Budding cells present. Fermentation absent. Aromatic compounds weakly or not utilised. Nitrate and nitrite utilised. Major CoQ system CoQ-8.
Type genus: Cystofilobasidium Oberw. & Bandoni.
Genus accepted: Cystofilobasidium.
Notes: The family Cystofilobasidiaceae was originally proposed within the order Filobasidiales mainly based on the morphological features of the holobasidia. Three genera Cystofilobasidium, Mrakia, and Xanthophyllomyces were included in this family (Wells & Bandoni 2001). Phylogenetic analyses suggest that this family represents an order independent of Filobasidiales (Fell et al., 1999, Fell et al., 2000, Scorzetti et al., 2002, Boekhout et al., 2011a, Weiss et al., 2014, Liu et al., 2015) and support the proposal of Cystofilobasidiales by Fell et al. (1999). The family Cystofilobasidiaceae is emended here to include only the Cystofilobasidium clade (Fig. 1). The species in this family produce much longer (up to 65 μm) holobasidia (Fell et al., 1999, Sampaio, 2011a, Weiss et al., 2014) than those (8–12 μm) produced by the teleomorphic species in the other family Mrakiaceae within Cystofilobasidiales.
Order Filobasidiales Jülich, Bibl. Mycol. 85: 324. 1981.
The order Filobasidiales was originally introduced to accommodate yeasts lacking basidiocarps and having holobasidia with passively released basidiospores (Jülich 1981). Three families, namely Filobasidiaceae, Cystofilobasidiaceae and Rhynchogastremaceae were included in this order by Wells & Bandoni (2001). As discussed above, the Cystofilobasidiaceae sensu Wells & Bandoni (2001) represents a distinct order and this family is emended in this study to include only the genus Cystofilobasidium. Rhynchogastremaceae was monotypic, including only Rhynchogastrema coronatum (Metzler et al. 1989). LSU rRNA sequence analyses assigned this dimorphic mycoparasite to the Tremellales as shown in Weiss et al. (2014) and this study (Fig. 5A). Molecular phylogenetic analyses showed that the order Filobasidiales without Cystofilobasidiaceae and Rhynchogastremaceae represents a monophyletic lineage, which includes species from the teleomorphic genera Filobasidium and Syzygospora and the anamorphic genus Cryptococcus (Fell et al., 2000, Scorzetti et al., 2002, Boekhout et al., 2011a, Millanes et al., 2011, Weiss et al., 2014, Liu et al., 2015). Three families, namely Filobasidiaceae, Syzygosporaceae and Christianseniaceae have been proposed in this order. However, they were not resolved as separate groups in the previous and present molecular phylogenetic analyses. Therefore, the families and genera within this order are recircumscribed in this study based on the phylogenetic analyses of the seven-genes (Fig. 1) and the LSU rRNA datasets (Fig. 2, Fig. 4), resulting in the recognition of two families and seven genera (Table 1).
Filobasidiaceae L.S. Olive, J. Elisha Mitchell Scient. Soc. 84: 261. 1968. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
This family is circumscribed to accommodate a monophyletic group containing the Filobasidium, albidus, gastricus, Cryptococcus arrabidensis and Syzygospora clades resolved in the trees from the seven-gene dataset (Liu et al. 2015) and the expanded LSU rRNA dataset (Fig. 1, Fig. 2).
Basidiocarps waxy to gelatinous in lichenicolous species, absent in cultures. Hyphae with clamp connection and haustorial branches may be present. Holobasidia tubular or long with terminally sessile basidiospores. Pseudohyphae and true hyphae occasionally produced. Sexual reproduction present in some species. Budding cells present. Ballistoconidia absent. Fermentation absent. Nitrate utilised by several species. The major CoQ system CoQ-9 or CoQ-10.
Type genus: Filobasidium L.S. Olive. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout
Genera accepted: Filobasidium, Goffeauzyma, Heterocephalacria, Naganishia, and Syzygospora.
Notes: The family Filobasidiaceae was proposed by Olive (1968) to accommodate the species Filobasidium floriforme. Three teleomorphic genera, namely Filobasidium, Filobasidiella and Cystofilobasidium, were then included in this family (Kwon-Chung 1987). These genera share the presence of long slender holobasidia, sessile terminal basidiospores and dolipore-like septa without parenthesomes (Kwon-Chung 1987). The genera Filobasidiella and Cystofilobasidium were transferred to the Tremellales and Cystofilobasidiales, respectively, based on molecular phylogenetic analyses (Fell et al., 2000, Scorzetti et al., 2002). Phylogenetic analysis of the expanded LSU rRNA dataset showed that four Syzygospora species were closely related to the taxa assigned to Filobasidiaceae (Fig. 4). Therefore, the Filobasidiaceae is emended here to include Filobasidium, Naganishia (i.e. the albidus clade), Goffeauzyma gen. nov. (i.e. the gastricus clade), Heterocephalacria (i.e. the Cryptococcus arrabidensis clade) and Syzygospora.
Filobasidium L.S. Olive, J. Elisha Mitchell Scient. Soc. 84: 261. 1986. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
This genus is emended to accommodate the species in the Filobasidium clade (floriforme clade sensu Fonseca et al. 2011) circumscribed by the phylogenetic analyses of seven genes (Liu et al. 2015).
Basidiocarps absent. Cultures often pigmented and pinkish-white. Sexual reproduction present in some species. Hyphae with clamp connections and haustorial branches may be present. Holobasidia tubular with terminal, sessile basidiospores. Pseudohyphae occasionally produced. Budding cells present. Fermentation absent. l-malic, saccharic, protocatechuic and p-hydroxybenzoic acids often utilised. Nitrate may or may not be utilised. Starch-like compounds usually produced. The major CoQ system CoQ-9 or CoQ-10.
Type species: Filobasidium floriforme L.S. Olive, J. Elisha Mitchell scient. Soc. 84: 261. 1968.
Notes: Filobasidium capsuligenum is excluded from this genus because it is located outside the Filobasidium clade and is closely related to the cylindricus clade (i.e. Piskurozyma gen. nov.). The genus Filobasidium currently contains nine species (Table 1).
New combinations for Filobasidium
Filobasidium chernovii (Á. Fonseca, Scorzetti & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812191.
Basionym: Cryptococcus chernovii Á. Fonseca et al., Can. J. Microbiol. 46: 20. 2000.
Filobasidium magnum (Lodder & Kreger-van Rij) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812192.
Basionym: Cryptococcus laurentii var. magnus Lodder & Kreger-van Rij, Yeast, a taxonomic study, [Edn 1]: 670. 1952.
Filobasidium oeirense (Á. Fonseca, Scorzetti & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812193.
Basionym: Cryptococcus oeirensis Á. Fonseca et al., Can. J. Microbiol. 46
Filobasidium stepposum (Golubev & J.P. Samp.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812194.
Basionym: Cryptococcus stepposus Golubev & J.P. Samp. Mycol. Res. 110: 960. 2006.
Filobasidium wieringae (Á. Fonseca, Scorzetti & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812195.
Basionym: Cryptococcus wieringae Á. Fonseca et al., Can. J. Microbiol. 46: 25. 2000.
Naganishia S. Goto, J. Ferment. Technology 41: 461. 1963. emend. A.M. Yurkov & T. Boekhout.
This genus is resurrected and emended here to accommodate the albidus clade comprised of 15 species that previously belonged to Cryptococcus (Fell et al., 2000, Scorzetti et al., 2002, Fonseca et al., 2011, Liu et al., 2015).
Basidiocarps absent. Hyphae and pseudohyphae usually absent. Sexual reproduction not observed. Budding cells present. Ballistoconidia absent. Fermentation absent. l-malic, vanillic, ferulic, caffeic, p-coumaric protocatechuic and hydroxybenzoic acids often utilised. Nitrate utilised. Starch-like compounds usually produced. Major CoQ system CoQ-9 or CoQ-10.
Type species: Naganishia globosa S. Goto, J. Ferment. Technology 41: 461. 1963. MycoBank MB335061.
= Cryptococcus saitoi Á. Fonseca, Scorzetti & Fell, Can. J. Microbiol. 46: 24. 2000. MycoBank MB464349.
Notes: The genus Naganishia was described to accommodate the yeast Naganishia globosus (Goto 1963), which was later synonymised with Cryptococcus saitoi based on rRNA sequence data (Fonseca et al. 2011). Some of the other species comprising this clade were described earlier than N. globosus, but they were originally classified in the genera Torula, Torulopsis and Rhodotorula. Since type species of these genera are placed outside Tremellomycetes, none of them can be used to reclassify the albidus clade. The generic name Naganishia also appears to be the oldest in the clade where the type species of the genus is specified and a type strain is preserved. Cryptococcus onofrii and C. vaughanmartiniae were recently described as cold-adaptive species in the albidus clade (Turchetti et al. 2015). Their affinity to the albidus clade was confirmed in this study by ML analysis using the expanded LSU rRNA gene sequence dataset (Fig. 4). Thus, these two Cryptococcus species are transferred to the genus Naganishia together with the Cryptococcus species included in the albidus clade in the seven-genes dataset (Liu et al. 2015). Fifteen species are presently accepted in the genus Naganishia (Table 1).
New combinations for Naganishia
Naganishia adeliensis (Scorzetti, I. Petrescu, Yarrow & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813139.
Basionym: Cryptococcus adeliensis Scorzetti et al., Antonie van Leeuwenhoek 77: 155. 2000.
Naganishia albidosimilis (Vishniac & Kurtzman) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813140.
Basionym: Cryptococcus albidosimilis Vishniac & Kurtzman, Int. J. Syst. Bacteriol. 42: 550. 1992.
Naganishia albida (Saito) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813140.
Basionym: Torula albida Saito, Jap. J. Bot. 1: 43. 1922.
Naganishia antarctica (Vishniac & Kurtzman) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813145.
Basionym: Cryptococcus antarcticus Vishniac & Kurtzman, Int. J. Syst. Bacteriol. 42: 548. 1992.
Naganishia bhutanensis (Goto & Sugiy.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813165.
Basionym: Cryptococcus bhutanensis Goto & Sugiy., Can. J. Bot. 48: 2097. 1970.
Naganishia cerealis (Passoth, A.-C. Andersson, Olstorpe, Theelen, Boekhout & Schnürer) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813167.
Basionym: Cryptococcus cerealis Passoth et al., Antonie van Leeuwenhoek 96: 641. 2009.
Naganishia diffluens (Zach) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813172.
Basionym: Torulopsis diffluens Zach, Wolfram & Zach, Arch. Derm. Syph. 170: 690. 1934.
Naganishia friedmannii (Vishniac) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813175.
Basionym: Cryptococcus friedmannii Vishniac, Mycologia 77: 150. 1985.
Naganishia liquefaciens (Saito & M. Ota) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813177.
Basionym: Torulopsis liquefaciens Saito & M. Ota, Jozogaku Zasshi 12: 167. 1934.
Naganishia onofrii (Turchetti, Selbmann & Zucconi) A.M. Yurkov, comb. nov. MycoBank MB813182.
Basionym: Cryptococcus onofrii Turchetti et al., Extremophiles 19: 149–159. 2015.
Naganishia randhawae (Z.U. Khan, S.O. Suh. Ahmad, F. Hagen, Fell, Kowshik, Chandy & Boekhout) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813246.
Basionym: Cryptococcus randhawai Z.U. Khan et al., Antonie van Leeuwenhoek 97: 256. 2010.
Naganishia uzbekistanensis (Á. Fonseca, Scorzetti & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813248.
Basionym: Cryptococcus uzbekistanensis Á. Fonseca et al., Can. J. Microbiol. 46: 25. 2000.
Naganishia vaughanmartiniae (Turchetti, Blanchette & Arenz) A.M. Yurkov, comb. nov. MycoBank MB813249.
Basionym: Cryptococcus vaughanmartiniae Turchetti et al., Extremophiles 19: 157. 2015.
Naganishia vishniacii (Vishniac & Hempfling) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813250.
Basionym: Cryptococcus vishniacii Vishniac & Hempfling, Int. J. Syst. Bacteriol. 29: 155. 1979.
Goffeauzyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813251.
Etymology: The genus is named in honour of the yeast biologist and geneticist André Goffeau, who was the initiator and coordinator of the genome sequence project of Saccharomyces cerevisiae.
The new genus is proposed for the gastricus clade recognised from the seven-genes tree (Liu et al. 2015), which consists of six species previously belonging to Cryptococcus.
Basidiocarps absent. True hyphae and pseudohyphae not observed. Sexual reproduction not observed. Budding cells present. Ballistoconidia absent. Fermentation absent. Low-weight aromatic compounds often not utilised. Nitrate usually not utilised. Major CoQ system CoQ-9.
Type species: Goffeauzyma gastrica (Reiersöl & di Menna) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout comb. nov.
Notes: The gastricus clade recognised in Liu et al. (2015) consists of C. gastricus and C. gilvescens isolated from soil (Fell et al., 2000, Boekhout et al., 2011a, Fonseca et al., 2011); C. aciditolerans, C. ibericus and C. metallitolerans in the ARD ecoclade that were obtained from acid rock drainage (Gadanho & Sampaio 2009); and C. agrionensis from acidic water of a volcanic environment (Russo et al. 2010). Though the species of the ARD ecoclade clustered together, they did not form a significantly supported clade in the seven-genes tree (Liu et al. 2015) and in the LSU rRNA gene tree (Fig. 4). The PRBO test indicated that the six species of the gastricus clade did not show significant deviation compared to the species in the reference clades (Table 2). Thus they are assigned to a single genus at present which currently contains six species (Table 1).
New combinations for Goffeauzyma
Goffeauzyma aciditolerans (Gadanho & J.P. Samp.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813252.
Basionym: Cryptococcus aciditolerans Gadanho & J.P. Samp., Int. J. Syst. Evol. Microbiol. 59: 2378. 2009.
Goffeauzyma agrionensis (Russo, Libkind, Samp. & van Broock) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813253.
Basionym: Cryptococcus agrionensis Russo et al., Int. J. Syst. Evol. Microbiol. 60: 998. 2010.
Goffeauzyma gastrica (Reiersöl & di Menna) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813254.
Basionym: Cryptococcus gastricus Reiersöl & di Menna, Antonie van Leeuwenhoek 24: 28. 1958.
Goffeauzyma gilvescens (Chernov & Babeva) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813255.
Basionym: Cryptococcus gilvescens Chernov & Babeva, Mikrobiologiya 57: 1032. 1988.
Goffeauzyma iberica (Gadanho & J.P. Samp.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813256.
Basionym: Cryptococcus ibericus Gadanho & J.P. Samp., Int. J. Syst. Evol. Microbiol. 59: 2378. 2009.
Goffeauzyma metallitolerans (Gadanho & J.P. Samp.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813257.
Basionym: Cryptococcus metallitolerans Gadanho & J.P. Samp., Int. J. Syst. Evol. Microbiol. 59: 2379. 2009.
Heterocephalacria Berthier, Mycotaxon 12: 114. 1980. emend. A. M. Millanes, M. Wedin & A.M. Yurkov.
This genus is resurrected and emended to accommodate the clade represented by Cryptococcus arrabidensis recognised in the seven-genes tree (Liu et al. 2015) and was expanded to include two Syzygospora and five undescribed Cryptococcus species in this study (Fig. 4). The type of Heterocephalacria, H. solida (Berthier 1980), is morphologically similar to these two lichenicolous “Syzygospora” species, viz. basidia with up to four sterigmata, spores obliquely attached to sterigmata, both spores and sterigmata refractive at the point of attachment, and most likely belongs here.
Basidiocarps waxy to gelatinous in lichenicolous species. Hyphae thin-walled with clamp connections and haustorial branches. Holobasidia tubular or long with terminal sessile basidiospores. Sexual reproduction not observed in culture. True hyphae and pseudohyphae absent in culture. Budding cells present. Fermentation absent. Nitrate utilised. Starch-like compounds usually not produced. Major CoQ system not known.
Type species: Heterocephalacria solida Berthier, Mycotaxon 12: 114. 1980.
≡ Syzygospora solida (Berthier) Ginns, Mycologia 78: 632. 1986.
Notes: The genus Heterocephalacria with one species, H. solida, was described by Berthier (1980) for a capitate clavarioid fungus with partially cruciately septate apices of the basidia and secondary spores (i.e. conidia). This genus was treated as a synonym of the genus Syzygospora by Ginns (1986) based on the morphological similarity of H. solida with Syzygospora species. The type species of Syzygospora, S. alba, is located in a separate clade (Fig. 4). The emended genus Heterocephalacria currently contains four described species (Table 1) and four additional sequences representing potential new species were obtained from public databases (Fig. 4).
New combinations for Heterocephalacria
Heterocephalacria arrabidensis (Á. Fonseca, Scorzetti & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813258.
Basionym: Cryptococcus arrabidensis Á. Fonseca et al., Can. J. Microbiol. 46: 20. 2000.
Heterocephalacria bachmannii (Diederich & M.S. Christ.) Millanes & Wedin, comb. nov. MycoBank MB813259.
Basionym: Syzygospora bachmannii Diederich & M.S. Christ., Bibl. Lichenol. 61: 30. 1996.
Heterocephalacria physciacearum (Diederich) Millanes & Wedin, comb. nov. MycoBank MB813260.
Basionym Syzygospora physciacearum Diederich, Bibl. Lichenol. 61: 38. 1996.
Syzygospora G.W. Martin, J. Wash. Acad. Sci. 27: 112. 1937.
= Christiansenia Hauerslev, Friesia 9: 43. 1969.
This genus is restricted to include the species in the clade represented by Syzygospora alba, the type species of the genus, as recognised in the tree drawn from the LSU D1/D2 dataset (Fig. 4).
Basidiocarps thin, gelatinous, subhyaline on the hymenial surface of the host or a cerebriform mass up to 12 cm diam. Hyphae with clamp connections present. Haustorial branches rather frequent, typically subglobose, arising from clamp connections. Basidia suburniform to cylindrical or clavate, 50–120 × 6–8 μm, holobasidiate or incompletely septate. Basidiospores broadly ellipsoid. Zygoconidia develop as two swellings, one on each side of the transverse septum in the conidiogenous cells.
Type species: Syzygospora alba G.W. Martin, J. Wash. Acad. Sci. 27: 112. 1937.
Notes: Syzygospora proved to be polyphyletic (Sampaio, 2004, Millanes et al., 2011). The phylogenetic analysis of the expanded LSU rRNA gene sequence dataset in this study showed that Syzygospora effibulata formed a strongly supported clade with Tremella polyporina in Tremellales, while the other species of Syzygospora studied occurred in three separate clades in the Filobasidiales. Syzygospora bachmannii and S. physciacearum were assigned to the emended genus Heterocephalacria as discussed above. Syzygospora sorana was located in a different clade together with Cryptococcus fildesensis and Filobasidium capsuligenum (see below). The type of the genus, S. alba, formed a well supported clade with Christiansenia (Syzygospora) pallida (Fig. 4). Both species form zygoconidia and are mycoparasitic on the hymenial surface of Corticiaceae (aphyllophoralean fungi) differing from the hosts of other Syzygospora species (Ginns 1986). Thus, the genus Syzygospora currently includes only S. alba and S. pallida (Table 1).
Piskurozymaceae X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, fam. nov. MycoBank MB812196.
Etymology: In reference to the name of the type genus Piskurozyma gen. nov.
This family is proposed for a strongly supported monophyletic lineage including the aerius (Solicoccozyma gen. nov.) and cylindricus (Piskurozyma gen. nov.) clades and a single-species clade Filobasidium capsuligenum in Filobasidiales resolved by the seven-gene dataset analysis (Fig. 1).
Basidiocarps absent. Pseudohyphae and true hyphae may be present. Clamp connections on dikaryotic hyphae occasionally present. Haustorial branches not present. Sexual reproduction with holobasidia. Budding cells present. Ballistoconidia absent. Fermentation usually absent. Nitrate usually utilised. The major CoQ system CoQ-10.
Type genus: Piskurozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
Genera accepted: Piskurozyma gen. nov. and Solicoccozyma gen. nov.
Piskurozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB812206.
Etymology: The genus is named in honour of the late Jure Piškur for his contribution to yeast genetics, physiology and evolutionary biology.
This genus is proposed for the cylindricus clade (Liu et al. 2015) containing three species previously classified in the genera Cryptococcus or Bullera as shown in the trees obtained from the seven-genes dataset. Phylogenetic analysis of the extended LSU rRNA gene dataset suggests close relationships with the teleomorphic species Filobasidium capsuligenum and Syzygospora sorana, and two recently described cryptococci, C. filicatus and C. fildensis (Fig. 4).
Basidiomes, if present, within the hymenium of the host. Dikaryotic hyphae with clamp connections. Haustoria not reported. In culture true hyphae occur occasionally. Pseudohyphae absent. Sexual reproduction observed for some species. Holobasidia slender with terminal sessile basidiospores. Budding cells present. Ballistoconidia may be present. Fermentation occasionally present. Nitrate may be utilised. Starch-like compounds usually produced. Major CoQ system CoQ-10.
Type species: Piskurozyma cylindrica (Á. Fonseca, Scorzetti & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
Notes: Filobasidium capsuligenum was regarded as a single-species clade because of its unique fermentative ability. However, the close relationship of this species with the cylindricus clade was resolved in the seven-genes tree with strong support (Liu et al. 2015). In the constrained ML tree from the expanded LSU rRNA sequence dataset, F. capsuligenum clustered together with a fungicolous species, Syzygospora sorana, and two recently published Cryptococcus species, namely C. filicatus (Golubev & Sampaio 2009) and C. fildesensis (Zhang et al. 2014). Therefore these species are assigned to the genus Piskurozyma gen. nov. This genus currently contains seven described species (Table 1) and two additional sequences representing potential new species were obtained from public databases (Fig. 4).
New combinations for Piskurozyma
Piskurozyma capsuligena (Fell, Statzell, I.L. Hunter & Phaff) A.M. Yurkov, comb. nov. MycoBank MB813122.
Basionym: Leucosporidium capsuligenum Fell et al., Antonie van Leeuwenhoek 35: 444. 1969.
Piskurozyma cylindrica (Á. Fonseca, Scorzetti & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812208.
Basionym: Cryptococcus cylindricus Á. Fonseca et al., Can. J. Microbiol. 46: 22. 2000.
Piskurozyma fildesensis (T. Zhang & L.-Y. Yu) A.M. Yurkov, comb. nov. MycoBank MB813124.
Basionym: Cryptococcus fildesensis T. Zhang & L.Y. Yu, Int. J. Syst. Evol. Microbiol. 64: 675–679. 2013.
Piskurozyma filicatus (Golubev & J.P. Samp.) Kachalkin, comb. nov. MycoBank MB814788.
Basionym: Cryptococcus filicatus Golubev & J.P. Sampaio, J. Gen. Appl. Microbiol. 55: 445. 2009.
≡ Cryptococcus filicatus var. filicatus Golubev & J.P. Sampaio, J. Gen. Appl. Microbiol. 55: 445. 2009.
≡ Cryptococcus filicatus var. pelliculosus Golubev & J.P. Sampaio, J. Gen. Appl. Microbiol. 55: 445. 2009.
Piskurozyma silvicola (Golubev & J.P. Samp.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812209.
Basionym: Cryptococcus silvicola Golubev & J.P. Samp., Antonie van Leeuwenhoek 89: 48. 2006.
Piskurozyma sorana (Hauerslev) A.M. Yurkov, comb. nov. MycoBank MB813129.
Basionym: Syzygospora sorana Hauerslev, Op. bot. 100: 113. 1989.
Piskurozyma taiwanensis (Nakase, Tsuzuki & M. Takash.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812207.
Basionym: Bullera taiwanensis Nakase et al., J. Gen. Appl. Microbiol. 48: 349. 2002.
Solicoccozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB812197.
Etymology: In reference to the ecological origin of the species in this genus that are mostly isolated from soils.
This genus is proposed for the aerius clade (Fonseca et al., 2011, Liu et al., 2015) containing six anamorphic species previously belonging to the genus Cryptococcus (Liu et al. 2015).
Basidiocarps absent. Sexual reproduction with holobasidia may be present. Pseudohyphae and true hyphae occasionally produced. Clamp connections may be present on dikaryotic hyphae. Haustorial branches not observed. Budding cells present. Ballistoconidia absent. Fermentation absent. Nitrate utilised. Starch-like compounds usually not produced. The major CoQ system CoQ-10.
Type species: Solicoccozyma aeria (Saito) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
Notes: The aerius clade on which the new genus is based was consistently resolved and strongly supported in the trees using different datasets (Fonseca et al., 2011, Liu et al., 2015, this study). The species of this genus are usually obtained from soils (Botha, 2006, Fonseca et al., 2011, Yurkov et al., 2012). They frequently produce thick polysaccharide capsules and accumulate substantial amount of lipids (Tanimura et al. 2014). They have a pronounced ability to assimilate aldaric acids and low-weight aromatic compounds, such as l-malic, l-tartaric, saccharic, mucic, caffeic, gentisic, p-coumaric, protocatechuic and hydroxybenzoic acids (Fonseca et al. 2011). This genus currently contains six described species (Table 1).
New combinations for Solicoccozyma
Solicoccozyma aeria (Saito) A.M. Yurkov, comb. nov. MycoBank MB812198.
Basionym: Torula aeria Saito, Jap. J. Bot. 1: 41. 1922.
Solicoccozyma fuscescens (Golubev) A.M. Yurkov, comb. nov. MycoBank MB812201.
Basionym: Cryptococcus fuscescens Golubev, J. Gen. Appl. Microbiol. 39: 428. 1984.
Solicoccozyma keelungensis (C.F. Chang & S.M. Liu) A.M. Yurkov, comb. nov. MycoBank MB812202.
Basionym: Cryptococcus keelungensis C.F. Chang & S.M. Liu, Int. J. Syst. Evol. Microbiol. 58: 2974. 2008.
Solicoccozyma phenolicus (Á. Fonseca, Scorzetti & Fell) A.M. Yurkov, comb. nov. MycoBank MB812203.
Basionym: Cryptococcus phenolicus Á. Fonseca et al., Can. J. Microbiol. 46: 24. 2000.
Solicoccozyma terreus (Di Menna) A.M. Yurkov, comb. nov. MycoBank MB812204.
Basionym: Cryptococcus terreus Di Menna, J. Gen. Microbiol. 11: 195. 1954.
= Cryptococcus himalayensis Goto & Sugiyama, Can. J. Bot. 48: 2099. 1970.
= Cryptococcus elinovii Golubev, Mikol. Fitopatol. 13: 466. 1979.
Solicoccozyma terricola (T.A. Pedersen) A.M. Yurkov, comb. nov. MycoBank MB812205.
Basionym: Cryptococcus terricola T.A. Pedersen, Bull. Jard. Bot. État Brux. 31: 101. 1958.
Order Holtermanniales Libkind, Wuczkowski, Turchetti & Boekhout, Int. J. Syst. Evol. Microbiol. 61: 685. 2011.
This order was proposed by Wuczkowski et al. (2011) to include the monotypic teleomorphic genus Holtermannia and the anamorphic genus Holtermanniella. The latter was proposed to accommodate four Cryptococcus species and a new anamorphic species closely related to Holtermannia corniformis (Wuczkowski et al. 2011). The order Holtermanniales was confirmed to be a distinct lineage in the trees from the seven-gene and the expanded LSU datasets with strong support values (Fig. 1, Fig. 2). The five anamorphic Holtermanniella species clustered together in a robust clade separate from the teleomorphic species Holtermannia corniformis (Fig. 1, Fig. 2). Therefore, the two genera Holtermannia with one species and Holtermanniella with five species are accepted here (Table 1).
Order Tremellales Fries, Syst. mycol. 1: 2. 1821.
Tremellales is the largest order in Tremellomycetes and six families, namely Cuniculitremaceae, Tremellaceae, Sirobasidiaceae, Tetragoniomycetaceae, Phragmoxenidiaceae and Rhynchogastremaceae containing yeast and yeast-like taxa have been proposed within this order (Metzler et al., 1989, Bandoni, 1995, Wells and Bandoni, 2001, Kirschner et al., 2001). This taxonomic concept included only teleomorphic species and was circumscribed based on basidial morphology, septal pore anatomy and ballistospore production (Bandoni 1995). The monotypic family Phragmoxenidiaceae was tentatively assigned to the Tremellales by Oberwinkler et al. (1990). The inclusion of this family in the order is still uncertain because molecular data are not available from this family (Bandoni, 1995, Wells and Bandoni, 2001). The Phragmoxenidiaceae species is therefore not included in this study. Previous phylogenetic analyses showed the paraphyletic nature of the families in Tremellales after the inclusion of anamorphic yeast and yeast-like taxa (Fell et al., 2001, Scorzetti et al., 2002, Sampaio, 2004). We re-circumscribed the families and genera in Tremellales based on the multigene phylogeny (Liu et al. 2015) and the PRBO test performed in this study. A total of 11 families and 28 genera are accepted in Tremellales here (Table 1, Fig. 1, Fig. 2, Fig. 5).
Bulleribasidiaceae X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, fam. nov. MycoBank MB812215.
Etymology: In reference to the name of the type genus Bulleribasidium.
This family is proposed to accommodate the monophyletic lineage containing the Bulleribasidium, Derxomyces, Dioszegia, Hannaella, melastomae and dimennae clades in the tree obtained from the seven genes (Liu et al. 2015).
Basidiocarps not present. Pseudohyphae and true hyphae may be present. Septal pore a dolipore with poorly developed parenthesome-like structures. Sexual reproduction observed in some species. Basidia cylindrical or occasionally globose, two-celled, with transverse and occasionally longitudinal septa, and globose basidiospores. Budding cells present. Ballistoconidia occasionally present. Fermentation absent. Nitration usually not utilised. Starch-like compounds usually produced. Major CoQ system CoQ-9 or CoQ-10.
Type genus: Bulleribasidium J.P. Sampaio. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
Genera accepted: Bulleribasidium, Derxomyces, Dioszegia, Hannaella, Nielozyma gen. nov., and Vishniacozyma gen. nov.
Notes: Except for one teleomorphic species, Bulleribasidium oberjochense, all the species included in this family are known from the asexual states only.
Bulleribasidium J.P. Sampaio., M. Weiss & R. Bauer, Mycologia 94: 874. 2002. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
This genus Bulleribasidium is emended to accommodate the Bulleribasidium clade containing B. oberjochense and ten anamorphic species previously belonging to Mingxiaea (Liu et al. 2015).
Basidiocarps not present. Pseudohyphae and true hyphae may be present. Septa with dolipores with poorly developed parenthesome-like structures. Sexual reproduction observed in some species. Basidia cylindrical or occasionally globose, two-celled, with transverse and occasionally longitudinal septa, with globose basidiospores. Budding cells present. Ballistoconidia rotationally symmetrical.
Type species: Bulleribasidium oberjochense J.P. Samp., Gadanho, M. Weiss & R. Bauer, Mycologia 94: 875. 2002.
Notes: The teleomorphic genus name Bulleribasidium, which was proposed in 2002 (Sampaio et al. 2002) has nomenclatural priority over the anamorphic genus name Mingxiaea proposed in 2011 (Wang et al. 2011). The species of the latter are, therefore, transferred to the genus Bulleribasidium. Eleven species are currently accepted in this genus (Table 1, Fig. 5E).
New combinations for Bulleribasidium
Bulleribasidium begoniae (Nakase, Tsuzuki, F.L. Lee & M. Takash.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812218.
Basionym: Bullera begoniae Nakase et al., Mycoscience 45: 290. 2004.
≡ Mingxiaea begoniae (Nakase et al.) Q.-M. Wang et al., Int. J. Syst. Evol. Microbiol. 61: 213. 2011.
Bulleribasidium foliicola (Q.M. Wang, F.Y. Bai, Boekhout & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB812232.
Basionym: Mingxiaea foliicola Q.M. Wang et al., Int. J. Syst. Evol. Microbiol. 61: 217. 2011.
Bulleribasidium hainanense (Q.M. Wang, F.Y. Bai, Boekhout & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813261.
Basionym: Mingxiaea hainanensis Q.M. Wang et al., Int. J. Syst. Evol. Microbiol. 61: 215. 2011.
Bulleribasidium panici (Fungsin, M. Takash. & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813262.
Basionym: Bullera panici Fungsin et al., Microbiol. Culture Coll. 19: 27. 2003.
≡ Mingxiaea panici (Fungsin et al.) Q.-M. Wang et al., Int. J. Syst. Evol. Microbiol. 61: 213. 2011.
Bulleribasidium pseudovariabile (F.Y. Bai, M. Takash. & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813263.
Basionym: Bullera pseudovariabilis F.Y. Bai et al., Antonie van Leeuwenhoek 83: 261. 2003.
≡ Mingxiaea pseudovariabilis (F.Y. Bai et al.) Q.-M. Wang et al., Int. J. Syst. Evol. Microbiol. 61: 214. 2011.
Bulleribasidium sanyaense (Q.M. Wang, F.Y. Bai, Boekhout & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813264.
Basionym: Mingxiaea sanyaensis Q.M. Wang et al., Int. J. Syst. Evol. Microbiol. 61: 214. 2011.
Bulleribasidium setariae (Nakase, Tsuzuki, F.L. Lee & M. Takash.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813265.
Basionym: Bullera setariae Nakase et al., Mycoscience 45: 292. 2004.
≡ Mingxiaea setariae (Nakase et al.) Q.-M. Wang et al., Int. J. Syst. Evol. Microbiol. 61: 213. 2011.
Bulleribasidium siamense (Fungsin, M. Takash. & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813266
Basionym: Bullera siamensis Fungsin et al., Microbiol. Culture Coll. 19: 29. 2003.
≡ Mingxiaea siamensis (Fungsin et al.) Q.-M. Wang et al., Int. J. Syst. Evol. Microbiol. 61: 214. 2011.
Bulleribasidium variabile (Nakase & M. Suzuki) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813267.
Basionym: Bullera variabilis Nakase & M. Suzuki, J. Gen. appl. Microbiol. 33: 350. 1987.
≡ Mingxiaea variabilis (Nakase & Suzuki) Q.-M. Wang et al., Int. J. Syst. Evol. Microbiol. 61: 214. 2011.
Bulleribasidium wuzhishanense (Q.M. Wang, F.Y. Bai, Boekhout & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813268.
Basionym: Mingxiaea wuzhishanensis Q.M. Wang et al., Int. J. Syst. Evol. Microbiol. 61: 218. 2011.
Nielozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813269.
Etymology: The genus is named in honour of C.B. van Niel for his pioneering contributions to the study of ballistoconidium-forming yeasts.
This genus is proposed for the melastomae clade recognised from the seven-genes phylogeny (Liu et al. 2015).
Basidiocarps not present. Colonies usually yellowish to brownish. Pseudohyphae and true hyphae not observed. Sexual reproduction not observed. Budding cells present. Ballistoconidia present. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-10.
Type species: Nielozyma melastomae (Nakase, Tsuzuki, F.L. Lee & M. Takash.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
Notes: Nielozyma gen. nov. is closely related to the genus Dioszegia (Nakase et al., 2004, Liu et al., 2015). Nielozyma species usually form yellowish to brownish colonies (Nakase et al. 2004), being distinct from those of Dioszegia species that usually are orange-coloured (Wang & Bai 2008). This genus currently contains two described species (Table 1, Fig. 5E).
New combinations for Nielozyma
Nielozyma formosana (Nakase, Tsuzuki, F.L. Lee & M. Takash.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813270.
Basionym: Bullera formosana Nakase et al., Syst. Appl. Microbiol. 27: 562. 2004.
Nielozyma melastomae (Nakase, Tsuzuki, F.L. Lee & M. Takash.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813271.
Basionym: Bullera melastomae Nakase et al., Syst. Appl. Microbiol. 27: 560. 2004.
Vishniacozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813272.
Etymology: The genus is named in honour of the American microbiologist Helen S. Vishniac for her contributions to the study of microbial diversity and yeast ecology, especially of cold-adapted yeasts.
This genus is proposed for the victoriae (sensu Fonseca et al. 2011) and dimennae clades recognised in Boekhout et al. (2011a) and Liu et al. (2015).
Basidiocarps gelatinous, mycoparasitic. Hyphae thin-walled with clamp connections. H-shaped conidia (e.g. zygoconidia) may be present. Haustoria not known. Basidia subglobose to ellipsoidal or clavate, with basal clamp connections, two- to four-celled, longitudinally or obliquely septa, with ellipsoidal basidiospores. Cultures are often pigmented, pinkish-white, orange-white or yellowish-white. Hyphae not observed in culture. Poorly developed pseudohyphae present. Sexual reproduction not observed in culture. Budding cells present. Ballistoconidia occasionally present. Fermentation absent. Nitrate not utilised. Starch-like compounds weakly produced or not produced. Major CoQ system CoQ-9 or CoQ-10.
Type species: Vishniacozyma carnescens (Verona & Luchetti) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
Notes: The victoriae clade (sensu Fonseca et al. 2011) coincides with the dimennae clade as recognised in Boekhout et al. (2011a) and Liu et al. (2015) and consists of six Cryptococcus and one Bullera species. In the tree derived from the expanded LSU rRNA gene sequence dataset, two teleomorphic mycoparasites Trimorphomyces nebularis and Trimorphomyces sp. CBS 200.94 (Takashima et al., 2003, Kirschner and Chen, 2008) and three recently published Cryptococcus species were also located in this clade (Fig. 5D). Yeasts of this genus are widespread eurybionts and are often found associated with plant material (Fonseca and Inácio, 2006, Fonseca et al., 2011, Yurkov et al., 2015). A possible mycoparasitic nature has been suggested for these yeasts, which may have sexual trimorphomyces-like morphs (Kirschner & Chen 2008). This genus currently contains eleven described species (Table 1) and four additional sequences representing potential new species were obtained from public databases (Fig. 5D).
New combinations for Vishniacozyma
Vishniacozyma globispora (B.N. Johri & Bandoni) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813273.
Basionym: Bullera globispora B.N. Johri & Bandoni, in Subramanian (ed.), Taxonomy of Fungi, (Proc. int. Symp. Madras, 1973) Part 2 (Madras) 2: 539. 1984.
Vishniacozyma carnescens (Verona & Luchetti) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813274.
Basionym: Torulopsis carnescens Verona & Luchetti, Boll. R. Istituto Superiore Agrario di Pisa 12: 280. 1936.
≡ Cryptococcus carnescens (Verona & Luchetti) M. Takash et al., Int. J. Syst. Evol. Microbiol. 53: 1192. 2003.
Vishniacozyma dimennae (Fell & Phaff) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813275.
Basionym: Cryptococcus dimennae Fell & Phaff, Antonie van Leeuwenhoek 33: 467. 1967.
Vishniacozyma foliicola (Q.M. Wang & F.Y. Bai) A.M. Yurkov, comb. nov. MycoBank MB813277.
Basionym: Cryptococcus foliicola Q.M. Wang & F.Y. Bai, J. Gen. Appl. Microbiol. 57: 287. 2011.
Vishniacozyma heimaeyensis (Vishniac) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813278.
Basionym: Cryptococcus heimaeyensis Vishniac, Can. J. Microbiol. 48: 464. 2002.
Vishniacozyma nebularis (Vishniac) A.M. Yurkov, comb. nov. MycoBank MB813279.
Basionym: Trimorphomyces nebularis R. Kirschner & Chee J. Chen, Nova Hedwigia 87: 448. 2008.
Vishniacozyma psychrotolerans (V. de García, Zalar, Brizzio, Gunde-Cim. & van Broock) A.M. Yurkov, comb. nov. MycoBank MB813281.
Basionym: Cryptococcus psychrotolerans V. de García et al., FEMS Microbiol. Ecol. 82: 535. 2012.
Vishniacozyma peneaus (Phaff, Mrak & O.B. Williams) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813282.
Basionym: Rhodotorula peneaus Phaff et al., Mycologia 44: 438. 1952.
≡ Cryptococcus peneaus (Phaff et al.) M. Takash. et al., Int. J. Syst. Evol. Microbiol. 53: 1192. 2003.
Vishniacozyma taibaiensis (Q.M. Wang & F.Y. Bai) A.M. Yurkov, comb. nov. MycoBank MB813283.
Basionym: Cryptococcus taibaiensis Q.M. Wang & F.Y. Bai, J. Gen. Appl. Microbiol. 57: 5. 2011.
Vishniacozyma tephrensis (Vishniac) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813284.
Basionym: Cryptococcus tephrensis Vishniac, Can. J. Microbiol. 48: 466. 2002.
Vishniacozyma victoriae (M.J. Montes, Belloch, Galiana, M.D. García, C. Andrés, S. Ferrer, Torr.-Rodr. & J. Guinea) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813285.
Basionym: Cryptococcus victoriae M.J. Montes et al., Syst. Appl. Microbiol. 22: 104. 1999.
Tremellaceae Fr., Syst. mycol. 1: lv. 1821.
This family is re-delimited to accommodate only the Tremella sensu stricto (= mesenterica) clade circumscribed by the phylogenetic analyses of seven genes (Liu et al. 2015) and consists of ten Tremella species previously included in the mesenterica and fuciformis groups as distinguished by Chen (1998). Additionally, Cryptococcus yokohamensis is included in this family based on previous phylogenetic results by Alshahni et al. (2011) and results from the analysis of the expanded LSU rRNA gene sequence dataset (Fig. 5D).
The diagnosis of the family is based on the description of the genus Tremella as recircumscribed below.
Type genus: Tremella Pers. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
Genus accepted: Tremella.
Notes: This family was created by Fries (1821) to include fungi with gelatinous fruitbodies and was later restricted by Patouillard (1900) to genera with tremelloid basidia. Bandoni (1984) made a major revision of this family based on the ultrastructure of the septal pore and restricted the Tremellaceae to the genera Holtermannia, Tremella and Trimorphomyces. Later, three other genera, Bulleromyces, Sirotrema and Xenolachne, were also included in this family (Bandoni, 1995, Wells and Bandoni, 2001). Phylogenetic analyses indicated that the genera assigned to the Tremellaceae did not form a monophyletic group but occurred scattered in different lineages (Bandoni, 1984, Fell et al., 2000, Scorzetti et al., 2002, Sampaio, 2004, Boekhout et al., 2011a, Millanes et al., 2011, Weiss et al., 2014, Liu et al., 2015). This family is, therefore, re-defined here to include only the Tremella sensu stricto clade as presented in Liu et al. (2015) and as shown in Fig. 1.
Tremella Pers., Neues Mag. Bot. 1: 111. 1794. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
This genus is re-delimited to include the species in the Tremella sensu stricto clade (= mesenterica clade) including the type species of the genus, T. mesenterica. This clade is circumscribed based on the seven-genes phylogeny (Liu et al. 2015).
Basidiocarps minute (0.3–0.5 cm in diameter) to large (to 5–10 cm high). Basidiocarps variable in colour, ranging from whitish-yellow, cream, yellowish-orange to brown, reddish or black. Clamp connections and haustorial branches present in dikaryotic hyphae. Basidia globose, subglobose, ellipsoid, oval, clavate or pyriform, two- or four-celled with longitudinal or oblique septa, occasionally with transverse septa. Basidiospores globose to ellipsoid. Budding cells originate from germinating basidiospores. Fermentation absent in yeast states. Nitrate not utilised. Major CoQ systems CoQ-9 or CoQ-10.
Type species: Tremella mesenterica (Schaeff.) Retz.
Notes: The genus Tremella as traditionally circumscribed is the largest polyphyletic genus in the order Tremellales (Weiss et al. 2014). Molecular phylogenetic analyses based on LSU rRNA gene sequences showed that Tremella species occurred in several distantly related clades (Fell et al., 2000, Scorzetti et al., 2002, Sampaio, 2004, Bandoni and Boekhout, 2011b, Boekhout et al., 2011a, Millanes et al., 2011, Weiss et al., 2014, Liu et al., 2015, also see Fig. 2 of this study) and should be re-classified. Here we re-define the genus Tremella to include only the species in the Tremella sensu stricto clade as recognised in the tree derived from seven genes (Liu et al. 2015). As discussed above, the Tremella sensu stricto clade showed a significant deviation from the reference thresholds in the PRBO analysis (Table 2) and two subclades, mesenterica and fuciformis, with distinguishable morphological characters could be identified within the clade as shown in Chen (1998) and Liu et al. (2015). These data imply that the Tremella sensu stricto clade probably can be reclassified into two genera in the future. At present, we prefer to keep them in the genus Tremella. The reclassification of the entire Tremella genus is unfeasible at present because even the D1/D2 LSU rRNA gene sequences are only available from less than half of the described Tremella species (Millanes et al., 2011, Weiss et al., 2014, Liu et al., 2015, this study).
An anamorphic yeast species, Cryptococcus yokohamensis, was recently described and showed a close relationship to the Tremella species in the Tremella sensu stricto clade (Alshahni et al. 2011). This species exhibits phenotypic characteristics similar to those of the yeast states of Tremella. Specifically, C. yokohamensis has two major coenzymes CoQ-9 and CoQ-10, being similar to T. mesenterica and T. coalescens (Alshahni et al. 2011). Thus, C. yokohamensis represents a yeast state of a species in the Tremella sensu stricto clade and is transferred to Tremella. The re-defined genus Tremella currently contains twelve species (Table 1, Fig. 5D).
New combination for Tremella
Tremella yokohamensis (Alshahni, Satoh & Makimura) A.M. Yurkov, comb. nov. MycoBank MB813286.
Basionym: Cryptococcus yokohamensis Alshahni et al., Int. J. Syst. Evol. Microbiol. 61: 3069. 2011.
Rhynchogastremataceae (Oberw. & B. Metzler) Syst. Appl. Microbiol. 12: 283. 1989. emend. X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov.
This family is emended to accommodate the well supported monophyletic lineage consisting of the aureus, Auriculibuller, Bandoniozyma, Papiliotrema, pseudoalba and laurentii clades as recognised in Liu et al. (2015).
Basidiocarps if present, minute. Sexual reproduction present in some species. Dikaryotic hyphae with clamp connections and haustorial branches. Basidia transversely septate, cylindrical, and in small clusters. Basidiospores globose or allantoid. In culture pseudohyphae and true hyphae occasionally present. Budding cells present. Ballistoconidia may be present. Fermentation present in some species. Nitrate not utilised. Starch-like compounds usually produced. Major CoQ system CoQ-10 as far as known.
Type genus: Rhynchogastrema B. Metzler & Oberw. emend. X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov
Genera accepted: Rhynchogastrema emend. and Papiliotrema emend.
Notes: The family Rhynchogastremataceae was originally proposed for a heterobasidiomycetous species Rhynchogastrema coronatum (Metzler et al. 1989). The expanded LSU rRNA gene sequence analysis located this species in the Bandoniozyma clade (Fig. 5A). Therefore, the Rhynchogastremataceae is expanded here to include the species in the Bandoniozyma and closely related clades that formed a 100 % supported monophyletic lineage (Liu et al. 2015) as shown in Fig. 1.
Papiliotrema J. P. Sampaio., M. Weiss & R. Bauer, Mycologia 94: 875. 2002. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, MycoBank MB813287.
This genus is emended to accommodate a well-supported monophyletic clade containing teleomorphic Papiliotrema and Auriculibuller species (Liu et al. 2015) as well as anamorphic yeasts including 16 Cryptococcus and three Bullera species (Table 1, Fig. 5A).
Basidiocarps, if present, minute, ca. 0.5 mm in diameter. Dikaryotic hyphae with clamp connections and haustorial branches. Basidia cylindrical and transversely septate. Cultures pale to brownish yellow. Pseudohyphae and true hyphae may be present. Budding cells present. Ballistoconidia may be present. Sexual reproduction observed in some species. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-10.
Type species: Papiliotrema bandonii J. P. Sampaio., Gadanho, M. Weiss & R. Bauer, Mycologia 94: 875. 2002.
Notes: The species assigned to the emended genus Papiliotrema here were located in the aureus, Auriculibuller, laurentii, Papiliotrema and pseudoalba clades in the seven-genes phylogenetic tree (Liu et al. 2015). In order to avoid creating genera for these small clades, we decided to combine them in a single genus. The PRBO test did not show significant deviation for the emended Papiliotrema generic concept (Table 2), supporting this taxonomic treatment. The genus Papiliotrema was proposed in 2002 (Sampaio et al. 2002), thus having nomenclature priority over Auriculibuller, which was proposed in 2004 (Sampaio et al. 2004). The genera Auriculibuller and Papiliotrema are morphologically similar and their sexual states form clavate basidia with transverse septa, differing clearly from those formed by Tremella species (Sampaio et al., 2002, Sampaio et al., 2004, Sampaio, 2011c). Twenty-two species are currently accepted in the emended genus Papiliotrema (Table 1, Fig. 5A).
New combinations for Papiliotrema
Papiliotrema anemochoreius (C.H. Pohl, Kock, P.W.J. van Wyk & Albertyn) Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813287.
Basionym: Cryptococcus anemochoreius C.H. Pohl et al., Int. J. Syst. Evol. Microbiol. 56: 2705. 2006.
Papiliotrema aurea (Saito) M. Takash., Sugita, Shinoda & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813288.
Basionym: Torula aurea Saito, Mitt. Jpn. J. Bot. 1: 44. 1922.
≡ Cryptococcus aureus (Saito) M. Takash. et al., Int. J. Syst. Evol. Microbiol. 53: 1192. 2003.
Papiliotrema aspenensis (K. Ferreira-Paim, T.B. Ferreira, L. Andrade-Silva, D.J. Mora, D.J. Springer, J. Heitman, F.M. Fonseca, D. Matos, M.S.C. Melhem & M.L. Silva-Vergara) X.Z. Liu, F.Y. Bai, A.M. Yurkov & Boekhout comb. nov. MycoBank MB814710.
Basionym: Cryptococcus aspenensis K. Ferreira-Paim et al., PloS ONE 9: e108633. 2014.
Papiliotrema baii (A.M. Yurkov, M.A. Guerreiro & Á. Fonseca) A.M. Yurkov, comb. nov. MycoBank MB813648.
Basionym: Cryptococcus baii A.M. Yurkov et al., PLoS One 10: e0120400. 2015.
Papiliotrema flavescens (Saito) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813289.
Basionym: Torula flavescens Saito, Jap. J. Bot. 1: 43. 1922.
≡ Cryptococcus flavescens (Saito) C.E. Skinner, Am. Midl. Nat. 43: 249. 1950.
Papiliotrema fonsecae (V. de García, Zalar, Braizzio, Gunde-Cim. & van Brollck) A.M. Yurkov, comb. nov. MycoBank MB813290.
Basionym: Cryptococcus fonsecae V. de García et al., FEMS Microbiol. Ecol. 82: 536. 2012.
Papiliotrema frias (V. de García, Zalar, Braizzio, Gunde-Cim. & van Brollck) A.M. Yurkov, comb. nov. MycoBank MB813292.
Basionym: Cryptococcus frias V. de García et al., FEMS Microbiol. Ecol. 82: 537. 2012.
Papiliotrema fuscus (J.P. Samp., J. Inácio, Fonseca & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813432.
Basionym: Auriculibuller fuscus J.P. Samp. et al., Int. J. Syst. Evol. Microbiol. 54: 989. 2004.
Papiliotrema hoabinhensis (D.T. Luong, M. Takash., Ty. Dung & Nakase) A.M. Yurkov, comb. nov. MycoBank MB813293.
Basionym: Bullera hoabinhensis D.T. Luong et al., J. Gen. Appl. Microbiol. 51: 340. 2005.
Papiliotrema japonica (J.P. Samp., Fonseca & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813294.
Basionym: Bullera japonica J.P. Samp. et al., Int. J. Syst. Evol. Microbiol. 54: 990. 2004.
Papiliotrema laurentii (Kuff.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813295.
Basionym: Torula laurentii Kuff., Ann. Soc. Roy. Sci. Méd. Natur. Brux. 74: 38. 1920.
≡ Cryptococcus laurentii (Kuff). C.E. Skinner, Am. Midl. Nat. 43: 249. 1950.
Papiliotrema mangalensis (Fell, Statzell & Scorzett) A.M. Yurkov, comb. nov. MycoBank MB813296.
Basionym: Cryptococcus mangalensis Fell et al., Antonie van Leeuwenhoek 99: 548. 2011.
Papiliotrema nemorosus (W.I. Golubev, Gadanho, J.P. Samp. & N.W. Golubev) X.Z. Liu, F.Y. Bai, A.M. Groenew. & Boekhout, comb. nov. MycoBank MB813298.
Basionym: Cryptococcus nemorosus W.I. Golubev et al., Int. J. Syst. Evol. Microbiol. 53: 907. 2003.
Papiliotrema perniciosus (W.I. Golubev, Gadanho, J.P. Samp. & N.W. Golubev) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813299.
Basionym: Cryptococcus perniciosus W.I. Golubev et al., Int. J. Syst. Evol. Microbiol. 53: 910. 2003.
Papiliotrema pseudoalba (Nakase & M. Suzuki) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813300.
Basionym: Bullera pseudoalba Nakase & M. Suzuki, J. Gen. Appl. Microbiol. 32: 131. 1986.
= Cryptococcus cellulolyticus Nakase et al., J. Gen. Appl. Microbiol 42: 9. 1996.
Papiliotrema ruineniae (A.M. Yurkov, M.A. Guerreiro & Á. Fonseca) A.M. Yurkov, comb. nov. MycoBank MB813649.
Basionym: Cryptococcus ruineniae A.M. Yurkov et al., PLoS One 10: e0120400. 2015.
Papiliotrema rajasthanensis (Saluja & G.S. Prasad) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813301.
Basionym: Cryptococcus rajasthanensis Saluja & G.S. Prasad, Int. J. Syst. Evol. Microbiol. 57: 417. 2007.
Papiliotrema taeanensis (K.S. Shin & Y.H. Park) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813302.
Basionym: Cryptococcus taeanensis K.S. Shin & Y.H. Park, Int. J. Syst. Evol. Microbiol. 55: 1367. 2005.
Papiliotrema terrestris (Crestani, Landell, Faganello, Vainstein, Vishniac & P. Valente) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813303.
Basionym: Cryptococcus terrestris Crestani et al., Int. J. Syst. Evol. Microbiol. 59: 635. 2009.
Papiliotrema wisconsinensis (K. Sylvester, Q.M. Wang & C.T. Hittinger) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB815284.
Basionym: Cryptococcus wisconsinensis K. Sylvester et al., FEMS Yeast Res. 15: 7. 2015.
Rhynchogastrema B. Metzler & Oberw., Syst. Appl. Microbiol. 12: 281. 1989. emend. X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov.
This genus is emended to include species classified in the anamorphic genus Bandoniozyma and is based on the phylogenetic analyses of the seven-gene (Liu et al. 2015) and the expanded LSU rRNA gene datasets (Fig. 5A).
Basidiocarps not present. Dikaryotic hyphae thin-walled with clamp connections and haustorial branches. Septa with dolipores and parenthesomes of cup-shaped vesicles of the Tremella-type. Sexual reproduction observed in some species. Basidium with basally swollen probasidium, elongated to subglobose, with apex of the neck partly cruciately septate. Budding cells present, globose, subglobose, ovoid or ellipsoidal. Ballistoconidia not produced. Pseudohyphae and true hyphae with clamp connections may be present in cultures. Fermentation of glucose usually present. Nitrate not utilised. Starch-like compounds are usually formed. Major CoQ system not known.
Type species: Rhynchogastrema coronatum B. Metzler & Oberw., Syst. Appl. Microbiol. 12: 281. 1989.
Notes: The species of the recently described anamorphic genus Bandoniozyma (Valente et al. 2012) formed a well-supported clade in the trees obtained from the analyses of the rRNA ITS (Valente et al. 2012) and the sequences of seven genes studied (Liu et al. 2015). Sequences of the D1/D2 region of the LSU rRNA gene of Rhynchogastrema coronatum, the type species of the teleomorphic genus (Metzler et al. 1989), were independently obtained for the type specimen BBA 65155 (GenBank KJ170152) by Weiss et al. (2014) and the ex-type culture DSM 28188 (deposited in DSMZ collection by Bertold Metzler) by Andrey Yurkov (GenBank LN870267). The sequences derived from both sources are identical. Phylogenetic analysis showed that Rhynchogastrema coronatum was nested within the genus Bandoniozyma with 99 % and 92 % bootstrap support in ML and MP analyses, respectively (Fig. 5A), suggesting that Bandoniozyma species represent yeast stages of Rhynchogastrema species. Based on the nomenclatural priority principle, the name Rhynchogastrema is selected for this genus. A fermentative species, Cryptococcus nanyangensis, was described by Hui et al. (2012). This species was closely related to a group of unnamed Cryptococcus strains, which were assigned to the genus Bandoniozyma by Valente et al. (2012). The ML tree from the expanded LSU rRNA dataset confirmed that C. nanyangensis clustered together with Bandoniozyma and Rhynchogastrema species (Fig. 5A). The former is therefore transferred to the genus Rhynchogastrema. Nine species are currently accepted in this genus (Table 1, Fig. 5A). The yeast states of most species of this genus can ferment glucose, which is an unusual trait among basidiomycetous yeasts.
New combinations for Rhynchogastrema
Rhynchogastrema aquatica (Brandao, Valente, Pimenta & Rosa) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov, comb. nov. MycoBank MB813305.
Basionym: Bandoniozyma aquatica Brandao et al., PLoS One 7: e46060. 2012.
Rhynchogastrema complexa (Landell, Pagnocca, Sette, Passarini, Garcia, Ribeiro, Lee, Brandao, Rosa & Valente) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov, comb. nov. MycoBank MB813306.
Basionym: Bandoniozyma complexa Landell et al., PLoS One 7: e46060. 2012.
Rhynchogastrema fermentans (Lee) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov, comb. nov. MycoBank MB813307.
Basionym: Bandoniozyma fermentans C.F. Lee, PLoS One 7: e46060. 2012.
Rhynchogastrema glucofermentans (S.O. Suh & Blackwell) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov, comb. nov. MycoBank MB813308.
Basionym: Bandoniozyma glucofermentans S.O. Suh & Blackwell, PLoS One 7: e46060. 2012.
Rhynchogastrema nanyangensis (F.L. Hui & Q.H. Niu) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov, comb. nov. MycoBank MB813309.
Basionym: Cryptococcus nanyangensis F.L. Hui & Q.H. Niu, Curr. Microbiol. 65: 619. 2012.
Rhynchogastrema noutii (Boekhout, Fell, Scorzett & Theelen) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov, comb. nov. MycoBank MB813304.
Basionym: Bandoniozyma noutii Boekhout et al., PLoS One 7: e46060. 2012.
Rhynchogastrema tunnelae (Boekhout, Fell, Scorzetti & Theelen) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov, comb. nov. MycoBank MB813310.
Basionym: Bandoniozyma tunnelae Boekhout et al., PLoS One 7: e46060. 2012.
Rhynchogastrema visegradensis (Peter & Dlauchy) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov, comb. nov. MycoBank MB813311.
Basionym: Bandoniozyma visegradensis Peter & Dlauchy, PLoS One 7: e46060. 2012.
Bulleraceae X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, fam. nov. MycoBank MB813328.
Etymology: In reference to the name of the type genus Bullera.
This family is proposed to accommodate a monophyletic lineage formed by the amylolyticus (i.e. Genolevuria gen. nov.), Bulleromyces and hannae (i.e. Bullera emend.), and moriformis clades and four single-species lineages, namely Cryptococcus allantoinivorans (Pseudotremella), C. mujuensis (Fonsecazyma), Sirobasidium intermedium and Tremella giraffa in the tree obtained from the seven-genes dataset (Liu et al. 2015).
Basidiocarps tremella-like, 1–2 cm in diameter. Dikaryotic hyphae with clamp connections and haustorial branches. Sexual reproduction observed in some species. Basidia transversely, obliquely or longitudinally septate. Basidiospores globose, fusiform to narrowly clavate. Budding cells present. Pseudohyphae occasionally present. Ballistoconidia may be present. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-10 as far as known.
Type genus: Bullera Derx emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout
Genera accepted: Bullera emend., Fonsecazyma gen. nov., Genolevuria gen. nov., and Pseudotremella (Table 1, Fig. 1).
Notes: In the constrained ML tree inferred from the expanded LSU rRNA gene dataset, the Tremella species in Clade I of Millanes et al. (2011) were assigned to the new family Bulleraceae though bootstrap support was lacking (Fig. 2, Fig. 5B). Cryptococcus allantoinivorans was incorporated into the moriformis clade together with two newly added species. A new genus Pseudotremella is proposed for this expanded clade (Fig. 5B). Cryptococcus mujuensis and the recently described species Cryptococcus tronadorensis and Kwoniella betulae formed a well supported clade for which a new genus Fonsecazyma is proposed (Fig. 5B). The Sirobasidium intermedium lineage was expanded to contain two more species, namely Sirobasidium brefeldianum and Tremella exigua, which correspond to Tremella clade I in Millanes et al. (2011). The Tremella giraffa lineage was expanded with eight Tremella species corresponding to Tremella clade III in Millanes et al. (2011). We recommend reclassifying Sirobasidium and Tremella species in this family, as well as Biatoropsis (Fig. 5B) when additional molecular data are available from all or the majority of the species of these three basidiocarp-forming genera. Cryptococcus cuniculi was described by Shin et al. (2006) as a member of the Kwoniella clade showing some relatedness to Kwoniella (Cryptococcus) heveanensis as well as Cryptococcus tronadorensis in de Garcia et al. (2012). These results were not supported in our analyses, and C. cuniculi clustered close to Tremella clade I in Millanes et al. (2011) in the expanded LSU rRNA gene analysis (Fig. 5B). In our opinion, this species should be re-classified together with respective species of Biatoropsis, Sirobasidium and Tremella. Currently, we propose to keep it unclassified as Cryptococcus cuniculi pro tem. until additional molecular data is available. Similarly, two Tremella species were not assigned to any clade in the expanded LSU rRNA gene analysis (Fig. 5B) as well as in the combined rRNA genes analysis (Millanes et al. 2011). Thus, we recommend keeping them unclassified as Tremella haematommatis pro tem. and Tremella ramalinae pro tem. until additional molecular data are available.
Bullera Derx, Annls Mycol. 28: 11. 1930. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
= Bulleromyces Boekhout & Á. Fonseca, Antonie van Leeuwenhoek 59: 91. 1991.
This genus is emended to include the species in the Bulleromyces and hannae clades recognised from the multigene phylogeny (Liu et al. 2015).
Basidiocarps absent. Sexual reproduction observed in some species. Dikaryotic hyphae with clamp connections and haustorial branches. Basidia subglobose, clavate or ovoid, and transversely, obliquely or longitudinally septated. Budding cells present. Ballistoconidia rotationally or bilaterally symmetrical. Pseudohyphae not observed. Fermentation absent. Nitrate not utilised. Starch-like compounds usually produced. Major CoQ system CoQ-10.
Type species: Bullera alba (W.F. Hanna) Derx.
Notes: The genus Bullera as circumscribed by Boekhout et al. (2011b) contained more than 40 species, which occurred distributed in many different clades of different orders (Fell et al., 2000, Scorzetti et al., 2002, Boekhout et al., 2011a, Weiss et al., 2014, Liu et al., 2015). The sexual state of the type species Bullera alba was discovered by Boekhout et al. (1991) and Bulleromyces albus as the teleomorph of this species was consequently described. According to the “One fungus = One name” principle implemented in the Code of Nomenclature for algae, fungi, and plants (McNeill et al. 2012), the genus name Bullera has nomenclatural priority over Bulleromyces. The genus Bullera emend. currently includes four species (Table 1, Fig. 5B), which were located into two closely related clades in the tree obtained from the seven-genes dataset (Liu et al. 2015). Though these species show a significant deviation from the reference threshold (Table 2), they are kept in the genus Bullera at present to accommodate their phenotypic similarity and close phylogenetic relationship and to minimise name changes.
Fonsecazyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813136.
Etymology: The genus is named in honour of the Portuguese mycologist Álvaro Fonseca, for his contributions to the study of diversity and systematics of basidiomycetous yeasts.
This genus is proposed for the clade represented by Cryptococcus mujuensis, which was resolved as a single-species lineage in the seven-genes tree (Liu et al. 2015) and was supplemented by two newly described species Cryptococcus tronadorensis (de Garcia et al. 2012) and Kwoniella betulae (Sylvester et al. 2015) in the LSU rRNA gene tree (Fig. 5C).
Basidiocarps unknown. Sexual reproduction has not been observed. Pseudohyphae and true hyphae have not been observed. Budding cells present. Ballistoconidia are not formed. Fermentation is absent. Nitrate is not utilised. Major CoQ system is CoQ-10.
Type species: Fonsecazyma mujuensis (K.S. Shin & Y.H. Park) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout comb. nov.
Notes: The phylogenetic analysis of the seven-gene dataset in Liu et al. (2015) indicated that Cryptococcus mujuensis was closely related to Sirobasidium intermedium. Cryptococcus tronadorensis was originally described as a new species related to the Kwoniella clade based on the LSU rRNA gene sequences similarity in de Garcia et al. (2012). Similarly, the recently described Kwoniella betulae (Sylvester et al. 2015) was also placed in this clade. Unfortunately, Cr. mujuensis was not employed in these studies. In the tree obtained from the expanded LSU rRNA gene dataset, Cr. mujuensis, Cr. tronadorensis and K. betulae formed a strongly supported clade, which was only distantly related to the Kwoniella and the S. intermedium clades (Fig. 5C). Thus, the new genus Fonsecazyma is proposed to accommodate these three species (Table 1). Other sequences obtained from public databases belong to a mislabelled K. heveanensis (GenBank AF406890) and three undescribed Cryptococcus species (Fig. 5C).
New combinations for Fonsecazyma
Fonsecazyma betulae (K. Sylvester, Q.M. Wang, C. T. Hittinger) A.M. Yurkov, A.V. Kachalkin & Boekhout, comb. nov. MycoBank MB814752.
Basionym: Kwoniella betulae K. Sylvester et al., FEMS Yeast Res. 15: 7. 2015.
Fonsecazyma mujuensis (K.S. Shin & Y.H. Park) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813341.
Basionym: Cryptococcus mujuensis K.S. Shin & Y.H. Park, Int. J. Syst. Evol. Microbiol. 56: 2243. 2006.
Fonsecazyma tronadorensis (V. De Garcia, Zalar, Brizzio, Gunde-Cim. & van Brook) A.M. Yurkov, comb. nov. MycoBank MB813342.
Basionym: Cryptococcus tronadorensis V. De García et al., FEMS Microbiology Ecology. 82: 536. 2012.
Genolevuria X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813329.
Etymology: The genus is named in honour of the French yeast genomics consortium “Genolevures” that produced high-quality yeast genome data.
This genus is proposed for the amylolyticus clade comprising four Cryptococcus species in the tree derived from the seven-genes dataset (Liu et al. 2015).
Basidiocarps not present. Colonies usually orange-coloured. Pseudohyphae occasionally present. True hyphae not observed. Budding cells present. Ballistoconidia not observed. Fermentation absent. Nitrate not utilised. Starch-like compounds weakly produced or not produced at all. Major CoQ system CoQ-10.
Type species: Genolevuria amylolytica (Á. Fonseca, J. Inácio & Spenc.-Mart.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout
Notes: The species of this genus have orange-coloured colonies (Inácio et al., 2005, Wang et al., 2007, Landell et al., 2009, Liu et al., 2015), which is not a common characteristic among tremellaceous yeasts (Fonseca et al. 2011). Four species are currently accepted in this genus (Table 1, Fig. 5B).
New combinations for Genolevuria
Genolevuria amylolytica (Á. Fonseca, J. Inácio & Spenc.-Mart.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813330.
Basionym: Cryptococcus amylolyticus Á. Fonseca et al., FEMS Yeast Res. 5: 1177. 2005.
Genolevuria armeniaca (Á. Fonseca & J. Inácio) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813331.
Basionym: Cryptococcus armeniacus Á. Fonseca & J. Inácio, FEMS Yeast Res. 5(12): 1177. 2005.
Genolevuria bromeliarum (Landell & P. Valente) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813332.
Basionym: Cryptococcus bromeliarum Landell & P. Valente, Int. J. Syst. Evol. Microbiol. 59: 911. 2009.
Genolevuria tibetensis (F.Y. Bai & Q.M. Wang) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813333.
Basionym: Cryptococcus tibetensis F.Y. Bai & Q.M. Wang, J. Gen. Appl. Microbiol., Tokyo 53: 282. 2007.
Pseudotremella X.Z. Liu, F.Y. Bai, A.M. Yurkov, M. Groenew. & Boekhout, gen. nov. MycoBank MB813334.
Etymology: In reference to the tremella-like basidiocarp and basidium morphology.
This genus is proposed for the moriformis lineage recognised by Chen (1998) and Liu et al. (2015), which was expanded with two Cryptococcus species in the tree obtained from the analysis of the LSU rRNA gene dataset (Fig. 5B).
Basidiocarps pustulate, erumpent or tuberculate on wood, pulvinate, hemispherical-moriform to cerebriform, white to pinkish or cream, amber to dark reddish-brown, drying brownish to black. Dikaryotic hyphae hyaline to brown or reddish, with clamp connections and occasionally haustorial branches. Basidia globose, ellipsoidal, obovoid, pyriform or capitate; four-celled with longitudinal to oblique septa. Budding cells sometimes present. Poorly developed pseudohyphae sometimes present. Fermentation absent. Nitrate not utilised. Major CoQ system unknown.
Type species: Pseudotremella moriformis (Berk.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
Notes: In the tree obtained from the seven-genes dataset, Tremella moriformis and Cryptococcus allantoinivorans clustered together with strong BP and PP support values, though they were regarded as representing single-species lineages (Liu et al. 2015). In the tree from the expanded LSU rRNA gene dataset (Fig. 5B), the Tremella species in the moriformis clade recognised in Chen (1998) and Cryptococcus allantoinivorans formed a monophyletic clade together with a recently described species, Cryptococcus lacticolor (Satoh et al. 2013). Thus, we propose the new genus Pseudotremella to accommodate this clade. Four species are currently accepted in this genus (Table 1, Fig. 5B).
It should be noted that a sequence of Tremella indecorata AM5 derived from specimen Santos s.n. was located in the moriformis clade, being in agreement with Millanes et al. (2011). However, three strains obtained from different specimens of the species from different origin and hosts, CBS 6976, originated from Salix sp., HBZ194 and DSM 5727 (both from Diatrype sp. growing on Sorbus aucuparia twigs, Germany), constituted a separate clade distantly related to the moriformis clade (Fig. 5A). However, strain CBS 6976 is placed with support closely related to Pseudotremella in an unconstrained LSU-based tree – together also with strains HBZ194 and DSM 5727 (data not shown). This represents a conflict with the placement of CBS 6976 in the seven-genes tree (Liu et al. 2015), where it is located in the aurantia clade (=Naematelia). This conflict should be further investigated in the future. Specimen AM5 and strain CBS 6976 are in any case not conspecific. Strain AM5 differs from strain HBZ194 by 7 % (42/613 bp) nucleotide divergence in the LSU rRNA gene sequence, which suggest that Tremella indecorata constitutes a species complex. A taxonomic revision for T. indecorata cannot be made until the species concept of this species and the strains and sequences belonging to it are verified. Thus, we keep this species unassigned to any lineage as Tremella indecorata pro tem. (Table 1).
New combinations for Pseudotremella
Pseudotremella allantoinivorans (Middelhoven) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813335.
Basionym: Cryptococcus allantoinivorans Middelhoven, Antonie van Leeuwenhoek 87: 103. 2004.
Pseudotremella lacticolor (Satoh & Makimura) A.M. Yurkov, comb. nov. MycoBank MB813337.
Basionym: Cryptococcus lacticolor Satoh & Makimura, Antonie van Leeuwenhoek 104: 90. 2013.
Pseudotremella moriformis (Berk.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813338.
Basionym: Tremella moriformis Berk., in Smith, Engl. Fl., Fungi (Edn 2) (London) 34: tab. 2446. 1836.
Pseudotremella nivalis (Chee J. Chen) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813340.
Basionym: Tremella nivalis Chee J. Chen, Bibl. Mycol. 174: 101. 1998.
Sirobasidiaceae Lindau, Engler & Prantl, Nat. Pflanzenfam., Teil. I (Leipzig) 1: 89. 1897.
The family Sirobasidiaceae was proposed by Möller (1895) and validated by Lindau to accommodate the genus Sirobasidium. Fibulobasidium was the second genus assigned to this family (Bandoni, 1979, Bandoni, 1995, Wells and Bandoni, 2001). Sirobasidiaceae are characterised by basidia arranged in linear chains (Sirobasidium) or in clusters (Fibulobasidium), which form passively released basidiospores. Phylogenetic studies indicated that the Fibulobasidium species formed a well-supported monophyletic clade while Sirobasidium species belonged to divergent lineages in the Tremellales (Bandoni et al., 2011, Liu et al., 2015). Sirobasidium magnum and S. japonicum formed a monophyletic lineage with the genus Fibulobasidium (Fig. 5B), whereas S. brefeldianum and S. intermedium grouped with Tremella exigua in the Bulleraceae (Millanes et al., 2011, Weiss et al., 2014, Liu et al., 2015, Fig. 5B). The phylogenetic position of the type species of Sirobasidium, S. sanguineum, is unknown because sequence data were not available for this species. Therefore, it is difficult to re-delimitate the genus Sirobasidium at present. We suggest to maintain the family Sirobasidiaceae for the monophyletic lineage formed by the genus Fibulobasidium and the two Sirobasidium species (Fig. 1, Fig. 5B).
Cuniculitremaceae J.P. Sampaio., R. Kirschner & M. Weiss, Antonie van Leeuwenhoek 80: 155. 2001
Type genus: Cuniculitrema J.P. Sampaio. & R. Kirshner, Antonie van Leeuwenhoek 80: 155. 2001 (synonym of Sterigmatosporidium).
Genera accepted: Fellomyces, Kockovaella and Sterigmatosporidium (Table 1, Fig. 1).
Notes: The family Cuniculitremaceae was proposed by Kirschner et al. (2001) to accommodate the teleomorphic genus Cuniculitrema and two anamorphic genera, Fellomyces and Kockovaella, based on their close phylogenetic relationship and the common morphological feature of producing conidia on stalks. Cuniculitrema was described for the teleomorph of Sterigmatosporidium (Kirschner et al. 2001). The latter was proposed by Kraepelin & Schulze (1982) and has nomenclatural priority over the former. Therefore, Cuniculitrema is treated as a synonym of Sterigmatosporidium here.
Fellomyces Y. Yamada & I. Banno, J. Gen. Appl. Microbiol., Tokyo 30: 524. 1984.
This genus is restricted to accommodate the Fellomyces clade as recognised in Liu et al. (2015) based on the analysis of the seven-genes dataset.
Basidiocarps unknown. Sexual reproduction not observed. Pseudohyphae and true hyphae may be present. Blastoconidia at the top of stalk-like conidiophores. Budding cells present. Ballistoconidia not present. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-10.
Type species: Fellomyces polyborus (D.B. Scott & Van der Walt) Y. Yamada & I. Banno, J. Gen. Appl. Microbiol. 30: 524. 1984.
Notes: This genus as previously delimited contained twelve species (Lopandic et al. 2011) but the Fellomyces clade recognised in Liu et al. (2015) included only four Fellomyces species. The support for the affinity of F. horovitzae to this clade in the seven-genes tree was weak (Liu et al. 2015). This species differs in physiological characteristics when compared to other Fellomyces species (Lopandic et al. 2011). We maintain this species in the emended genus Fellomyces for the time being to avoid creating a monotypic genus. This genus currently contains four species (Table 1, Fig. 5D).
Kockovaella Nakase, I. Banno & Y. Yamada, J. Gen. Appl. Microbiol., Tokyo 37: 178. 1991. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
This genus is emended to accommodate the Kockovaella clade as recognised in Liu et al. (2015), which includes non-ballistoconidium-forming species previously belonging to the genus Fellomyces.
Basidiocarps unknown. Sexual reproduction not observed. Pseudohyphae absent or poorly developed. True hyphae not observed. Blastoconidia separate from the parental cells at the distal end of stalk-like conidiophores. Budding cells present. Ballistoconidia may be present. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-10.
Type species: Kockovaella thailandica Nakase, I. Banno & Y. Yamada, J. Gen. Appl. Microbiol. 37: 178. 1991.
Notes: The Kockovaella clade recognised in Liu et al. (2015) includes eight Fellomyces species and all currently recognised eleven Kockovaella species (Takashima & Nakase 2011). As previously circumscribed, the genus Kockovaella was differentiated from the genus Fellomyces only by the production of ballistoconidia (Takashima & Nakase 2011), a property that has since long been shown to be an unstable character (Nakase et al. 1993). The PRBO test, however, indicated that the genus is significantly more divergent than the genera that fit into the optimal boundaries (Table 2), discouraging us to merge this clade with the Fellomyces clade into a single genus, which will significantly expand the deviation of the genus further. Nineteen species are currently accepted in the genus Kockovaella (Table 1, Fig. 5D).
New combinations for Kockovaella
Kockovaella chinensis (Prillinger, G. Kraep. & Lopandic) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813343.
Basionym: Fellomyces chinensis Prillinger et al., Syst. Appl. Microbiol. 20: 579. 1997.
Kockovaella distylii (Hamam., Kuroy. & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813344.
Basionym: Fellomyces distylii Hamam. et al., Int. J. Syst. Bacteriol. 48: 290. 1998.
Kockovaella fuzhouensis (J.Z. Yue) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813345.
Basionym: Sterigmatomyces fuzhouensis J.Z. Yue, Acta Mycol. Sin. 1: 81. 1982.
≡ Fellomyces fuzhouensis (J.Z. Yue) Y. Yamada & I. Banno, J. Gen. Appl. Microbiol. 34: 506. 1988.
Kockovaella lichenicola (Prillinger, G. Kraep. & Lopandic) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813346.
Basionym: Fellomyces lichenicola Prillinger et al., Syst. Appl. Microbiol. 20: 582. 1997.
Kockovaella mexicana (Lopandic, O. Molnár & Prillinger) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813347.
Basionym: Fellomyces mexicanus Lopandic et al., Microbiol. Res. 160: 8. 2005.
Kockovaella ogasawarensis (Hamam., Kuroy. & Nakase) X.Z. Liu, F.Y. Bai, Groenew. & Boekhout, comb. nov. MycoBank MB813348.
Basionym: Fellomyces ogasawarensis Hamam. et al., Int. J. Syst. Bacteriol. 48: 289. 1998.
Kockovaella sichuanensis (Prillinger, G. Kraep. & Lopandic) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813349.
Basionym: Fellomyces sichuanensis Prillinger et al., Syst. Appl. Microbiol. 20: 582. 1997.
Kockovaella prillingeri X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, sp. nov. MycoBank MB813350 Holotype: CBS 7552.
≡ Fellomyces thailandicus Prillinger et al., Syst. Appl. Microbiol. 20: 583. 1997.
Notes: The name Kockovaella thailandica, which is the type species of the genus Kockovaella, already exists. Thus a new name Kockovaella prillingeri is proposed for Fellomyces thailandicus as this species has to be transferred to the genus Kockovaella and a new combination using the epithet 'thailandicus' is not possible in this genus. The new name is proposed in honour of Hansjörg Prillinger who discovered and described the species Fellomyces thailandicus.
Sterigmatosporidium G. Kraep. & U. Schulze, Antonie van Leeuwenhoek 48: 479. 1983.
= Cuniculitrema J. P. Sampaio & R. Kirschner, Antonie van Leeuwenhoek 80: 155. 2001.
Notes: The name Sterigmatosporidium was described before Cuniculitrema and thus has nomenclatural priority. The genus has only one species, S. polymorphum with the name C. polymorpha listed as synonym (Table 1).
Naemateliaceae X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, fam. nov. MycoBank MB813351.
Etymology: In reference to the name of type genus Naematelia.
This family is proposed to accommodate a monophyletic lineage consisting of the aurantia clade formed by four Tremella species and a single-species clade represented by Cryptococcus cistialbidi as resolved in the seven-genes tree (Liu et al. 2015).
Basidiocarps when formed caespitose, tuberculate or spherical to thick foliaceous or crumpled, invory, yellow, bright orange, reddish. Basidia subglobose, globose to ovoid, four-celled and with vertical or occasionally transverse septa. Orange-pigmented colonies may be present. True hyphae with clamp connections and haustorial branches sometimes present. Budding cells present. Ballistoconidia sometimes present. Fermentation absent. Nitrate and nitrite not utilised. Major CoQ system CoQ-10.
Type genus: Naematelia Fr.
Genera accepted: Naematelia and Dimennazyma.
Notes: The species Bullera arundinariae branched before the aurantia clade and Cryptococcus cistialbidi in the seven-genes Bayesian tree, but this phylogenetic relationship was not supported (Liu et al. 2015). Thus, it is not included in the new family Naemateliaceae. C. cistialbidi occurred at different positions in the ML and NJ trees obtained from the seven-genes dataset (Liu et al. 2015). Possible relationship of the Tremella Clade II species (e.g. T. cladoniae, T. leptogii, T. phaeophysciae) with the aurantia clade was shown in the analyses of rRNA genes (Millanes et al., 2011, Liu et al., 2015, Fig. 5A in this study), but the affinity lacked support in rRNA gene analyses. The taxonomic assignment of the Clade II species remains to be determined. A strain representing T. indecorata in Liu et al. (2015) was placed close to Clade II species in the expanded LSU rRNA gene tree (Fig. 5A).
Naematelia Fr., Obs. Myc. 2: 370. 1818.
This genus is resurrected to accommodate species in the aurantia clade (Chen, 1998, Millanes et al., 2011, Liu et al., 2015).
Basidiocarps caespitose, tuberculate or spherical to thick foliaceous or crumpled, invory, yellow, bright orange, reddish. Basidia subglobose, globose to ovoid, four-celled and with vertical or occasionally transverse septa. True hyphae with clamp connections and haustorial branches present. Budding cells present. Ballistoconidia sometimes present. Fermentation absent. Nitrate and nitrite not utilised. Major CoQ system CoQ-10.
Type species: Naematelia encephala (Pers.) Fr., Observ. mycol. (Havniae) 2: 370. 1818.
Basionym: Tremella encephala Pers., Syn. meth. fung. (Göttingen) 2: 623. 1801.
Notes: The genus contains four species previously classified as Tremella (Chen, 1998, Millanes et al., 2011, Liu et al., 2015). These species formed a strongly supported monophyletic clade in the trees from the seven-genes (Liu et al. 2015) and expanded LSU datasets (Fig. 5A), and also in an unconstrained LSU tree (data not shown).
New combinations for Naematelia
Naematelia aurantialba (Bandoni & M. Zang) Millanes & Wedin, comb. nov. MycoBank MB813352.
Basionym: Tremella aurantialba Bandoni & M. Zang, Mycologia 82: 270. 1990.
Naematelia microspora (Lloyd) Millanes & Wedin, comb. nov. MycoBank MB813357.
Basionym: Tremella microspora Lloyd, Mycol. Writ. 6 (Letter 64): 991. 1920.
Dimennazyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813339.
Etymology: The genus is named in honour of M. E. di Menna for her pioneering work on yeast ecology and diversity.
This genus is proposed to accommodate the single-species lineage Cryptococcus cistialbidi in the tree derived from the seven-genes dataset (Liu et al. 2015).
Basidiocarps not present. Colonies usually orange-coloured. Pseudohyphae occasionally present. True hyphae not observed. Budding cells present. Ballistoconidia not observed. Fermentation absent. Nitrate not utilised. Starch-like compounds weakly produced or not produced. Major CoQ system CoQ-10.
Type species: Dimennazyma cistialbidi (Á. Fonseca, J. Inácio & Spenc.-Mart.) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout comb. nov. MycoBank MB814753.
Basionym: Cryptococcus cistialbidi Á. Fonseca et al., FEMS Yeast Res. 5: 1177. 2005.
Notes: The species Cryptococcus cistialbidi formed a single-species branching before the aurantia clade (i.e. Naematelia gen. nov.) with strong support in the seven-genes tree (Liu et al. 2015). This species produces orange-coloured colonies as emphasised by Inácio et al. (2005), which is different from the yeast stage of the Naematelia species that have yellowish brown colonies. Thus C. cistialbidi is not assigned to Naematelia. The colony colour of C. cistialbidi is similar to those produced by the genus Genolevuria (i.e. the amylolyticus clade in Fonseca et al. 2011). However, C. cistialbidi was distantly related to Genolevuria in the seven-genes tree (Liu et al. 2015). Therefore, we propose a new genus for this single-species lineage.
Carcinomycetaceae Oberw. & Bandoni, Nordic JI Bot. 2: 507. 1982. emend. A.M. Yurkov.
Basidiocarps mycoparasitic, forming a thin layer on the surface of host. Basidia subglobose or cylindrical, longitudinally or cruciately septated, with four sterigmata. Budding cells present. Ballistoconidia occasionally present. Pseudohyphae and true hyphae not observed in culture. Sexual reproduction not observed in culture. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-10.
Type genus: Carcinomyces Oberw. & Bandoni.
Genus accepted: Carcinomyces.
Notes: Oberwinkler & Bandoni (1982) proposed the family Carcinomycetaceae to accommodate three mycoparasitic genera Carcinomyces, Christiansenia and Syzygospora. While the type genus of this family is placed in the order Tremellales, two other genera are phylogenetically related to Filobasidiales (Millanes et al. 2011, Fig. 2 of this study). A close relationship between the genus Carcinomyces (Syzygospora effibulata) and Tremella polyporina was shown previously in the analysis of the combined rRNA dataset (Millanes et al. 2011). In the present study a well-supported clade formed by Bullera arundinariae, Syzygospora effibulata and Tremella polyporina was resolved in the expanded LSU rRNA gene tree (Fig. 5D). The species B. arundinariae was the only member of this clade included in the seven-genes phylogenetic analysis in Liu et al. (2015). B. arundinariae branched before the aurantia clade (i.e. Naematelia) and Cryptococcus cistialbidi (i.e. Dimennazyma) in the seven-genes Bayesian tree, but this phylogenetic relationship was not supported (Liu et al. 2015). Thus, we prefer to classify these genera into two different families, Naemateliaceae (Naematelia and Dimennazyma) and Carcinomycetaceae (Carcinomyces).
Carcinomyces Oberw. & Bandoni, Nordic JI Bot. 2: 507. 1982. emend. A.M. Yurkov.
This genus is resurrected and emended to accommodate a well-supported clade formed by Bullera arundinariae, Syzygospora effibulata and Tremella polyporina in the expanded LSU rRNA gene tree (Fig. 2, Fig. 5D). The diagnosis of Carcinomyces is based on the description of the family Carcinomycetaceae.
Type species: Carcinomyces effibulatus (Ginns & Sunhede) Oberw. & Bandoni, Nordic Journal of Botany 2: 509. 1982.
≡ Christiansenia effibulata Ginns & Sunhede, Bot. Notiser 131: 168. 1978.
≡ Syzygospora effibulata (Ginns & Sunhede) Ginns. Mycologia 78: 626. 1986.
Notes: Carcinomyces was proposed by Oberwinkler & Bandoni (1982) to accommodate two species transferred from Christiansenia, namely Ch. mycetophila and Ch. effibulata, with the latter being the type species. Ginns (1986) treated Carcinomyces, Christiansenia and Heterocephalacria as synonyms of Syzygospora. Molecular phylogenetic analyses showed that the genus Syzygospora sensu Ginns (1986) was polyphyletic (Millanes et al. 2011, also see Fig. 2 of this study). As discussed above, S. alba (type of Syzygospora) and Ch. pallida (type of Christiansenia) were located in a clade within the Filobasidiales, while S. effibulata (= Ch. effibulata) was located in a well supported clade together with Bullera arundinariae and Tremella polyporina in the Tremellales (Fig. 2). Therefore, the genus Carcinomyces is resurrected here for this clade. The taxonomic assignment of the genus Carcinomyces in Tremellales remains to be determined as discussed above. The affinity of Carcinomyces (Christiansenia) mycetophila with this genus needs to be confirmed using molecular data.
New combinations for Carcinomyces
Carcinomyces arundinariae (Fungsin, M. Takash. & Nakase) A.M. Yurkov, comb. nov. MycoBank MB813361.
Basionym: Bullera arundinariae Fungsin et al., Microbiol. Culture Coll. 18: 86. 2002.
Carcinomyces polyporina (D.A. Reid) A.M. Yurkov, comb. nov. MycoBank MB813362.
Basionym: Tremella polyporina D.A. Reid, Trans. Br. Mycol. Soc. 55: 416. 1970.
Trimorphomycetaceae X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, fam. nov. MycoBank MB813363.
Etymology: In reference to the name of the type genus Trimorphomyces.
This family is proposed for a well-supported monophyletic lineage including the flavus clade and Trimorphomyces papilionaceus, Bullera miyagiana and B. sakaeratica (Liu et al. 2015).
Basidiocarps minute, pustulate up to 2 mm diameter, watery-gelatinous, milky to faintly greenish. H-shaped dikaryotic conidia form dikaryotic hyphae with clamp connections and haustorial branches. Alternatively, these conidia germinate with paired blastogenous conidia. Basidia tremella-like, narrowly clavate, pyriform to stalked capitate, and four-celled. Basidiospores bud or form ballistoconidia. In culture, true hyphae may form extensively. Budding cells present, with polar or multilateral budding. Ballistoconidia occasionally present. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-10.
Type genus: Trimorphomyces Bandoni & Oberw.
Genera accepted: Trimorphomyces, Carlosrosaea, Saitozyma and Sugitazyma.
Notes: The species assigned to Trimorphomycetaceae also formed a monophyletic lineage in the tree obtained from the expanded LSU rRNA gene dataset, but bootstrap support was lacking (Fig. 5D). Two lichenicolous Tremella species (T. diploschistina and T. parmeliarum) are only loosely connected with other species and we therefore refrain from including them in any family until additional molecular data are available. Contrarily, Bullera ninhbinhensis clustered together with Cryptococcus podzolicus with strong support (Fig. 5E) and we thus suggest to include this species in the family. In the original description Bullera vrieseae (Landell et al. 2015) was placed together with Bullera sakaeratica and B. miyagiana in a well-supported clade, which is resolved in this study as members of the family Trimorphomycetaceae (Fig. 5E). Relationships between the three Bullera species were not supported in our analyses. Bullera sakaeratica was closely related to Trimorphomyces papilionaceus with strong support, while B. miyagiana and B. vrieseae were located in significantly diverged branches with unresolved positions (Fig. 5E), suggesting that they represent independent lineages in the family Trimorphomycetaceae. Thus, we propose new genera for B. vrieseae and B. miyagiana. The phylogenetic relationships of Tremella diploschistina and T. parmeliarum with other species were also not resolved in our analyses. We do not reclassify the two sexual species and leave them temporary as T. diploschistina pro tem and T. parmeliarum pro tem.
Trimorphomyces Bandoni & Oberw., Syst. Appl. Microbiol. 4: 106. 1983. emend. X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov.
This genus is emended to include an anamorphic yeast species Bullera sakaeratica, which is closely related to the type species of Trimorphomyces (Liu et al. 2015, Fig. 5E in this study).
Basidiocarps mycoparasitic, small, watery-gelatinous. Hyphae with clamp connections and haustorial branches may be present. Basidia narrow clavate, pyriform to capitate, four-celled, longitudinally or obliquely septa. Basidiospores subglobose. H-shaped zygoconidia may be present. Budding cells present. Ballistoconidia occasionally present. Sexual reproduction not observed in culture. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-10.
Type species: Trimorphomyces papilionaceus Bandoni & Oberw, Syst. Appl. Microbiol. 4: 106. 1983.
Notes: The close relationship between T. papilionaceus and B. sakaeratica was shown in different studies (Boekhout et al., 2011a, Weiss et al., 2014, Liu et al., 2015). As discussed above, these species were treated as two single-species lineages in Liu et al. (2015) due to the consideration that the former is morphologically unique (Bandoni & Boekhout 2011a). The separation of Trimorphomyces species in distantly related clades and the close relationships of some Trimorphomyces species with some Cryptococcus and Bullera species as shown in the tree obtained from the expanded LSU rRNA gene dataset (Fig. 5D, E) suggested that the phenotypically circumscribed genus Trimorphomyces is polyphyletic and that Trimorphomyces species may have a common yeast stage. Therefore, we transfer B. sakaeratica to the genus Trimorphomyces here. Another described Trimorphomyces species, T. nebularis (Kirschner & Chen 2008), is transferred to the genus Vishniacozyma gen. nov. Trimorphomyces papilionaceus forms zygoconidia from a single conidiogenous cell, differing from T. nebularis (Vishniacozyma nebularis comb. nov.) which produces zygoconidia from two neighbouring conidiogenous cells (Bandoni and Oberwinkler, 1983, Kirschner and Chen, 2008). The genus Trimorphomyces currently contains two species (Table 1, Fig. 5E).
New combinations for Trimorphomyces
Trimorphomyces sakaeraticus (Fungsin, M. Takash. & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew., Boekhout & A.M. Yurkov, comb. nov. MycoBank MB813365.
Basionym: Bullera sakaeratica Fungsin et al., Microbiol. Culture Coll. 19: 37. 2003.
Saitozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813367.
Etymology: The genus is named in honour of the Japanese yeast taxonomist Masuyoshi Saito.
This genus is proposed for the flavus clade containing three species previously assigned to the genus Cryptococcus (Liu et al. 2015).
Basidiocarps not known. Cultures can be pigmented and pale to brownish-yellow in colour. Pseudohyphae and true hyphae may be present. Sexual reproduction not observed in culture. Budding cells present. Ballistoconidia occasionally present. Fermentation absent. Nitrate not utilised. Starch-like compounds weakly produced or not formed. Major CoQ system CoQ-10.
Type species: Saitozyma flava (Saito) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813368.
Basionym: Torula flavus Saito, Jap. J. Bot. 1: 45. 1922.
Notes: The flavus clade delimited in Liu et al. (2015) based on multiple genes is equivalent to the podzolicus clade in Boekhout et al. (2011a), which was based on the analysis of LSU rRNA gene (D1/D2 domains) sequences, even though this clade received no bootstrap support. In the tree from the expanded LSU rRNA gene dataset, the close relationship between C. podzolicus and Bullera ninhbinhensis was confirmed, but they did not form a monophyletic clade with the other two species, C. flavus and C. paraflavus, in the flavus clade (Fig. 5E). Since the affinity of C. podzolicus with the flavus clade was strongly supported in the trees obtained both from the seven genes and the three rRNA gene datasets (Liu et al. 2015), we assign these species to Saitozyma gen. nov. Four species are currently accepted in this new genus (Table 1) and an additional sequence representing a potential new species was obtained from public databases (Fig. 5E).
New combinations for Saitozyma
Saitozyma ninhbinhensis (Luong, M. Takash., Dung & Nakase) A.M. Yurkov, comb. nov. MycoBank MB813370.
Basionym: Bullera ninhbinhensis Luong et al., J. Gen. Appl. Microbiol. 6: 335–342. 2002.
Saitozyma paraflava (Golubev & J.P. Samp.) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813372.
Basionym: Cryptococcus paraflavus Golubev & J.P. Samp., J. Gen. Appl. Microbiol. 50: 68. 2004.
Saitozyma podzolica (Babeva & Reshetova) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813373.
Basionym: Candida podzolica Babeva & Reshetova, Mikrobiologiya, Seriya B 44: 333. 1975.
Sugitazyma A. M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB814754.
Etymology: The genus is named in honour of the Japanese yeast taxonomist Takashi Sugita.
This genus is proposed for the single-species lineage Bullera miyagiana resolved in the tree inferred from seven genes (Liu et al. 2015) and the expanded LSU rRNA gene dataset (Fig. 5E).
Basidiocarps not known. Pseudohyphae and true hyphae may be present. Sexual reproduction not observed in culture. Budding cells present. Ballistoconidia may be present. Fermentation absent. Nitrate not utilised. Starch-like compounds produced. Major CoQ system CoQ-10.
Type species: Sugitazyma miyagiana (Nakase, Itoh, Takem. & Bandoni) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb nov. Mycobank MB814755.
Basionym: Bullera miyagiana Nakase et al., J. Gen. Appl. Microbiol. 36: 35. 1990.
Notes: Relationships between Bullera miyagiana and other Trimorphomycataceae remained unclear in the expanded analysis of the LSU rRNA gene sequences (Fig. 5E). We propose to erect a new genus for this single-species lineage to restrict the genus Bullera to the type lineage in the family Bulleraceae. The genus Sugitazyma currently contains one species (Table 1).
Carlosrosaea A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB814756.
Etymology: The genus is named in honour of the Brazilian yeast taxonomist and ecologist Carlos A. Rosa.
This genus is proposed for the single-species lineage Bullera vrieseae resolved in the tree inferred from the expanded LSU rRNA gene dataset (Fig. 5E).
Basidiocarps not known. Pseudohyphae and true hyphae not observed in culture. Sexual reproduction not observed in culture. Budding cells present. Ballistoconidia may be present. Fermentation absent. Nitrate sometimes utilised. Starch-like compounds not produced. Major CoQ system unknown.
Type species: Carlosrosaea vrieseae (Landell, Brandão, Safar, Gomes, Félix, Santos, Pagani, Ramos, Broetto, Mott, Valente & Rosa) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb nov. Mycobank MB814757.
Basionym: Bullera vrieseae Landell et al., Int. J. Syst. Evol. Microbiol. 65: 2469. 2015.
Notes: Relationships between Bullera vrieseae and other Trimorphomycataceae remained unclear in the expanded analysis of the LSU rRNA gene sequences (Fig. 5E). B. vrieseae was distantly related to B. miyagiana (Sugitazyma gen. nov.) and showed additionally distinct phenotypic characters (Landell et al. 2015). We propose to erect a new genus for this single-species lineage to restrict the genus Bullera emend. to the type lineage in the family Bulleraceae. The genus Carlosrosaea contains currently one species (Table 1) and an additional sequence representing a potential new species (LSU: GenBank KJ690943) was found in public databases.
Cryptococcaceae Kütz. ex Castell. & Chalm., Man. Trop. Med., 3rd Edn: 1070. 1919. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
This family is emended here to accommodate the monophyletic lineage consisting of the Cryptococcus and Kwoniella clades (Fig. 1) as recognised in Liu et al. (2015).
Basidiocarps not known. Pseudohyphae and true hyphae present or not, but present after mating. Basidia one-celled, cylindrical, clavate, capitate with four chains of acropetal basidiospores, or globose to ovoid with longitudinal and transverse septa. Basidiospores passively released, variably shaped, smooth or somewhat rough bacilliform, ellipsoid, globose, oblong-pentagonal or obpyriform to cylindrical. Budding cells usually present, but may be absent. Fermentation absent. Nitrate not utilised. Starch-like compounds usually produced. Major CoQ system CoQ-9 or CoQ-10.
Type genus: Cryptococcus Vuillemin
Genera accepted: Cryptococcus and Kwoniella.
Notes: The family Cryptococcaceae was originally established to contain anamorphic tremellomycetous yeasts, including the genera Bullera, Cryptococcus, Fellomyces, Kockovaella, Trichosporon, Tschuchiyaea and Udeniomyces (Kützing, 1833, Van der Walt, 1987). Molecular phylogenetic studies indicated that this family and most of the genera assigned to it are not monophyletic (Wells, 1994, Fell et al., 2000, Scorzetti et al., 2002, Sampaio, 2004, Liu et al., 2015). Based on the seven-genes phylogeny (Liu et al. 2015), the family Cryptococcaceae is emended here to accommodate only two emended genera Cryptococcus and Kwoniella (Table 1, Table 2, Fig. 1).
Cryptococcus Vuillemin, Rev. Gén. Sci. Pures Appl. 12: 741. 1901. Nomen Cons. (McNeil et al. 2006), emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
= Filobasidiella Kwon-Chung, Mycologia 67: 1198. 1975.
= Tsuchiyaea Yamada, Kawasaki, Itoh, Banno et Nakase, J. Gen. Appl. Microbiol. 34: 509. 1988.
This genus is emended to accommodate the Cryptococcus clade described in Findley et al., 2009, Liu et al., 2015 and the LSU rRNA gene tree (Fig. 5C), and includes the species belonging to the C. neoformans/C. gattii complex, Filobasidiella depauperata and F. lutea.
Basidiocarps not present. True hyphae may be present, but present after mating. Basidia one-celled, slender, cylindrical sphaeropedunculate. Basidiospores in four long basipetal chains, smooth, or somewhat rough bacilliform, ellipsoid, globose, oblong-pentagonal or obpyriform. Haustorial branches present or not. Budding cells usually present, but in some species also absent. Thick polysaccharide capsules may be present. Fermentation absent. Nitrate and nitrite not utilised. Starch-like compounds usually produced. Major CoQ system CoQ-9 or CoQ-10.
Type species: Cryptococcus neoformans (Sanfelice) Vuillemin, Rev. Gén. Sci. Pures Appl. 12: 747–750. 1901.
Notes: The generic name Cryptococcus was originally created by Kützing (1833) for an organism placed among the algae and only one species C. mollis was included, but the description of this species was not presented. Vuillemin (1901) recircumscribed Cryptococcus to include pathogenic yeasts and transferred Saccharomyces neoformans to this genus. The genus Cryptococcus has been conserved with C. neoformans as the type species as proposed by Fell et al. (1989). Many molecular phylogenetic studies have shown that Cryptococcus as circumscribed until now (Fonseca et al. 2011) is a highly artificial genus that contains species distributed in all five major lineages of Tremellomycetes (Takashima and Nakase, 1999, Fell et al., 2000, Scorzetti et al., 2002, Sampaio, 2004, Fonseca et al., 2011, Weiss et al., 2014, Liu et al., 2015). The teleomorphic genus Filobasidiella was described by Kwon-Chung (1975) for the perfect state of C. neoformans. Four species are currently accepted in the genus Filobasidiella (Kwon-Chung 2011). Filobasidiella bacillispora was described for the sexual stage of serotypes B and C of C. neoformans (Kwon-Chung 1976). Two other Filobasidiella species, F. depauperata and F. lutea, are mycoparasites but lack ontogenetic yeast states. The affinity of F. depauperata with the Cryptococcus/Filobasidiella clade has been shown in different studies (Sivakumaran et al., 2002, Findley et al., 2009, Rodriguez-Carres et al., 2010, Boekhout et al., 2011a, Weiss et al., 2014, Liu et al., 2015). Sequence analyses of the ITS regions and SSU rRNA gene showed that F. lutea belongs to the Cryptococcus/Filobasidiella clade (Sivakumaran et al. 2002). Unfortunately, the sequence data from this species are neither available from GenBank, nor from other databases, nor from these authors. Filobasidiella xianghuijun is the fifth species described in the genus (Zang 1999), but the taxonomic and phylogenetic position of this species remains to be confirmed by molecular means (Kwon-Chung 2011). Recently, a sexual cycle with a tetrapolar mating system was found in C. amylolentus (Findley et al. 2012). So far, all the species in the Cryptococcus/Filobasidiella clade have been shown to have similar sexual forms. The C. neoformans/C. gattii complex was recently revised and presently contains seven species (Hagen et al. 2015). In that paper it was also suggested that the anamorph name Cryptococcus should have priority over the teleomorph name Filobasidiella (Kwon-Chung 1975) as the former has nomenclatural priority, is much more commonly used and, importantly, refers to the name of an important infectious disease, cryptococcosis, that is caused by species of this genus (Heitman et al. 2011). Thus, the Filobasidiella species without a yeast state are transferred here to Cryptococcus. Fonseca et al. (2011) considered Tsuchiyaea wingfieldii (basionym Sterigmatomyces wingfieldii) a synonym of C. amylolentus because of similar SSU, ITS and LSU D1/D2 sequences. This point of view is supported by similar sequences of four protein-coding genes (Liu et al. 2015). Phylogenetic analysis of four MAT related genes, STE3, STE20, SXI1α and LPD1 indicated minor differences between T. wingfieldii and C. amylolentus (Findley et al. 2012). Thus, further studies are needed to fully understand the taxonomic relationship between these two species. Ten species are currently accepted in this genus (Table 1).
New combinations for Cryptococcus
Cryptococcus depauperatus (Petch) Boekhout, Liu, Bai & M. Groenew., comb. nov. MycoBank MB813376.
Basionym: Aspergillus depauperatus Petch, Trans. Br. Mycol. Soc. 16: 245. 1932 (‘1931’).
≡ Filobasidiella depauperata (Petch) Samson et al., Antonie van Leeuwenhoek 49: 454. 1983.
= Filobasidiella arachnophila Malloch et al., Can. J. Bot. 56: 1823. 1978.
Notes: The close relatedness between this species and C. neoformans (cited as F. neoformans) was shown by SSU rRNA gene sequence analysis (Kwon-Chung et al. 1995). Multilocus phylogenetic analysis using ITS, TEF-1 alpha and RPB1 sequences corroborated these conclusions and found the species related to C. neoformans (cited as var. grubii), C. deneoformans (cited as var. neoformans), C. gattii (cited as VGI) and C. deuterogattii (cited as VGII) (Rodriguez-Carres et al. 2010). Another study using six genes (the above with addition of RPB2, mitSSU and the D1/D2 domains of LSU rDNA) showed that also Tsuchiyaea wingfieldii and Cryptococcus amylolentus were phylogenetically related (Findley et al. 2009).
Cryptococcus luteus (Roberts) Boekhout, Liu, Bai & M. Groenew., comb. nov. MycoBank MB814758.
Basionym: Filobasidiella lutea Roberts, Mycotaxon 60, 198. 1997.
Notes: This species could not be studied by molecular means due to the absence of a culture. The close relatedness between this species, C. neoformans (cited as Filobasidiella neoformans) and C. depauparatus (cited as Filobasidiella depauparata) was shown by sequence analysis of ITS and SSU rRNA gene fragments (Sivakumaran et al. 2002).
Kwoniella Statzell-Tallman & Fell, FEMS Yeast Res. 8: 107. 2008. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
This genus is emended to include the species of the Kwoniella clade containing K. mangroviensis and six anamorphic species previously classified in Bullera and Cryptococcus (Liu et al. 2015).
Basidiocarps unknown. Sexual reproduction with tetrapolar mating system observed in some species. Basidia polymorphic, lageniform, globose to ovoid with longitudinal and transverse septa. Basidiospores passively released, globose, ovoid to cylindrical. Pseudohyphae and true hyphae may be present. Budding cells present. Ballistoconidia occasionally present. Fermentation absent. Nitrate not utilised. Starch-like compounds are usually produced. Major CoQ system CoQ-10.
Type species: Kwoniella mangroviensis Statzell, Belloch & Fell, FEMS Yeast Res. 8: 107. 2008.
Notes: The genus Kwoniella was proposed for a teleomorphic yeast K. mangroviensis isolated from mangrove habitats (Statzell-Tallman et al. 2008). The second species of the genus, K. heveanensis, was proposed for the teleomorph of Cryptococcus heveanensis (Sun et al. 2011). Recently, two teleomorphic species, K. europaea and K. botswanensis, closely related to K. mangroviensis were described (Guerreiro et al. 2013). All sexual species displayed a tetrapolar mating system (Guerreiro et al. 2013). Since the use of a dual nomenclature for classification of sexual and asexual morphs of fungi was recently terminated, three asexual species were described in the genus Kwoniella, K. shandongensis (Chen et al. 2012), K. newhampshirensis and K. betulae (Sylvester et al. 2015). The Kwoniella clade was consistently resolved in different studies (Findley et al., 2009, Boekhout et al., 2011a, Guerreiro et al., 2013, Weiss et al., 2014, Liu et al., 2015). However, the affinity of Cryptococcus cuniculi to this clade remains controversial. Boekhout et al. (2011a) and Weiss et al. (2014) showed that this species belonged to the Kwoniella clade according to LSU rRNA gene sequence analyses, but Liu et al. (2015) showed that C. cuniculi branched before the Cryptococcus and Kwoniella clades in an analysis of combined SSU, ITS and LSU rRNA gene sequences. In the tree obtained from the expanded LSU rRNA gene dataset in this study, C. cuniculi was located in a branch distantly related to the Kwoniella clade (Fig. 5B, C). Thus, the final phylogenetic position of C. cuniculi remains to be determined. The recently described K. betulae was also placed outside the genus Kwoniella in a well-supported clade (i.e. Fonsecazyma gen. nov.) containing C. mujuensis and C. tronadorensis (Fig. 5C). Eleven species are accepted in Kwoniella emend. in this study (Table 1) and three sequences representing potential new species were obtained from public databases (Fig. 5C). Kwoniella species are usually saprobic and differ from the closely aligned pathogenic yeasts in the genus Cryptococcus emend. by their inability to grow at 37 °C (Findley et al. 2009).
New combinations for Kwoniella
Kwoniella dendrophila (Van der Walt & D.B. Scott) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813381.
Basionym: Bullera dendrophila Van der Walt & D.B. Scott, Antonie van Leeuwenhoek 36: 384. 1970.
Kwoniella bestiolae (Thanh, Hai & Lachance) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813383.
Basionym: Cryptococcus bestiolae Thanh et al., FEMS Yeast Res. 6: 301. 2006.
Kwoniella dejecticola (Thanh, Hai & Lachance) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813384.
Basionym: Cryptococcus dejecticola Thanh et al., FEMS Yeast Res. 6: 303. 2006.
Kwoniella pini (Golubev & Pfeiffer) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813387.
Basionym: Cryptococcus pinus Golubev & Pfeiffer, Int. J. Syst. Evol. Microbiol. 58: 1970. 2007.
Kwoniella shivajii (S.R. Ravella, S.A. James, C.J. Bond, I.N. Roberts, K. Cross, Retter & P.J. Hobbs) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813389.
Basionym: Cryptococcus shivajii S.R. Ravella et al., Curr. Microbiol. 60: 14. 2010.
Phaeotremellaceae A.M. Yurkov & Boekhout, fam. nov. MycoBank MB813390.
Etymology: In reference to the name of the type genus Phaeotremella.
This family is proposed to accommodate the foliacea clade and a single-species lineage, Cryptococcus spencermartinsiae (Liu et al. 2015).
Basidiocarps, if present, foliaceous with caespitose lobes, gelatinous, tan to cinnamon or light brown when fresh, drying dark brown to black. Hyphae with clamp connections; haustorial branches occasionally present. Basidia globose, subglobose, ovoid or ellipsoidal, four-celled and with longitudinal to diagonal or transverse septa. Basidiospores subglobose to broadly ellipsoidal. Budding cells present in culture. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-10.
Type genus: Phaeotremella Rea
Genera accepted: Phaeotremella and Gelidatrema.
Notes: The foliacea clade was resolved as early branching Tremellales in the seven-genes Bayesian tree without PP support; however, in the seven-genes ML and NJ trees, this clade was branching prior to the Tremellales and Trichosporonales lineages (Liu et al. 2015). The foliacea clade was also resolved as a well-supported monophyletic group in the trees constructed from rRNA genes datasets, but its position varied. In the tree from the combined SSU, ITS-5.8S and LSU datasets, the foliacea clade is branching before the Tremellales and Trichosporonales lineages in Millanes et al. (2011), but between these two lineages in Liu et al. (2015). In the trees from the LSU rRNA gene datasets containing more filamentous fruitbody-forming taxa, this clade located within the Tremellales (Boekhout et al., 2011a, Weiss et al., 2014), but as a group (equivalent to Phaeotremella) branching before the order in this study (Fig. 1). The affinity of Cryptococcus spencermartinsiae to Phaeotremellaceae fam. nov. remains to be confirmed. It branches before the foliacea clade with strong to moderate support in the Bayesian and ML trees obtained from the seven-genes dataset, but it occurred in a branch distantly related to this clade in the combined rRNA genes tree (Liu et al. 2015). In the tree from the expanded LSU dataset, C. spencermartinsiae branched more early than the foliacea (Phaeotremella) clade, but without bootstrap support (Fig. 5C). Furthermore, this species is not able to grow above 25 °C in contrast to the other species in the foliacea clade (Phaeotremella gen. nov.) that can grow at 30 °C. Thus, we propose to accommodate it in the new genus Gelidatrema gen. nov.
Phaeotremella Rea, Trans. Br. Mycol. Soc. 3: 377. 1912. emend. A.M. Yurkov & Boekhout.
Type species: Phaeotremella pseudofoliacea Rea, Trans. Br. Mycol. Soc. 3: 377. 1912.
= Tremella foliacea Pers., Observationes mycologicae 2: 98. 1800.
This genus is resurrected and emended to accommodate the foliacea clade (Chen, 1998, Boekhout et al., 2011a, Millanes et al., 2011, Weiss et al., 2014, Liu et al., 2015, also see Fig. 5B).
Basidiocarps, if present, foliaceous with caespitose lobes, gelatinous, tan to cinnamon or light brown when fresh, drying dark brown to black. Hyphae with clamp connections; haustorial branches occasionally present. Basidia globose, subglobose, ovoid or ellipsoidal, four-celled and with longitudinal to diagonal or transverse septa. Basidiospores subglobose to broadly ellipsoidal. In culture budding cells sometimes present. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-10.
Notes: Phaeotremella was described by Rea (1912) including only Phaeotremella pseudofoliacea, which was considered a synonym of Tremella foliacea (Roberts 1999). As discussed above, the foliacea clade was consistently resolved as a well supported clade in different studies using various datasets, though its phylogenetic position varied (Chen, 1998, Boekhout et al., 2011a, Millanes et al., 2011, Weiss et al., 2014, Liu et al., 2015, this study Fig. 1). Seven species, including two Cryptococcus and five Tremella species were included in this clade with 100 % bootstrap support (Fig. 1) and therefore are transferred to Phaeotremella (Table 1).
New combinations for Phaeotremella
Phaeotremella fagi (Middelhoven & Scorzetti) A.M. Yurkov & T. Boekhout, comb. nov. MycoBank MB813391.
Basionym: Cryptococcus fagi Middelhoven & Scorzetti, Antonie van Leeuwenhoek 90: 63. 2006.
Phaeotremella simplex (H.S. Jacks. & G.W. Martin) Millanes & Wedin, comb. nov. MycoBank MB813392.
Basionym: Tremella simplex H.S. Jacks. & G.W. Martin, Mycologia 32: 687. 1940.
Phaeotremella skinneri (Phaff & Carmo Souza) A.M. Yurkov & T. Boekhout, comb. nov. MycoBank MB813393.
Basionym: Cryptococcus skinneri Phaff & Carmo Souza, Antonie van Leeuwenhoek 28: 205. 1962.
Phaeotremella mycetophiloides (Kobayasi) Millanes & Wedin, comb. nov. MycoBank MB813394.
Basionym: Tremella mycetophiloides Kobayasi, Sci. Rep. Tokyo Bunrika Daig., Sect. B 4: 13. 1939.
Phaeotremella mycophaga (G.W. Martin) Millanes & Wedin, comb. nov. MycoBank MB813395.
Basionym: Tremella mycophaga G.W. Martin, Mycologia 32: 686. 1940.
Phaeotremella neofoliacea (Chee J. Chen) Millanes & Wedin, comb. nov. MycoBank MB813396.
Basionym: Tremella neofoliacea Chee J. Chen, Bibl. Mycol. 174: 135. 1998.
Gelidatrema A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB814759.
Etymology: In reference to the origin of this psychrophilic yeast and its phylogenetic relatedness to the order Tremellales.
This genus is proposed for the single-species lineage Cryptococcus spencermartinsiae as resolved in the seven-genes tree (Liu et al. 2015) and the expanded LSU rRNA gene tree (Fig. 5C).
Basidiocarps not know. Pseudohyphae and true hyphae not observed in culture. Sexual reproduction not observed in culture. Budding cells present. Ballistoconidia absent. Fermentation absent. Nitrate not utilised. Starch-like compounds not produced. Major CoQ system unknown.
Type species: Gelidatrema spencermartinsiae (Garcia, Brizzio, Boekhout, Theelen, Libkind & van Broock) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb nov. MycoBank MB814760.
Basionym: Cryptococcus spencermartinsiae Garcia et al., Int. J. Syst. Evol. Microbiol. 60: 710. 2010.
Notes: Relationships between Cryptococcus spencermartinsiae and Phaeotremella remained unclear in phylogenetic analyses. Cryptococcus spencermartinsiae branched earlier than the foliacea (Phaeotremella) clade, but without bootstrap support in the expanded LSU rRNA gene tree (Fig. 5C) and with moderate support in the tree inferred from seven genes (Liu et al. 2015). Additionally, C. spencermartinsiae showed phenotypic characters distinct from other members of the foliacea clade (de García et al. 2010). Thus, we propose to erect a new genus for this single-species lineage. The genus Gelidatrema currently contains one species (Table 1), but a few additional sequences representing potential new species (ITS: GenBank KC455886, DQ242634) were found in public databases.
Order Trichosporonales Boekhout & Fell, FEMS Yeast Res. 1: 265–270. 2001.
The order Trichosporonales was introduced to accommodate arthroconidia-producing Trichosporon species that formed a clade in a sequence analysis of the D1/D2 domains of LSU rRNA gene (Fell et al. 2000). The distinction of this order from the order Tremellales was supported by the seven-genes phylogeny (Liu et al. 2015). Several Bullera and Cryptococcus species were included in this order although they do not produce arthroconidia (Takashima et al., 2001, Fungsin et al., 2006, Boekhout et al., 2011a, Weiss et al., 2014, Liu et al., 2015). In view of physiological, morphological and molecular data, two families, namely Trichosporaceae emend. and Tetragoniomycetaceae emend. and ten genera, including one emended and four new genera are accepted in this order (Table 1).
Trichosporonaceae Nann. [as ‘Trichosporaceae’], Repert. mic. uomo: 285. 1934. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout.
= Asterotremellaceae Prillinger et al., J. Gen. Appl. Microbiol. 53: 174. 2007.
This family is emended to accommodate the monophyletic lineage comprising the brassicae/gracile, cutaneum, haglerorum, porosum, Trichosporon and Vanrija clades, and four single-species lineages comprising three Cryptocococcus and one Trichosporon species as recognised in multigene phylogenies (Liu et al. 2015).
Basidiocarps unknown. Sexual reproduction not known. Septate hyphae with arthroconidia usually present. Pseudohyphae may be present. Budding cells usually present. Fermentation absent. Nitrate not utilised. This family has a pronounced ability to assimilate aromatic compounds. Major CoQ system CoQ-9 or CoQ-10.
Type genus: Trichosporon Behrend.
Genera accepted: Trichosporon, Apiotrichum emend., Cutaneotrichospon gen. nov., Effuseotrichosporon gen. nov., Haglerozyma gen. nov., and Vanrija.
Notes: Trichosporonaceae was introduced to include the Trichosporon species that were assigned to Geotrichoides and Proteomyces (Nannizzi 1934). The scope of this family has been expanded by the discovery of more Trichosporon species (Sugita and Nakase, 1998, Colombo et al., 2011), but was also reduced by the exclusion of the genus Geotrichum, an ascomycetous genus that also forms arthroconidia (Weijman 1979). Some Cryptococcus species also belonged to this clade. Thus, this family is emended to accommodate a monophyletic lineage consisting of the brassicae/gracile, cutaneum, haglerorum, humicola (i.e. Vanrija pro parte), porosum, and Trichosporon clades, as well as four single-species clades of three Cryptocococcus species and one Trichosporon species as recognised in Liu et al. (2015).
Trichosporon Behrend, Berliner Klin. Wochenschr. 21: 464. 1890.
This genus is re-defined to accommodate only the species in the Trichosporon clade recognised in the seven-genes phylogeny (Liu et al. 2015).
Basidiocarps not known. Septate hyphae with arthroconidia usually present. Sexual reproduction not observed. Budding cells present. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-9.
Type species: Trichosporon ovoides Behrend
Notes: Pleurococcus beigelii Küchenmeister & Rabenhorst (Rabenhorst 1867) was probably the first fungus representing the current genus Trichosporon. The name Pleurococcus was rejected by Guého et al. (1992a) as it concerned a poorly described species and no material was preserved. The name Trichosporon was introduced to include only one species, T. ovoides, isolated from an infected moustache hair from Germany (Behrend 1890). Guého et al. (1992a) selected a neotype for T. ovoides, CBS 7556 isolated from capital white piedra occurring on a Caucasian, a habitat that agreed with the original description of the species by Behrend. These authors also rejected the neotypification of T. beigelii with ATCC 28592 (= CBS 2466) as proposed by McPartland & Goff (1991), but they selected this strain as neotype for T. cutaneum when using a more narrow species concept for this taxon. A major contribution to the taxonomy of the genus Trichosporon was made by Guého et al. (1992b) by recircumscribing several species using rRNA sequence analysis, nutritional physiology, electron microscopy of septal pores, and the analysis of the G+C content of the DNA and DNA–DNA hybridisation experiments. Using sequence analysis of the D2 domain of the 26S rRNA, five clusters of species were observed: 1) T. sporotrichoides, T. dulcitum, T. gracile, T. multisporum, T. laibachii, and T. loubieri; 2) T. cutaneum, T. jirovecii, T. mucoides, and T. moniliiforme; 3) T. coremiiforme, T. aquatile, T. faecale, T. asahii, T. asteroids and Fissurella filamenta; 4) T. ovoides and T. inkin; and 5) T. brassicae and T. montevideense (Guého et al. 1992b). It is interesting to note that this clustering is fully congruent with our results from the analyses of multiple genes.
Recently, many new Trichosporon species have been described or have been transferred to this genus from other genera (Guého et al., 1998, Colombo et al., 2011, Sugita, 2011). According to the latest monographic treatment (Sugita 2011) the genus contains 37 species. The Trichosporon species are distributed over five clades, namely brassicae, cutaneum, gracile, porosum and Trichosporon (Middelhoven et al., 2004, Sugita et al., 2004, Sugita, 2011), which are similar to those observed before in analyses of partial LSU rRNA sequences (Guého et al. 1992b). Serological characteristics correlate well with these clades and therefore were applied as a taxonomic marker in this group (Tsuchiya et al., 1974, Sugita, 2011). The genus is restricted here to accommodate only the Trichosporon clade, including the neotype species T. ovoides (Liu et al. 2015). Species belonging to this newly circumscribed genus Trichosporon have serotype II. Twelve species are accepted in this genus (Table 1, Fig. 6).
Cutaneotrichosporon X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813397.
Etymology: In reference to the specific epithet of the type species in this genus.
This genus is proposed for the well supported monophyletic group consisting of the cutaneum and haglerorum clades together with Trichosporon guehoae, Cryptococcus curvatus, Cr. cyanovorans and Cr. daszewskae (Liu et al. 2015, Fig. 6 of this study).
Basidiocarps not known. Sexual reproduction not observed. True hyphae disarticulate into ovoid, cylindrical or cubic arthroconidia. Pseudohyphae abundant or not. Budding cells present. Fermentation absent. Nitrate not utilised, nitrite utilised or not. Major CoQ system CoQ-10.
Type species: Cutaneotrichosporon cutaneum (Beurm., Gougerot & Vaucher bis) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. Mycobank MB13398.
Notes: Close relationships between the cutaneum and haglerorum clades were supported in recent phylogenetic studies (Sugita, 2011, Liu et al., 2015, Takashima et al., 2015, Fig. 6 of this study). The next well-supported node clustered with cutaneum and haglerorum clades with T. guehoae, Cr. curvatus, Cr. cyanovorans and Cr. daszewskae (Fig. 6). Since this expanded clade received strong support in multi-gene studies by Liu et al. (2015) and Takashima et al. (2015) as well as the extended LSU rRNA gene phylogeny (Fig. 6), we propose to classify the cutaneum and haglerorum clades together with T. guehoae, Cr. curvatus, Cr. cyanovorans and Cr. daszewskae (Fig. 6) in a single genus Cutaneotrichosporon gen. nov., in order to avoid proposing several small genera with only one or two species. The genus Cutaneotrichosporon currently contains 15 species (Table 1).
New combinations for Cutaneotrichosporon
Cutaneotrichosporon arboriformis (Sugita, M. Takash., Sano, Nishim., Kinebuchi, S. Yamag. & Osanai) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813407.
Basionym: Cryptococcus arboriformis Sugita et al., Microbiol. Immunol. 51: 544. 2007.
Cutaneotrichosporon curvatus (Diddens & Lodder) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB814761.
Basionym: Candida heveanensis Diddens & Lodder var. curvata Diddens & Lodder, Beitr. Monogr. Hefenarten. II Teil. Die anaskosporogene Hefen: 310. 1942.
≡ Cryptococcus curvatus (Diddens & Lodder) Golubev, Mikol. Fitopatol. 15: 467. 1981.
Cutaneotrichosporon cutaneum (de Beurmann, Gougerot & Vaucher) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813398.
Basionym: Oidium cutaneum de Beurmann et al., Bull. Mém. Soc. Méd. Hôpit. Paris 18: 52. 1910.
≡ Trichosporon cutaneum (de Beurmann et al.) M. Ota, Annls Parasit. hum. comp. 4: 12. 1926.
Cutaneotrichosporon cyanovorans (Motaung, Albertyn, J. L. F. Kock et Pohl) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB814762.
Basionym: Cryptococcus cyanovorans Motaung et al., Int. J. Syst. Evol. Microbiol. 62: 1211. 2012.
Cutaneotrichosporon daszewskae (M. Takash., Sugita, Shinoda & Nakase) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB814763.
Basionym: Cryptococcus daszewskae M. Takash. et al., Int. J. Syst. Evol. Microbiol. 51: 2204, 2001.
Cutaneotrichosporon debeurmannianum (Sugita, M. Takash., Nakase & Shinoda) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813399.
Basionym: Trichosporon debeurmannianum Sugita et al., Int. J. Syst. Evol. Microbiol. 51: 1221–1228. 2001.
Cutaneotrichosporon dermatis (Sugita, M. Takash., Nakase, Ichikawa, Ikeda & Shinoda) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813400.
Basionym: Trichosporon dermatis Sugita et al., Int. J. Syst. Evol. Microbiol. 51: 1221. 2001.
Cutaneotrichosporon guehoae (Middelhoven, Scorzettii & Fell) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB814764.
Basionym: Trichosporon guehoae Middelhoven et al., Can. J. Microbiol. 45: 687. 1999.
Cutaneotrichosporon jirovecii (Frágner) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813401.
Basionym: Trichosporon jirovecii Frágner, Česká Mykol. 23: 160. 1969.
Cutaneotrichosporon haglerorum (Middelhoven, Á. Fonseca, S.C. Carreiro, Pagnocca & O.C. Bueno) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813408.
Basionym: Cryptococcus haglerorum Middelhoven et al., Antonie van Leeuwenhoek 83: 168. 2003.
Cutaneotrichosporon moniliiforme (Weigmann & A. Wolff) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813403.
Basionym: Oidium moniliiforme Weigmann & A. Wolff, Zentbl. Bakt. ParasitKde, Abt. II 22: 668. 1909.
≡ Trichosporon moniliiforme (Weigmann & A. Wolff) E. Guého & M.T. Smith, Antonie van Leeuwenhoek 61: 309. 1992.
Cutaneotrichosporon mucoides (E. Guého & M.T. Smith) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813402.
Basionym: Trichosporon mucoides E. Guého & M.T. Smith, Antonie van Leeuwenhoek 61: 312. 1992.
Cutaneotrichosporon oleaginosus (J.J. Zhou, S.O. Suh & Gujjari) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB815305.
Basionym: Trichosporon oleaginosus J.J. Zhou et al., Mycologia 103: 1115. 2011.
Cutaneotrichosporon smithiae (Middelhoven, Scorzetti, Sugita & Fell) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813404.
Basionym: Trichosporon smithiae Middelhoven et al., Int. J. Syst. Evol. Microbiol. 54: 979. 2004.
Cutaneotrichosporon terricola (Sugita, M. Takash. & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813405.
Basionym: Trichosporon terricola Sugita et al., J. Gen. Appl. Microbiol. 48: 295. 2002.
Haglerozyma X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB813406.
Basionym: Trichosporon chiarellii Pagnocca, Legaspe, Rodrigues & Ruivo, Int. J. Syst. Evol. Microbiol. 60: 1457. 2010.
Etymology: The genus is named in honour of Allen N. Hagler for his work on yeast ecology and physiology.
This genus is proposed for the single-species lineage Trichosporon chiarellii, which showed some phylogenetic relatedness to the genus Cutaneotrichosporon in the seven-genes tree (Liu et al. 2015).
Basidiocarps not known. Sexual reproduction not observed. True hyphae sometimes disarticulate into arthroconidia. Pseudohyphae abundant or not. Budding cells present. Fermentation absent. Nitrate and nitrite not utilised. The major CoQ system unknown.
Type species: Haglerozyma chiarellii (Pagnocca, Legaspe, Rodrigues & Ruivo) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. Mycobank MB814765.
Notes: Trichosporon chiarellii was branching earlier than the genus Cutaneotrichosporon in the seven-genes tree with strong posterior probability support from Bayesian analysis but without bootstrap support from ML and NJ analyses (Liu et al. 2015). In the expanded LSU rRNA gene analysis, T. chiarellii was located in the same position without statistical support (Fig. 6), suggesting the species represents a distinct lineage. The genus currently contains one species (Table 1).
Apiotrichum Stautz, Phytopath. Z. 3: 209. 1931. emend. A.M. Yurkov & Boekhout.
= Hyalodendron Diddens, Centbl. Bakt, Parasitkde, Abt. II 90: 316. 1934.
= Protendomycopsis Windisch, Beitr. Biol. Pfl. 41: 355, 1965.
The genus Apiotrichum is resurrected and emended here for the gracile/brassicae and porosum clades containing 18 Trichosporon species (Middelhoven et al., 2004, Sugita, 2011, Liu et al., 2015). Close relationships between these clades were resolved in multi-gene studies by Liu et al. (2015) and Takashima et al. (2015) as well as an extended LSU rRNA gene phylogeny (Fig. 6). Several genus names are available for species contained in this clade. The genus Apiotrichum was described for Apiotrichum (Trichosporon) porosum by Stautz (1931) and Hyalodendron was proposed by Diddens (1934) to accommodate Hyalodendron (Trichosporon) lignicola. The former name has nomenclature priority over the latter. Therefore, Hyalodendron is treated as a synonym of Apiotrichum.
Basidiocarps not known. Sexual reproduction not observed. Septate and branched hyphae present that form arthroconidia. Catenate or lateral conidia occasionally present. Pseudohyphae present. Budding cells present. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-9.
Type species: Apiotrichum porosum Stautz, Phytopath. Z. 3: 209. 1931
Notes: The genus Apiotrichum was proposed with A. porosum as the type species (Stautz 1931). Though this species could not produce fragmenting mycelium, viz. arthroconidia, it was transferred to the genus Trichosporon based on rRNA gene sequence analysis and a similar physiology with other Trichosporon species (Middelhoven et al. 2001). Species belonging to this genus have serotype I and III, and their major coenzyme Q system is Q-9 (Sugita, 2011, Takashima et al., 2015). Close relationships between these three clades received strong support in multi-gene studies by Liu et al. (2015) and Takashima et al. (2015) as well as the extended LSU rRNA gene phylogeny (Fig. 1). For reasons of conservatism we combine the three clades into a single genus Apiotrichum. The genus currently contains 20 species (Table 1).
New combinations for Apiotrichum
Apiotrichum brassicae (Nakase) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813409.
Basionym: Trichosporon brassicae Nakase, J. Gen. Appl. Microbiol. 17: 417. 1971.
Apiotrichum cacaoliposimilis (J.L. Zhou, S.O. Suh & Gujjari) Kachalkin, A.M. Yurkov & Boekhout, comb. nov. MycoBank MB814766.
Basionym: Trichosporon cacaoliposimilis J.L. Zhou et al., Mycologia 103: 1114. 2011.
Apiotrichum dehoogii (Middelhoven, Scorzetti & Fell) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813410.
Basionym: Trichosporon dehoogii Middelhoven et al., Int. J. Syst. Evol. Microbiol. 54: 979. 2004.
Apiotrichum domesticum (Sugita, A. Nishikawa & Shinoda) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813411.
Basionym: Trichosporon domesticum Sugita et al., J. Gen. Appl. Microbiol. 41: 431. 1995.
Apiotrichum dulcitum (Berkhout) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813412.
Basionym: Oospora dulcita Berkhout, Consid. Vég. Vosges: 50–51 + pl. 3. 1923.
≡ Trichosporon dulcitum (Berkhout) Weijman, Antonie van Leeuwenhoek 45: 126. 1979.
Apiotrichum gamsii (Middelhoven, Scorzetti, Sigler & Fell) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813413.
Basionym: Trichosporon gamsii Middelhoven et al., Int. J. Syst. Evol. Microbiol. 54: 982. 2004.
Apiotrichum gracile (Weigmann & A. Wolff) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813414.
Basionym: Oidium gracile Weigmann & A. Wolff, Zentbl. Bakt. ParasitKde, Abt. II 22: 668. 1909.
≡ Trichosporon gracile (Weigmann & A. Wolff) E. Guého & M.T. Smith, Antonie van Leeuwenhoek 61: 307. 1992.
Apiotrichum laibachii (Windisch) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813415.
Basionym: Endomyces laibachii Windisch, Beitr. Biol. Pfl. 41: 356. 1965.
≡ Trichosporon laibachii (Windisch) E. Guého & M.T. Smith, Antonie van Leeuwenhoek 61: 302. 1992.
Apiotrichum lignicola (Diddens) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813416.
Basionym: Hyalodendron lignicola Diddens, Zentbl. Bakt. ParasitKde, Abt. II 90: 317. 1934.
≡ Trichosporon lignicola (Diddens) Fell & Scorzetti, Int. J. Syst. Evol. Microbiol. 54: 997. 2004.
Apiotrichum loubieri (Morenz) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813417.
Basionym: Geotrichum loubieri Morenz, Mykologische Schriftenreihe, Leipzig 2: 46. 1964.
≡ Trichosporon loubieri (Morenz) Weijman, Antonie van Leeuwenhoek 45: 126. 1979.
Apiotrichum montevideense (L.A. Queiroz) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813418.
Basionym: Endomycopsis montevideensis L.A. Queiroz, Mycopath. Mycol. Appl. 51: 311. 1973.
≡ Trichosporon montevideense (L.A. Queiroz) E. Guého & M.T. Smith, Antonie van Leeuwenhoek 61: 301. 1992.
Apiotrichum mycotoxinivorans (O. Molnár, Schatzm. & Prillinger) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813420.
Basionym: Trichosporon mycotoxinivorans O. Molnár et al., Syst. Appl. Microbiol. 27: 664. 2004.
Apiotrichum siamense (Nakase, Jindam., Sugita & H. Kawas.) Kachalkin, A.M. Yurkov & Boekhout, comb. nov. MycoBank MB814767.
Basionym: Trichosporon siamense Nakase et al., Mycoscience 47: 107. 2006.
Apiotrichum scarabaeorum (Middelhoven, Scorzetti & Fell) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813421.
Basionym: Trichosporon scarabaeorum Middelhoven et al., Int. J. Syst. Evol. Microbiol. 54: 981. 2004.
Apiotrichum sporotrichoides (van Oorschot) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813422.
Basionym: Trichosporiella sporotrichoides van Oorschot, Stud. Mycol. 20: 66. 1980.
Apiotrichum vadense (Middelhoven, Scorzetti & Fell) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813423.
Basionym: Trichosporon vadense Middelhoven et al., Int. J. Syst. Evol. Microbiol. 54: 976. 2004.
Apiotrichum veenhuisii (Middelhoven, Scorzetti & Fell) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813424.
Basionym: Trichosporon veenhuisii Middelhoven et al., Int. J. Syst. Evol. Microbiol. 50: 382. 2000.
Apiotrichum wieringae (Middelhoven) A.M. Yurkov & Boekhout, comb. nov. MycoBank MB813425.
Basionym: Trichosporon wieringae Middelhoven, Antonie van Leeuwenhoek 86: 331. 2004.
Apiotrichum xylopini (S.O. Suh, Lee, Gujjari & Zhou) Kachalkin, A.M. Yurkov & Boekhout, comb. nov. MycoBank MB814768.
Basionym: Trichosporon xylopini S.O. Suh et al., Int. J. Syst. Evol. Microbiol. 61: 2540. 2010.
Vanrija R.T. Moore, Bot. Mar. 23: 367. 1980.
= Asterotremella H.J. Prillinger, K. Lopandic, K. Sterflinger, E. Metzger & R. Bauer, ScienceWeek@Austria: 11. 1993. Nomen nudum. (Note: The name Asterotremella is introduced as a nomen nudum as a description, a diagnosis, and information on a type were not provided).
= Asterotremella Prillinger, Lopandic & Sugita, J. Gen, Appl. Microbiol. 53: 171. 2007.
The concept of this genus as emended by Weiss et al. (2014) is accepted.
Basidiocarps not known. Sexual reproduction not observed. Pseudohyphae and true hyphae present. Arthroconidia usually not present. Budding cells present. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-9 or CoQ-10.
Type species: Vanrija humicola (Dasz.) R.T. Moore
Notes: The genus Vanrija was proposed with V. humicola as the type strain (Moore 1980). Following suggestions made by Okoli et al. (2007) and Fonseca et al., 2011, Weiss et al., 2014 emended this genus to accommodate the humicola clade, including five Cryptococcus species, namely C. ramirezgomezianus (= Vanrija albida, basionym Sporobolomyces albidus), C. humicola, C. longa, C. musci, and C. pseudolongus (Takashima et al. 2001), but they did not present an emended diagnosis for the genus. The genus Asterotremella and family Asterotremellaceae based on the study of yeasts isolated from Asterophora basidiocarps has been introduced by Prillinger et al. (1993) and later described (Prillinger et al. 2007), indicating in both cases close relationships with the species C. humicola, the type species of the genus Vanrija (Moore 1980). However, Vanrija has nomenclatural priority over Asterotremella. Two species, which were described as Asterotremella, namely A. meifongana and A. nantouana were placed in the humicola clade in the expanded LSU rRNA gene tree with high bootstrap support (Fig. 6). Cryptococcus fragicola, which did not cluster in the Vanrija clade in Weiss et al. (2014), was shown to be closely related to this clade in the Bayesian and ML trees based on the seven-genes dataset with strong PP and BP support (Liu et al. 2015) as well as in the combined rRNA genes analysis in Wang & Wang (2015). Cryptococcus thermophilus was not included in the phylogenetic analysis performed by Weiss et al. (2014) and clustered loosely with the Vanrija core group in the LSU rRNA gene tree in Takashima et al. (2015) and Wang & Wang (2015). This species, however, clustered inside the clade delimited with Cr. fragicola in Takashima et al. (2015) and in the expanded LSU rRNA gene tree (Fig. 6). Therefore, we transfer these four species to the genus Vanrija. The genus currently contains nine species (Table 1).
New combinations for Vanrija
Vanrija fragicola (M. Takash., Sugita, Shinoda & Nakase) X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB813426.
Basionym: Cryptococcus fragicola M. Takash. et al., Int. J. Syst. Evol. Microbiol. 51: 2205. 2001.
Vanrija meifongana (C.F. Lee) Kachalkin, A.M. Yurkov & Boekhout, comb. nov. MycoBank MB814769.
Basionym: Asterotremella meifongana C.F. Lee, Antonie van Leeuwenhoek 99: 647. 2011.
Vanrija nantouana (C.F. Lee) Kachalkin, A.M. Yurkov & Boekhout, comb. nov. MycoBank MB814770.
Basionym: Asterotremella nantouana C.F. Lee, Antonie van Leeuwenhoek 99: 648. 2011.
Vanrija thermophila (Vogelmann, Chaves & Hertel) Kachalkin, A.M. Yurkov & Boekhout, comb. nov. MycoBank MB814772.
Basionym: Cryptococcus thermophilus Vogelmann et al., Int. J. Syst. Evol. Microbiol. 62: 1719. 2012.
Effuseotrichosporon A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB814773.
Etymology: In reference to the widespread distribution (Lat. effuse) of the species.
This genus is proposed for the single-species lineage formed by Trichosporon vanderwaltii in Trichosporonaceae (Fig. 2, Fig. 6).
Basidiocarps not known. Sexual reproduction not observed. True hyphae disarticulate into arthroconidia. Pseudohyphae abundant or not. Budding cells present. Fermentation absent. Nitrate and nitrite sometimes utilised. The major CoQ system unknown.
Type species: Effuseotrichosporon vanderwaltii (Motaung, Albertyn, Kock, C.F. Lee, S.O. Suh, M. Blackwell & C.H. Pohl) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MB814774.
Basionym: Trichosporon vanderwaltii Motaung et al., Antonie van Leeuwenhoek. 103: 315. 2013.
Notes: Trichosporon vanderwaltii was described for a group of strains from different sources (Motaung et al. 2013). These strains formed a sister clade to the brassicae/porosum clades (i.e. Apiotrichum gen. nov.) in the tree drawn from the ITS and LSU D1/D2 sequences but statistical support for this phylogeny was lacking (Motaung et al. 2013). The affinity of T. vanderwaltii to the family Trichosporonaceae was resolved in this study but its phylogenetic relationships to other clades of the family remained unclear (Fig. 6). This species showed wide geographic distribution and we expect more species to be described in the future. Currently the genus contains one species (Table 1).
Tetragoniomycetaceae Oberw. & Bandoni, Can. J. Bot. 59: 1039. 1981. emend. X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout
This family is emended to accommodate the monophyletic lineage comprising Tetragoniomyces, Takashimella (i.e. the formosensis clade), Cryptocotrichosporon and Cryptococcus marinus recognised in Liu et al. (2015) and the present study (Fig. 1, Fig. 2, Fig. 6).
Basidiocarps not known or growing as a thin film on sclerotia of a basidiomycetous host. Sexual reproduction unknown or by basidia that occur terminally on hyphae; initially thin-walled, ellipsoid to globose, becoming thick-walled and four-celled; mature basidia deciduous, rhomboidal; germination by dikaryotic hyphae or germ tubes that may conjugate. In culture pseudohyphae and true hyphae present or not. Budding cells present or absent. Ballistoconidia bilaterally symmetrical. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-10.
Type genus: Tetragoniomyces Oberw. & Bandoni
Genera accepted: Bandonia, Cryptotrichosporon, Takashimella and Tetragoniomyces.
Notes: The family Tetragoniomycetaceae was established for a single species, Tetragoniomyces uliginosus (Oberwinkler & Bandoni 1981). This species produces unusual deciduous basidia that germinate directly into a filamentous phase and does not have a unicellular yeast phase. Molecular phylogenetic analysis indicated that T. uliginosus is an early-branching member of Trichosporonales with loose connection to Cryptocotrichosporon anacardii (Millanes et al., 2011, Weiss et al., 2014). The latter is branching before the formosensis clade (i.e. Takashimella) with strong support in the Bayesian and ML trees obtained from the seven-genes dataset (Liu et al. 2015). The tree inferred from the expanded LSU rRNA gene dataset confirmed this phylogenetic relationship (Fig. 6). A new Cryptotrichosporon species, C. tibetense that is able to form ballistoconidia was described recently and the genus description emended consequently (Wang & Wang 2015). The close phylogenetic relationship between Cryptotrichosporon and Tetragoniomyces was supported in Wang & Wang (2015). The genus Tetragoniomyces remains monotypic and the genera Cryptotrichosporon (Okoli et al. 2007) and Takashimella (Wang & Wang 2015) contain two and four species, respectively (Table 1). Cryptococcus marinus was branching more early than this family in the Bayesian and ML trees derived from the seven-genes dataset, but its position was not resolved in the seven-genes NJ tree and the combined rRNA genes tree (Liu et al. 2015). In all molecular phylogenetic studies performed to date this species occurred as a single species long-branch lineage (Fell et al., 2000, Scorzetti et al., 2002). Below we propose a new genus Bandonia to accommodate this species.
Bandonia A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, gen. nov. MycoBank MB814775.
Etymology: In honour of Robert J. Bandoni for his valuable contribution to the studies of Tremellomycetes.
The genus is proposed to accommodate the single-species lineage Cryptococcus marinus, which was placed in Tetragoniomycetaceae (Liu et al. 2015, Fig. 6 of this study).
Basidiocarps not known. Sexual reproduction not observed. Pseudohyphae present or not. Budding cells present or absent. Ballistoconidia not observed. Fermentation absent. Nitrate not utilised. Major CoQ system CoQ-10.
Type species: Bandonia marina (van Uden & Zobell) A.M. Yurkov, X.Z. Liu, F.Y. Bai, M. Groenew. & Boekhout, comb. nov. MycoBank MB814776.
Basionym: Candida marina van Uden & Zobell, Antonie van Leeuwenhoek. 28: 278. 1962.
Notes: The phylogenetic position of Cryptococcus marinus in Tremellomycetes remained unclear (e.g. Fonseca et al. 2011) before the seven-genes phylogenetic analysis (Liu et al. 2015). The latter analysis placed the species in Trichosporonales with some affinity to the genera Cryptotrichosporon and Takashimella. The species is known from a single strain only and is characterised by several unusual physiological properties (Fonseca et al. 2011).
Acknowledgements
We thank Walter Gams and David Hawksworth for their nomenclatural advice, and the former also for his help on the correct writing of the names. Masako Takashima helped in finding some original publications in Japanese. Roland Kirschner, Berthold Metzler, Franz Oberwinkler and Hansjoerg Prillinger also provided us some original literature for which they are acknowledged. This study was supported by grants No. 31010103902, No. 30700001 and No. 30970013 from the National Natural Science Foundation of China (NSFC), KSCX2-YW-Z-0936 from the Knowledge Innovation Program of the Chinese Academy of Sciences and grant No. 10CDP019 from the Royal Netherlands Academy of Arts and Sciences (KNAW).
MW acknowledges support from the Swedish Taxonomy Initiative, and from the Swedish Research Council (VR 621-2006-3760, 621-2009-5372, 621-2012-3990). AM acknowledges a grant from the Ministerio de Economía y Competitividad, Spain (CGL2012-40123). AY acknowledges a grant from the Fundação para a Ciência e a Tecnologia, Portugal (grant number PTDC/BIA-BIC/4585/2012). AK acknowledges a grant from the Russian Scientific Foundation, Russia (grant number 14-50-00029). TB is supported by grant NPRP 5-298-3-086 of Qatar Foundation. The authors are solely responsible for the content of this manuscript.
Footnotes
Peer review under responsibility of CBS-KNAW Fungal Biodiversity Centre.
Contributor Information
T. Boekhout, Email: t.boekhout@cbs.knaw.nl.
F.-Y. Bai, Email: baify@im.ac.cn.
References
- Alshahni M.M., Makimura K., Satoh K. Cryptococcus yokohamensis sp. nov., a basidiomycetous yeast isolated from trees and a Queensland koala kept in a Japanese zoological park. International Journal of Systematic and Evolutionary Microbiology. 2011;61:3068–3071. doi: 10.1099/ijs.0.027144-0. [DOI] [PubMed] [Google Scholar]
- Avise J.C., Johns G.C. Proposal for a standardized temporal scheme of biological classification for extant species. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:7358–7363. doi: 10.1073/pnas.96.13.7358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise J.C., Liu J.X. On the temporal inconsistencies of Linnean taxonomic ranks. Biological Journal of the Linnean Society. 2011;102:707–714. [Google Scholar]
- Bab'eva I.P. Tausonia pamirica gen. nov. sp. nov., a psychrophilic yeast-like micromycete from the soils of Pamir. Mikrobiologiia. 1998;67:189–194. [Google Scholar]
- Bab'eva I.P., Lisichkina G.A., Reshetova L.S. Mrakia curviuscula sp. nov.: a new psychrophilic yeast from forest substrates. Microbiology (Moscow) 2002;71:449–454. (translated from Mikrobiologiia71: 526–632) [PubMed] [Google Scholar]
- Bandoni R.J. Fibulobasidium: a new genus in the Sirobasidiaceae. Canadian Journal of Botany. 1979;57:264–268. [Google Scholar]
- Bandoni R.J. The Tremellales and Auriculariales: an alternative classification. Transactions of the Mycological Society of Japan. 1984;25:489–530. [Google Scholar]
- Bandoni R.J. Dimorphic heterobasidiomcetes: taxonomy and parasitism. Studies in Mycology. 1995;38:13–28. [Google Scholar]
- Bandoni R.J., Boekhout T. Trimorphomyces Bandoni & Oberwinkler. In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1591–1594. [Google Scholar]
- Bandoni R.J., Boekhout T. Tremella Persoon (1794) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1567–1590. [Google Scholar]
- Bandoni R.J., Oberwinkler F. On some species of Tremella described by Alfred Möller. Mycologia. 1983;75:854–863. [Google Scholar]
- Bandoni R.J., Sampaio J.P., Boekhout T. Sirobasidium de Lagerheim & Patouillard (1982) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1545–1548. [Google Scholar]
- Barraclough T.G., Humphreys A.M. The evolutionary reality of species and higher taxa in plants: a survey of post-modern opinion and evidence. New Phytologist. 2015;207:291–296. doi: 10.1111/nph.13232. [DOI] [PubMed] [Google Scholar]
- Bauer R., Begerow D., Sampaio J.P. The simple-septate basidiomycetes: a synopsis. Mycological Progress. 2006;5:41–66. [Google Scholar]
- Behrend G. Über Trichomycosis nodosa (Juhel-Rénoy): piedra (Osorio) Berlin Klinische Wochenschrift. 1890;27:464–467. [Google Scholar]
- Berthier J. Une nouvelle tremellale clavarioide Heterocephalacria solida gen. et sp. nov. Mycotaxon. 1980;12:111–116. [Google Scholar]
- Boekhout T., Bai F.Y., Nakashi N. Bullera Derx (1930) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1635–1648. [Google Scholar]
- Boekhout T., Fonseca A., Batenburg-van der Vegte W.H. Bulleromyces genus novum (Tremellales), a teleomorph for Bullera alba, and the occurrence of mating in Bullera variabilis. Antonie van Leeuwenhoek. 1991;59:81–93. doi: 10.1007/BF00445652. [DOI] [PubMed] [Google Scholar]
- Boekhout T., Fonseca A., Sampaio J.P. Classification of heterobasidiomycetous yeasts: characteristics and affiliation of genera to higher taxa of Heterobasidiomycetes. Canadian Journal of Microbiology. 1993;39:276–290. doi: 10.1139/m93-040. [DOI] [PubMed] [Google Scholar]
- Boekhout T., Fonseca Á., Sampaio J.P. Discussion of teleomorphic and anamorphic basidiomycetous yeasts. In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1356–1367. [Google Scholar]
- Botha . Yeasts in soil. In: Peter G., Rosa C.A., editors. Biodiversity and ecophysiology of yeasts. Springer; Berlin: 2006. pp. 21–240. [Google Scholar]
- Castresana J. Cytochrome b phylogeny and the taxonomy of great apes and mammals. Molecular Biology and Evolution. 2001;18:465–471. doi: 10.1093/oxfordjournals.molbev.a003825. [DOI] [PubMed] [Google Scholar]
- Celio G.J., Padamsee M., Dentinger B.T. Assembling the Fungal Tree of Life: constructing the structural and biochemical database. Mycologia. 2006;98:850–859. doi: 10.3852/mycologia.98.6.850. [DOI] [PubMed] [Google Scholar]
- Chen C.J. Morphological and molecular studies in the genus Tremella. Bibliotheca Mycologica. 1998;174:1–225. [Google Scholar]
- Chen R., Jiang Y.-M., Wei S.-C. Kwoniella shandongensis sp. nov., a basidiomycetous yeast isolated from soil and bark from an apple orchard. International Journal of Systematic and Evolutionary Microbiology. 2012;62:2774–2777. doi: 10.1099/ijs.0.039172-0. [DOI] [PubMed] [Google Scholar]
- Colombo A.L., Padovan A.C.B., Chaves G.M. Current knowledge of Trichosporon spp. and Trichosporonosis. Clinical Microbiology Reviews. 2011;24:682–700. doi: 10.1128/CMR.00003-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Palma M., Libkind D., Sampaio J.P. Global distribution, diversity hot spots and niche transitions of an astaxanthin-producing eukaryotic microbe. Molecular Ecology. 2014;23:921–932. doi: 10.1111/mec.12642. [DOI] [PubMed] [Google Scholar]
- de García C., Brizzio S., Russo G. Cryptococcus spencermartinsiae sp. nov., a basidiomycetous yeast isolated from glacial waters and apple fruits. International Journal of Systematic and Evolutionary Microbiology. 2010;60:707–711. doi: 10.1099/ijs.0.013177-0. [DOI] [PubMed] [Google Scholar]
- de Garcia V., Zalar P., Brizzio S. Cryptococcus species (Tremellales) from glacial biomes in the southern (Patagonia) and northern (Svalbard) hemispheres. FEMS Microbiology Ecology. 2012;82:523–539. doi: 10.1111/j.1574-6941.2012.01465.x. [DOI] [PubMed] [Google Scholar]
- Diddens H.A. Eine neue Pilzgattung, Hyalodendron. Zentralblatt für Bakteriologie und Parasitenkunde Abteilung II. 1934;90:315–319. [Google Scholar]
- Farris J.S. Phylogenetic classification of fossils with recent species. Systematic Biology. 1976;25:271–282. [Google Scholar]
- Fell J.W. Mrakia Y. Yamada & Komagata (1987) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1503–1510. [Google Scholar]
- Fell J.W., Blatt G. Separation of strains of the yeasts Xanthophyllomyces dendrorhous and Phaffia rhodozyma based on rDNA IGS and ITS sequence analysis. Journal of Industrial Microbiology & Biotechnology. 1999;21:677–681. doi: 10.1038/sj.jim.2900681. [DOI] [PubMed] [Google Scholar]
- Fell J.W., Boekhout T., Fonseca A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. International Journal of Systematic and Evolutionary Microbiology. 2000;50:1351–1371. doi: 10.1099/00207713-50-3-1351. [DOI] [PubMed] [Google Scholar]
- Fell J.W., Boekhout T., Fonseca A. Basidiomycetous yeasts. In: McLaughlin D.J., McLaughlim E.J., Lemke P., editors. Springer-Verlag; Berlin: 2001. pp. 3–35. (The mycota. Vol. VII. Part B., Systematics and evolution). [Google Scholar]
- Fell J.W., Guého-Kellermann E. Guehomyces Fell & Scorzetti (2004) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1773–1775. [Google Scholar]
- Fell J.W., Johnson E.A., Scorzetti G. Xanthophyllomyces Golubev (1995) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1595–1599. [Google Scholar]
- Fell J.W., Kurtzman C.P., Kwon-Chung K.J. Proposal to conserve Cryptococcus (Fungi) Taxon. 1989;38:151–152. [Google Scholar]
- Fell J.W., Roeijmans H., Boekhout T. Cystofilobasidiales, a new order of basidiomycetous yeasts. International Journal of Systematic and Evolutionary Microbiology. 1999;49:907–913. doi: 10.1099/00207713-49-2-907. [DOI] [PubMed] [Google Scholar]
- Fell J.W., Scorzetti G. Reassignment of the basidiomycetous yeasts Trichosporon pullulans to Guehomyces pullulans gen. nov., comb. nov. and Hyalodendron lignicola to Trichosporon lignicola comb. nov. International Journal of Systematic and Evolutionary Microbiology. 2004;54:995–998. doi: 10.1099/ijs.0.03017-0. [DOI] [PubMed] [Google Scholar]
- Fell J.W., Scorzetti G., Statzell-Tallman A. Molecular diversity and intragenomic variability in the yeast genus Xanthophyllomyces: the origin of Phaffia rhodozyma? FEMS Yeast Research. 2007;7:1399–1408. doi: 10.1111/j.1567-1364.2007.00297.x. [DOI] [PubMed] [Google Scholar]
- Findley K., Rodriguez-Carres M., Metin B. Phylogeny and phenotypic characterization of pathogenic Cryptococcus species and closely related saprobic taxa in the Tremellales. Eukaryotic Cell. 2009;8:353–361. doi: 10.1128/EC.00373-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findley K., Sun S., Fraser J.A. Discovery of a modified tetrapolar sexual cycle in Cryptococcus amylolentus and the evolution of MAT in the Cryptococcus species complex. PLoS Genetics. 2012;8:e1002528. doi: 10.1371/journal.pgen.1002528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca Á., Boekhout T., Fell J.W. Cryptococcus Vuillemin (1901) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1661–1737. [Google Scholar]
- Fonseca Á., Inácio J. Phylloplane yeasts. In: Péter G., Rosa C.A., editors. Biodiversity and ecophysiology of yeasts. Springer; Berlin: 2006. pp. 263–301. [Google Scholar]
- Fries E.M. Systema Mycologicum. 1821;I:lv. [Google Scholar]
- Fungsin B., Takashima M., Sugita T. Bullera koratensis sp. nov. and Bullera lagerstroemiae sp. nov., two new ballistoconidium-forming yeast species in the Trichosporonales clade isolated from plant leaves in Thailand. The Journal of General Applied Microbiology. 2006;52:73–81. doi: 10.2323/jgam.52.73. [DOI] [PubMed] [Google Scholar]
- Gadanho M., Sampaio J.P. Cryptococcus ibericus sp. nov., Cryptococcus aciditolerans sp. nov. and Cryptococcus metallitolerans sp. nov., a new ecoclade of anamorphic basidiomycetous yeast species from an extreme environment associated with acid rock drainage in São Domingos pyrite mine, Portugal. International Journal of Systematic and Evolutionary Microbiology. 2009;59:2375–2379. doi: 10.1099/ijs.0.008920-0. [DOI] [PubMed] [Google Scholar]
- Ginns J. The genus Syzygospora (Heterobasidiomycetes, Syzygosporaceae) Mycologia. 1986;78:619–636. [Google Scholar]
- Göker M., García-Blázquez G., Voglmayr H. Molecular taxonomy of phytopathogenic fungi: a case study in Peronospora. PLoS One. 2009;4:e6319. doi: 10.1371/journal.pone.0006319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göker M., Grimm G.W., Auch A.F. A clustering optimization strategy for molecular taxonomy applied to planktonic foraminifera SSU rDNA. Evolutionary Bioinformatics Online. 2010;6:97–112. doi: 10.4137/ebo.s5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goloboff P.A., Farris J.S., Nixon K.C. TNT, a free program for phylogenetic analysis. Cladistics. 2008;24:774–786. [Google Scholar]
- Golubev W.I. Perfect state of Rhodomyces dendrorhous (Phaffia rhodozyma) Yeast. 1995;11:101–110. doi: 10.1002/yea.320110202. [DOI] [PubMed] [Google Scholar]
- Golubev W., Sampaio J. New filobasidiaceous yeasts found in the phylloplane of a fern. Journal of General and Applied Microbiology. 2009;55:441–446. doi: 10.2323/jgam.55.441. [DOI] [PubMed] [Google Scholar]
- Goto S. On a new yeast genus Naganishia. Journal of Fermentation Technology. 1963;41:459–462. [Google Scholar]
- Guého E., de Hoog G.S., Smith Mth. Neotypification of the genus Trichosporon. Antonie van Leeuwenhoek. 1992;61:285–288. doi: 10.1007/BF00713937. [DOI] [PubMed] [Google Scholar]
- Guého E., Smith MTh, de Hoog G.S. Contributions to a revision of the genus Trichosporon. Antonie van Leeuwenhoek. 1992;61:289–316. doi: 10.1007/BF00713938. [DOI] [PubMed] [Google Scholar]
- Guého E., Smith MTh, de Hoog G.S. Trichosporon Behrend. In: Kurtzman C.P., Fell J.W., editors. The yeasts, a taxonomic study. 4th edn. Elsevier; Amsterdam: 1998. pp. 854–872. [Google Scholar]
- Guerreiro M.A., Springer D.J., Rodrigues J.A. Molecular and genetic evidence for a tetrapolar mating system in the basidiomycetous yeast Kwoniella mangrovensis and two novel sibling species. Eukaryotic Cell. 2013;12:746–760. doi: 10.1128/EC.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen F., Khayhan K., Theelen B. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genetics and Biology. 2015;78:16–48. doi: 10.1016/j.fgb.2015.02.009. [DOI] [PubMed] [Google Scholar]
- Heitman J., Kozel T.R., Kwon-Chung K.J. ASM Press; Washington, DC: 2011. Cryptococcus: from human pathogen to model yeast; pp. 3–620. [Google Scholar]
- Hennig W. Univ. Illinois Press; Urbana: 1966. Phylogenetic systematics. [Google Scholar]
- Hibbett D.A., Binder M., Bischoff J.F. A high-level phylogenetic classification of the Fungi. Mycological Research. 2007;3:509–547. doi: 10.1016/j.mycres.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Holt B.G., Jonsson K.A. Reconciling hierarchical taxonomy with molecular phylogenies. Systematic Biology. 2014;63:1010–1017. doi: 10.1093/sysbio/syu061. [DOI] [PubMed] [Google Scholar]
- Hui F.L., Niu Q.H., Ke T. Cryptococcus nanyangensis sp. nov., a new basidiomycetous yeast isolated from the gut of wood-boring larvae. Current Microbiology. 2012;65:617–621. doi: 10.1007/s00284-012-0203-7. [DOI] [PubMed] [Google Scholar]
- Humphreys A.M., Barraclough T.G. The evolutionary reality of higher taxa in mammals. Proceedings of the Royal Society B-Biological Sciences. 2014;281:2013–2750. doi: 10.1098/rspb.2013.2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inácio J., Portugal L., Spencer-Martins I. Phylloplane yeasts from Portugal: seven novel anamorphic species in the Tremellales lineage of the Hymenomycetes (Basidiomycota) producing orange-coloured colonies. FEMS Yeast Research. 2005;5:1167–1183. doi: 10.1016/j.femsyr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- James T.Y., Kauff F., Schoch C.L. Reconstructing the early evolution of the fungi using a six gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- Johns G.C., Avise J.C. A comparative summary of genetic distances in the vertebrates from the mitochondrial cytochrome b gene. Molecular Biology and Evolution. 1998;15:1481–1490. doi: 10.1093/oxfordjournals.molbev.a025875. [DOI] [PubMed] [Google Scholar]
- Jülich W. Bibliotheca Mycologica. 1981;85:1–485. [Google Scholar]
- Kirk P.M., Cannon P.E., Minter D.W. 10th edn. CAB International; Wallingford: 2008. Ainsworth & Bisby's Dictionary of the Fungi. [Google Scholar]
- Kirschner R., Chen C.J. A new species of Trimorphomyces (Basidiomycota, Tremellales) from Taiwan. Nova Hedwigia. 2008;87:445–455. [Google Scholar]
- Kirschner R., Sampaio J.P., Gadanho M. Cuniculitrema polymorpha (Tremellales, gen. nov. and sp. nov.), a heterobasidiomycete vectored by bark beetles, which is the teleomorph of Sterigmatosporidium polymorphum. Antonie van Leeuwenhoek. 2001;80:149–161. doi: 10.1023/a:1012275204498. [DOI] [PubMed] [Google Scholar]
- Knapp S., McNeill J., Turland N. Changes to publication requirements made at the XVIII International Botanical Congress in Melbourne – what does e-publication mean for you? MycoKeys. 2011;1:21–27. doi: 10.1186/1471-2148-11-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraepelin G., Schulze U. Sterigmatosporidium gen. n., a new heterothallic basidiomycetous yeast, the perfect state of a new species of Sterigmatomyces fell. Antonie van Leeuwenhoek. 1982;48:471–483. doi: 10.1007/BF00448420. [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P. Phylogenetic circumscription of Saccharomyces, Kluyveromyces and other members of the Saccharomycetaceae, and the proposal of the new genera Lachancea, Nakaseomyces, Naumovia, Vanderwaltozyma and Zygotorulaspora. FEMS Yeast Research. 2003;4:233–245. doi: 10.1016/S1567-1356(03)00175-2. [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P., Albertyn J., Basehoar-Powers E. Multigene phylogenetic analysis of the Lipomycetaceae and the proposed transfer of Zygozyma species to Lipomyces and Babjevia anomala to Dipodascopsis. FEMS Yeast Research. 2007;7:1027–1034. doi: 10.1111/j.1567-1364.2007.00246.x. [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. [Google Scholar]
- Kurtzman C.P., Fell J.W., Boekhout T. Methods for isolation, phenotypic characterization and maintenance of yeasts. In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 87–110. [Google Scholar]
- Kurtzman C.P., Robnett C.J. Phylogenetic relationships among yeasts of the ‘Saccharomyces complex’ determined from multigene sequence analyses. FEMS Yeast Research. 2003;3:417–432. doi: 10.1016/S1567-1356(03)00012-6. [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P., Robnett C.J. Multigene phylogenetic analysis of the Trichomonascus, Wickerhamiella and Zygoascus yeast clades, and the proposal of Sugiyamaella gen. nov. and 14 new species combinations. FEMS Yeast Research. 2007;7:141–151. doi: 10.1111/j.1567-1364.2006.00157.x. [DOI] [PubMed] [Google Scholar]
- Kurtzman C.P., Robnett C.J., Basehoar-Powers E. Phylogenetic relationships among species of Pichia, Issatchenkia and Williopsis determined from multigene sequence analysis, and the proposal of Barnettozyma gen. nov., Lindnera gen. nov. and Wickerhamomyces gen. nov. FEMS Yeast Research. 2008;8:939–954. doi: 10.1111/j.1567-1364.2008.00419.x. [DOI] [PubMed] [Google Scholar]
- Kützing F.T. 1833. Algarum aquae dulcis Germanicarum, Decas III. [Google Scholar]
- Kwon-Chung K.J. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia. 1975;67:1197–1200. [PubMed] [Google Scholar]
- Kwon-Chung K.J. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia. 1976;68:942–946. [PubMed] [Google Scholar]
- Kwon-Chung K.J. Filobasidiaceae – a taxonomic survey. Studies in Mycology. 1987;30:75–85. [Google Scholar]
- Kwon-Chung K.J. Filobasidiella Kwon-Chung (1975) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1443–1455. [Google Scholar]
- Kwon-Chung K.J., Chang Y.C., Bauer R. The characteristics that differentiate Filobasidiella depauperata from Filobasidiella neoformans. Studies in Mycology. 1995;38:67–79. [Google Scholar]
- Landell M.F., Brandão L.R., Safar S.V. Bullera vrieseae sp. nov., a tremellaceous yeast species isolated from bromeliads. International Journal of Systematic and Evolutionary Microbiology. 2015;65:2466–2471. doi: 10.1099/ijs.0.000285. [DOI] [PubMed] [Google Scholar]
- Landell M.F., Inácio J., Fonseca Á. Cryptococcus bromeliarum sp. nov., an orange-coloured basidiomycetous yeast isolated from bromeliads in Brazil. International Journal of Systematic and Evolutionary Microbiology. 2009;59:910–913. doi: 10.1099/ijs.0.005652-0. [DOI] [PubMed] [Google Scholar]
- Lindgren H., Diederich P., Goward T. The phylogenetic analysis of fungi associated with lichenized ascomycete genus Bryoria reveals new lineages in the Tremellales including a new species Tremella huuskonenii hyperparasitic on Phacopsis huuskonenii. Fungal Biolology. 2015;119:844–856. doi: 10.1016/j.funbio.2015.06.005. [DOI] [PubMed] [Google Scholar]
- Liu X.Z., Wang Q.M., Theelen B. Phylogeny of tremellomycetous yeasts and related dimorphic and filamentous basidiomycetes reconstructed from multiple gene sequence analyses. Studies in Mycology. 2015;81:1–26. doi: 10.1016/j.simyco.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder J., Kreger-van Rij N.J.W. North-Holland; Amsterdam: 1952. The yeasts, a taxonomic study. [Google Scholar]
- Lopandic K., Prillinger H., Wuczkowski M. Fellomyces Y. Yamada & Banno (1984) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1759–1772. [Google Scholar]
- Lumbsch H.T. How objective are genera in euascomycetes? Perspectives in Plant Ecology Evolution and Systematics. 2002;5:91–101. [Google Scholar]
- Margesin R., Fell J.W. Mrakiella cryoconiti gen. nov., sp. nov., a psychrophilic, anamorphic, basidiomycetous yeast from alpine and arctic habitats. International Journal of Systematic and Evolutionary Microbiology. 2008;58:2977–2982. doi: 10.1099/ijs.0.2008/000836-0. [DOI] [PubMed] [Google Scholar]
- McLaughlin D.J., Frieders E.M., Lü H. A microscopist's view of heterbasidiomycete phylogeny. Studies in Mycology. 1995;38:91–109. [Google Scholar]
- McNeil L.K., Barrie F.R., Burdet H.M. 2006. International Code of Botanical Nomenclature (Vienna Code) pp. 1–568. Regnum Veg. 146 (XVI) [Google Scholar]
- McNeill J., Barrie F.R., Buck W.R. A.R.G. Gantner Verlag KG; Ruggell, Liechtenstein: 2012. International Code of Nomenclature for algae, fungi, and plants (Melbourne Code)http://www.iapt-taxon.org/nomen/main.php Regnum Vegetabile 154. Available at: [Google Scholar]
- McPartland M.M., Goff J.P. Neotypification of Trichosporon beigelii: morphological, pathological and taxonomic consideration. Mycotaxon. 1991;41:173–178. [Google Scholar]
- Meier-Kolthoff J.P., Auch A.F., Klenk H.-P. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff J.P., Klenk H.P., Göker M. Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. International Journal of Systematic and Evolutionary Microbiology. 2014;64:352–356. doi: 10.1099/ijs.0.056994-0. [DOI] [PubMed] [Google Scholar]
- Metzler B., Oberwinkler F., Petzold H. Rhynchogastrema gen. nov. and Rhynchogastremaceae fam. nov. (Tremellales) Systematic and Applied Microbiology. 1989;12:280–287. [Google Scholar]
- Middelhoven W.J., Scorzetti G., Fell J.W. Trichosporon porosum comb. nov., an anamorphic basidiomycetous yeast inhabiting soil, related to the loubieri/laibachii group of species that assimilate hemicelluloses and phenolic compounds. FEMS Yeast Research. 2001;1:15–22. doi: 10.1111/j.1567-1364.2001.tb00009.x. [DOI] [PubMed] [Google Scholar]
- Middelhoven W.J., Scorzetti G., Fell J.W. Systematics of the anamorphic basidiomycetous yeast genus Trichosporon Behrend with the description of five novel species: Trichosporon vadense, T. smithiae, T. dehoogii, T. scarabaeorum and T. gamsii. International Journal of Systematic and Evolutionary Microbiology. 2004;54:975–986. doi: 10.1099/ijs.0.02859-0. [DOI] [PubMed] [Google Scholar]
- Millanes A.M., Diederich P., Ekman S. Phylogeny and character evolution in the jelly fungi (Tremellomycetes, Basidiomycota, Fungi) Molecular Phylogenetics and Evolution. 2011;61:12–28. doi: 10.1016/j.ympev.2011.05.014. [DOI] [PubMed] [Google Scholar]
- Miller M.W., Yoneyama M., Soneda M. Phaffia, a new yeast genus in the Deuteromycotina (Blastomycetes) International Journal of Systematic Bacteriology. 1976;26:286–291. [Google Scholar]
- Möller A. Protobasidiomyceten. In: Schimper A.F.W., editor. Vol. 8. Gustav Fischer; Jena: 1895. pp. 1–179. (Botanische Mitteilungen aus den Tropen). [Google Scholar]
- Monaghan M.T., Wild R., Elliot M. Accelerated species inventory on Madagascar using coalescent-based models of species delineation. Systematic Biology. 2009;58:298–311. doi: 10.1093/sysbio/syp027. [DOI] [PubMed] [Google Scholar]
- Moore R.T. Taxonomic proposals for the classification of marine yeasts and other yeast-like fungi including the smuts. Botanica Marina. 1980;23:361–373. [Google Scholar]
- Motaung T.E., Albertyn J., Kock J.L. Trichosporon vanderwaltii sp. nov., an asexual basidiomycetous yeast isolated from soil and beetles. Antonie van Leeuwenhoek. 2013;103:313–319. doi: 10.1007/s10482-012-9811-2. [DOI] [PubMed] [Google Scholar]
- Nakase T., Jan-ngam H., Tsuzuki S. Two new ballistoconidium-forming yeast species, Bullera melastomae and Bullera formosana, found in Taiwan. Systematic and Applied Microbiology. 2004;27:558–564. doi: 10.1078/0723202041748118. [DOI] [PubMed] [Google Scholar]
- Nakase T., Takematsu A., Yamada Y. Molecular approaches to the taxonomy of ballistosporous yeasts based on the analysis of the partial nucleotide sequences of 18S ribosomal ribonucleic acids. The Journal of General and Applied Microbiology. 1993;39:107–134. [Google Scholar]
- Nannizzi A. Vol. 4. Siena; 1934. pp. 1–557. (Repertorio sistematico dei miceti dell' uomo e degli animali). [Google Scholar]
- Nguyen T.H., Fleet G.H., Rogers P.L. Composition of the cell walls of several yeast species. Applied Microbiology and Biotechnology. 1998;50:206–212. doi: 10.1007/s002530051278. [DOI] [PubMed] [Google Scholar]
- Niwata Y., Takashima M., Tornai-Lehoczki J. Udeniomyces pannonicus sp. nov., a ballisto-conidium-forming yeast isolated from leaves of plants in Hungary. International Journal of Systematic and Evolutionary Microbiology. 2002;52:1887–1892. doi: 10.1099/00207713-52-5-1887. [DOI] [PubMed] [Google Scholar]
- Oberwinkler F., Bandoni R.J. Tetragoniomyces gen. nov. and Tetragoniomycetaceae fam. nov. (Tremellales) Canadian Journal of Botany. 1981;59:1034–1040. [Google Scholar]
- Oberwinkler F., Bandoni R.J. Carcinomycetaceae: a new family in the Heterobasidiomycetes. Nordic Journal of Botany. 1982;2:501–516. [Google Scholar]
- Oberwinkler F., Bauer R., Schneller J. Phragmoxenidium mycophilum sp. nov., an unusual mycoparasitic heterobasidiomycete. Systematic and Applied Microbiology. 1990;13:186–191. [Google Scholar]
- Okoli I., Oyeka C.A., Kwon-Chung K.J. Cryptotrichosporon anacardii gen. nov., sp. nov., a new Trichosporonoid capsulate basidiomycetous yeast from Nigeria that is able to form melanin on niger seed agar. FEMS Yeast Research. 2007;7:339–350. doi: 10.1111/j.1567-1364.2006.00164.x. [DOI] [PubMed] [Google Scholar]
- Olive L.S. An unusual heterobasidiomycete with Tilletia-like basidia. Journal of the Elisha Mitchell Scientific Society. 1968;84:261–266. [Google Scholar]
- Paradis E. Springer; New York: 2006. Analysis of phylogenetics and evolution with R. [Google Scholar]
- Patouillard N.T. Lucien Declure; Lons-le-Saunier: 1900. Essai Taxonomique sur les Familles et les Genres des Hyménomycètes; pp. 17–20. (in French) [Google Scholar]
- Pattengale N.D., Alipour M., Bininda-Emonds O.R.P. How many bootstrap replicates are necessary? Lecture Notes in Computer Science. 2009;5541:184–200. [Google Scholar]
- Pons J., Barraclough T.G., Gomez-Zurita J. Sequence-based species delimitation for the DNA taxonomy of undescribed insects. Systematic Biology. 2006;55:595–609. doi: 10.1080/10635150600852011. [DOI] [PubMed] [Google Scholar]
- Prillinger H., Lopandic K., Sugita T. Asterotremella gen. nov. albida, an anamorphic tremelloid yeast isolated from the agarics Asterophora lycoperdoides and Asterophora parasitica. The Journal of General and Applied Microbiology. 2007;53:167–175. doi: 10.2323/jgam.53.167. [DOI] [PubMed] [Google Scholar]
- Prillinger H., Oberwinkler F., Umile C. Analysis of cell wall carbohydrates (neutral sugars) from ascomycetous and basidiomycetous yeasts with and without derivatization. The Journal of General and Applied Microbiology. 1993;39:1–34. [Google Scholar]
- Rabenhorst L. Zwei Parasiten an den todten haaren der Chignons. Hedwigia. 1867;6:1–49. [Google Scholar]
- R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2015. R: A language and environment for statistical computing.https://www.R-project.org/ [Google Scholar]
- Rea C. New and rare British fungi. Transactions of the British Mycological Society. 1912;3:376–380. [Google Scholar]
- Roberts P. British Tremella species II: T. encephala, T. steidleri & T. foliacea. Mycologist. 1999;13:127–131. [Google Scholar]
- Rodriguez-Carres M., Findley K., Sun S. Morphological and genomic characterization of Filobasidiella depauperata: a homothallic sibling species of the pathogenic Cryptococcus species complex. PLoS One. 2010;5:e9620. doi: 10.1371/journal.pone.0009620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo G., Libkind D., Ulloa R.J. Cryptococcus agrionensis sp. nov., a basidiomycetous yeast of the acidic rock drainage ecoclade, isolated from an acidic aquatic environment of volcanic origin. International Journal of Systematic and Evolutionary Microbiology. 2010;60:996–1000. doi: 10.1099/ijs.0.012534-0. [DOI] [PubMed] [Google Scholar]
- Sampaio J.P. Diversity, phylogeny and classification of basidiomycetous yeasts. In: Agerer R., Piepenbring M., Blanz P., editors. Frontiers in basidiomycote mycology. IHW-Verlag; Eching: 2004. pp. 49–80. [Google Scholar]
- Sampaio J.P. Cystofilobasidium Oberwinkler & Bandoni (1983) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1423–1432. [Google Scholar]
- Sampaio J.P. Tausonia Bab'eva (1998) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1999–2001. [Google Scholar]
- Sampaio J.P. Auriculibuller Sampaio (2004) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1379–1381. [Google Scholar]
- Sampaio J.P., Fonseca A. Physiological aspects in the systematic of heterobasidiomycetous yeasts. Studies in Mycology. 1995;38:29–46. [Google Scholar]
- Sampaio J.P., Inacio J., Fonseca A. Auriculibuller fuscus gen. nov., sp. nov. and Bullera japonica sp. nov., novel taxa in the Tremellales. International Journal of Systematic and Evolution Microbiology. 2004;54:987–993. doi: 10.1099/ijs.0.02970-0. [DOI] [PubMed] [Google Scholar]
- Sampaio J.P., Wei β M., Gadanho M. New taxa in the Tremellales: Bulleribasidium oberjochense gen. et sp. nov., Papiliotrema bandonii gen. et sp. nov. and Fibulobasidium murrhardtense sp. nov. Mycologia. 2002;94:873–887. [PubMed] [Google Scholar]
- Sanderson M.J. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Molecular Biology and Evolution. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Satoh K., Maeda M., Umeda Y. Cryptococcus lacticolor sp. nov. and Rhodotorula oligophaga sp. nov., novel yeasts isolated from the nasal smear microbiota of Queensland koalas kept in Japanese zoological parks. Antonie van Leeuwenhoek. 2013;104:83–93. doi: 10.1007/s10482-013-9928-y. [DOI] [PubMed] [Google Scholar]
- Scorzetti G., Fell J.W., Fonseca A. Systematics of basidiomycetous yeasts: a comparison of large subunit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Research. 2002;2:495–517. doi: 10.1111/j.1567-1364.2002.tb00117.x. [DOI] [PubMed] [Google Scholar]
- Shin K.S., Oh H.M., Park Y.H. Cryptococcus mujuensis sp. nov. and Cryptococcus cuniculi sp. nov., basidiomycetous yeasts isolated from wild rabbit faeces. International Journal of Systematic and Evolutionary Microbiology. 2006;56:2241–2244. doi: 10.1099/ijs.0.64353-0. [DOI] [PubMed] [Google Scholar]
- Sivakumaran S., Bridge P., Roberts P. Genetic relatedness among Filobasidiella species. Mycopathologia. 2002;156:157–162. doi: 10.1023/a:1023309311643. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Systematic Biology. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Statzell-Tallman A.1, Belloch C., Fell J.W. Kwoniella mangroviensis gen. nov., sp. nov. (Tremellales, Basidiomycota), a teleomorphic yeast from mangrove habitats in the Florida Everglades and Bahamas. FEMS Yeast Research. 2008;8:103–113. doi: 10.1111/j.1567-1364.2007.00314.x. [DOI] [PubMed] [Google Scholar]
- Stautz W. Beiträge zur Schleimflussfrage. Phytopathologische Zeitschrift. 1931;3:163–229. [Google Scholar]
- Stielow B., Bratek Z., Orczán A.K.I. Species delimitation in taxonomically difficult fungi: the case of Hymenogaster. PLoS One. 2011;6:e15614. doi: 10.1371/journal.pone.0015614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S., Nakase T. Molecular phylogenetic study of the basidiomycetous anamorphic yeast genus Trichosporon and related taxa based on small subunit ribosomal DNA sequences. Mycoscience. 1998;39:7–13. [Google Scholar]
- Sugita T. Trichosporon Behrend (1890) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 2015–2061. [Google Scholar]
- Sugita T., Ikeda R., Nishikawa A. Analysis of Trichosporon isolates obtained from the houses of patients with summer-type hypersensitivity pneumonitis. Journal of Clinical Microbiology. 2004;42:5467–5471. doi: 10.1128/JCM.42.12.5467-5471.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Metin B., Findley K. Validation of Kwoniella heveanensis, teleomorph of the basidiomycetous yeast Cryptococcus heveanensis. Mycotaxon. 2011;116:227–229. [Google Scholar]
- Sylvester K., Wang Q.M., James B. Temperature and host preferences drive the diversification of Saccharomyces and other yeasts: a survey and the discovery of eight new yeast species. FEMS Yeast Research. 2015 doi: 10.1093/femsyr/fov002. Epub 2015 Mar 4. [DOI] [PubMed] [Google Scholar]
- Takashima M., Hamamoto M., Nakase T. Taxonomic significance of fucose in the class Urediniomycetes: distribution of fucose in cell wall and phylogeny of urediniomycetous yeasts. Systematic and Applied Microbiology. 2000;23:63–70. doi: 10.1016/S0723-2020(00)80047-8. [DOI] [PubMed] [Google Scholar]
- Takashima M., Manabe R., Iwasaki W. Selection of orthologous genes for construction of a highly resolved phylogenetic tree and clarification of the phylogeny of Trichosporonales species. PLoS One. 2015;10:e0131217. doi: 10.1371/journal.pone.0131217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima M., Nakase T. Molecular phylogeny of the genus Cryptococcus and related species based on the sequences of SSU rDNA and internal transcribed spacer regions. Microbiology and Culture Collection. 1999;15:35–47. [Google Scholar]
- Takashima M., Nakase T. Kockovaella Nakase, Banno & Y. Yamada (1991) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The yeasts: a taxonomic study. Elsevier; Amsterdam: 2011. pp. 1781–1794. [Google Scholar]
- Takashima M., Sugita T., Shinoda T. Reclassification of the Cryptococcus humicola complex. International Journal of Systematic and Evolutionary Microbiology. 2001;51:2199–2210. doi: 10.1099/00207713-51-6-2199. [DOI] [PubMed] [Google Scholar]
- Takashima M., Sugita T., Shinoda T. Three new combinations from the Cryptococcus laurentti complex: Cryptococcus aureus, Cryptococcus carnescens and Cryptococcus peneaus. International Journal of Systematic and Evolutionary Microbiology. 2003;53:1187–1194. doi: 10.1099/ijs.0.02498-0. [DOI] [PubMed] [Google Scholar]
- Talavera G., Lukhtanov V.A., Pierce N.E. Establishing criteria for higher-level classification using molecular data: the systematics of Polyommatus blue butterflies (Lepidoptera, Lycaenidae) Cladistics. 2013;29:166–192. doi: 10.1111/j.1096-0031.2012.00421.x. [DOI] [PubMed] [Google Scholar]
- Tanimura A., Takashima M., Sugita T. Cryptococcus terricola is a promising oleaginous yeast for biodiesel production from starch through consolidated bioprocessing. Scientific Reports. 2014;4:4776. doi: 10.1038/srep04776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau T., Atkinson B., Ripley B. 2015. rpart: Recursive Partitioning and Regression Trees.http://CRAN.R-project.org/package=rpart R package version 4.1–10. [Google Scholar]
- Tindall B.J., Rosselló-Móra R., Busse H.J. Notes on the characterization of prokaryote strains for taxonomic purposes. International Journal of Systematic and Evolutionary Microbiology. 2010;60:249–266. doi: 10.1099/ijs.0.016949-0. [DOI] [PubMed] [Google Scholar]
- Tsuchiya T., Fukazawa Y., Taguchi M. Serologic aspects on yeast classification. Mycopathologia et Mycologia Applicata. 1974;53:77–91. doi: 10.1007/BF02127199. [DOI] [PubMed] [Google Scholar]
- Turchetti B., Selbmann L., Blanchette R.A. Cryptococcus vaughanmartiniae sp. nov. and Cryptococcus onofrii sp. nov.: two new species isolated from worldwide cold environments. Extremophiles. 2015;19:149–159. doi: 10.1007/s00792-014-0692-3. [DOI] [PubMed] [Google Scholar]
- Valente P., Boekhout T., Landell M.F. Bandoniozyma gen. nov., a genus of germentative and non-fermentative tremellaceous yeast species. PLoS One. 2012;7:e46060. doi: 10.1371/journal.pone.0046060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Walt J.P. Criteria and methods used in classification. In: Lodder J., editor. The yeasts, a taxonomic study. 2nd edn. North-Holland; Amsterdam: 1970. pp. 34–113. [Google Scholar]
- Van der Walt J.P. The yeasts – a conspectus. In: de Hoog G.S., Smith MTh, Weijman A.C.M., editors. The expanding realm of yeast-like fungi. Elsevier; Amsterdam: 1987. pp. 19–32. (Stud. Mycol. 30, 19–31) [Google Scholar]
- Van der Walt J.P., Yarrow D. Methods for the isolation, maintenance, classification and identification of yeasts. In: Kreger-van Rij N.J.W., editor. The yeasts, a taxonomic study. 3rd edn. Elsevier; Amsterdam: 1984. pp. 45–104. [Google Scholar]
- Vences M., Guayasamin J.M., Miralles A. To name or not to name: criteria to promote economy of change in Linnaean classification schemes. Zootaxa. 2013;3636:201–244. doi: 10.11646/zootaxa.3636.2.1. [DOI] [PubMed] [Google Scholar]
- Vogelmann S.A., Chaves S., Hertel C. Cryptococcus thermophilus sp. nov., isolated from cassava sourdough. International Journal of Systematic and Evolutionary Microbiology. 2012;62:1715–1720. doi: 10.1099/ijs.0.032748-0. [DOI] [PubMed] [Google Scholar]
- Vuillemin P. Les blastomycètes pathogènes. Revue Générale des Sciences Pures et Appliquées. 1901;12:732–751. [Google Scholar]
- Wang L., Wang Q.M. Molecular phylogenetic analysis of ballistoconidium-forming yeasts in Trichosporonales (Tremellomycetes): a proposal for Takashimella gen. nov. and Cryptotrichosporon tibetense sp. nov. PLoS One. 2015;10:e0132653. doi: 10.1371/journal.pone.0132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q.M., Bai F.Y. Molecular phylogeny of basidiomycetous yeasts in the Cryptococcus luteolus lineage (Tremellales) based on nuclear rRNA and mitochondrial cytochrome b gene sequence analyses: proposal of Derxomyces gen. nov. and Hannaella gen. nov., and description of eight novel Derxomyces species. FEMS Yeast Research. 2008;8:799–814. doi: 10.1111/j.1567-1364.2008.00403.x. [DOI] [PubMed] [Google Scholar]
- Wang Q.M., Bai F.Y., Fungsin B. Proposal of Mingxiaea gen. nov. for the anamorphic basidiomycetous yeast species in the Bulleribasidium clade (Tremellales) based on molecular phylogenetic analysis, with six new combinations and four novel species. International Journal of Systematic and Evolutionary Microbiology. 2011;61:210–219. doi: 10.1099/ijs.0.019299-0. [DOI] [PubMed] [Google Scholar]
- Wang Q.M., Wang S.A., Jia J.H. Cryptococcus tibetensis sp. nov., a novel basidiomycetous anamorphic yeast species isolated from plant leaves. The Journal of General and Applied Microbiology. 2007;53:281–285. doi: 10.2323/jgam.53.281. [DOI] [PubMed] [Google Scholar]
- Weijman A.C. Carbohydrate composition and taxonomy of Geotrichum, Trichosporon and allied genera. Antonie Van Leeuwenhoek. 1979;45:119–127. doi: 10.1007/BF00400785. [DOI] [PubMed] [Google Scholar]
- Weiss M., Bauer R., Begerow D. Spotlights on Heterobasidiomycetes. In: Agerer R., Blanz P., Piepenbring M., editors. Frontiers in basidiomycote mycology. IHW-Verlag; Eching: 2004. pp. 7–48. [Google Scholar]
- Weiss M., Bauer R., Sampaio J.P. Tremellomycetes and related groups. In: McLaughlin D.J., Spatafora J.W., editors. Springer-Verlag; Berlin: 2014. pp. 349–350. (Systematics and evolution, The mycota VII Part A). [Google Scholar]
- Wells K. Jelly fungi, then and now! Mycologia. 1994;86:18–48. [Google Scholar]
- Wells K., Bandoni R.J. Heterobasidiomycetes. In: McLaughlin D.J., McLaughlin E.G., Lemke P.A., editors. Springer-Verlag; Berlin: 2001. pp. 85–120. (The Mycota VII, Systematics and Evolution, Part B). [Google Scholar]
- Wiley E., Lieberman B. John Wiley & Sons Inc.; Hoboken (New Jersey): 2011. Phylogenetics: theory and practice of phylogenetic systematics. [Google Scholar]
- Wuczkowski M., Passoth V., Turchetti B. Description of Holtermanniella takashimae sp. nov., Holtermanniella gen. nov. and proposal of the order Holtermanniales to accommodate Tremellomycetous yeasts of the Holtermanniales clade. Internatonal Journal of Systematic and Evolutionary Microbiology. 2011;61:680–689. doi: 10.1099/ijs.0.019737-0. [DOI] [PubMed] [Google Scholar]
- Yarrow D. Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman C.P., Fell J.W., editors. The yeasts, a taxonomic study. 4th edn. Elsevier; Amsterdam: 1998. pp. 77–100. [Google Scholar]
- Yarza P., Yilmaz P., Pruesse E. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nature Review Microbiology. 2014;12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- Yurkov A.M., Inácio J., Chernov I.Y. Yeast biogeography and the effects of species recognition approaches: the case study of widespread basidiomycetous species from brich forests in Russia. Current Microbiology. 2015;70:587–601. doi: 10.1007/s00284-014-0755-9. [DOI] [PubMed] [Google Scholar]
- Yurkov A.M., Kemler M., Begerow D. Assessment of yeast diversity in soils under different management regions. Fungal Ecology. 2012;5:24–35. [Google Scholar]
- Zang M. A new taxon, Filobasidiella xianghuijun Zang, associated with Tremella fuciformis. Edible Fungi of China. 1999;18:43–44. [Google Scholar]
- Zhang T., Zhang Y.Q., Liu H.Y. Cryptococcus fildesensis sp. nov. a psychrophilic basidiomycetous yeast isolated from Antarctic moss. International Journal of Systematic and Evolutionary Microbiology. 2014;64:675–679. doi: 10.1099/ijs.0.054981-0. [DOI] [PubMed] [Google Scholar]