Abstract
Unesterified cholesterol accumulates in late endosomes in cells expressing the misfolded cystic fibrosis transmembrane conductance regulator (CFTR). CFTR misfolding in the endoplasmic reticulum (ER) or general activation of ER stress led to dynein-mediated clustering of cholesterol-loaded late endosomes at the Golgi region, a process regulated by ER-localized VAMP-associated proteins (VAPs). We hypothesized that VAPs serve as intracellular receptors that couple lipid homeostasis through interactions with two phenylalanines in an acidic track (FFAT) binding signals (found in lipid sorting and sensing proteins, LSS) with proteostasis regulation. VAPB inhibited the degradation of ΔF508-CFTR. The activity was mapped to the ligand-binding major sperm protein (MSP) domain, which was sufficient in regulating CFTR biogenesis. We identified mutations in an unstructured loop within the MSP that uncoupled VAPB-regulated CFTR biogenesis from basic interactions with FFAT. Using this information, we defined functional and physical interactions between VAPB and proteostasis regulators (ligands), including the unfolded protein response sensor ATF6 and the ER degradation cluster that included FAF1, VCP, BAP31, and Derlin-1. VAPB inhibited the degradation of ΔF508-CFTR in the ER through interactions with the RMA1-Derlin-BAP31-VCP pathway. Analysis of pseudoligands containing tandem FFAT signals supports a competitive model for VAP interactions that direct CFTR biogenesis. The results suggest a model in which VAP-ligand binding couples proteostasis and lipid homeostasis leading to observed phenotypes of lipid abnormalities in protein folding diseases.
Keywords: amyotrophic lateral sclerosis (ALS) (Lou Gehrig disease), cholesterol regulation, cystic fibrosis transmembrane conductance regulator (CFTR), ER-associated degradation, proteostasis
Introduction
Perturbation of cellular cholesterol homeostasis is seen in cystic fibrosis (CF)2 and other protein folding diseases (1–5). Expression of ΔF508-CFTR, which fails to fold in the ER and is removed by ER-associated degradation (ERAD), leads to the accumulation of unesterified cholesterol in late endosomes (LE) (2) and to defects in the traffic of internalized (labeled) cholesterol from LE (6). The accumulation of unesterified cholesterol in LE is accompanied by a reduction in cellular cholesterol esters and by defects in the traffic of glycosphingolipids (2). These outcomes are reminiscent of cholesterol traffic defects observed in Niemann Pick disease type C (NPC) (7). In NPC, cholesterol traffic from LE to the ER, where it is esterified, is inhibited. Cholesterol accumulation in ΔF508-CFTR-expressing cells or NPC fibroblasts is alleviated by the overexpression of Rab9 (2, 8, 9). Rab9 can either function directly, augmenting the traffic of sterols from LE to the trans-Golgi network, or indirectly, by facilitating trafficking of proteins required for intracellular cholesterol transport. The lack of cholesterol supply to the ER is accompanied by the activation of SREBP (sterol regulatory element-binding protein) and endogenous cholesterol synthesis both in model CF cells (6) and mice that contain an in-frame deletion in the CFTR gene (10). Importantly, these cellular lipid phenotypes are somewhat mirrored in CF patients that present deficiencies in essential fatty acids as well as low plasma LDL and HDL levels (5).
Cholesterol accumulation in LE is not specific to the ΔF508-CFTR mutation, does not require CFTR function, and is observed in cells expressing CFTR-related mutant transporters (2) or unrelated misfolded proteins that are processed in the ER (3, 4). Overall, these observations suggest that protein misfolding in the ER leads to global effects on lipid homeostasis. Indeed aberrant cholesterol distribution is observed during general perturbation of protein folding in the ER; drugs that perturb ER N-glycosylation or oxidative folding similarly lead to cholesterol accumulation in LE (11–13). The mechanisms that enable the coupling of protein biogenesis in the ER with cholesterol flow remain undefined. Changes to transcription programs involved in cholesterol synthesis and alterations in cholesterol uptake and intracellular traffic may both play a role in altering cholesterol levels and distribution. These changes might be utilized to address protein misfolding in the ER. Introduction of free cholesterol or long chain saturated fatty acids in the ER membrane leads to activation of ER stress (14). Therefore, it may be that when ER-associated folding or degradation machineries are perturbed or overloaded, the traffic of free cholesterol to the ER is reduced to minimize the derived biosynthetic and processing load. This reduction would be functionally homologous to the general inhibition of protein synthesis during the development of ER stress. The ER unfolded protein response (UPR) couples the adjustments to protein folding and degradation mechanisms with phospholipid and sterol synthesis (15). However, mechanisms that couple the intracellular cholesterol distribution with protein folding remain largely undefined.
The global nature of lipid effects observed in CF patients and the relation to protein folding in the ER led us to hypothesize that VAMP-associated proteins (VAPs) might be involved in regulated coupling of protein folding with lipid homeostasis. VAPs are evolutionarily conserved ER-localized type II membrane proteins that function in both protein and lipid homeostasis (16–18). The proteins serve as anchors that physically link the ER with other organelles forming membrane contact sites (MCS). VAPs are anchored in the ER membrane via the C terminus to present a coiled-coil dimerization domain and an N-terminal major sperm protein (MSP) domain in the cytosol. The MSP domain binds FFAT signals (two phenylalanine residues in an acidic track) containing ligands, mostly involved in lipid sorting and sensing activities (16). LSS include members of the oxysterol-binding protein-related proteins (ORPs) in mammals or the oxysterol-binding Saccharomyces cerevisiae homologs in yeast (19). LSS function at MCS that connect the ER (through FFAT binding) with Golgi, endosomes, and the plasma membranes. ORPs function directly in sterol and phospholipid transfer, using an oxysterol regulatory domain to bind and mediate bidirectional inter-membrane exchange of monophosphorylated phosphoinositides with sterols. ORPs may also function as sensors that report on lipid environments. ER-anchored ORPs function in a VAP-independent manner to exchange phosphoinositide with phosphatidylserine. In addition, a number of lipid-modifying and transfer enzymes interact with the ER using FFAT-VAP binding and function in ceramide traffic, phosphoinositide, and phosphatidic acid exchange and regulated phosphoinositide phosphorylation and dephosphorylation cycles that provide directionality for cellular lipid flow (20–25). Regulation of MCS assembly also allows for lipid-modifying enzymes to function in trans and regulate organelle positioning within the cell. A role for VAPs in regulating hepatitis C virus replication complexes has been shown, and this role is likely related to regulatory activities that control the lipid composition of these membranous structures (26, 27). Overall, a role for VAPs as central elements in general lipid homeostasis is well established.
VAPs are also implicated in the regulation of proteostasis. Yeast VAP (Scs2, suppressor of choline sensitivity) couples Opi1 regulation with UPR activity to control ER membrane expansion during ER stress (28). Opi1 regulates Ino1 activity, a key element in phospholipid synthesis. Scs2 also interacts with the ER-localized proteostasis regulators Yet1p-Yet3p (29). The relation between VAP, UPR, and proteostasis regulation is conserved in mammals. VAPs interact with and regulate the activity of the mammalian UPR sensors Ire1 and ATF6 (30, 31). ATF6 regulates UPR-mediated membrane expansion, linking the regulation of lipid biosynthesis with protein folding in the ER (32).
The physiological role of VAP in regulating proteostasis is undefined. However, the significance of such regulation is exemplified by the role of VAPB mutants in the development of familial amyotrophic lateral sclerosis (ALS) (31, 33–36). A P56S mutation in the MSP domain of VAPB destabilizes the domain and supports the aggregation of VAPB with VAPA leading to a general loss of function. Deletion of Drosophila dVAP is manifested as a proteostasis disease, leading to the accumulation of protein aggregates and the build up of ER quality control compartments characteristic of inhibited protein degradation (37–39). The cytosolic MSP domain of dVAP is cleaved and secreted (using an unconventional secretion pathway) serving as an activating ligand for Eph receptors (40, 41). The P56S mutation inhibits cleavage and signaling activities. Therefore, the loss of VAP signaling may lead to the development of ALS. However, the loss of dVAP can be partially compensated by the expression of ORP8, an FFAT-independent ER-anchored ORP (36). Thus VAP-FFAT binding and lipid transfer activities are likely involved in regulating cellular proteostasis and might contribute to the development of ALS (38). We therefore hypothesize that VAPs function as ER-localized receptors, which couple proteostasis with lipid homeostasis.
We now demonstrate that VAPs (or isolated MSP domains) inhibit the degradation of misfolded ΔF508-CFTR in the ER. We defined mutations in the MSP domain that uncouple this proteostasis activity from VAP expression and defined selective interactions of VAP-MSP with proteostasis ligands regulating UPR and ER-associated degradation of CFTR. We provide evidence to suggest a model in which competitive ligand binding by VAP-MSP regulates proteostasis and lipid homeostasis coupling.
Experimental Procedures
Cells, Chemicals, and Antibodies
HeLa, HEK293, and ΔF508-CFTR stably expressing HEK293 cells and the RNF5/RMA1 knockdown HeLa cell lines (provided by Dr. Z. Ronai, Burnham Institute, La Jolla, CA) were maintained in Dulbecco's modified Eagle's medium (Thermo Scientific Hyclone) supplemented with 10% fetal bovine serum (FBS, Serum Source International and Thermo Scientific Hyclone) and 1% penicillin/streptomycin solution (Mediatech) at 37 °C with 5% CO2. In some experiments, cells were grown in 10% delipidated FBS (Cocalico Biologicals). U18666A was purchased from BIOMOL Research Labs. Cycloheximide and tunicamycin were purchased from Sigma. Antibodies used in this study include anti-calnexin (StressGen), anti-Lamp2 (gift from Dr. G. Apodaca and the Developmental Studies Hybridoma Bank), anti-HA (Invitrogen and Covance), anti-GFP (Invitrogen and Polysciences, Inc.), anti-FLAG M2 (Sigma), anti-p97/VCP (Research Diagnostics, Inc.), anti-RMA1 (Abgent), anti-β-actin (Abcam), and anti-BAP31 (gift from Dr. G. Shore, McGill University, Montreal Canada).
Plasmids
Mammalian expression vectors encoding HA-tagged human VAPA and VAPB were kindly provided by Dr. C. Hoogenraad (Utrecht University, Utrecht, The Netherlands). The VAPA construct was a variant that harbors a substitution of a conserved Glu to Gly at position 185. The substitution resides outside of the proteostasis-active MSP domain of VAPA and in agreement did not affect VAPA activity in our assays. Importantly, site-directed mutagenesis generated several variants used in the structure-function analysis of VAPs. The N-terminal MSP domain was expressed by mutagenizing amino acids 132 and 125 in VAPA and VAPB, respectively, to stop codons. The HA-VAPA-MSP construct was generated using the forward primer 5′-CCAAATTGAGATGCGTATTTGAATAGCCCAATGAAAATGATAAATTGAATGATATGG-3′ and reverse primer 5′-CCATATCATTCAATTTATCATTTTCATTGGGCTATTCAAATACGCATCTCAATTTGG-3′, and the HA-VAPB-MSP construct was generated using the forward primer 5′-GGATTCAAAACTTAGATGTGTGTTTGAATAGCCAGCAGAGAATGATAAACC-3′ and reverse primer 5′-GGTTTATCATTCTCTGCTGGCTATTCAAACACACATCTAAGTTTTGAATCC-3′.
Conversion of amino acids 201 and 196 in VAPA and VAPB, respectively, to stop codons produced truncations after the VAP coiled-coil domain (HA-VAPA-MSP+CC andHA-VAPB-MSP+CC). The VAPA-specific primers used were 5′-CGGCACCTGAGAGATGAAGGTTTATAACTCAGAAAGGTAGCACATTCGG-3′ and 5′-CCGAATGTGCTACCTTTCTGAGTTATAAACCTTCATCTCTCAGGTGCCG-3′, and the VAPB-specific primers used were 5′-GCAGTTCAAGGAAGAAGATGGACTGTAAATGAGGAAGACAGTGCAGAGCAACAGC-3′ and 5′-GCTGTTGCTCTGCACTGTCTTCCTCATTTACAGTCCATCTTCTTCCTTGAACTGC-3′.The ALS-causing mutation in VAPB, P56S, was generated using the 5′-CGTAGGTACTGTGTGAGGTCCAACAGCGGAATCATCGATGC-3′ and 5′-GCATCGATGATTCCGCTGTTGGACCTCACACAGTACCTACG-3′ primers. Sequence alignment of VAPA and VAPB identified proline 63 as the ALS-related residue in VAPA, which was mutated to serine 63 using the 5′-GCCGGTACTGTGTGAGGTCCAACAGTGGAATTATTGACC-3′ and 5′-GGTCAATAATTCCACTGTTGGACCTCACACAGTACCGGC-3′ primers.
VAP double point mutations within the MSP domain, as identified previously (42), prevent FFAT binding, and these constructs were generated in VAPA using the primers 5′-CCGAATGAAAAGAGTAAACACGACTTTGACGTACAGACAATTTTTGCTCCACC-3′ and 5′-GGTGGAGCAAAAATTGTCTGTACGTCAAAGTCGTGTTTACTCTTTTCATTCGG-3′ (HA-VAPA-K95D/M97D).
In VAPB, the double point mutations within the MSP domain were generated using the 5′-CCCAATGAGAAAAGTAAACACGACTTTGACGTTCAGTCTATGTTTGCTCC-3′ and 5′-GGAGCAAACATAGACTGAACGTCAAAGTCGTGTTTACTTTTCTCATTGGG-3′ primers (HA-VAPB-K87D/M89D).
Analysis of the VAP-MSP domain identified exposed acidic residues within an unstructured loop, which were mutagenized to generate the 2DK mutants. Two reverse-charge point mutations in VAPA (D84K/D86K) and VAPB (D77K/D79K), which were termed the “2DK” mutants, were generated in VAPA using the primers 5′-GCTACAGCCCTTTAAGTATAAGCCGAATGAAAAGAGTAAACACAAGTTTATGG-3′ and 5′-CCATAAACTTGTGTTTACTCTTTTCATTCGGCTTATACTTAAAGGGCTGTAGC-3′ and in VAPB using the primers5′-GTGATGTTACAGCCTTTCAAGTATAAGCCCAATGAGAAAAGTAAACACAAGTTTATGG-3′ and 5′-CCATAAACTTGTGTTTACTTTTCTCATTGGGCTTATACTTGAAAGGCTGTAACATCAC-3′.
The GFP-3×FFAT ligand was generated by subcloning two Myc-tagged FFAT fragments (rabbit OSBP residues 347–372) into a plasmid containing a GFP-tagged FFAT fragment (human Nir2 residues 344–360). The Myc-FFAT (43) and GFP-FFAT (44) constructs were kindly provided by Dr. J. Ngsee (Ottawa Health Research Institute, University of Ottawa, Canada) and Dr. C. Hoogenraad (Utrecht University, Utrecht, The Netherlands), respectively. To generate the GFP-3×FFAT plasmid, an internal BamHI restriction enzyme site within the Myc-FFAT construct was first removed by site-directed mutagenesis using the forward primer 5′-GGTACCGAGCTCGGTTCCTCTGGCAAAGG-3′ and reverse primer 5′-CCTTTGCCAGAGGAACCGAGCTCGGTACC-3′. PCR amplificationof the Myc-FFAT construct using the forward primer 5′-AGTCACGGATCCGAGCAAAAGCTCATTTCTGAGGAAGATCTCC-3′ and reverse primer 5′-CACTGGTCTAGATTCAGGCATGGTGATGATCTCAGGGGC-3′ generated a fragment containing the Myc-tagged FFAT region with 5′-BamHI and 3′-XbaI sites that were used to insert a second FFAT fragment into the GFP-FFAT plasmid (GFP-2×FFAT). An additional Myc-FFAT fragment was cloned into the GFP-2×FFAT plasmid using 5′-XhoI and 3′-EcoRI sites added by PCR amplification with the forward primer 5′-AGTCAGCTCGAGCTGAGCAAAAGCTCATTTCTGAGGAAGATCTCC-3′ and reverse primer 5′-CACTGGGAATTCGATTCAGGCATGGTGATGATCTCAGGGGC-3′ (GFP-3×FFAT). When indicated, the FFAT fragment originally found within the GFP-FFAT plasmid was replaced in the GFP-3×FFAT plasmid with a Myc-FFAT fragment using 5′-EcoRI and 3′-BamHI sites added by PCR amplification with the forward primer 5′-CGTCAGGAATTCAGAGCAAAAGCTCATTTCTGAGGAAGATCTCC-3′ and reverse primer 5′-CATTGAGGATCCTTCAGGCATGGTGATGATCTCAGGGGC-3′ (GFP-3×FFAT-pure).
The p3×FLAG-ATF6 expression vector was kindly provided Dr. R. Prywes (Addgene plasmid 11975) and encodes a 3×FLAG-tagged human ATF6 protein. This plasmid was altered by site-directed mutagenesis using the forward primer 5′-CCAGAGGCTTAAAGTCCCTAGTTGAAAGCGAAGAGTTGTCTGTGTGATGATAGTATTGGC-3′ and reverse primer 5′-GCCAATACTATCATCACACAGACAACTCTTCGCTTTCAACTAGGGACTTTAAGCCTCTGG-3′ to generate a stop codon at amino acid 374 (P374X) and produce the N-terminal nuclear form of ATF6 (3×FLAG-nATF6).
The FLAG-tagged human FAF1 expression vector was obtained from Dr. K.-J. Lee (Ewha Womans University, Seoul, South Korea). The GFP-tagged human ORP1L expression construct was donated by Dr. V. Olkkonen (Minerva Foundation Institute for Medical Research, Helsinki, Finland). The GFP-tagged dynamitin expression vector was obtained from Dr. T. Schroer (The Johns Hopkins University, Baltimore, MD). The FLAG-tagged BAP31 expression vector was obtained from Dr. G. C. Shore (McGill University, Montreal, Canada).
ATF6 Luciferase Assay
HEK293 cells seeded in 24-well dishes were transfected with the p5×ATF6-GL3 luciferase reporter (300 ng, Addgene plasmid 11976), the pGL4.75 Renilla control vector (15 ng), and either pEGFP-C1 or HA-VAPB constructs (100 ng). 24 hours post-transfection, cells were washed with PBS and incubated with media with 2 μg/ml tunicamycin for 6 or 12 h. Cell lysates were collected in 100 μl of lysis buffer, and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega) per the manufacturer's protocol and detected by the Synergy H1 Hybrid Microplate Reader (BioTek). ATF6-derived luciferase luminescence was normalized to the Renilla control luminescence, and responses to tunicamycin treatment of VAPB-transfected cells were compared with GFP-transfected cells. Data were analyzed using Student's t tests.
Co-immunoprecipitations
HeLa cells were co-transfected with HA-VAPB (full-length or MSP) and various GFP-FFAT DNA constructs using Lipofectamine 2000 (Invitrogen), according to the manufacturer's protocol. 24 h after transfections, cells were washed with PBS and lysed in 50 mm Tris-HCl, pH 7.5, 150 NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100 (EM Sciences), 1% sodium deoxycholate (MP Biomedicals), 1× Complete protease inhibitor mixture (Roche Applied Science), and 1 mm PMSF. Lysates were incubated with anti-HA antibodies overnight with rotation at 4 °C and collected on protein G-agarose (Pierce). After extensive washing, protein complexes were analyzed by Western blotting using HA and GFP antibodies.
GST Pulldowns
Both the wild type and 2DK-MSP domains of VAPB were PCR-amplified with the forward primer 5′-CATAGGATCCATGGCGAAGGTGGAACAGG-3′ and the reverse primer 5′-GCGCGAATTCCTATTCAAACACACATCTAAGTTTTGAATCC-3′ to add 5′-BamHI and 3′-EcoRI sites that were cloned into the pGEX-4T-1 expression vector. The constructs were transformed into BL21 (DE3)-competent Escherichia coli cells (Invitrogen) and induced with isopropyl 1-thio-β-d-galactopyranoside to express GST-tagged VAPB MSP domains (GST-VAPB-WT-MSP and GST-VAPB-2DK-MSP), and proteins were purified on glutathione-Sepharose 4B beads using standard protocol (GE Healthcare). For pulldown experiments, GST-VAPB-MSP proteins (250 μg for interaction analysis and 50 μg for binding competition analysis) were pre-bound to beads, incubated with HeLa cell lysates (prepared in 25 mm Hepes, pH 7.2, 125 mm KOAc, 5 mm MgOAc, 2 mm EDTA, 2 mm EGTA, 1 mm DTT, 1% Triton X-100, 1 mm PMSF, 1 mm orthovanadate), collected by centrifugation, washed, and analyzed by Western blots with selective antibodies or mass spectrometry.
Immunostaining
Cells were seeded on cover glass (Fisher) in 24-well dishes and fixed in 4% formaldehyde in PBS. Fixed samples were blocked with 5% goat serum (Sigma) in PBS and 0.05% saponin (Sigma), incubated with selective primary antibodies, stained with Alexa Fluor 488 or 594 dye conjugates secondary antibodies (Invitrogen) diluted in PBS-saponin, washed, and mounted on slides (Fisher) with Fluoromount G (eBioscience) containing DAPI (5 μg/ml). Filipin complex (Sigma) in PBS was incubated with cells after immunostaining to detect free cholesterol in cells and mounted without DAPI. Images were acquired on an Olympus Fluoview 1000 confocal system using an inverted microscope (IX-81 Olympus) and ×60 NA 1.42 PLAPON objective. Images were processed using FV10-ASW Version 02.00.03.10 (Olympus Corp.) and Adobe Photoshop CS3 (Adobe Photoshop Version 10.0.1 (Adobe)).
CFTR Expression Analysis
Human wild type and ΔF508-CFTR expression constructs were co-transfected with various HA-VAPs and GFP-FFAT DNA constructs using the Effectene reagent (Qiagen) following the manufacturer's protocol. After transfection, cells were washed with PBS, lysed in PBS containing 1% Triton X-100 (EM Sciences), 0.1% SDS (Bio-Rad), and 0.5% sodium deoxycholate (MP Biomedicals). Lysates were mixed with SDS sample buffer and denatured by incubating and shaking at 37 °C for 30 min. Samples were separated on 5 or 8% SDS-polyacrylamide gels and transferred overnight onto Protran BA83 nitrocellulose membranes (GE Healthcare). CFTR expression was detected using CFTR-specific antibodies (217 and 596) purchased from Cystic Fibrosis Foundation Therapeutics (University of North Carolina at Chapel Hill) and goat anti-mouse HRP-conjugated secondary antibodies (Pierce). To assess protein loading, samples were assayed for β-actin expression levels (Abcam). Quantitative densitometry was conducted using the Quantity One software (Bio-Rad).
Results
Misfolding of CFTR Leads to Cholesterol Accumulation and LE Clustering
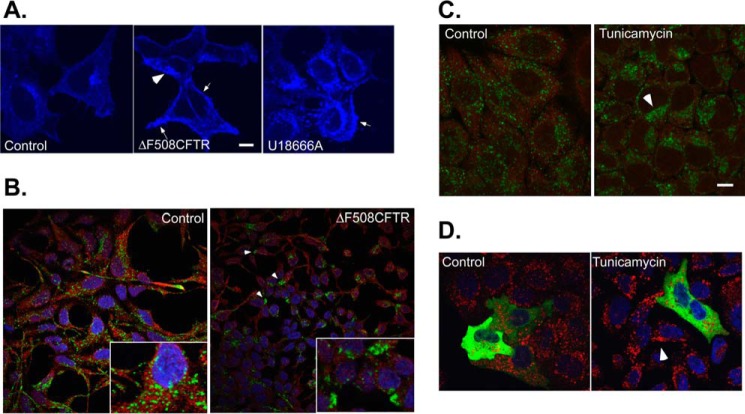
Dietary cholesterol taken by cells is released from lipoprotein particles in LE and lysosome compartments generating free cholesterol that can be transported back to the ER where it is further processed. Cells derived from CF patients or ones that express exogenous ΔF508-CFTR were previously shown to accumulate abnormal levels of free endosomal cholesterol (2, 6), and we verified these observations in HEK293 cells. HEK293 stably expressing ΔF508-CFTR stained with the cholesterol dye filipin revealed high levels of free cholesterol as observed in cells treated with U18666A, a cationic drug that inhibits the flow of cholesterol from LE to the ER (Fig. 1A). ER-LE MCS are regulated by cholesterol levels in LE and are abrogated in high cholesterol conditions leading to dynein-mediated LE clustering at the microtubule-organizing center (MToC) (45). We examined cells stably expressing ΔF508-CFTR proteins for LE localization. Although Golgi (gpp130) and ER (calnexin) and early endosome (Rab5) morphologies were not affected by the expression of ΔF508-CFTR (data not shown), LE (marked by Lamp2) exhibited a shift from scattered morphology to robust clustering at the juxtanuclear area near the MToC in ΔF508-CFTR-expressing cells (Fig. 1B). Previous studies have shown that induction of general protein misfolding in the ER and activation of the UPR lead to accumulation of free cholesterol in LE (11). We therefore analyzed whether activation of UPR also leads to LE clustering as observed in ΔF508-CFTR-expressing cells. We inhibited general ER folding in cells using tunicamycin, a drug that inhibits N-glycosylation in the ER leading to the activation of UPR. Tunicamycin treatment led to clustering of LE at the MToC (Fig. 1C).
FIGURE 1.
Cellular distribution of late endosomes is regulated by protein misfolding in the ER. A, HEK293 cells (control) treated or untreated with U18666A (3 μg/ml for 6 h) as indicated or HEK293 cell stably expressing ΔF508-CFTR were fixed and stained with filipin to detected cholesterol distribution. B, HEK293 cells (control) or HEK293 cell stably-expressing ΔF508-CFTR were stained for ER (red, calnexin) and late endosomes (LE, green, Lamp2). Insets are a magnification of cells from the above fields. C, control or tunicamycin-treated HeLa cells (2 μg/ml for 24 h as indicated) were fixed and stained for ER (calnexin, red) and LE (Lamp2, green). D, as in C. HeLa cells were transfected with GFP-p50 dynamitin and treated with control or tunicamycin (4 μg/ml for 6 h, as indicated). The localization of LE (Lamp2, red) was determined by IF. Bars, 5 μm.
Increased cholesterol levels in LE can be sensed by ORP1L (45–48). In low cholesterol conditions, ORP1L establishes MCS between LE and ER membranes by using its FFAT signal to bind VAPs on the ER and ankyrin and pleckstrin homology domains to bind endosomal Rab7-RILP (16, 45, 49). Increased cholesterol concentrations in LE leads to conformational change that occludes the FFAT motif. The resulting release of ORP1L from VAP-ER binding promotes the binding of the dynactin-dynein motor complex on LE, leading LE transport to the MToC. In agreement, clustering of LE at the MToC in cells treated with tunicamycin was inhibited by the expression of dynamitin p50, which uncouples the dynein motor from cargo organelles (Fig. 1D). Because cholesterol sensing and LE position in the cell are regulated by the ER-localized VAP receptors, we hypothesize that VAP receptors link MCS assembly and cholesterol transfer with protein biogenesis in the ER.
VAP Proteins Regulate the Stability of CFTR
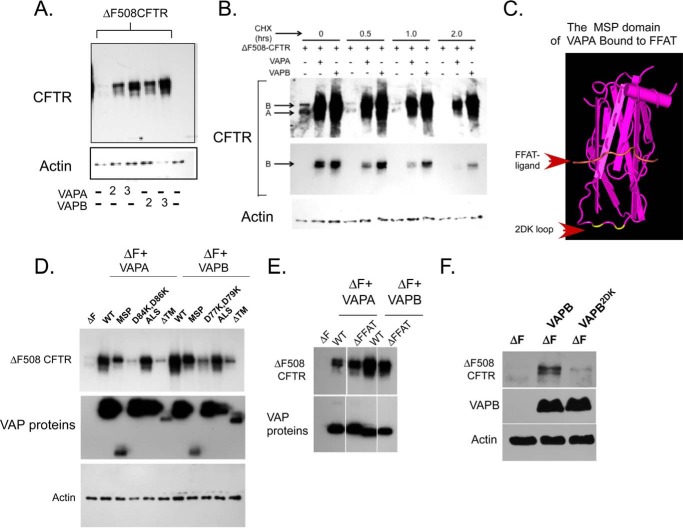
As a first test to examine whether VAPs, which are involved in LE-ER interactions, participate in coupling sterol transfer to proteostasis, we examined whether VAPs affect CFTR biogenesis. Overexpression of VAPB (or VAPA-E185G, see under “Experimental Procedures”) led to a robust increase in ΔF508-CFTR expression (Fig. 2A). The F508 phenylalanine deletion in CFTR is causative in 90% of CF patients. The mutation destabilizes CFTR, which is re-routed for rapid ER-associated degradation (50). The observed stabilization of ΔF508-CFTR upon co-expression with VAPs might be the result of increased synthesis and thus physiologically insignificant. Alternatively, VAPs may lead to reduced degradation rates of ΔF508-CFTR. To distinguish between these possibilities, we analyzed the stability of CFTR during a cycloheximide chase. As can be seen in Fig. 2B, ΔF508-CFTR stability was enhanced by the expression of VAPs. The results suggest that VAPs regulate CFTR biogenesis in the ER.
FIGURE 2.
VAPs regulate ΔF508-CFTR biogenesis. A, HEK293 cells were transfected with plasmids expressing ΔF508-CFTR and increasing amounts of HA-tagged VAPA or VAPB DNA (2× or 3× over CFTR DNA, as indicated). Cell lysates were collected and analyzed for CFTR and actin expression as indicated. Actin provides a loading control. B, HEK293 cells were transfected with ΔF508-CFTR together with VAPA or VAPB (in a DNA ratio of 1:3, respectively) as indicated. Cycloheximide (CHX) (25 μg/ml) was added for the indicated time, and the expression of CFTR was defined by Western blot. Arrows point to ER-localized immature bands A and B of CFTR as indicated. Middle panel is a lower exposure of the blot shown on the top to emphasize the difference in CFTR expression between VAP-expressing and control cells. Actin provides a loading control. Similar results were recorded in three independent experiments. C, structure of an FFAT-bound MSP domain of rat VapA (42). Arrows indicate the positions of the 2DK loop and the FFAT peptide as indicated. The positions of the D84K and D86K mutations in the 2DK loop are highlighted in yellow. D, structure-function analysis of VAPA and VAPB in CFTR stabilization. ΔF508-CFTR was expressed either with control (GFP) or together with VAPA and VAPB constructs, full-length (WT), isolated MSP fragments (VAPA(1–132); VAPB(1–124)), full-length VAPA-D84K, D86K, and VAPB-D77K, D79K (2DK mutants), ALS VAP mutants (VAPA-P63S and VAPB-P56S), and trans-membrane domain deleted mutants (ΔTM) (all at a 1 (ΔF508-CFTR) to 3 (tested proteins) DNA ratio) as indicated. Cell lysates were analyzed for VAP expression (using HA antibodies), CFTR, and actin (as a loading control). E, ΔF508-CFTR was expressed without or with VAPA-K95D/M97D or VAPB-K87D/M89D (both deficient in FFAT binding, ΔFFAT) and analyzed as in D. F, bronchial epithelial cells (cystic fibrosis bronchial epithelial cells) were transfected with VAPB or VAPB-2DK mutant as in D, and the expression of CFTR, HA-VAP, and actin was determined using Western blots.
Mutations in VAP-MSP Functionally Uncouple VAP Proteins from CFTR Stabilization
To explore the mechanisms by which VAPs might regulate CFTR biogenesis, we analyzed the molecular basis for the observed activity. VAPB is a type II membrane protein, anchored in the ER membrane through a C-terminal transmembrane domain (18). The protein contains a central coiled-coil domain that supports VAP oligomerization (51) and an N-terminal MSP domain, which functions in FFAT binding (42). The structure of the MSP domain of VAPA in complex with FFAT was previously defined (Fig. 2C). We prepared structure-guided mutants of VAPB and truncated fragments and analyzed their effect on CFTR biogenesis (Fig. 2, D and E). Expression of the MSP domain in isolation as a cytosolic soluble protein was sufficient in stabilizing CFTR. Therefore, membrane anchor or the coiled-coil domains of VAPB are not required for the observed stabilization. A mutation within the MSP domain (P56S in VAPB) destabilizes the protein and leads to the development of ALS (31). The mutant VAP retains activity in FFAT binding or UPR interactions, yet it exhibits higher propensity to oligomerize, an activity dependent on the coiled-coil domain and accelerated by the MSP mutation (51). In agreement with these results, VAPs harboring the P56S (or equivalent in VAPA-P63S) mutation had no effect on VAP-induced ΔF508-CFTR stabilization (Fig. 2D). It could be that interactions with FFAT containing LSS proteins regulate CFTR biogenesis. However, mutations that inhibit FFAT binding (K87D, M89D, and VAPA-K95D, M97D) for the most part had a marginal effect on the stabilization activity of VAPs (Fig. 2E). Although the MSP domain in isolation was sufficient in promoting ΔF508-CFTR stability, extending the fragment to include the CC domain inhibited this activity (Fig. 2D). It may be that uncontrolled assembly of the MSP domain driven by CC interactions may inhibit the stabilization activity.
Inspection of the MSP structure revealed an unstructured loop that extends from the β-sheet core of the FFAT binding domain. The structural information suggests that the loop may not affect direct FFAT peptide binding, although regulatory activities cannot be excluded. Within this loop, we identified a pair of acidic residues (Asp-84 and Asp-86 in VAPA and Asp-77 and Asp-79 in VAPB, see Fig. 2C, residue positions highlighted in yellow) and reversed their charge (replacement with lysine, resulting mutant termed 2DK, Fig. 2C). As anticipated from the VAPA MSP structure (42), the mutation did not affect expression or localization of the resulting mutants and did not affect protein stability (Fig. 2, D and F, and data not shown). However, and surprisingly, the mutations inhibited the ΔF508-CFTR stabilization activities of VAPA or VAPB. Moreover, the mutations further inhibited the stabilization activity of the isolated MSP domains (data not shown). Thus, an additional interaction interface on VAPs is required for the regulation of ΔF508-CFTR stability. Having established a structural determinant that uncoupled VAP expression from its effect on CFTR biogenesis effects, we analyzed whether VAPs effects on ΔF508-CFTR stability are also exerted in a more physiological setting for CFTR expression by using cystic fibrosis bronchial epithelial cells. As observed in HEK cells, VAPB stabilized ΔF508-CFTR, transiently expressed in cystic fibrosis bronchial epithelial cells, whereas the 2DK mutant, expressed at similar levels, failed to affect CFTR stability (Fig. 2F). Overall, the results established a requirement for VAP activity in CFTR stabilization that resides in the soluble MSP domain and requires acidic charges on an unstructured loop within the domain. The analysis further established the physiological relevance in bronchial epithelia.
ATF6 Regulation Does Not Contribute to CFTR Stabilization by VAPB
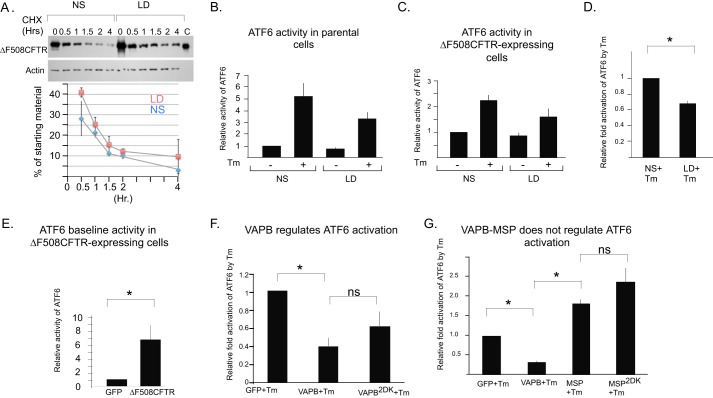
VAPB has been previously implicated in the regulation of UPR by interactions with ATF6 (30). Yeast two-hybrid analysis demonstrated that VAPB interacts with ATF6, and a similar interaction was detected in cells using a fluorescence complementation assay. The interaction inhibits ATF6 during UPR activation. Depletion of ATF6α was shown to enhance ΔF508-CFTR stability and to regulate CFTR expression (52, 53). ATF6 selectively controls the expression of a defined set of genes involved in ERAD providing a plausible mechanism for the stabilization of ΔF508-CFTR by VAPB (54). We tested whether ATF6 activities are regulated by sterol homeostasis and whether VAPB-ATF6 interactions are sensitive to the 2DK mutations that inhibit ΔF508-CFTR stabilization. The modulation of cholesterol homeostasis through the incubation of cells in lipoprotein-deficient (LD) media and/or treated with β-methyl cyclodextrin can lead to partial correction of folding and trafficking of misfolded proteins from the ER (4, 55). In ΔF508-CFTR-expressing cells, a reduction in the NPC phenotype of cholesterol accumulation was observed following an acute treatment with β-methyl cyclodextrin. The observed reduction in unesterified cholesterol is accompanied by functional rescue of ΔF508-CFTR, derived in part by the arrival and increased retention of ΔF508-CFTR at the plasma membrane (56). Misfolded glucocerebrosidase mutants that are arrested in the ER also lead to the accumulation of unesterified cholesterol in LE in fibroblasts derived from Gaucher disease patients. The incubation of such fibroblasts in LD reduces the accumulation of unesterified cholesterol, inhibits ER-associated degradation, and induces the traffic and functional rescue of ER-retained glucocerebrosidase mutants (4). We did observe a ΔF508-CFTR rescue phenotype by incubating ΔF508-CFTR-expressing cells in LD alone or LD supplemented with β-methyl cyclodextrin, although such a phenotype was infrequent (data not shown). The reasons for this variability is unknown and yet likely reflects variable ΔF508-CFTR expression levels that can lead to leakage of stabilized ΔF508-CFTR from the ER in LD-treated cells. However, we did observe a consistent delay in the ΔF508-CFTR degradation rate in LD-treated cells when analyzed using a cycloheximide chase (Fig. 3A). Importantly, sterol homeostasis regulates VAP-controlled MCS, so we analyzed whether ATF6 activities are modulated under these conditions. We analyzed the activation of endogenous ATF6 in parental and ΔF508-CFTR-expressing cells using a luciferase-based transcription reporter as reported previously (57). Treatment of cells with tunicamycin for 6 h led to ∼6-fold activation of ATF6-driven transcription in parental HEK293 cells (Fig. 3B). However, activation was markedly reduced in ΔF508-CFTR-expressing cells (2-fold, Fig. 3C). This result is likely derived from desensitization of ATF6 activation in ΔF508-CFTR-expressing cells. Indeed, transient expression of ΔF508-CFTR in parental HEK293 cells led by itself to 6-fold elevation of basal ATF6 activity when compared with cells expressing GFP (Fig. 3E). Thus, ΔF508-CFTR expression might have led to chronic activation and desensitization of UPR activities. Importantly, incubation of cells for 24 h in LD led to overall inhibition of Tm-induced ATF6 activation in both parental and ΔF508-CFTR-expressing cells (Fig. 3, B–D). Short incubations in LD (4 h) did not affect ATF6 activity suggesting that an adaptation period is required for a new homeostasis to develop (data not shown). The results suggest that ΔF508-CFTR stabilization may correlate with ATF6 activity. We thus first examined the physical interactions of ATF6 with VAPB. Because the CFTR stabilization activity resided in the MSP domain, we produced the MSP domain of VAPB as a GST-tagged protein for the analysis of protein-protein interactions using glutathione-Sepharose beads in pulldown assays with cell lysates (Fig. 4A). As control, we analyzed the interactions of the domains with the FFAT-containing ligand EGFP-ORP1L, transiently expressed in HeLa cells. Both VAPB-MSP and VAPB-MSP-2DK interacted with EGFP-ORP1L in HeLa cell lysates, confirming that both WT and mutated domains are folded and the FFAT-binding site is largely intact (Fig. 4B). The results are in agreement with the structurally defined minimal FFAT-binding site, which resides away from the 2DK-containing loop (42). We prepared lysates from cells expressing FLAG-tagged ATF6 for analysis. GST-MSP interacted with FLAG-ATF6 (Fig. 4B). Activation of ATF6 involves a traffic step from the ER to the Golgi where two Golgi resident proteases, site 1 protease and site 2 protease, release the N terminus of ATF6 from membranes for transcription regulation (58). GST-MSP interacted with the active FLAG-nATF6 fragment (Fig. 4B). Importantly, the 2DK mutation partially inhibited yet did not abolish these interactions. The results are in agreement with previous analysis of VAPB-ATF6 interactions using yeast two-hybrid and fluorescent complementation assays (30). The inhibition of VAPB-MSP interactions by the 2DK mutation supports a role of ATF6 in ΔF508-CFTR stabilization. We therefore analyzed how VAPB regulates ATF6 activation. Expression of VAPB led to elevated basal activity of ATF6 (data not shown), which may reflect an ER overload response. When UPR was induced by tunicamycin (Tm) and the magnitude of ATF6 activation above basal activity was analyzed, VAPB expression led to a 50% inhibition of Tm fold activation as reported previously (30). The 2DK mutant that inhibits VAPB interactions with nATF6 was less effective in inhibiting Tm-induced ATF6 activation, although the difference was not statistically significant (Fig. 3F). The results may still suggest that the ATF6 arm of the UPR is a target, which can regulate CFTR stability. Because expression of VAPB, an ER membrane protein by itself, led to activation of ATF6, we further tested whether the soluble, isolated MSP domain, which is functional in CFTR stabilization, is similarly active in ATF6 regulation. Expression of either the MSP or the MSP-2DK domain failed to elevate basal ATF6 activity. Importantly, Tm-induced activation of ATF6 was not inhibited by the expression of both MSP domains (Fig. 3G). The results collectively suggest that VAPB expression and sterol homeostasis can regulate ATF6 activity. However, given the lack of regulation by the soluble MSP domain, which is functional in CFTR stabilization, ATF6 regulation is not a primary target by which VAPB affects ΔF508-CFTR degradation. Indeed, the inhibition of ΔF508-CFTR degradation is an acute effect unlikely derived from UPR-controlled proteostasis readjustments.
FIGURE 3.
ATF6 activities in VAPB-mediated ΔF508-CFTR stabilization. A, HEK293 cells stably expressing ΔF508-CFTR were incubated in the presence of normal (NS) or lipoprotein-deficient (LD) serum for 24 h, and the stability of ΔF508-CFTR during cycloheximide (CHX) chase (time points as indicated) was determined by Western blots with actin as a loading control. Lane C contains ΔF508-CFTR-expressing ER microsomes. A representative experiment is shown, and the average of two experiments normalized to actin loading is presented in the bottom panel with bars representing S.E. The trend of slower degradation in LD incubations was maintained in all experiments, yet variability was observed in overall ΔF508-CFTR degradation rates. B, control or tunicamycin (Tm, 2 μg/ml for 6 h)-induced ATF6 activation (normalized to non induced control condition) was measured in HEK293 cells and incubated in the presence of normal or LD serum for 24 h as indicated. ATF6 activity was determined as described under “Experimental Procedures.” C, Tm-induced ATF6 activation was measured in HEK293 cells stably-expressing ΔF508-CFTR as in B, in the presence of normal or LD serum as indicated. D, compilation of the fold stimulation of ATF6 activity by Tm in cells incubated in normal (defined as 1) or LD media. E, basal ATF6 activity was measured in HEK293 cells transfected with GFP or ΔF508-CFTR as indicated. F, Tm-induced ATF6 activation was measured as in B, in cells transiently expressing GFP, VAPB, or VAPB-2DK as indicated. Fold activation normalized to the Tm-induced activation recorded in cells expressing GFP (where activation is arbitrary set as 1) is shown. G, Tm-induced (2 μg/ml, 12 h) ATF6 activity was measured as in F, in cells transiently expressing GFP, VAPB, VAPB-MSP or VAPB-MSP-2DK mutant. Note that the MSP domain, which stabilizes ΔF508-CFTR, did not inhibit ATF6 activation. Bars in B–G are S.E.-derived from at least three independent experiments; * indicates statistically significant differences between groups (p < 0.05), as determined by using unpaired two-tailed Student's t test.
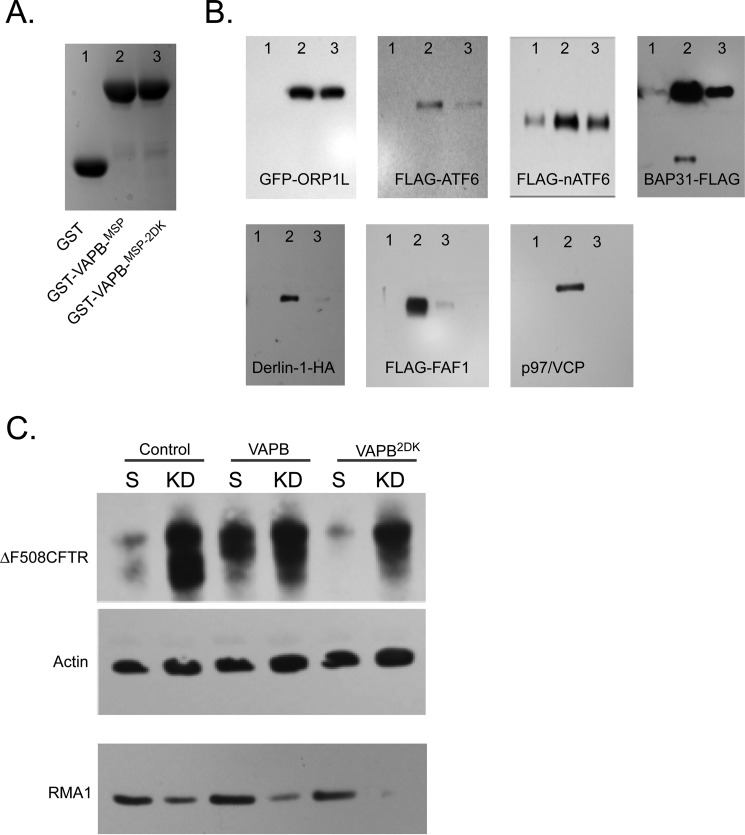
FIGURE 4.
Interactions between VAPB-MSP and proteostasis regulators can control ΔF508-CFTR biogenesis. A, Coomassie Blue-stained gel showing GST (lane 1), GST-VAPB-MSP (lane 2), or GST-VAPB-MSP-2DK (lane 3) proteins that were isolated in pulldown assays as utilized for the following analysis. B, analysis of interactions of GST (lane 1) and GST-VapB-MSP (lane 2) or GST-VAPB-MSP-2DK (lane 3) domains shown in A using HeLa cell lysates transiently expressing GFP-ORP1L, FLAG-ATF6, FLAG-nATF6, Derlin-1-HA, FLAG-FAF1, and BAP31-FLAG (detected with anti-tag antibodies using Western blots) as indicated. Endogenous p97/VCP was detected with selective antibodies. C, HeLa cells stably expressing scrambled (S) or RMA1-directed shRNA (KD) were transfected with ΔF508-CFTR together with GFP (control), HA-VAPB, or HA-VAPB-2DK as indicated, and the expression of ΔF508-CFTR, RMA1 (KD control), and actin (loading control) was determined by Western blots.
VAPB Interactions with ERAD Components Regulate ΔF508-CFTR Degradation
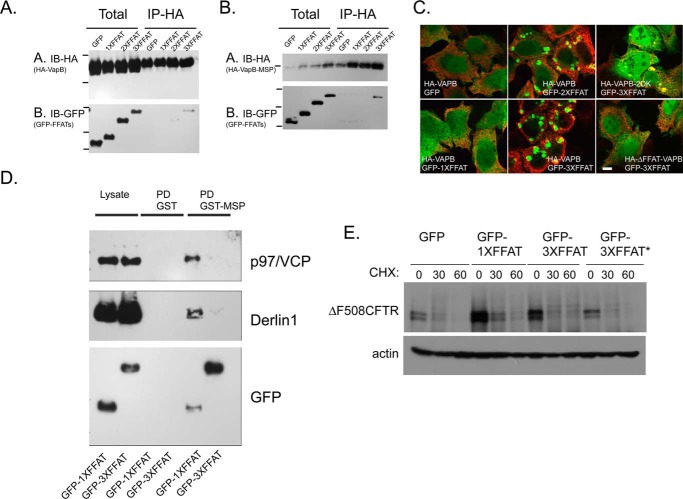
Having defined mutations that uncouple VAP expression from regulation of CFTR biogenesis, we used the information to analyze proteostasis-active VAPB interactions. Because the MSP domain is localized in the cytosol, we conducted a qualitative preliminary screen to identify cytosolic proteins that interact with GST-VAPB-MSP or GST-VAPB-MSP-2DK using the rat liver cytosol fraction, pulldown on GS beads, and mass spectrometry (MS), yielding a limited set of hits that were somewhat sensitive to the 2DK mutation (data not shown). Importantly, some of our hits were recently reported in quantitative MS analyses of VAPB interactions, including ORP and proteostasis proteins (59, 60). We therefore considered the possibility that the 2DK loop may regulate selective FFAT-driven interactions. As reported previously, we also detected interactions between VAP-MSP and FAF1 (61), a protein implicated in Parkinson disease (62). FAF1 utilizes a non-canonical FFAT motif to interact with VAPB, and this interaction is further required to support VCP-VAPB binding. We analyzed the interactions of FAF1 and VCP (a FAF1-binding protein) to determine whether the interactions are sensitive to the DK mutation. For analysis, we used VAPB-MSP domains in pulldown assays from lysates of cells transiently expressing FLAG-tagged FAF1. FAF1, a regulator of ERAD, contains a UBA domain that binds ubiquitinated proteins, a UAS domain that binds long chain fatty acids, and a UBX domain that interacts with p97/VCP exclusively when it is bound to UFD1/NPL4 and is thus committed to ERAD activities (63–65). Both FLAG-FAF1 and endogenous VCP were effectively pulled down with the MSP domain, and the interactions were inhibited by the proteostasis-uncoupling 2DK mutations to suggest that the 2DK loop regulates FFAT-mediated interactions (Fig. 4B). Overexpression of FAF1 by itself failed to affect ΔF508-CFTR degradation (data not shown). We therefore analyzed VAPB-MSP for interactions with BAP31 and Derlin-1; both interact with p97/VCP to support ERAD of ΔF508-CFTR (66). Derlin-1 is a rhomboid pseudoprotease that is required for protein dislocation from ER membranes (67). Overexpression of Derlin-1 leads to accelerated proteasome-dependent degradation of ΔF508-CFTR, whereas its depletion by shRNA leads to protein stabilization (68). Similarly, depletion of BAP31 stabilizes and further supports functional correction of ΔF508-CFTR (69, 70). VCP, BAP31, and Derlin-1 are found in complex with ΔF508-CFTR that controls dislocation and ER degradation (69). BAP31 and Derlin-1 (both endogenous and exogenously expressed tagged proteins) interacted with GST-MSP in cell lysates, but not with GST, and these interactions were inhibited by the 2DK mutations (Fig. 4B and data not shown). Studies in yeast suggested that the yeast homolog of BAP31-BAP29, the Yet1-Yet3 complex, interacts with yeast VAP-Scs2; therefore, these interactions are evolutionarily conserved (29). Overall, our analysis and recently published MS data highlighted two main groups of VAPB-MSP (direct or indirect)-binding proteins, one composed of components of the UPR and a cluster of ERAD and cytosolic protein degradation machinery (ATF6, FAF1, VCP, BAP31, and Derlin-1). The second group contained FFAT-carrying lipid sorting and sensing proteins and proteins that may regulate MCS (ORPs or Rab3-GAP1/2) supporting a role of VAP regulating the coupling between sterol traffic and proteostasis. The identity of primary (direct) interacting partners and the detailed map of the VAP-interactome remain to be determined.
VAPB Regulates ΔF508-CFTR Degradation via the RMA1 Pathway
BAP31 and Derlin-1 participate in a degradation pathway of ΔF508-CFTR that is controlled by the activity of the E3 ligase RMA1 (66, 71). BAP31 and Derlin-1 are in complex with RMA1, which preferentially identifies structural instabilities in NBD1, including ΔF508, leading to ubiquitination and BAP31-Derlin-1-mediated delivery to proteasomal degradation (69). To test whether VAPB-induced stabilization of ΔF508-CFTR utilizes the BAP31-Derlin-1-RMA1 pathway, we analyzed the effects of VAPB on CFTR stability in cells stably expressing scrambled or RNF5/RMA1 targeting shRNA (Fig. 4C). Although the cells only exhibited partial depletion of RMA1, this depletion led to stabilization of ΔF508-CFTR (Fig. 4C) as reported previously (71, 72). VAPB, which stabilizes ΔF508-CFTR, did not modify the effect of RMA1-mediated stabilization. The results suggest that VAPB functions to regulate the RMA1-BAP31-Derlin1 degradation pathway of ΔF508-CFTR.
Ligand Binding (FFAT or Proteostasis Ligands) Controls Functional VAP Interactions
Multiple interactions between VAP and FFAT-containing ligands have been described, and for many, a vital cellular function has been suggested. VAPs may function as a passive anchor point on the ER membranes to link FFAT ligands with the ER. In a passive mechanism, VAPs are found in large excess to their binding partners and thus may not play a regulatory role. However, previous functional data as well as the data presented here suggest an alternative model, in which VAP-ligand binding provides a regulation point in cell physiology as seen here with CFTR. How can multiple interactions of VAPB regulate CFTR proteostasis and sterol homeostasis and are these involved in coupling of the two activities? One simple mechanism is competition; binding of one ligand can inhibit interactions and thus regulation of others. To test this hypothesis, we produced artificial VAP ligands and tested these for VAP receptor binding and for the ability to functionally compete with other VAP ligands. VAPs bind FFAT signals in a mechanism that most likely involves coincidence-based recognition. Such binding is assisted by added interactions with lipids and protein determinants. To overcome this, we adopted a strategy of increasing FFAT binding through increased avidity. We generated tandem repeats of FFAT signals tagged with GFP for analysis. A similar strategy was utilized to follow FFAT binding in yeast, where ligand containing a single FFAT signal remains cytosolic, yet the ones that contain tandem FFAT repeats became localized with Scs2 on ER membranes (73). To examine the binding of tandem repeats to VAP, we co-expressed the HA-tagged VAPB or the VAPB-MSP domain together with GFP-nxFFAT reporters and monitored interactions by co-immunoprecipitation (Fig. 5, A and B). Although GFP and GFP-1×FFAT did not co-precipitate with VAPB, the 2×FFAT and more pronouncedly the 3×FFAT precipitated with HA-VAPB, suggesting that increased avidity can indeed overcome low affinity to support receptor binding (Fig. 5A). Similar results were observed when HA-VAPB-MSP-FFAT binding was analyzed, yet these interactions were more robust (Fig. 5B). To further monitor receptor binding, we followed the localization of HA-VAPB and FFAT ligands co-expressed in transfected cells (Fig. 5C). A recent study utilized a similar approach to demonstrate VAPA clustering at the Golgi region upon expression of an FFAT ligand appended to a Golgi-selective lipid-binding module (23). All transiently expressed GFP-FFAT ligands exhibited diffused cytosolic staining (Fig. 5C and data not shown). To examine receptor binding, we co-expressed HA-tagged VAPB with 1×, 2×, and 3×FFAT for morphological inspection. Strikingly, although GFP-1×FFAT remained largely cytosolic and HA-VAPB remained in a typical reticular ER pattern, the 2×FFAT and more pronouncedly the 3×FFAT ligand formed large ER aggregates in the presence of expressed VAPB (Fig. 5C), and these aggregates contained other ER membrane proteins (data not shown). ER aggregation was not observed when an FFAT binding-deficient HA-VAPB mutant was co-expressed suggesting that VAP-FFAT binding is reported morphologically through this membrane aggregation. The VAPB-2DK mutant also aggregated with the 3×FFAT ligand, in agreement with functional FFAT binding recorded in GST pulldown assays (Figs. 5C and 4B). It is interesting to note that we have not seen FFAT aggregates in the absence of exogenous co-expressed HA-VAPB. It may be that the concentration of endogenous VAP is insufficient to drive global rearrangements of ER membranes. Alternatively, isolated FFAT signals are effectively outcompeted by endogenous FFAT-LSS interactions that might be assisted by regulated co-incidence binding.
FIGURE 5.
Receptor-ligand interactions can regulate ΔF508-CFTR biogenesis. A, HeLa cells were co-transfected with HA-VAPB and GFP or GFP-tagged with 1×, 2×, or 3× copies of FFAT signals as indicated. HA-VAPB was isolated by immunoprecipitation (IP-HA) from cell lysates (Total) and probed for HA and GFP. Note interactions of 3×FFAT with VAPB. IB, immunoblot. B, similar co-immunoprecipitation analysis was conducted from cells expressing HA-VAPB-MSP domain and GFP-FFATs as indicated. C, HeLa cells were co-transfected with HA-VAPB, HA-VAPB-2DK, or HA-VAPB-ΔFFAT together with GFP, GFP-1×FFAT, GFP-2×FFAT, or GFP-3×FFAT, as indicated, and the localization of HA (red) or GFP (green) was determined by indirect (red) and direct (green) fluorescence. D, lysates prepared from cells transiently expressing GFP-1×FFAT or GFP-3×FFAT were subjected to pulldown analysis using GST or GST-VAPB-MSP proteins and analyzed for interactions with p97/VCP, Derlin1, and GFP-tagged FFATs on Western blots. E, cells expressing ΔF508-CFTR together with GFP, 1×, or 3× FFATs were analyzed for ΔF508-CFTR stability using cycloheximide (CHX) chase for the indicated times (actin serves as a loading control). 3×FFAT contained two copies of Myc-tagged FFAT signals of rabbit OSPB (residues 347–372) and one copy of an FFAT signal from human Nir2 (residues 344–360). 3×FFAT* is a tandem repeat of three copies of the OSBP-FFAT signal.
However, having established receptor binding by tandem FFAT ligands, we analyzed whether these ligands can also outcompete endogenous proteostasis ligands for VAP binding and whether such competition functionally modulates CFTR biogenesis. For analysis, we expressed the GFP-1×FFAT or 3×FFAT ligands in cells and prepared cell lysates. We then followed interactions of FFAT and proteostasis ligands using the GST pulldown assay. We titrated the GST-MSP and determined minimal concentrations that allowed for robust detection of binding with p97/VCP and Derlin-1 (data not shown). In this experimental setup, interactions between the 1×FFAT and VAPB-MSP were observed, yet these were not as robust as the interactions observed with the 3×FFAT ligand, corroborating the co-immunoprecipitation-based analysis in cells (Fig. 5D). Importantly, although p97/VCP or Derlin-1 interactions were detected in lysates expressing 1×FFAT, these interactions were abolished in lysates containing 3×FFAT ligands, supporting a competitive ligand-binding mode for VAP receptors (Fig. 5D). VAPs binding to proteostasis ligands may inhibit their activity. In the simplest model, VAP interactions with proteostasis regulators can be competed by the expression of artificial FFAT ligands thus accelerating ΔF508-CFTR degradation. We expressed FFAT ligands together with ΔF508-CFTR and analyzed CFTR stability using cycloheximide chase (Fig. 5E). Expression of FFAT ligands (1× and 3×FFAT) led to enhanced ΔF508-CFTR degradation. Therefore, ligand competition may direct VAP regulatory activities.
Discussion
Abnormalities in the distribution and levels of sterols and ceramides are documented in patients with protein folding diseases suggesting that coupling exists between proteostasis and lipid homeostasis (1, 2, 4, 5). However, the molecular basis for these abnormalities is unknown. VAPs bind LSS and proteostasis ligands and can regulate both processes. We therefore hypothesized that VAPs function as ER receptors that couple both processes. During protein misfolding in the ER, VAPs inhibit membrane contacts to slow lipid flow back to the ER, thus reducing biosynthetic load to accommodate the developing stress (Fig. 6).
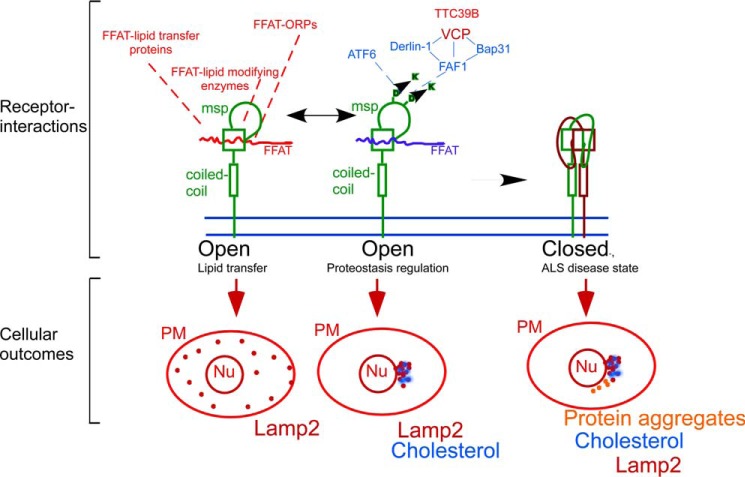
FIGURE 6.
Model for VAP receptor activities in coupling proteostasis with lipid homeostasis. The VAP receptor (green) utilizes its MSP domain (as marked) to interact with FFAT-containing ligands. Interactions are governed by competition between lipid-regulated LSS ligands (red) and proteostasis-regulated ligands (blue). The balance between the interactions can either lead to effective lipid flow or clustering of lamp2 containing late endosomes at the MToC and the accumulation of free cholesterol in the cells (see cellular outcomes depicted in the lower panel). VAP receptors can dimerize and oligomerize, and this activity regulates ligand binding and cellular outcomes (Nu, nucleus; PM, plasma membrane, red dots, late endosomes; blue dots, cholesterol; orange dots, protein aggregates). ALS-causing mutation P56S favors oligomerization, leading to the adaptation of closed receptor conformation, which does not bind ligands, leading to the deregulation of both proteostasis and lipid homeostasis.
Our studies support this hypothesis as follows. 1) VAP regulated membrane contact sites between ER and LE detached in response to protein misfolding in the ER (including that of CFTR) leading to the accumulation of free cholesterol in LE (Fig. 1). 2) VAPs, which regulate LE-ER MCS and sterol traffic, also regulated ΔF508-CFTR degradation using the MSP FFAT-binding domain (Fig. 2). 3) The functional interactions of VAPs with ATF6 (Fig. 3) and ERAD components of the RMA1-BAP31-Derlin-1-VCP pathway correlate with the regulation of ΔF508-CFTR biogenesis (Fig. 4); selective MSP mutations (2DK) inhibited the binding of VAP-MSP to ERAD ligands and the regulation of CFTR biogenesis by VAPs (Figs. 2–4). 4) Cellular sterol homeostasis regulates ATF6 activity and ERAD of ΔF508-CFTR (Fig. 3). 5) Competitive VAP-ligand binding (which can be defined in our experiments as a form of biological inactivation) can regulate CFTR biogenesis (Fig. 5). The results support a role for the ER-anchored VAPs in coupling between protein folding in the ER and lipid transfer and homeostasis (Fig. 6).
The mechanism of VAP-mediated regulation remains to be defined. In our experiments, VAPs regulate protein biogenesis by binding and sequestration of proteostasis regulators resulting in their inhibition. These regulators include components of the ERAD machinery and stress signaling elements ATF6 (Fig. 3) (30) and Ire1 (31). VAP is known to support directional intracellular lipid transfer. In yeast, VAPs directly regulate lipid synthesis during the activation of ER stress. How then is the coupling between the two activities achieved? A simple model suggests that the coupling activity is determined by ligand availability and VAP occupancy. In such a model, the occupation of the receptor by proteostasis ligands leads to inhibition of lipid flow and the observed accumulation of free cholesterol in late endosomes and the plasma membrane as observed in protein folding diseases (Fig. 6). However, the mechanism by which ligand interactions with VAP are regulated remains to be defined. For known LSS, availability is defined by lipid binding. The binding of 25-hydroxycholesterol by OSBP leads to stabilization of VAP-mediated Golgi-ER MCS (23, 74). In contrast, the binding of cholesterol by ORP1L leads to conformational change and inhibition of VAP binding (45). ORPs are utilized to sense and report lipid homeostasis in different organelles. In our model, LSS function to survey the membrane lipid composition of intracellular compartments using their lipid-binding modules and convey this information through conformational changes that regulate binding to VAPs in the ER. The observed complex regulation (with both inhibitory and stimulatory roles for lipids in regulation of VAP interactions) is likely utilized to optimize the coordination between proteostasis and overall cellular lipid traffic and homeostasis.
Similar regulation of ligand binding may apply for the interactions of proteostasis regulators with VAPs. We identified and verified FAF1 as a VAPB-interacting protein. FAF1 oligomerization is regulated by the interactions of its UAS domain with long chain unsaturated fatty acids (usFA) while linking to the p97-supported degradation machinery using its UBX domain (63–65). Thus, it might be that usFA regulate FAF1-Vap-VCP binding, although the source of usFA remains to be defined. In that respect, it is interesting to note that FAF2, a homolog of FAF1, is localized to lipid droplets (75), and a role for lipid droplets in the regulation of ERAD has been proposed (76). Thus, lipid droplets may provide regulatory usFAs during protein degradation. TTC39B was further identified as a VAPB binding partner. TTC39B is an evolutionarily conserved protein that is genetically linked to the development of dyslipidemia in humans (77–79). TTC39B contains a tetratricopeptide domain, possibly linking it to the protein chaperone system. The S. cerevisiae homolog of TTC39B/C Iml2, is localized with protein inclusions (that are characteristic of protein folding diseases) and is involved in physically linking those with lipid droplets. TTC39B/Iml2 activity is required for a lipid droplet-assisted clearance of cytosolic protein inclusions (80).
While our study was prepared for publication, mass spectrometry analyses of VAPB interactions using immunoprecipitation also identified FAF1 and VCP as binding partners for VAPB (60, 61). Importantly, the studies defined a non-canonical FFAT motif in FAF1 that was required for the interactions of both p97/VCP and FAF1 with VAPB. Future studies should further explore the possibility that FAF1-TTC39B and VAPs function on the same VAPB-RMA1-p97/VCP-Derlin-BAP31 ERAD pathway (Figs. 2 and 4), whereas lipid droplets provide the required environment for protein extraction from the ER and degradation. If so, VAPB binding may be utilized to coordinate lipid signals derived from lipid droplets with protein degradation in the ER.
Signaling pathways further control ligand availability and VAP interactions. OSBP phosphorylation enhances its VAP-mediated interactions with ER membranes (81, 82), and other LSS and proteostasis ligands may also be modified to regulate VAP binding and activity in response to signals that couple cells with organism physiology.
In our model (Fig. 6), the receptor itself is a passive element. However, ligand binding is also controlled by the status of the receptor. Given the sensitivity of FAF1 and VCP binding to the 2DK mutation (Fig. 4), the results suggest that the 2DK-containing loop regulates selective interactions of FFAT with the MSP domain and might play a role in the regulation of receptor assembly. VAPs dimerize and oligomerize using their coiled-coil (CC) cytosolic domain, and structural studies suggest that assembly regulates ligand binding (42).
A mutation in the MSP domain of VAPB (P56S) leads to the development of ALS, and it increases the propensity of VAPB to oligomerize, an activity mediated by the CC domain (51). Thus, the ligand-binding MSP domain regulates receptor assembly. In ALS, the VAPB P56S mutation can occlude MSP binding through induced oligomerization, trapping the receptor in a “closed” conformation that will inactivate FFAT ligand binding (Fig. 6). Thus, in its closed state, VAP will not support lipid flow between the ER and cellular membranes leading to abnormal distribution of cellular lipids. Under such conditions, proteostasis will also be deregulated (Fig. 6). In agreement, abnormal cholesterol and ceramide levels as well as protein inclusions are observed in relevant samples from sporadic ALS patients and importantly in defined ALS mouse models as is predicted by a loss of proteostasis and lipid homeostasis coupling (1). Future studies should examine the role of VAP homo- and hetero-assembly in physiological ligand binding and the role of ligand binding in controlling receptor assemblies. Importantly, the proposed ligand-receptor regulatory mode of activity for VAP receptors offers an avenue for the development of future therapeutics. VAP ligands might be utilized to selectively modulate cellular proteostasis outcomes in CF and other protein-folding diseases.
Author Contributions
M. A. designed the studies. W. L. E., K. S., X. G., and C. C. W. performed experiments and analyzed data. M. A., R. A. F., X. G., and W. L. E. analyzed and summarized the data. M. A. wrote the manuscript with input and editing from R. A. F. and W. L. E.
Acknowledgments
We thank Drs. C. Hoogenraad, J. Ngsee, K.-J. Lee, V. Olkkonen, T. Schroer, Z. Ronai, G. Shore, and G. Apodaca for providing valuable reagents.
This work was supported by Cystic Fibrosis Foundation (CFF) Grant FRIZZE05XX0 (to R. A. F.), National Institutes of Health Grants 5R01DK092807 (to M. A.) and 5P30DK072506 (to R. A. F.), and CFF Postdoctoral Fellowship ERNST10F0 (to W. L. E.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- CF
- cystic fibrosis
- LSS
- lipid sensing and sorting proteins
- LE
- late endosomes
- ER
- endoplasmic reticulum
- MSP
- major sperm protein
- CFTR
- cystic fibrosis transmembrane conductance regulator
- VAP
- VAMP-associated protein
- ALS
- amyotrophic lateral sclerosis
- OSBP
- oxysterol-binding protein
- ORP
- OSBP-related protein
- UPR
- unfolded protein response
- ERAD
- ER-associated degradation
- NPC
- Niemann Pick disease type C
- CC
- coiled-coil
- usFA
- unsaturated fatty acid
- Tm
- tunicamycin
- MCS
- membrane contact site
- FFAT
- two phenylalanines in an acidic track
- LD
- lipoprotein-deficient
- MToC
- microtubule-organizing center.
References
- 1. Cutler R. G., Pedersen W. A., Camandola S., Rothstein J. D., and Mattson M. P. (2002) Evidence that accumulation of ceramides and cholesterol esters mediates oxidative stress-induced death of motor neurons in amyotrophic lateral sclerosis. Ann. Neurol. 52, 448–457 [DOI] [PubMed] [Google Scholar]
- 2. Gentzsch M., Choudhury A., Chang X. B., Pagano R. E., and Riordan J. R. (2007) Misassembled mutant ΔF508 CFTR in the distal secretory pathway alters cellular lipid trafficking. J. Cell Sci. 120, 447–455 [DOI] [PubMed] [Google Scholar]
- 3. Ron I., and Horowitz M. (2005) ER retention and degradation as the molecular basis underlying Gaucher disease heterogeneity. Hum. Mol. Genet. 14, 2387–2398 [DOI] [PubMed] [Google Scholar]
- 4. Ron I., and Horowitz M. (2008) Intracellular cholesterol modifies the ERAD of glucocerebrosidase in Gaucher disease patients. Mol. Genet. Metab. 93, 426–436 [DOI] [PubMed] [Google Scholar]
- 5. Worgall T. S. (2009) Lipid metabolism in cystic fibrosis. Curr. Opin. Clin. Nutr. Metab. Care 12, 105–109 [DOI] [PubMed] [Google Scholar]
- 6. White N. M., Jiang D., Burgess J. D., Bederman I. R., Previs S. F., and Kelley T. J. (2007) Altered cholesterol homeostasis in cultured and in vivo models of cystic fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 292, L476–L486 [DOI] [PubMed] [Google Scholar]
- 7. Cianciola N. L., Carlin C. R., and Kelley T. J. (2011) Molecular pathways for intracellular cholesterol accumulation: common pathogenic mechanisms in Niemann-Pick disease type C and cystic fibrosis. Arch. Biochem. Biophys. 515, 54–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Narita K., Choudhury A., Dobrenis K., Sharma D. K., Holicky E. L., Marks D. L., Walkley S. U., and Pagano R. E. (2005) Protein transduction of Rab9 in Niemann-Pick C cells reduces cholesterol storage. FASEB J. 19, 1558–1560 [DOI] [PubMed] [Google Scholar]
- 9. Choudhury A., Dominguez M., Puri V., Sharma D. K., Narita K., Wheatley C. L., Marks D. L., and Pagano R. E. (2002) Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J. Clin. Invest. 109, 1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xu Y., Tertilt C., Krause A., Quadri L. E., Crystal R. G., and Worgall S. (2009) Influence of the cystic fibrosis transmembrane conductance regulator on expression of lipid metabolism-related genes in dendritic cells. Respir. Res. 10, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Colgan S. M., Tang D., Werstuck G. H., and Austin R. C. (2007) Endoplasmic reticulum stress causes the activation of sterol regulatory element binding protein-2. Int. J. Biochem. Cell Biol. 39, 1843–1851 [DOI] [PubMed] [Google Scholar]
- 12. Kim A. J., Shi Y., Austin R. C., and Werstuck G. H. (2005) Valproate protects cells from ER stress-induced lipid accumulation and apoptosis by inhibiting glycogen synthase kinase-3. J. Cell Sci. 118, 89–99 [DOI] [PubMed] [Google Scholar]
- 13. Sage A. T., Walter L. A., Shi Y., Khan M. I., Kaneto H., Capretta A., and Werstuck G. H. (2010) Hexosamine biosynthesis pathway flux promotes endoplasmic reticulum stress, lipid accumulation, and inflammatory gene expression in hepatic cells. Am. J. Physiol. Endocrinol. Metab. 298, E499–E511 [DOI] [PubMed] [Google Scholar]
- 14. Pineau L., Colas J., Dupont S., Beney L., Fleurat-Lessard P., Berjeaud J. M., Bergès T., and Ferreira T. (2009) Lipid-induced ER stress: synergistic effects of sterols and saturated fatty acids. Traffic 10, 673–690 [DOI] [PubMed] [Google Scholar]
- 15. Ron D., and Walter P. (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell. Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 16. Loewen C. J., and Levine T. P. (2005) A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J. Biol. Chem. 280, 14097–14104 [DOI] [PubMed] [Google Scholar]
- 17. Skehel P. A., Fabian-Fine R., and Kandel E. R. (2000) Mouse VAP33 is associated with the endoplasmic reticulum and microtubules. Proc. Natl. Acad. Sci. U.S.A. 97, 1101–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Skehel P. A., Martin K. C., Kandel E. R., and Bartsch D. (1995) A VAMP-binding protein from Aplysia required for neurotransmitter release. Science 269, 1580–1583 [DOI] [PubMed] [Google Scholar]
- 19. Weber-Boyvat M., Zhong W., Yan D., and Olkkonen V. M. (2013) Oxysterol-binding proteins: functions in cell regulation beyond lipid metabolism. Biochem. Pharmacol. 86, 89–95 [DOI] [PubMed] [Google Scholar]
- 20. Olkkonen V. M. (2013) OSBP-related proteins: liganding by glycerophospholipids opens new insight into their function. Molecules 18, 13666–13679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Litvak V., Dahan N., Ramachandran S., Sabanay H., and Lev S. (2005) Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat. Cell Biol. 7, 225–234 [DOI] [PubMed] [Google Scholar]
- 22. Kim Y. J., Guzman-Hernandez M. L., Wisniewski E., and Balla T. (2015) Phosphatidylinositol-phosphatidic acid exchange by Nir2 at ER-PM contact sites maintains phosphoinositide signaling competence. Dev. Cell 33, 549–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mesmin B., Bigay J., Moser von Filseck J., Lacas-Gervais S., Drin G., and Antonny B. (2013) A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell 155, 830–843 [DOI] [PubMed] [Google Scholar]
- 24. Chung J., Torta F., Masai K., Lucast L., Czapla H., Tanner L. B., Narayanaswamy P., Wenk M. R., Nakatsu F., and De Camilli P. (2015) Intracellular transport. PI4P/phosphatidylserine countertransport at ORP5- and ORP8-mediated ER-plasma membrane contacts. Science 349, 428–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hanada K., Kumagai K., Tomishige N., and Yamaji T. (2009) CERT-mediated trafficking of ceramide. Biochim. Biophys. Acta 1791, 684–691 [DOI] [PubMed] [Google Scholar]
- 26. Kukihara H., Moriishi K., Taguwa S., Tani H., Abe T., Mori Y., Suzuki T., Fukuhara T., Taketomi A., Maehara Y., and Matsuura Y. (2009) Human VAP-C negatively regulates hepatitis C virus propagation. J. Virol. 83, 7959–7969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hamamoto I., Nishimura Y., Okamoto T., Aizaki H., Liu M., Mori Y., Abe T., Suzuki T., Lai M. M., Miyamura T., Moriishi K., and Matsuura Y. (2005) Human VAP-B is involved in hepatitis C virus replication through interaction with NS5A and NS5B. J. Virol. 79, 13473–13482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brickner J. H., and Walter P. (2004) Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol. 2, e342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilson J. D., Thompson S. L., and Barlowe C. (2011) Yet1p-Yet3p interacts with Scs2p-Opi1p to regulate ER localization of the Opi1p repressor. Mol. Biol. Cell 22, 1430–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gkogkas C., Middleton S., Kremer A. M., Wardrope C., Hannah M., Gillingwater T. H., and Skehel P. (2008) VAPB interacts with and modulates the activity of ATF6. Hum. Mol. Genet. 17, 1517–1526 [DOI] [PubMed] [Google Scholar]
- 31. Suzuki H., Kanekura K., Levine T. P., Kohno K., Olkkonen V. M., Aiso S., and Matsuoka M. (2009) ALS-linked P56S-VAPB, an aggregated loss-of-function mutant of VAPB, predisposes motor neurons to ER stress-related death by inducing aggregation of co-expressed wild-type VAPB. J. Neurochem. 108, 973–985 [DOI] [PubMed] [Google Scholar]
- 32. Maiuolo J., Bulotta S., Verderio C., Benfante R., and Borgese N. (2011) Selective activation of the transcription factor ATF6 mediates endoplasmic reticulum proliferation triggered by a membrane protein. Proc. Natl. Acad. Sci. U.S.A. 108, 7832–7837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kuijpers M., van Dis V., Haasdijk E. D., Harterink M., Vocking K., Post J. A., Scheper W., Hoogenraad C. C., and Jaarsma D. (2013) Amyotrophic lateral sclerosis (ALS)-associated VAPB-P56S inclusions represent an ER quality control compartment. Acta Neuropathol. Commun. 1, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuijpers M., Yu K. L., Teuling E., Akhmanova A., Jaarsma D., and Hoogenraad C. C. (2013) The ALS8 protein VAPB interacts with the ER-Golgi recycling protein YIF1A and regulates membrane delivery into dendrites. EMBO J. 32, 2056–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moumen A., Virard I., and Raoul C. (2011) Accumulation of wildtype and ALS-linked mutated VAPB impairs activity of the proteasome. PLoS ONE 6, e26066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Moustaqim-Barrette A., Lin Y. Q., Pradhan S., Neely G. G., Bellen H. J., and Tsuda H. (2014) The amyotrophic lateral sclerosis 8 protein, VAP, is required for ER protein quality control. Hum. Mol. Genet. 23, 1975–1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chai A., Withers J., Koh Y. H., Parry K., Bao H., Zhang B., Budnik V., and Pennetta G. (2008) hVAPB, the causative gene of a heterogeneous group of motor neuron diseases in humans, is functionally interchangeable with its Drosophila homologue DVAP-33A at the neuromuscular junction. Hum. Mol. Genet. 17, 266–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Forrest S., Chai A., Sanhueza M., Marescotti M., Parry K., Georgiev A., Sahota V., Mendez-Castro R., and Pennetta G. (2013) Increased levels of phosphoinositides cause neurodegeneration in a Drosophila model of amyotrophic lateral sclerosis. Hum. Mol. Genet. 22, 2689–2704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sanhueza M., Zechini L., Gillespie T., and Pennetta G. (2014) Gain-of-function mutations in the ALS8 causative gene VAPB have detrimental effects on neurons and muscles. Biol. Open 3, 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tsuda H., Han S. M., Yang Y., Tong C., Lin Y. Q., Mohan K., Haueter C., Zoghbi A., Harati Y., Kwan J., Miller M. A., and Bellen H. J. (2008) The amyotrophic lateral sclerosis 8 protein VAPB is cleaved, secreted, and acts as a ligand for Eph receptors. Cell 133, 963–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Han S. M., Tsuda H., Yang Y., Vibbert J., Cottee P., Lee S. J., Winek J., Haueter C., Bellen H. J., and Miller M. A. (2012) Secreted VAPB/ALS8 major sperm protein domains modulate mitochondrial localization and morphology via growth cone guidance receptors. Dev. Cell 22, 348–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaiser S. E., Brickner J. H., Reilein A. R., Fenn T. D., Walter P., and Brunger A. T. (2005) Structural basis of FFAT motif-mediated ER targeting. Structure 13, 1035–1045 [DOI] [PubMed] [Google Scholar]
- 43. Prosser D. C., Tran D., Gougeon P. Y., Verly C., and Ngsee J. K. (2008) FFAT rescues VAPA-mediated inhibition of ER-to-Golgi transport and VAPB-mediated ER aggregation. J. Cell Sci. 121, 3052–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Teuling E., Ahmed S., Haasdijk E., Demmers J., Steinmetz M. O., Akhmanova A., Jaarsma D., and Hoogenraad C. C. (2007) Motor neuron disease-associated mutant vesicle-associated membrane protein-associated protein (VAP) B recruits wild-type VAP into endoplasmic reticulum-derived tubular aggregates. J. Neurosci. 27, 9801–9815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rocha N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., and Neefjes J. (2009) Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7-RILP-p150 Glued and late endosome positioning. J. Cell Biol. 185, 1209–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lehto M., Laitinen S., Chinetti G., Johansson M., Ehnholm C., Staels B., Ikonen E., and Olkkonen V. M. (2001) The OSBP-related protein family in humans. J. Lipid Res. 42, 1203–1213 [PubMed] [Google Scholar]
- 47. Im Y. J., Raychaudhuri S., Prinz W. A., and Hurley J. H. (2005) Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature 437, 154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vihervaara T., Uronen R. L., Wohlfahrt G., Björkhem I., Ikonen E., and Olkkonen V. M. (2011) Sterol binding by OSBP-related protein 1L regulates late endosome motility and function. Cell. Mol. Life Sci. 68, 537–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Loewen C. J., Roy A., and Levine T. P. (2003) A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22, 2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Riordan J. R. (2005) Assembly of functional CFTR chloride channels. Annu. Rev. Physiol. 67, 701–718 [DOI] [PubMed] [Google Scholar]
- 51. Kim S., Leal S. S., Ben Halevy D., Gomes C. M., and Lev S. (2010) Structural requirements for VAP-B oligomerization and their implication in amyotrophic lateral sclerosis-associated VAP-B(P56S) neurotoxicity. J. Biol. Chem. 285, 13839–13849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bartoszewski R., Rab A., Twitty G., Stevenson L., Fortenberry J., Piotrowski A., Dumanski J. P., and Bebok Z. (2008) The mechanism of cystic fibrosis transmembrane conductance regulator transcriptional repression during the unfolded protein response. J. Biol. Chem. 283, 12154–12165 [DOI] [PubMed] [Google Scholar]
- 53. Kerbiriou M., Le Drévo M. A., Férec C., and Trouvé P. (2007) Coupling cystic fibrosis to endoplasmic reticulum stress: differential role of Grp78 and ATF6. Biochim. Biophys. Acta 1772, 1236–1249 [DOI] [PubMed] [Google Scholar]
- 54. Adachi Y., Yamamoto K., Okada T., Yoshida H., Harada A., and Mori K. (2008) ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct. Funct. 33, 75–89 [DOI] [PubMed] [Google Scholar]
- 55. Cholon D. M., O'Neal W. K., Randell S. H., Riordan J. R., and Gentzsch M. (2010) Modulation of endocytic trafficking and apical stability of CFTR in primary human airway epithelial cultures. Am. J. Physiol. Lung Cell. Mol. Physiol. 298, L304–L314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lim C. H., Bijvelds M. J., Nigg A., Schoonderwoerd K., Houtsmuller A. B., de Jonge H. R., and Tilly B. C. (2007) Cholesterol depletion and genistein as tools to promote F508delCFTR retention at the plasma membrane. Cell. Physiol. Biochem. 20, 473–482 [DOI] [PubMed] [Google Scholar]
- 57. Wang Y., Shen J., Arenzana N., Tirasophon W., Kaufman R. J., and Prywes R. (2000) Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem. 275, 27013–27020 [DOI] [PubMed] [Google Scholar]
- 58. Haze K., Yoshida H., Yanagi H., Yura T., and Mori K. (1999) Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hantan D., Yamamoto Y., and Sakisaka T. (2014) VAP-B binds to Rab3GAP1 at the ER: its implication in nuclear envelope formation through the ER-Golgi intermediate compartment. Kobe J. Med. Sci. 60, E48–E56 [PubMed] [Google Scholar]
- 60. Huttlin E. L., Ting L., Bruckner R. J., Gebreab F., Gygi M. P., Szpyt J., Tam S., Zarraga G., Colby G., Baltier K., Dong R., Guarani V., Vaites L. P., Ordureau A., Rad R., et al. (2015) The BioPlex network: a systematic exploration of the human interactome. Cell 162, 425–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Baron Y., Pedrioli P. G., Tyagi K., Johnson C., Wood N. T., Fountaine D., Wightman M., and Alexandru G. (2014) VAPB/ALS8 interacts with FFAT-like proteins including the p97 cofactor FAF1 and the ASNA1 ATPase. BMC Biol. 12, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sul J. W., Park M. Y., Shin J., Kim Y. R., Yoo S. E., Kong Y. Y., Kwon K. S., Lee Y. H., and Kim E. (2013) Accumulation of the parkin substrate, FAF1, plays a key role in the dopaminergic neurodegeneration. Hum. Mol. Genet. 22, 1558–1573 [DOI] [PubMed] [Google Scholar]
- 63. Lee J. J., Park J. K., Jeong J., Jeon H., Yoon J. B., Kim E. E., and Lee K. J. (2013) Complex of FAF1 with VCP-Npl4-Ufd1 and polyubiquitinated proteins promotes endoplasmic reticulum-associated degradation (ERAD). J. Biol. Chem. 288, 6998–7011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim H., Zhang H., Meng D., Russell G., Lee J. N., and Ye J. (2013) UAS domain of Ubxd8 and FAF1 polymerizes upon interaction with long-chain unsaturated fatty acids. J. Lipid Res. 54, 2144–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kim K. H., Kang W., Suh S. W., and Yang J. K. (2011) Crystal structure of FAF1 UBX domain in complex with p97/VCP N domain reveals a conformational change in the conserved FcisP touch-turn motif of UBX domain. Proteins 79, 2583–2587 [DOI] [PubMed] [Google Scholar]
- 66. Younger J. M., Chen L., Ren H. Y., Rosser M. F., Turnbull E. L., Fan C. Y., Patterson C., and Cyr D. M. (2006) Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 126, 571–582 [DOI] [PubMed] [Google Scholar]
- 67. Greenblatt E. J., Olzmann J. A., and Kopito R. R. (2011) Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant α-1 antitrypsin from the endoplasmic reticulum. Nat. Struct. Mol. Biol. 18, 1147–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sun F., Zhang R., Gong X., Geng X., Drain P. F., and Frizzell R. A. (2006) Derlin-1 promotes the efficient degradation of the cystic fibrosis transmembrane conductance regulator (CFTR) and CFTR folding mutants. J. Biol. Chem. 281, 36856–36863 [DOI] [PubMed] [Google Scholar]
- 69. Wang B., Heath-Engel H., Zhang D., Nguyen N., Thomas D. Y., Hanrahan J. W., and Shore G. C. (2008) BAP31 interacts with Sec61 translocons and promotes retrotranslocation of CFTRΔF508 via the derlin-1 complex. Cell 133, 1080–1092 [DOI] [PubMed] [Google Scholar]
- 70. Lambert G., Becker B., Schreiber R., Boucherot A., Reth M., and Kunzelmann K. (2001) Control of cystic fibrosis transmembrane conductance regulator expression by BAP31. J. Biol. Chem. 276, 20340–20345 [DOI] [PubMed] [Google Scholar]
- 71. Delaunay A., Bromberg K. D., Hayashi Y., Mirabella M., Burch D., Kirkwood B., Serra C., Malicdan M. C., Mizisin A. P., Morosetti R., Broccolini A., Guo L. T., Jones S. N., Lira S. A., Puri P. L., et al. (2008) The ER-bound RING finger protein 5 (RNF5/RMA1) causes degenerative myopathy in transgenic mice and is deregulated in inclusion body myositis. PLoS ONE 3, e1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Morito D., Hirao K., Oda Y., Hosokawa N., Tokunaga F., Cyr D. M., Tanaka K., Iwai K., and Nagata K. (2008) Gp78 cooperates with RMA1 in endoplasmic reticulum-associated degradation of CFTRΔF508. Mol. Biol. Cell 19, 1328–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mikitova V., and Levine T. P. (2012) Analysis of the key elements of FFAT-like motifs identifies new proteins that potentially bind VAP on the ER, including two AKAPs and FAPP2. PLoS ONE 7, e30455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ridgway N. D., Dawson P. A., Ho Y. K., Brown M. S., and Goldstein J. L. (1992) Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J. Cell Biol. 116, 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Krahmer N., Hilger M., Kory N., Wilfling F., Stoehr G., Mann M., Farese R. V. Jr., and Walther T. C. (2013) Protein correlation profiles identify lipid droplet proteins with high confidence. Mol. Cell. Proteomics 12, 1115–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Olzmann J. A., Richter C. M., and Kopito R. R. (2013) Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc. Natl. Acad. Sci. U.S.A. 110, 1345–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kathiresan S., Willer C. J., Peloso G. M., Demissie S., Musunuru K., Schadt E. E., Kaplan L., Bennett D., Li Y., Tanaka T., Voight B. F., Bonnycastle L. L., Jackson A. U., Crawford G., Surti A., Guiducci C., et al. (2009) Common variants at 30 loci contribute to polygenic dyslipidemia. Nat. Genet. 41, 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., Johansen C. T., Fouchier S. W., Isaacs A., Peloso G. M., Barbalic M., et al. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Myocardial Infarction Genetics Consortium, Kathiresan S., Voight B. F., Purcell S., Musunuru K., Ardissino D., Mannucci P. M., Anand S., Engert J. C., Samani N. J., Schunkert H., Erdmann J., Reilly M. P., Rader D. J., Morgan T., et al. (2009) Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 41, 334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Moldavski O., Amen T., Levin-Zaidman S., Eisenstein M., Rogachev I., Brandis A., Kaganovich D., and Schuldiner M. (2015) Lipid droplets are essential for efficient clearance of cytosolic inclusion bodies. Dev. Cell 33, 603–610 [DOI] [PubMed] [Google Scholar]
- 81. Nhek S., Ngo M., Yang X., Ng M. M., Field S. J., Asara J. M., Ridgway N. D., and Toker A. (2010) Regulation of oxysterol-binding protein Golgi localization through protein kinase D-mediated phosphorylation. Mol. Biol. Cell 21, 2327–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Goto A., Liu X., Robinson C. A., and Ridgway N. D. (2012) Multisite phosphorylation of oxysterol-binding protein regulates sterol binding and activation of sphingomyelin synthesis. Mol. Biol. Cell 23, 3624–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]