Abstract
Actin-interacting protein 1 (AIP1) is a conserved WD repeat protein that promotes disassembly of actin filaments when actin-depolymerizing factor (ADF)/cofilin is present. Although AIP1 is known to be essential for a number of cellular events involving dynamic rearrangement of the actin cytoskeleton, the regulatory mechanism of the function of AIP1 is unknown. In this study, we report that two AIP1 isoforms from the nematode Caenorhabditis elegans, known as UNC-78 and AIPL-1, are pH-sensitive in enhancement of actin filament disassembly. Both AIP1 isoforms only weakly enhance disassembly of ADF/cofilin-bound actin filaments at an acidic pH but show stronger disassembly activity at neutral and basic pH values. However, a severing-defective mutant of UNC-78 shows pH-insensitive binding to ADF/cofilin-decorated actin filaments, suggesting that the process of filament severing or disassembly, but not filament binding, is pH-dependent. His-60 of AIP1 is located near the predicted binding surface for the ADF/cofilin-actin complex, and an H60K mutation of AIP1 partially impairs its pH sensitivity, suggesting that His-60 is involved in the pH sensor for AIP1. These biochemical results suggest that pH-dependent changes in AIP1 activity might be a novel regulatory mechanism of actin filament dynamics.
Keywords: actin, cofilin, cytoskeleton, pH regulation, severing, AIP1, actin dynamics, actin turnover, depolymerization
Introduction
Regulated disassembly of actin filaments is often required for reorganization of the actin cytoskeleton (1, 2). Spontaneous fragmentation and depolymerization of actin filaments are too rare or slow to account for rapid filament turnover or drastic remodeling of the actin cytoskeleton. The actin depolymerizing factor (ADF)/cofilin2 family of actin regulatory proteins is one of the major groups of proteins that promote severing and depolymerization of actin filaments and are involved in a number of cellular events that require enhanced turnover of actin filaments (1). ADF/cofilin binds to the side of actin filaments by cross-bridging two longitudinal actin subunits, and ADF/cofilin-bound actin filaments are structurally different from bare actin filaments (3–7). Filament severing occurs preferentially at a junction between ADF/cofilin-bound and bare segments within a filament (8, 9). Therefore, when actin filaments are fully decorated by ADF/cofilin, severing occurs less frequently than when actin filaments are occupied by ADF/cofilin at a low density (10, 11).
Actin-interacting protein 1 (AIP1), a conserved WD repeat protein, promotes disassembly of ADF/cofilin-bound actin filaments (12), and its activity appears to be maximal when actin filaments are fully decorated by ADF/cofilin (13–16). Although AIP1 has been reported previously to enhance filament disassembly by capping the barbed ends of ADF/cofilin-severed actin filaments (17, 18), recent studies have shown that highly purified AIP1 lacks capping activity and still enhances severing of ADF/cofilin-bound filaments (16, 19–21). AIP1 further shortens ADF/cofilin-severed actin filaments (14) and contributes to the maintenance of high concentrations of actin monomers in cells (22). Therefore, AIP1 and ADF/cofilin are important players in the actin disassembly mechanism in cytoskeletal remodeling (1, 2).
Several regulatory mechanisms for ADF/cofilin are known to be important for spatial and temporal regulation of actin dynamics (23), but the mechanism that regulates AIP1 remains unknown. Because the effects of AIP1 on actin are highly dependent on ADF/cofilin, regulation of ADF/cofilin can indirectly modulate AIP1 activity. AIP1 is very potent in the disassembly of ADF/cofilin-bound actin filaments (17, 24). Therefore, an inhibitory mechanism for AIP1 would have to be operated when actin filaments need to be stabilized.
Intracellular pH is one of the regulatory factors for the dynamics of the actin cytoskeleton (25). Several actin-regulatory proteins exhibit pH-dependent changes in their properties and act as pH sensors for cytoskeletal reorganization (26). For example, talin, a focal adhesion protein (27, 28), and hisactophilin, a histidine-rich protein in Dictyostelium (29), bind to actin with higher affinity at low pH. Mammalian cyclase-associated protein 1 severs actin filaments at acidic pH (30). Gelsolin and adseverin, two related actin-severing and capping proteins, are constitutively active below pH 6.0 independently of calcium (31, 32). Fascin, an actin-bundling protein, is more susceptible to inhibitory phosphorylation by protein kinase C at lower pH (33, 34). Perhaps the most well characterized pH-dependent actin regulator is ADF/cofilin. Most ADF/cofilin proteins are sensitive to pH in vitro and exhibit higher activity to promote actin filament disassembly at higher pH (35–39). Colocalization of ADF/cofilin with actin filaments in cells is enhanced when the intracellular pH is lowered (40). Binding of ADF/cofilin (mammalian cofilin 1) with phosphoinositides is enhanced at lower pH under some in vitro conditions, with His-133 near the C terminus acting as a pH sensor (41, 42). Furthermore, binding of ADF/cofilin with cortactin is reduced at elevated pH (43). Either phosphoinositides or cortactin inhibits the interaction between ADF/cofilin and actin, and these pH sensitivities contribute to actin-dependent membrane protrusions in migratory cells (41, 43). However, the precise mechanism of pH-dependent cytoskeletal regulation and other pH sensors that mediate this process remains largely unknown.
In the nematode Caenorhabditis elegans, two ADF/cofilin isoforms, UNC-60A and UNC-60B, are expressed from the unc-60 gene by alternative splicing (44) and have genetically distinct functions (45–47). UNC-60A has very weak actin filament-severing and strong monomer-sequestering activities, whereas UNC-60B has strong actin filament-severing and nearly no monomer-sequestering activities (48, 49). Both of them show only weak pH sensitivity in actin filament disassembly when filament severing and depolymerization are examined in solution (49). In this study, we found two C. elegans AIP1 isoforms, UNC-78 and AIPL-1 (50–52), have pH sensitivity in disassembly of UNC-60B-bound actin filaments. Therefore, AIP1 may function as a novel pH sensor for the regulation of actin filament dynamics.
Experimental Procedures
Proteins and Materials
Rabbit muscle actin was purified from acetone powder (Pel-Freeze Biologicals) as described previously (53). UNC-60B was expressed in Escherichia coli and purified as described previously (48). GST alone was expressed from pGEX-2T (GE Healthcare Life Sciences) and purified using glutathione Uniflow resin (Clontech).
Mutagenesis, Expression, and Purification of Recombinant UNC-78 and AIPL-1 Proteins
GST-tagged UNC-78 and AIPL-1 were expressed in E. coli and purified as described previously (52, 54). H60K and H60A mutations of AIPL-1 were introduced by site-directed mutagenesis of the expression vector for AIPL-1 (pGEX-AIPL-1) using a QuikChange site-directed mutagenesis kit. The mutations and other coding sequence for AIPL-1 were verified by DNA sequencing. Both mutant AIPL-1 proteins were expressed as GST fusion proteins and purified with the same procedures as GST-AIPL-1 (wild-type).
F-actin Sedimentation Assays
Assays were performed in F buffer (0.1 m KCl, 2 mm MgCl2, and 0.2 mm DTT) at various pH values containing 20 mm MES (pH 6.0), 20 mm HEPES (pH 7.0), 20 mm HEPES (pH 7.5), 20 mm Tris (pH 8.0), or 20 mm Tris (pH 8.5). F-actin (10 μm) in F buffer was incubated with or without 20 μm UNC-60B and various concentrations of GST, GST-UNC-78, or GST-AIPL-1 for 30 min at room temperature. Then the mixtures were ultracentrifuged at 80,000 rpm for 15 min at 20 °C using a Beckman TLA-100 rotor. Supernatants and pellets were adjusted to the same volumes and subjected to SDS-PAGE (12% acrylamide gel) and staining with Coomassie Brilliant Blue R-250 (National Diagnostics). Gels were scanned by an Epson Perfection V700 photo scanner at 300 dpi, and band intensity was quantified using ImageJ.
Light-scattering Assays
F-actin (4 μm) in F buffer at various pH values was mixed with 5 μm UNC-60B and 0–0.5 μm GST, GST-UNC-78, or GST-AIPL-1, and light scattering at an angle of 90° and a wavelength of 400 nm (5-nm slit width for both excitation and emission) was measured over time with an F-4500 fluorescence spectrophotometer (Hitachi High Technologies).
Direct Observation of Actin Filament Severing by Fluorescence Microscopy
Rhodamine- and biotin-labeled actin filaments (20% rhodamine- and 1% biotin labels) were tethered to anti-biotin antibody-coated coverslips and observed with a Nikon TE2000-U epifluorescence microscope equipped with a high numerical aperture (NA) lens (Plan Apo TIRF; ×60; numerical aperture, 1.45; Nikon) and an ORCA Flash D4 complementary metal-oxide-semiconductor (CMOS) camera (Hamamatsu Photonics) as described previously (55). The pH of F buffer (containing 10 mm ascorbic acid as an anti-bleaching agent) was adjusted by 10 mm HEPES (pH 6.7 or 7.5) or 20 mm Tris-HCl (pH 8.5). Images were recorded for 200 s at a rate of 1 s/frame. Filament-severing events were counted, and the frequency (rate) of filament severing was calculated as the number of severings per micrometer per micromolar UNC-60B per second. Direct observation of actin filaments, as shown in Fig. 3, D and E, was performed as described previously (24) using DyLight 549-labeled actin (20% labels) (56), a Nikon TE2000 epifluorescence microscope, and a SPOT RT Monochrome charge-coupled device (CCD) camera (Diagnostic Instruments).
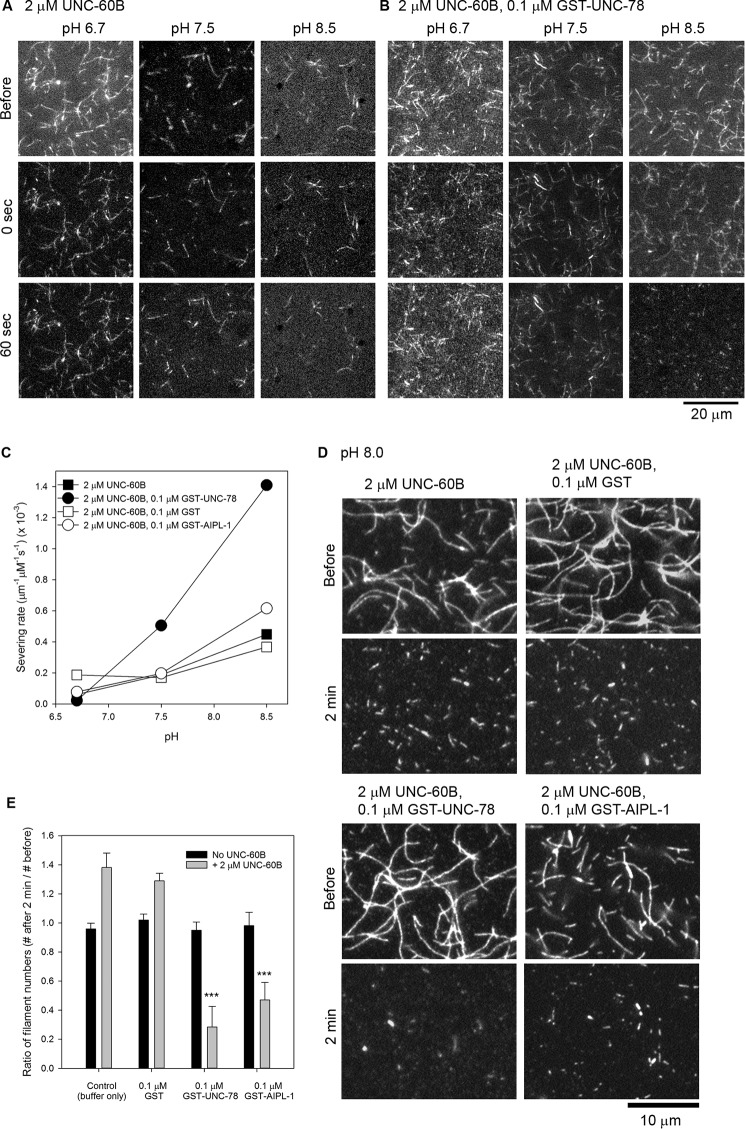
FIGURE 3.
pH-dependent severing of actin filaments by UNC-60B (ADF/cofilin) and AIP1 isoforms. A and B, rhodamine-labeled actin filaments were attached to a glass surface and observed by time-lapse imaging using epifluorescence microscopy. Representative images of actin filaments before and at 0 and 60 s after adding 2 μm UNC-60B (A) or 2 μm UNC-60B and 0.1 μm GST-UNC-78 (B) at pH 6.7, 7.5, or 8.5 are shown. C, severing rates were analyzed quantitatively from time-lapse movies in the initial period of 15–30 s and plotted as a function of pH. The data are average rates of severing. Total accumulated filament lengths of 290–1400 μm for each condition were analyzed. D and E, DyLight 549-labeled actin filaments were attached to a glass surface, and snapshot images of the filaments before and 2 min after treatments at pH 8.0 with or without UNC-60B, GST, GST-UNC-78, or GST-AIPL-1 were examined. Representative images of actin filaments are shown in D. The numbers of actin filaments regardless of length in a 20 × 20 μm image field were counted before (F1) and 2 min after treatments (F2). The ratios of the number of filaments were calculated by dividing F2 by F1. Data are mean ± S.D. (n = 5). ***, p < 0.001 as compared with control.
Results
C. elegans AIP1s Enhance Disassembly of ADF/Cofilin-bound Actin Filaments in a pH-dependent Manner
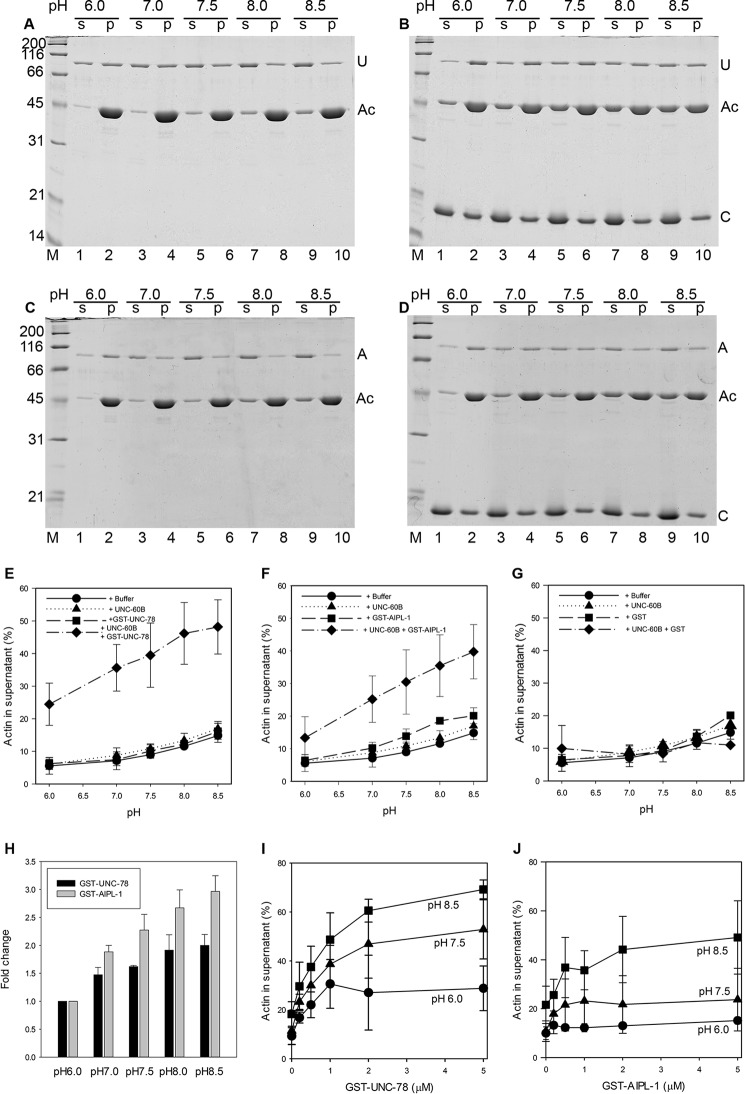
The pH sensitivity of UNC-78 and AIPL-1, the two C. elegans AIP1 isoforms, was characterized by F-actin sedimentation assays to estimate the extent of filament disassembly in a range of pH values (6.0–8.5). UNC-78 and AIPL-1 were used as GST fusion proteins (52, 54), and GST alone was used in control experiments. UNC-60B (a muscle-specific C. elegans ADF/cofilin) or GST-UNC-78 alone had only minor effects on the amounts of pelletable actin at the examined pH values (Fig. 1, A and E). Slight increases in unpelletable actin at high pH were also detected by actin alone to similar extents (Fig. 1E, circles), which is consistent with a previous report of suppressed actin polymerization at high pH (57). In these experiments, an excessive concentration of UNC-60B (20 μm) was added to F-actin (10 μm). Under these conditions, although UNC-60B weakly severs actin filaments, it does not sequester actin monomers, and UNC-60B-severed filaments are still long enough to be sedimented by ultracentrifugation (49).
FIGURE 1.
pH-dependent disassembly of actin filaments by UNC-60B (ADF/cofilin) and AIP1 isoforms as determined by sedimentation assays. A–D, F-actin (10 μm) was incubated with 2 μm GST-UNC-78 (A), 20 μm UNC-60B and 2 μm GST-UNC-78 (B), 2 μm GST-AIPL-1 (C), or 20 μm UNC-60B and 2 μm GST-AIPL-1 (D) at various pH values, as indicated on each panel, for 30 min. The mixtures were ultracentrifuged and fractionated into supernatants (s) and pellets (p), which were analyzed by SDS-PAGE. The positions of the bands of actin (Ac), GST-UNC-78 (U), GST-AIPL-1 (A), and UNC-60B (C) are indicated at the right in all panels. Molecular weight markers in kilodalton (M) are shown at the left in panels A and C. The same markers were used in B and D. E–H, quantitative analysis of the sedimentation assays. Sedimentation assays with 10 μm F-actin, as shown in A–D, were performed with 2 μm GST-UNC-78 (E), 2 μm GST-AIPL-1 (F), or 2 μm GST (G), and the percentages of actin in the supernatants were quantified by densitometry and plotted as a function of pH. The same data for controls with buffer only (circles) and 20 μm UNC-60B (triangles) were used in E–G. The data for GST-UNC-78, GST-AIPL-1, or GST in the absence (squares) or presence of 20 μm UNC-60B (diamonds) are shown in each panel. For the results of GST-UNC-78 and GST-AIPL-1 in the presence of 20 μm UNC-60B, the -fold changes in actin in the supernatants at various pH levels compared with those at pH 6.0 were calculated and are shown in H. Results are mean ± S.D. of three independent experiments. I and J, quantitative results of the sedimentation assays with 10 μm F-actin, 20 μm UNC-60B, and various concentrations of GST-UNC-78 (I) or GST-AIPL-1 (J) at pH 6.0 (circles), 7.5 (triangles), or 8.5 (squares).
In the presence of both UNC-60B and GST-UNC-78, actin in the supernatants was increased, and more actin remained in the supernatants with increasing pH (Fig. 1, B and E). On the basis of the shifts of actin into the supernatant fractions, actin filament disassembly by GST-UNC-78 was estimated to be enhanced 2-fold at pH 8.5 compared with pH 6.0 (Fig. 1H). GST alone did not have detectable effects on actin disassembly (Fig. 1G). Because UNC-60B by itself did not show significant pH sensitivity in this assay, we concluded that UNC-78 had pH-sensitive activity to disassemble UNC-60B-bound actin filaments.
Another C. elegans AIP1 isoform, AIPL-1, also showed pH-dependent enhancement of actin disassembly in the F-actin sedimentation assays (Fig. 1, C, D, and F). Although GST-AIPL-1 had slightly weaker activity than GST-UNC-78, it showed greater pH sensitivity than GST-UNC-78 (Fig. 1, C, D, and F). At pH 6.0, GST-AIPL-1 only slightly enhanced disassembly of UNC-60B-bound actin filaments (Fig. 1D, lanes 1 and 2), but this activity was greatly enhanced with increasing pH (Fig. 1, D and F), resulting in a 3-fold increase in actin disassembly at pH 8.5 compared with pH 6.0 (Fig. 1H). Both GST-UNC-78 and GST-AIPL-1 showed pH-sensitive effects in a wide range of concentrations, with maximum activities above 1–2 μm of GST-UNC-78 (Fig. 1I) or GST-AIPL-1 (Fig. 1J).
AIP1-dependent actin disassembly was also examined by light-scattering assays at different pH values (Fig. 2). C. elegans AIP1s are most effective in filament disassembly when binding of UNC-60B to F-actin is saturated (15, 52). Under these conditions, UNC-60B nearly completely quenches the fluorescence of pyrene-labeled actin filaments (49). Therefore, we used light scattering to monitor the kinetics of filament disassembly. At pH 6.5 and 8.0, UNC-60B (5 μm) instantaneously increased light scattering of F-actin (4 μm) because of F-actin binding that increased the mass of the filaments, which was followed by very slow decreases, most likely because of weak actin depolymerization or filament severing (Fig. 2, A and B, red curves). Therefore, UNC-60B alone did not show pH sensitivity in this assay, as reported previously (49). GST did not have significant effects on the UNC-60B-induced changes in light scattering (Fig. 2, A and B, cyan curves). Both GST-UNC-78 and GST-AIPL-1 decreased light scattering of UNC-60B-bound actin filaments at pH 6.5 or 8.0 (Fig. 3A, green and blue curves, respectively). At pH 8.0, both GST-UNC-78 and GST-AIPL-1 induced faster and greater drops in light scattering than at pH 6.5, indicating stronger activities of GST-UNC-78 and GST-AIPL-1 at basic pH (compare Fig. 2, A and B, green and blue curves, respectively), which is consistent with the results of the F-actin sedimentation assays. GST-AIPL-1 consistently induced slower disassembly than GST-UNC-78 in three independent experiments (representative data are shown in Fig. 2, A and B). Therefore, these kinetics results are consistent with the F-actin sedimentation assays (Fig. 1) in which GST-UNC-78 showed stronger activity than GST-AIPL-1 under all examined conditions. The results provide qualitative evidence that disassembly of UNC-60B-bound actin filaments by UNC-78 or AIPL-1 is enhanced at basic pH.
FIGURE 2.
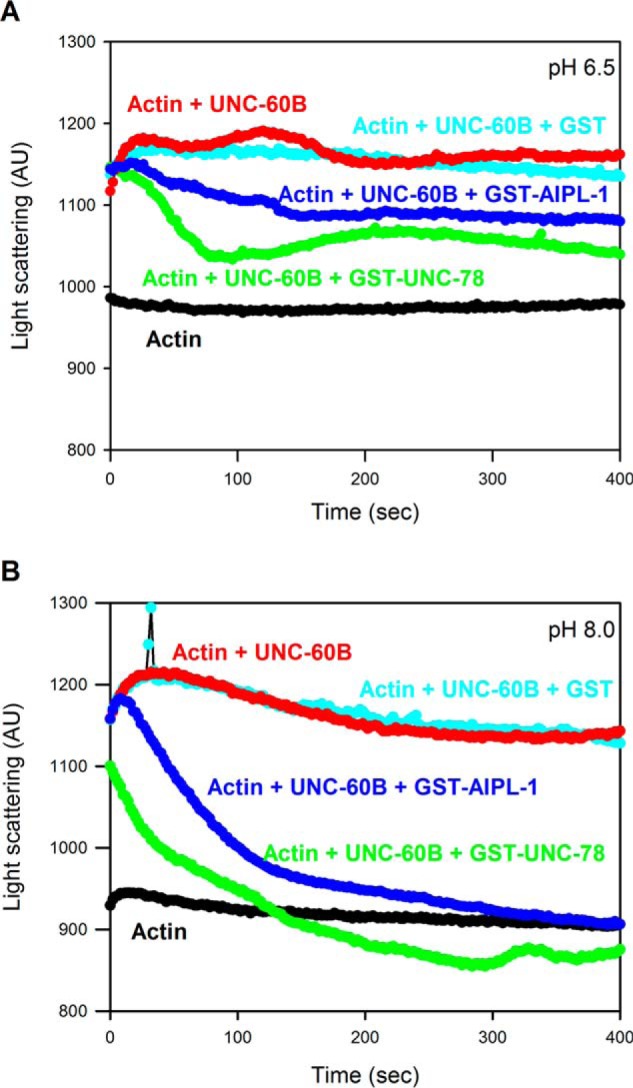
pH-dependent disassembly of actin filaments by UNC-60B (ADF/cofilin) and AIP1 isoforms as determined by light-scattering assays. F-actin (4 μm) was incubated alone (black) or in a mixture with 5 μm UNC-60B (red), 5 μm UNC-60B and 0.5 μm GST (cyan), 5 μm UNC-60B and 0.5 μm GST-UNC-78 (green), or 5 μm UNC-60B and 0.5 μm GST-AIPL-1 (blue) at pH 6.5 (A) or 8.0 (B), and light scattering (arbitrary units (AU)) at a wavelength of 400 nm was monitored over time.
Recent studies by multiple groups have clearly demonstrated that enhanced filament severing by AIP1 is one of the mechanisms to promote filament disassembly (16, 19–21). Therefore, we examined effects of pH on actin filament severing by AIP1 using time-lapse imaging of single actin filaments (Fig. 3). The severing rates (Fig. 3C) were quantified within initial incubation periods of 15–30 s because the length of total filaments did not decrease significantly, which allowed us to accurately estimate the severing frequency. UNC-60B by itself exhibited modest pH sensitivity in severing surface-immobilized actin filaments (Fig. 3, A and C). This was unexpected because UNC-60B by itself has only very weak pH sensitivity in F-actin sedimentation and light-scattering assays (Figs. 1 and 2) (49). C. elegans AIP1 isoforms also showed pH-dependent severing activities. At pH 6.7, GST, GST-UNC-78, or GST-AIPL-1 did not enhance severing of actin filaments in the presence of UNC-60B (Fig. 3C). As the pH increased, GST-UNC-78 strongly enhanced severing (Fig. 3, B and C), whereas GST-AIPL-1 modestly enhanced severing only at pH 8.5 (Fig. 3C).
To determine whether GST-AIPL-1 enhances filament severing during a prolonged incubation period, we also analyzed snapshot images before and 2 min after incubation at pH 8.0 (Fig. 3C). At this time point, precise counting of severing events was difficult because of significant shortening and loss of filaments. Therefore, we counted the numbers of remaining actin filaments regardless of their length and calculated -fold changes in the numbers of actin filaments before and after the treatments. In the absence of UNC-60B, there were no major changes in the numbers of filaments (fold change, ∼1.0; Fig. 3E). In the presence of UNC-60B with or without GST, the number of filaments increased to 1.3- to 1.4-fold because of severing (Fig. 3E), whereas it was decreased to 0.3-fold by GST-UNC-78 or 0.5-fold by GST-AIPL-1 in the presence of UNC-60B (Fig. 3E), indicating that actin filament disassembly was enhanced by UNC-78 or AIPL-1 by promoting filament severing or depolymerization at pH 8.0. All of these results suggest that enhancement of actin filament severing is one of the mechanisms of pH-dependent promotion of actin filament disassembly by AIP1 isoforms.
Binding of AIP1 to Cofilin Actin Filaments Is Independent of pH
To understand the mechanism of pH sensitivity of AIP1, we examined whether binding of AIP1 to ADF/cofilin-bound actin filaments is affected by pH. In the F-actin sedimentation assays, both UNC-78 and AIPL-1 co-sedimented with UNC-60B-bound F-actin at pH 6.0 (Fig. 1, A–D), where the activity to disassemble filaments was weak. The amounts of sedimented UNC-78 or AIPL-1 was apparently decreased as pH increased (Fig. 1, A–D). However, sedimented actin was also decreased in the presence of UNC-60B, and estimation of F-actin binding was not precise under these conditions. To uncouple filament binding from filament disassembly, we used a mutant form of UNC-78 with defective actin-severing activity. We have reported previously that UNC-78 with four point mutations (E126A, D168A, F182A, and F192A), designated UNC-78(4X), binds to UNC-60B-bound actin filaments but does not sever the filaments (58). In F-actin co-sedimentation assays in the absence of UNC-60B, GST-UNC-78(4X) co-sedimented with F-actin, but co-sedimentation was decreased with increasing pH (Fig. 4, A, lanes 1–6, and B, circles). In the presence of UNC-60B, co-sedimentation of GST-UNC-78(4X) with F-actin was enhanced but remained at similar levels at the examined pH values (Fig. 4, A, lanes 7–12, and B, squares). In addition, GST-UNC-78(4X) did not enhance actin filament severing in the presence of UNC-60B in the microscope assay.3 These results strongly suggest that binding of AIP1 to ADF/cofilin-bound actin filaments is pH-independent and that the subsequent filament severing process by AIP1 is presumably pH-dependent. These experiments also show that UNC-78 retained its property to bind to UNC-60B-bound actin filaments and was not simply denatured under low pH conditions.
FIGURE 4.
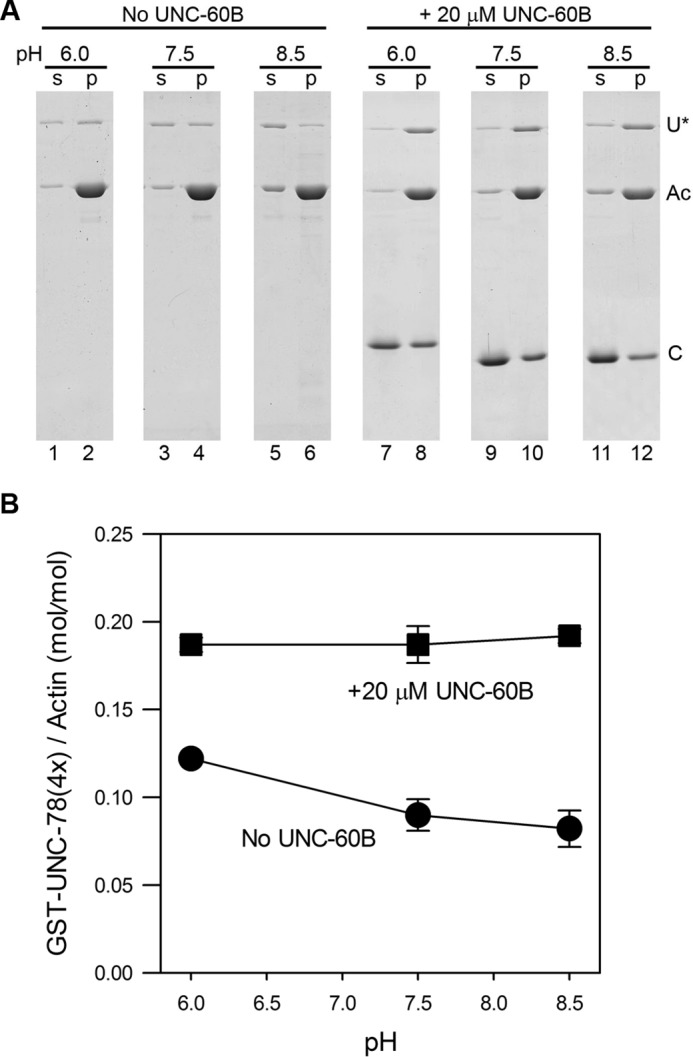
pH-insensitive binding of a severing-defective mutant of UNC-78 (AIP1) to UNC-60B (ADF/cofilin)-bound actin filaments. A, sedimentation assays were performed with F-actin (10 μm) and 2 μm GST-UNC-78(4X) (a severing-defective mutant) in the absence (lanes 1–6) or presence of 20 μm UNC-60B (lanes 7–12) at pH 6.0, 7.5, or 8.5. The positions of the bands of GST-UNC-78(4X) (U*), actin (Ac), and UNC-60B (C) are shown at the right. Note that the results were obtained from different gels and that the position of each protein is somewhat variable. s, supernatant; p, pellet. B, quantitative results of molar ratios of GST-UNC-78(4X) and actin in pellets as a function of pH. Results are mean ± S.D. of three independent experiments.
A Conserved Histidine Residue of AIP1 Is Involved in pH Sensing
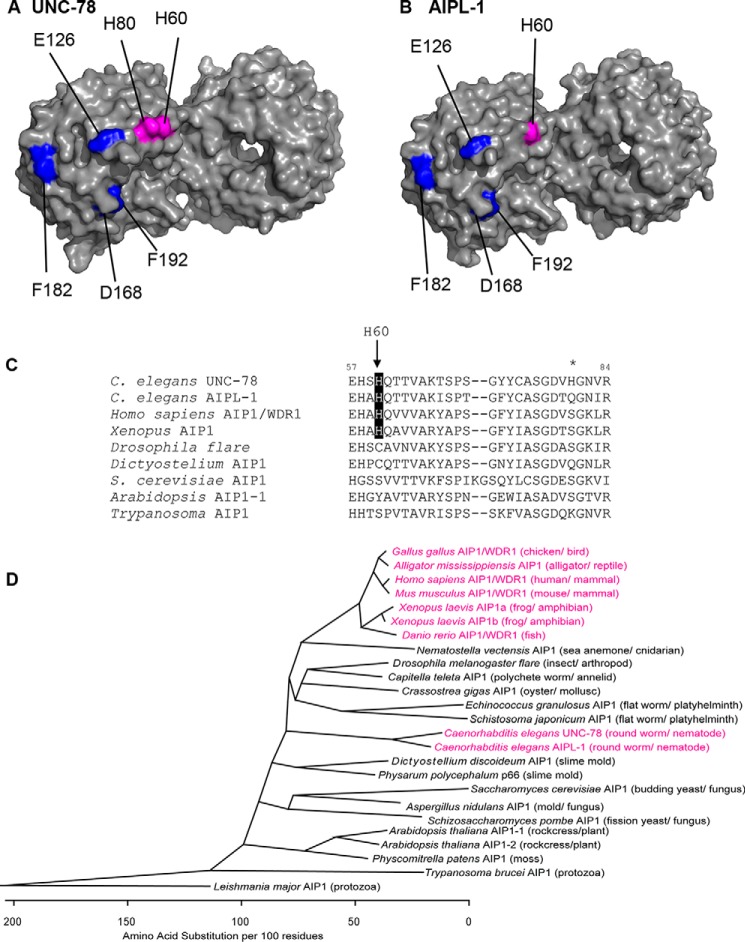
To understand how AIP1 senses pH in the surrounding solution, we tested whether any histidine residue is involved in pH sensitivity. A histidine side chain (pKa, ∼ 6.5) is charged positively by protonation at acidic pH but uncharged at basic pH (26). UNC-78 and AIPL-1 have 20 and 19 histidine residues, respectively. However, many of them are buried as integral parts of β propellers within a consensus sequence of WD repeats (59). By comparing histidine residues exposed on the surface of UNC-78 and AIPL-1, we found that His-60 is common to both UNC-78 and AIPL-1 and located on the concave surface of UNC-78 and AIPL-1, where ADF/cofilin is predicted to bind (60) (Fig. 5, A and B), suggesting that ionization states of this residue can influence the activity of AIP1. Among known AIP1 proteins, histidine is conserved at this position in AIP1s in nematodes and vertebrates (mammals, birds, reptiles, amphibians, and fish) but not in invertebrates other than nematodes, plants, protozoans, and fungi (Fig. 5, C and D). UNC-78 has another exposed histidine (His-80) that is located close to His-60 (Fig. 5A), but AIPL-1 has only one histidine residue at this location (Fig. 5B). By comparing AIP1 sequences, His-80 is unique to UNC-78 (Fig. 5C). To simplify the analysis, we used AIPL-1 and replaced His-60 with Ala (H60A) to remove an ionization group or Lys (H60K) to add a pH-independent positive charge. AIPL-1(H60A) is uncharged at this position and expected to mimic a high pH state of AIPL-1. This mutant had much weaker activity than the wild type across the examined pH values and failed to mimic an uncharged high pH state of AIPL-1 (Fig. 6A, squares). Although the activity of AIPL-1(H60A) did not suggest its involvement in pH sensing, it indeed suggests that His-60 is important for the activity of AIPL-1.
FIGURE 5.
Mapping of histidine residues on the surface of the AIP1 isoforms UNC-78 and AIPL-1. A and B, surface models of the crystal structure of UNC-78 (PDB code 1PEV) (54) and a homology model of AIPL-1 (52). Residues that are essential for severing activity (Glu-126, Asp-168, Phe-182, and Phe-192) are shown in blue. These are mutated in the severing-defective UNC-78(4X) mutant. Histidine residues on the surface are shown in magenta. Molecular graphics were generated by PyMOL 1.7 (Schrödinger, LLC.). C, alignment of sequences around His-60 of AIP1s from various species. The position of His-80 of UNC-78 is indicated by the asterisk. D, phylogenetic analysis of AIP1 sequences. AIP1 sequences from various species were aligned by the ClustalW method (62), and AIP1s that have histidine at positions equivalent to His-60 of C. elegans AIP1s are shown in magenta.
FIGURE 6.
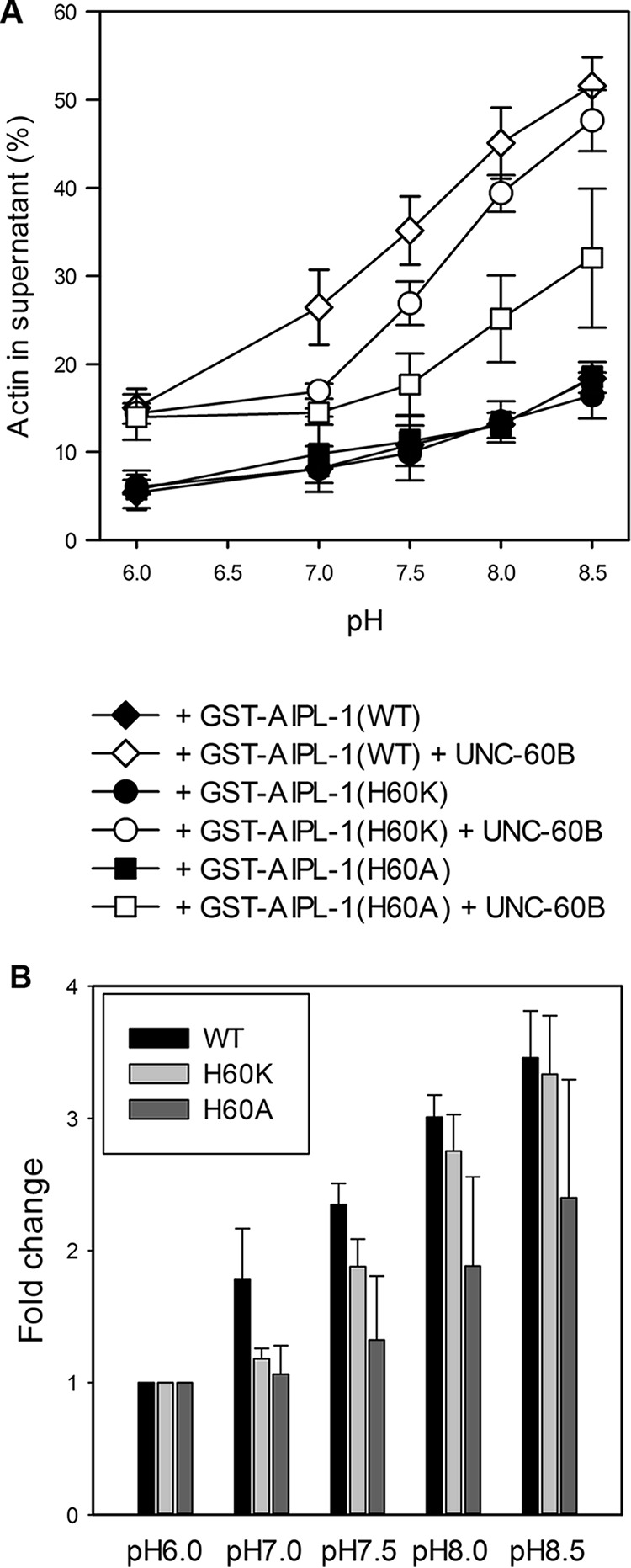
Effects of mutations of His-60 on the pH sensitivity of AIPL-1 (AIP1). A, actin sedimentation assays were performed with 10 μm F-actin and 2 μm GST-AIPL-1 (wild-type, diamonds), 2 μm GST-AIPL-1(H60K) (circles), or 2 μm GST-AIPL-1(H60A) (squares) in the absence (black symbols) or presence of 20 μm UNC-60B (white symbols), and the percentages of actin in the supernatants were plotted as a function of pH. B, for the results in the presence of 20 μm UNC-60B, the -fold changes in actin in the supernatants at various pH levels compared with those at pH 6.0 were calculated. Results are mean ± S.D. of three independent experiments.
AIPL-1(H60K) is charged positively constitutively at this position and expected to mimic a low pH state of AIPL-1. This mutant had significantly lower activity than wild-type AIPL-1 at pH 7.0 and 7.5 (Fig. 6A, compare diamonds for the wild type and circles for H60K). However, at pH 8.0 and 8.5, AIPL-1(H60K) showed nearly equivalent activity to wild-type AIPL-1 (Fig. 6, A and B). Therefore, a constantly positive charge at position 60 of AIPL-1 prevents its activity enhancement at ∼pH 7.0, suggesting that uncharging of His-60 at neutral and basic pH by deprotonation is partly important for pH sensing by AIPL-1. The observation that the activity of AIPL-1(H60K) can be enhanced fully at ∼pH 8.0 (Fig. 6A) suggests that additional residue(s) of AIPL-1 is/are involved in pH sensing.
Discussion
In this study, we demonstrated that two C. elegans AIP1 isoforms, UNC-78 and AIPL-1, possess pH-sensitive activity to enhance disassembly of UNC-60B (ADF/cofilin)-bound actin filaments. Both UNC-78 and AIPL-1 enhanced actin disassembly more strongly at basic pH than at acidic pH. Association of a severing-defective UNC-78 mutant with UNC-60B-bound actin filaments was not pH-sensitive, suggesting that filament disassembly is the pH-sensitive process. Mutations at a conserved histidine (His-60) residue in AIPL-1 altered its pH sensitivity, suggesting that this histidine is involved in the pH-sensitive activity of AIP1. Therefore, a change in intracellular pH may be a mechanism to regulate AIP1-dependent actin filament dynamics.
To date, pH-dependent modulation of the AIP1 activity, as demonstrated in this study, is the only known regulatory mechanism for AIP1. Because actin filament disassembly by AIP1 is dependent on ADF/cofilin, modulation of ADF/cofilin could indirectly influence AIP1-mediated actin dynamics. Nonetheless, direct regulation of AIP1 could be an another important mechanism to control actin dynamics. AIP1 is highly effective in actin filament disassembly when F-actin is saturated by ADF/cofilin (14, 15). However, ADF/cofilin is not effective in filament severing at high concentrations but is optimal in filament severing at substoichiometric concentrations (8, 11). Therefore, when active ADF/cofilin is present at high concentrations, alterations in AIP1 activity could trigger rapid changes in actin dynamics without modulation of ADF/cofilin. In vertebrates, ADF/cofilins are relatively strongly pH-sensitive and exhibit higher activities to disassemble actin filaments at basic pH than at neutral and acidic pH (35–38). If vertebrate AIP1 (also known as WDR1) is also pH-sensitive, then a combination of ADF/cofilin and AIP1 could be a highly sensitive pH-dependent regulatory system for actin dynamics. In C. elegans, the two ADF/cofilin isoforms, UNC-60A and UNC-60B, are only weakly pH-sensitive in solution, and phosphoregulation of ADF/cofilin has not been demonstrated because homologs of vertebrate ADF/cofilin kinases (LIM (Lin-11/Isl-1/Mec-3) kinases and testis-specific kinases) and phosphatases (Slingshot phosphatases) are absent in the C. elegans genome. Therefore, pH regulation of AIP1 might be one important mechanism to regulate actin filament disassembly. However, the significance of pH changes in cytoskeleton reorganization in C. elegans is currently unknown and will be an important subject for future research.
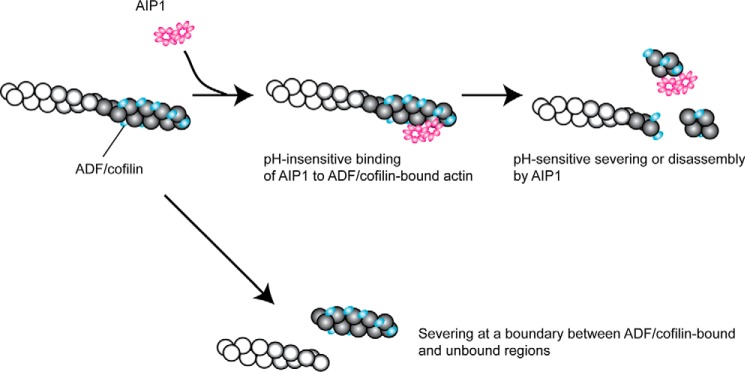
On the basis of these results, we hypothesize a two-step mechanism for AIP1-mediated severing or disassembly of ADF/cofilin-bound actin filaments (Fig. 7). We have reported previously that the UNC-78(4X) mutant can bind to ADF/cofilin-bound actin filaments but does not sever them (58). This indicates that filament binding and severing/disassembly are two separate processes that can be uncoupled by a mutation. This study further supports this view by demonstrating that filament binding by UNC-78(4X) is pH-insensitive but severing/disassembly is pH-sensitive, which strongly suggest that these are two separate processes. If the pH regulation of AIP1 is important in cells, then this two-step mechanism can provide the machinery for rapid actin reorganization. At low pH, AIP1 can be in a “standby mode” by binding to ADF/cofilin-bound actin filaments without inducing disassembly. When the pH is raised, these AIP1- and ADF/cofilin-bound actin filaments can be disassembled rapidly. In addition, AIP1 remains bound transiently to severed filaments (Fig. 7, top right) and contributes to capping when coronin is present (20), but the effect of pH on this process remains to be investigated.
FIGURE 7.
A two-step model of severing or disassembly of ADF/cofilin-bound actin filaments by AIP1. ADF/cofilin binds preferentially to ADP-actin (as shown by shaded actin subunits) in a cooperative manner, and severing of the filament occurs frequently at a boundary between ADF/cofilin-bound and -unbound segments (bottom). In the presence of AIP1, AIP1 first binds to an ADF/cofilin-bound region in the filament in a pH-insensitive manner (top center) and then promotes severing or disassembly of the filament in a pH-sensitive manner (top right).
Our mutational analysis of AIPL-1 indicates that His-60 is involved in pH sensing. However, mutations at His-60 did not completely abolish the pH sensitivity of AIPL-1, suggesting that other residues are also involved in pH sensing. Histidines are part of a consensus sequence of WD repeats (59), and most histidines within WD repeats of AIP1 are buried as integral parts of β propellers (54). We cannot exclude the possibility that some of these histidines are exposed under certain conditions and act as pH sensors. We have previously characterized a H535Y mutation of UNC-78 and found that this mutation significantly impairs the activity of UNC-78 and reduces protein levels of UNC-78 in vivo (58). This histidine is a part of a WD repeat and buried in the crystal structure of UNC-78, suggesting that the H535Y mutation disrupted the conformation of UNC-78. Therefore, additional mutagenesis on other histidines is expected to yield similar results or impairment in the solubility of the protein. To identify additional pH sensors in AIP1, other unbiased non-invasive methods, such as nuclear magnetic resonance, might be better to identify pH-dependent changes in AIP1.
Whether pH sensitivity is common to AIP1s in other organisms remains unknown. Interestingly, His-60 is conserved in vertebrate AIP1s and nematode AIP1s (Fig. 5, C and D) but not in AIP1s in other organisms. However, as discussed above, additional unknown pH sensors are likely involved, and we cannot predict which AIP1s are pH-sensitive. AIP1s in Drosophila (flare) and Dictyostelium have cysteine at a position equivalent to His-60 of C. elegans AIP1s (Fig. 5C). Because the pKa value of a cysteine side chain is ∼8.0, the cysteine could function as a pH sensor at high pH. Therefore, it would be interesting to determine whether AIP1s from different organisms have different pH sensitivities. Both C. elegans UNC-78 and AIPL-1 have Cys-74 near His-60 (Fig. 5C). However, Cys-74 is buried in the molecule and unlikely to participate in the pH regulation. pH sensing mechanisms and pH-dependent regulation of the actin cytoskeleton are known to be important in the migration of mammalian cells (25, 26, 61), and ADF/cofilin and AIP1 could function together to enhance the actin dynamics at the leading edge of migrating cells. However, the importance of pH on cytoskeletal control in other organisms is poorly understood. Accumulation of biochemical information on pH-sensitive actin regulators and future cell biological and genetic studies may reveal novel links between pH sensing and actin-dependent cellular events.
Author Contributions
K. N. designed and performed the experiments and analyzed the data shown in Figs. 1, 2, and 4–6. K. H. designed and performed the severing assays by microscopy and analyzed the data shown in Fig. 3, A–C. H. T. designed and performed the severing assays by microscopy, analyzed the data shown in Fig. 3, A–C, and edited the paper. S. O. developed the study, designed experiments, performed the experiments shown in Fig. 3, D and E, analyzed the data, and wrote the paper.
Acknowledgment
We thank Dr. Masahiro Sokabe for helpful discussions.
This work was supported by National Institutes of Health Grant R01 AR048615 (to S. O.) and Grant-in-Aid 15K07025 from the Ministry of Education, Culture, Sports, Science, and Technology Japan (to H. T.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
K. Nomura, K. Hayakawa, H. Tatsumi, and S. Ono, unpublished data.
- ADF
- actin-depolymerizing factor
- AIP1
- actin-interacting protein 1.
References
- 1. Ono S. (2007) Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int. Rev. Cytol. 258, 1–82 [DOI] [PubMed] [Google Scholar]
- 2. Brieher W. (2013) Mechanisms of actin disassembly. Mol. Biol. Cell 24, 2299–2302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McGough A., Pope B., Chiu W., and Weeds A. (1997) Cofilin changes the twist of F-actin: implications for actin filament dynamics and cellular function. J. Cell Biol. 138, 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Galkin V. E., Orlova A., VanLoock M. S., Shvetsov A., Reisler E., and Egelman E. H. (2003) ADF/cofilin use an intrinsic mode of F-actin instability to disrupt actin filaments. J. Cell Biol. 163, 1057–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ono S., McGough A., Pope B. J., Tolbert V. T., Bui A., Pohl J., Benian G. M., Gernert K. M., and Weeds A. G. (2001) The C-terminal tail of UNC-60B (ADF/cofilin) is critical for maintaining its stable association with F-actin and is implicated in the second actin-binding site. J. Biol. Chem. 276, 5952–5958 [DOI] [PubMed] [Google Scholar]
- 6. Kudryashov D. S., Galkin V. E., Orlova A., Phan M., Egelman E. H., and Reisler E. (2006) Cofilin cross-bridges adjacent actin protomers and replaces part of the longitudinal F-actin interface. J. Mol. Biol. 358, 785–797 [DOI] [PubMed] [Google Scholar]
- 7. Ressad F., Didry D., Xia G. X., Hong Y., Chua N. H., Pantaloni D., and Carlier M. F. (1998) Kinetic analysis of the interaction of actin-depolymerizing factor (ADF)/cofilin with G- and F-actins: comparison of plant and human ADFs and effect of phosphorylation. J. Biol. Chem. 273, 20894–20902 [DOI] [PubMed] [Google Scholar]
- 8. Suarez C., Roland J., Boujemaa-Paterski R., Kang H., McCullough B. R., Reymann A. C., Guérin C., Martiel J. L., De la Cruz E. M., and Blanchoin L. (2011) Cofilin tunes the nucleotide state of actin filaments and severs at bare and decorated segment boundaries. Curr. Biol. 21, 862–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McCullough B. R., Grintsevich E. E., Chen C. K., Kang H., Hutchison A. L., Henn A., Cao W., Suarez C., Martiel J. L., Blanchoin L., Reisler E., and De La Cruz E. M. (2011) Cofilin-linked changes in actin filament flexibility promote severing. Biophys. J. 101, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pavlov D., Muhlrad A., Cooper J., Wear M., and Reisler E. (2007) Actin filament severing by cofilin. J. Mol. Biol. 365, 1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Andrianantoandro E., and Pollard T. D. (2006) Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24, 13–23 [DOI] [PubMed] [Google Scholar]
- 12. Ono S. (2003) Regulation of actin filament dynamics by actin depolymerizing factor/cofilin and actin-interacting protein 1: new blades for twisted filaments. Biochemistry 42, 13363–13370 [DOI] [PubMed] [Google Scholar]
- 13. Rodal A. A., Tetreault J. W., Lappalainen P., Drubin D. G., and Amberg D. C. (1999) Aip1p interacts with cofilin to disassemble actin filaments. J. Cell Biol. 145, 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Okada K., Obinata T., and Abe H. (1999) XAIP1: a Xenopus homologue of yeast actin interacting protein 1 (AIP1), which induces disassembly of actin filaments cooperatively with ADF/cofilin family proteins. J. Cell Sci. 112, 1553–1565 [DOI] [PubMed] [Google Scholar]
- 15. Mohri K., and Ono S. (2003) Actin filament disassembling activity of Caenorhabditis elegans actin-interacting protein 1 (UNC-78) is dependent on filament binding by a specific ADF/cofilin isoform. J. Cell Sci. 116, 4107–4118 [DOI] [PubMed] [Google Scholar]
- 16. Nadkarni A. V., and Brieher W. M. (2014) Aip1 destabilizes cofilin-saturated actin filaments by severing and accelerating monomer dissociation from ends. Curr. Biol. 24, 2749–2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Okada K., Blanchoin L., Abe H., Chen H., Pollard T. D., and Bamburg J. R. (2002) Xenopus actin-interacting protein 1 (XAip1) enhances cofilin fragmentation of filaments by capping filament ends. J. Biol. Chem. 277, 43011–43016 [DOI] [PubMed] [Google Scholar]
- 18. Michelot A., Grassart A., Okreglak V., Costanzo M., Boone C., and Drubin D. G. (2013) Actin filament elongation in Arp2/3-derived networks is controlled by three distinct mechanisms. Dev. Cell 24, 182–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chen Q., Courtemanche N., and Pollard T. D. (2015) Aip1 promotes actin filament severing by cofilin and regulates constriction of the cytokinetic contractile ring. J. Biol. Chem. 290, 2289–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jansen S., Collins A., Chin S. M., Ydenberg C. A., Gelles J., and Goode B. L. (2015) Single-molecule imaging of a three-component ordered actin disassembly mechanism. Nat. Commun. 6, 7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gressin L., Guillotin A., Guérin C., Blanchoin L., and Michelot A. (2015) Architecture dependence of actin filament network disassembly. Curr. Biol. 25, 1437–1447 [DOI] [PubMed] [Google Scholar]
- 22. Okreglak V., and Drubin D. G. (2010) Loss of Aip1 reveals a role in maintaining the actin monomer pool and an in vivo oligomer assembly pathway. J. Cell Biol. 188, 769–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Van Troys M., Huyck L., Leyman S., Dhaese S., Vandekerkhove J., and Ampe C. (2008) Ins and outs of ADF/cofilin activity and regulation. Eur. J. Cell Biol. 87, 649–667 [DOI] [PubMed] [Google Scholar]
- 24. Ono S., Mohri K., and Ono K. (2004) Microscopic evidence that actin-interacting protein 1 actively disassembles actin-depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 279, 14207–14212 [DOI] [PubMed] [Google Scholar]
- 25. Damaghi M., Wojtkowiak J. W., and Gillies R. J. (2013) pH sensing and regulation in cancer. Front. Physiol. 4, 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schönichen A., Webb B. A., Jacobson M. P., and Barber D. L. (2013) Considering protonation as a posttranslational modification regulating protein structure and function. Annu. Rev. Biophys. 42, 289–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goldmann W. H., Hess D., and Isenberg G. (1999) The effect of intact talin and talin tail fragment on actin filament dynamics and structure depends on pH and ionic strength. Eur. J. Biochem. 260, 439–445 [DOI] [PubMed] [Google Scholar]
- 28. Schmidt J. M., Zhang J., Lee H. S., Stromer M. H., and Robson R. M. (1999) Interaction of talin with actin: sensitive modulation of filament crosslinking activity. Arch. Biochem. Biophys. 366, 139–150 [DOI] [PubMed] [Google Scholar]
- 29. Scheel J., Ziegelbauer K., Kupke T., Humbel B. M., Noegel A. A., Gerisch G., and Schleicher M. (1989) Hisactophilin, a histidine-rich actin-binding protein from Dictyostelium discoideum. J. Biol. Chem. 264, 2832–2839 [PubMed] [Google Scholar]
- 30. Normoyle K. P., and Brieher W. M. (2012) Cyclase-associated protein (CAP) acts directly on F-actin to accelerate cofilin-mediated actin severing across the range of physiological pH. J. Biol. Chem. 287, 35722–35732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lamb J. A., Allen P. G., Tuan B. Y., and Janmey P. A. (1993) Modulation of gelsolin function: activation at low pH overrides Ca2+ requirement. J. Biol. Chem. 268, 8999–9004 [PubMed] [Google Scholar]
- 32. Lueck A., Yin H. L., Kwiatkowski D. J., and Allen P. G. (2000) Calcium regulation of gelsolin and adseverin: a natural test of the helix latch hypothesis. Biochemistry 39, 5274–5279 [DOI] [PubMed] [Google Scholar]
- 33. Yamakita Y., Ono S., Matsumura F., and Yamashiro S. (1996) Phosphorylation of human fascin inhibits its actin binding and bundling activities. J. Biol. Chem. 271, 12632–12638 [DOI] [PubMed] [Google Scholar]
- 34. Ono S., Yamakita Y., Yamashiro S., Matsudaira P. T., Gnarra J. R., Obinata T., and Matsumura F. (1997) Identification of an actin binding region and a protein kinase C phosphorylation site on human fascin. J. Biol. Chem. 272, 2527–2533 [DOI] [PubMed] [Google Scholar]
- 35. Yonezawa N., Nishida E., and Sakai H. (1985) pH control of actin polymerization by cofilin. J. Biol. Chem. 260, 14410–14412 [PubMed] [Google Scholar]
- 36. Hayden S. M., Miller P. S., Brauweiler A., and Bamburg J. R. (1993) Analysis of the interactions of actin depolymerizing factor with G- and F-actin. Biochemistry 32, 9994–10004 [DOI] [PubMed] [Google Scholar]
- 37. Hawkins M., Pope B., Maciver S. K., and Weeds A. G. (1993) Human actin depolymerizing factor mediates a pH-sensitive destruction of actin filaments. Biochemistry 32, 9985–9993 [DOI] [PubMed] [Google Scholar]
- 38. Abe H., Ohshima S., and Obinata T. (1989) A cofilin-like protein is involved in the regulation of actin assembly in developing skeletal muscle. J. Biochem. 106, 696–702 [DOI] [PubMed] [Google Scholar]
- 39. Chen H., Bernstein B. W., Sneider J. M., Boyle J. A., Minamide L. S., and Bamburg J. R. (2004) In vitro activity differences between proteins of the ADF/cofilin family define two distinct subgroups. Biochemistry 43, 7127–7142 [DOI] [PubMed] [Google Scholar]
- 40. Bernstein B. W., Painter W. B., Chen H., Minamide L. S., Abe H., and Bamburg J. R. (2000) Intracellular pH modulation of ADF/cofilin proteins. Cell Motil. Cytoskeleton 47, 319–336 [DOI] [PubMed] [Google Scholar]
- 41. Frantz C., Barreiro G., Dominguez L., Chen X., Eddy R., Condeelis J., Kelly M. J., Jacobson M. P., and Barber D. L. (2008) Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J. Cell Biol. 183, 865–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhao H., Hakala M., and Lappalainen P. (2010) ADF/cofilin binds phosphoinositides in a multivalent manner to act as a PIP2-density sensor. Biophys. J. 98, 2327–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Magalhaes M. A., Larson D. R., Mader C. C., Bravo-Cordero J. J., Gil-Henn H., Oser M., Chen X., Koleske A. J., and Condeelis J. (2011) Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J. Cell Biol. 195, 903–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. McKim K. S., Matheson C., Marra M. A., Wakarchuk M. F., and Baillie D. L. (1994) The Caenorhabditis elegans unc-60 gene encodes proteins homologous to a family of actin-binding proteins. Mol. Gen. Genet. 242, 346–357 [DOI] [PubMed] [Google Scholar]
- 45. Ono S., Baillie D. L., and Benian G. M. (1999) UNC-60B, an ADF/cofilin family protein, is required for proper assembly of actin into myofibrils in Caenorhabditis elegans body wall muscle. J. Cell Biol. 145, 491–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ono K., Parast M., Alberico C., Benian G. M., and Ono S. (2003) Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J. Cell Sci. 116, 2073–2085 [DOI] [PubMed] [Google Scholar]
- 47. Ono K., Yamashiro S., and Ono S. (2008) Essential role of ADF/cofilin for assembly of contractile actin networks in the C. elegans somatic gonad. J. Cell Sci. 121, 2662–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ono S., and Benian G. M. (1998) Two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins, encoded by the unc-60 gene, differentially regulate actin filament dynamics. J. Biol. Chem. 273, 3778–3783 [DOI] [PubMed] [Google Scholar]
- 49. Yamashiro S., Mohri K., and Ono S. (2005) The two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins differently enhance actin filament severing and depolymerization. Biochemistry 44, 14238–14247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ono K., and Ono S. (2014) Two actin-interacting protein 1 isoforms function redundantly in the somatic gonad and are essential for reproduction in Caenorhabditis elegans. Cytoskeleton 71, 36–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ono S. (2001) The Caenorhabditis elegans unc-78 gene encodes a homologue of actin-interacting protein 1 required for organized assembly of muscle actin filaments. J. Cell Biol. 152, 1313–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ono S., Nomura K., Hitosugi S., Tu D. K., Lee J. A., Baillie D. L., and Ono K. (2011) The two actin-interacting protein 1 genes have overlapping and essential function for embryonic development in Caenorhabditis elegans. Mol. Biol. Cell 22, 2258–2269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pardee J. D., and Spudich J. A. (1982) Purification of muscle actin. Methods Enzymol. 85, 164–181 [DOI] [PubMed] [Google Scholar]
- 54. Mohri K., Vorobiev S., Fedorov A. A., Almo S. C., and Ono S. (2004) Identification of functional residues on Caenorhabditis elegans actin-interacting protein 1 (UNC-78) for disassembly of actin depolymerizing factor/cofilin-bound actin filaments. J. Biol. Chem. 279, 31697–31707 [DOI] [PubMed] [Google Scholar]
- 55. Hayakawa K., Sakakibara S., Sokabe M., and Tatsumi H. (2014) Single-molecule imaging and kinetic analysis of cooperative cofilin-actin filament interactions. Proc. Natl. Acad. Sci. U.S.A. 111, 9810–9815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Liu Z., Klaavuniemi T., and Ono S. (2010) Distinct roles of four gelsolin-like domains of Caenorhabditis elegans gelsolin-like protein-1 in actin filament severing, barbed end capping, and phosphoinositide binding. Biochemistry 49, 4349–4360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tsuboi K. K., Markel R. P., and Tomita J. (1965) The pH-dependent nature of actin transformation. Arch. Biochem. Biophys. 112, 82–88 [DOI] [PubMed] [Google Scholar]
- 58. Mohri K., Ono K., Yu R., Yamashiro S., and Ono S. (2006) Enhancement of actin-depolymerizing factor/cofilin-dependent actin disassembly by actin-interacting protein 1 is required for organized actin filament assembly in the Caenorhabditis elegans body wall muscle. Mol. Biol. Cell 17, 2190–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Smith T. F., Gaitatzes C., Saxena K., and Neer E. J. (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24, 181–185 [DOI] [PubMed] [Google Scholar]
- 60. Clark M. G., Teply J., Haarer B. K., Viggiano S. C., Sept D., and Amberg D. C. (2006) A genetic dissection of Aip1p's interactions leads to a model for Aip1p-cofilin cooperative activities. Mol. Biol. Cell 17, 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martin C., Pedersen S. F., Schwab A., and Stock C. (2011) Intracellular pH gradients in migrating cells. Am. J. Physiol. Cell Physiol. 300, C490–495 [DOI] [PubMed] [Google Scholar]
- 62. Thompson J. D., Higgins D. G., and Gibson T. J. (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]