Abstract
Flavonols are a group of secondary metabolites that affect diverse cellular processes. They are considered putative negative regulators of the transport of the phytohormone auxin, by which they influence auxin distribution and concomitantly take part in the control of plant organ development. Flavonols are accumulating in a large number of glycosidic forms. Whether these have distinct functions and diverse cellular targets is not well understood. The rol1-2 mutant of Arabidopsis thaliana is characterized by a modified flavonol glycosylation profile that is inducing changes in auxin transport and growth defects in shoot tissues. To determine whether specific flavonol glycosides are responsible for these phenotypes, a suppressor screen was performed on the rol1-2 mutant, resulting in the identification of an allelic series of UGT89C1, a gene encoding a flavonol 7-O-rhamnosyltransferase. A detailed analysis revealed that interfering with flavonol rhamnosylation increases the concentration of auxin precursors and auxin metabolites, whereas auxin transport is not affected. This finding provides an additional level of complexity to the possible ways by which flavonols influence auxin distribution and suggests that flavonol glycosides play an important role in regulating plant development.
Keywords: Arabidopsis thaliana, auxin, flavonoid, metabolism, transport, UGT89C1, auxin transport, auxin turnover, flavonols, rol1-2, flavonol glycosides, rhamnose*
Introduction
Flavonoids are secondary metabolites produced via the phenylpropanoid pathway (1), and they serve different functions such as protection from UV irradiation (2), plant-microbe interaction (3), regulation of levels of reactive oxygen species (4), inhibition of cell cycle, and control of cell growth (5, 6). In addition, they are also thought to influence transcriptional and signaling processes (7–9). A major role of flavonols, a subgroup of flavonoids, appears to be the modification of auxin-related processes due to their property of modulating auxin transport (10–14). In some tissues, auxin is transported from cell to cell in a polarized fashion, operated mainly via auxin transporters of the ABCB, AUX1/LAX, and PIN families (15, 16). Flavonoids bind to and inhibit the auxin transport proteins ABCB1 and ABCB19 (17) and interfere with the interaction of these two proteins with the immunophilin-like protein, FKBP42/TWISTED DWARF1 (18). Recent data show that the pinoid kinase, which influences polar localization of PIN proteins (19) and ABCB transport activity, is a likely target of flavonol action (20). The accumulation of flavonols is altered in the pin2 mutant, the agravitropic phenotype of which can be partially complemented by exogenous flavonols due to their ability to modify expression of PIN genes (21–23). Hence, flavonols and auxin are able to mutually influence each other, confirming a functional interdependence in their biological activity.
Flavonoid biosynthesis is well characterized, and a number of mutants affected in the enzymes committed to the different steps have been identified in Arabidopsis thaliana (1) (Fig. 1). These lines frequently show a pale yellow seed coat due to the absence of proanthocyanidins and were thus named transparent testa (tt) mutants (24). Flavonols are produced from dihydroflavonols via the activity of the flavonol synthase, and thus represent a side branch of the flavonoid biosynthesis pathway. In Arabidopsis, the flavonols kaempferol, quercetin, and isorhamnetin are glycosylated by one or several sugars, mainly Glc, Rha, and rarely Ara, at the C3 and C7 position of the flavonol backbone (25, 26) through the function of UDP-dependent glycosyltransferases (UGTs),4 some of which have been identified (8, 26–29). The biological relevance of flavonol glycosylation remains controversial. In vitro experiments show activity of flavonol aglycones, suggesting that these are the compounds active in modulating polar auxin transport (10, 17, 18, 20). However, the identification of mutant phenotypes induced by changes in the flavonol glycosylation profile suggests function for at least some flavonol glycosides (30, 31).
FIGURE 1.
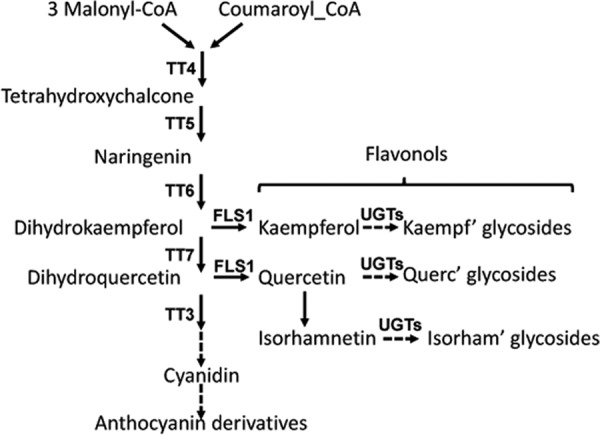
Overview of flavonoid biosynthesis. The phenylpropanoid pathway leads to the synthesis of flavonoids. These encompass a number of different compounds, not all of which are indicated in this scheme. Enzymes leading to the synthesis of flavonols (kaempferol, quercetin, isorhamnetin) are indicated. TT (transparent testa) proteins were identified based on the pale brown phenotype of seeds of corresponding mutant lines. FLS1: flavonol synthase 1, the predominant FLS protein in Arabidopsis. UGTs: UDP-dependent glycosyltransferases needed for the glycosylation of flavonols.
Of the major auxin (indole-3-acetic acid; IAA) produced by plants, only a minor fraction of ∼1% exists in this active form. Auxin can be conjugated to amino acids and/or sugars (mainly Glc in Arabidopsis) for storage in an inactive form or as an initial step for degradation (32, 33). Indole-3-acetyl-alanine (IAA-Ala) and indole-3-acetyl-leucine (IAA-Leu) are readily hydrolyzable, resulting in active free IAA, whereas indole-3-acetyl-aspartate (IAA-Asp) and indole-3-acetyl-glutamate (IAA-Glu) remain largely stable, suggesting that the dynamics of the reversible (de-)conjugation are decisive in regulating the pool of active auxin. Auxin homeostasis can be affected by mutations in loci involved in auxin (de-)conjugation, and multiple mutations in these loci can induce auxin-related phenotypes ((Ref. 34) and references therein). In Arabidopsis, the most abundant degradation products of auxin are produced via oxidation, resulting predominantly in oxIAA (2-oxindole-3-acetic acid) and oxIAA-Glc (2-oxindole-3-acetyl-glucose, glucosyl-ester of oxIAA) (35, 36). This oxidation contributes to the formation of auxin gradients (37) that are crucial for auxin-driven organ development (38–41). oxIAA levels are increased in the flavonoid-deficient tt4 mutant but decreased in the flavonol over-accumulator tt3 (42). This correlation suggests that the anti-oxidant activity of flavonols (4) influences auxin degradation.
The rol1-2 (repressor of lrx1) mutant is affected in one of three RHM genes, RHM1, encoding rhamnose synthase, which converts UDP-Glc to UDP-Rha (43). The rol1-2 mutant was identified in a screen for alterations in cell wall structures and shows modifications in the Rha-rich cell wall component pectin (43). In addition, rol1-2 also shows changes in the flavonol glycosylation profile, mainly a reduction in the degree of rhamnosylation, whereas the overall quantity of flavonols is not affected (30, 44). This confirms the importance of RHM1, which is co-expressed with genes important for flavonol biosynthesis (26) and for flavonol glycosylation. rol1-2 seedlings are characterized by shorter roots and root hairs than in the wild type. The rol1-2 seedling shoot develops deformed trichomes and hyponastic cotyledons with brick-shaped pavement cells, whereas the wild type develops regular trichomes and epinastic cotyledons with puzzle-shaped jigsaw-like pavement cells (30, 43). There is no obvious growth defect detectable in rol1-2 adult plants, presumably due to the functional redundancy among the three RHM genes (45).
The short root phenotype of the rol1-2 mutant is most likely induced by the changes in pectin structures (43). By contrast, the aberrant shoot phenotype of the rol1-2 mutant is connected to the altered flavonol composition. Mutations affecting steps in flavonoid biosynthesis (Fig. 1) upstream of flavonols such as tt4 or tt6, mutations in the flavonol synthase (FLS1), or mutations in a positive regulator of flavonol biosynthesis encoded by MYB111 all suppress rol1-2, resulting in wild type-like shoot development, whereas root development is comparable with the rol1-2 single mutant (30, 44). A mutation in TT7, blocking quercetin accumulation, or any step farther downstream in the pathway has no effect on the rol1-2 shoot phenotype (30, 44). Together, these findings suggest that the rol1-2 phenotype is induced by the accumulation of flavonol glycosides that interfere with proper plant development and that kaempferols are sufficient to induce this defect. Yin et al. (31) have shown that the dwarf growth phenotype of the flavonoid 3-O-glucosyltransferase mutant ugt78d2 correlates with the over-accumulation of 3-O-7-O-rhamnosylated kaempferol (K-R-3-R-7), and interfering with K-R-3-R-7 biosynthesis suppresses the growth defect of the ugt78d2 mutant. By contrast, the flavonol species inducing the rol1-2 phenotype is most likely not K-R-3-R-7, because this compound is present in lower amounts than in the wild type. Thus, it is likely that several flavonol glycosides can have an effect on plant development. Both rol1-2 and ugt78d2 show changes in auxin concentration or transport activity. For rol1-2, it was shown that auxin transport is reverted to wild-type levels when blocking flavonol accumulation (44), thus indicating that flavonols do interfere with auxin distribution.
Aiming at the identification of flavonol glycosides that induce the rol1-2 phenotype, EMS-induced suppressor mutants of rol1-2 were selected for specific changes in flavonol glycosylation. Several alleles of the 7-O-rhamnosyltransferase locus UGT89C1 were identified. The auxin transport activity in rol1-2 is not changed by a ugt89c1 mutation, but the levels of auxin conjugates and catabolites are strongly increased in the ugt89c1 mutant background. This indicates that flavonols affect not only auxin transport but also auxin turnover, and in this way modify auxin homeostasis.
Experimental Procedures
Plant Material, Growth Conditions, EMS Mutagenesis, and Mutant Screen
All lines described in this study are in the Col-0 genetic background. The rol1-2 allele and fls1-3 allele used in this study are described elsewhere (43, 44). For all analyses described, the ugt89c1-3 nonsense allele was used. Seeds were surface-sterilized with 1% sodium hypochlorite, 0.03% Triton X-100, plated on half-strength Murashige and Skoog medium containing 0.6% Phytagel, 2% sucrose, 100 μg/ml myo-inositol, stratified for 4 days at 4 °C, and grown in a vertical orientation in a 16-h light, 8-h dark cycle at 22 °C. For propagation and crossings, 10-day-old plants were transferred to soil, grown in a 16-h light, 8-h dark cycle at 22 °C, and irradiated with white light (Biolux, Osram). The EMS screen was performed as described (44), and 75,000 M2 seedlings were screened for a suppressed rol1-2 mutant phenotype. All ugt89c1 alleles were backcrossed at least three times to Col-0 and rol1-2 prior to analysis. Plant transformation was done as described (43), and transgenic plants were selected with BASTA (10 μg/ml).
DNA Constructs, Plant Transformation, and Molecular Markers
For the UGT89C1 complementation construct, the UGT89C1 genomic clone was PCR-amplified with the primers F7RT_PC 5′-GGCGCGCCAGACTACAGTTTGGCTAACCAG-3′ and F7RT_R3C 5′-TGAACCGCGTGTGTAATGTATC-3′, encompassing 1.5-kb promoter, 1.3-kb coding sequence, and 0.35-kb terminator sequence. The resulting fragment (UGT89C1:UGT89C1) was cloned into pGEM-T Easy (Promega) for sequencing. For the GFP fusion construct, a BamHI site was introduced into UGT89C1:UGT89C1 clone by PCR 3′ of the ATG (N-terminal fusion). PCR was performed with the primers F7RT_R4_NGFP 5′-GGATCCCATGATTGATGTTTTTTTCTTTC-3′ and F7RT_F6_NGFP 5′-GGATCCACAACAACAACAACGAAGAAGC-3′. A previously produced GFP construct flanked by BamHI sites (46) was cloned into this BamHI site, resulting in UGT89C1:GFP-UGT89C1. This construct was cloned into the binary vector pBART (47) by NotI.
The molecular markers for the rol1-2 and fls1-3 mutations are described elsewhere (43, 44). For detection of ugt89c1-3, a PCR was performed using the primers F7RT_F4 5′-TGATGCTTTCTCTATTAAGTCCAT-3′ and F7RT_R5_CGFP 5′-GGATCCCAAACACATCTCTGCAACGAG-3′. The PCR product was cut by ApaI (cuts wild type) and run on a 1.5% agarose gel. For detection of ugt89c1-11, PCR was performed using the primers F7RT_F4 5′-TGATGCTTTCTCTATTAAGTCCAT-3′ and F7RT_R5_CGFP 5′-GGATCCCAAACACATCTCTGCAACGAG-3′. PCR was performed at 55 °C annealing temperature, 1.5 mm MgCl2, and 1 min of elongation time, with 35 cycles. The PCR product was cut by BglII (cuts mutant) and run on a 1.5% agarose gel. All other ugt89c1 alleles were confirmed by sequencing.
Microscopic Analysis
Phenotypic screening for and analysis of ugt89c1 rol1-2 was performed using a binocular microscope. GFP fluorescence was photographed using a Leica DM6000 stereomicroscope. Gel prints of epidermal cells were produced following an established protocol (48) and observed by differential interference contrast microscopy using a Leica DMR microscope.
Flavonol Content Analysis
The analysis of the flavonol accumulation profile was done as described (44). Seedlings were grown in a vertical orientation for 6 days as described. One hundred intact seedlings were cut in the hypocotyl region, and roots and shoots were pooled separately, frozen in liquid nitrogen, and lyophilized to determine the dry weight. The dried material was incubated in 500 μl of 80% methanol overnight at 4 °C and subsequently macerated with a pestle, followed by vigorous vortexing. After pelleting the cell debris by centrifugation, the supernatant was transferred to a fresh tube and evaporated in a SpeedVac centrifuge, with the temperature being limited to a maximum of 42 °C. After evaporation, the pellet was resuspended in 100 μl of fresh 80% methanol and used for analysis. HPLC-ESI-MS and MS/MS experiments were performed on an ACQUITY UPLC (Waters) connected to a Bruker maXis high-resolution quadrupole time-of-flight mass spectrometer (Bruker Daltonics). An ACQUITY BEH C18 HPLC column (1.7 μm, 2.1 × 100 mm fitted with a 2 × 2-mm guard column) was used with a gradient of solvent A (H2O, 0.1% (v/v) HCOOH) and solvent B (CH3CN, 0.1% (v/v) HCOOH), at 0.45-ml flow rate and with a linear gradient from 5 to 95% B within 30 min.
The mass spectrometer was operated in the negative electrospray ionization mode. MS acquisitions were performed in the full scan mode in the mass range from m/z 50 to 2,000 at 25,000 resolution (full width at half-maximum) and two scans per second. The MS instrument was optimized for maximum signal intensities of quercitrine at m/z 447. Masses were calibrated with a 2 mm solution of sodium formate between m/z 180 and 1,472 prior to analysis. The lock mass signal of hexakis (1H,1H,2H-perfluoroethoxy)phosphazene at m/z 556.00195 was further used as lock mass during the HPLC run. Flavonols were identified by the molecular mass determined by MS and by interpretation of fragments obtained by MS/MS, by comparison of both fragmentation patterns and retention times with previous analyses (30), and by their absence in flavonol-less rol1-2 fls1-3 mutants (44). Purification of each flavonol glycoside in sufficient quantities for producing standard curves was not possible. Therefore, the content of each individual flavonol was expressed as the peak area of the corresponding high-resolution extracted ion chromatogram. The total flavonol content was calculated by summing up all flavonol peak areas, divided by the weight of plant material used for extraction. Either the total or the individual flavonol contents were compared between different experiments.
Auxin Transport Experiments and Quantification of Derivatives
Arabidopsis mesophyll protoplasts were prepared as described (49) from rosette leaves of plants grown on soil under 100 μm m−2 s−1 white light, 8-h light, 16-h dark cycle at 22 °C. Intact protoplasts were isolated as described (50) and loaded by incubation with 1-[4-3H]naphthalene acetic acid (25 Ci mmol−1; American Radiolabeled Chemicals) on ice. External radioactivity was removed by separating protoplasts using a 50–30-5% Percoll gradient. Samples were incubated at 25 °C, and efflux was halted by silicon oil centrifugation. Effluxed radioactivity was determined by scintillation counting of aqueous phases and is presented as the relative efflux of the initial efflux (efflux prior to incubation), which was set to zero. Efflux experiments were performed with 3–5 independent protoplast preparations with four replicas for each time point. Basipetal (shoot-ward) auxin transport was measured according to Ref. 51. Radioactive auxin ([3H]IAA) was applied to the root tips of 15 seedlings (n = 4), and 5–10-mm segments were cut after 18 h and counted. Segments in the wild-type Col were set as 100%, and others were calculated accordingly.
Measuring the concentration of auxin, auxin conjugates, and auxin degradation products was done as follows. The determination of IAA and its metabolites included extraction and purification (according to Ref. 52) followed by quantitation using LC-MS/MS (as described in Ref. 53). Briefly, a sample (about 100 mg of fresh weight) was homogenized in liquid nitrogen. The homogenate was supplied with 500 μl of cold extraction buffer (methanol/water/formic acid, 15/10/5, v/v/v, −20 °C) and with a mixture of stable isotope-labeled internal standards: [13C6]IAA (Cambridge Isotope Laboratories) and [2H5-15N1]IAA-Asp, [2H5-15N1]IAA-Glu, and [2H2]OxIAA (OlChemIm, Olomouc, Czech Republic); 10 pmol each. After incubation for 30 min at −20 °C, the extract was centrifuged at 28,000 × g (centrifuge Eppendorf 5430 R, Eppendorf AG, Hamburg, Germany), and pellet was re-extracted once. Pooled supernatants were evaporated in vacuum concentrator (Alpha RVC, Martin Christ, Osterode am Harz, Germany), and then a sample residue was dissolved into 0.1 m formic acid and applied to mixed mode reversed phase-cation exchange SPE column (Oasis-MCX, Waters). Auxin and its metabolites were eluted with methanol. Eluate was evaporated to dryness in a vacuum concentrator and dissolved into 30 μl of 15% acetonitrile. An aliquot was analyzed on an HPLC (Ultimate 3000, Dionex) coupled to a hybrid triple quadrupole/linear ion trap mass spectrometer (3200 Q TRAP, Applied Biosystems) set in selected reaction monitoring mode. Quantification of hormones was done using the isotope dilution method with multilevel calibration curves (r2 > 0.99). Data processing was carried out with Analyst 1.5 software (Applied Biosystems).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBankTM/European Molecular Biology Laboratory (EMBL) databases under the following accession numbers: FLS1, At5g08640; ROL1/RHM1, At1g78570; UGT89C1, At1g06000.
Results
Altered Flavonol Glycosylation Suppresses the rol1-2 Shoot Growth Defect
The rol1-2 shoot phenotype is characterized by hyponastic cotyledons, brick-shaped adaxial pavement cells of cotyledons, and deformed trichomes (Fig. 2A). Mutations in a glycosyltransferase involved in producing the rol1-2 phenotype-inducing flavonol(s) would be expected to revert the rol1-2 phenotype and act as suppressor(s) of the rol1-2 phenotype. Thus, to identify possible rol1-2 phenotype-inducing flavonols, rol1-2 seeds were mutagenized with ethyl methanesulfonate. After mutagenesis, the seeds were propagated, and seedlings of the M2 generation were screened for a wild-type shoot phenotype that is characterized by epinastic cotyledons with puzzle-shaped pavement cells and upright, branched trichomes (Fig. 2A). The identified lines were propagated and confirmed in the M3 generation. Positive lines were backcrossed with rol1-2, and only those with an F2 segregation pattern indicating recessive suppressor mutations were further analyzed. Several lines were identified that suppressed the rol1-2 phenotype, yet showed accumulation of flavonols to levels comparable with the wild type. Among these lines, only one group exhibited the lack of one specific type of flavonols, namely those rhamnosylated at the C7 position. Based on this finding, the previously identified flavonol 7-rhamnosyltransferase gene UGT89C1 (29) was identified as a potential candidate suppressor gene, and sequencing of the UGT89C1 locus At1g06000 indeed led to the identification of a mutation in this gene in all these mutants. In total, nine different ugt89c1-alleles were identified. Because two T-DNA insertion alleles of ugt89c1 were already described, the newly identified EMS alleles were numbered ugt89c1-3 to ugt89c1-11. In total, six missense and three nonsense mutations were identified (Table 1). The ugt89c1-3 and ugt89c1-6 alleles are affected in the same codon, but the resulting nonsense mutations originate from two different mutation events. Hence, the ugt89c1-3 and ugt89c1-6 lines represent two independent alleles with the same effect at the protein level. All mutant ugt89c1 alleles identified in this study lead to a comparable suppression of the rol1-2 shoot phenotype, resulting in seedlings with epinastic cotyledons, jigsaw puzzle-shaped pavement cells, and wild type-like trichomes (Fig. 2A).
FIGURE 2.
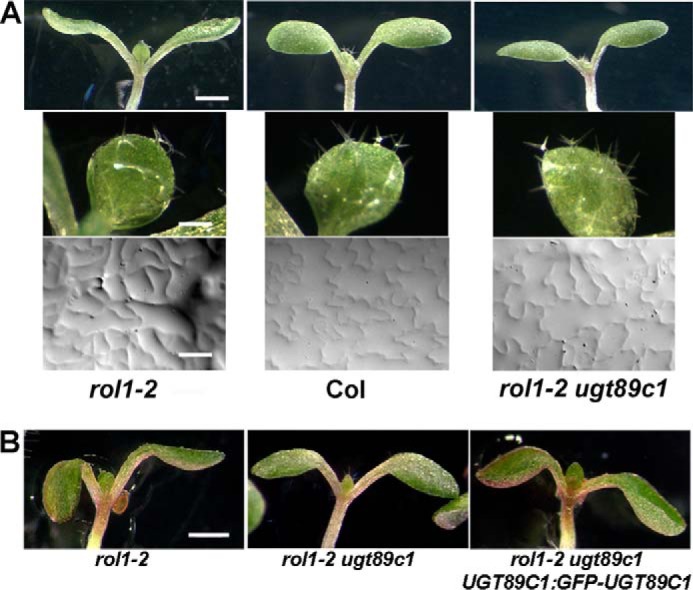
The rol1-2 mutant shoot cell growth phenotype is suppressed by mutations in UGT89C1. A, the rol1-2 mutant shows hyponastic bending of cotyledons, distorted trichomes, and brick-shaped adaxial pavement cells, whereas the wild type (Col) shows epinastic bending of cotyledons, normal shaped trichomes, and a jigsaw puzzle-like structure of adaxial pavement cells. All ugt89c1 alleles identified in this study suppress the rol1-2 shoot phenotype, i.e. rol1-2 ugt89c1 double mutants develop comparable with the wild type. Pictures of the rol1-2 ugt89c1-3 line are shown that are representative for all other ugt89c1 lines. B, the UGT89C1:GFP-UGT89C1 construct complements the ugt89c1 mutation, and hence encodes a functional UGT89C1 protein, resulting in a rol1-2 mutant phenotype. Bars = 1 mm for shoot (A, upper panels, and B); 0.2 mm for trichomes (A, middle panels); and 40 μm for pavement cells (A, lower panels).
TABLE 1.
List of ugt89c1 alleles identified in this screen
| Allele name | Mutation |
|---|---|
| ugt89c1-3 | Stop 314a |
| ugt89c1-4 | G123R |
| ugt89c1-5 | S19F |
| ugt89c1-6 | Stop 314a |
| ugt89c1-7 | G249V |
| ugt89c1-8 | Stop335 |
| ugt89c1-9 | D356N |
| ugt89c1-10 | T4I |
| ugt89c1-11 | R253W |
a The mutations producing the stop codon are not identical.
ugt89c1 Mutants Specifically Lack 7-Rhamnosylated Flavonols
A detailed analysis of the flavonol content of rol1-2 ugt89c1 mutants was performed by HPLC-MS. Individual flavonols were identified by comparing the data with previous studies where flavonols were identified by HPLC-MS/MS, correlation with reference compounds, and the absence in a flavonol-deficient flavonol synthase1 (fls1) mutant background (30, 44). For these and all subsequent analyses, the ugt89c1-3 nonsense allele was used. The quantification of flavonol accumulation using the sum of areas of all identified flavonol-peaks in HPLC-MS analyses confirmed that the overall flavonol content of the rol1-2 ugt89c1-3 line is comparable with the wild type or the rol1-2 mutant (Fig. 3A). As a negative control, the rol1-2 fls1-3 double mutant was included that shows a strongly reduced flavonol content due to a lesion in the flavonol synthase 1 gene (44). The quantification of individual flavonol peaks confirmed the absence of all identifiable 7-O-rhamnosylated flavonols in rol1-2 ugt89c1 mutants. Other flavonol glycoside derivatives were still present, some at levels higher than in the wild type or rol1-2 single mutant, as shown for the kaempferol-3-O-glucoside (Fig. 3B). Flavonols of root and shoot tissue are affected, confirming the ubiquitous importance of UGT89C1 for flavonol 7-O-rhamnosylation. These data indicate a lack of genetic redundancy in Arabidopsis with respect to 7-O-rhamnosylation of flavonols. Hence, UGT89C1 appears to be the only enzyme that is capable of catalyzing this reaction to a significant extent. The lack of 7-O-rhamnosylated flavonols in rol1-2 ugt89c1-3 suggests an important role of these compounds in the induction of the rol1-2 shoot phenotype.
FIGURE 3.
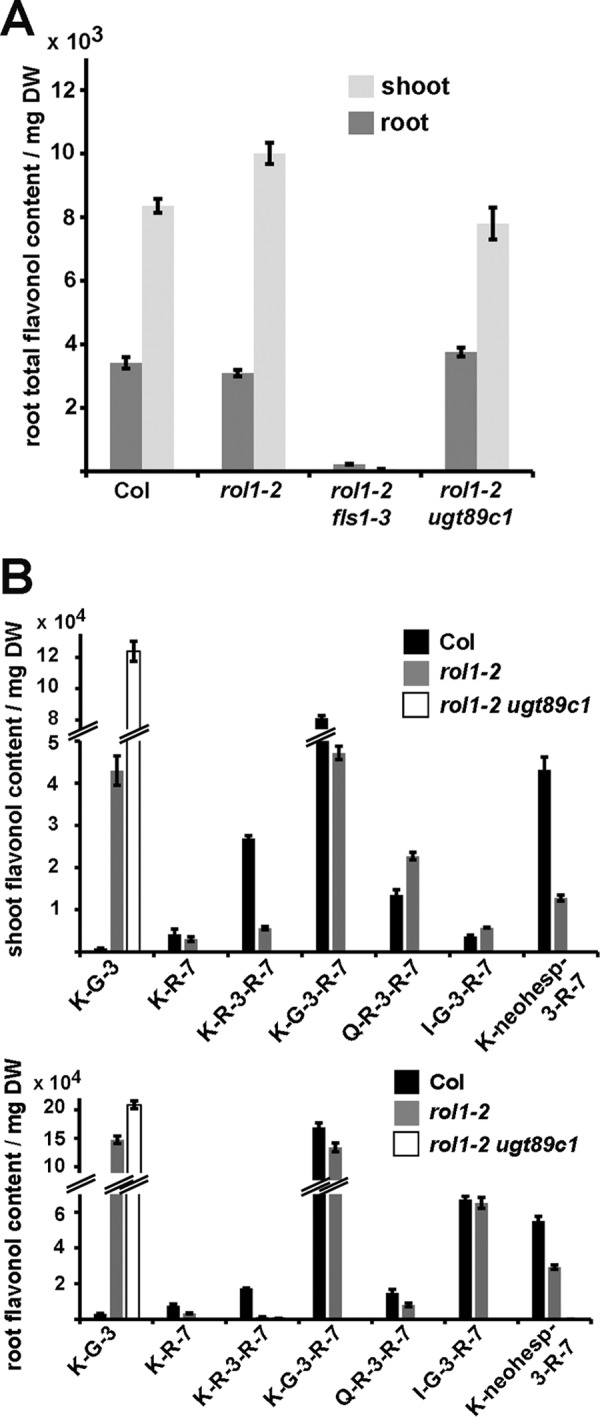
Content of total flavonols and of 7-O-rhamnosylated flavonols. A, total flavonol accumulation is comparable in the wild type, rol1-2, and rol1-2 ugt89c1-3. Values of rol1-2 fls1-3, which fail to accumulate flavonols, serve as negative control. DW, dry weight. B, the rol1-2 ugt89c1-3 line shows a loss of 7-rhamnosylated flavonols, whereas e.g. 3-O-glucosylated flavonols are still present and accumulate both in the shoot (upper graph) and in the root (lower graph). Only a sub-selection of flavonols is shown. For quantification, the area under each peak of the HPLC elution profile was used as an arbitrary unit. K-G-3, kaempferol-3-O-glucoside; K-R-7, kaempferol-7-O-rhamnoside; K-G-3-R-7, kaempferol-3-O-glucoside-7-O-rhamnoside; Q-R-3-R-7, quercetin-3-O-7-O-rhamnoside; I-G-3-R-7, isorhamnetin-3-O-glucoside-7-O-rhamnoside; K-neoh-3-R-7, kaempferol-3-O-neohesperidoside-7-O-rhamnoside. Error bars indicate mean ± S.E.
Different Protein Domains Are Affected in the ugt89c1 Alleles
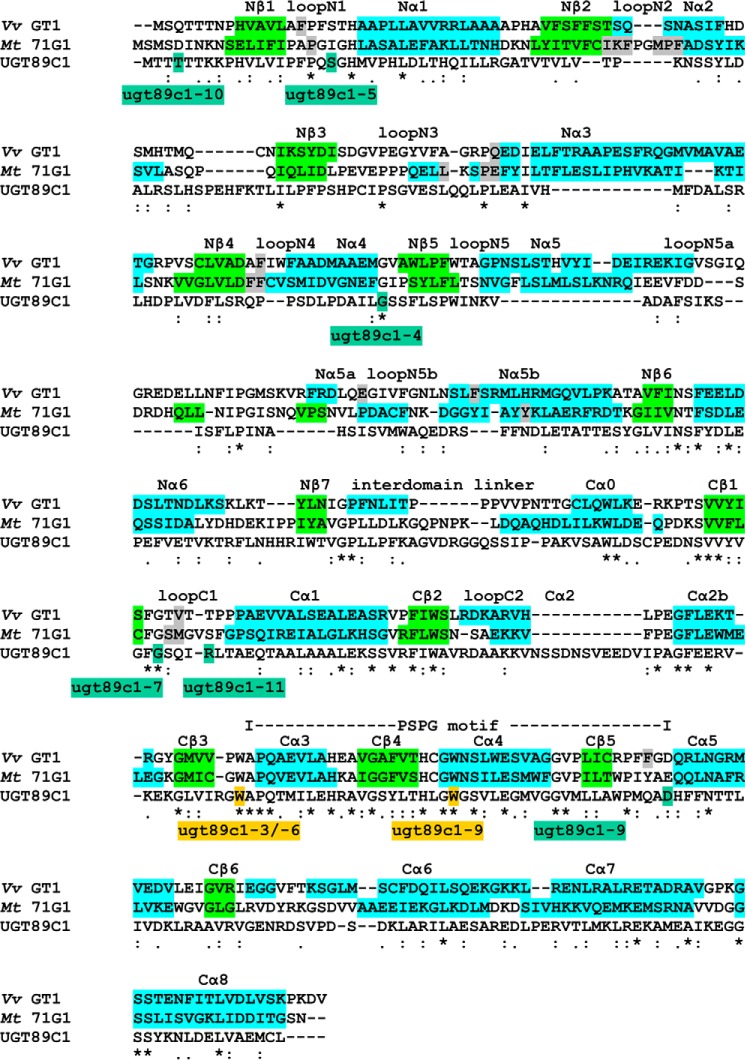
Analysis of the UGT89C1 protein sequence was chosen as a strategy to elucidate the effect of the different mutations on the secondary and tertiary protein structure. UGT89C1 was compared with the flavonol-specific UGTs VvGT1 from Vitis vinifera and Mt71G1 of Medicago truncatula whose secondary and tertiary structures are known (54, 55). VvGT1 and Mt71G1 accept flavonols as donor substrates and therefore were selected for sequence and structural comparison. Previous analysis of four plant UDP-dependent glycosyltransferases revealed a low overall conservation on the protein sequence level, but a high conservation on the level of secondary and tertiary protein structure (56). The alignment (Fig. 4) shows only one short region of 45 amino acids, the PSPG domain that is relatively well conserved and important for binding of the UDP-sugar substrate. All three nonsense alleles and the missense allele ugt89c1-9, introducing an D356N substitution, affect residues in this PSPG domain. Mutational analysis has shown that the Asp residue affected in ugt89c1-9 is essential for function of VvGT1 (55). Of the remaining five alleles, four alter the N1- or C1-loops (ugt89c1-5, -10, and ugt89c1-7, -11, respectively) that are involved in forming the sugar donor binding pocket (56). Altogether, protein sequence analysis revealed that a vast majority of mutations that revert the rol1-2 phenotype are directly related to sugar donor binding on the UGT89C1 enzyme.
FIGURE 4.
Structural alignment of primary sequences of different UGTs with UGT89C1. The crystal structure of VvGT1 (V. vinifera (Vv)) and Mt71G1 (Medicago truncatula (Mt)) was already solved, and different domains are marked with colors. α-Helices are highlighted in blue, and β-strands are in light green. The structurally important loops are labeled. Nonsense mutations identified in this screen are marked in beige, and missense mutations are in dark green. The dashed line indicates the PSPG motif important for UDP-sugar donor specificity. Residues reported to be involved in acceptor pocket formation are highlighted in gray. Asterisks mark identity, colons mark high similarities, single dots mark lower similarities, and blank positions indicate no similarities between the amino acids in the compared sequences. Adopted from Ref. 56.
UGT89C1 Expression Profile
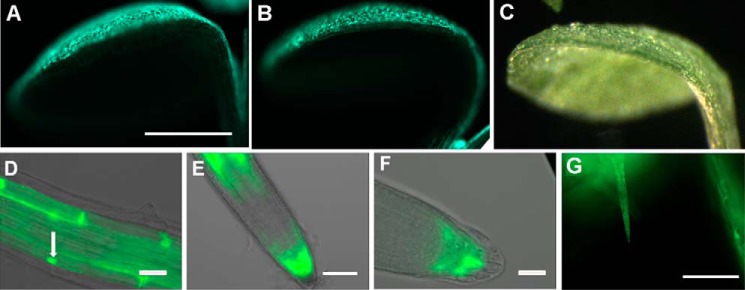
The rol1-2 cell growth phenotype in cotyledons is characterized by brick-shaped pavement cells on the adaxial side. Previous studies revealed that the flavonol synthase FLS1 is expressed asymmetrically on the adaxial side (44), providing an explanation for the limitation of the flavonol-related cell growth defect to this side of the cotyledons. To investigate the localization of UGT89C1, a GFP-UGT89C1 construct under the control of the UGT89C1 promoter was produced and transformed into Arabidopsis. Complementation of the rol1-2 ugt89c1-3 mutant was used to assess the impact of GFP on UGT89C1 protein activity. In the T2 generation, transgenic plants developed the typical rol1-2 shoot growth phenotype (Fig. 2B), confirming complementation and thus activity of the GPF-UGT89C1 fusion protein. To assess protein localization, the UGT89C1:GFP-UGT89C1 construct was transformed into wild-type Arabidopsis and GFP fluorescence was analyzed in seedlings of the T2 generation of several independent transgenic lines. GFP fluorescence could be detected on the adaxial side of cotyledons and emerging leaves (Fig. 5A, C), comparable with the previously reported pattern observed in FLS1:GFP-FLS1 (Fig. 5B) and RHM1:RHM1-GFP (RHM1 is identical to ROL1) transgenic lines (44). Trichomes also showed a strong GFP fluorescence (Fig. 5G). In roots, GFP fluorescence was predominant in columella cells and in the late elongation/early differentiation zone, correlating with zones of high auxin concentrations. Subcellularly, GFP-UGT89C1 localizes to the cytoplasm and the nucleus (Fig. 5, D–F). The expression pattern of GFP-UGT89C1 is similar to FLS1 (44) confirming the previous finding that UGT89C1 is co-expressed with other genes coding for enzymes of the flavonoid biosynthesis pathway (29).
FIGURE 5.
Localization of GFP-UGT89C1. A and B, in cotyledon of 6-day-old seedlings, fluorescence induced by UGT89C1:GFP-UGT89C1 (A) is restricted to the adaxial side, very similar to seedlings containing FLS1:FLS1-GFP (B). C, bright field image of a cotyledon corresponding to those shown in A and B. In roots, fluorescence of GFP-UGT89C1 was observed in the cortex and vascular cell layers of the differentiation zone. D, on the subcellular level, a fraction of the fusion protein localizes to the nucleus (arrow). E, in root tips, fluorescence occurred around the meristematic and later elongation zone. F, detailed analysis of root tips revealed GFP-UGT89C1 expression in columella and lateral root cap cells. G, GFP-UGT89C1 induced fluorescence in trichomes. Bars = 1 mm (A–C); 15 μm (D and F); 30 μm (E); and 100 μm (G).
Auxin Distribution but Not Auxin Transport Is Modified by the ugt89c1 Mutations
Flavonols have been shown to modify auxin-related processes (13, 14). rol1-2 mutant seedlings indeed show an altered auxin distribution and transport activity, as determined by auxin concentration measurements and transport assays (30, 44). To determine whether ugt89c1 functions as a suppressor of rol1-2 by influencing auxin distribution, auxin export was measured in the wild type, rol1-2, and rol1-2 ugt89c1 mutant lines. Due to the strong growth defect of rol1-2 mutant seedlings, which might interfere with interpretation of auxin transport in a whole-seedling assay, rosette leaf mesophyll protoplasts were used following an established protocol (44). In a first step, flavonols were extracted from wild-type, rol1-2, and rol1-2 ugt89c1 protoplasts to confirm that the mutations have an effect on flavonol accumulation in this cell type. Comparable with flavonols of seedlings, flavonols of rol1-2 protoplasts revealed a glycosylation profile that is distinct from wild-type protoplasts in that they accumulate fewer rhamnosylated and more glucosylated flavonols (Fig. 6A) (30). rol1-2 ugt89c1 protoplasts did not accumulate 7-O-rhamnosylated flavonols. Hence, protoplasts of the different genotypes showed the expected flavonol glycosylation profile, making them suitable to investigate the effect of the two mutations on auxin transport. Auxin export rates were determined in protoplasts using the synthetic auxin, 1-NAA, and revealed that the rol1-2 mutant shows a modified transport efficiency as compared with the wild type. rol1-2 ugt89c1-3 double mutant protoplasts, however, show a transport rate that is not different from the one of rol1-2 (Fig. 6B). Hence, mutations in ugt89c1 suppress the rol1-2 phenotype by an auxin transport-independent manner. This is particularly interesting because the flavonol-less rol1-2 fls1 mutant suppresses rol1-2 by changing auxin transport back to wild-type levels (44). Hence, the mode of action of fls1 and ugt89c1 is different. To sustain cellular auxin transport data and to back up measurements performed with synthetic auxin 1-NAA, we measured basipetal (shoot-ward) IAA transport in roots. Comparable with 1-NAA transport in protoplasts, root basipetal transport of IAA was revealed to be changed in rol1-2 as compared with the wild type. The rol1-2 ugt89c1 double mutant shows transport activity similar to rol1-2, again indicating that ugt89c1 does not modify the rol1-2-induced alteration in auxin transport (Fig. 6C). To investigate whether ugt89c1 changes auxin levels in a different way, concentrations of free auxin and auxin-related metabolites were determined in wild type, rol1-2, and rol1-2 ugt89c1 mutant seedlings. Here, two additional lines were included: the ugt89c1 single mutant to confirm effects found for rol1-2 ugt89c1; and the rol1-2 fls1-3 double mutant to obtain indications of whether ugt89c1 and fls1-3 indeed suppress rol1-2 by different means. The levels of the prevailing auxin of Arabidopsis, IAA, was subject to variation between several biologically independent experiments. As a consequence, the increase in auxin concentration in rol1-2 as compared with the wild type was not as pronounced as reported previously (30) and did not fulfill the criteria of statistical significance (t test, p < 0.05). The reason for the observed variation is not clear at this point. Quantification of the auxin precursor indole-3-acetonitrile (IAN) and auxin conjugates and degradation products, on the other hand, gave very consistent results: ugt89c1 shows increased accumulation of IAN as well as of auxin conjugates and degradation intermediates such as IAA-Glc or oxIAA-Glc (Fig. 7). This effect of ugt89c1 is observed not only in the context of the rol1-2 mutation that causes aberrant accumulation of flavonols, but also in the ugt89c1 single mutant as compared with the wild type, suggesting that the accumulation of auxin precursor and derivatives is genuinely affected by flavonol glycosides.
FIGURE 6.
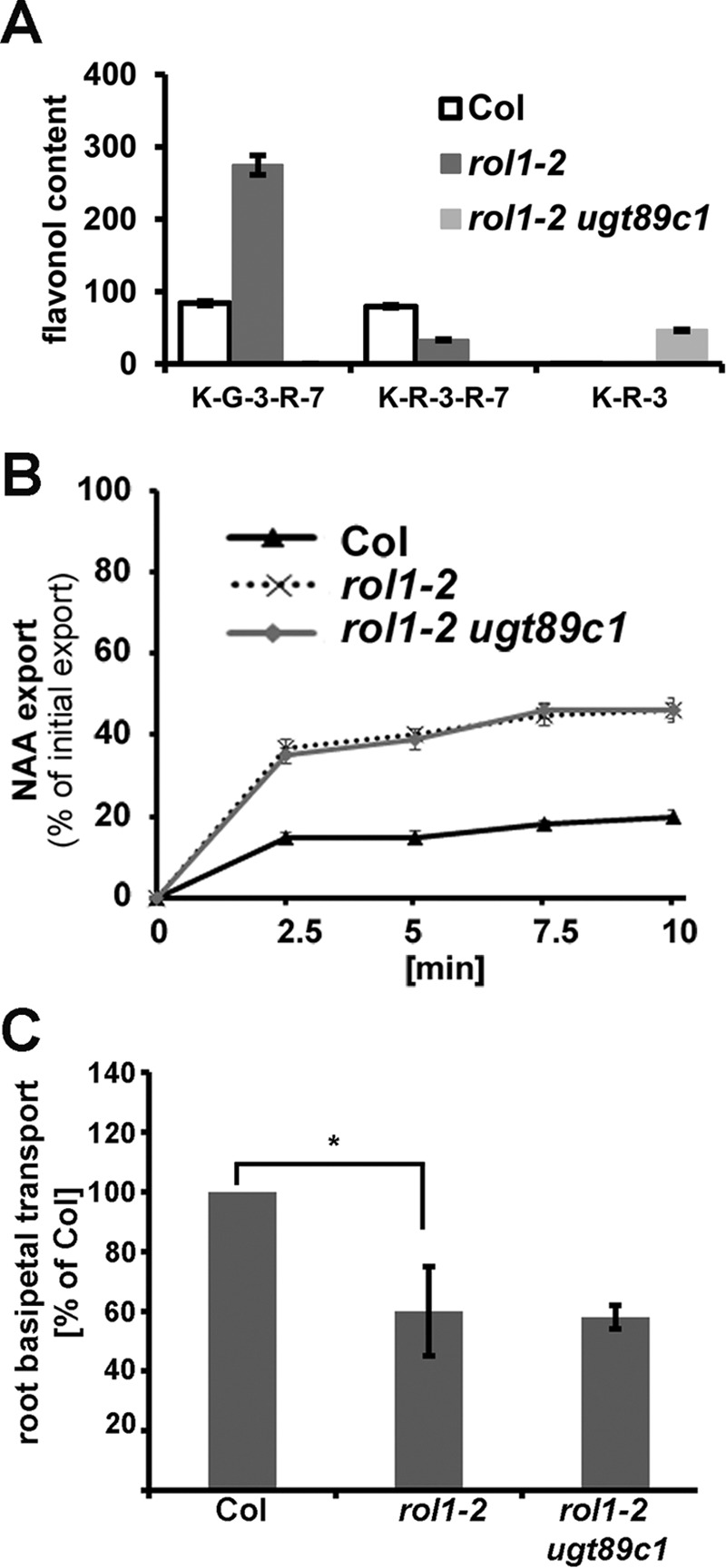
Auxin transport in rol1-2 plants is not affected by ugt89c1. A, flavonol content analysis of protoplasts revealed the same effects of rol1-2 and ugt89c1 mutations as in intact plants. The area under each peak of the HPLC elution profile was used to determine flavonol content. K-G-3-R-7, kaempferol-3-O-glucoside-7-O-rhamnoside; K-R-3, kaempferol-3-O-rhamnoside. B, auxin (1-NAA) export from protoplasts is affected in rol1-2 as compared with the wild type (Col) but not altered by the ugt89c1 mutation. C, as compared with the wild type (Col), rol1-2 and rol1-2 ugt89c1 show similar, reduced basipetal IAA transport in roots. Asterisks indicate statistically significant differences (t test, p < 0.05). Error bars indicate mean ± S.E.
FIGURE 7.
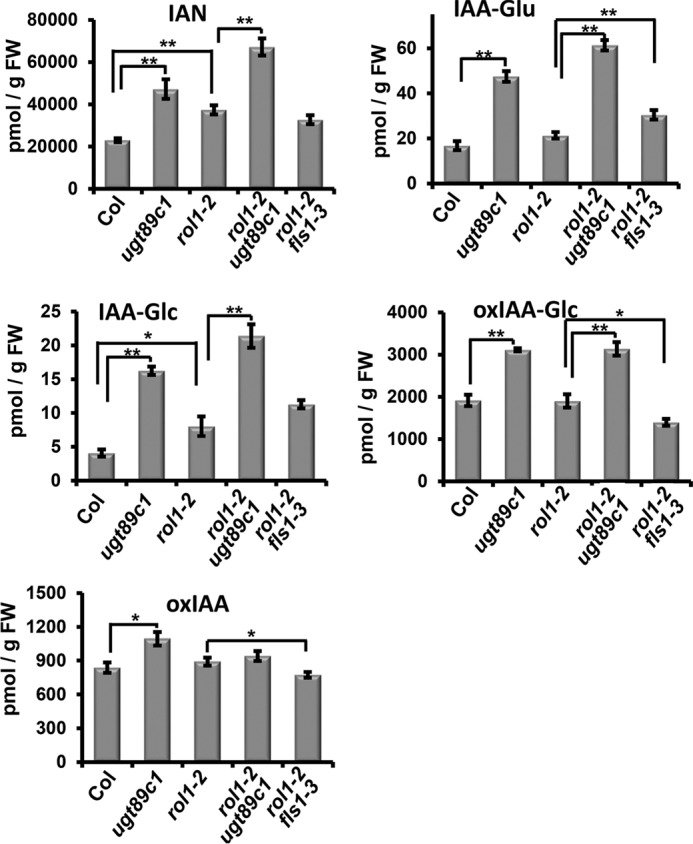
ugt89c1 affects levels of auxin precursor and metabolites. Quantification of the auxin conjugates IAA-Glc and IAA-Glu, the auxin precursor IAN, and the degradation products oxIAA and oxIAA-Glc is shown for wild type (Col), rol1-2, and ugt89c1 single mutants, and for rol1-2 ugt89c1 and rol1-2 fls1-3 double mutants. FW, fresh weight. Statistically significant differences (t test) with p < 0.05 and p < 0.01 are indicated with one and two asterisks, respectively. Error bars indicate mean ± S.E.
Discussion
Flavonoids are a group of phenylpropanoic secondary metabolites known to influence cellular processes. They are of interest as nutrition additives due to their health-promoting effect (57). However, the knowledge of the effects of flavonoids on cellular processes in either animals or plants is still limited. The Arabidopsis rol1-2 shoot phenotype, characterized by defects in cell growth and development, is induced by flavonols, a subgroup of flavonoids (30, 44), and thus represents a model to characterize the active flavonol(s) and their mode of action.
Flavonol Glycosides Influence Plant Development
The study presented here identifies 7-O-rhamnosylated flavonols as being important for inducing the growth defects in rol1-2. The enzyme UGT89C1, conjugating rhamnose onto flavonols (29), is encoded by a single gene in Arabidopsis, and mutations in UGT89C1 were identified as suppressors of the rol1-2 phenotype. In seedling shoots, UGT89C1 expression is limited to the adaxial side of cotyledons, emerging leaves, and trichomes. The same protein accumulation pattern has been found for ROL1/RHM1 and FLS1 (44), which explains the asymmetry in the flavonol-induced growth phenotype that is limited in cotyledons to the adaxial side. The co-localization is also in line with previous findings that genes involved in flavonoid biosynthesis (including FLS1, RHM1, and UGT89C1) are co-regulated (26). The data suggest that flavonol glycosides are the active flavonol species and that 7-O-rhamnosylated flavonols are involved in inducing the rol1-2 phenotype. Evidence for an active role of glycosylated flavonols in plant development was also provided by the analysis of the 3-O-glucosyltransferase mutant ugt78d2, which shows a dwarfed growth phenotype. This mutant contains increased levels of 3-O-7-O-rhamnosylated flavonols, and mutating either the 3-O-rhamnosyltransferases or the 7-O-rhamnosyltransferases ugt78d1 or ugt89c1, respectively, suppresses the dwarfed phenotype of ugt78d2. Hence, 3-O-7-O-rhamnosylated flavonols appear to induce the growth defects developed by the ugt78d2 mutant (31). Both the ugt78d2 and the rol1-2 mutant phenotypes are also found in combination with the tt7 mutation (30, 31) that prevents synthesis of quercetin and causes over-accumulation of kaempferol (Fig. 1). Thus, accumulation of the flavonol kaempferol appears to be sufficient to interfere with proper plant development. Similarly, the tt7 single mutant was reported to have increased inhibition of auxin transport, again suggesting that kaempferol and its glycosylated forms are particularly active flavonols (13). The mechanism of flavonol-induced growth defects, however, is not the same in rol1-2 and ugt72d2. First, in contrast to ugt78d2, there is a reduction of 3-O-7-O-rhamnosylated flavonols in rol1-2 as compared with the wild type (30). Second, although introducing the ugt89c1 mutation suppresses both ugt78d2 and rol1-2, the introduction of the 3-O-rhamnosyl transferase mutation ugt78d1 only suppresses ugt72d2 but not rol1-2 (30, 31). This raises the question as to how suppression of the ugt78d2 and rol1-2 mutant phenotypes is induced. Although the mechanism of suppression of the ugt78d2 mutant by ugt78d1 and ugt89c1 has not yet been investigated (31), the work presented here suggests that ugt89c1 suppresses the rol1-2 phenotype by modulating levels of the auxin precursor IAN and auxin metabolites rather than auxin transport.
Flavonols Influence Auxin Homeostasis in Several Ways
The suppression of rol1-2 by tt4, tt6, and fls1 (30, 44) fit the picture of flavonols being negative regulators of auxin transport (11). Indeed, interfering with flavonol biosynthesis in rol1-2 by an fls1 mutation reverts the modified auxin transport to wild-type levels and suppresses the rol1-2 phenotype. In contrast to the fls1 mutation, however, ugt89c1 does not seem to interfere with auxin transport. As compared with the wild type, the rol1-2 mutant shows an increase in auxin efflux in the protoplast assay but a decrease in auxin transport in the root basipetal transport. This discrepancy can be explained by the very different experimental setup in the assays. In the protoplast assay, primarily auxin efflux is measured from a non-polarized, cellular system after loading with radioactive auxin. In contrast, the root basipetal transport assay is performed on the entire root, so that the resulting value comes from the complex auxin transport machinery present in the root tissues. However, and importantly, the absence of any effect of the additional ugt89c1 mutation is found in both systems. This unexpected finding can be explained by different flavonol species having different functions in terms of regulating auxin levels. Although some interfere with auxin transport, others seem to influence auxin metabolism. Kaempferols are important for the development of the ugt78d2 and rol1-2 phenotypes or for pollen development in maize (6, 30, 31), whereas other studies point at quercetin as an important flavonol (58, 59). In the rol1-2 fls1 double mutant, all flavonols are absent, resulting in wild type-like auxin transport, whereas a defect in 7-O-rhamnosylation only interferes with auxin metabolism but not transport. An indication for multiple modes of action of flavonols was also obtained from analyzing the rol1-2 shoot phenotype in more detail. In seedlings, a correlation between increased auxin levels and growth defects was observed. However, phenocopying the increased auxin levels in wild-type seedlings by exposing them to the auxin efflux inhibitor 1-naphthylphthalamic acid resulted in hyponastic cotyledons comparable with the rol1-2 shoot phenotype, whereas the defects in pavement cell shape and trichome development were not observed (30). Hence, negatively regulating auxin transport is likely not the only way by which flavonols in the rol1-2 mutant interfere with plant development. The results presented here suggest that UGT89C1 influences the activity of auxin biosynthesis and its conjugation/degradation. Although oxIAA and oxIAA-Glc are irreversible degradation intermediates, IAA-amino acid derivatives can mostly be converted back to active, free auxin (32–34, 60). Interfering with the activity of amidohydrolases that convert IAA-amino acid conjugates to free IAA affects free auxin levels and induces auxin-related phenotypes (61, 62). A mutation in ugt89c1 causes an increase in the level of auxin precursor and auxin metabolites, the only exception being oxIAA accumulation in rol1-2 ugt89c1 that is comparable with rol1-2. Both low-abundant (such as IAA-Glu or IAA-Glc) as well as highly abundant (such as the auxin precursor IAN or the auxin degradation product oxIAA-Glc) auxin derivatives are affected to a similar extent. However, the low-abundant IAA derivatives are reversible conjugates that contribute to the pool of all IAA metabolites only very little (<1%). The accumulation of the precursor IAN in the ugt89c1 and ugt89c1 rol1-2 mutants suggests that conversion of this molecule to free IAA is inhibited. On the other hand, according to the levels of oxIAA-Glc, irreversible oxidative degradation is increased in those mutants. Thus, UGT89C1 seems to influence the production as well as the degradation of auxin. Considering the large amount of the precursor IAN in the entire pool of auxin-related metabolites, the regulation of the last step of auxin biosynthesis appears to be a main target process of the activity of UGT89C1. At this point, it is not clear why these changes in the pool of auxin and its derivatives are not reflected in a clear change in the level of free auxin. Several repetitions of auxin measurements gave somewhat varying results with an increase in rol1-2 as compared with the wild type, whereas rol1-2 ugt89c1 reduced auxin levels again. However, the differences were not severalfold, did not fulfill the criteria of statistical significance, and therefore cannot be considered in the interpretation. Such variations may reflect fine-tuning of levels of free auxin as the only auxin-active form in relation to subtle variations in developmental stages of plant material used to analyze auxin content in biologically independent samples. It can be assumed that the level of free auxin is under strict control and may be modified only temporarily and locally by control mechanisms that are functional in the absence of UGT89C1. Blocking flavonol biosynthesis in the rol1-2 fls1-3 mutant has comparatively little effect on the auxin derivatives. In three out of five auxin-related compounds, the changes induced by fls1-3 are the opposite of those induced by ugt89c1, i.e. oxIAA-Glc is reduced in rol1-2 fls1-3 as compared with rol1-2, but increased in rol1-2 ugt89c1. These data indicate again that changes in the flavonol glycosylation pattern do not have the same effect on auxin levels as blocking altogether flavonol synthesis. Presumably, the balance between different glycosylated forms of flavonols including 7-O-rhamnosylated flavonols is critical for auxin homeostasis, and thus, for proper plant development. The mechanism by which flavonols influence auxin conjugation and catabolism remains to be uncovered. The level of oxIAA has been shown to be increased in the flavonol-deficient tt4 mutant and reduced in the flavonol over-accumulator tt3, an observation that is attributed to the reactive oxygen species-scavenging activity of flavonols (9, 42). However, mutations in ugt89c1 do not affect the overall accumulation of flavonols. This suggests that different flavonol glycosides might have different reactive oxygen species-scavenging activities, possibly because of distinct subcellular localization. Alternatively, ugt89c1 mutations might not influence auxin oxidation via modulated reactive oxygen species-scavenging activity. It has been shown that changing flavonol glycosylation affects gene expression (8), and thereby, genes involved in auxin metabolism might also be affected. Formally, it cannot be excluded that UGT89C1 has another biochemical activity in addition to its function as a rhamnosyltransferase. However, UGT89C1 appears to recruit specifically Rha as the substrate and flavonols as acceptor for the glycosyltransferase reaction, and its gene expression is tightly linked with other genes involved in flavonol biosynthesis (26, 29), making such a scenario less likely.
Together, the findings of this study reveal the effect of flavonol glycosides on auxin metabolic turnover. In addition to the known effect of flavonols on auxin transport, this represents a new mechanism for flavonols to influence auxin homeostasis.
Author Contributions
B. M. K., S. E., R. B., P. D., and M. G. performed experiments; L. B., E. Z., and C. R. designed experiments; and C. R. designed the project and wrote the paper.
This work was supported the Swiss National Science Foundation (Grants 122157 and 138472) and by the Forschungskredit of the University of Zürich (to B. M. K., S. E., and C. R.); by the Czech Science Foundation (Grant P305/11/0797) (to P. D. and E. Z.); and the Swiss National Science Foundation (Grant 31003A_144223) and the Novartis foundation (to M. G.). The authors declare that they have no conflicts of interest with the contents of this article.
- UGT
- UDP-dependent glycosyltransferases
- K-R-3-R-7
- kaempferol-3-O-7-O-rhamnoside
- IAA
- indole-3-acetic acid
- IAA-Glc
- IAA-glucose
- IAA-Asp
- IAA-aspartate
- IAA-Glu
- IAA-glutamate
- oxIAA
- 2-oxindole-3-acetic acid
- oxIAA-Glc
- oxIAA-glucose
- IAN
- indole-3-acetonitrile
- EMS
- ethyl methanesulfonate
- ESI
- electrospray ionization
- 1-NAA
- 1-naphthalene acetic acid
- FLS
- flavonol synthase.
References
- 1. Lepiniec L., Debeaujon I., Routaboul J. M., Baudry A., Pourcel L., Nesi N., and Caboche M. (2006) Genetics and biochemistry of seed flavonoids. Annu. Rev. Plant Biol. 57, 405–430 [DOI] [PubMed] [Google Scholar]
- 2. Costa D., Galvao A. M., Di Paolo R. E., Freitas A. A., Lima J. C., Quina F. H., and Macanita A. L. (2015) Photochemistry of the hemiketal form of anthocyanins and its potential role in plant protection from UV-B radiation. Tetrahedron 71, 3157–3162 [Google Scholar]
- 3. Zhang J., Subramanian S., Stacey G., and Yu O. (2009) Flavones and flavonols play distinct critical roles during nodulation of Medicago truncatula by Sinorhizobium meliloti. Plant J. 57, 171–183 [DOI] [PubMed] [Google Scholar]
- 4. Brown J. E., Khodr H., Hider R. C., and Rice-Evans C. A. (1998) Structural dependence of flavonoid interactions with Cu2+ ions: implications for their antioxidant properties. Biochem. J. 330, 1173–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taylor L. P., and Grotewold E. (2005) Flavonoids as developmental regulators. Curr. Opin. Plant Biol. 8, 317–323 [DOI] [PubMed] [Google Scholar]
- 6. Mo Y. Y., Nagel C., and Taylor L. P. (1992) Biochemical complementation of chalcone synthase mutants defines a role for flavonols in functional pollen. Proc. Natl. Acad. Sci. U.S.A. 89, 7213–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Peer W. A., and Murphy A. S. (2006) Flavonoids as signal molecules. in The Science of Flavonoids (Grotewold E. ed), pp. 239–268, Springer-Verlag, Berlin [Google Scholar]
- 8. Yin R., Messner B., Faus-Kessler T., Hoffmann T., Schwab W., Hajirezaei M.-R., von Saint Paul V., Heller W., and Schäffner A. R. (2012) Feedback inhibition of the general phenylpropanoid and flavonol biosynthetic pathways upon a compromised flavonol-3-O-glycosylation. J. Exp. Bot. 63, 2465–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nakabayashi R., Yonekura-Sakakibara K., Urano K., Suzuki M., Yamada Y., Nishizawa T., Matsuda F., Kojima M., Sakakibara H., Shinozaki K., Michael A. J., Tohge T., Yamazaki M., and Saito K. (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 77, 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jacobs M., and Rubery P. H. (1988) Naturally-occurring auxin transport regulators. Science 241, 346–349 [DOI] [PubMed] [Google Scholar]
- 11. Buer C. S., and Muday G. K. (2004) The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 16, 1191–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brown D. E., Rashotte A. M., Murphy A. S., Normanly J., Tague B. W., Peer W. A., Taiz L., and Muday G. K. (2001) Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 126, 524–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Peer W. A., Bandyopadhyay A., Blakeslee J. J., Makam S. N., Chen R. J., Masson P. H., and Murphy A. S. (2004) Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell 16, 1898–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peer W. A., Blakeslee J. J., Yang H., and Murphy A. S. (2011) Seven things we think we know about auxin transport. Mol. Plant 4, 487–504 [DOI] [PubMed] [Google Scholar]
- 15. Zažímalová E., Murphy A. S., Yang H., Hoyerová K., and Hosek P. (2010) Auxin transporters: why so many? Cold Spring Harb. Perspect. Biol. 2, a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Remmer J., and Murphy A. S. (2014) Intercellular transport of auxin. in Auxin and its role in plant development (Zažímalová E., Petrášek J., and Benková E. eds.), pp. 75–100, Springer, Vienna [Google Scholar]
- 17. Bouchard R., Bailly A., Blakeslee J. J., Oehring S. C., Vincenzetti V., Lee O. R., Paponov I., Palme K., Mancuso S., Murphy A. S., Schulz B., and Geisler M. (2006) Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J. Biol. Chem. 281, 30603–30612 [DOI] [PubMed] [Google Scholar]
- 18. Bailly A., Sovero V., Vincenzetti V., Santelia D., Bartnik D., Koenig B. W., Mancuso S., Martinoia E., and Geisler M. (2008) Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J. Biol. Chem. 283, 21817–21826 [DOI] [PubMed] [Google Scholar]
- 19. Michniewicz M., Zago M. K., Abas L., Weijers D., Schweighofer A., Meskiene I., Heisler M. G., Ohno C., Zhang J., Huang F., Schwab R., Weigel D., Meyerowitz E. M., Luschnig C., Offringa R., and Friml J. (2007) Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell 130, 1044–1056 [DOI] [PubMed] [Google Scholar]
- 20. Henrichs S., Wang B., Fukao Y., Zhu J., Charrier L., Bailly A., Oehring S. C., Linnert M., Weiwad M., Endler A., Nanni P., Pollmann S., Mancuso S., Schulz A., and Geisler M. (2012) Regulation of ABCB1/PGP1-catalysed auxin transport by linker phosphorylation. EMBO J. 31, 2965–2980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen R., Hilson P., Sedbrook J., Rosen E., Caspar T., and Masson P. H. (1998) The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Proc. Natl. Acad. Sci. U.S.A. 95, 15112–15117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luschnig C., Gaxiola R. A., Grisafi P., and Fink G. R. (1998) EIR1, a root-specific protein involved in auxin transport, is required for gravitropism in Arabidopsis thaliana. Genes Dev. 12, 2175–2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Santelia D., Henrichs S., Vincenzetti V., Sauer M., Bigler L., Klein M., Bailly A., Lee Y., Friml J., Geisler M., and Martinoia E. (2008) Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. J. Biol. Chem. 283, 31218–31226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Koornneef M. (1990) Mutations affecting the testa color in Arabidopsis. Arab. Inf. Serv. 19, 113–115 [Google Scholar]
- 25. Routaboul J. M., Kerhoas L., Debeaujon I., Pourcel L., Caboche M., Einhorn J., and Lepiniec L. (2006) Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 224, 96–107 [DOI] [PubMed] [Google Scholar]
- 26. Yonekura-Sakakibara K., Tohge T., Matsuda F., Nakabayashi R., Takayama H., Niida R., Watanabe-Takahashi A., Inoue E., and Saito K. (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20, 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jones P., Messner B., Nakajima J., Schäffner A. R., and Saito K. (2003) UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. J. Biol. Chem. 278, 43910–43918 [DOI] [PubMed] [Google Scholar]
- 28. Tohge T., Nishiyama Y., Hirai M. Y., Yano M., Nakajima J., Awazuhara M., Inoue E., Takahashi H., Goodenowe D. B., Kitayama M., Noji M., Yamazaki M., and Saito K. (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J. 42, 218–235 [DOI] [PubMed] [Google Scholar]
- 29. Yonekura-Sakakibara K., Tohge T., Niida R., and Saito K. (2007) Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 282, 14932–14941 [DOI] [PubMed] [Google Scholar]
- 30. Ringli C., Bigler L., Kuhn B. M., Leiber R. M., Diet A., Santelia D., Frey B., Pollmann S., and Klein M. (2008) The modified flavonol glycosylation profile in the Arabidopsis rol1 mutants results in alterations in plant growth and cell shape formation. Plant Cell 20, 1470–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yin R., Han K., Heller W., Albert A., Dobrev P. I., Zažímalová E., and Schäffner A. R. (2014) Kaempferol 3-O-rhamnoside-7-O-rhamnoside is an endogenous flavonol inhibitor of polar auxin transport in Arabidopsis shoots. New Phytol. 201, 466–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ostin A., Kowalyczk M., Bhalerao R. P., and Sandberg G. (1998) Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol. 118, 285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tam Y. Y., Epstein E., and Normanly J. (2000) Characterization of auxin conjugates in Arabidopsis: low steady-state levels of indole-9-acetyl-aspartate indole-3-acetyl-glutamate, and indole-3-acetyl-glucose. Plant Physiol. 123, 589–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Korasick D. A., Enders T. A., and Strader L. C. (2013) Auxin biosynthesis and storage forms. J. Exp. Bot. 64, 2541–2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Novák O., Hényková E., Sairanen I., Kowalczyk M., Pospíšil T., and Ljung K. (2012) Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 72, 523–536 [DOI] [PubMed] [Google Scholar]
- 36. Kai K., Horita J., Wakasa K., and Miyagawa H. (2007) Three oxidative metabolites of indole-3-acetic acid from Arabidopsis thaliana. Phytochemistry 68, 1651–1663 [DOI] [PubMed] [Google Scholar]
- 37. Pencík A., Simonovik B., Petersson S. V., Henyková E., Simon S., Greenham K., Zhang Y., Kowalczyk M., Estelle M., Zažímalová E., Novák O., Sandberg G., and Ljung K. (2013) Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the indole-3-acetic acid catabolite 2-oxindole-3-acetic acid. Plant Cell 25, 3858–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Benjamins R., and Scheres B. (2008) Auxin: the looping star in plant development. Annu. Rev. Plant Biol. 59, 443–465 [DOI] [PubMed] [Google Scholar]
- 39. Vanneste S., and Friml J. (2009) Auxin: a trigger for change in plant development. Cell 136, 1005–1016 [DOI] [PubMed] [Google Scholar]
- 40. Habets M. E. J., and Offringa R. (2014) PIN-driven polar auxin transport in plant developmental plasticity: a key target for environmental and endogenous signals. New Phytol. 203, 362–377 [DOI] [PubMed] [Google Scholar]
- 41. Runions A., Smith R., and Prusinkiewicz P. (2014) Computational models of auxin-driven development. in Auxin and its role in plant development (Zažímalová E., Petrášek J., and Benková E. eds.), Springer, Vienna: pp 315–357 [Google Scholar]
- 42. Peer W. A., Cheng Y., and Murphy A. S. (2013) Evidence of oxidative attenuation of auxin signalling. J. Exp. Bot. 64, 2629–2639 [DOI] [PubMed] [Google Scholar]
- 43. Diet A., Link B., Seifert G. J., Schellenberg B., Wagner U., Pauly M., Reiter W. D., and Ringli C. (2006) The Arabidopsis root hair cell wall formation mutant lrx1 is suppressed by mutations in the RHM1 gene encoding a UDP-l-rhamnose synthase. Plant Cell 18, 1630–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kuhn B. M., Geisler M., Bigler L., and Ringli C. (2011) Flavonols accumulate asymmetrically and affect auxin transport in Arabidopsis. Plant Physiol. 156, 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Reiter W. D., and Vanzin G. F. (2001) Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant. Mol. Biol. 47, 95–113 [PubMed] [Google Scholar]
- 46. Leiber R.-M., John F., Verhertbruggen Y., Diet A., Knox J. P., and Ringli C. (2010) The TOR pathway modulates the structure of cell walls in Arabidopsis. Plant Cell 22, 1898–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Stintzi A., and Browse J. (2000) The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. U.S.A. 97, 10625–10630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Horiguchi G., Fujikura U., Ferjani A., Ishikawa N., and Tsukaya H. (2006) Large-scale histological analysis of leaf mutants using two simple leaf observation methods: identification of novel genetic pathways governing the size and shape of leaves. Plant J. 48, 638–644 [DOI] [PubMed] [Google Scholar]
- 49. Geisler M., Blakeslee J. J., Bouchard R., Lee O. R., Vincenzetti V., Bandyopadhyay A., Titapiwatanakun B., Peer W. A., Bailly A., Richards E. L., Ejenda K. F. K., Smith A. P., Baroux C., Grossniklaus U., Müller A., Hrycyna C. A., Dudler R., Murphy A. S., and Martinoia E. (2005) Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 44, 179–194 [DOI] [PubMed] [Google Scholar]
- 50. Geisler M., Kolukisaoglu H. U., Bouchard R., Billion K., Berger J., Saal B., Frangne N., Koncz-Kalman Z., Koncz C., Dudler R., Blakeslee J. J., Murphy A. S., Martinoia E., and Schulz B. (2003) TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Biol. Cell 14, 4238–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lewis D. R., and Muday G. K. (2009) Measurement of auxin transport in Arabidopsis thaliana. Nat. Protoc. 4, 437–451 [DOI] [PubMed] [Google Scholar]
- 52. Dobrev P. I., and Kamínek M. (2002) Fast and efficient separation of cytokinins from auxin and abscisic acid and their purification using mixed-mode solid-phase extraction. J. Chromatogr. A 950, 21–29 [DOI] [PubMed] [Google Scholar]
- 53. Dobrev P. I., and Vankova R. (2012) Quantification of abscisic acid, cytokinin, and auxin content in salt-stressed plant tissues. Methods Mol. Biol. 913, 251–261 [DOI] [PubMed] [Google Scholar]
- 54. Shao H., He X., Achnine L., Blount J. W., Dixon R. A., and Wang X. (2005) Crystal structures of a multifunctional triterpene/flavonoid glycosyltransferase from Medicago truncatula. Plant Cell 17, 3141–3154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Offen W., Martinez-Fleites C., Yang M., Kiat-Lim E., Davis B. G., Tarling C. A., Ford C. M., Bowles D. J., and Davies G. J. (2006) Structure of a flavonoid glucosyltransferase reveals the basis for plant natural product modification. EMBO J. 25, 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Osmani S. A., Bak S., and Møller B. L. (2009) Substrate specificity of plant UDP-dependent glycosyltransferases predicted from crystal structures and homology modeling. Phytochemistry 70, 325–347 [DOI] [PubMed] [Google Scholar]
- 57. Butelli E., Titta L., Giorgio M., Mock H.-P., Matros A., Peterek S., Schijlen E. G. W. M., Hall R. D., Bovy A. G., Luo J., and Martin C. (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308 [DOI] [PubMed] [Google Scholar]
- 58. Buer C. S., Kordbacheh F., Truong T. T., Hocart C. H., and Djordjevic M. A. (2013) Alteration of flavonoid accumulation patterns in transparent testa mutants disturbs auxin transport, gravity responses, and imparts long-term effects on root and shoot architecture. Planta 238, 171–189 [DOI] [PubMed] [Google Scholar]
- 59. Lewis D. R., Ramirez M. V., Miller N. D., Vallabhaneni P., Ray W. K., Helm R. F., Winkel B. S. J., and Muday G. K. (2011) Auxin and ethylene induce flavonol accumulation through distinct transcriptional networks. Plant Physiol. 156, 144–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Woodward A. W., and Bartel B. (2005) Auxin: Regulation, action, and interaction. Ann. Bot. 95, 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rampey R. A., LeClere S., Kowalczyk M., Ljung K., Sandberg G., and Bartel B. (2004) A family of auxin-conjugate hydrolases that contributes to free indole-3-acetic acid levels during Arabidopsis germination. Plant Physiol. 135, 978–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Spiess G. M., Hausman A., Yu P., Cohen J. D., Rampey R. A., and Zolman B. K. (2014) Auxin input pathway disruptions are mitigated by changes in auxin biosynthetic gene expression in Arabidopsis. Plant Physiol. 165, 1092–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]