Abstract
The available structural information on LeuT and structurally related transporters suggests that external loop 4 (eL4) and the outer end of transmembrane domain (TM) 10′ participate in the reversible occlusion of the outer pathway to the solute binding sites. Here, the functional significance of eL4 and the outer region of TM10′ are explored using the sodium/proline symporter PutP as a model. Glu-311 at the tip of eL4, and various amino acids around the outer end of TM10′ are identified as particularly crucial for function. Substitutions at these sites inhibit the transport cycle, and affect in part ligand binding. In addition, changes at selected sites induce a global structural alteration in the direction of an outward-open conformation. It is suggested that interactions between the tip of eL4 and the peptide backbone at the end of TM10′ participate in coordinating conformational alterations underlying the alternating access mechanism of transport. Together with the structural information on LeuT-like transporters, the results further specify the idea that common design and functional principles are maintained across different transport families.
Keywords: amino acid transport, electron paramagnetic resonance (EPR), membrane protein, protein chemistry, protein conformation, PutP, proline, secondary transport, solute/sodium symport, spin labeling
Introduction
Secondary transporters harness the energy stored in (electro)chemical gradients of a solute or ion (e.g. proton or sodium ion) to drive transport of a second solute across cell membranes. All transporters are proposed to follow an alternating access mechanism in which the binding sites for the ligands are alternately exposed to one or the other side of the membrane (1). This mechanism is supported by crystal structures of transporters in different conformations as well as protein chemical and spectroscopic analyses (e.g. Refs. 2–6). A recurring feature in transporter structures is the presence of an internal pseudo-symmetry of sets of transmembrane domains (TMs)3 (7). Structural rearrangements that may include collective movements of groups of TMs and displacements of partially unwound α-helices can cause an alternate exposure of the usually central ligand binding sites to either side of the membrane (2, 7, 8). In addition, there is evidence that also external and internal loop regions participate in the reciprocal opening and closing of outer and inner pathways (9–13). For example, the external gate in LeuT is closed through interactions between TM1b and TM10, eL4 and TM10, and TM6b and TM11 (13).
The successful determination of structures of secondary transporters revealed a class of sequence-unrelated symporters and antiporters that share the same structural fold with the founding member, the bacterial sodium-dependent amino acid transporter LeuT (neurotransmitter/sodium symporter family, TC 2.A.22) (8, 14–16). A core of 10 TMs that are arranged in two five-helix inverted repeats characterize the fold (8, 14, 15). Two crystal structures of the sodium/galactose symporter of Vibrio parahemeolyticus vSGLT (solute/sodium symporter family, SSS, TC 2.A.21) suggest that also SSS transporters possess a core structure with a LeuT-like fold (17, 18).
We use the sodium/proline symporter PutP (SSS family) as a model to explore molecular principles of sodium-coupled transport by SSS proteins. PutP and other proline-specific SSS family members are found in bacteria and archaea (19), where they are responsible for the uptake of the amino acid as a nutrient, or are involved in proline transport during adaptation to osmotic stress (20–22). Based on these and other functions, PutP is of significance for various bacteria host interactions including the virulence of human pathogens (21, 23–25).
PutP contains 13 TMs (26, 27) of which TMs 2 to 11 correspond to TMs 1 to 10 of the LeuT core (in the following designated as TMs 1′ to 10′)4 (28, 29). Based on the structure of vSGLT (17), a homology model of PutP was generated (28) and advanced based on experimentally determined secondary structure and distance restraints (29). The restraint-based homology model is in agreement with the idea of a LeuT-like structural fold for SSS members. Based on Cys accessibility analyses, PutP seems to be most stable in an inward-open conformation (28, 30–32). An extracellular pathway has not yet been identified. Structure-function analyses suggest that amino acids of TMs 1′ and 8′ form a sodium binding site corresponding to the Na2 site in LeuT (28, 30–32). Proline binding presumably involves amino acids of TMs 1′, 3′, 6′, and 8′ (28, 30, 33). In addition to the central binding site, recent computational and binding analyses provide evidence for the existence of a second, more externally localized proline binding site formed by amino acids of TMs 1′, 2′, 6′, and 10′ (34). The function of the second binding site in PutP has yet to be elucidated.
Recently, we reported on the role of the external loop 4 (eL4) as extracellular gate in PutP (29). The results of a complete spin-labeling site scan combined with electron paramagnetic resonance spectroscopy suggest that eL4 is involved in transmitting ligand-induced conformational changes at the binding sites to adjacent TMs, leading to closure of the extracellular gate. Thereby, Phe-314 in eL4b anchors the loop by means of hydrophobic contacts to TM1′ close to the ligand binding sites. The scanning mutagenesis approach identified also Glu-311 in eL4 as particularly important for transport, but its role in the transport cycle remained enigmatic (29). Evidence for a role of eL4 in gating comes also from crystallographic and electron paramagnetic resonance spectroscopic analyses of the bacterial transporters LeuT and the sodium/hydantoin transporter Mhp1 (nucleobase/sodium symporter 1 family, 2.A.39) (10, 12, 13, 35). eL4 is also of functional significance in eukaryotic representatives of LeuT-like transporters, e.g. for the serotonin transporter SERT (36), the γ-aminobutyric acid transporter GAT-1 (37), and the glycine transporters GlyT1 and GlyT2 (38).
Based on the interactions between the tip of eL4 and the outer end of TM10′ in the crystal structures of the inward-open conformations of LeuT (13) and vSGLT (17), we set out to gain a better understanding of the functional significance of eL4 and the region around the outer end of TM10′ in SSS proteins. Using PutP of Escherichia coli as a model we found that a negative charge at position 311 (tip of eL4) is crucial for transport, whereas there is relatively little influence on ligand binding. The restraint-based homology model presented here suggests interactions (i.e. H-bond formation) of the side chain carboxyl with the peptide backbone close to the outer end of TM10′. A screen of the latter region identified two clusters of amino acids as particularly crucial for function. Analysis of the accessibility of strategically placed Cys residues on the inner and outer side of PutP suggests that substitutions at positions 311 or 404 support a conformation that is more open to the outside than the wild-type. This conclusion is further supported by a distance analysis of spin-labeled PutP using electron paramagnetic resonance spectroscopy. Taken together, the results further support the idea that eL4 is involved in the formation of the extracellular gate in PutP and potentially other members of the SSS family. Thereby, interactions of Glu-311 at the tip of eL4 probably with the outer end of TM10′ are required for the transmission of conformational changes underlying the alternating access mechanism.
Experimental Procedures
Bacterial Strains and Plasmids
E. coli DH5α (F− φ80d lacZ ΔM15 Δ(lacZYA-argF) U169 deoR recA1 endA1 hsd R17 (rk−,mk+) phoA supE44 λ− thi-1 gyrA96 relA1) was used as carrier for plasmids. E. coli WG170 (F− trp lacZ rpsL thi Δ(putPA)101 proP219) (39) harboring given plasmids was used for expression of the putP gene and biochemical assays. The following plasmids, derivatives of pT7-5 (40), containing the lac promoter/operator for expression of the putP gene were used for all gene manipulations: pT7-5/putP and pT7-5/putP(ΔC), each of which harboring a cassette version of the putP gene encoding PutP-wild-type and an engineered transporter devoid of all five native Cys residues (PutP(ΔC)), respectively, and a C-terminal attached amino acid sequence resembling the FLAG epitope and a His6 tag (41). Vector pTrc99a (42) was used for overexpression.
Site-directed Mutagenesis
Desired nucleotide substitutions in putP were generated by PCR with Phusion DNA polymerase using plasmid pT7-5/putP or pT7-5/putP(ΔC) as a template and synthetic mutagenic oligonucleotides. Resulting PCR fragments were cloned in plasmid pT7-5/putP or pT7-5/putP(ΔC). The plasmid DNA was verified by sequencing using an ABI 3730 capillary sequencer.
Transport Assay
Active transport of l-[U-14C]proline was measured in E. coli WG170 harboring plasmids pT7-5/putP encoding PutP with given amino acid replacements as described (43). Initial rates of transport were calculated from the initial linear portion of the time course, and steady-state levels of l-[U-14C]proline accumulation were taken from time points after leveling off of the uptake curve. Standard deviations were calculated from at least triplicate determinations.
Immunological Analysis
Relative amounts of PutP with given amino acid replacements in membranes of E. coli WG170 harboring plasmid pT7-5/putP were estimated by Western blot analysis with HRP-linked mouse anti-FLAG IgG directed against the FLAG epitope at the C terminus of each PutP variant as described (44).
Cys Accessibility to Sulfhydryl Reagents
[14C]N-Ethylmaleimide ([14C]NEM) labeling and the impact of methanethiosulfonate ethylammonium (MTSEA), methyl methanethiosulfonate (MMTS), and methanethiosulfonate ethylsulfonate (MTSES) on the labeling reaction were analyzed with right-side out membrane vesicles (45) prepared from E. coli WG170 transformed with pTrc99a/putP(ΔC) harboring given mutations essentially as described (31). The accessibility of Cys to fluorescein-5-maleimide (FM) was determined with randomly oriented membrane vesicles prepared from the same E. coli strain following the protocol of Raba et al. (32). In short, vesicles were incubated with 200 μm FM at 25 °C for defined periods of time; then labeling reactions were stopped by addition of 10 mm β-mercaptoethanol. PutP was purified as described (41), and equal amounts of protein were subjected to SDS-PAGE. Fluorescent bands of PutP were detected, and fluorescence was normalized to the total amount of PutP.
Proline Binding Assays
Cells of E. coli WG170 transformed with pTrc99a/putP with given mutations were disrupted using a high-pressure cell disruption system (Constant Systems) at 1.35 kbar. Membrane vesicles of E. coli WG170 harboring pTrc99a served as negative control. Membrane vesicles were preincubated with 50 mm NaCl, 20 μm carbonyl cyanide m-chlorophenylhydrazone, and 5 μm monensin at 25 °C for 30 min. For all binding assays, 100 mm potassium phosphate buffer, pH 7.5, 2 mm β-mercaptoethanol was used.
For the differential radial capillary action of ligand assay (DRaCALA), the protocol of Roelofs et al. (46) was followed. l-[U-14C]Proline (final concentration 1 μm, 55 μCi/μmol) was added to the preincubated membrane vesicles containing 9 mg/ml of total protein, and samples were incubated at 25 °C for 10 min. Five-μl aliquots were subsequently pipetted onto dry nitrocellulose (GE Healthcare) in triplicates. The nitrocellulose was exposed to a Storage Phosphor Screen, and a Typhoon Trio Imager (Amersham Biosciences) was used for detection of radioactivity.
For quantitative binding assays, 200-μl aliquots of the preincubated membrane vesicles containing 12.5 mg/ml of total protein were filled into the sample chamber of a Thermo Scientific Single-Use RED Plate with RED inserts (8K MWCO). The adjacent chamber contained 350 μl of buffer with 50 mm NaCl and l-[U-14C]proline (55 μCi/μmol) yielding final concentrations in both chambers of 0.2 to 50 μm. For PutP variants for which saturation was not reached, concentrations of up to 350 μm l-[U-14C]proline (20 μCi/μmol) were used. The plate was sealed using self-sticking tape and incubated at 4 °C in an Eppendorf Thermomixer comfort with a mixing frequency of 750 rpm for 22 h. Then, 50-μl aliquots were removed from each chamber, ECOLUMETM liquid scintillation mixture (MP Biomedicals) was added and radioactivity was measured using a PerkinElmer Tri-Carb® 2910TR Liquid Scintillation Analyzer. The kd values were determined using the Enzyme Kinetics module of SigmaPlot 12.5.
Site-directed Spin Labeling and Double Electron Electron Resonance (DEER) Spectroscopy
Membranes of E. coli WG170 transformed with pTrc99a/putP(ΔC) were prepared and double-Cys PutP variants were purified and spin-labeled with 1 mm (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate (MTSSL, Toronto Research Chemicals) as described (47). For reconstitution into liposomes (41), a lipid to protein ratio of 40:1 (w/w) was adjusted. To all samples, 10% d8-glycerol was added before freezing. Q-band DEER measurements were performed as described by Raba et al. (29). The frequency offset between pump and observer pulses was 100 MHz in all measurements and in some of the measurements a power-upgraded (200 W) Bruker ElexsysII E580 spectrometer was used instead of the home-built spectrometer used in Ref. 29.
Determination of Sodium
Sodium concentrations in buffers used for transport assays were determined with a VARIAN AA240 atomic absorption spectrometer.
Protein Determination
Determination of protein was performed according to a modified Lowry method (48) for total membrane protein, and according to Bradford (49) for detergent-solubilized protein.
Molecular Graphics and Analyses
Molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (50).
Results
PutP Model
For structural information, our previous homology and restraint-based model of PutP (29) was further advanced by including the experimentally determined distance between positions 298 and 326 (see DEER analysis below) (Fig. 1, A and B). The resulting model represents PutP in an inward-open conformation similar to previous models (28, 29), and consistent with previous Cys accessibility analyses (28, 30, 32). The model matches well all nine DEER-based distances used as restraints (positions 50–304 (cross membrane), 91–371, 190–371 (cytoplasmic side), 294–326, 294–446, 298–326, 298–446, 299–318, 326–446 (periplasmic side); root mean square deviation for all distance restraints = 0.72 Å).
FIGURE 1.
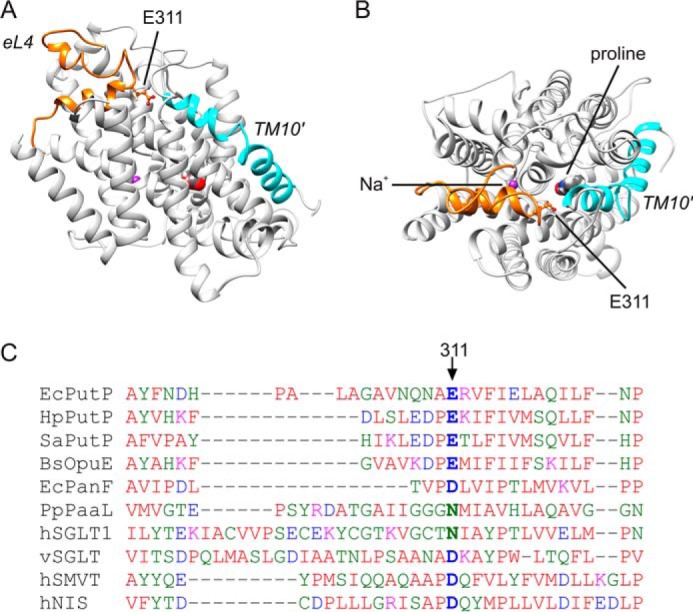
Structural model of PutP. A and B, homology and restraint-based model of PutP in an inward-open conformation with eL4 highlighted in orange and TM10′ in turquoise (A, top view; B, side view). The model represents an advancement of earlier models (28, 29), and matches nine DEER-based distances (root mean square deviations for all distance restraints = 0.72 Å). The side chain of Glu-311 is shown in ball-and-stick representation. The indicated putative locations of the binding sites for l-proline and sodium were taken from Olkhova et al. (28). C, sequence alignment of amino acids of eL4 of SSS family members. The alignment was performed with Clustal Omega (53).
Glu-311 is localized at the tip of the helix-turn-helix motif of eL4 of PutP (Fig. 1, A and B). The residue is conserved in SSS family members with proline specificity, and can be replaced by Asp or Asn in pro- and eukaryotic family members of different substrate specificity (Fig. 1C).
Substitution of Glu-311 Impacts Transport Kinetics
To learn more about the functional significance of the amino acid side chain at position 311, the native Glu was replaced by polar (Cys, Gln, Ser), apolar (Ala), and other charged amino acids (Asp, Arg). Under standard test conditions (50 mm NaCl, 10 μm l-[14C]proline), PutP-E311C and -E311D showed 1 and 10%, respectively, of the proline uptake activity of the wild-type. All other substitutions (E311A, E311Q, E311R, E311S) reduced the activity to values <0.5% of the wild-type (Fig. 2A). Analysis of transport kinetics of PutP-E311C and -E311D according to Michaelis-Menten revealed highly reduced maximum rates of transport (Vmax) (0.5 and 9% of the wild-type value). By contrast, the sodium concentration causing half-maximum stimulation of transport (K0.5(Na+)) and the proline concentration at Vmax/2 (Km(Pro)) were not or only slightly altered (Table 1). The Michaelis-Menten parameters could not be determined for PutP-E311A, -E311Q, -E311R, and -E311S due to very low activity or inactivity. All activity parameters were normalized to the amount of PutP in the membrane, and therefore the observed deviations of kinetic parameters from wild-type values were not due to fluctuations of the amount of transporter in the membrane. In fact, none of the substitutions had a significant impact on the amount of PutP (not shown). The results indicate that length and negative charge of the side chain at position 311 are of pivotal significance for the transport cycle of PutP.
FIGURE 2.
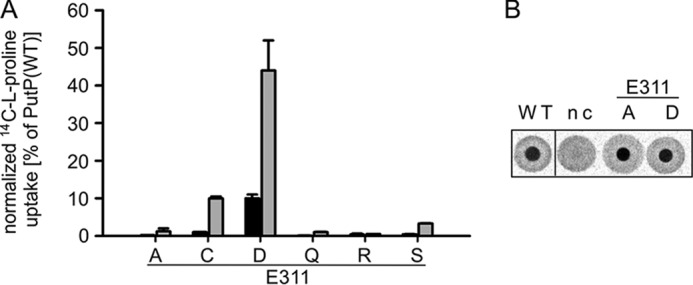
Impact of the substitution of Glu-311 in eL4 on sodium-coupled proline uptake and binding. A, normalized initial rates of transport (black) and maximum accumulation of proline inside the cells (gray). Transport of 10 μm l-[14C]proline into E. coli WG170 was assayed in the presence of 50 mm NaCl and 20 mm d-lactate (sodium salt) at 25 °C under aerobic conditions using a rapid filtration method. Results are shown as percentage of wild-type values, and transport activities are normalized to the amount of PutP based on Western blot analysis. Standard deviations were calculated from triplicate determinations of a representative experiment. B, l-[14C]proline binding to membrane vesicles containing given PutP variants detected by DRaCALA. E. coli WG170 membrane vesicles were incubated with 50 mm NaCl, 20 μm carbonyl cyanide p-chlorophenylhydrazone, and 5 μm monensin at 25 °C for 30 min before adding 1 μm l-[14C]proline for 10 min. Aliquots were subsequently pipetted onto dry nitrocellulose in triplicates. nc, negative control with membranes of E. coli WG170 transformed with pTrc99a without putP. All DRaCALA spots are from the same assay.
TABLE 1.
Uptake kinetics and proline binding of PutP bearing replacements of given amino acids
l-[14C]Proline uptake by E. coli WG170 producing either PutP (wild-type) or PutP with given replacements was measured in the presence of 50 mm NaCl and 20 mm d-lactate (Na+ salt) at proline concentrations from 0.2 to 250 μm. For determination of K0.5(Na+), transport of 10 μm proline was measured in the presence of 0.005 to 100 mm NaCl at 25 °C. Binding of l-[14C]proline to E. coli WG170 membranes was measured at proline concentrations from 0.2 to 50 μm; if saturation was not reached, concentrations up to 350 μm proline were used. The resulting data were plotted according to Michaelis-Menten, Lineweaver-Burk, and Eadie-Hofstee using the kinetic module of the SigmaPlot software.
| Substitution | Km(Pro) | Vmaxa | K0.5(Na+) | Kd(Pro) |
|---|---|---|---|---|
| μm | nmol min−1 mg−1 | μm | ||
| Wild-type | 2.2 ± 0.4 | 64.6 ± 1.7 | 19.1 ± 2.4 | 11.4 ± 1.0 |
| E311A | –b | – | – | 9.3 ± 0.6 |
| E311C | 1.6 ± 0.6 | 0.3 ± 0.02 | 17.9 ± 2.9 | NDc |
| E311D | 4.0 ± 0.3 | 6.0 ± 0.1 | 17.8 ± 1.4 | 5.5 ± 1.2 |
| E311Q | – | – | – | ND |
| E311R | – | – | – | ND |
| E311S | – | – | – | ND |
| A404C | 6.1 ± 2.0 | 19.8 ± 1.6 | 19.5 ± 2.5 | 4.1 ± 0.6 |
| A404E | – | – | – | ND |
| A404G | 77.0 ± 8.5 | 134.5 ± 6.2 | 81.5 ± 14.3 | 148.2 ± 23.3 |
| A406C | 18.9 ± 3.4 | 21.5 ± 1.1 | 56.0 ± 4.8 | ND |
a Vmax values were normalized to the amount of PutP in the membrane.
b Transport activities were below 1% of PutP(wild-type) preventing reliable determination of kinetic parameters.
c ND, not determined.
To further evaluate the role of Glu-311 in the transport cycle, proline binding was initially tested using DRaCALA. A semi-quantitative analysis revealed that active (PutP-E311D) and inactive (PutP-E311A) transporter variants bind proline similar as the wild-type (Fig. 2B). For a more quantitative estimation of the binding properties of PutP, the dissociation constant for proline (Kd(Pro)) was determined by equilibrium dialysis (Table 1). The binding data of the wild-type were well fitted by a single binding site model and yielded a Kd(Pro) of 11.4 ± 1.0 μm. The latter value was relatively high compared with a Km(Pro) of 2.2 ± 0.4 μm. The difference may result from overlapping binding kinetics of a high affinity binding site and a lower affinity binding site whose existence was previously suggested (34). Substitution of Glu-311 reduced Kd(Pro) to different degrees toward the value of Km(Pro) (Table 1). Together with the unaffected kinetic parameters K0.5(Na+) and Km(Pro) of PutP-E311D and -E311C the results suggest that Glu-311 does not play a pivotal role in binding of sodium or proline.
Cys-311 Is Accessible from the Aqueous Phase
To gain information on the location of position 311, we analyzed rates of the modification of PutP(ΔC)-E311C by sulfhydryl reagents of different polarity and size (Fig. 3A). Cys-311 readily reacted with [14C]NEM, and this reaction was efficiently inhibited by preincubation with the neutral and negatively charged sulfhydryl-specific reagents MMTS and MTSES, respectively. Also the positively charged MTSEA inhibited the reaction albeit less efficiently than the other methanethiosulphonate reagents (Fig. 3B). Finally, individual or simultaneous addition of the PutP ligands sodium and l-proline did not significantly affect the labeling of Cys-311 (not shown). These results indicate that Cys-311 is accessible from the aqueous phase. The observation does not agree with our model of PutP in an inward-open conformation, in which Glu-311 is buried in the protein interior similar as its counterpart Asp-336 in vSGLT (Protein Data Bank code 3DH4) (17). The discrepancy may reflect the dynamics of the transporter or an altered opening probability induced by the placement of Cys at position 311.
FIGURE 3.
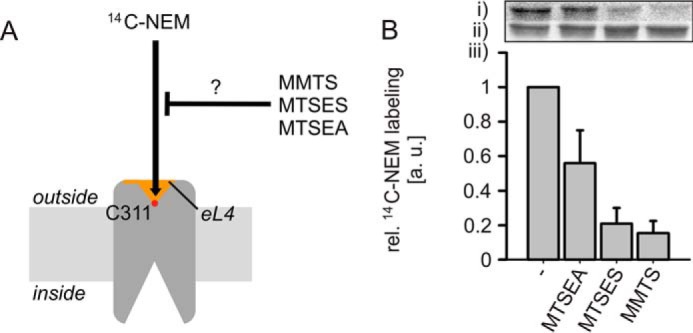
Accessibility of Cys-311 in PutP(ΔC) to sulfhydryl reagents. A, scheme of the experimental procedure. The efficiency of Cys-311 labeling by MTSEA, MTSES, or MMTS was estimated by analyzing the impact of these compounds on the subsequent labeling with [14C]N-ethylmaleimide ([14C]NEM). B, results of the labeling experiment. Right-side out membrane vesicles of E. coli WG170 containing given PutP derivatives were preincubated with 500 μm MTSEA, MTSES, or MMTS at 25 °C for 10 min if indicated. Then, labeling with 500 μm [14C]NEM was performed at 25 °C for 5 min. Reactions were stopped by addition of 2 mm dithiothreitol. After labeling, PutP was purified and subjected to SDS-PAGE. Radioactivity was detected with a PhosphorImager (i), and total amounts of protein were estimated based on Coomassie Blue staining (ii). The radioactivity was normalized to the amount of protein, and relative labeling yields were determined by arbitrary setting the [14C]NEM labeling yield in the absence of other sulfhydryl compounds to 1 (iii). Standard deviations were calculated from three independent experiments, whereas a representative PhosphorImager-generated autoradiograph and Coomassie-stained bands are shown.
Arg Residues in the Proximity of Glu-311 Are Unimportant for Function
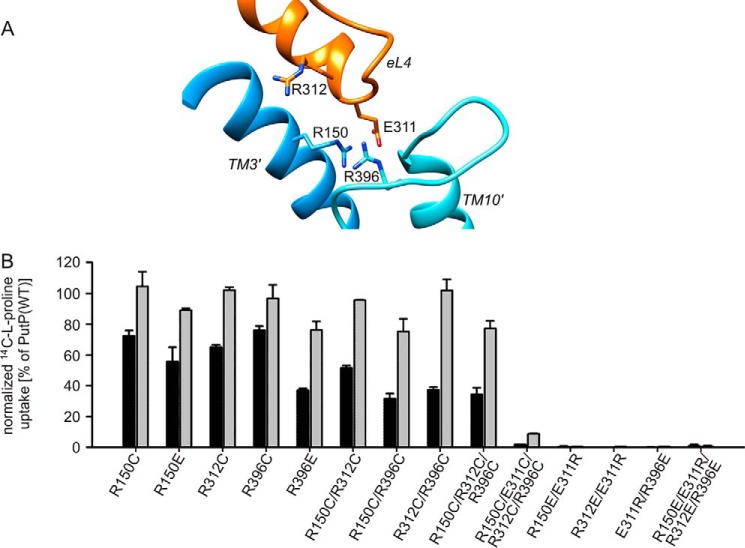
Next, we set out to identify interactions of Glu-311 that are of functional relevance. Based on the above demonstrated requirement of a negative charge at position 311 for PutP activity, we first considered electrostatic interactions. Indeed an inspection of our PutP model identified three positively charged amino acids in the immediate proximity of Glu-311 (Fig. 4A). However, an amino acid substitution analysis revealed that none of the Arg residues was particularly crucial for transport (Fig. 4B). Even a transporter with all threeArg residues replaced (PutP-R312C/R150C/R396C) catalyzed transport with 35 and 80% of the initial rate and maximum level of proline accumulation, respectively, of the wild-type. Furthermore, all combinations with Glu-311 substitutions (e.g. PutP-E311R/R396E) were inactive as the corresponding Glu-311 substitution alone. In conclusion, evidence for a particular functional significance of electrostatic interactions of Glu-311 with positively charged amino acids in its proximity could not be obtained.
FIGURE 4.
Role of Arg residues near Glu-311. A, zoom into the PutP model shown in Fig. 1 highlighting eL4 and Arg-150 (TM3′, blue), Arg-312 (eL4, orange), and Arg-396 (loop between TMs 9′ and 10′, turquoise) in the vicinity of Glu-311 (eL4, orange). B, impact of the substitution of the Arg residues on sodium-coupled proline uptake. Transport was analyzed as described in the legend of Fig. 2. Normalized initial rates of transport (black) and maximum accumulation of proline inside the cells (gray) are shown as percentage of wild-type values. Transport activities are normalized to the amount of PutP based on Western blot analysis. Standard deviations were calculated from triplicate determinations of a representative experiment.
Amino Acids Close to the Periplasmic End of TM10′ Are of Functional Significance
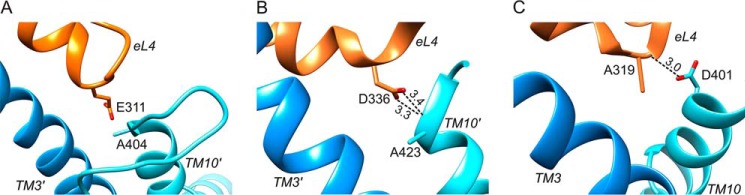
Further inspection of our PutP model revealed a location of the side chain carboxyl group of Glu-311 close to the protein backbone at the periplasmic end of TM10′ (Fig. 5A). The closest distance was ∼2.6 Å (in our model between Glu-311(Oδ) and Ala-404(N)) supporting the idea of a H-bond formation between the carboxyl group and the peptide backbone at the end of TM10′. Further support for this idea comes from the crystal structures of inward-open conformations of vSGLT (Protein Data Bank code 3DH4) (17) and LeuT (Protein Data Bank code 3TT3) (13). In vSGLT, the side chain carboxyl group of Asp-336 at the tip of eL4 is in H-bond distance to the protein backbone at the periplasmic end of TMD10′ (Ala-423(N)) (Fig. 5B). In the inward-open conformation of LeuT (Protein Data Bank code 3TT3) (13), Ala-319(N) in eL4 and Asp-401(Oγ) at the outer end of TMD10 are in H-bond distance (Fig. 5C).
FIGURE 5.
Interactions of eL4 with TM10′ in transporters with a LeuT-fold. A, detail of the PutP model shown in Fig. 1 highlighting eL4 with Glu-311 and TM10′ with Ala-404. B, detail of the crystal structure of vSGLT (Protein Data Bank 3DH4 (17)) highlighting the interaction between Asp-336 in eL4 and Ala-423 in TM10′. C, detail of the crystal structure of LeuT in an inward-open conformation (Protein Data Bank 3TT3 (13)) highlighting the interaction between Ala-319 in eL4 and Asp-401 in TM10. Distances highlighted by dotted lines are in Å.
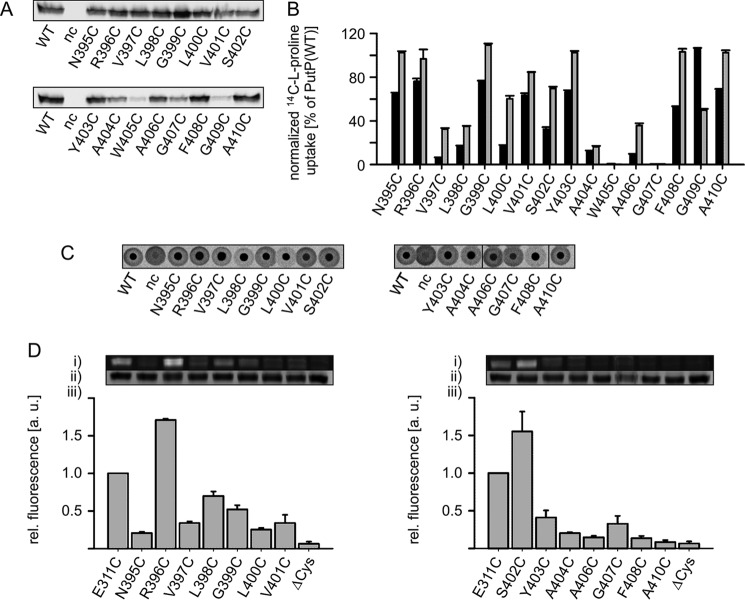
Based on these observations we analyzed the functional significance of amino acids in the region around the outer end of TM10′. We performed a Cys scanning analysis of amino acid positions 395 to 410. Substitution of Ala-404, Trp-405, Gly-407, and Gly-409 led to reduced amounts of PutP in the membrane (Fig. 6A). Only in the case of PutP-G409C was the reduced amount of PutP (11% of wild-type) fully accounted for by its reduced transport activity (12% of wild-type). When normalized to the amount of PutP in the membrane, the activity of PutP-G409C was similar to wild-type (Fig. 6B). The normalized transport activity pattern revealed two clusters of positions of particular functional importance: Val-397, Leu-398, Leu-400, and Ala-404, Trp-405, Ala-406, Gly-407 (Fig. 6B). Semi-quantitative analysis of proline binding by DRaCALA demonstrated that the PutP variants tested were able to bind proline (Fig. 6C). Because the proline concentration in the assay (1 μm) was below Kd(Pro) of the wild-type (Table 1), and strong differences in spot intensity were not detected, the individual substitutions did apparently not lead to larger changes in substrate affinity. The concentrations of PutP-W405C and -G409C in the membrane were too low to yield a reliable signal in the DRaCALA assay.
FIGURE 6.
Functional significance and accessibility of the region around the end of TM10′. A, relative amounts of PutP with given amino acid replacementsin E. coli WG170 membranes as determined by Western blot analysis. B, normalized initial rates of transport (black) and maximum accumulation of proline in E. coli WG170 (gray). Transport was analyzed as described in the legend of Fig. 2. Transport activities are normalized to the amount of PutP based on Western blot analysis. C, l-proline binding to membrane vesicles containing PutP variants detected by DRaCALA. Binding was analyzed as described in the legend of Fig. 2. All DRaCALA spots are from the same assay. D, accessibility of TM10′ residues to FM. Membrane vesicles of E. coli WG170 transformed with pTrc99a/putP(ΔC) containing a single Cys residue were incubated with 200 μm FM at 25 °C for 10 min. Labeling reactions were stopped by addition of 10 mm β-mercaptoethanol, PutP was purified as described (41), and equal amounts of protein were subjected to SDS-PAGE. (i) fluorescent bands of PutP; (ii) total amounts of PutP based on Coomassie staining; (iii) FM fluorescence normalized to the total amount of PutP. The yield of Cys labeling with the native Glu at position 311 was arbitrarily set to 1 in each experiment. Standard deviations were calculated from three independent experiments, whereas representative fluorescent and Coomassie-stained bands are shown. All bands of an individual experiment come from the same gel.
To learn more about the topology of this region of PutP, the accessibility of the individual positions to fluorescein-5-maleimid was analyzed. For this purpose, all Cys substitutions were placed into PutP(ΔC). Labeling was performed with randomly oriented membrane vesicles, labeled protein was purified, and relative labeling ratios were determined. PutP(ΔC)-E311C was used for comparison, and its relative labeling ratio was arbitrarily set to 1 (Fig. 6D). The analysis revealed that the positions of Arg-396 and Ser-402 are highly accessible from the aqueous phase suggesting a location at the surface of the protein, i.e. in an exposed loop region. The positions of Leu-398 and Gly-399 showed an intermediate accessibility, whereas positions 397, 400, 402, and 403 to 410 appeared to be buried in the protein or located in an apolar environment. The results support our model of PutP (Fig. 1, A and B) and place position 402 at the outer boarder of TM10′.
Alterations at Positions 404 and 406 Impact Transport Kinetics
Because a previous investigation already suggested Leu-398 to be part of an external proline binding site (34), we set out to obtain more detailed information on the significance of Ala-404 and Ala-406 in the above identified second cluster of functionally important amino acids on transport kinetics. Kinetic analyses of PutP-A404C and -A406C revealed reduced normalized Vmax for both PutP variants (30% of the wild-type value). However, both substitutions differ in the effect on the apparent affinity parameters. PutP-A404C showed only slightly altered values for Km(Pro) and K0.5(Na+), whereas Cys in place of Ala-406 caused an about 10- and 3-fold increase of Km(Pro) and K0.5(Na+), respectively (Table 1).
To learn more about the significance of the side chain at position 404, the native Ala was replaced also with Gly and Glu (Fig. 7). Reduced amounts of PutP in the membrane were observed only upon substitution by Glu (Fig. 7B). The normalized initial rate of proline uptake by PutP-A404G was highly reduced compared with the wild-type, and PutP-A404E was inactive. Reversal of amino acids at positions 311 and 404 (PutP-E311A/A404E) did not restore activity (Fig. 7A). Further kinetic analyses of PutP-A404C revealed a reduced normalized Vmax (30% of the wild-type value), and only slightly altered values for K0.5(Na+) and Km(Pro) (Table 1). On the contrary, placement of glycine at position 404 stimulated Vmax (210% of the wild-type value), and led to increased values for K0.5(Na+) (4-fold increase) and Km(Pro) (35fold increase) (Table 1). Due to a transport activity below 1% of the wild-type value, kinetic parameters could not be determined for PutP-A404E. However, the DRaCALA assay demonstrated the capability to bind proline for PutP-A404C, -A404E, -A404G, and -E311A/A404E (Fig. 7C). Binding to PutP-A404G was impaired compared with the other PutP variants considering the amount of PutP in the membrane (Fig. 7C). Equilibrium dialysis revealed that the Kd(Pro) of PutP-A404C was reduced to a value close to Km(Pro) similar as observed for the Glu-311 substitutions. On the contrary, Kd(Pro) of PutP-A404G was increased 13-fold compared with wild-type (Table 1).
FIGURE 7.
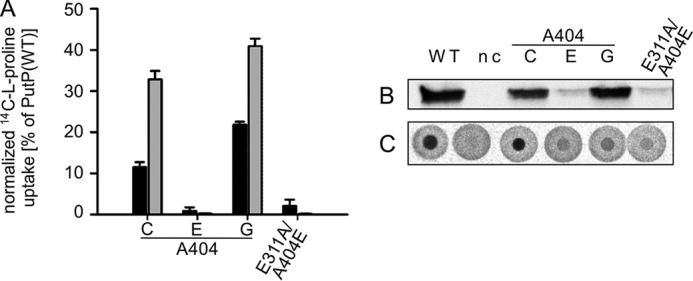
Impact of the substitution of Ala-404 on PutP function. A, normalized initial rates of transport (black) and maximum accumulation of proline in E. coli WG170 (gray). Transport was analyzed as described in the legend of Fig. 2. Transport activities are normalized to the amount of PutP based on Western blot analysis. B, relative amounts of PutP with given amino acid replacements in E. coli WG170 membranes as determined by Western blot analysis. C, l-proline binding to membrane vesicles containing PutP variants detected by DRaCALA. Binding was analyzed as described in the legend of Fig. 2.
Taken together, the kinetic analyses confirm that Ala-404 and Ala-406 are particularly crucial for transport. The kinetic data are consistent with the idea that Ala-404 is crucial for tertiary interactions during the transport cycle (e.g. external gate closure). Gly at position 404 stimulates the transport cycle and affects binding. Ala-406 directly or indirectly influences proline binding.
Alteration of Glu-311 or Ala-404 Induces a Global Conformational Change
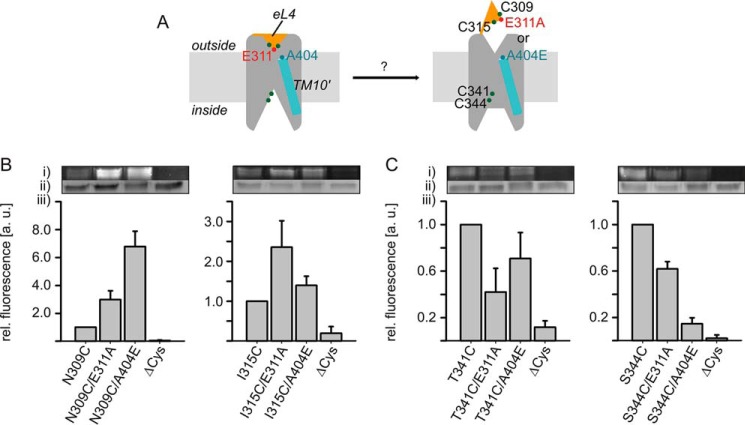
According to previous Cys accessibility analyses (28, 30, 32), PutP is most stable in an inward-open conformation. Conditions triggering a structure more open to the outside have not been identified yet. Because in LeuT transient interactions between the tip of eL4 and the outer end of TM10 contribute to gate closure (13), we speculated that a disruption of the corresponding interaction in PutP may induce an opening of the outer gate. Furthermore, changes at the outer gate might also affect the inwardly oriented pathway of PutP. To test these ideas, we analyzed the influence of individual substitutions E311A and A404E on the accessibility of strategically placed Cys residues in the putative outer gate and in the putative inner pathway to FM (Fig. 8A). In the experiments we observed a significant stimulation of the labeling rates of Cys placed at positions 309 (close to the tip of eL4) and 315 (in helix eL4b) in PutP-E311A and -A404E compared with wild-type (Fig. 8B). On the contrary, labeling of Cys placed at positions 341 or 344 (cytoplasmic half of TM8′ lining an inwardly oriented pathway) was inhibited in both PutP variants relative to wild-type (Fig. 8C). These results suggest that the placement of Ala at position 311 or Glu at position 404 can induce a global conformational change that involves an increase of the opening probability of the extracellular gate and a decrease of the opening probability of the inner pathway.
FIGURE 8.
Impact of the E311A and A404E substitutions on the putative inner and outer pathways to the substrate binding sites. A, schematic presentation of the experiment. Cys residues (green circles) were placed into the putative pathways, and modification by FM was analyzed in PutP(ΔC), PutP(ΔC)-E311A, and -A404E containing the given Cys substitutions. B, impact of the substitution of E311A or A404E on the accessibility of Cys-309 and Cys-315 in eL4. C, impact of the substitution of E311A or A404E on the accessibility of Cys-341 and Cys-344 lining the putative inner pathway. Membrane vesicles of E. coli WG170 transformed with pTrc99a/putP(ΔC) containing the given substitutions were incubated with 200 μm FM at 25 °C for 1 min (Cys-309 and Cys-315) or 0.5 min (Cys-341 and Cys-344). Sample preparation for SDS-PAGE was performed as described in the legend of Fig. 6. (i) Fluorescent bands of PutP; (ii) total amounts of PutP based on Coomassie staining; (iii) FM fluorescence normalized to the total amount of PutP. The yield of Cys labeling with the native Glu at position 311 and the native Ala at position 404 was arbitrary set to 1 in each experiment. Standard deviations were calculated from three independent experiments, whereas representative fluorescent and Coomassie-stained bands are shown. All bands of an individual experiment come from the same gel.
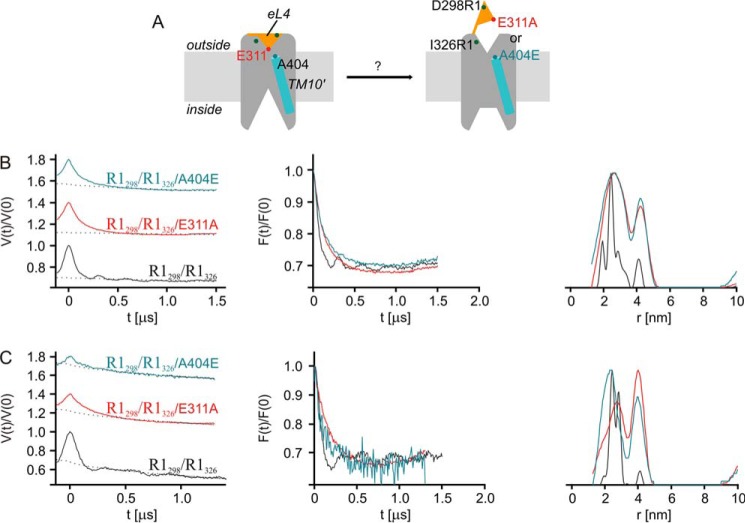
DEER Analysis
For a further evaluation of the proposed opening of the extracellular gate, we applied site-directed spin labeling and DEER spectroscopy (51). For this purpose, we placed a pair of Cys residues at positions 298 (eL4a) and 326 (outer end of TM8′) in PutP(ΔC) (Fig. 9A). These positions were chosen because (i) they appeared to be suitable for detection of a possible relocation of eL4 relative to the membrane integral part of PutP based on an analysis of structural transitions of transporters with a LeuT-fold (8); (ii) the amino acids at these positions were shown not to be of functional significance (29, 32); and (iii) they turned out in our experiments to yield well defined interspin distances in the otherwise wild-type background.
FIGURE 9.
DEER analysis of the impact of Glu-311 and Ala-404 substitutions on (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate (MTSSL)-labeled PutP(ΔC)-R1298/R1326. A, schematic presentation of the site-directed spin labeling experiment highlighting the sites of labeling. B, DEER analysis of PutP(ΔC)-R1298/R1326 in detergent solution. C, DEER analysis of PutP(ΔC)-R1298/R1326 reconstituted into liposomes. Shown are the echo amplitude in the DEER experiment as a function of dipolar evolution time and normalized to the value at zero time (left column) with background fits (dashed black lines), the background-corrected and renormalized echo amplitude (form factor) scaled to the same depth of the dipolar modulation (middle column), and distance distributions computed by Tikhonov regularization with regularization factor of 10 and normalized to the maximum amplitude of probability density P(r) of finding distance r (right column). In the left column, data for E311A and A404E are vertically shifted for better visibility. Data were analyzed with DeerAnalysis (54).
Purified PutP(ΔC)-R1298/R1326 (R1 represents a nitroxide spin label attached to Cys) was subjected to DEER analysis in its n-dodecyl-β-d-maltoside-solubilized state as well as reconstituted into proteoliposomes. Both conditions yielded a short distance with a narrow distance distribution (26 ± 2.6 Å) (Fig. 9, B and C) as is visible in the primary data (left column) and form factor (middle column) by a fast initial decay (short distance) and several oscillations after the initial decay (narrow distribution). The distance distributions obtained by Tikhonov regularization (right column) are consistent with these observations. Upon substitution of Glu-311 the oscillations after the initial decay disappeared (left and middle columns), indicating a broadening of the distance distribution that was indeed observed in the data obtained by Tikhonov regularization (right column). The mean distance increased, as is also seen by the slower initial decay of the echo signal (left and middle column). Very similar results were obtained upon replacement of Ala-404 by Glu (Fig. 9, B and C). The lower signal strength with the latter PutP variant was due to lower protein concentrations of the preparation, whereas the lower DEER modulation depth may indicate lower accessibility of the labeling sites in PutP-A404E. These results can be explained by a higher conformational flexibility of the spin labels at the reporter positions and/or a longer distance between positions 298 (eL4a) and 326 (TM8′).
Discussion
The available structural information on LeuT and transporters with a related fold suggests that eL4 participates in the reversible occlusion of a pathway open to the outside and leading to the central solute binding site (10, 12, 13, 35). Specifically, crystallization of LeuT in different conformations implies that the tip of eL4 interacts with the outer end of TM10 upon gate closure (13). Here we address the functional significance of amino acids at the tip of eL4 and the region around the outer end of TM10′ using PutP as a model. We demonstrate that a negative charge at position 311 at the tip of the hairpin structure of eL4 of PutP is particularly crucial for transport, whereas binding of sodium and proline to the binding sites is only marginally affected. Positively charged amino acids in the proximity of Glu-311 proved irrelevant for transport. On the contrary, our Cys scanning mutagenesis identified two clusters of functional important amino acids in the region around the outer end of TM10′: Val-397, Leu-398, Leu-400, and Ala-404, Trp-405, Ala-406, Gly-407.
What is the role of these in part small aliphatic amino acids in PutP? Even the drastic substitutions of Glu-311 by Ala and Ala-404 by Glu do not significantly impair binding of either proline or sodium. These substitutions must therefore affect other steps in the transport cycle. Considering the role of the respective sites in LeuT (13) (Fig. 5C), our results support the idea of an involvement of both sites in closing the outer gate. The idea is further fostered by the observation that side chain alterations at positions 311 or 404 appear to induce a global conformational alteration in the direction of closing of the previously proposed inner cavity (28, 30, 32), and opening of an outer pathway. This conclusion is supported by alterations of the accessibility of Cys placed in the inner and outer ligand pathways. For the outer pathway, evidence for gate opening comes also from our DEER-based distance analysis. A safe statement on the extent of the conformational change cannot be made. However, because the measurements are performed with PutP in its native environment, the membrane, and there is only little influence on ligand binding, we expect the induced state to resemble a conformation close to a native state occurring during the transition to an outward-open conformation. Previously investigated ligand-induced changes in PutP involve transitions only between an inward-open and an occluded state (28, 30, 32). Alterations of the opening probability of the inner and outer gates have been observed also upon substitution of amino acids involved in gating in LeuT (6).
In the model of PutP presented here, the side chain of Glu-311 is in H-bond distance to the backbone at position 404 (Fig. 5A). However, our functional analysis cannot exclude that Glu-311 interacts with an amino acid at a different position. Candidates for the interaction site come from the more N-terminal cluster of functionally important amino acids including Val-397, Leu-398, and Leu-400. In fact, due to an uncertain local alignment of the TMs 10′ of PutP and vSGLT, there is a register shift between the alignment originally used for PutP modeling (28) and the alignment of Faham et al. (17) that locates Leu-398 at the outer end of TM10′. The Cys accessibility analysis presented here places position 402 at the outer boarder of TM10′ thereby supporting our model of PutP (Figs. 5A and 6D).
Besides being involved in gate closure, are there other potential functions of the amino acids around the outer end of TM10′? Our results indicate that alterations also of Ala-406 affect proline binding. Furthermore, differing from the Cys substitution, Gly at position 404 increases the apparent affinity parameters Km(Pro) and Kd(Pro) by more than 1 order of magnitude and to a lesser extent also K0.5(Na+) (Table 1). Furthermore, the significantly stimulated Vmax of PutP-A404G suggests that the smallest and achiral amino acid glycine at this position facilitates conformational alterations associated with the transport cycle and underpins the significance of TM10′ for the alternating access mechanism. The results are somewhat reminiscent of the impact of the substitution of the outer gate residue Phe-453 in TM10′ of hSGLT1 (52). The authors conclude that the gate side chains are not simply barriers but are important for coordinating conformational changes, and thus efficient coupling of Na+ and sugar transport by hSGLT1 (52).
Leu-398 of PutP was previously proposed to participate in proline binding to a second, more external site (34). If indeed eL4 interacts with the peptide at or close to position 398, one may speculate on a role of the proposed external binding site in regulating the dynamics of gating. In fact, in the corresponding region of vSGLT, Ala-423 interacts with the tip of eL4 (17), and Phe-424 is suggested to be part of a more external substrate binding site (34).
What is the role of eL4 and TM10′ in PutP under physiological conditions, i.e. sodium-coupled uptake of proline? Previously, we suggested that upon substrate binding a pulling force arising from interactions with amino acids above binding site residues in TM1′ is exerted on Phe-314 in eL4b anchoring eL4 into the outer pathway of PutP (29). The anchoring causes an inward movement of TM7b′ and a displacement of TM8′ (29). Here we extend the model by implicating interactions between Glu-311 at the tip of eL4 and the peptide backbone at the end of TM10′ in external gate closure and stabilization of an inward-open conformation. The significance of both structural elements for coordinating conformational alterations may arise from the fact that both eL4 and TM10′ connect groups of TMs moving relative to each other during the transport cycle as identified in the crystal structures of different conformations of LeuT and Mhp1 (3, 8, 13).
Taken together with the described interactions between eL4 and the periplasmic end of TM10′ in vSGLT (between Asp-336 and Ala-423) and LeuT (between Ala-319 and Asp-401), our observations further specify the idea that common design and functional principles are maintained across different transport families with similar structure.
Author Contributions
S. B. generated, purified, labeled all mutants, and performed cysteine accessibility and functional analyses; K. G. and Y. P. performed electron paramagnetic resonance measurements and spectral analyses; G. J. performed the distance restraint-based homology modeling; M. B. participated in mutant generation and characterization; S. D. and S. F. participated in the cysteine accessibility analyses; G. J. and H. J. planned and supervised the experiments; and H. J., S. B., and G. J. wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
We thank Michelle Eder for excellent technical assistance, and Emil Tonon who participated in the analyses in a lab research course (H. J. group). We also thank the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, supported by NIGMS P41-GM103311.
This work was supported by Deutsche Forschungsgemeinschaft Grants JU333/3-2 and JU333/4-2 (to H. J.) and Schweizer Nationalfonds Grant 200021E_129360 (to G. J.). The authors declare that they have no conflicts of interest with the contents of this article.
TMs 2 to 11 of PutP correspond to the core TMs 1 to 10 of LeuT, and are numbered 1′ to 10′ in the text.
- TM
- transmembrane domain
- DEER
- double electron electron resonance
- DRaCALA
- differential radial capillary action of ligand assay
- eL
- external loop
- FM
- fluorescein-5-maleimide
- MMTS
- methyl methanethiosulfonate
- MTSEA
- methanethiosulfonate ethylammonium
- MTSES
- methanethiosulfonate ethylsulfonate
- MTSSL
- (1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate
- NEM
- N-ethylmaleimide
- SSS
- solute/sodium symporter.
References
- 1. Jardetzky O. (1966) Simple allosteric model for membrane pumps. Nature 211, 969–970 [DOI] [PubMed] [Google Scholar]
- 2. Krishnamurthy H., Piscitelli C. L., and Gouaux E. (2009) Unlocking the molecular secrets of sodium-coupled transporters. Nature 459, 347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shimamura T., Weyand S., Beckstein O., Rutherford N. G., Hadden J. M., Sharples D., Sansom M. S., Iwata S., Henderson P. J., and Cameron A. D. (2010) Molecular basis of alternating access membrane transport by the sodium-hydantoin transporter Mhp1. Science 328, 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perez C., Koshy C., Yildiz O., and Ziegler C. (2012) Alternating-access mechanism in conformationally asymmetric trimers of the betaine transporter BetP. Nature 490, 126–130 [DOI] [PubMed] [Google Scholar]
- 5. Kaback H. R. (2015) A chemiosmotic mechanism of symport. Proc. Natl. Acad. Sci. U.S.A. 112, 1259–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhao Y., Terry D., Shi L., Weinstein H., Blanchard S. C., and Javitch J. A. (2010) Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature 465, 188–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Forrest L. R. (2013) Structural biology: (pseudo-)symmetrical transport. Science 339, 399–401 [DOI] [PubMed] [Google Scholar]
- 8. Jeschke G. (2013) A comparative study of structures and structural transitions of secondary transporters with the LeuT fold. Eur. Biophys. J. 42, 181–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kazmier K., Sharma S., Quick M., Islam S. M., Roux B., Weinstein H., Javitch J. A., and McHaourab H. S. (2014) Conformational dynamics of ligand-dependent alternating access in LeuT. Nat. Struct. Mol. Biol. 21, 472–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Claxton D. P., Quick M., Shi L., de Carvalho F. D., Weinstein H., Javitch J. A., and McHaourab H. S. (2010) Ion/substrate-dependent conformational dynamics of a bacterial homolog of neurotransmitter:sodium symporters. Nat. Struct. Mol. Biol. 17, 822–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Forrest L. R., and Rudnick G. (2009) The rocking bundle: a mechanism for ion-coupled solute flux by symmetrical transporters. Physiology 24, 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Weyand S., Shimamura T., Yajima S., Suzuki S., Mirza O., Krusong K., Carpenter E. P., Rutherford N. G., Hadden J. M., O'Reilly J., Ma P., Saidijam M., Patching S. G., Hope R. J., Norbertczak H. T., Roach P. C., Iwata S., Henderson P. J., and Cameron A. D. (2008) Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science 322, 709–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krishnamurthy H., and Gouaux E. (2012) X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature 481, 469–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abramson J., and Wright E. M. (2009) Structure and function of Na+-symporters with inverted repeats. Curr. Opin. Struct. Biol. 19, 425–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Khafizov K., Staritzbichler R., Stamm M., and Forrest L. R. (2010) A study of the evolution of inverted-topology repeats from LeuT-fold transporters using AlignMe. Biochemistry 49, 10702–10713 [DOI] [PubMed] [Google Scholar]
- 16. Yamashita A., Singh S. K., Kawate T., Jin Y., and Gouaux E. (2005) Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature 437, 215–223 [DOI] [PubMed] [Google Scholar]
- 17. Faham S., Watanabe A., Besserer G. M., Cascio D., Specht A., Hirayama B. A., Wright E. M., and Abramson J. (2008) The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321, 810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Watanabe A., Choe S., Chaptal V., Rosenberg J. M., Wright E. M., Grabe M., and Abramson J. (2010) The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature 468, 988–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jung H. (2002) The sodium/substrate symporter family: structural and functional features. FEBS Lett. 529, 73–77 [DOI] [PubMed] [Google Scholar]
- 20. Zaprasis A., Bleisteiner M., Kerres A., Hoffmann T., and Bremer E. (2015) Uptake of amino acids and their metabolic conversion into the compatible solute proline confers osmoprotection to Bacillus subtilis. Appl. Environ. Microbiol. 81, 250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schwan W. R., Lehmann L., and McCormick J. (2006) Transcriptional activation of the Staphylococcus aureus putP gene by low-proline-high osmotic conditions and during infection of murine and human tissues. Infect. Immun. 74, 399–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jung H., Hilger D., and Raba M. (2012) The Na+/l-proline transporter PutP. Front. Biosci. 17, 745–759 [DOI] [PubMed] [Google Scholar]
- 23. Rivera-Ordaz A., Bracher S., Sarrach S., Li Z., Shi L., Quick M., Hilger D., Haas R., and Jung H. (2013) The sodium/proline transporter PutP of Helicobacter pylori. PLoS ONE 8, e83576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vílchez S., Molina L., Ramos C., and Ramos J. L. (2000) Proline catabolism by Pseudomonas putida: cloning, characterization, and expression of the put genes in the presence of root exudates. J. Bacteriol. 182, 91–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Crawford J. M., Kontnik R., and Clardy J. (2010) Regulating alternative lifestyles in entomopathogenic bacteria. Curr. Biol. 20, 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jung H., Rübenhagen R., Tebbe S., Leifker K., Tholema N., Quick M., and Schmid R. (1998) Topology of the Na+/proline transporter of Escherichia coli. J. Biol. Chem. 273, 26400–26407 [DOI] [PubMed] [Google Scholar]
- 27. Wegener C., Tebbe S., Steinhoff H. J., and Jung H. (2000) Spin labeling analysis of structure and dynamics of the Na+/proline transporter of Escherichia coli. Biochemistry 39, 4831–4837 [DOI] [PubMed] [Google Scholar]
- 28. Olkhova E., Raba M., Bracher S., Hilger D., and Jung H. (2011) Homology model of the Na+/proline transporter PutP of Escherichia coli and its functional implications. J. Mol. Biol. 406, 59–74 [DOI] [PubMed] [Google Scholar]
- 29. Raba M., Dunkel S., Hilger D., Lipiszko K., Polyhach Y., Jeschke G., Bracher S., Klare J. P., Quick M., Jung H., and Steinhoff H. J. (2014) Extracellular loop 4 of the proline transporter PutP controls the periplasmic entrance to ligand binding sites. Structure 22, 769–780 [DOI] [PubMed] [Google Scholar]
- 30. Pirch T., Landmeier S., and Jung H. (2003) Transmembrane domain II of the Na+/proline transporter PutP of Escherichia coli forms part of a conformationally flexible, cytoplasmic exposed aqueous cavity within the membrane. J. Biol. Chem. 278, 42942–42949 [DOI] [PubMed] [Google Scholar]
- 31. Hilger D., Böhm M., Hackmann A., and Jung H. (2008) Role of Ser-340 and Thr-341 in transmembrane domain IX of the Na+/proline transporter PutP of Escherichia coli in ligand binding and transport. J. Biol. Chem. 283, 4921–4929 [DOI] [PubMed] [Google Scholar]
- 32. Raba M., Baumgartner T., Hilger D., Klempahn K., Härtel T., Jung K., and Jung H. (2008) Function of transmembrane domain IX in the Na+/proline transporter PutP. J. Mol. Biol. 382, 884–893 [DOI] [PubMed] [Google Scholar]
- 33. Pirch T., Quick M., Nietschke M., Langkamp M., and Jung H. (2002) Sites important for Na+ and substrate binding in the Na+/proline transporter of Escherichia coli, a member of the Na+/solute symporter family. J. Biol. Chem. 277, 8790–8796 [DOI] [PubMed] [Google Scholar]
- 34. Li Z., Lee A. S., Bracher S., Jung H., Paz A., Kumar J. P., Abramson J., Quick M., and Shi L. (2015) Identification of a second substrate-binding site in solute-sodium symporters. J. Biol. Chem. 290, 127–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kazmier K., Sharma S., Islam S. M., Roux B., and Mchaourab H. S. (2014) Conformational cycle and ion-coupling mechanism of the Na+/hydantoin transporter Mhp1. Proc. Natl. Acad. Sci. U.S.A. 111, 14752–14757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mitchell S. M., Lee E., Garcia M. L., and Stephan M. M. (2004) Structure and function of extracellular loop 4 of the serotonin transporter as revealed by cysteine-scanning mutagenesis. J. Biol. Chem. 279, 24089–24099 [DOI] [PubMed] [Google Scholar]
- 37. Zomot E., and Kanner B. I. (2003) The interaction of the γ-aminobutyric acid transporter GAT-1 with the neurotransmitter is selectively impaired by sulfhydryl Modification of a conformationally sensitive cysteine residue engineered into extracellular loop IV. J. Biol. Chem. 278, 42950–42958 [DOI] [PubMed] [Google Scholar]
- 38. Carland J. E., Handford C. A., Ryan R. M., and Vandenberg R. J. (2014) Lipid inhibitors of high affinity glycine transporters: identification of a novel class of analgesics. Neurochem. Int. 73, 211–216 [DOI] [PubMed] [Google Scholar]
- 39. Stalmach M. E., Grothe S., and Wood J. M. (1983) Two proline porters in Escherichia coli K-12. J. Bacteriol. 156, 481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tabor S., and Richardson C. C. (1985) A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. U.S.A. 82, 1074–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jung H., Tebbe S., Schmid R., and Jung K. (1998) Unidirectional reconstitution and characterization of purified Na+/proline transporter of Escherichia coli. Biochemistry 37, 11083–11088 [DOI] [PubMed] [Google Scholar]
- 42. Amann E., Ochs B., and Abel K. J. (1988) Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69, 301–315 [DOI] [PubMed] [Google Scholar]
- 43. Quick M., and Jung H. (1997) Aspartate 55 in the Na+/proline permease of Escherichia coli is essential for Na+-coupled proline uptake. Biochemistry 36, 4631–4636 [DOI] [PubMed] [Google Scholar]
- 44. Quick M., Tebbe S., and Jung H. (1996) Ser57 in the Na+/proline permease of Escherichia coli is critical for high-affinity proline uptake. Eur. J. Biochem. 239, 732–736 [DOI] [PubMed] [Google Scholar]
- 45. Kaback H. R. (1971) Bacterial membranes. Methods Enzymol. 22, 99–120 [Google Scholar]
- 46. Roelofs K. G., Wang J., Sintim H. O., and Lee V. T. (2011) Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. Proc. Natl. Acad. Sci. U.S.A. 108, 15528–15533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hilger D., Polyhach Y., Jung H., and Jeschke G. (2009) Backbone structure of transmembrane domain IX of the Na+/proline transporter PutP of Escherichia coli. Biophys. J. 96, 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peterson G. L. (1977) A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83, 346–356 [DOI] [PubMed] [Google Scholar]
- 49. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 50. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., and Ferrin T. E. (2004) UCSF chimera: a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 51. Jeschke G., and Polyhach Y. (2007) Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys. Chem. Chem. Phys. 9, 1895–1910 [DOI] [PubMed] [Google Scholar]
- 52. Sala-Rabanal M., Hirayama B. A., Loo D. D., Chaptal V., Abramson J., and Wright E. M. (2012) Bridging the gap between structure and kinetics of human SGLT1. Am. J. Physiol. Cell Physiol. 302, C1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sievers F., Wilm A., Dineen D., Gibson T. J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., Thompson J. D., and Higgins D. G. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jeschke G., Chechik V., Ionita P., Godt A., Zimmermann H., Banham J., Timmel C. R., Hilger D., and Jung H. (2006) DeerAnalysis2006—a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Res. 30, 473–498 [Google Scholar]