Abstract
Positive findings from pre-clinical and clinical studies involving depletion or supplementation of microRNA (miRNA) engender optimism about miRNA–based therapeutics. However, off-target effects must be considered. Predicting these effects is complicated. Each miRNA may target many gene transcripts, and the rules governing imperfectly complementary miRNA:target interaction are incompletely understood. Several databases provide lists of the relatively small number of experimentally confirmed miRNA:target pairs. Although incomplete, this information might allow assessment of at least some of the off-target effects. We evaluated the performance of four databases of experimentally validated miRNA:target interactions (miRWalk 2.0, miRTarBase, miRecords, and TarBase 7.0) using a list of 50 alphabetically consecutive genes. We examined the provided citations to determine the degree to which each interaction was experimentally supported. To assess stability, we tested at the beginning and end of a five month period. Results varied widely by database. Two of the databases changed significantly over the course of five months. Most reported evidence for miRNA:target interactions was indirect or otherwise weak, and relatively few interactions were supported by more than one publication. Some returned results appear to arise from simplistic text searches that offer no insight into the relationship of the search terms, may not even include the reported gene or miRNA, and may thus be invalid. We conclude that validation databases provide important information, but not all information in all extant databases is up-to-date or accurate. Nevertheless, the more comprehensive validation databases may provide useful starting points for investigation of off-target effects of proposed small RNA therapies.
Keywords: microRNA, validation, interaction, database, off-target effect
INTRODUCTION
MicroRNAs (miRNAs) are therapeutically targetable small non-coding RNA molecules that regulate gene expression. Binding to imperfectly complementary miRNA recognition elements (MREs) within messenger RNAs (mRNAs), preferentially to sites in the 3’ untranslated region of the transcript, miRNAs bring mRNAs and the RNA-induced silencing complex (RISC) into proximity, effecting target gene silencing [Bartel 2009]. Aberrant expression of miRNAs has been reported in association with various diseases, both neoplastic and non-neoplastic. As such, miRNAs are candidate biomarkers, alone or in combination with other factors. Protected in biological fluids within proteins and double-membraned vesicles [Arroyo et al. 2011; Tosar et al. 2015; Turchinovich and Burwinkel 2012; Turchinovich et al. 2011], extracellular miRNAs may also be one important component of a “liquid biopsy” that reflects the state of cells and tissues of origin without incurring the risk of tissue biopsy. Nevertheless, developing accurate miRNA-based diagnostics and prognostics has proven challenging [Haider et al. 2014; Leidner et al. 2013; Witwer 2015].
As important as biomarker status is the potential role of miRNA in therapy. In cancers, for example, an aberrantly overexpressed miRNA that silences tumor suppressors could be silenced by an antisense inhibitor. A depleted miRNA that keeps proliferation-related genes in check could be supplemented by supplying a precursor molecule or a mature miRNA mimic. An overexpressed gene could be targeted by supplying an excess of one or more known targeting miRNAs, while expression of a suppressed gene could be rescued with antisense inhibitors of targeting miRNAs.
A successful example of a miRNA-targeted therapy is miravirsen, an antisense inhibitor of hsa-miR-122-5p, commonly referred to as miR-122 [Janssen et al. 2013]. In liver cells, where it is highly enriched, miR-122 may comprise up to two thirds of all miRNA. In contrast with canonical miRNA-mediated suppression, miR-122 enhances hepatitis C virus (HCV) replication by binding to two sites in the HCV 5’ UTR. miR-122 is thus required for optimal replication of HCV. Miravirsen is an injectable next generation oligonucleotide with a stability-conferring backbone. Despite the abundance of miR-122 in liver, Phase I trials of miravirsen indicated no excessive adverse side effects, and Phase II trials revealed a significant and prolonged knockdown of HCV viral load (as well as reduced circulating cholesterol) [Janssen et al. 2013; Lieberman and Sarnow 2013]. Trials of small RNA drugs that are designed to supplement deficient miRNAs are also underway. For example, MRX34 has been developed to target liver cancers and entered trials in 2013 [Ling et al. 2013].
In the development of any therapeutic, potential off-target effects must be evaluated. Several types of off-target effects of RNA-targeted therapies are known or have been postulated [Jackson and Linsley 2010]. Innate immune responses can be triggered by exposure to double-stranded RNA of certain lengths [Co et al. 2011; Dixit and Kagan 2013; Kawai and Akira 2008]. Excess RNA might saturate the RNA silencing machinery, preventing normal regulation of endogenous genes by endogenous small RNAs [Grimm 2011; Grimm et al. 2006; Jackson and Linsley 2010]. These types of non-specific effects generally require extremely high doses and will not be treated further here. Instead, we wish to examine sequence-specific off-target effects. By “off-target,” we refer to a molecular, functional interaction with any target but the desired therapeutic target. These undesired targets may be important to evaluate, since most mRNAs are targets of miRNAs, and each miRNA may regulate many transcripts [Friedman et al. 2009].
Bioinformatics methods are a useful approach to predicting off-target binding. However, this approach is imperfect. Some well-characterized miRNA:target interactions include poor seed region binding that would not necessarily be predicted by targeting algorithms [Lal et al. 2009]. The opposite is also true. Just because a site is a high-likelihood target does not mean that functional regulation occurs, or that it occurs at measurable, biologically consequential levels (i.e., beyond the range of normal expression variation [Seitz 2009]). Some apparently strong predicted interactions have yielded no experimental evidence. The idea that bona fide target sites will be conserved across species is sound; however, only seed-binding site conservation can be investigated easily in this context. Sites predicted by multiple targeting programs are only as accurate as the algorithms themselves, most of which are very similar. A recently published method for direct target identification by cross-linking, ligation, and sequencing of hybrid miRNA:target molecules (CLASH) [Helwak et al. 2013] has added more evidence to the notion that many interactions may rely only partly or not at all on the seed region, reinforcing doubts about the utility of bioinformatics-only approaches.
Experimental identification and verification of miRNA:target interactions can be conducted in several ways [Clancy et al. 2007; Wang et al. 2007]. Reporter assays introduce a construct in which a sequence encoding a reporter gene such as luciferase is followed by a putative target sequence, often from the 3’ UTR of a transcript of interest. When a miRNA binds to the target, the reporter signal is diminished compared with controls. The MRE(s) may then be mutated and the experiment repeated to confirm direct binding. Because of the confirmation of direct binding, reporter assays are often considered to be the strongest form of evidence for direct interaction. Other assays used to detect changes in putative targets include measures of abundance of specific targets such as transcripts (qPCR, Northern blotting) or protein products (western blot, ELISA). Finally, microarrays, high throughput sequencing, and mass-spectrometry proteomics provide indirect, correlative evidence of miRNA targets.
While only a vanishingly small percentage of genuine miRNA:target interactions have been identified and verified experimentally by mutation-based confirmation—and very few of these by more than one research group or in multiple cell or tissue types—databases of verified targets might be useful in gauging the likelihood that essential genes could be disrupted by proposed RNA-targeted therapeutics. If known target genes are not expressed in a target cell type; if the proposed RNA therapy affects the levels of these genes only slightly (because of weak interactions or low concentration of therapeutic molecules); or if altering expression of these genes does not harm the target cell, the researcher would have at least some evidence for or against expected off-target effects. Indeed, several such databases are freely available online.
We conducted an evaluation of four validated target databases: miRWalk [Dweep et al. 2011], miRTarBase [Hsu et al. 2011; Hsu et al. 2014], miRecords [Xiao et al. 2009], and TarBase 7.0 [Vlachos et al. 2015]. These databases allow the user to input a single interaction partner or multiple partners, and return a list of miRNAs and/or gene transcripts, along with the PubMed ID of the article(s) supporting each miRNA:target interaction. Some databases also catalogue the experimental method underlying the evidence (e.g., luciferase assay, gene expression microarray, etc.) and other information, such as “prediction scores” returned by targeting algorithms for each miRNA:MRE interaction. We evaluated each database at two time points, five months apart. Using a list of 50 alphabetically consecutive genes for each database, we were surprised to observe substantial discordance between databases. However, at the final time point, the most comprehensive database, TarBase 7.0, contained most non-erroneous entries from other databases as well as many others. Although databases cannot take the place of experimentation, comprehensive platforms like TarBase 7.0 may provide good starting-off points for investigation of the literature and of potential off-target effects of miRNAs.
METHODS
Selection of Genes
To standardize our search and to compare findings across four platforms (see below), the same gene transcripts were searched on each platform. 50 alphabetically consecutive genes were chosen from an alphabetized list from GenBank (http://www.ncbi.nlm.nih.gov/genbank/). GeneCards (http://www.genecards.org/) was searched, as needed, for aliases of these genes. The GenBank designations were: A1BG, A1CF, A2M, A2ML1, A3GALT2, A4GALT, A4GNT, AAAS, AACS, AADAC, AADACL2, AADACL3, AADACL4, AADAT, AAED1, AAGAB, AAK1, AAMDC, AAMP, AANAT, AAR2, AARD, AARS, AARS2, AARSD1, AASDH, AASDHPPT, AASS, AATF, AATK, ABAT, ABCA1, ABCA10, ABCA12, ABCA13, ABCA2, ABCA3, ABCA4, ABCA5, ABCA6, ABCA7, ABCA8, ABCA9, ABCB1, ABCB10, ABCB11, ABCB4, ABCB5, ABCB6, and ABCB7. Corresponding ENSEMBL designations for TarBase were: ENSG00000121410, ENSG00000148584, ENSG00000175899 ENSG00000166535, ENSG00000184389, ENSG00000128274, ENSG00000118017, ENSG00000094914, ENSG00000081760, ENSG00000114771, ENSG00000197953, ENSG00000188984, ENSG00000204518, ENSG00000109576, ENSG00000158122, ENSG00000103591, ENSG00000115977, ENSG00000087884, ENSG00000127837, ENSG00000129673, ENSG00000131043, ENSG00000205002, ENSG00000090861, ENSG00000124608, ENSG00000266967, ENSG00000157426, ENSG00000149313, ENSG00000008311, ENSG00000108270, ENSG00000181409, ENSG00000183044, ENSG00000165029, ENSG00000154263, ENSG00000144452, ENSG00000179869, ENSG00000107331, ENSG00000167972, ENSG00000198691, ENSG00000154265, ENSG00000154262, ENSG00000064687, ENSG00000141338, ENSG00000154258, ENSG00000085563, ENSG00000135776, ENSG00000073734, ENSG00000005471, ENSG00000004846, ENSG00000115657, and ENSG00000131269
Database Searching and Literature Validation
miRNA partners of the 50 selected genes were queried in miRecords, miRTarbase, miRWalk, and TarBase. Searches were performed using an online database interface (miRWalk, TarBase) or comprehensive spreadsheet data downloaded from the internet platform (miRecords, miRTarBase). Additional details about each database can be found in the Results section (below). The search was done in January, 2015 and again in May, 2015 to assess changes to the platform. As needed to resolve differences, spot checks were performed again in June, 2015.
The minimal relevant outputs for each database (Table 1) were: 1) genes that were putatively targeted by 2) a specific miRNA according to 3) a peer-reviewed publication, represented by a PubMed ID (PMID). Some databases additionally provided information on the technique(s) used to establish the interaction. Where this information was unavailable, we attempted to find it by consulting the primary literature. Articles corresponding to PMIDs were obtained through PubMed, PubMed Central, or journal websites, accessed through the Welch Library of The Johns Hopkins University. Abstracts and methods sections were scanned for confirmation of miRNA and gene names. If the interaction was not located immediately, we used text search (“control-f”) to find the miRNA or gene name in the article. In the case of no returns, GeneCards (http://www.genecards.org/) was used to find gene aliases that were then also queried. Finally, we downloaded and searched supplemental material if the interaction could not be found in the main text or figures. Searches were first performed by VK and YJL, repeated by DM, and checked by KWW.
Table 1.
Minimal relevant outputs for each database
| Target Gene | miRNA | PMID | Experiments | Evidence? |
|---|---|---|---|---|
| ABCA1 | hsa-miR-33a | 20466882 | Luciferase | Strong |
| ABCA1 | hsa-miR-33b | 20466882 | Luciferase | Strong |
| ABCA1 | hsa-miR-33a | 20466885 | Luciferase | Strong |
| ABCA1 | hsa-miR-33a | 20566875 | Luciferase | Strong |
| ABCA1 | hsa-miR-33a | 20732877 | Luciferase | Strong |
| ABCA1 | hsa-miR-758 | 21885853 | Luciferase | Strong |
| ABCA1 | hsa-miR-33a | 22011750 | N/A | Strong |
| ABCA1 | hsa-miR-33b | 22011750 | N/A | Strong |
| ABCA1 | hsa-miR-33a | 22315319 | Luciferase | Strong |
| AACS | hsa-miR-122* | 19296470 | Luciferase | Weak or none |
| AADAC | hsa-miR-34b | 18519671 | Microarray | Weak or none |
| AADAC | hsa-miR-34b* | 18519671 | Microarray | Weak or none |
| AADAC | hsa-miR-34c-3p | 18519671 | Microarray | Weak or none |
| AADAC | hsa-miR-34c-5p | 18519671 | Microarray | Weak or none |
| AADAC | hsa-miR-129-3p | 21547903 | qRT-PCR | Weak or none |
| AADAC | hsa-miR-129-5p | 21547903 | qRT-PCR | Weak or none |
| AADAC | hsa-miR-203 | 21547903 | qRT-PCR | Weak or none |
| AADAC | hsa-miR-34a | 21547903 | qRT-PCR | Weak or none |
| AADAC | hsa-miR-34a* | 21547903 | qRT-PCR | Weak or none |
| AADAC | hsa-miR-34b | 21547903 | qRT-PCR | Weak or none |
| AADAC | hsa-miR-34b* | 21547903 | qRT-PCR | Weak or none |
| AADAC | hsa-miR-34c-3p | 21547903 | qRT-PCR | Weak or none |
| AADAC | hsa-miR-34c-5p | 21547903 | qRT-PCR | Weak or none |
| AADAC | hsa-miR-424 | 21547903 | qRT-PCR | Weak or none |
| AADAC | hsa-miR-424* | 21547903 | qRT-PCR | Weak or none |
| AADAC | hsa-miR-373 | 21785829 | Microarray | Weak or none |
| AADAC | hsa-miR-373* | 21785829 | Microarray | Weak or none |
| AADAC | hsa-miR-34b | 22052540 | Microarray | Weak or none |
| AADAC | hsa-miR-34b* | 22052540 | Microarray | Weak or none |
| AARSD1 | hsa-let-7b | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-let-7b* | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-1 | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-155 | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-155* | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-16 | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-30a | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-30a* | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-30b | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-30b* | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-30c | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-30c-1* | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-30c-2* | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-30d | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-30d* | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-30e | 18668040 | Proteomics | Weak or none |
| AARSD1 | hsa-miR-30e* | 18668040 | Proteomics | Weak or none |
| AASDHPPT | hsa-miR-1274a | 20818168 | Sequencing | Weak or none |
| AASDHPPT | hsa-miR-1274b | 20818168 | Sequencing | Weak or none |
| AATF | hsa-miR-149 | 19082544 | Western blot, PCR | Weak or none |
| AATF | hsa-miR-149* | 19082544 | Western blot, PCR | Weak or none |
| AATK | hsa-miR-338-3p | 18684991 | qRT-PCR, Western blot | Weak or none |
| AATK | hsa-miR-338-5p | 18684991 | qRT-PCR, Western blot | Weak or none |
| AATK | hsa-miR-338-3p | 22064487 | Microarray | Weak or none |
| AATK | hsa-miR-338-5p | 22064487 | Microarray | Weak or none |
| ABCA1 | hsa-miR-33a* | 20466882 | Luciferase | Weak or none |
| ABCA1 | hsa-miR-33b* | 20466882 | Luciferase | Weak or none |
| ABCA1 | hsa-miR-33a* | 20466885 | Luciferase | Weak or none |
| ABCA1 | hsa-miR-33a* | 20566875 | Luciferase | Weak or none |
| ABCA1 | hsa-miR-33a* | 20732877 | Luciferase | Weak or none |
| ABCA1 | hsa-miR-216a | 21764575 | Microarray | Weak or none |
| ABCA1 | hsa-miR-216b | 21764575 | Microarray | Weak or none |
| ABCA1 | hsa-miR-302a* | 21764575 | Microarray | Weak or none |
| ABCA1 | hsa-miR-33a* | 22011750 | N/A | Weak or none |
| ABCA1 | hsa-miR-33b* | 22011750 | N/A | Weak or none |
| ABCA1 | hsa-miR-33a* | 22315319 | Luciferase | Weak or none |
| ABCB1 | hsa-miR-27a* | 18619946 | RT-PCR | Weak or none |
| ABCB1 | hsa-miR-27a | 20624637 | PCR, Western blot | Weak or none |
| ABCB1 | hsa-miR-27a* | 20624637 | PCR, Western blot | Weak or none |
| ABCB1 | hsa-miR-451 | 21948564 | PCR, Northern blot | Weak or none |
| ABCB1 | hsa-miR-212 | 22241070 | Array | Weak or none |
| ABCB1 | hsa-miR-328 | 22241070 | Array | Weak or none |
| ABCB1 | hsa-miR-519c-3p | 22887864 | PCR, Northern blot | Weak or none |
| ABCB1 | hsa-miR-519c-5p | 22887864 | PCR, Northern blot | Weak or none |
| ABCB1 | hsa-miR-520h | 22887864 | PCR, Northern blot | Weak or none |
| ABCB4 | hsa-miR-15a | 21563233 | Array | Weak or none |
| ABCB4 | hsa-miR-15a* | 21563233 | Array | Weak or none |
| ABCB4 | hsa-miR-16 | 21563233 | Array | Weak or none |
| ABCB4 | hsa-miR-16-1* | 21563233 | Array | Weak or none |
| AACS | hsa-miR-122 | 19296470 | Luciferase | Weak or none |
| ABCA1 | hsa-miR-302a | 21764575 | Microarray | Weak or none |
| ABCB1 | hsa-miR-27a | 18619946 | RT-PCR | Weak or none |
| ABCB1 | hsa-miR-451 | 18619946 | RT-PCR | Weak or none |
Evidence Classification
Evidence for miRNA:target interaction was classified as “strong” or “weak or none” (Table 1). Some databases provided their own assessment of evidence strength. Experimental evidence of a direct interaction of a miRNA with its target—most often in the form of results of 3’ UTR reporter assays in which the putative miRNA recognition element (MRE) was also mutated, and combined or not with measurements of mRNA and/or protein abundance—was classified as “strong” evidence. Indirect evidence of interaction, including correlative ‘omics’ approaches such as RNA sequencing, microarrays, and proteomics, was considered “weak.” In some cases, we were unable to find any mention of the reported miRNA and/or its target in the cited literature, leading to a “none” designation. Out of an abundance of caution, considering that we may have missed evidence contained in difficult-to-access or missing data outside the articles and their supplemental materials, we elected to give the databases the benefit of the doubt and combine weak and none into one category.
Analysis
Data were entered into Google Sheets and Microsoft Excel and manipulated in the latter using common functions and searches to compare the different databases and query time points. An Excel workbook with summarized data is provided as Table 1.
RESULTS
miRecords (http://mirecords.biolead.org/)
miRecords consists of two components: The Validated Targets component is a large database of experimentally validated miRNA targets and the Predicted Targets component of miRecords is an integration of predicted miRNA targets produced by 11 miRNA target prediction programs. To conduct our search, the validated targets dataset was downloaded, and data corresponding to our 50 genes were selected.
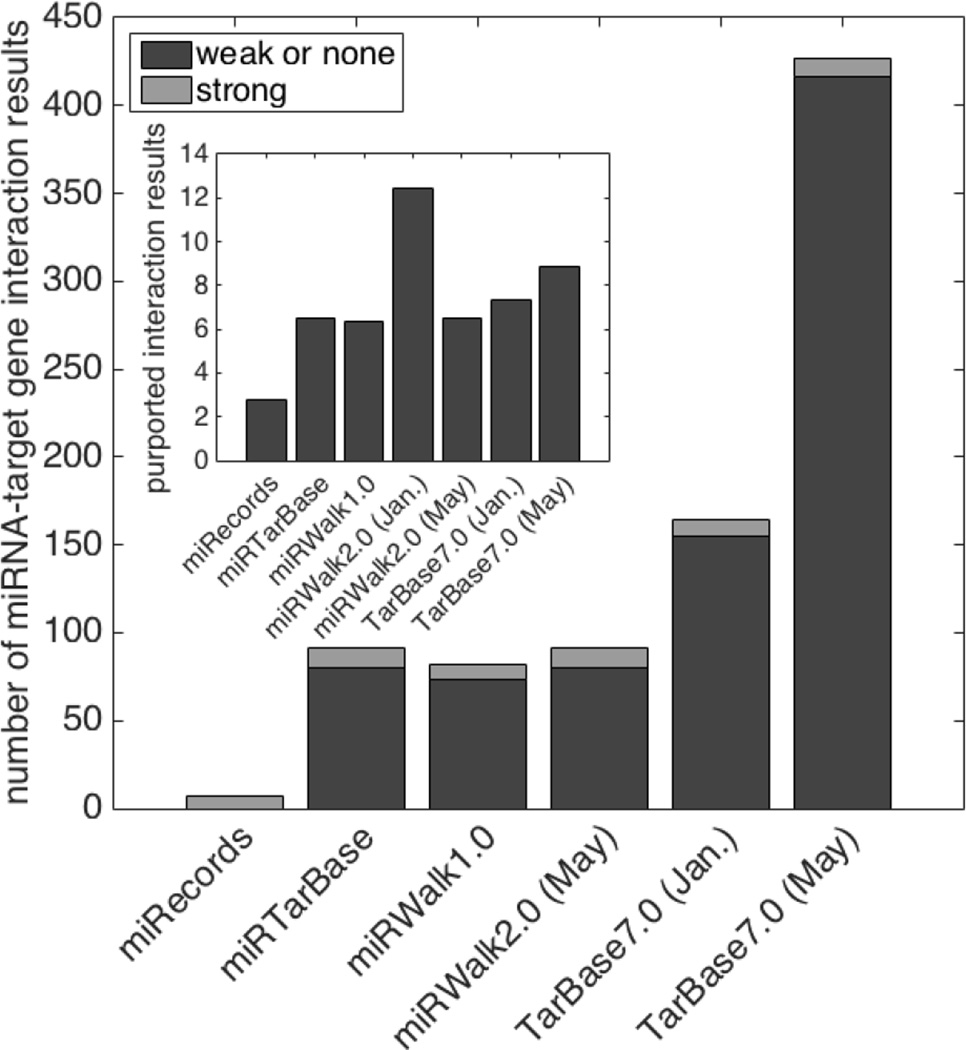
miRecords returned seven interactions of six miRNAs with three genes (including one pair with two apparent sites; Table 2). All were based on solid evidence including luciferase reporter assays (Figure 1, Table 2). Unsurprisingly, since the last update to the platform was in May, 2013, results did not change across the study period.
Table 2. Database comparisons.
Summary of results.
| Database | Genes | miRNAs | Associations | Strong | Weak or none |
Percent strong |
2×+ support |
Publications |
|---|---|---|---|---|---|---|---|---|
| miRecords | 3 | 6 | 7 | 7 | 0 | 100% | 0 | 5 |
| miRTarBase | 33 | 54 | 91 | 11 | 80 | 12.09% | 2 | 19 |
| miRWalk 1.0 | 9 | 58 | 82 | 9 | 73 | 10.98% | ND | 24 |
| miRWalk 2.0 (Jan 2015) | 35 | 594 | 5468 | 21 | 5447 | 0.38% | ND | 48 |
| miRWalk 2.0 (May 2015) | 33 | 54 | 91 | 11 | 80 | 12.09% | 2 | 19 |
| TarBase 7.0 (Jan 2015) | 36 | 110 | 164 | 9 | 155 | 5.49% | 5 | 27 |
| TarBase 7.0 (May 2015) | 32 | 209 | 476 | 18 | 458 | 2.65% | >30 | >35 |
Shown are results of queries performed through each indicated platform in January and May, 2015. For databases/versions that did not change between January and May of 2015, only one entry is given. Columns are as follows: genes (out of 50 queried genes); total number of miRNAs reported; associations (unique miRNA:target interactions); associations with strong evidence or weak evidence, and percent strong evidence; miRNA:target interactions with support from two or more publications; and total number of publications on which the reported interactions are based. Note that miRTarBase and the May 2015 query of miRWalk 2.0 returned the same results.
Figure 1. Number of miRNA:target interactions reported by each database.
Experimental evidence of direct interaction was classified as “strong," while that of indirect interaction was classified as “weak or none” along with no support. For database returns that did not change during our study period, only one set of data are shown. Apparently spurious miRWalk 2.0 results from January are not included in the main graph. Inset: Total number (log(2) scale) of interaction results returned by all database searches, including the January returns for miRWalk 2.0.
Advantage(s)
miRecords is the most conservative of the databases we queried, with strong evidence for all returned interactions.
Disadvantage(s)
The platform has not been updated since 2013. Numerous well-supported interactions are not returned.
miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/search.php)
The miRTarBase platform allows the user to search by miRNA or target gene. Methods used to validate the miRNA-target interaction are ranked as strong or less strong. Target site reporter assays, alone or combined with target abundance measurements such as Western blot or qPCR, are considered to be strong evidence. Correlations provided by microarray, NGS, pSILAC, and others are considered to be less strong evidence. We selected data corresponding to our 50 genes from the miRTarBase Homo sapiens catalog (file name: hsa_MTI.xls), which is available in the “download” section of the website. The spreadsheet includes miRTarBase ID, miRNA, target gene, experiments, support type, and references (PMID).
miRTarbase returned results for 33/50 genes, based on evidence from 19 publications. 91 interactions, including 54 miRNAs, were identified (Table 1), with 11 interactions supported by strong evidence (Figure 1, Table 2). Interactions with ABCA1 were reported for two miRNAs by more than one publication.
Advantage(s)
miRTarBase offers an easily downloaded spreadsheet and is relatively stable.
Disadvantage(s)
Numerous well-supported interactions are not returned.
miRWalk (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/custom.html)
The web-interface of miRWalk2.0 is broadly classified into the Predicted Target Module (PTM) and the Validated Target Module (VTM). PTM hosts miRNA:target interaction information compiled from multiple prediction algorithms and includes the nuclear and mitochondrial transcriptomes of human, mouse and rat. VTM hosts experimentally verified miRNA interaction information associated with genes, pathways, organs, diseases, cell lines, OMIM disorders and literature on miRNAs. To search VTM, we selected “gene-miRNA targets.” The list of 50 genes was copied and pasted into the search box after selecting the species (human), database (gene) and input identifier type (official symbol). A link for validated gene-miRNA interactions leads the user to a new window with a table listing the target genes, miRNA, EntrezID, MIMATID and PMID. This table was downloaded for viewing with Microsoft Excel. We also queried the 1.0 version of the database for comparison.
miRWalk initially returned by far the largest number of associations of the four databases: 5468 associations involving 594 miRNAs and 35 of our 50 genes (Table 2). However, 4,742 of these interactions were listed as being supported by one paper, the CLASH study of Helwak, et al., (2013) and only 22 (0.4%) were deemed to be supported by strong evidence. As a frame of reference, we consulted the previous version of the database, miRWalk 1.0, which remains accessible and returned only 82 interactions: 58 miRNAs and 9 of our 50 transcripts, based on 24 publications. In miRWalk 1.0, 9 of the 82 interactions (11%) were supported by strong evidence (Figure 1 and Table 1). At the end of our study period, the search returns differed drastically from those of both the miRWalk 1.0 and the previous 2.0 query. Now, only 91 associations were returned, featuring 54 miRNAs and 33 genes (Table 2). We noticed with some surprise that these results were identical to those of the stable miRTarBase returns (Table 2).
Advantage(s)
At our last query, the platform seemed to return identical results to miRTarBase, implying either perfect agreement of independent search strategies or mirroring.
Disadvantage(s)
miRWalk appears to have started as a simple text search in version 1.0, with a high percentage of erroneous results, and this problem persisted into version 2.0. During the course of our study, 2.0 seems to have undergone a radical transformation, and, at least according to our results, now appears to be a mirror of miRTarBase. These changes do not seem to be well documented on the website, and in light of past problems, users should be careful to check current or future results with different databases and the literature.
TarBase 7.0 (http://snf-152837.vm.okeanos.grnet.gr/projects/dianauniverse/index.php?r=tarbase)
The TarBase web-interface includes a search bar at the top of the page. We copied our list of 50 genes into the search bar. The platform returned a page asking the user to select the species of interest for each gene if “more than one gene is found with this name,” and we selected the appropriate H. sapiens gene for each entry. The TarBase data return identifies miRNA, target, the utilized experimental methodology, and a “prediction score” column for selected interactions. Clickable features allow the user to obtain a link to PubMed for supporting literature, then (depending on user-specified filters) direction of regulation, site information, experimental conditions including cell or tissue type and treatment, and more.
Our results show that TarBase v7.0 was updated between January and May of 2015, growing from 164 associations involving our 50 genes to 476, and from nine strongly supported interactions to at least 18 (Table 1, Figure 1, Table 2). Furthermore, interactions attributed to more than one publication grew from five to at least 30. Approximately ten apparently erroneous entries were also removed from the first to the second time point, some of them still found in other databases. Several methods corrections or alterations were also made. For example, erroneous references to reporter assays were replaced with the correct “IP” (immunoprecipitation) category, but CLASH evidence was occasionally replaced with “Other”. Some of the increase in association numbers was due to repeats or multiple MREs in the same sequence, as we found numerous instances in which a miRNA:target interaction was listed multiple times for the same publication. No source article identifier(s) are immediately returned with the results, necessitating tedious manual checking of each entry to reveal sources as clickable links to PubMed. Because of this, it is possible that we missed some multiply supported interactions, and we present some of our findings as minimums.
Advantage(s)
TarBase is the most comprehensive database we tested. At our final query time point, it included almost all non-error associations found in other databases, and many more besides (Figure 1, Table 2). TarBase shows evidence of careful curation, since several erroneous entries were removed during the course of our study. It also grew, as one might expect with expansion of the literature and annotation thereof. Detailed information about methods used to identify each interaction was provided.
Disadvantage(s)
The need to click on entries to obtain references, then to click again for positional and other information, was tedious. A minority of miRNA:target interactions appeared to be listed multiple times for the same publication. Although this may have been due in some cases to multiple target sites, positional information was (understandably) often "unknown". A downloadable version is currently unavailable.
Comparison of Database Results
Comparing results from miRecords, miRTarBase, and TarBase (we exclude miRWalk 2.0, which seems to have become a mirror of miRTarBase), we found that 43 /91 miRNA:target associations from miRTarBase (and corresponding PMIDs) were shared by TarBase at the first time point. Up to 121 of 164 associations reported by TarBase were not shared with miRTarBase (although a small number of these may have been duplicates). Approximately two-thirds of the associations that were unique to miRTarBase came from CLASH; between January and May, TarBase appears to have added many or perhaps all of these associations through a more inclusive approach to the CLASH data. Other additions were also made to TarBase (including some duplications), and there were several deletions of apparently erroneous material. As a result of these changes, the last queried version of TarBase returned 476 associations (again, encompassing numerous duplications). These now include 86 of the 91 miRTarBase associations; among the remaining five unique miRTarBase interactions were several apparent errors, so presumably TarBase, which now also includes miRTarBase results, has also been carefully curated. Of the six associations reported by miRecords (one association includes two MREs in the same transcript), only one was shared by all three databases in January, while three were uniquely shared by miRecords and miRTarBase. By June, however, five of the six interactions were shared by miRecords and TarBase.
DISCUSSION
To summarize our results, we find that 1) miRecords is accurate but incomplete; 2) miRTarBase is more expansive but still incomplete; 3) miRWalk in its various versions has a history of unreliability and as of this writing appears to mirror miRTarBase instead; and 4) TarBase is the most comprehensive of the four databases and shows the best evidence of curation.
Several weaknesses of our analysis should be mentioned. Firstly, we did not attempt to construct our own database and cannot estimate how comprehensive the databases may be in comparison with the literature. It was not our intent to search the literature exhaustively to find entries that may have been missed by the databases we examined. We have simply compared databases with each other, not with the entire literature.
Secondly, for the most part, we relied in our reporting on the strength of evidence provided by the individual databases. Because of the difficulties in checking all article versions and databases (in some cases, the latter were even unavailable), we chose to give the databases the benefit of the doubt even when we could not find evidence of a specific interaction. For this reason, we did not report separate “no evidence” conclusions. However, we are aware that some entries, particularly older entries in miRWalk, are completely unsupported and appear to result from simple text searches. For example, our January, 2015 query of miRWalk 2.0 suggested that ABCB1 was regulated by miRs-27a-5p, -30c-5p, and -27a-3p, citing a 2012 publication [Padmanabhan et al. 2012]. In fact, the article investigates regulation of ABCG2 by two other miRNAs and mentions ABCB1 in the abstract and introduction merely as another example of a multidrug transporter. The three miRNAs in question are not mentioned at all, but the numbers “27” and “30” are mentioned several times as catalog numbers, or in references or lengths of time. Clearly, databases that rely on such searches are unreliable, and it is possible that our findings are overly optimistic.
Thirdly, like the databases that reported strength of evidence, we placed weight on reporter assays that can incorporate site-specific mutations. But is this appropriate? Reporter assays are considered to be the best evidence of direct regulation. At the same time, these assays are highly artificial. Typically, the reporter gene is followed by a nucleic acid sequence corresponding, e.g., to an MRE, tandem MREs, or a partial or full 3’ UTR (or other sequence). That is, the full nucleic acid context of the native mRNA is not present. Perhaps more importantly, the stoichiometry of miRNA and target is often greatly distorted, especially when both are transfected at molar excess into cells, in essence making these molecules the “only game in town” for the suppression machinery. Most such assays are also performed in cell lines, which contain an overrepresentation of “active” miRNA-containing complexes [La Rocca et al. 2015]. Likely because of this, most research using reporter assays curiously describes miRNA-mediated downregulation much greater than the fractional fold changes revealed by ‘omics’ studies. Of course, such tiny changes cannot easily be measured with qPCR and other technologies. Perhaps the best approach is not luciferase assays, at least not on their own, but rather precipitation studies (especially CLASH) [Helwak et al. 2013] that seek to sequence or otherwise detect miRNAs and their targets that are associated with the RNA silencing machinery.
In summary, miRNA:target validation databases should be used with caution when seeking confirmed interactions of miRNAs and their MREs in target transcripts. The user is encouraged to check the strength of the evidence carefully with the primary literature, especially when planning follow-up experiments. Of the databases we reviewed, Diana-TarBase v7.0 is clearly the most comprehensive and up-to-date. However, given the relative lack of information in the literature, the widely varying quality of evidence between studies, and the dearth of multi-study confirmation of individual interactions, results from even the best validation databases are, in our opinion, of questionable value as input for “pathway” analyses. Instead, databases are perhaps most useful to highlight multiply supported interactions, which could serve as a starting-off point for investigations of off-target interactions of proposed miRNA-based therapies.
Supplementary Material
Acknowledgments
The authors thank other members of the Witwer lab for helpful comments and would like to acknowledge funding from several sources. Both KWW and YJL have received support from the Johns Hopkins University Center for AIDS Research (P30AI094189). DCM is supported by T32 OD011089. KWW is supported in part by R01 DA040385.
REFERENCES
- Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy JL, Nousch M, Humphreys DT, Westman BJ, Beilharz TH, Preiss T. Methods to analyze microRNA-mediated control of mRNA translation. Methods Enzymol. 2007;431:83–111. doi: 10.1016/S0076-6879(07)31006-9. [DOI] [PubMed] [Google Scholar]
- Co JG, Witwer KW, Gama L, Zink MC, Clements JE. Induction of Innate Immune Responses by SIV In Vivo and In Vitro: Differential Expression and Function of RIG-I and MDA5. J Infect Dis. 2011;204(7):1104–1114. doi: 10.1093/infdis/jir469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Kagan JC. Intracellular pathogen detection by RIG-I-like receptors. Adv Immunol. 2013;117:99–125. doi: 10.1016/B978-0-12-410524-9.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by "walking" the genes of three genomes. J Biomed Inform. 2011;44(5):839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D. The dose can make the poison: lessons learned from adverse in vivo toxicities caused by RNAi overexpression. Silence. 2011;2:8. doi: 10.1186/1758-907X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441(7092):537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Haider BA, Baras AS, McCall MN, Hertel JA, Cornish TC, Halushka MK. A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLoS One. 2014;9(2):e89565. doi: 10.1371/journal.pone.0089565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153(3):654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39(Database issue):D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42(Database issue):D78–D85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Linsley PS. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov. 2010;9(1):57–67. doi: 10.1038/nrd3010. [DOI] [PubMed] [Google Scholar]
- Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- La Rocca G, Olejniczak SH, Gonzalez AJ, Briskin D, Vidigal JA, Spraggon L, DeMatteo RG, Radler MR, Lindsten T, Ventura A, et al. In vivo, Argonaute-bound microRNAs exist predominantly in a reservoir of low molecular weight complexes not associated with mRNA. Proc Natl Acad Sci U S A. 2015;112(3):767–772. doi: 10.1073/pnas.1424217112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, O'Day E, Chowdhury D, Dykxhoorn DM, Tsai P, Hofmann O, et al. miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to "seedless" 3'UTR microRNA recognition elements. Mol Cell. 2009;35(5):610–625. doi: 10.1016/j.molcel.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidner RS, Li L, Thompson CL. Dampening enthusiasm for circulating microRNA in breast cancer. PLoS One. 2013;8(3):e57841. doi: 10.1371/journal.pone.0057841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman J, Sarnow P. Micromanaging hepatitis C virus. N Engl J Med. 2013;368(18):1741–1743. doi: 10.1056/NEJMe1301348. [DOI] [PubMed] [Google Scholar]
- Ling H, Fabbri M, Calin GA. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov. 2013;12(11):847–865. doi: 10.1038/nrd4140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirna Therapeutics I. A Multicenter Phase I Study of MRX34, MicroRNA miR-RX34 Liposome Injectable Suspension. 2013 [Google Scholar]
- Padmanabhan R, Chen KG, Gillet JP, Handley M, Mallon BS, Hamilton RS, Park K, Varma S, Mehaffey MG, Robey PG, et al. Regulation and expression of the ATP-binding cassette transporter ABCG2 in human embryonic stem cells. Stem Cells. 2012;30(10):2175–2187. doi: 10.1002/stem.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H. Redefining microRNA targets. Curr Biol. 2009;19(10):870–873. doi: 10.1016/j.cub.2009.03.059. [DOI] [PubMed] [Google Scholar]
- Tosar JP, Gambaro F, Sanguinetti J, Bonilla B, Witwer KW, Cayota A. Assessment of small RNA sorting into different extracellular fractions revealed by high-throughput sequencing of breast cell lines. Nucleic Acids Res. 2015;43(11):5601–5616. doi: 10.1093/nar/gkv432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A, Burwinkel B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol. 2012;9(8):1066–1075. doi: 10.4161/rna.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39(16):7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos IS, Paraskevopoulou MD, Karagkouni D, Georgakilas G, Vergoulis T, Kanellos I, Anastasopoulos IL, Maniou S, Karathanou K, Kalfakakou D, et al. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43(Database issue):D153–D159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Doench JG, Novina CD. Analysis of microRNA effector functions in vitro. Methods. 2007;43(2):91–104. doi: 10.1016/j.ymeth.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Witwer KW. Circulating MicroRNA Biomarker Studies: Pitfalls and Potential Solutions. Clin Chem. 2015;61(1):56–63. doi: 10.1373/clinchem.2014.221341. [DOI] [PubMed] [Google Scholar]
- Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37(Database issue):D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.