Abstract
Recombination between homologous chromosomes is required for the faithful meiotic segregation of chromosomes and leads to the generation of genetic diversity. The conserved meiosis-specific Dmc1 recombinase catalyzes homologous recombination triggered by DNA double strand breaks through the exchange of parental DNA sequences. Although providing an efficient rate of DNA strand exchange between polymorphic alleles, Dmc1 must also guard against recombination between divergent sequences. How DNA mismatches affect Dmc1-mediated DNA strand exchange is not understood. We have used fluorescence resonance energy transfer to study the mechanism of Dmc1-mediated strand exchange between DNA oligonucleotides with different degrees of heterology. The efficiency of strand exchange is highly sensitive to the location, type, and distribution of mismatches. Mismatches near the 3′ end of the initiating DNA strand have a small effect, whereas most mismatches near the 5′ end impede strand exchange dramatically. The Hop2-Mnd1 protein complex stimulates Dmc1-catalyzed strand exchange on homologous DNA or containing a single mismatch. We observed that Dmc1 can reject divergent DNA sequences while bypassing a few mismatches in the DNA sequence. Our findings have important implications in understanding meiotic recombination. First, Dmc1 acts as an initial barrier for heterologous recombination, with the mismatch repair system providing a second level of proofreading, to ensure that ectopic sequences are not recombined. Second, Dmc1 stepping over infrequent mismatches is likely critical for allowing recombination between the polymorphic sequences of homologous chromosomes, thus contributing to gene conversion and genetic diversity.
Keywords: DNA-binding protein, DNA damage, DNA recombination, homologous recombination, meiosis, DNA mismatches, DNA strand exchange, Dmc1, Hop2-Mnd1, heterologous DNA sequences
Introduction
Homologous recombination (HR)2 is a major, conserved pathway for the repair of DNA double strand breaks (DSBs). During meiosis, HR establishes physical linkages between homologous chromosomes from maternal and paternal origins as pairs. These linkages produce chiasmata, which are the cytological manifestation of the crossover products of HR. Together with sister chromatid cohesion, these chromosome crossovers ensure the orderly segregation of each chromosome pair in the first meiotic division (1, 2). The cells derived from the meiotic cell division have half the number of chromosomes as their parent, which is an essential feature of sexual reproduction involving the generation of haploid gametes. Errors in meiotic chromosome segregation lead to miscarriages and Down, Klinefelter, Edwards, and Turner syndromes in humans, which are all characterized by aneuploidy stemming from a deficiency in meiotic chromosome segregation (3–5).
Meiotic HR is initiated by cleavage of chromosomal DNA at multiple sites by the Spo11 protein to generate DSBs (6). These DSBs are processed by nucleases to generate 3′ single-stranded DNA (ssDNA) tails (7, 8). A recombinase polymerizes on the ssDNA tails to form a helical nucleoprotein filament, termed the presynaptic filament, capable of searching for, interacting with, and invading a homologous double-stranded DNA (dsDNA) target in a reaction named homologous pairing (9–11). The heteroduplex region formed by homologous pairing is expanded in the next step, referred to as DNA strand exchange. In most eukaryotes, homologous DNA pairing and strand exchange are catalyzed by Dmc1 and Rad51 proteins, which are structural and functional homologues of the bacterial recombinase RecA. Whereas Dmc1 acts specifically in meiosis, Rad51 functions in both meiosis and mitotic cells.
Previous studies have suggested that, similar to Rad51 and RecA, the recognition of DNA homology by human Dmc1, which is strongly influenced by helix stability and mismatched base pairs, requires the preferential breathing of A:T base pairs (12–14). Although bacterial RecA and eukaryotic Rad51 proteins have been well characterized, less is known about the mechanism by which Dmc1 promotes the recognition of homology and discriminates between homeologous DNA sequences.
Notably, in vitro, eukaryotic recombinases usually exhibit a less robust strand exchange activity and higher tolerance to interruptions of homology than RecA protein (15). At present, it is not clear if the ability of the eukaryotic recombinases to tolerate DNA mismatches during the catalysis of homologous DNA pairing and strand exchange is subject to regulation by their accessory factors. Indeed, the proper function of Dmc1 and Rad51 requires their interactions with several accessory proteins (see Refs. 9, 11, and 16 and citations within). Hop2 and Mnd1 are two conserved proteins essential for HR completion via their interactions with Dmc1 and Rad51 (17–22). In Hop2−/− and Mnd1−/− mouse spermatocytes, Dmc1 or Rad51 is loaded onto the resected ends of DSBs, but further progression of recombination is impaired (23, 24). Biochemical studies have shown that Hop2 and Mnd1 are associated in a heterodimeric complex that stimulates the DNA strand invasion activity of Rad51 and Dmc1, and it acts by stabilizing Dmc1/Rad51 nucleoprotein filaments on ssDNA, and by facilitating the condensation and capture of dsDNA in conjunction with the presynaptic filament (17, 25). In this study, we have analyzed the effect of Hop2-Mnd1 in the Dmc1-catalyzed strand exchange reaction using homologous or mismatch-containing oligonucleotide substrates.
Considering that homologous chromosomes diverge in their primary sequence (26), the ability of Dmc1 to tolerate mismatches in the heteroduplex DNA joints may be crucial for successful HR between parental chromosomes. However, to ensure that HR occurs only between homologs, the Dmc1-mediated DNA strand exchange reaction must also exhibit a certain limit of tolerance of the number and types of DNA mismatches in carrying out DNA pairing and strand exchange.
The lack of information in this important aspect of meiotic recombination has prompted us to study the effect of DNA mismatches on the efficiency of Dmc1-mediated DNA strand exchange. Specifically, we have carried out a systematic study of Dmc1-catalyzed DNA strand exchange with different mismatched substrates using fluorescence resonance energy transfer (FRET) techniques. Our results show that Dmc1-mediated strand exchange is highly sensitive to the location, type, and distribution of mismatches. The presence of a few mismatches at specific locations of the incoming strand can inhibit strand exchange considerably. This rejection of the incoming strand, which cannot be explained merely by changes in the stability of the duplex DNA, suggests a mechanism that uses ATP hydrolysis to amplify the fidelity of homology search, being akin to kinetic proofreading (27, 28). Furthermore, addition of the Hop2-Mnd1 heterodimer enhances the efficiency of DNA strand exchange with the mismatched substrates. A DNA binding site present on Mnd1 is needed for this Hop2-Mnd1 attribute.
Experimental Procedures
DNA Substrates
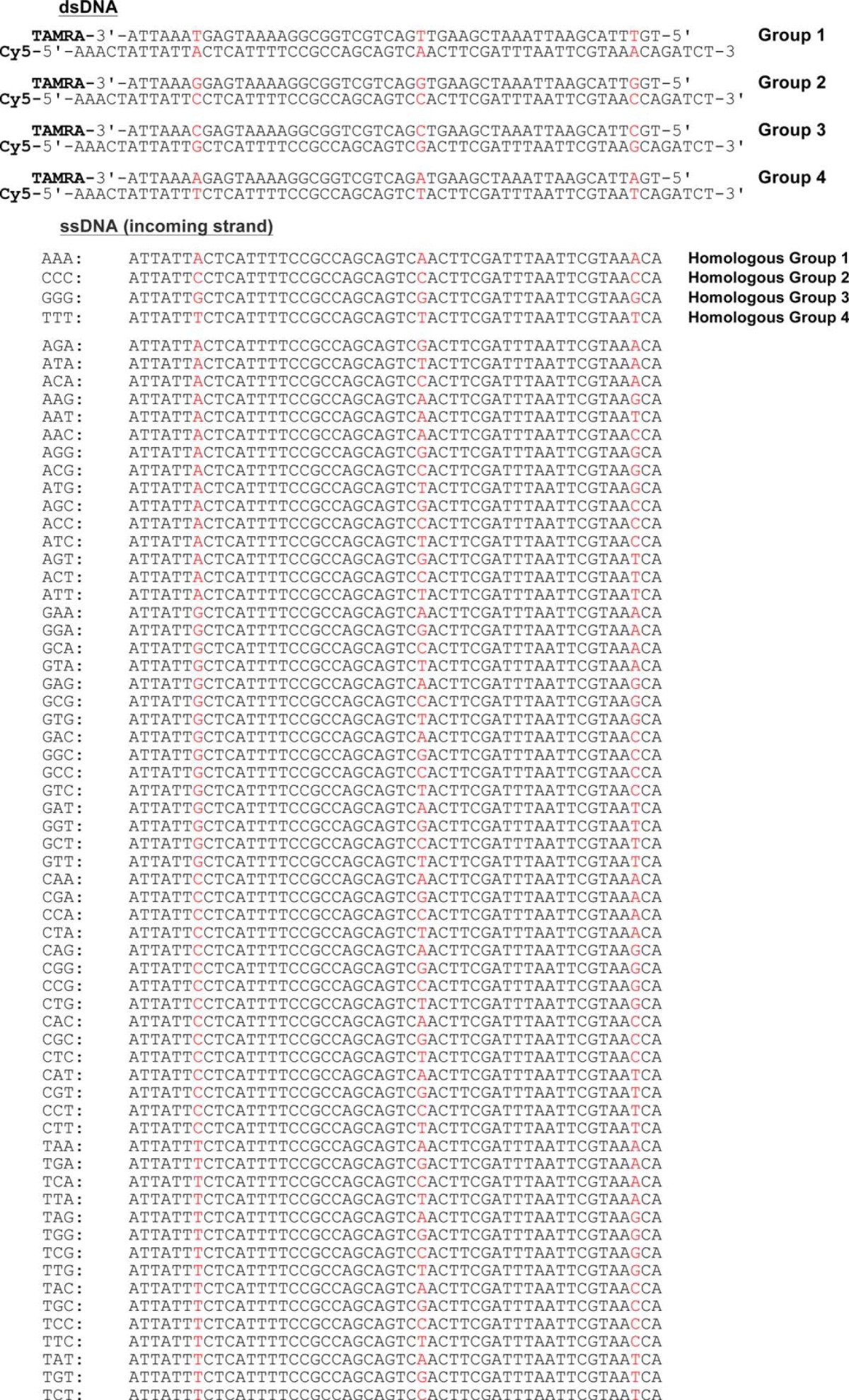
All the oligonucleotides were purchased from Integrated DNA Technologies (IDT, Coralville, IA). Selected oligonucleotides were fluorescently labeled at the 3′ end with tetramethylrhodamine (TAMRA) or at the 5′ end with Cy5 and were purified by HPLC and PAGE. Unlabeled oligonucleotides were purified by PAGE. To prepare dsDNA, the complementary strands were mixed in a 1:1 molar ratio in 10 mm Tris-HCl (pH 7.5), 100 mm NaCl, 10 mm MgCl2, and hybridized by cooling from 90 °C to room temperature over 2.5 h. The sequences of the oligonucleotides are shown in Table 1. The concentration of ssDNA and dsDNA was determined by absorbance at 260 nm and is expressed as molar concentration of nucleotides or base pairs for ssDNA or dsDNA.
TABLE 1.
Sequences of dsDNA and invading ssDNA used in this study
Variable nucleotides are highlighted in color.
Proteins
Human Dmc1 was purified as described previously (29) with modifications. Briefly, the protein was expressed in Escherichia coli BL21 (DE3)pLysS carrying a pET-16b-Dmc1 plasmid using the INDUCERTM (moleculA, VA 20166) and purified by consecutive chromatographic steps on nickel-nitrilotriacetic acid (Thermo Fisher Scientific), reactive blue (Bio-Rad), heparin-Sepharose, and MonoQ (GE Healthcare) columns. Mouse wild type and mutant Hop2-Mnd1 complexes were expressed in the codon plus PR strain (Stratagene) carrying the pET15b-Hop2-Mnd1 plasmid and purified as described previously (19, 22). Briefly, the protein complex was purified by co-expressing His-tagged Hop2 and untagged Mnd1 in the same expression system and employing consecutive chromatographic steps on nickel-nitrilotriacetic acid, MonoQ, Macro hydroxyapatite (Bio-Rad), heparin-Sepharose, and MonoS columns. The concentration of proteins was determined by the Bradford assay. E. coli RecA protein was purchased from New England Biolabs Inc.
DNA Strand Exchange Reactions
Nucleoprotein complexes were formed by incubating Dmc1 protein (3 μm) with unlabeled 50-mer ssDNA (10 μm nucleotides) for 8 min at 37 °C in buffer (250 μl, final volume) containing 20 mm Tris-HCl (pH 7.5), 50 mm KCl, 2 mm MgCl2, 3 mm ATP, and 100 μg/ml of BSA. The strand exchange reaction was initiated by adding labeled dsDNA (5 μm base pairs) and incubated at 37 °C for 40 min. Variation in the fluorescence emission (FE) of TAMRA was recorded every 1.25 s at 580 nm on excitation at 556 nm. FRET measurements were performed using an ISS-PC1 photon counting spectrofluorometer (ISS Inc.).
For DNA strand exchange reactions done with Hop2-Mnd1, Dmc1 (3 μm) was incubated with ssDNA in the same buffer for 5 min at 37 °C, followed by the addition of wild type or mutant Hop2-Mnd1 and incubated for an additional 5 min. Then, the strand exchange reaction was initiated by the addition of labeled dsDNA. Dmc1 and Hop2-Mnd1 were used at a 4:1 molar ratio. RecA-ssDNA filaments were formed by incubating RecA (0.4 μm) with unlabeled ssDNA (10 μm nucleotides) for 2 min at 37 °C. The DNA and buffer conditions were identical as described for the Dmc1 reaction, except that ATP was present at 1 mm and an ATP-regenerating system (7.5 mm creatine phosphate and 30 units ml−1 creatine kinase) was added. The reaction was carried out for 22 min.
FRET Measurements and Analysis
Fig. 1A illustrates our experimental design for measuring DNA strand exchange catalyzed by the recombinases (see also Results). The fraction of dsDNA having undergone strand exchange (x) was calculated using the following equation: d2.x + d1.(1-x) = d; where “d1” is the fluorescence emission from the donor (TAMRA) measured in a sample containing fully hybridized labeled duplexes, “d2” is the emission from the donor measured in a sample containing non-hybridized labeled oligonucleotides, and “d” is the measured emission of the donor, respectively. The relative strand exchange efficiency was calculated as the increase of FE at 580 nm at the end of the reaction (tf) relative to the value obtained with the homologous ssDNA (which was set as 1 for each oligonucleotide group): (FEtf − FEt0) ssDNA/(FEtf − FEt0) homologous.
FIGURE 1.
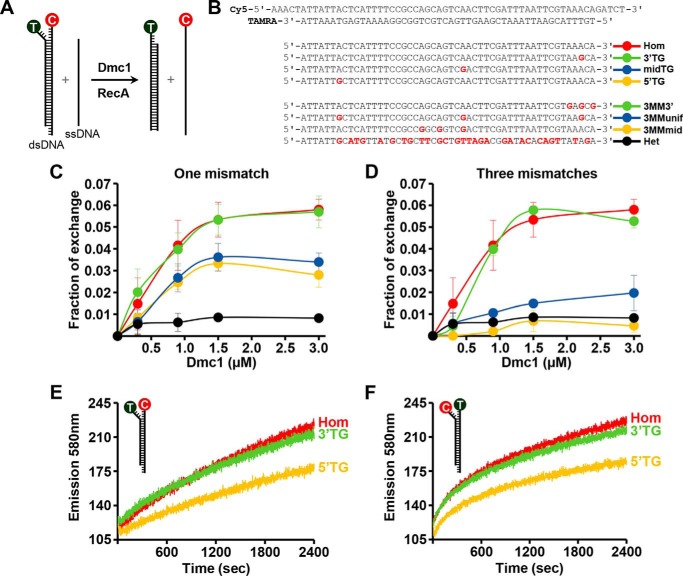
Effect of mismatches on Dmc1-promoted strand exchange. A, experimental scheme to study Dmc1-catalyzed recombination in vitro. T, TAMRA (λ excitation: 556 nm; λ emission: 580 nm). C, Cy5 (λ excitation: 649 nm; λ emission: 665 nm). B, labeled duplex and the corresponding invading unlabeled ssDNA sequences used in the experiment. Variable nucleotides are highlighted in red. Hom, 100% homologous oligonucleotide; Het, heterologous; 60% of divergence; 5′TG, midTG, and 3′TG refers to a single TG mismatch at 5′ end, middle, or 3′ end, respectively, of the incoming strand; 3MMunif, three mismatches uniformly distributed; 3MM3′ and 3MMmid, three mismatches clustered on the 3′ end or near the central region of the incoming strand. C and D, fraction of exchange as a function of Dmc1 concentration for one TG mismatch in different positions or three TG mismatches with different distributions. Reactions shown in panels C and D were carried out for 40 min. Each data point represents an average of three independent determinations. Error bars are the S.D. E and F, effect of reporter dye placement on Dmc1 strand exchange between homologous DNA substrates or containing one mismatch at the 5′ or 3′ end.
Results
Assays of DNA Strand Exchange by FRET
An essential attribute of recombinases is the ability to search for a homologous sequence amid an excess of heterologous DNA, and to discriminate against target sequences that are not completely homologous to the initiating ssDNA. During DNA homology search, the Dmc1-ssDNA presynaptic filament engages a duplex DNA molecule and samples segments of the duplex DNA until homology is found. However, for successful recombination between chromosome homologs to occur, the presynaptic filament must be able to tolerate a certain level of heterology between the recombining DNA molecules. These characteristics for Dmc1-mediated strand exchange reactions are poorly understood. We have examined the influence of DNA heterologies on Dmc1-promoted HR by conducting a systematic study of DNA strand exchange between oligonucleotides with different degrees of heterology using a FRET-based method. The scheme shown in Fig. 1A illustrates our experimental design, which follows procedures described previously (27). Briefly, linear dsDNA was formed by annealing complementary oligonucleotides each labeled with a fluorescent dye that constitutes a donor (3′TAMRA)-acceptor (5′Cy5) pair (Fig. 1A). The labeled dsDNA was mixed with linear unlabeled ssDNA complementary to the TAMRA strand and that was preincubated with Dmc1. In the absence of strand exchange, the donor-acceptor pair yields high transfer efficiency due to their close proximity. Fluorescence emission of the donor (TAMRA) at 580 nm becomes greatly enhanced upon DNA strand exchange because of the replacement of the Cy5-oligonucleotide by the unlabeled DNA. In experiments to be described in the following sections, the FRET assay was used to study the effects of the position, number, and type of DNA mismatches on the efficiency of strand exchange catalyzed by human Dmc1 and E. coli RecA.
Location-specific Effect of DNA Mismatches on Dmc1-mediated DNA Strand Exchange
In initial experiments, we examined ssDNA substrates that differ from the 50-bp duplex target by a single TG mismatch or three such mismatches being located at different positions (Fig. 1B). Fig. 1, C and D, show the fraction of duplex DNA that undergoes exchange as a function of Dmc1 concentration. Under the experimental conditions used, Dmc1 binds to all the ssDNA oligonucleotides used in our study (not shown). With the fully homologous DNA substrates, the fraction of the duplex substrate that had undergone exchange was 0.07 with 3 μm Dmc1 (Fig. 1, C and D), reaching a maximum of 0.15 with 6 μm Dmc1 (not shown). The presence of one TG mismatch near the 3′ end of the initiating ssDNA (3′ TG) did not affect strand exchange appreciably, but a mismatch either in the middle or the 5′ end of the ssDNA (midTG and 5′ TG, respectively) had a significant negative impact in this regard (Fig. 1, B and C). With the substrates that harbor 3 mismatches, the location of the mismatches also had a distinct effect on strand exchange efficiency. Specifically, whereas little negative effect occurred with the ssDNA wherein all 3 mismatches are located near the 3′ end of the DNA (3MM3′), a large reduction in strand exchange efficiency was seen when the mismatches were either uniformly distributed over the substrate (3MMunif) or clustered within the middle of the substrate (3MMmid) (Fig. 1, B and D). With the heterologous ssDNA (28 mismatches, 60% of divergence), DNA strand exchange was reduced to the background level. We note that the use of oligonucleotides with slightly different sequences did not apparently influence the effect of the number and position of mismatches in DNA strand exchange (oligonucleotides Group 1–4, see below).
We next asked whether the placement of the reporter dyes would affect the outcome of strand exchange. We prepared another set of substrates with the locations of the fluorophores being reversed (i.e. TAMRA at the 5′ end and Cy5 at the 3′ end) in the duplex substrate. Importantly, we again observed a similar bias in strand exchange, with the 5′ mismatch affecting the efficiency of the reaction much more than the 3′ mismatch (Fig. 1, E and F).
Taken together, the results show that the presence of a single mismatch in 50-mer substrates can affect Dmc1-mediated DNA strand exchange significantly, although the position of the mismatch appears to be a critical factor in this regard. The general trend is that strand exchange efficiency decreases with the mismatch being proximal to the 5′ end of the initiating ssDNA strand.
The Effect of DNA Mismatches Is Unrelated to Strand Exchange Directionality or Stability of the Duplex Product
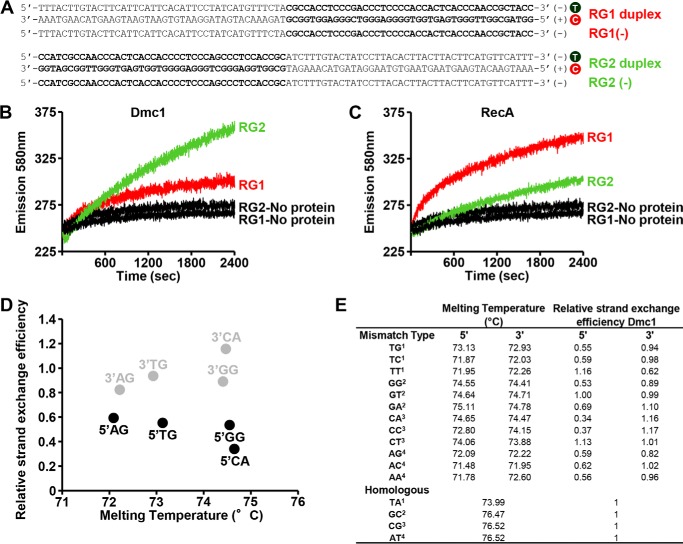
The strong influence of mismatches located at the 5′ versus 3′ end of the initiating ssDNA could stem from the directionality in the Dmc1-mediated strand exchange process. Previous studies showed that DNA strand exchange efficiency is attenuated when the GC content of the substrates increases (30). Therefore, strand exchange was assayed using oligonucleotide substrates with a GC-rich region present at either the 5′ or 3′ end of the ssDNA (Fig. 2A). The presynaptic filament was formed on the oligonucleotide RG1(−) (3′, 71% GC content, 42-mer) or RG2(−) (5′, 71% GC content, 42-mer) and paired with the corresponding duplex. Dmc1 could mediate DNA strand exchange with both pairs of substrates, but more efficiently with the ssDNA that harbors high GC content at its 5′ end (Fig. 2B). The strand exchange bias observed is consistent with strand exchange being propagated in the 3′-5′ direction with respect to the initiating ssDNA. As a control, we examined the action of RecA protein, which is known to promote exchange in the 5′-3′ direction (30). In contrast to what was observed for Dmc1, the RecA reaction occurred more rapidly with the ssDNA substrate in which the GC-rich region lies within the 3′ end of the substrate (Fig. 2C). The findings reported here show that in short range interactions involving the exchange of a few dozen base pairs, Dmc1 promotes strand exchange in a 3′-5′ direction, which is opposite to that of RecA. Thus, the strong influence of 5′ mismatches on Dmc1 may be unrelated to the directionality of the strand exchange reaction.
FIGURE 2.
The directionality of strand exchange and stability of the dsDNA product of strand exchange do not influence Dmc1 strand exchange efficiency. A, chimeric DNA substrates used to study the directionality of the strand exchange. Nucleotides in bold show the 71% GC-rich region. T, TAMRA; C, Cy5. Sequences of the single-strand oligonucleotides, either RG1(−) (3′, 71% GC-rich) or RG2(−) (5′, 71% GC-rich), are 100% complementary to RG1(+)C or RG2(+)C, respectively. B and C, the kinetics of strand exchange promoted by Dmc1 or RecA with substrates shown in A. D and E, comparative analysis between the melting temperature of dsDNA formed by the Dmc1 strand exchange reaction (homologous or containing single mismatches) and relative strand exchange efficiency obtained during the strand exchange reaction promoted by Dmc1. Melting temperature for each duplex was calculated using the software “MELTING 5” (45). The superscripts in E indicate the corresponding Group (see Fig. 3A).
Alternatively, inhibition of strand exchange by mismatches located at the 5′ end of the initiating ssDNA may be explained by differences in the stability of the dsDNA product of the reaction. In this case, lower duplex DNA melting temperature may favor the reverse reaction of strand exchange. However, a comparative analysis indicates that the melting temperature of duplex products containing specific single mismatches at either the 5′ or 3′ end of the initiating ssDNA is similar (Fig. 2, D and E), but the relative efficiency of strand exchange involving these substrates is clearly dependent on the location and type of the mismatch. In summary, there is no correlation between the melting temperature of the duplex product of strand exchange and the strength of inhibition of the strand exchange reaction by a DNA mismatch.
Differential Effect of Type and Position of Mismatches on DNA Strand Exchange
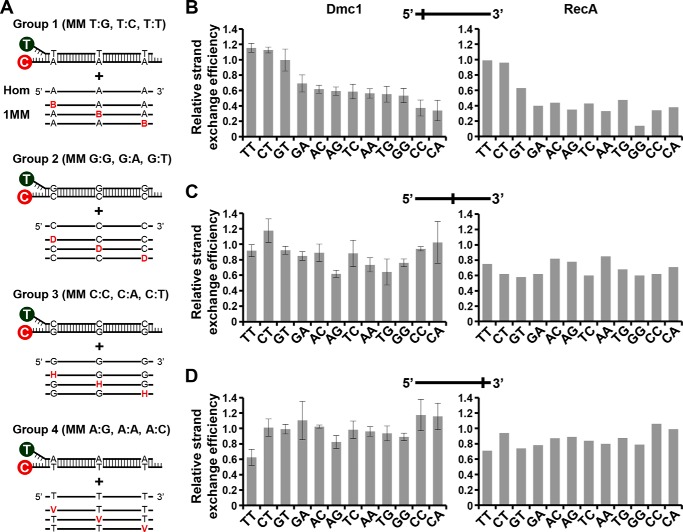
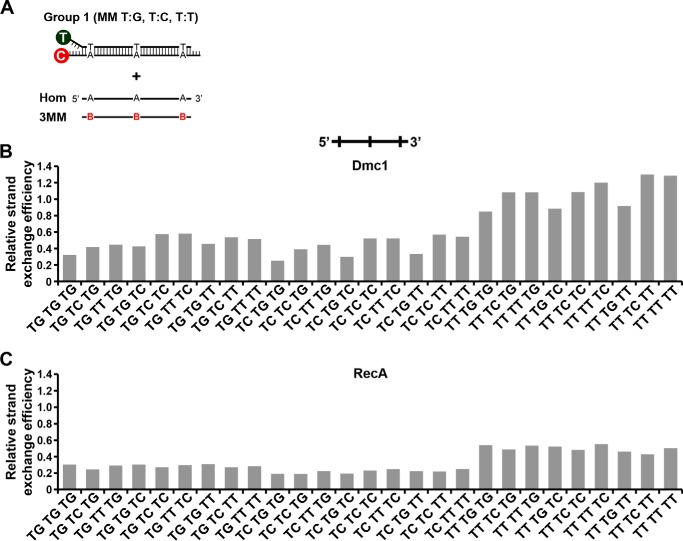
We asked whether the DNA mismatch type exert an effect on the strand exchange efficiency. For this, we constructed four different duplex substrates with each differing by 1 base pair in three fixed positions to each ssDNA substrates that we used alongside the control that is completely homologous to the duplex (Fig. 3A). This allowed us to study the effect of all 12 types of single mismatches at three different positions between the recombining DNA molecules (near the 5′ end, in the middle, or near 3′ end). The results from this extensive series of experiments are shown in Fig. 3, B–D. The general trend is that Dmc1 strand exchange is more sensitive to different types of mismatches when they are located at the 5′ end of the initiating ssDNA, with the following order of severity of inhibition: CA ≈ CC > GG ≈ TG ≈ AA ≈ TC ≈ AG ≈ AC ≈ GA > GT ≈ CT ≈ TT. Some mismatches located in the center of the ssDNA substrate, namely, TG, GG, AG and AA, also resulted in an inhibition of strand exchange, but most mismatches located at the 3′ end did not show any inhibitory effect. Interestingly, contrary to that observed when a TT mismatch was located at the 5′ end, a TT mismatch at the 3′ end resulted in a large reduction of Dmc1 strand exchange efficiency. We also note that, whereas TG and TC mismatches at the 5′ end inhibited strand exchange considerably, GT and CT mismatches had no effect.
FIGURE 3.
Effect of type and position of a single mismatch on Dmc1 and RecA strand exchange efficiency. A, schematic representation of mismatch type and position used in this study. 12 types of mismatches at three different positions of the invading ssDNA (near the 5′ end, in the middle, or near the 3′ end) were generated using four labeled dsDNA (Groups 1, 2, 3, and 4). The three types of mismatches generated for each group are indicated. Variable nucleotides in the incoming ssDNA strand are highlighted in color. Nucleotide codes are: B: G, C, or T; D: G, A, or T; H: C, A, or T; and V: G, A, or C. For each group, we used a homologous (Hom) and 9 different ssDNA to generate a single mismatch. Oligonucleotide sequences of the labeled dsDNA and the invading ssDNA are shown in Table 1. B–D, relative strand exchange efficiency of Dmc1 and RecA with the ssDNA oligonucleotide containing different types of single mismatches located near the 5′ end (B), middle (C), or 3′ end (D) of the incoming strand. The homologous ssDNA was arbitrary considered as 1 for each Group. Values are the mean ± S.D. from three independent experiments. For RecA, results from duplicate experiments are shown.
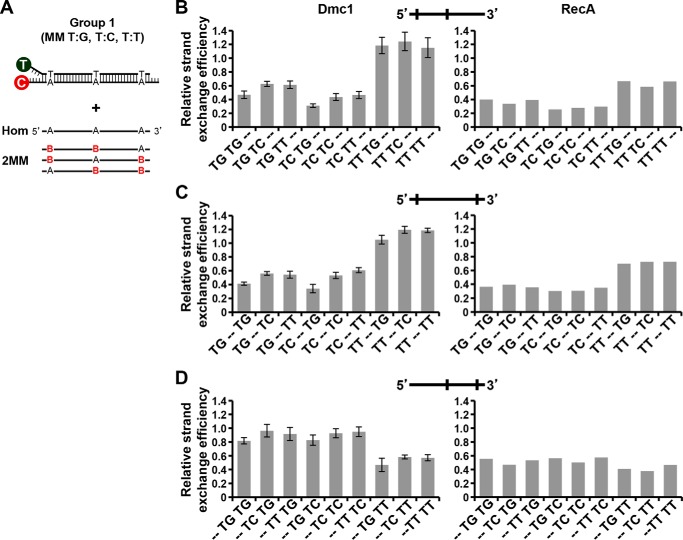
We proceeded to analyze the effect on Dmc1-catalyzed strand exchange of oligonucleotides containing two and three mismatches. The results obtained with these substrates (oligonucleotides of Group 1) are shown in Figs. 4 and 5, respectively. We observed that the efficiency of strand exchange mainly depends on the type of mismatch present at the 5′ position, whereas the frequency of mismatches has less influence. For example, the presence of a TG or TC mismatch at the 5′ position (in oligonucleotides containing two or three mismatches) led to a strong inhibition of strand exchange, independently of the mismatch type present in the middle or in the 3′ end (Figs. 4, B and C, and 5B). Otherwise, the presence of a 5′ TT mismatch resulted in relatively low inhibition of strand exchange. However, the presence of a 3′ TT mismatch resulted in an increased inhibitory effect when another mismatch was located in the middle (Fig. 4D). The inhibitory positioning effect of a given mismatch may be explained by changes in structure and stability, which are largely dependent on the nucleotide sequence context. Overall, except for specific mismatch types, the general trend of inhibition observed for single mismatches is the same as that observed when we increased the frequency of mismatches, for both Group 1 (Figs. 4 and 5) and Group 2 (Table 1) substrates.
FIGURE 4.
Effect of type and position of two mismatches on Dmc1 strand exchange efficiency. A, scheme of mismatches type and position used in this study. Variable nucleotides in ssDNA are highlighted in color. B stands for G, C, or T. The homologous (Hom) and a set of 27 different ssDNA substrates were tested. Oligonucleotide sequences of the labeled dsDNA and invading ssDNA are shown in Table 1. B–D, relative strand exchange efficiency of Dmc1 and RecA with ssDNA oligonucleotides containing two mismatches in different positions of the incoming strand. Type of mismatches are indicated in the x axis labels. Dashes indicate the absence of mismatch.
FIGURE 5.
Effect of three mismatches on Dmc1-mediated strand exchange. A, scheme of mismatches type and position used in this study. Variable nucleotides in ssDNA are highlighted in color. B stands for G, C, or T. The homologous (Hom) and a set of 27 different ssDNA substrates were tested. Oligonucleotide sequences of the labeled dsDNA and invading ssDNA are shown in Table 1. B and C, relative strand exchange efficiency of Dmc1 and RecA with ssDNA oligonucleotides containing three mismatches in all possible combinations.
Using the same sets of mismatched DNA substrates, we tested the strand exchange reaction catalyzed by RecA and the results are shown in Figs. 3–5. In general, RecA-mediated DNA strand exchange has a lower tolerance for DNA mismatches than the equivalent reaction catalyzed by Dmc1. Thus, our results reveal that both Dmc1 and RecA are sensitive to the mismatch type as well as its location, distribution, and frequency during the catalysis of DNA strand exchange.
Hop2-Mnd1 Stimulate Strand Exchange of a Single 5′ TG Mismatch
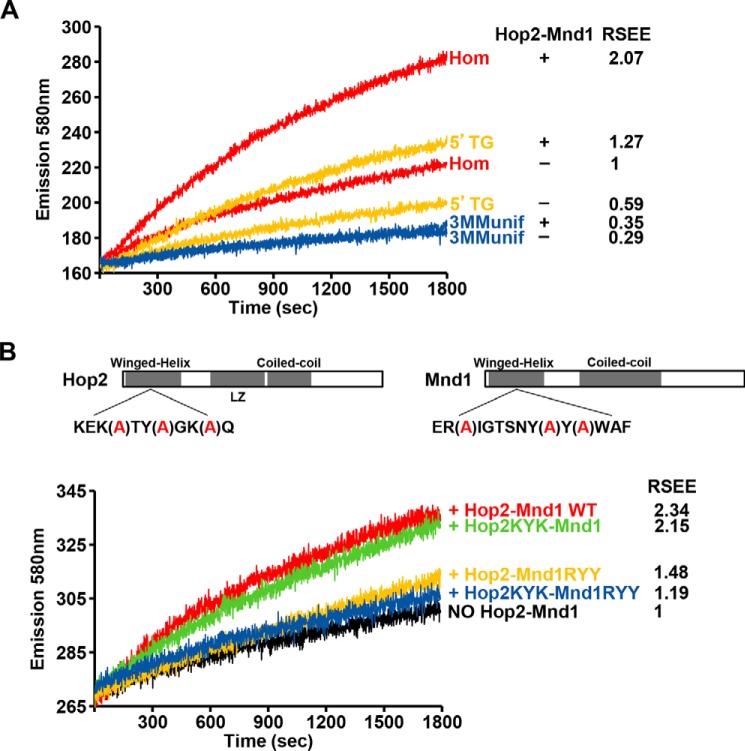
The efficiency of Dmc1-mediated DNA strand exchange is enhanced by the heterodimeric Hop2-Mnd1 complex (18, 25, 31, 32), which acts in two critical steps of the exchange process (17, 25). First, Hop2-Mnd1 stabilizes the Dmc1-ssDNA presynaptic filament, and second, it acts in conjunction with the presynaptic filament to mediate the capture of dsDNA. Here, we asked whether Hop2-Mnd1 by stimulating Dmc1 would help overcome the inhibitory effects of DNA mismatches in Dmc1-mediated DNA strand exchange. We examined in real-time the kinetics of Dmc1-mediated strand exchange between homologous DNA substrates and for those containing mismatches in the presence or absence of Hop2-Mnd1 (Fig. 6A). Addition of Hop2-Mnd1 resulted in 2-fold enhancement of Dmc1 strand exchange with the homologous ssDNA. As expected, Hop2-Mnd1 did not enhance the RecA-mediated strand exchange reaction, indicating its specificity for Dmc1 (results not shown). Notably, Hop2-Mnd1 facilitated strand exchange with the oligonucleotide containing a single TG mismatch either at the 5′ (Fig. 6A) or 3′ end (results not shown). However, the stimulatory effect of Hop2-Mnd1 becomes limited by the presence of an increasing number of mismatches, as it was unable to overcome the presence of three DNA mismatches (Fig. 6A).
FIGURE 6.
Effect of Hop2-Mnd1 in the strand exchange reaction catalyzed by Dmc1. A, real-time analysis of Dmc1 strand exchange between homologous DNA substrates or those containing mismatches in the presence or absence of Hop2-Mnd1. B, analysis of wild type or mutant variants of Hop2-Mnd1 on Dmc1-catalyzed strand exchange between homologous DNA. The relative strand exchange efficiency (RSEE) is shown for each reaction.
To understand the function of Hop2-Mnd1 in more depth, we analyzed mutants of the complex that harbor a compound point mutation in the winged-helix DNA binding domain of either Hop2 (the KYK mutation) or Mnd1 (the RYY mutation), and also a mutant that harbors both mutations (19, 32). The Hop2 KYK mutation partially compromises the ability of Hop2-Mnd1 to function in the Dmc1-mediated D-loop reaction, whereas the Mnd1 RYY mutation and Hop2 KYK-Mnd1 RYY double mutation have a much stronger effect in this regard (19). Comparative analysis revealed that the increase in fluorescence emission observed with Hop2-Mnd1 and either homologous DNA substrates or oligonucleotides containing a 5′ TG mismatch is indistinguishable to that observed when the Hop2 KYK-Mnd1 mutant was used. Notably, Hop2-Mnd1 RYY and Hop2 KYK-Mnd1 RYY nearly abolished the strand exchange stimulatory activity of Hop2-Mnd1 with all of the DNA substrates tested (Fig. 6B). In summary, our results indicate the requirement of the Mnd1 DNA binding activity in Dmc1 strand exchange stimulation.
Discussion
The tolerance of a recombinase to interruptions of homology must be closely regulated to maintain the fine balance between gene divergence and fidelity of repair. Excessive tolerance will result in an increase in recombination between nonallelic sequences, which may be detrimental to the cell. In this regard, we have observed that Dmc1-mediated DNA strand exchange is nearly abolished with oligonucleotides containing three mismatches. This suggests that Dmc1 may constitute the first barrier against recombination of substantially divergent DNA sequences. In this context, the DNA mismatch repair system may provide a second level of proofreading to ensure that heterologous DNA sequences do not recombine. We can imagine that in situations in which the mismatch repair system is either down-regulated or inactivated, the determinant mechanism to ensure approximate homology must be provided by the recombination process itself. However, a high discrimination imposed by a recombinase against DNA mismatches may prevent effective DNA strand exchange between homologous chromosomes in meiosis.
Although the ability of Dmc1 to bypass DNA mismatches is considered a critical aspect of meiotic recombination, no systematic study on the effect of mismatches on the activity of Dmc1 has been conducted. Here, we have determined the effects of the types, number, and position of DNA mismatches DNA strand exchange promoted by Dmc1. Our results show that Dmc1 can tolerate infrequent mismatches between the invading DNA and the homologous target DNA. This is consistent with the role of Dmc1 in promoting recombination between polymorphic maternal and paternal homologous chromosomes during meiosis. The ability of Dmc1 to step over certain DNA mismatches, together with subsequent correction of the mismatched base pairs, will lead to changes in the repaired DNA sequences, thus contributing to allelic variation (33). In accordance with our results, a recent report has provided evidence that RecA, Rad51, and Dmc1 all stabilize strand exchange intermediates in three nucleotide steps, and a single mismatch impacts upon the recognition of an entire base triplet. Importantly, among the three recombinases, only Dmc1 can stabilize mismatched triplets (34). This distinctive characteristic of Dmc1 may also be important for shielding mismatched DNA intermediates from premature dissolution by the mismatch repair machinery and may help guide homologous versus sister chromatic template choice. The differences observed between RecA and Dmc1 are in agreement with our results showing an increased level of tolerance to mismatches of Dmc1 when compared with RecA, and may explain why in our assay Dmc1-mediated DNA strand exchange is insensitive to certain types and positions of mismatches.
What is the origin of the differential sensitivity among recombinases to DNA mismatches? The changes in the efficiency of DNA strand exchange with oligonucleotides substrates containing different mismatches at the 5′ end may be explained by variations in the structure of the mismatches. In this sense, several studies have demonstrated that some mismatches have properties that are different from those of Watson-Crick base pairs (Refs. 35–37 and citations within). For example, the GA (anti-syn) conformation causes little local or global distortion of the B-DNA helix, whereas the AG (anti-syn) mismatch has poor base stacking within the helix, which perturbs the DNA backbone (35, 38). However, for both Dmc1 and RecA, we have observed that mismatches located toward the 5′ end of the initiating ssDNA have, in general, a stronger inhibitory effect than those located at the 3′ end of the DNA. This effect cannot be explained solely by differences in the stability (i.e. melting temperature) of the dsDNA product generated during strand exchange. It seems possible that Dmc1 is more sensitive to the specific conformation of a mismatch (39), which is influenced by the local sequence context and related to stacking and H-bonding interactions (37). This may also help explain differences between our results and those from previous work on RecA showing that 5′ mismatches had a smaller negative impact than 3′ mismatches on DNA strand exchange efficiency (27). If this is the case, then more work will be needed to define how DNA sequence context affects the sensitivity of RecA and other recombinases to a given mismatch.
We note that the sensitivity of Dmc1 to a mismatch may also be influenced by the free energy associated with the bending of DNA, which is lower for mismatch-containing heteroduplexes (40). In the case of homoduplexes, DNA bending is observed to occur via smooth deformations, whereas for heteroduplexes, kinks are observed at the mismatch site during strong bending. It is possible that some mismatches are more adept at inducing DNA bending, which may affect the recognition of different heteroduplex types by a recombinase (40). In agreement with this idea, RecA binds to dsDNA containing some single mismatches with higher efficiency than to perfectly paired duplex DNA (41). Moreover, the binding affinity is dependent on the conformation of the bulged bases, the kinking angles produced by the bulges, the type of mismatch, and the flanking sequences (41). Another relevant factor to consider is that the presence of mismatches may alter the ability of the recombinase to stretch DNA, which has been considered a hallmark of homology recognition mediated by the RecA family of proteins. This is, under conditions where the recombinase promotes homologous pairing, recombinase-ssDNA filaments are formed in which the ssDNA within the filament is stretched resulting in unstacking of the DNA bases (25, 42–44).
The DNA strand-exchange activity of Dmc1 is stimulated by Hop2-Mnd1. We have shown that Hop2-Mnd1 stimulates Dmc1-mediated DNA strand exchange even when there is a single TG mismatch. This result is consistent with the fact that Hop2-Mnd1 acts in a DNA homology independent fashion through prolonging the lifetime of duplex DNA bound within the Dmc1 presynaptic filament (17, 25), Hop2-Mnd1 helps alleviate the restriction imposed by a 5′ mismatch. Thus, our results support the premise that Hop2-Mnd1 acts by enhancing an earlier homology independent stage of the recombination reaction (25).
Author Contributions
R. J. P. conceived the study. M. V. B. and R. J. P. designed the study and conducted the study. M. V. B., R. J. P., M. R. M., C. E. A, W. Z., and P. S. analyzed the data. W. Z. and P. S. provided essential material for the study. M. V. B. and R. J. P. wrote the manuscript. All authors reviewed the results, edited the manuscript, and approved the final version of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants ES015252 (to P. S.) and GM103636 and March of Dimes Grant FY14–256 (to R. J. P.). The authors declare that they have no conflicts of interest with the contents of this article.
- HR
- homologous recombination
- DSBs
- double-strand breaks
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- TAMRA
- tetramethylrhodamine
- FE
- fluorescence emission.
References
- 1. Kleckner N. (1996) Meiosis: how could it work? Proc. Natl. Acad. Sci. U.S.A. 93, 8167–8174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Roeder G. S. (1997) Meiotic chromosomes: it takes two to tango. Genes Dev. 11, 2600–2621 [DOI] [PubMed] [Google Scholar]
- 3. Hassold T. J., and Jacobs P. A. (1984) Trisomy in man. Annu. Rev. Genet. 18, 69–97 [DOI] [PubMed] [Google Scholar]
- 4. Hassold T., and Matsuyama A. (1979) Origin of trisomies in human spontaneous abortions. Hum. Genet. 46, 285–294 [DOI] [PubMed] [Google Scholar]
- 5. Hunt P. A., and Hassold T. J. (2008) Human female meiosis: what makes a good egg go bad? Trends Genet. 24, 86–93 [DOI] [PubMed] [Google Scholar]
- 6. Keeney S., Giroux C. N., and Kleckner N. (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 [DOI] [PubMed] [Google Scholar]
- 7. Neale M. J., Pan J., and Keeney S. (2005) Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature 436, 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Garcia V., Phelps S. E., Gray S., and Neale M. J. (2011) Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature 479, 241–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Neale M. J., and Keeney S. (2006) Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442, 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. San Filippo J., Sung P., and Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 11. Sansam C. L., and Pezza R. J. (2015) Connecting by breaking and repairing: mechanisms of DNA strand exchange in meiotic recombination. FEBS J. 282, 2444–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gupta R. C., Folta-Stogniew E., O'Malley S., Takahashi M., and Radding C. M. (1999) Rapid exchange of A:T base pairs is essential for recognition of DNA homology by human Rad51 recombination protein. Mol. Cell 4, 705–714 [DOI] [PubMed] [Google Scholar]
- 13. Gupta R. C., Golub E., Bi B., and Radding C. M. (2001) The synaptic activity of HsDmc1, a human recombination protein specific to meiosis. Proc. Natl. Acad. Sci. U.S.A. 98, 8433–8439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bazemore L. R., Folta-Stogniew E., Takahashi M., and Radding C. M. (1997) RecA tests homology at both pairing and strand exchange. Proc. Natl. Acad. Sci. U.S.A. 94, 11863–11868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Volodin A. A., Bocharova T. N., Smirnova E. A., and Camerini-Otero R. D. (2009) Reversibility, equilibration, and fidelity of strand exchange reaction between short oligonucleotides promoted by RecA protein from Escherichia coli and human Rad51 and Dmc1 proteins. J. Biol. Chem. 284, 1495–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sung P. (1997) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem. 272, 28194–28197 [DOI] [PubMed] [Google Scholar]
- 17. Chi P., San Filippo J., Sehorn M. G., Petukhova G. V., and Sung P. (2007) Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev. 21, 1747–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pezza R. J., Petukhova G. V., Ghirlando R., and Camerini-Otero R. D. (2006) Molecular activities of meiosis-specific proteins Hop2, Mnd1, and the Hop2-Mnd1 complex. J. Biol. Chem. 281, 18426–18434 [DOI] [PubMed] [Google Scholar]
- 19. Zhao W., Saro D., Hammel M., Kwon Y., Xu Y., Rambo R. P., Williams G. J., Chi P., Lu L., Pezza R. J., Camerini-Otero R. D., Tainer J. A., Wang H. W., and Sung P. (2014) Mechanistic insights into the role of Hop2-Mnd1 in meiotic homologous DNA pairing. Nucleic Acids Res. 42, 906–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Uanschou C., Ronceret A., Von Harder M., De Muyt A., Vezon D., Pereira L., Chelysheva L., Kobayashi W., Kurumizaka H., Schlögelhofer P., and Grelon M. (2013) Sufficient amounts of functional HOP2/MND1 complex promote interhomolog DNA repair but are dispensable for intersister DNA repair during meiosis in Arabidopsis. Plant Cell 25, 4924–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen Y. K., Leng C. H., Olivares H., Lee M. H., Chang Y. C., Kung W. M., Ti S. C., Lo Y. H., Wang A. H., Chang C. S., Bishop D. K., Hsueh Y. P., and Wang T. F. (2004) Heterodimeric complexes of Hop2 and Mnd1 function with Dmc1 to promote meiotic homolog juxtaposition and strand assimilation. Proc. Natl. Acad. Sci. U.S.A. 101, 10572–10577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Petukhova G. V., Pezza R. J., Vanevski F., Ploquin M., Masson J. Y., and Camerini-Otero R. D. (2005) The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 12, 449–453 [DOI] [PubMed] [Google Scholar]
- 23. Petukhova G. V., Romanienko P. J., and Camerini-Otero R. D. (2003) The Hop2 protein has a direct role in promoting interhomolog interactions during mouse meiosis. Dev. Cell 5, 927–936 [DOI] [PubMed] [Google Scholar]
- 24. Pezza R. J., Voloshin O. N., Volodin A. A., Boateng K. A., Bellani M. A., Mazin A. V., and Camerini-Otero R. D. (2014) The dual role of HOP2 in mammalian meiotic homologous recombination. Nucleic Acids Res. 42, 2346–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pezza R. J., Voloshin O. N., Vanevski F., and Camerini-Otero R. D. (2007) Hop2/Mnd1 acts on two critical steps in Dmc1-promoted homologous pairing. Genes Dev. 21, 1758–1766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Månér S., Massa H., Walker M., Chi M., Navin N., Lucito R., Healy J., Hicks J., Ye K., Reiner A., Gilliam T. C., Trask B., Patterson N., Zetterberg A., and Wigler M. (2004) Large-scale copy number polymorphism in the human genome. Science 305, 525–528 [DOI] [PubMed] [Google Scholar]
- 27. Sagi D., Tlusty T., and Stavans J. (2006) High fidelity of RecA-catalyzed recombination: a watchdog of genetic diversity. Nucleic Acids Res. 34, 5021–5031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Urena D. E., Zhang Z., Tsai Y. C., Wang Y. Z., and Chen J. (2011) From strand exchange to branch migration; bypassing of non-homologous sequences by human Rad51 and Rad54. J. Mol. Biol. 405, 77–91 [DOI] [PubMed] [Google Scholar]
- 29. Masson J. Y., Davies A. A., Hajibagheri N., Van Dyck E., Benson F. E., Stasiak A. Z., Stasiak A., and West S. C. (1999) The meiosis-specific recombinase hDmc1 forms ring structures and interacts with hRad51. EMBO J. 18, 6552–6560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gupta R. C., Golub E. I., Wold M. S., and Radding C. M. (1998) Polarity of DNA strand exchange promoted by recombination proteins of the RecA family. Proc. Natl. Acad. Sci. U.S.A. 95, 9843–9848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pezza R. J., Camerini-Otero R. D., and Bianco P. R. (2010) Hop2-Mnd1 condenses DNA to stimulate the synapsis phase of DNA strand exchange. Biophys. J. 99, 3763–3772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moktan H., Guiraldelli M. F., Eyster C. A., Zhao W., Lee C. Y., Mather T., Camerini-Otero R. D., Sung P., Zhou D. H., and Pezza R. J. (2014) Solution Structure and DNA-binding properties of the winged helix domain of the meiotic recombination HOP2 protein. J. Biol. Chem. 289, 14682–14691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pratto F., Brick K., Khil P., Smagulova F., Petukhova G. V., and Camerini-Otero R. D. (2014) DNA recombination. Recombination initiation maps of individual human genomes. Science 346, 1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee J. Y., Terakawa T., Qi Z., Steinfeld J. B., Redding S., Kwon Y., Gaines W. A., Zhao W., Sung P., and Greene E. C. (2015) DNA recombination: base triplet stepping by the Rad51/RecA family of recombinases. Science 349, 977–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brown J., Brown T., and Fox K. R. (2003) Cleavage of fragments containing DNA mismatches by enzymic and chemical probes. Biochem. J. 371, 697–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Patel D. J., Kozlowski S. A., Ikuta S., and Itakura K. (1984) Dynamics of DNA duplexes containing internal G.T, G.A, A.C, and T.C pairs: hydrogen exchange at and adjacent to mismatch sites. Fed. Proc. 43, 2663–2670 [PubMed] [Google Scholar]
- 37. Peyret N., Seneviratne P. A., Allawi H. T., and SantaLucia J. Jr. (1999) Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A, C.C, G.G, and T.T mismatches. Biochemistry 38, 3468–3477 [DOI] [PubMed] [Google Scholar]
- 38. Webster G. D., Sanderson M. R., Skelly J. V., Neidle S., Swann P. F., Li B. F., and Tickle I. J. (1990) Crystal structure and sequence-dependent conformation of the A.G mispaired oligonucleotide d(CGCAAGCTGGCG). Proc. Natl. Acad. Sci. U.S.A. 87, 6693–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Allawi H. T., and SantaLucia J. Jr. (1998) Nearest-neighbor thermodynamics of internal A.C mismatches in DNA: sequence dependence and pH effects. Biochemistry 37, 9435–9444 [DOI] [PubMed] [Google Scholar]
- 40. Sharma M., Predeus A. V., Mukherjee S., and Feig M. (2013) DNA bending propensity in the presence of base mismatches: implications for DNA repair. J. Phys. Chem. B 117, 6194–6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Y. H., Bortner C. D., and Griffith J. (1993) RecA binding to bulge- and mismatch-containing DNAs: certain single base mismatches provide strong signals for RecA binding equal to multiple base bulges. J. Biol. Chem. 268, 17571–17577 [PubMed] [Google Scholar]
- 42. Voloshin O. N., Wang L., and Camerini-Otero R. D. (1996) Homologous DNA pairing promoted by a 20-amino acid peptide derived from RecA. Science 272, 868–872 [DOI] [PubMed] [Google Scholar]
- 43. Benson F. E., Stasiak A., and West S. C. (1994) Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 13, 5764–5771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nishinaka T., Ito Y., Yokoyama S., and Shibata T. (1997) An extended DNA structure through deoxyribose-base stacking induced by RecA protein. Proc. Natl. Acad. Sci. U.S.A. 94, 6623–6628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dumousseau M., Rodriguez N., Juty N., and Le Novère N. (2012) MELTING, a flexible platform to predict the melting temperatures of nucleic acids. BMC Bioinformatics 13, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]