Abstract
Chronic kidney disease (CKD) is associated with an increase in serum retinol; however, the underlying mechanisms of this disorder are poorly characterized. Here, we found that the alteration of hepatic metabolism induced the accumulation of serum retinol in 5/6 nephrectomy (5/6Nx) mice. The liver is the major organ responsible for retinol metabolism; accordingly, microarray analysis revealed that the hepatic expression of most CYP genes was changed in 5/6Nx mice. In addition, D-box-binding protein (DBP), which controls the expression of several CYP genes, was significantly decreased in these mice. Cyp3a11 and Cyp26a1, encoding key proteins in retinol metabolism, showed the greatest decrease in expression in 5/6Nx mice, a process mediated by the decreased expression of DBP. Furthermore, an increase of plasma transforming growth factor-β1 (TGF-β1) in 5/6Nx mice led to the decreased expression of the Dbp gene. Consistent with these findings, the alterations of retinol metabolism and renal dysfunction in 5/6Nx mice were ameliorated by administration of an anti-TGF-β1 antibody. We also show that the accumulation of serum retinol induced renal apoptosis in 5/6Nx mice fed a normal diet, whereas renal dysfunction was reduced in mice fed a retinol-free diet. These findings indicate that constitutive Dbp expression plays an important role in mediating hepatic dysfunction under CKD. Thus, the aggravation of renal dysfunction in patients with CKD might be prevented by a recovery of hepatic function, potentially through therapies targeting DBP and retinol.
Keywords: circadian clock, hepatocyte, renal physiology, retinol, transforming growth factor beta (TGF-beta)
Introduction
Chronic kidney disease (CKD)3 affects the functions of various organs (1, 2) and can cause cardiovascular dysfunction and heart and liver failure (3–5), thus reducing quality of life. Impaired renal function inhibits the production of various metabolites in the body and aggravates renal fibrosis in CKD. The uremic toxins that accumulate during the early stages of CKD promote renal cell apoptosis via transforming growth factor-β1 (TGF-β1) (6). Renal dysfunction in CKD is further promoted by signaling via the renin-angiotensin-aldosterone system (7). Because inhibition of these signaling pathways is insufficient for renal protection, the development of novel therapies is needed for treating renal fibrosis and dysfunctions.
Circadian rhythms have been observed in many biological systems and numerous physiological functions (8, 9). In mammals, the master pacemaker controlling circadian rhythms is the hypothalamic suprachiasmatic nucleus. Recent molecular dissection of the circadian clock system has revealed that the core circadian oscillator consists of a transcription-translation feedback loop. CLOCK and BMAL1 form a heterodimer and activate transcription of the period (PER) and cryptochrome (CRY) genes. Once the PER and CRY proteins reach a critical concentration, they attenuate CLOCK/BMAL1 transactivation, thereby generating circadian oscillation through their own transcription (9, 10). The clock genes constituting the core oscillation loop control downstream events by regulating the rhythmic expression of clock-controlled genes (11). The existence of peripheral clocks in the liver, kidney, lung, and other organs is important for regulating various biological functions (12, 13). Thus, the efficacies and toxicities of many drugs vary depending on dosing times associated with the 24-h rhythms of biochemical, physiological, and behavioral processes under the control of the circadian clock (14). Recently, several clock genes have been found to control the circadian rhythms of cytochrome P450 (CYP) genes (14). We confirmed that expression of the CYP3A4 gene, encoding an important enzyme mediating xenobiotic metabolism in humans, exhibits a circadian rhythm in HepG2 cells (14). Disruption of these molecular clockworks has been linked to the onset of diseases such as cancer, metabolic syndrome, and diabetes in both humans and animal models (15, 16).
Recently, metabolic dysfunction has been also demonstrated in patients with CKD (17–19). This is related, in part, to the fact that CKD affects not only the renal clearance of drugs but also the metabolism of xenobiotics and bioactive compounds by inhibiting key enzymatic systems within the liver. Indeed, several pharmacokinetics studies have demonstrated that the non-renal clearance of multiple drugs is reduced in patients with CKD (20, 21). In addition, the serum concentration of retinol, which in bioactive compounds is a fat-soluble vitamin A, is also elevated in patients with CKD (22–24); however, the influences of excessive retinol on CKD are poorly understood. Furthermore, almost all retinol is ingested via food and is metabolized primarily by the liver. Several CYPs are involved in retinol metabolism (25–27), but little is known regarding the relationship between CYPs and retinol in CKD. In the hepatic dysregulation observed in patients with CKD, patient serum has been shown to contain a mediator that can decrease CYPs in hepatocytes (28–34). However, the mechanisms and factors that decrease hepatic CYP gene expression under CKD remain to be elucidated.
In this study, we aimed to identify the increase mechanisms of serum retinol and to provide evidence that the amelioration of hepatic dysfunction would be beneficial to reduce the symptoms of CKD. We utilized wild-type 5/6 nephrectomy (5/6Nx) CKD model mice to examine the expression and regulatory pathways of proteins involved in liver metabolism and their regulation by the local circadian clock.
Experimental Procedures
Mouse Experiments
Four-week-old male ICR mice (Charles River Laboratories Japan, Inc., Yokohama, Japan) were housed in a light-controlled room (lights on from ZT0 to ZT12) at 24 ± 1 °C and 60 ± 10% humidity, with food and water available ad libitum. Mice were synchronized to the lighting conditions for 2 weeks before surgery. Male ICR mice (5 weeks old) were purchased from Charles River Laboratories Japan, Inc. This study was performed using Id2-knock-out mice of mixed genetic background (ICR) and wild-type ICR mice. We prepared mouse models of CKD by performing 5/6Nx operations in two stages under inhalation anesthesia (Baxter). In the first surgical procedure (at 7 weeks of age), two-thirds of the left kidney was removed by cutting off both poles. Seven days later, the right kidney was completely removed. After the second operation, mice were housed for 8 weeks (until they were 16 weeks old) in order to achieve CKD. Sham-operated mice were subjected to laparotomy on the same days as the procedure in the 5/6Nx mice.
A mouse monoclonal anti-TGF-β1 antibody (1d11; R&D Systems, Minneapolis, MN) was reconstituted in PBS and administered to mice via intraperitoneal injection three times a week at a dose of 5 mg/kg body weight. Treatment was initiated 6 weeks after the 5/6Nx operation and continued for 2 weeks. IgG (Zymed Laboratories Inc., San Francisco, CA) was reconstituted in PBS and administered at a dose of 5 mg/kg body weight as a control. All animals used in this study were treated in accordance with the guidelines stipulated by the Animal Care and Use Committee of Kyushu University.
Cell Culture
Hepatocytes were isolated from male mice using a collagenase perfusion method (32). Cells were plated in Williams' medium (Sigma-Aldrich) containing 5% FBS (SAFC Bioscience, Kansas City, MO), 0.1 μm insulin, 0.1 μm dexamethasone, and 2% penicillin/streptomycin and incubated at 37 °C in a humidified environment (5% CO2, 95% air). Following a 4-h attachment period, the culture medium was replaced with serum-free hepatocyte maintenance medium (Lonza, Walkersville, MD) containing 0.1 μm insulin, 0.1 μm dexamethasone, 50 μg/ml gentamicin, and 50 ng/ml amphotericin B. Serum samples were obtained from 5/6Nx mice. TGF-β1 (10 ng/ml) was dissolved in DMSO. After preincubation, the medium was changed to DMEM (Sigma-Aldrich) containing 10% serum from 5/6Nx mice (0% FBS) or TGF-β1 (10 ng/ml; 10% FBS), and the mouse hepatocytes were grown for another 24 h. The mouse hepatocytes were exposed to 20 nm SD208 (R&D Systems; dissolved in DMSO) for 1 h before the 5/6Nx serum or TGF-β1 treatment began.
Measurement of Mouse Body Weight
Body weight was monitored at 8 weeks after operation using the Sartorius CPA22025 (Sartorius, Göttingen, Germany).
Serum Blood Urea Nitrogen (BUN) Measurements
Serum BUN levels were determined using a manufactured kit (Wako Pure Chemical Industries Co. Ltd., Osaka, Japan).
TUNEL Staining
Kidneys were sliced into 10-μm sections using HM550-OMP (Thermo Scientific Microm). Sliced samples were labeled by TdT-mediated dUTP using an In Situ Apoptosis Detection Kit (TaKaRa, Otsu, Japan). Renal sections were observed by fluorescence microscopy.
Determination of Caspase-3/7 Activity
Apoptosis was determined using an Apo-One caspase-3/7 reagent kit (Promega).
Microarray Gene Expression Analysis
The mouse liver was collected at ZT6 in 8 weeks after the operation. Total RNA was isolated with RNAiso (TaKaRa). The quality of extracted RNA was analyzed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). The cRNA was amplified and labeled using a Low Input Quick Amp Labeling Kit (Agilent Technologies, Inc., Loveland, CO). Labeled cRNA was hybridized to a 44K Agilent 60-mer oligomicroarray (Whole Mouse Genome Microarray Kit version 2.0). To identify down-regulated or up-regulated genes, we calculated Z-scores and ratios (non-log-scaled -fold change) from the normalized signal intensities of each probe. We set criteria for down-regulated genes as follows: Z-score ≤ −2.0 and ratio ≤ 0.6. Functional analysis of the decreased gene expression was performed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database on the DAVID system. Raw microarray data were submitted to Gene Expression Omnibus GEO in NCBI (accession numbers GSE35135 and GSE57799).
Quantitative RT-PCR Analysis
Complementary DNA (cDNA) was synthesized by reverse transcription using the ReverTra Ace qPCR RT kit (Toyobo, Osaka, Japan). Diluted cDNA samples were analyzed by real-time PCR using THUNDERBIRD SYBR qPCR Mix (Toyobo) and the 7500 real-time PCR system (Applied Biosystems). Sequences for PCR primers are listed in Table 1.
TABLE 1.
Quantitative RT-PCR analysis PCR primers
| Gene symbol | GenBankTM accession number | Sequence |
|---|---|---|
| Clock | NM_007715 | 5′-TTGCTCCACGGGAATCCTT-3′ (forward) |
| 5′-GGAGGGAAAGTGCTCTGTTGTAG-3′ (reverse) | ||
| Per2 | NM_011066 | 5′-ATCTCCAGGCGGTGTTGAAG-3′ (forward) |
| 5′-AGGGTTACGTCTGGGCCTCT-3′ (reverse) | ||
| Cry1 | NM_007771 | 5′-AGGGAACCCCATCTGTGTTC-3′ (forward) |
| 5′-TGGTGCATTCCAAGGATCGT-3′ (reverse) | ||
| Per1 | NM_011065 | 5′-CCAGGCCCGGAGAACCTTTTT-3′ (forward) |
| 5′-CGAAGTTTGAGCTCCCGAAGTG-3′ (reverse) | ||
| Dec1 | NM_011498 | 5′-GGATTTTGCCCACATGTACC-3′ (forward) |
| 5′-TCAACGTAAGCTCCAGAACC-3′ (reverse) | ||
| Dbp | NM_016974 | 5′-CCGTGGAGGTGCTAATGACCT-3′ (forward) |
| 5′-CCTCTGAGAAGCGGTGTCT-3′ (reverse) | ||
| E4bp4 | NM_017373 | 5′-TTTTGTGGACGAGCATGAGCCTG-3′ (forward) |
| 5′-TGGGAGTAAGTGGAGAAAGAGC-3′ (reverse) | ||
| Cyp3a11 | NM_007818 | 5′-GAACTTCTCCTTCCAGCCTT-3′ (forward) |
| 5′-ACCAGGCATCAAAACCAATC-3′ (reverse) | ||
| Cyp26a1 | NM_007811 | 5′-ACATTGCAGATGGTGCTTCA-3′ (forward) |
| 5′-TCACCTCGGGGTAGACCA-3′ (reverse) | ||
| Id2 | NM_010496 | 5′-ACTCGCATCCCACTATCGTCAG-3′ (forward) |
| 5′-AGACACCGCTTCTCAGAGG-3′ (reverse) | ||
| Tcf7l2 | NM_001142918 | 5′-GAACTCGGAAAACTCCTCGGC-3′ (forward) |
| 5′-CTCGGAAACTTTCGGAGCGA-3′ (reverse) | ||
| Pxr | NM_010936 | 5′-GATGGAGGTCTTCAAATCTGCC-3′ (forward) |
| 5′-GGCCCTTCTGAAAAACCCCT-3′ (reverse) | ||
| Car | NM_009803 | 5′-CCCTGACAGACCCGGAGTTA-3′ (forward) |
| 5′-GCCGAGACTGTTGTTCCATAAT-3′ (reverse) | ||
| Hnf1α | NM_009327 | 5′-TCTACAACTGGTTTGCCAACC-3′ (forward) |
| 5′-GGCCTCTGTGCTCAGCAGGC-3′ (reverse) | ||
| Hnf4α | NM_008261 | 5′-GGGAGAATGCGACTCTCTAA-3′ (forward) |
| 5′-GTTGGCAATCTTCTTTGCCC-3′ (reverse) | ||
| β-actin | NM_007393 | 5′-GACGGCCAGGTCATCACTATT-3′ (forward) |
| 5′-TACCACCAGACAGCACTGTGTT-3′ (reverse) |
Western Blotting
Total cell lysate proteins were isolated with CelLyticTM MT Cell Lysis Reagent (Sigma-Aldrich). Cytoplasmic and nuclear proteins were isolated with the NE-PER nuclear protein extraction kit (Pierce). Total cell lysate, cytosol, and nuclear proteins were separated by SDS-PAGE and transferred to PVDF membranes. The membranes were reacted with primary antibodies against mouse DBP (Aviva Systems Biology, San Diego, CA), ID2 (Santa Cruz Biotechnology, Inc.), TGF-βR1 (Abcam, Cambridge, UK), TGF-βR2 (Abcam), phospho-SMAD3 (Ser-423/425) (Santa Cruz Biotechnology), TCF7L2 (Cell Signaling Technology, Inc., Boston, MA), β-actin (Santa Cruz Biotechnology), or Pol II (Santa Cruz Biotechnology). The immunocomplexes were reacted with anti-rat, anti-mouse, and anti-rabbit IgG secondary antibodies and Chemi-Lumi One reagent (Nacalai Tesque, Inc., Kyoto, Japan). The membranes were photographed, and the density of each band was analyzed with an ImageQuant LAS 3000 mini (Fuji Film).
Luciferase Reporter Assay
The 5′-flanking region of the mouse Cyp3a11 gene (from bp −501 to −1) was amplified with the Elongase Enzyme mix (Invitrogen) using PCR. The PCR products were purified and ligated into a pGL4.12 Basic vector (Cyp3a11-Luc). The sequence of the D-box (AGCATATAA) on Cyp3a11-Luc was mutated to KpnI (GGTACCTAA) using a QuikChange site-directed mutagenesis kit (Stratagene). The 5′-flanking region of the mouse Cyp26a1 gene (from bp −5096 to −3900; +1 indicates the transcription start site) was amplified with the Elongase Enzyme mix using PCR. The PCR products were purified and ligated into a pGL4.12 Basic vector (Cyp26a1-Luc). The sequence of the D-box (site 1, ATTCTGCAAT; site 2, ATTATTTAAT) on Cyp26a1-Luc was mutated to XhoI (site 1, CTCGAGCAAT) or KpnI (site 2, GGTACCAAT) using a QuikChange site-directed mutagenesis kit (Stratagene). The promoter of the mouse Dbp gene (from bp +625 to +1044; +1 indicates the transcription start site) was amplified using the Elongase Enzyme using PCR. The PCR products were purified and ligated into a pGL4.12 Basic vector (Dbp-Luc). The sequence of the E-box (CACATG) on Dbp-Luc was mutated to BglII (AGATCT) using a QuikChange site-directed mutagenesis kit (Stratagene). The promoter of the mouse Tcf7l2 gene (from bp −547 to +139; +1 indicates the transcription start site) was amplified using the Elongase Enzyme using PCR. The PCR products were purified and ligated into a pGL4.12 Basic vector (Tcf7l2-Luc). Sequences for PCR primers are listed in Table 2.
TABLE 2.
Luciferase reporter assay PCR primers
| Gene symbol | Sequence |
|---|---|
| CYP3a11 promoter (from bp −501 to −1) | 5′-ATACTCGAGCTTACAGCTCACCAGTGAAG-3′ (forward) |
| 5′-ATAAGATCTGAAGGGGATTATGTCTCAAC-3′ (reverse) | |
| CYP26a1 promoter (from bp −5096 to −3900) | 5′-ACGGCTAGCTCTTTCTCAGCAGTGAGTGG-3′ (forward) |
| 5′-ACGTAAGCTTTGCCCACTCTGCTGCACTG-3′ (reverse) | |
| Dbp promoter (from bp +625 to +1044) | 5′-ATACTCGAGTTAGGTCACTTAGGTCT-3′ (forward) |
| 5′-ATAAGATCTCGAGGATGGAAGTTTAGAA-3′ (reverse) | |
| Tcf7l2 promoter (from bp −547 to +139) | 5′-ATACTCGAGTAGATGGCCCAGCCTCTCTG-3′ (forward) |
| 5′-ATAAGATCTGTGCCGCTCGGATTTGAGTG-3′ (reverse) | |
| CYP3a11 mutation promoter | 5′-TTCTCCCGGTACCTAATTACTGC-3′ (forward) |
| 5′-GCAGTAATTAGGTACCGGGAGAA-3′ (reverse) | |
| CYP26a1 mutation promoter (D-box site 1) | 5′-AAGGAGCTCGAGCAATCTTCTG-3′ (forward) |
| 5′-CAGAAGATTGCTCGAGCTCCTT-3′ (reverse) | |
| CYP26a1 mutation promoter (D-box site 2) | 5′-TCATGAAGGTACCAATATCCTCA-3′ (forward) |
| 5′-TGAGGATATTGGTACCTTCATGA-3′ (reverse) |
Expression vectors for mouse Dbp, E4bp4, Clock, Bmal1, and Id2 were constructed as described previously (37–39). NIH3T3 was transfected with 100 ng/well reporter vector and 250 ng/well expression vector using Lipofectamine LTX reagent (Life Technologies Japan Ltd., Tokyo, Japan). In addition, 0.5 ng/well phRL-TK vector (Promega, Tokyo, Japan) was co-transfected as an internal control reporter. At 24 h after transfection, cell extracts were prepared with passive lysis buffer (Promega), and the extracts were used for assays of firefly luciferase and Renilla luciferase activity by luminometry. The ratio of firefly luciferase activity (expressed from reporter plasmids) to Renilla luciferase activity (expressed from phRL-TK) in each sample served as a measure of normalized luciferase activity.
Quantification of Serum Retinol Concentration
Serum retinol concentrations were measured by the Alliance HPLC system (Nihon Waters, Tokyo, Japan) (40). Serum was added to acetonitrile following acidification with 2 n acetic acid. After vortex mixing, water and an extraction mixture consisting of an n-hexane and 2-propanol (6.5:1.5, v/v) were added. The sample was extracted by vortex mixing for 2 min. After centrifugation (1000 × g, 10 min, 4 °C), the organic layer was removed and evaporated in a solvent evaporator at 15 °C. The residue was dissolved in mobile phase solvent A by vortex mixing. Following centrifugation at 300 × g for 3 min, the supernatant was transferred into an autosampler vial and injected. The solvent system consisted of methanol-water (3:1, v/v) containing 10 mm ammonium acetate and 0.025% acetic acid (solvent A) and of methanol/dichloromethane (4:1, v/v) (solvent B). A linear gradient from 100% solvent A to 100% solvent B was applied over a period of 30 min, followed by isocratic elution with solvent B for an additional 15 min (Table 3). Between each run, the analytical column was equilibrated with 100% solvent A for 15 min (total run time 60 min). The flow rate was set at 0.4 ml/min, and the detection wavelength was set between 320 and 380 nm (from 18 to 40 min). Monitoring wavelength was set at 350 nm.
TABLE 3.
Quantification of serum retinol concentration
| Time | Solvent A | Solvent B |
|---|---|---|
| min | % | % |
| 0.0 | 100 | 0 |
| 16.0 | 47 | 53 |
| 30.0 | 0 | 100 |
| 45.0 | 0 | 100 |
| 45.1 | 100 | 0 |
| 60.0 | 100 | 0 |
Construction of Dbp-knockdown Cells and Serum Shock Procedures
Serum shock was performed as follows (37, 38). Hepa1-6 cells were grown in DMEM supplemented with 10% FBS. On the day of serum shock, 50% FBS was added for 2 h, and then cells were transferred back into starvation medium. Cells were harvested for RNA every 4 h from 0 to 52 h after serum treatment. Stealth RNA of DBP (5′-UUC AAA GGU CAU UAG CAC CUC CAC G-3′) and the control (5′-UUC CCA GGA AUA CUC GAU UCC AAC G-3′) were designed by Life Technologies Japan. DBP short interfering RNA (siRNA) or control siRNA (50 pmol) was transfected into the cells using Lipofectamine RNAiMAX (Life Technologies Japan). The siRNA was transfected 24 h after serum treatment.
Transfection of Plasmid DNA in Mouse Liver
Mice (sham-operated or 5/6Nx mice) were injected with the suspensions (0.05 ml) containing pcDNA3.1-Dbp expression plasmid DNA or pcDNA3.1 (5 μg) complexes with SAFETRANS (Lac-α-CDE) (Cosmo Bio, Tokyo, Japan) 2 times/week at ZT6 via intravenous injection (41).
Statistical Analysis
The significance of the 24-h variations in each parameter was tested by ANOVA. p values of <0.05, determined from comparisons of the residuals before and after cosine curve fitting, indicated the detection of a rhythm. All data are the mean ± S.E. of at least three individual experiments. For all experiments in which three or more test groups were compared, we used one-way analysis of variance with Tukey's post hoc tests to evaluate differences resulting from the operation, drug, or antibody application and circadian time points tested. p values of <0.05 were considered significant.
Results
Down-regulation of Hepatic Cyp3a11 and Cyp26a1 Affects the Disruption of Retinol Metabolism in 5/6Nx Mice
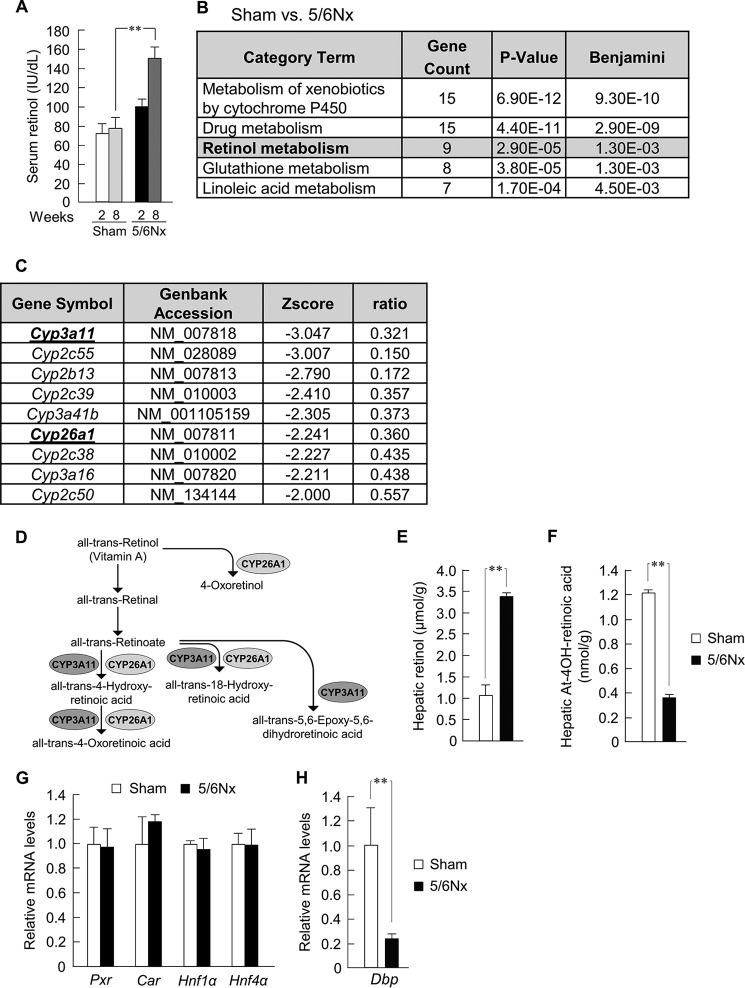
We quantified the serum concentration of retinol as our initial observation. In 5/6Nx mice, regarding the concentration profiles of retinol after 5/6Nx operation, we found a marked increase at 8 weeks (Fig. 1A). Although the mechanism of serum retinol elevation is unknown in rodents and humans following kidney dysfunction (34–36), several studies have demonstrated that serum retinol is controlled by hepatic metabolism (25, 26). To identify the genes whose hepatic expressions are down-regulated during the development of CKD, we performed microarray analysis by using RNA isolated from the livers of the sham-operated and 5/6Nx mice (Fig. 1, B and C). The expression of a large number of genes was repressed by disrupted renal function. We performed functional analysis of the genes by using the KEGG database (35) and found that five biological pathways were enriched in a statistically significant manner (Fig. 1B). Among these pathways, we focused on the retinol metabolism pathway. Among the down-regulated genes in the retinol metabolism pathway, Cyp3a11, which is responsible for the metabolism of over 50% of commonly prescribed drugs (42), showed the greatest decrease in expression in the 5/6Nx mice (Fig. 1C). Furthermore, Cyp26a1 has been identified as a key enzyme metabolizing retinol. Both Cyp3a11 and Cyp26a1 play multiple roles in retinol metabolism (Fig. 1D). Hepatic retinol levels significantly increased in 5/6Nx mice (Fig. 1E), and the hepatic expression of all-trans-4-hydroxyretinoic acid, one of the metabolites of retinol (Fig. 1D), also significantly decreased in 5/6Nx mice (Fig. 1F). These results suggest an overall decrease in retinol metabolism in CKD. Therefore, we further focused on Cyp3a11 and Cyp26a1 to investigate their down-regulation metabolisms during the CKD development.
FIGURE 1.
Down-regulation of hepatic Cyp3a11 and Cyp26a1 affects the disruption of retinol metabolism in 5/6Nx mice. A, serum concentration profiles of retinol at ZT6 at 2 and 8 weeks after the second operation in 5/6Nx mice. B, functional analysis of the decreased gene expression in the livers of 5/6Nx mice using the KEGG database on the DAVID system. The Benjamini false discovery test was used. C, decrease in the expression of genes involved in retinol metabolism in the 5/6Nx mouse liver. D, schematic representation of the retinol metabolism pathway. E and F, hepatic concentration profiles of retinol (E) and all-trans-4-Hydroxy-retinoic acid (F) of sham-operated (Sham) and 5/6Nx mice at ZT6. G and H, expression profiles of the general transcription factors of Cyps (G) and Dbp (H) mRNA in the liver of sham-operated and 5/6Nx mice at ZT6. Values are the means ± S.E. (error bars) for all experiments (n = 3–6). A, E, F, and H, **, p < 0.01.
Selected isoforms of hepatic CYPs are reduced in CKD secondary to a decrease in gene expression (28–34). We therefore investigated whether the expression of the general transcription factors of Cyp3a11 were changed in the liver of 5/6Nx mice. Contrary to our expectations, the mRNA levels of pregnane X receptor (Pxr), constitutive androstane receptor (Car), hepatocyte nuclear factor (Hnf-1α), and Hnf-4α in the liver of 5/6Nx mice were unchanged (Fig. 1G). Other transcription factors of Cyp genes, such as arylhydrocarbon receptor and peroxisome proliferator-activated receptor α, also remained at the same levels (data not shown); therefore, we next investigated other transcription factors. In research on the related genes by microarray analysis, the participation of D-box-binding protein (DBP) was suggested. The mRNA levels of Dbp in the liver of 5/6Nx mice were decreased (Fig. 1H).
Down-regulation of Hepatic DBP Affects the Disruption of Retinol Metabolism in 5/6Nx Mice
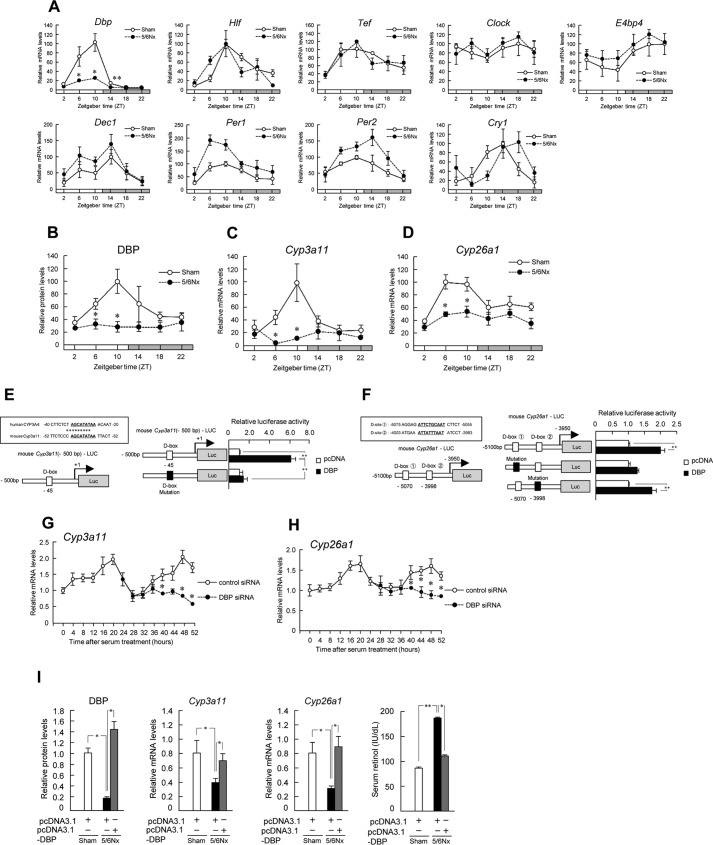
Expressions of Dbp exhibit circadian rhythms (43). Therefore, we quantified the temporal expression profiles of hepatic clock genes. Expression of Dbp was decreased at zeitgeber times (ZT; where ZT0 is the light on time, and ZT12 is the light off time) 6, 10, and 14 in 5/6Nx mice (Fig. 2A). The mRNA levels of Dbp oscillated over 24 h in both sham-operated and 5/6Nx mice. This result was the same as that for DBP protein levels (Fig. 2B). The mRNA levels of other clock genes and the expression of Per1 and Per2 tended to be increased (Fig. 2A).
FIGURE 2.
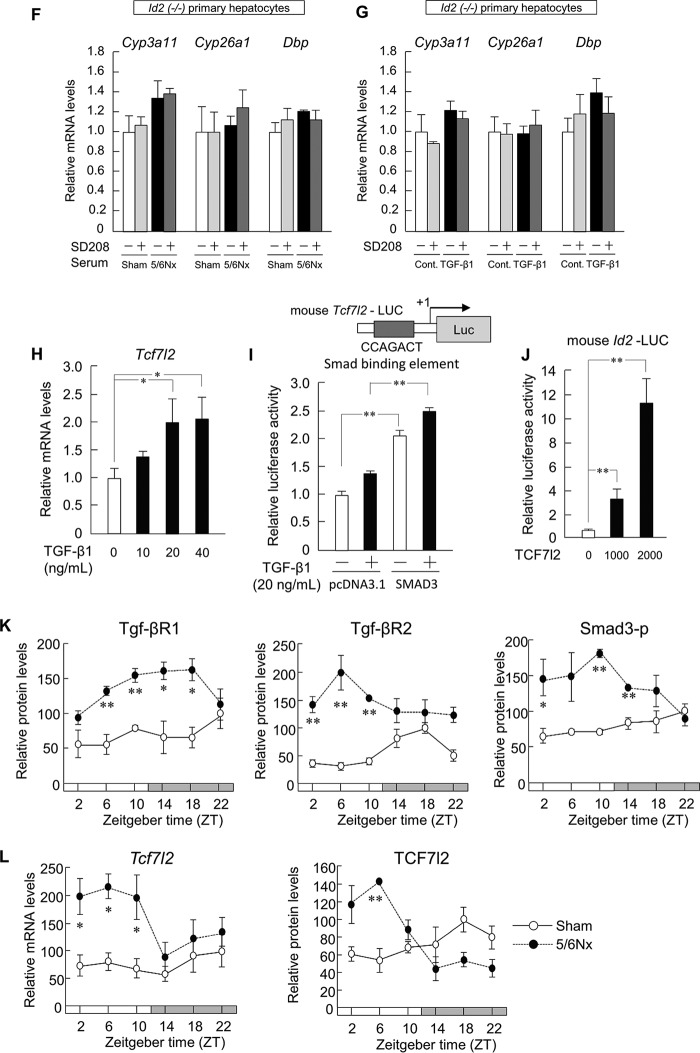
Down-regulation of hepatic DBP affects the disruption of retinol metabolism in 5/6Nx mice. A, temporal expression profiles of Dbp, Hlf, Tef, Clock, E4bp4, Dec1, Per1, Per2, and Cry1 mRNA levels in the livers of sham-operated (Sham) and 5/6Nx mice (Dbp, E4bp4, Dec1, Per2, and Cry1 in sham-operated and 5/6Nx mice; p < 0.05, ANOVA). B, temporal expression profiles of DBP protein levels in the livers of sham-operated and 5/6Nx mice (DBP in sham-operated and 5/6Nx mice; p < 0.05, ANOVA). C and D, temporal expression profiles of Cyp3a11 (C) and Cyp26a1 (D) mRNA in the livers of sham-operated and 5/6Nx mice (Cyp3a11 and Cyp26a1 in sham-operated mice; p < 0.05, ANOVA). E, left, computer-aided analysis identified the D-box in the promoter region of mouse Cyp3a11. Right, luciferase activities in Hepa1-6 cells after co-transfection with constructs encoding DBP and/or a reporter vector encoding the mouse Cyp3a11 promoter region (−500 to −1) and influence of D-box mutation on luciferase activity induction by DBP treatment. F, left, computer-aided analysis identified the D-box in the promoter region of mouse Cyp26a1. Right, luciferase activities in Hepa1-6 cells after co-transfection with constructs encoding DBP and/or a reporter vector encoding the mouse Cyp26a1 promoter region (−5100 to −3950) and influence of D-box mutation on luciferase activity induction by DBP treatment. G, temporal expression profiles of Cyp3a11 mRNA after serum shock in control or DBP siRNA-transfected cells. Hepa1-6 cells were transfected with the indicated siRNAs for 24 h after serum treatment. Significant time-dependent differences were observed in Cyp3a11 expression between control and DBP siRNA-transfected cells (p < 0.05, ANOVA). H, temporal expression profiles of Cyp26a1 mRNA after serum shock in control siRNA or DBP siRNA-transfected cells. Significant time-dependent differences in Cyp26a1 expression were observed between control and DBP siRNA-transfected cells (p < 0.05, ANOVA). I, hepatic expression profiles of DBP protein levels, Cyp3a11 and Cyp26a1 mRNA levels, and serum retinol concentration profiles in 5/6Nx mice by transfection with the Dbp expression vector. Values are the means ± S.E. (error bars) for all experiments (n = 3–6). A–D, G, and H, *, p < 0.05; **, p < 0.01, compared with the values at corresponding time points. E, F, and I, *, p < 0.05; **, p < 0.01.
Because the circadian expression of several human and mouse Cyp genes is controlled by molecular clocks (15), we quantified the temporal expression profiles of Cyp3a11 and Cyp26a1 in 5/6Nx mouse liver (Fig. 2, C and D). In 5/6Nx mice, the mRNA levels of Cyp3a11 and Cyp26a1 decreased at ZT6 and ZT10 (Fig. 2, C and D). The mRNA levels of Cyp3a11 and Cyp26a1 significantly oscillated over 24 h in sham-operated mice but not in 5/6Nx mice. Next, we investigated the expression mechanism of Cyp3a11 and Cyp26a1 genes (Fig. 2, E and F). The so-called “D-box,” which is the DNA element to which DBP binds, is present in the promoter regions of mouse Cyp3a11 and Cyp26a1. Luciferase reporter assays revealed that Dbp expression led to significantly increased transcriptional activity of both Cyp genes, whereas expression of the antagonist E4-binding protein 4 (E4bp4) significantly reversed this effect (Fig. 2, E and F). Furthermore, promoter activation by Dbp was reduced by D-box mutation (Fig. 2, E and F). Synchronization of the circadian clock in Hepa1-6 cells by treating with a high concentration of serum induced 24-h oscillation in the expression of Cyp3a11 and Cyp26a1 genes. Transfection of Hepa1-6 cells with Dbp siRNA significantly decreased the mRNA levels of Cyp3a11 and Cyp26a1 genes (Fig. 2, G and H). Consistent with this, the levels of DBP protein decreased as well in 5/6Nx mouse liver (Fig. 2B). Therefore, we attempted to rescue the decreased Cyp3a11 and Cyp26a1 expression by injection of a Dbp expression vector using Lac-α-CDE, novel hepatocyte-specific carriers. This experiment resulted in the recovery of hepatic Cyp3a11 and Cyp26a1 expression levels in 5/6Nx mice (Fig. 2I). Additionally, the accumulation of serum retinol in 5/6Nx mice was ameliorated (Fig. 2I).
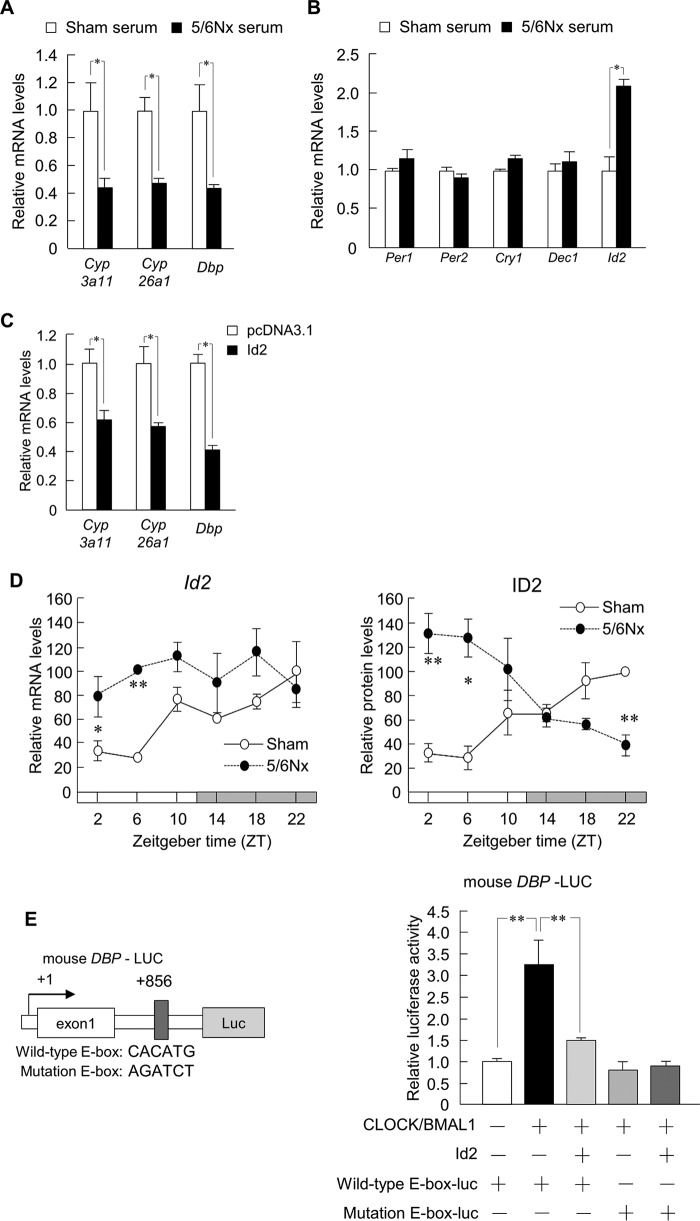
Aberrant Id2 Expression Is Associated with Hepatic Dysfunction in 5/6Nx Mice
To uncover additional transcriptional alterations associated with hepatic dysfunction, we determined whether treatment of wild-type primary hepatocytes with serum from 5/6Nx mice could decrease Cyp3a11, Cyp26a1, and Dbp expression. As shown in Fig. 3A, such decreases were observed. To identify candidate inhibitors of Dbp expression present in 5/6Nx serum, we surveyed clock genes that are known to suppress Dbp (Fig. 3B). Transcription of Dbp has been shown to be positively regulated by CLOCK-BMAL1, which binds to an E-box in its promoter (44). Conversely, CLOCK-BMAL1-dependent transcription is antagonized by inhibitor of DNA binding 2 (ID2), which prevents the complex from binding to E-boxes (45, 46). In agreement with these findings, we found that among the mRNAs studied, only Id2 mRNA levels were increased in 5/6Nx serum (Fig. 3B). Furthermore, Cyp3a11, Cyp26a1, and Dbp expression decreased in cells transfected with an Id2 expression vector (Fig. 3C). Therefore, we analyzed the expression profile of Id2 and found that the levels of both its transcript and protein were increased in 5/6Nx mouse livers (Fig. 3D). Luciferase reporter assays were performed to investigate the influence of Id2 on Dbp promoter activity and demonstrated that Dbp promoter activation by Clock/Bmal1 was inhibited by co-overexpression of Id2 (Fig. 3E) and that Dbp promoter activation by Clock/Bmal1 was reduced by E-box mutation (Fig. 3E).
FIGURE 3.
Aberrant ID2 expression is associated with hepatic dysfunction in 5/6Nx mice. A and B, expression profiles of Cyp3a11, Cyp26a1, and Dbp (A) and clock suppressor genes of DBP (Per1, Per2, Cry1, Dec1, and Id2) (B) in primary hepatocytes of wild-type mice, following incubation with serum from sham-operated (Sham) or 5/6Nx mice. C, expression profiles of Cyp3a11, Cyp26a1, and Dbp in Hepa1-6 cells incubated with pcDNA3.1 and an Id2 expression plasmid. D, temporal expression profiles of Id2 mRNA levels and protein levels in the livers of sham-operated and 5/6Nx mice at 8 weeks after the second 5/6Nx operation. E, left, computer-aided analysis identified the E-box in the promoter region of mouse Dbp. Right, influence of E-box mutation on luciferase activity induction by CLOCK/BMAL1 and Id2 treatment. Values are the means ± S.E. (error bars) for all experiments (n = 3–6). D, *, p < 0.05; **, p < 0.01, compared with the values at the corresponding time points. A–C and E, *, p < 0.05; **, p < 0.01.
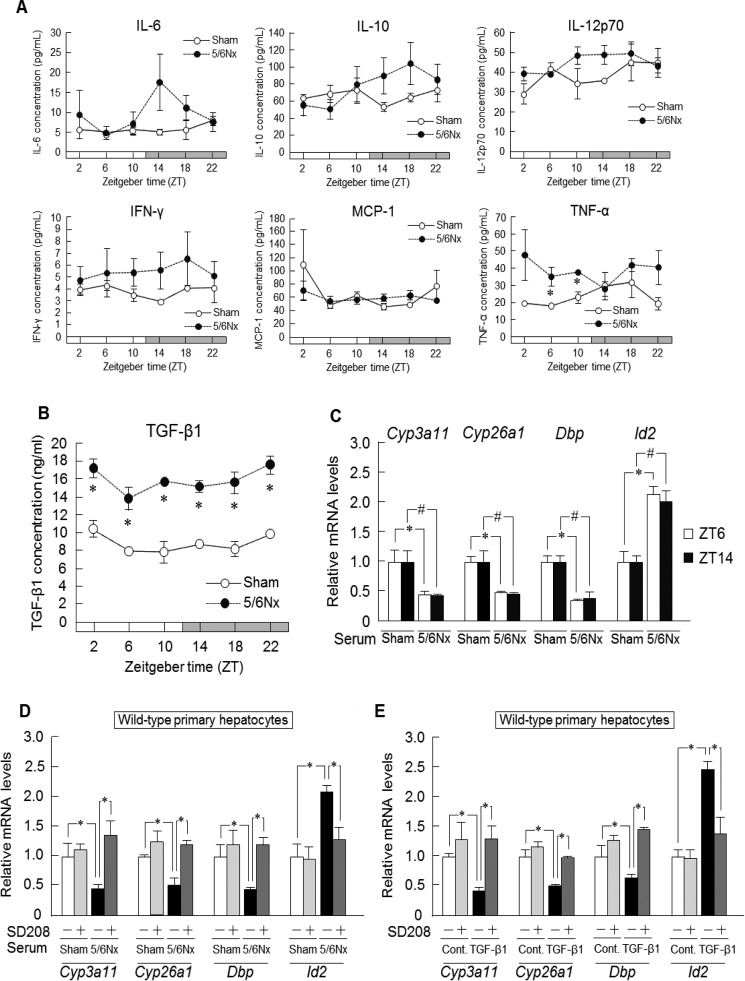
Increased Expression of TCF7L2 via TGF-β1 Signaling-induced Id2
In the serum component, a factor is present in the fraction containing proteins of molecular mass between 10 and 30 kDa (34); we therefore focused on cytokines. To identify the inducer of Id2 in serum, we investigated serum cytokine concentrations using a cytometric bead array or ELISA. We examined the variation in serum IL-6, IL-10, IL-12p70, IFN-γ, MCP-1, and TNF-α (Fig. 4A) and plasma Tgf-β1 levels over 24 h (Fig. 4B). TGF-β1 is present in platelet granules and is released upon platelet activation. Therefore, to measure the circulating levels of Tgf-β1, platelet-poor plasma was collected for measurements. Plasma Tgf-β1 increased throughout the day in 5/6Nx mice (Fig. 4B), and serum Tgf-β1 also increased (data not shown). In contrast, TNF-α increased only at ZT6 and ZT10 in 5/6Nx mice, whereas other cytokine concentrations remained unchanged throughout the day (Fig. 4A).
FIGURE 4.
Increased TCF7L2 expression via TGF-β1 signaling-induced Id2. A, temporal serum concentration profiles of IL-6, IL-10, IL-12p70, IFN-γ, MCP-1, and TNF-α of sham-operated (Sham) and 5/6Nx mice at 8 weeks after the second 5/6Nx operation. B, temporal plasma concentration profiles of TGF-β1 in sham-operated and 5/6Nx mice. C, expression profiles of Cyp3a11, Cyp26a1, Dbp, and Id2 in wild-type mouse primary hepatocytes incubated with serum from sham-operated and 5/6Nx mice at ZT6 or ZT14. D and E, expression profiles of Cyp3a11, Cyp26a1, Dbp, and Id2 in wild-type mouse primary hepatocytes incubated with serum from sham-operated or 5/6Nx mice (D) or incubated with TGF-β1 (E). The primary hepatocytes were pretreated with SD208 prior to incubation with serum or incubated with TGF-β1. F and G, expression profiles of Cyp3a11, Cyp26a1, and Dbp incubated with serum from wild-type sham-operated or wild-type 5/6Nx mice and pretreated with SD208 (F) or incubated with TGF-β1 and pretreated with SD208 (G) in Id2 (−/−) mouse primary hepatocytes. H, expression profiles in Hepa1-6 cells of Tcf7l2 mRNA levels after incubation with TGF-β1 (0, 10, 20, or 40 ng/ml) for 4 h. I, top, computer-aided analysis identified the Smad-binding protein in the promoter region of mouse Tcf7l2. Bottom, influence of Smad-binding protein on luciferase activity induction by TGF-β1 and SMAD3 treatment. J, TCF7L2 overexpression and mouse Id2-luciferase reporter activity in Hepa1-6 cells. K, temporal expression profiles of TGF-βR1, TGF-βR2, or SMAD-3p protein levels in the livers of sham-operated or 5/6Nx mice at 8 weeks after operation (TGF-βR2 in sham-operated and 5/6Nx mice; p < 0.05, ANOVA). L, temporal expression profiles of Tcf7l2 mRNA levels and protein levels in the livers of sham-operated and 5/6Nx mice at 8 weeks after the second 5/6Nx operation. Values are the means ± S.E. (error bars) for all experiments (n = 3–6). A, B, K, and L, *, p < 0.05; **, p < 0.01, compared with the values at the corresponding time points. C–E and H–J, *, p < 0.05; **, p < 0.01; #, p < 0.05.
To determine whether Tnf-α or Tgf-β1 down-regulated the expression of Cyp3a11, Cyp26a1, and Dbp, wild-type mouse primary hepatocytes were incubated with serum from 5/6Nx mice obtained at ZT6 or ZT14. Incubation with serum obtained at ZT6 increased the expression of both Tgf-β1 and Tnf-α; however, incubation with serum obtained at ZT14 increased only Tgf-β1 expression (Fig. 4C). We found that the mRNA levels of Cyp3a11, Cyp26a1, and Dbp were significantly decreased to the same degree not only in serum from 5/6Nx mice obtained at ZT6 but also in serum obtained at ZT14 (Fig. 4C). In addition, following incubation with serum from 5/6Nx mice and pretreatment with the TGF-β1 receptor 1 selective inhibitor SD208, the mRNA levels of Cyp3a11, Cyp26a1, and Dbp were substantively restored to the control level (Fig. 4D). Furthermore, when the cells were exposed to Tgf-β1, the mRNA levels of Cyp3a11, Cyp26a1, and Dbp decreased significantly (Fig. 4E). This down-regulation recovered following incubation with SD208 (Fig. 4E). Thus, we demonstrated that Tgf-β1 in 5/6Nx mice down-regulated Cyp3a11, Cyp26a1, and Dbp mRNA expression in primary hepatocytes. On the other hand, the reduction of Cyp3a11, Cyp26a1, and Dbp expression by Tgf-β1 was limited by knock-out of Id2 (Fig. 4, F and G).
We next investigated the mechanism of Id2 induction. Pathway analysis of microarray data identified the Tef/Tcf-β-catenin signal. Analysis of the Id2 promoter sequence and microarray analysis revealed a binding site for transcription factor 7-like 2 (Tcf7l2) (Fig. 4I), and Tcf7l2 has been shown to be regulated by TGF-β1 signaling (47). To examine the influence of Tgf-β1 on Tcf7l2 expression, Tcf712 expression in Hep1–6 cells was studied using dose-response experiments with Tgf-β1. The results showed that Tcf7l2 mRNA levels were induced by incubation with Tgf-β1 (Fig. 4H). Furthermore, Tcf7l2 promoter activity was activated by transfection of a Smad3 (small phenotype mothers against decapentaplegic 3) vector and treatment with Tgf-β1 (Fig. 4I). In addition, Id2 promoter-driven luciferase activity was increased by TCF7L2 overexpression (Fig. 4J).
Based on these results, we next investigated Tgf-β1 signaling in the liver. The binding of TGF-β1 to the type 1 TGF-β receptor (TGF-βr1) leads to transactivation of the type 2 TGF-β1 receptor (TGF-βr2) and increased phosphorylation of its downstream targets, SMAD2 and SMAD3 (48). In 5/6NX mice, hepatic Tgf-βR1 and Tgf-βR2 protein levels increased significantly (Fig. 4K), and Smad3 phosphorylation (Smad3-p) was induced during the light phase in these mice (Fig. 4K). Furthermore, the amounts of both TCF-7L2 protein and mRNA increased compared with those of sham-operated mice (Fig. 4L).
TGF-β1 Induces Metabolic Dysfunction in 5/6Nx Mouse Livers
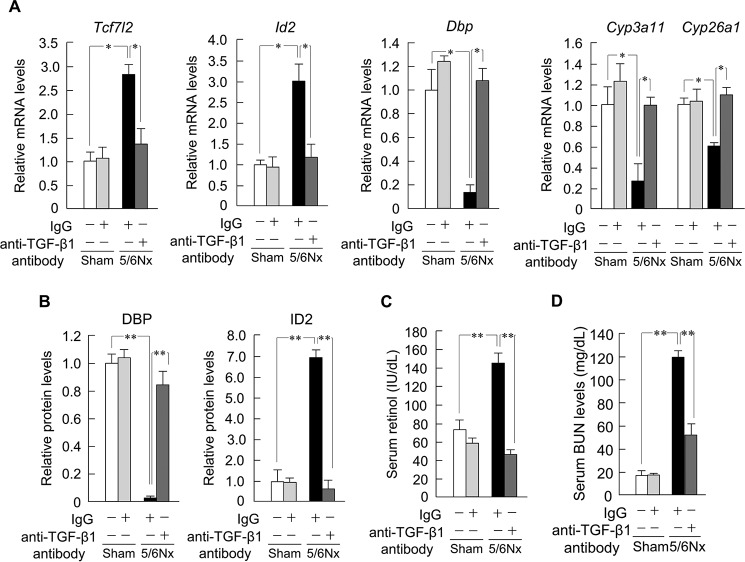
To examine the mechanism underlying the observed change in the periodicity of hepatic Tcf7l2 expression in 5/6Nx mice, an anti-TGF-β1 antibody was administrated via intraperitoneal injection from 7 to 8 weeks after the operation. This abrogated the influence of TGF-β1 on Tcf7l2, Id2, Dbp, Cyp3a11, and Cyp26a1 mRNA expression in the liver (Fig. 5A). In agreement, following anti-TGF-β1 administration, DBP and ID2 protein expression levels in 5/6Nx mice also reverted to those observed in sham-operated mice (Fig. 5B), and the levels of serum retinol decreased as well (Fig. 5C), as did the levels of serum BUN, an indicator of renal dysfunction (Fig. 5D).
FIGURE 5.
TGF-β1 induces metabolic dysfunction in 5/6Nx mouse livers. A, hepatic mRNA expression profiles of Tcf7l2, Id2, Dbp, Cyp3a11, and Cyp26a1 in 5/6Nx mice treated with an anti-TGF-β1 antibody. B, hepatic protein expression profiles of DBP or ID2 in 5/6Nx mice treated with an anti-TGF-β1 antibody. C, serum retinol concentration profiles in 5/6Nx mice treated with an anti-TGF-β1 antibody. D, serum concentration profiles of BUN in 5/6Nx mice treated with an anti-TGF-β1 antibody. Values are the means ± S.E. (error bars) for all experiments (n = 3–6). *, p < 0.05; **, p < 0.01.
Influence of the Accumulation of Serum Retinol on Renal Pathology in 5/6Nx Mice
Retinol is a fat-soluble vitamin A, and serum retinol concentration is raised in CKD (22–24) (Fig. 1A); however, the influence of retinol accumulation on renal function in CKD is not clear. Therefore, we finally focused on the role of retinol in CKD.
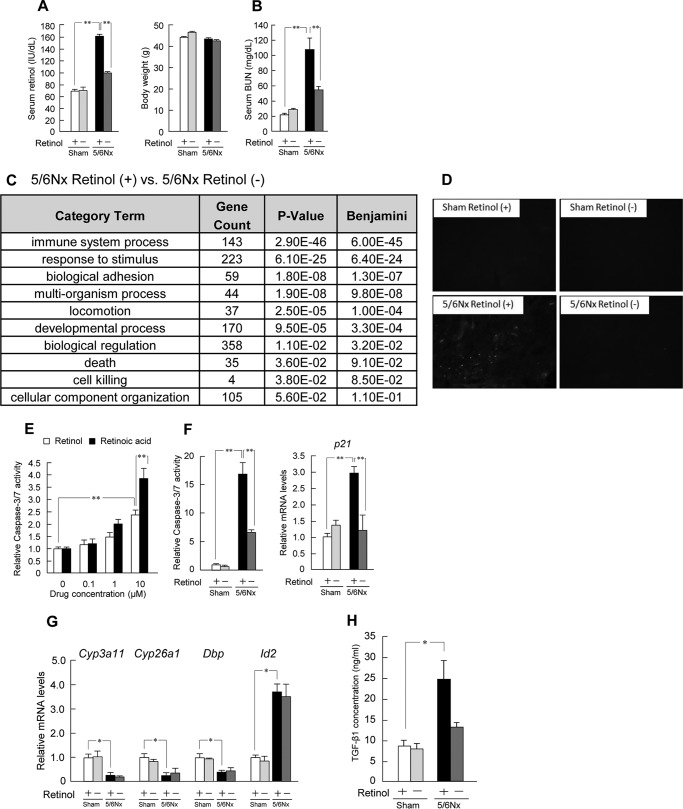
Because almost all retinol is ingested via food, we investigated the effects of retinol-free feeding (retinol (−)) in this model. In 5/6Nx retinol (−) mice, average body weight was unchanged (Fig. 6A), whereas the serum concentration of retinol decreased (Fig. 6A). Notably, BUN was significantly decreased in 5/6Nx retinol (−) mice (Fig. 6B).
FIGURE 6.
Retinol aggravates renal apoptosis in wild-type 5/6Nx mice. A, serum concentration profiles of retinol and body weight in wild-type sham-operated (Sham) and 5/6Nx mice with retinol-free feeding. B, serum concentration profiles of BUN in wild-type sham-operated and 5/6Nx mice with retinol-free feeding. C, functional analysis of the recovered expression of genes by retinol-free feeding in 5/6Nx mice, using the KEGG database on the DAVID system. D, fluorescence images of TUNEL staining in renal sections observed by fluorescence microscopy. E, influence on relative caspase-3/7 activity in response to treatment with retinol or retinoic acid in Renca. F, left, influence on relative caspase-3/7 activity in wild-type sham-operated and 5/6Nx mice with retinol-free feeding. Right, expression profiles of renal p21 mRNA levels in wild-type sham-operated and 5/6Nx mice with retinol-free feeding. G, mRNA expression profiles of hepatic Cyp3a11, Cyp26a1, Dbp, and Id2 in wild-type sham-operated and 5/6Nx mice with retinol-free feeding. H, plasma concentration profiles of TGF-β1 in wild-type sham-operated and 5/6Nx mice with retinol-free feeding. Values are the means ± S.E. (error bars) for all experiments (n = 3–6). A, B, and E–H, *, p < 0.05; **, p < 0.01.
To investigate the pathway underlying the amelioration of renal function associated with decreased retinol levels, we compared the results of microarray analysis of renal gene expression between 5/6Nx retinol (+) and (−) mice using functional analysis (Fig. 6C). This analysis, which utilized the KEGG database (35), identified biological pathways involved in the immune system process, response to stimulus, and death as being involved in the amelioration (Fig. 6C). It is well known that the apoptotic death of renal cells is induced in CKD (36) and that renal apoptosis is induced by retinol (49). Accordingly, following incubation of the murine kidney carcinoma cell line, Renca, with either retinoic acid or retinol, caspase-3/7 activities, which are major indicators of apoptosis, increased significantly and dose-dependently for both factors (Fig. 6E). TUNEL staining and caspase-3/7 activities in 5/6Nx retinol (+) mice showed marked increases as well (Fig. 6, D and F). These activations were significantly ameliorated in 5/6Nx retinol (-) mice (Fig. 6, D and F). In addition, p21 is an apoptosis-related gene, serves as an apoptosis marker in the kidney, and is regulated by retinol (50). The levels of renal p21 in 5/6Nx retinol (−) mice were also reduced compared with those in 5/6Nx retinol (+) mice (Fig. 6F). In the liver, there were no effects of the mRNA levels of Cyp3a11, Cyp26a1, Dbp, and Id2 in 5/6Nx retinol (−) mice (Fig. 6G). Finally, serum concentrations of TGF-β1 decreased slightly (Fig. 6H). Taken together, these results suggest that the liver-kidney axis by retinol aggravates the renal function in CKD.
Discussion
This study unveils a new aspect of the coupling between the kidney and liver by clock genes in CKD. The results of these studies provide insight into the mechanism of metabolic dysfunction, which involves the accumulation of serum retinol in CKD livers, and suggest that hepatic expression of Cyp3a11 and Cyp26a1 might serve as a key regulator of retinol metabolism. Furthermore, the results demonstrate that the abnormal signal transduction caused by elevated Tgf-β1 signaling under chronic renal dysfunction leads to high expression of hepatic Tcf7l2, which in turn induces Id2, resulting in the disruption of the 24-h Dbp rhythm and concomitant decreased expression of Cyp3a11 and Cyp26a1. We also showed that increased serum retinol levels affect renal apoptosis in CKD (Fig. 6C) and that renal dysfunction was ameliorated by the reduction of serum retinol (Fig. 6, C, D, and F).
In this study, the levels of serum retinol were elevated by 5/6Nx in mice (Fig. 1A). Almost all retinol is ingested as food, suggesting that the influence of retinol metabolism in the liver is important for endogenous retinol levels. Several hepatic CYPs have been shown to be involved in retinol metabolism (25–27), and microarray analysis revealed alterations in these retinol acid metabolism pathways (Fig. 1B). Specifically, the expression of Cyp3a11 and Cyp26a1, major retinoic metabolic CYPs, decreased in 5/6Nx mice (Fig. 1C). Therefore, to study potential alterations in the expression of general transcriptional factors involved in the metabolism of various biological chemicals in the livers of CKD mice, we quantified their expression profiles in sham-operated or 5/6Nx mouse livers. However, we found that the expression levels of Pxr, Car, Hnf-1α, and Hnf-4α in CKD livers were unchanged (Fig. 1G). On the other hand, the molecular clock is also an important mediator of various biological functions (8–10). The PAR domain basic leucine zipper (PAR bZip) transcription factors DBP, TEF, and HLF accumulate in a highly circadian manner in several peripheral tissues, especially in the liver, and, in turn, DBP plays a role in circadian rhythm regulation (39). In 5/6Nx mice, we found that hepatic Dbp expression was decreased, whereas the expression levels of Hlf and Tef were unchanged (Fig. 2A).
The selective alteration of DBP might be explained in part by the number of E-box elements contained therein. DBP has seven E-boxes (44), and the binding of BMAL1 and CLOCK to multiple extra- and intragenic E-boxes parallels the circadian transcription cycle. Furthermore, DBP is more highly expressed in the liver than are HLF and TEF (43, 44) and is therefore more affected in CKD. Comparatively, the expression of almost all clock genes was unchanged in 5/6Nx mouse livers, but the expression of Per1 and Per2 tended to be increased in 5/6Nx mice (Fig. 2A). This finding might be explained by the presence of a cAMP-response element in the Per1 and Per2 promoter region (51). Previous microarray analysis results showed that expression of activating transcription factor 5 (Atf5), which binds to cAMP-response element, was increased in 5/6Nx mice (51), which might increase Per1 and Per2 expression. Nevertheless, in vitro experiments showed that Per1 and Per2 expressions were unchanged (Fig. 3B), so the influence of Per1 and Per2 is slight in the down-regulation of hepatic Cyp genes. Computer-aided analysis identified a putative D-box in the promoter regions of mouse Cyp3a11 and Cyp26a1, and the expression levels of Cyp3a11 and Cyp26a1 were shown to be regulated by DBP. Previous studies with PAR bZip triple-knockdown mice showed that the expression of many metabolic genes was altered in the liver (44). These results suggest that decreased expression of hepatic DBP in CKD alters hepatic metabolism.
In this study, 5/6Nx mouse serum induced the reduction of Cyp3a11, Cyp26a1, and Dbp expression and the induction of Id2 expression (Fig. 3, A and B). ID2 is a rhythmically expressed helix-loop-helix transcriptional repressor that suppresses the transactivation of genes that positively regulate clock-controlled gene activity (46). In 5/6Nx mice, we found that hepatic Id2 was increased (Fig. 3D). Time-dependent difference of ID2 protein expression was shown. Recently, 24-h rhythm in posttranscriptional modification was identified (52). Therefore, Id2 mRNA may be modified by abnormal 24-h rhythm of posttranscriptional modification in CKD. The function of Id2 was elucidated only under Id2-deficient conditions; however, we found that the forced expression of Id2 can regulate Dbp transcription (Fig. 3, C and E). In addition, transfection with Id2 vector decreased the mRNA levels of Per1 and Per2 (data not shown). Thus, Id2 can suppress all of the positive elements of CCG activity; however, only the mRNA levels of Dbp decreased in 5/6Nx mice. This also may be partly explained by the number of E-box elements and the amount present in the liver, as described above. Thus, Id2 might be able to suppress the mRNA levels of Dbp in 5/6Nx mice. To determine whether Id2 participates in the decreased expression of Dbp in 5/6Nx mice, we attempted to create a mouse model of CKD by knocking out Id2 but failed because Id2−/− mice were viable for only about 10 weeks (40). In addition, the young Id2−/− mice did not endure 5/6Nx conditions. Therefore, we used primary Id2−/− hepatocyte cultures and found that Id2 mediates the inhibitory effects on Dbp expression (Fig. 4, F and G). These findings suggest that Id2 can influence Cyp3a11 and Cyp26a1 mRNA levels.
A serum factor that decreases Cyp gene expression has been found in the fraction containing proteins of molecular mass between 10 and 30 kDa (34). Indeed, several studies have demonstrated that the expression of Cyp genes is reduced by cytokines in in vitro experiments (53), suggesting the involvement of cytokines. Together, these findings suggest that Tgf-β1 can influence Id2 mRNA levels. In 5/6Nx mice, we showed that Tgf-β1 induces hepatic Id2 expression through Smad3 and Tcf7l2 (Fig. 4, H and I) (54); however, several reports showed that TGF-β1 repressed the expression of the Id2 gene family in in vitro experiments (55). This may be partly explained by the difference in the cell lines. TGF-β1 is known to have functional differences in each organ. In addition, it shows different reactions between transient and chronic conditions. These findings suggest that this difference in organs can influence Id2 mRNA levels. TCF7L2 has been recently shown to be significantly associated with human CKD progression and with schizophrenic behavior in mice (56, 57). In our study, Id2 expression was induced by Tcf7l2 overexpression in NIH3T3 fibroblast cells (Fig. 4J). Furthermore, the hepatic expression of both Tcf7l2 protein and mRNA was significantly affected in 5/6Nx mice (Fig. 4L). Thus, hepatic Id2 expression in 5/6Nx mice might be regulated by Tcf7l2. Additionally, we further demonstrated that anti-TGF-β1 antibody administration could prevent the decreased expression of Cyp3a11, Cyp26a1, and Dbp or the increased expression of Id2, serum retinol, and serum BUN induced by Tgf-β1 in 5/6Nx mice. These inhibitory effects of TGF-β1 signaling might be important for the development of therapeutic strategies in CKD (Fig. 5).
The concentration of serum retinol, a fat-soluble vitamin A, is raised in CKD (Fig. 1A); however, the influences of excessive retinol on endogenous functions are poorly understood. We found that the greatest decrease in expression of hepatic Cyp3a11 and Cyp26a1, encoding key proteins in retinol metabolism, induced the accumulation of serum retinol and renal apoptosis in 5/6Nx mice fed a normal diet, whereas renal dysfunction was reduced in mice fed a retinol-free diet. With retinol (−) feeding, the serum concentration of retinol was not zero because the half-life of retinol is about 200 days. In addition, as apoptosis-related genes, p21 is an apoptosis marker in the kidney and is regulated by retinol (50). The levels of renal p21 were also reduced in 5/6Nx retinol (−) mice (Fig. 6F), but they were not changed in the liver (data not shown). Therefore, there was no effect on the liver because apoptosis did not occur. This is related to the mRNA levels of hepatic Cyp3a11, Cyp26a1, Dbp, and Id2 being unchanged in 5/6Nx retinol (−) mice (Fig. 6G). We assume that the influence of retinol-free feeding has renal-specific effects, because the mRNA levels of renal Tgf-β1 decreased in 5/6Nx retinol (−) mice (data not shown); however, plasma concentrations of TGF-β1 decreased slightly (Fig. 6H). Therefore, we assume that there is no influence on liver metabolism and hepatic clock genes.
In conclusion, we found that increased TGF-β1 levels are involved in the regulation of metabolic activity in the liver, which is mediated by DBP and TCF7L2. As a transcription factor, DBP is widely expressed in tissues throughout the body, including in blood vessels, and regulates the expression of numerous genes. Decreased expression of DBP might affect not only drug metabolism but also CKD pathologies, such as renal function and cardiovascular or cerebrovascular disease. Our findings might be applicable to the prevention of the aggravation of hepatic dysfunctions, adverse drug side effects, and pathologies in patients with CKD.
Author Contributions
K. H. and N. M. participated in the design of the study, conducted the experiments, analyzed the data, and drafted the manuscript. E. I., H. I., K. T., K. I., Y. F., Y. Y., and M. M. conducted the experiments. K. Y. and A. D. analyzed microarray data. Y. Y. provided Id2 (−/−) mice. H. K. provided the serum from CKD patients. T. O., H. A., and Y. I. provided technical assistance. S. K. and S. O. conceived the study, participated in its design and coordination, and wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgment
We thank the Research Support Center, Graduate School of Medical Sciences, Kyushu University, for technical support.
This study was supported in part by Grant-in-Aid for Scientific Research A 25253038 (to S. O.), Scientific Research on Innovative Areas Grant 25136716 (to S. O.), Challenging Exploratory Research Grant 25670079 (to S. O.), a grant from the Uehara Memorial Foundation (to S. O.), Scientific Research C Grant 24590196 (to N. M.), a grant from the Japan Research Foundation for Clinical Pharmacology (to N. M.), Grant-in-Aid for JSPS Fellows 25-4175 (to K. H.) from the Japan Society for the Promotion of Science, and a grant from the Fukuoka Foundation for Sound Health. The authors declare that they have no conflicts of interest with the contents of this article.
- CKD
- chronic kidney disease
- CYP
- cytochrome P450
- 5/6Nx
- 5/6 nephrectomy
- KEGG
- Kyoto Encyclopedia of Genes and Genomes
- ANOVA
- analysis of variance
- ZT
- zeitgeber time(s)
- TEF
- thyrotroph embryonic factor
- HLF
- hepatic leukemia factor.
References
- 1. Barsoum R. S. (2006) Chronic kidney disease in the developing world. N. Engl. J. Med. 354, 997–999 [DOI] [PubMed] [Google Scholar]
- 2. Hedayati S. S., Bosworth H. B., Briley L. P., Sloane R. J., Pieper C. F., Kimmel P. L., and Szczech L. A. (2008) Death or hospitalization of patients on chronic hemodialysis is associated with a physician-based diagnosis of depression. Kidney Int. 74, 930–936 [DOI] [PubMed] [Google Scholar]
- 3. Murray A. M. (2008) Cognitive impairment in the aging dialysis and chronic kidney disease populations: an occult burden. Adv. Chronic Kidney Dis. 15, 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Enomoto M., Inoue Y., Namba K., Munezawa T., and Matsuura M. (2008) Clinical characteristics of restless legs syndrome in end-stage renal failure and idiopathic RLS patients. Mov. Disord. 23, 811–816; quiz 926. [DOI] [PubMed] [Google Scholar]
- 5. Krishnan A. V., and Kiernan M. C. (2007) Uremic neuropathy: clinical features and new pathophysiological insights. Muscle Nerve 35, 273–290 [DOI] [PubMed] [Google Scholar]
- 6. Codreanu I., Perico N., and Remuzzi G. (2005) Dual blockade of the renin-angiotensin system: the ultimate treatment for renal protection? J. Am. Soc. Nephrol. 16, S34–S38 [DOI] [PubMed] [Google Scholar]
- 7. Rüster C., and Wolf G. (2006) Renin-angiotensin-aldosterone system and progression of renal disease. J. Am. Soc. Nephrol. 17, 2985–2991 [DOI] [PubMed] [Google Scholar]
- 8. Vitaterna M. H., King D. P., Chang A. M., Kornhauser J. M., Lowrey P. L., McDonald J. D., Dove W. F., Pinto L. H., Turek F. W., and Takahashi J. S. (1994) Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science 264, 719–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kume K., Zylka M. J., Sriram S., Shearman L. P., Weaver D. R., Jin X., Maywood E. S., Hastings M. H., and Reppert S. M. (1999) mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell 98, 193–205 [DOI] [PubMed] [Google Scholar]
- 10. Gekakis N., Staknis D., Nguyen H. B., Davis F. C., Wilsbacher L. D., King D. P., Takahashi J. S., and Weitz C. J. (1998) Role of the CLOCK protein in the mammalian circadian mechanism. Science 280, 1564–1569 [DOI] [PubMed] [Google Scholar]
- 11. Ripperger J. A., Shearman L. P., Reppert S. M., and Schibler U. (2000) CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 14, 679–689 [PMC free article] [PubMed] [Google Scholar]
- 12. Panda S., Antoch M. P., Miller B. H., Su A. I., Schook A. B., Straume M., Schultz P. G., Kay S. A., Takahashi J. S., and Hogenesch J. B. (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320 [DOI] [PubMed] [Google Scholar]
- 13. Ueda H. R., Chen W., Adachi A., Wakamatsu H., Hayashi S., Takasugi T., Nagano M., Nakahama K., Suzuki Y., Sugano S., Iino M., Shigeyoshi Y., and Hashimoto S. (2002) A transcription factor response element for gene expression during circadian night. Nature 418, 534–539 [DOI] [PubMed] [Google Scholar]
- 14. Ohdo S., Koyanagi S., and Matsunaga N. (2010) Chronopharmacological strategies: intra- and inter-individual variability of molecular clock. Adv. Drug Deliv. Rev. 62, 885–897 [DOI] [PubMed] [Google Scholar]
- 15. Ohdo S., Koyanagi S., Matsunaga N., and Hamdan A. (2011) Molecular basis of chronopharmaceutics. J. Pharm. Sci. 100, 3560–3576 [DOI] [PubMed] [Google Scholar]
- 16. Bass J. (2012) Circadian topology of metabolism. Nature 491, 348–356 [DOI] [PubMed] [Google Scholar]
- 17. Singh A. K., and Kari J. A. (2013) Metabolic syndrome and chronic kidney disease. Curr. Opin. Nephrol. Hypertens. 22, 198–203 [DOI] [PubMed] [Google Scholar]
- 18. Dreisbach A. W., Japa S., Gebrekal A. B., Mowry S. E., Lertora J. J., Kamath B. L., and Rettie A. E. (2003) Cytochrome P4502C9 activity in end-stage renal disease. Clin. Pharmacol. Ther. 73, 475–477 [DOI] [PubMed] [Google Scholar]
- 19. Pichette V., and Leblond F. A. (2003) Drug metabolism in chronic renal failure. Curr. Drug Metab. 4, 91–103 [DOI] [PubMed] [Google Scholar]
- 20. Touchette M. A., and Slaughter R. L. (1991) The effect of renal failure on hepatic drug clearance. DICP 25, 1214–1224 [DOI] [PubMed] [Google Scholar]
- 21. Talbert R. L. (1994) Drug dosing in renal insufficiency. J. Clin. Pharmacol. 34, 99–110 [DOI] [PubMed] [Google Scholar]
- 22. Popper H., Steigmann F., and Dyniewicz H. A. (1945) Plasma vitamin A level in renal diseases. Am. J. Clin. Pathol. 15, 272–277 [Google Scholar]
- 23. Smith F. R., and Goodman D. S. (1971) Effects of diseases of the liver, thyroid and kidneys on the transport of vitamin A in human plasma. J. Clin. Invest. 50, 2426–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yatzidis H., Digenis P., and Fountas P. (1975) Hypervitaminosis A accompanying advanced chronic renal failure. Br. Med. J. 3, 352–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ross A. C., and Zolfaghari R. (2011) Cytochrome P450s in the regulation of cellular retinoic acid metabolism. Annu. Rev. Nutr. 31, 65–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fujii H., Sato T., Kaneko S., Gotoh O., Fujii-Kuriyama Y., Osawa K., Kato S., and Hamada H. (1997) Metabolic inactivation of retinoic acid by a novel P450 differentially expressed in developing mouse embryos. EMBO J. 16, 4163–4173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Luu L., Ramshaw H., Tahayato A., Stuart A., Jones G., White J., and Petkovich M. (2001) Regulation of retinoic acid metabolism. Adv. Enzyme Regul. 41, 159–175 [DOI] [PubMed] [Google Scholar]
- 28. Leber H. W., and Schütterle G. (1972) Oxidative drug metabolism in liver microsomes from uremic rats. Kidney Int. 2, 152–158 [DOI] [PubMed] [Google Scholar]
- 29. Van Peer A. P., and Belpaire F. M. (1977) Hepatic oxidative drug metabolism in rats with experimental renal failure. Arch. Int. Pharmacodyn. Ther. 228, 180–183 [PubMed] [Google Scholar]
- 30. Leber H. W., Gleumes L., and Schutterle G. (1978) Enzyme induction in the uremic liver. Kidney Int. Suppl. S43–S48 [PubMed] [Google Scholar]
- 31. Patterson S. E., and Cohn V. H. (1984) Hepatic drug metabolism in rats with experimental chronic renal failure. Biochem. Pharmacol. 33, 711–716 [DOI] [PubMed] [Google Scholar]
- 32. Uchida N., Kurata N., Shimada K., Nishimura Y., Yasuda K., Hashimoto M., Uchida E., and Yasuhara H. (1995) Changes of hepatic microsomal oxidative drug metabolizing enzymes in chronic renal failure (CRF) rats by partial nephrectomy. Jpn. J. Pharmacol. 68, 431–439 [DOI] [PubMed] [Google Scholar]
- 33. Leblond F., Guévin C., Demers C., Pellerin I., Gascon-Barré M., and Pichette V. (2001) Downregulation of hepatic cytochrome P450 in chronic renal failure. J. Am. Soc. Nephrol. 12, 326–332 [DOI] [PubMed] [Google Scholar]
- 34. Michaud J., Dubé P., Naud J., Leblond F. A., Desbiens K., Bonnardeaux A., and Pichette V. (2005) Effects of serum from patients with chronic renal failure on rat hepatic cytochrome P450. Br. J. Pharmacol. 144, 1067–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kanehisa M., Goto S., Kawashima S., and Nakaya A. (2002) The KEGG databases at GenomeNet. Nucleic Acids Res. 30, 42–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Megyesi J., Safirstein R. L., and Price P. M. (1998) Induction of p21WAF1/CIP1/SDI1 in kidney tubule cells affects the course of cisplatin-induced acute renal failure. J. Clin. Invest. 101, 777–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matsunaga N., Ikeda M., Takiguchi T., Koyanagi S., and Ohdo S. (2008) The molecular mechanism regulating 24-hr rhythm of CYP2E1 expression in the mouse liver. Hepatology 48, 240–251 [DOI] [PubMed] [Google Scholar]
- 38. Okazaki F., Matsunaga N., Okazaki H., Utoguchi N., Suzuki R., Maruyama K., Koyanagi S., and Ohdo S. (2010) Circadian rhythm of transferrin receptor 1 gene expression controlled by c-Myc in colon cancer-bearing mice. Cancer Res. 70, 6238–6246 [DOI] [PubMed] [Google Scholar]
- 39. Takiguchi T., Tomita M., Matsunaga N., Nakagawa H., Koyanagi S., and Ohdo S. (2007) Molecular basis for rhythmic expression of CYP3A4 in serum-shocked HepG2 cells. Pharmacogenet. Genomics 17, 1047–1056 [DOI] [PubMed] [Google Scholar]
- 40. Ali R., Campos B., Dyckhoff G., Haefeli W. E., Herold-Mende C., and Burhenne J. (2012) Quantification of retinoid concentrations in human serum and brain tumor tissues. Anal. Chim. Acta 725, 57–66 [DOI] [PubMed] [Google Scholar]
- 41. Arima H., Yamashita S., Mori Y., Hayashi Y., Motoyama K., Hattori K., Takeuchi T., Jono H., Ando Y., Hirayama F., and Uekama K. (2010) In vitro and in vivo gene delivery mediated by lactosylated dendrimer/α-cyclodextrin conjugates (G2) into hepatocyte. J. Control. Release 146, 106–117 [DOI] [PubMed] [Google Scholar]
- 42. Wang D., Guo Y., Wrighton S. A., Cooke G. E., and Sadee W. (2011) Intronic polymorphism in CYP3A4 affects hepatic expression and response to statin drugs. Pharmacogenomics J. 11, 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gachon F., Olela F. F., Schaad O., Descombes P., and Schibler U. (2006) The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 4, 25–36 [DOI] [PubMed] [Google Scholar]
- 44. Ripperger J. A., and Schibler U. (2006) Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat. Genet. 38, 369–374 [DOI] [PubMed] [Google Scholar]
- 45. Hou T. Y., Ward S. M., Murad J. M., Watson N. P., Israel M. A., and Duffield G. E. (2009) ID2 (inhibitor of DNA binding 2) is a rhythmically expressed transcriptional repressor required for circadian clock output in mouse liver. J. Biol. Chem. 284, 31735–31745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ward S. M., Fernando S. J., Hou T. Y., and Duffield G. E. (2010) The transcriptional repressor ID2 can interact with the canonical clock components CLOCK and BMAL1 and mediate inhibitory effects on mPer1 expression. J. Biol. Chem. 285, 38987–39000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nakano N., Itoh S., Watanabe Y., Maeyama K., Itoh F., and Kato M. (2010) Requirement of TCF7L2 for TGF-β-dependent transcriptional activation of the TMEPAI gene. J. Biol. Chem. 285, 38023–38033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wrana J. L., Attisano L., Wieser R., Ventura F., and Massagué J. (1994) Mechanism of activation of the TGF-β receptor. Nature 370, 341–347 [DOI] [PubMed] [Google Scholar]
- 49. Chen C. H., Hsieh T. J., Lin K. D., Lin H. Y., Lee M. Y., Hung W. W., Hsiao P. J., and Shin S. J. (2012) Increased unbound retinol-binding protein 4 concentration induces apoptosis through receptor-mediated signaling. J. Biol. Chem. 287, 9694–9707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yu Z., Li W., Lu Q., Wang L., Zhang X., Han P., Chen P., and Pei Y. (2008) p21 is required for atRA-mediated growth inhibition of MEPM cells, which involves RAR. J. Cell Biochem. 104, 2185–2192 [DOI] [PubMed] [Google Scholar]
- 51. Tischkau S. A., Mitchell J. W., Tyan S. H., Buchanan G. F., and Gillette M. U. (2003) Ca2+/cAMP response element-binding protein (CREB)-dependent activation of Per1 is required for light-induced signaling in the suprachiasmatic nucleus circadian clock. J. Biol. Chem. 278, 718–723 [DOI] [PubMed] [Google Scholar]
- 52. Lim C., Allada R. (2013) Emerging roles for post-transcriptional regulation in circadian clocks. Nat. Neurosci. 16, 1544–1550 [DOI] [PubMed] [Google Scholar]
- 53. Aitken A. E., and Morgan E. T. (2007) Gene-specific effects of inflammatory cytokines on cytochrome P450 2C, 2B6 and 3A4 mRNA levels in human hepatocytes. Drug Metab. Dispos. 35, 1687–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cao Y., Liu X., Zhang W., Deng X., Zhang H., Liu Y., Chen L., Thompson E. A., Townsend C. M. Jr., and Ko T. C. (2009) TGF-β repression of Id2 induces apoptosis in gut epithelial cells. Oncogene 28, 1089–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kon N., Hirota T., Kawamoto T., Kato Y., Tsubota T., and Fukada Y. (2008) Activation of TGF-β/activin signalling resets the circadian clock through rapid induction of Dec1 transcripts. Nat. Cell Biol. 10, 1463–1469 [DOI] [PubMed] [Google Scholar]
- 56. Köttgen A., Hwang S. J., Rampersaud E., Coresh J., North K. E., Pankow J. S., Meigs J. B., Florez J. C., Parsa A., Levy D., Boerwinkle E., Shuldiner A. R., Fox C. S., and Kao W. H. (2008) TCF7L2 variants associate with CKD progression and renal function in population-based cohorts. J. Am. Soc. Nephrol. 19, 1989–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Savic D., Distler M. G., Sokoloff G., Shanahan N. A., Dulawa S. C., Palmer A. A., and Nobrega M. A. (2011) Modulation of Tcf7l2 expression alters behavior in mice. PLoS One 6, e26897. [DOI] [PMC free article] [PubMed] [Google Scholar]