Abstract
Atrial fibrillation (AF) is the most common cardiac arrhythmia with well-established clinical and genetic risk components. Genome-wide association studies (GWAS) have identified 17 independent susceptibility signals for AF at 14 genomic regions, but the mechanisms through which these loci confer risk to AF remain largely undefined. This problem is not unique to AF, as the field of functional genomics, which attempts to bridge this gap from genotype to phenotype, has only uncovered the mechanisms for a handful of GWAS loci. Recent functional genomic studies have made great strides towards translating genetic discoveries to an underlying mechanism, but the large-scale application of these techniques to AF has remain limited. These advances, as well as the continued unresolved challenges for both common variation in AF and the functional genomics field in general, will be the subject of the following review.
Keywords: Atrial fibrillation, Genetics, Mechanism, Epigenetics, SNP
1. Introduction
Among all cardiac arrhythmias, atrial fibrillation (AF) is by far the most prevalent, with over 3 million current cases in the USA and 30 million cases worldwide. This number is expected to greatly increase over the next several decades as the population ages, an event that will further exacerbate the societal and monetary burden caused by this arrhythmia.
Clinical risk factors for AF are numerous and include advanced age, hypertension, obesity, and heart disease, but a significant hereditary component for AF risk has also been well described.1–4 This genetic contribution is observed as a markedly increased risk for individuals with a first-degree relative with AF.1,3–5 At present, estimates of the heritability of AF have been found to be as high as 62%, of which only a small portion has been mechanistically defined.4,5 This is evidenced by an inability for adjustment of clinical risk factors or common genetic variation to substantially alter the observed 40% increased AF risk in those with familial AF.4
Together, the total genetic contribution to AF risk can be broadly divided into three components:
rare coding variation with a large effect size such as that seen in familial forms of AF;
common variation identified by genome-wide association studies (GWAS) that confers a smaller, but additive, risk of disease;
undiscovered variation (common or rare variation, inherited or acquired epigenetic modifications, somatic variation, and mosaicism).
With respect to the rare coding variation, there have been numerous reports from individuals or families with AF with associated mutations in a variety of cardiac ion channels, signalling molecules, structural proteins, and transcription factors.6 These include mutations that are not germline, but instead somatic in nature, as has been described for the gap junction gene GJA5.7 It is likely that the decreasing cost of exome and genome sequencing will aid in the discovery of additional monogenic or polygenic variants that confer a large risk of disease, although primary tissue samples for identifying somatic causes will be difficult to obtain and analyse. In total, the utility in these studies is likely to be family-specific and may not be useful in the ultimate identification of generalizable pathways that may be therapeutic targets in the population as a whole.
With respect to the role of undiscovered genetic variation in AF, there continues to be a disconnect between the heritability of AF and the AF-associated common or rare variants described to date. This so-called ‘missing heritability’ can potentially be attributed to many potential aetiologies including common variants that have not been captured in current GWAS arrays, epigenetics, rare variants with strong effects, large or small copy number variants, and mosaicism that is poorly captured with current techniques.
Instead, more common risk-conferring AF variants, typically found at >1% frequency in the population, may provide valuable insights into targetable pathways. However, determining the mechanism through which these variants perturb disease risk has been challenging and is the subject of the following review.
2. Current synopsis of common gene variants conferring AF risk
In the mid-2000s, platforms for genotyping hundreds of thousands of common variants in an individual, combined with the collection of large numbers of patient samples, led to the rapid expansion of GWAS. In brief, these studies examine the relative frequency of individual common variants in cases vs. controls and identify genomic variants that are more or less commonly observed in the cases. GWAS have been broadly used to identify the genetic basis of a wide range of traits and diseases. The first such GWAS for AF was published in 2007, in which a region on 4q25, upstream of the transcription factor gene PITX2, was found to be significantly associated with AF.8
As the effectiveness of GWAS relies greatly on the sample size and resolution of the assay, efforts were made to improve both of these in subsequent analyses. In 2009, a pair of groups identified an additional AF-associated signal at 16q22,9,10 which was intronic to another transcription factor, ZFHX3. In a separate analysis focused on individuals with early onset AF, a third signal intronic to the KCNN3 gene on chromosome 1 was identified.11
Large increases in sample size and increased density of genotyping led to a trio of studies that identified many additional AF risk loci. In 2012, a meta-analysis in individuals of European and Japanese descent uncovered an additional six loci for AF.12 Next, in 2014, the combination of fine-mapping and conditional analyses revealed that there was not one but four independent loci at 4q25.13 Finally, also in 2014, a meta-analysis of over 13 000 cases and 70 000 controls identified a further 5 loci, one of which was exclusive to those of Asian descent.14 At present, there are 14 genomic regions of susceptibility for AF with 17 independent signals at these loci (Table 1).
Table 1.
Genetic loci associated with AF
| Genomic locus | Sentinel SNP | Relative risk | P-value | Proximal gene | eQTL by GTeX15 | Relative location to proximal gene |
|---|---|---|---|---|---|---|
| 4q25 | rs6817105 | 1.64 | 1.8 × 10−74 | PITX2 | None | 150 kb upstream |
| 4q25 | rs4400058 | 1.31 | 4.1 × 10−20 | PITX2 | None | 150 kb upstream |
| 4q25 | rs6838973 | 1.13 | 7.1 × 10−12 | PITX2 | None | 200 kb upstream |
| 4q25 | rs1448818 | 1.12 | 2.0 × 10−8 | PITX2 | None | Intronic |
| 16q22 | rs2106261 | 1.24 | 3.2 × 10−16 | ZFHX3 | None | Intronic |
| 1q21 | rs6666258 | 1.18 | 2.0 × 10−14 | KCNN3 | None | Intronic |
| 1q24 | rs3903239 | 1.14 | 8.4 × 10−14 | PRRX1 | RP1-79C4.4, skin | 46 kb upstream |
| 7q31 | rs3807989 | 0.90 | 3.6 × 10−12 | CAV1 | None | Intronic |
| 14q23 | rs1152591 | 1.13 | 5.8 × 10−13 | SYNE2 | ESR2, skin | Intronic |
| 9q22 | rs10821415 | 1.11 | 4.2 × 10−11 | C9orf3 | None | Intronic |
| 15q24 | rs7164883 | 1.19 | 2.8 × 10−17 | HCN4 | None | Intronic |
| 10q22 | rs10824026 | 0.87 | 4.0 × 10−9 | MYOZ1 | None | 20 kb upstream |
| 10q24 | rs12415501 | 1.18 | 6.5 × 10−16 | NEURL | None | Intronic |
| 12q24 | rs10507248 | 1.12 | 8.5 × 10−11 | TBX5 | None | Intronic |
| 3p25 | rs4642101 | 1.10 | 9.8 × 10−9 | CAND2 | CAND2, skeletal muscle | Intronic |
| 6q22 | rs13216675 | 1.10 | 2.2 × 10−8 | GJA1 | None | 600 kb downstream |
| 12q24 | rs6490029 | 1.22 | 3.9 × 10−9 | CUX2 | None | Intronic |
Although there have been rapid advances in the discovery of new genetic loci for AF, the clinical utility of GWAS variants remains unclear. For example, a genetic risk score consisting of the top 12 AF GWAS loci allows for a stratification of the population into a five-fold gradient of AF risk.14 However, a similar risk score resulted in only a modest increase in the prediction of incident AF beyond other known risk factors for arrhythmia.16 Furthermore, the effect of genotype at the top AF risk loci has also been examined in relation to the response to treatment, where AF genotype has been reported to predict response to cardioversion,17 pulmonary vein isolation,18–20 and anti-arrhythmic medications.21 In contrast, a single study in a Korean cohort found no association between AF risk alleles and response to AF ablations.22
Importantly, most of the studies relating genetic variants to AF treatments or outcomes have been of modest size and only considered the top few variants for AF. Thus, it will be interesting to re-examine each of these questions as we develop a more comprehensive picture of the AF genetics and gain access to larger patient populations. In future, it will also be interesting to determine whether AF genetic risk scores can be used to identify high-risk patients with heart failure or cryptogenic stroke who would benefit from more intensive monitoring or anticoagulation.
In the upcoming years, it is clear that increased resources will be applied to this endeavour, and we can expect GWAS for AF with massively increased sample sizes. Such an approach has been taken for other common diseases including coronary artery disease23 and schizophrenia,24 and what emerges is a comprehensive picture of the common genetic underpinnings of a disease. Larger GWASs also improve and refine the clinical risk prediction models of AF and will enable sufficiently powered analyses in cohorts stratified for AF outcomes such as stroke or heart failure.
Despite these anticipated advances, there remains a great knowledge gap in ‘how’ common variants at these loci confer AF risk. This void must be filled if we are to maximally utilize these discovery efforts to understand the molecular pathways underlying AF.
3. Challenges in defining common variant mechanisms
Although GWAS can be a powerful tool, this method is blind to the divergent pathways through which AF can arise, and it does not directly help to understand the basis for AF pathogenesis. For example, does the genotype at an AF SNP alter atrial fibrosis? Increase atrial diameter? Decrease atrial conduction time? Affect ion channel remodelling? Alter the positioning or development of the myocardial sleeves? Perturb autonomic tone? These and other possibilities25–27 will ultimately need to be considered when evaluating each locus. Such genotype–phenotype correlations are a daunting but important task that will provide insight for translational biology to develop novel, and hopefully personalized, treatments for AF.
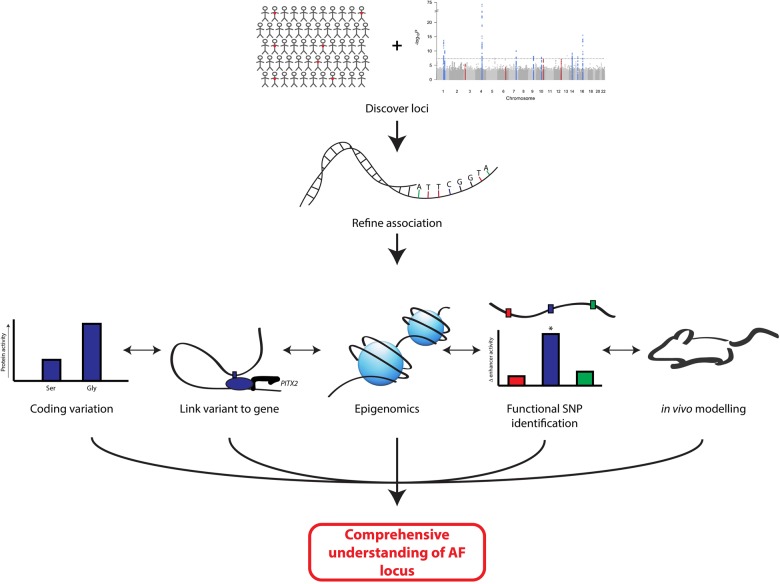
For most AF loci, the steps from an association to the phenotype are entirely unknown. AF-associated SNPs mark a general region of the genome without the implication that an individual SNP is causative. The definition of the functional SNP(s) will be key for identifying a transcriptional network for AF, and an integrated approach spanning from genetics to animal modelling will be necessary (Figure 1). First, it will be essential to refine the genetics association at a locus. Second, for coding variants, the challenge is relatively straightforward and will involve the determination of an amino acid substitution on protein function. However, as with most GWAS, the vast majority of AF loci reside in non-coding regions. Therefore, a third approach will be to link the AF-associated SNP to a gene in the region. Fourth, we will need to refine the epigenetic landscape at a given locus. Fifth, it will be necessary to develop higher throughput methods to identify functional variants, and finally, appropriate modelling of the disease-related gene can be undertaken. In practice, it will be essential to combine these different approaches to develop a comprehensive view of how a locus contributes to disease.
Figure 1.
Model for the functional definition of a GWAS association locus. GWAS and associated analyses identify a general genomic region of AF association, which is refined by additional genotyping, sequencing, or bioinformatics analyses. For coding variation, evaluation of altered protein function in vitro and in vivo for risk/protective alleles is conducted. For non-coding variation, variants are linked with altered expression of genes through eQTL or chromatin conformation capture. Concurrently, data sets with epigenomic information are mined for markers of regulatory DNA, which may contain associated SNPs. When candidates are found, functional variant(s) should be defined and validated with reporter assays. Translation of information on directionality of altered expression, amplitude of altered expression, and tissue type of interest inform modelling of AF mechanisms in animal models.
4. Difficulties in identifying functionally relevant variants
The challenge behind identification of functional non-coding variation is rooted in the linkage structure of the human genome, in which relatively large regions or haplotype blocks of many SNPs are inherited together. The ‘sentinel’ SNPs identified by GWAS serve as a marker for a larger region of SNPs, all with a similar degree of association with disease. Thus, it is important to note that the sentinel SNP is not necessarily the functional variant at a locus. Further complicating these efforts, it is also possible that a common SNP may only serve as a tag for a coding variant, rare variant, or a variation in copy number such as an insertion or deletion.
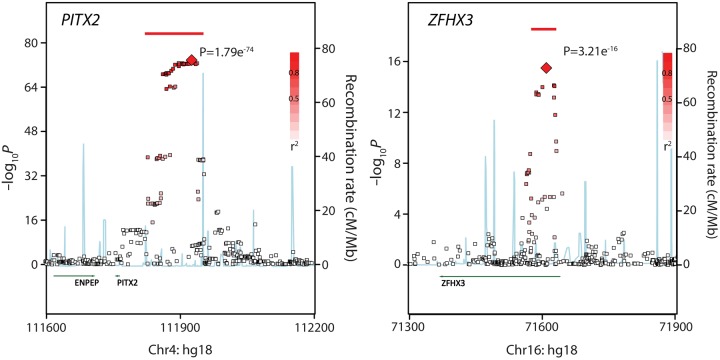
A convenient way to display the results of an association at a given locus is with a regional plot (Figure 2). The data are plotted as the –log of the P-value for the association with AF vs. chromosomal distance. Each point represents a distinct SNP that is tested for an association with AF, and the red colour provides the degree of linkage disequilibrium to the sentinel SNP at the locus. The two plots for PITX2 and ZFHX3 illustrate some of the challenges in deciphering the mechanism of the AF-associated variants. For example, in the plot of the PITX2 region (Figure 2, left), there are a large number of non-coding, common variants that span a nearly 75 kb region (illustrated by the red bar) that are all essentially equivalent to the top signal. The region is much narrower at the ZFHX3 locus (Figure 2, right), so one could expect that the identification of a functional variant would be more straightforward, yet there are still many potential variants to consider even at a relatively simple locus.
Figure 2.
Regional plot for AF association signals on chromosomes 4 and 16. Each point represents a single SNP with association level displayed on the left Y-axis. Red bars indicate the top linkage dissociation block that is approximately equivalently associated with altered risks of AF. Blue lines indicate peaks of recombination, with strength being displayed on the right Y-axis. Note that the region associated with AF in (A) (indicated by the red SNPs and bar) is quite large and at a great distance upstream of the closest gene, PITX2. In contrast, in (B), the AF-associated SNPs/region is considerably smaller and is located within the gene ZFHX3.
5. Refining the association signal
Given the challenges of the aforementioned studies, as a first step after the identification of an AF locus, it is helpful to refine the association signal. Potential approaches include performing conditional analyses, imputation to newer haplotype maps, genotyping with alternate arrays to capture exonic, copy number, or epigenetic markers, and targeted or whole genome sequencing studies.
In a conditional analysis, one corrects for the top SNPs in a region to determine whether there is just one or potentially multiple independent association signals at the locus. This technique allowed the discovery of an additional three loci at the 4q25 locus, which, when combined with that at the main 4q25 signal, could be used to predict a five-fold graded risk score in individuals of European or Japanese descent.14 It is possible that other AF emerging loci may also contain multiple independent signals.
In addition to fine-mapping, one may improve the resolution at a locus by re-imputing previous data to newer reference map of the genome. Imputation is a strategy whereby known genotype at several SNPs in a region can be used to predict those not directly genotyped. Initially, this prediction was based upon reference maps from the HapMap consortium consisting of approximately 2 million markers.28 In recent years, these maps have been supplanted by those built from the 1000 Genomes Project,29 wherein hundreds of genomes have been sequenced and assembled. With 8–10 million variants, imputation to the 1000 genomes data set will greatly increase the resolution of any GWAS locus, as has been the case with coronary artery disease.23
The greatest resolution of an association signal at a locus can be provided by whole-genome sequencing. Although genome sequencing was once far too costly, with the continued improvements in sequencing cost, quality, and turnaround times, such studies are increasingly feasible. Genome sequencing will permit the identification of all common genetic variants and uncover any previously unidentified causative rare or copy number variation that was previously tagged by common variants.
Although each of these approaches will improve the resolution at a given locus, they will not solve the challenge of defining a functional regulatory variant. Further prioritization and functional analysis would be necessitated to define which variant(s) underlies association.
6. Epigenomics to guide functional variant discovery
Even if genome sequencing data were readily available, there will still be dozens, if not hundreds, of possible candidate SNPs which may be a functional variant(s) at a given locus. Although it is possible that some may be coding variants, the vast majority of GWAS loci reside in non-coding regions (Figure 2, left, PITX2). For these loci, it is expected that functional variants act by differentially regulating the expression of nearby genes through enhancer or promoter activities. As comprehensively testing each of these with reporter assays, such as luciferase or EGFP expression, is an expensive and time-consuming endeavour, previous studies have attempted to limit the number of SNPs tested through prioritization schemes. The most widely used approach is to examine epigenetic data from the ENCODE project,30 including studies of DNase hypersensitivity and histone post-translational modification marks obtained from a series of immortalized cell lines and human primary samples.30 Together, these signals observed from these data sets allow researchers to hone in on regions of active chromatin that may be acting as enhancers or promoters in a given tissue. In theory, this can allow one to test far fewer SNPs as potential candidates.
While impressive for its time, the ENCODE data were limited in its scope and depth and have been integrated with the more comprehensive Roadmap Epigenomics Project data sets.31 These data, obtained from 111 epigenomes of human tissues, include additional CHIP-seq data for histone modifications, DNA methylation status, DNase hypersensitivity, and RNA sequencing data for relative expression. Using these data in combination, the Roadmap Epigenomics Project31 has released a series of models which predict the state of chromatin in a given tissue, a valuable resource for those looking to follow up on the functional basis for GWAS loci. As a template, in initial studies, these data were used to generate predictive models of where functional variants for 58 studies resided, including those for lipids, PR interval, blood pressure, and aortic root size.
Although the next release of the Roadmap data will be the result of more than 180 biological samples, its usefulness for the study of AF is limited by the lack of left atrial samples, nodal tissue, myocardial sleeves, or pulmonary vasculature. Inclusion of these samples in future epigenomic analyses will be important for the data set as a tool for functional follow-up for AF loci. Additionally, it should be noted that for human primary samples, the epigenomic map is a product of a mixed cell population from the tissue of interest. Therefore, if the molecular basis for an AF locus was the modification of enhancer activity in a minor cell population of the heart, it would be unlikely to be marked as ‘active chromatin’ in the aggregate analyses. A refinement of maps which take into account tissue heterogeneity will be an important improvement to watch for in future iterations of epigenetic modelling.
7. Linking AF SNPs to nearby genes
As regions of chromatin can form long-range regulatory interactions that can extend as far as a megabase, it is important to consider all genes in a wide region around an AF GWAS signal as potential candidate genes. Sometimes, there are relatively few candidate genes in a region, and the potential association with AF is clear, for example, the association between AF and the GJA1 or connexin 43 locus.14 However, in most other loci, there are many potential candidate genes with a number of plausible biological mechanisms. In these cases, determining which of these genes are regulated differentially based on genotype is of paramount importance prior to any further functional evaluation. Current approaches include expression quantitative trait analyses (eQTL) or the measurement of the physical interactions between regulatory regions and proximal promoters.
In an eQTL analysis, samples isolated from primary tissue are genotyped for a sentinel SNP at an AF locus, and then transcription of genes in the region is quantitated. If there is a correlation between the AF SNP and expression of a nearby gene, then that gene is highly likely be the causative one for AF. On a small scale, surgical samples from left atrial appendage tissue have been used to determine that the AF locus on chromosome 10q22 alters expression of a nearby gene (MYOZ1) and not the gene closest to the AF SNP (SYNPO2L).32 Similar analyses can also be performed more globally by performing genome-wide genotyping and RNA sequencing in relevant tissues.14,33 Large-scale projects for examining gene expression using RNA sequencing are currently underway in the Genotype Tissue Expression (GTEx) project.15 However, at present, there are 190 samples from the left atrial appendage, a number that is still underpowered for subtle effects of most GWAS functional variants. Future evaluations by the GTEx Consortium or other investigators will hopefully provide well-powered analysis of gene expression in the upcoming year.
A complimentary approach to find which genes are affected by GWAS SNPs is to examine the physical interaction between a potential enhancer(s) in a non-coding GWAS locus and the promoter of the genes in the region using chromatin conformation capture.34 Current models of distal enhancer action suggest that physical interactions occur between factors binding enhancers and the transcription initiation complexes at proximal promoters. Thus, by physically linking these enhancer: promoter interactions and examining the frequency with which chromosomally distant DNA regions are physically near to one another by sequencing or targeted polymerase chain reaction, one can develop a interaction map of the locus. One iteration of this approach is 4C, works from a limited number of reference points, and can provide a high-resolution interaction map of a reference point with all other regions at a given locus. Alternatively, Hi-C examines the interaction between all points in the genome with all other points in the genome. However, the latter currently provides low-resolution maps due to limited sequencing depth and is only available in a small number of cell and tissue types. Investment to improve both these characteristics in Hi-C data will greatly aid in the usefulness of these data to AF and other cardiovascular diseases.
Although eQTL signals or chromatin conformation capture provides strong evidence to implicate a given gene, the absence of either is uninformative. Both these strategies rely on homogeneous tissue samples for best resolution, and myocardial samples used for the creation of current maps are a mixture of several distinct cell types. Therefore, if an important interaction were to occur in a minor subpopulation, it would be unlikely to be observed above the noise of the assay. To address this, evaluation in cell subpopulations will be made feasible as quantitative in situ RNAseq techniques such as FISSEQ continue to mature.35,36 In the meantime, despite general technical limitations, generation of publically available eQTL and Hi-C maps for the left atrium, pulmonary vasculature, or nodal tissues would be a great advance for the functional genomic analysis of AF and should be a focus of subsequent efforts.
8. Limitations of modelling AF disease genes
Knowledge of functional variation is necessary to provide insight into genes that are linked with the development of AF and the ‘when and where’ of the altered regulation, but modelling these functional consequences is critical for comprehensive understanding of mechanism, secondary pharmaceutical screens, and therapeutic translation.
A definite functional analysis in the context of a complex and multifactorial disease such as AF needs a suitable animal model. So far, however, there is no ideal animal model for AF available.37,38 Large animals such as pigs have a cardiac anatomy similar to that of humans and would allow a comprehensive in vivo evaluation of the cardiovascular system including three-dimensional mapping, voltage mapping, or left atrial electrophysiology (EP) studies.38 Unfortunately, genetic manipulation to generate large transgenic animals is still not widely available, and housing and experimental costs would be prohibitive.39 Genetic engineering is routinely available in small animal models such as zebrafish and mouse models.40,41 Although zebrafish models can be used to evaluate basic EP parameters such as the action potential duration, they are limited by their small size and by the lack of a four-chamber heart.42,43 Mouse models seem to be a good compromise as transgenic mice are available, and performing an in vivo EP study including induction and evaluation of arrhythmias is possible.44,45 However, the dramatically different heart rates and the action potential duration in a mouse significantly limit the use of the model for studies of AF. A final approach would be to perform gene knockout or overexpression of candidates in induced pluripotent stem-cell-derived cardiomyocytes.46,47 These cells have the advantage of being human in origin, but have limitations related to unknown chamber specificity and various levels of maturity. Therefore, it is difficult for this model in its current state to recapitulate all of the features of the mature human myocardium.
9. Need for higher throughput methods to identify functional variation
Despite the many challenges outlined earlier, there have been some important successes in identifying functional variants at the SORT1 locus for myocardial infarction (MI)/low-density lipoprotein (LDL),48 NOS1AP in the QT interval,49 SCN10A/5A for the PR interval,50 BCL11A for foetal haemoglobin,51 and MEIS1 for restless legs syndrome,52 among others. However, the effort required for these analyses is immense and time-consuming, problems best displayed by the vast disconnect between the thousands of known GWAS loci and handful which are defined mechanistically.
It is also important to note that these studies have relied on important advantages, which are largely not available for most other GWAS loci. For example, the SORT1 locus for MI/LDL encompassed a 6 kb region which contained only six SNPs to be functionally analysed. This is notable when compared to the PITX2 locus for AF on 4q25 (Figure 2), which is ∼75 kb in length and incorporates nearly 100 associated SNPs. Similar smaller regions of associations have been observed in all mechanistically defined loci, but this advantageous size is the exception rather than the rule for GWAS loci. Therefore, integration of additional refinement methods is an important pursuit which will increase the rate at which functional variants are described and defined.
In addition to the prioritization schemes using fine-mapping and epigenomic marks, the use of massively parallel reporter assays53–56 will also aid in this rate of functional variant discovery. These assays, which are capable of profiling hundreds or thousands of variants simultaneously, have been applied to the study of transcription factor base preference and to the study of enhancer organization, but their potential for the study of variants identified by GWAS has remained untapped.
Finally, following discovery of a functional SNP, the definition of mechanism for altered activity, presumably through differential transcription factor binding, has become standard. Although this has not been demonstrated at any locus for AF, CHIP-seq studies have aided discovery efforts for other GWAS loci. In those studies, CHIP-seq signals obtained from large-scale pulldown and mapping efforts were used to discover the altered transcription factor binding site. In other cases in which CHIP-seq data might not exist, databases such as JASPAR,57 Haploreg,58 PROMO,59 Transfac,60 or Uniprobe61 may be useful for predicting differential binding of transcription factors. Efforts can also be made to determine transcription factor de novo, although nuclear lysates from appropriate tissues and comprehensive follow-up in vitro and in vivo would be necessary for the establishment of a convincing mechanism.
10. Conclusions
To move forward in the dissection of AF loci will require an integrated approach with collaborative teams of investigators with a diverse set of skills to enable each of the studies outlined earlier. Progress in dissecting AF and other common disease loci has remained challenging due to the complex nature of AF disease pathogenesis, but the advent of newly designed analysis tools, release of public data sets for linking locus to gene modulation, increased ease of genetic modulation of gene expression, and improvements in sequencing technology should aid in the rate of discovery in the near future. Ultimately, only through a shared understanding of the genetics, epigenomics, three-dimensional chromatin organization, and gene modelling studies, we will reach our goal of more completely understanding the link between genetic variation and the causative mechanisms for AF.
Conflict of interest: P.T.E. is the principal investigator on a grant from Bayer HealthCare to the Broad Institute related to the genetics and therapeutics of atrial fibrillation.
Funding
This work was supported by grants from the National Institutes of Health to P.T.E. (1RO1HL092577, R01HL128914, and K24HL105780). S.C. was supported by a Marie Curie International Outgoing Fellowship within the 7th European Community Framework Programme (PIOF-GA-2012-328352) and by the German Centre for Cardiovascular Research (DZHK; 81X2600210, 81X2600204). P.T.E. is also supported by an Established Investigator Award from the American Heart Association (13EIA14220013) and by the Fondation Leducq (14CVD01).
References
- 1.Arnar DO, Thorvaldsson S, Manolio TA, Thorgeirsson G, Kristjansson K, Hakonarson H, Stefansson K. Familial aggregation of atrial fibrillation in Iceland. Eur Heart J 2006;27:708–712. [DOI] [PubMed] [Google Scholar]
- 2.Ellinor PT, Yoerger DM, Ruskin JN, MacRae CA. Familial aggregation in lone atrial fibrillation. Hum Genet 2005;118:179–184. [DOI] [PubMed] [Google Scholar]
- 3.Fox CS, Parise H, D'Agostino RB, Lloyd-Jones DM, Vasan RS, Wang TJ, Levy D, Wolf PA, Benjamin EJ. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. JAMA 2004;291:2851–2855. [DOI] [PubMed] [Google Scholar]
- 4.Lubitz SA, Yin X, Fontes JD, Magnani JW, Rienstra M, Pai M, Villalon ML, Vasan RS, Pencina MJ, Levy D, Larson MG, Ellinor PT, Benjamin EJ. Association between familial atrial fibrillation and risk of new-onset atrial fibrillation. JAMA 2010;304:2263–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christophersen IE, Ravn LS, Budtz-Joergensen E, Skytthe A, Haunsoe S, Svendsen JH, Christensen K. Familial aggregation of atrial fibrillation: a study in Danish twins. Circ Arrhythm Electrophysiol 2009;2:378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tucker NR, Ellinor PT. Emerging directions in the genetics of atrial fibrillation. Circ Res 2014;114:1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gollob MH, Jones DL, Krahn AD, Danis L, Gong X-Q, Shao Q, Liu X, Veinot JP, Tang ASL, Stewart AFR, Tesson F, Klein GJ, Yee R, Skanes AC, Guiraudon GM, Ebihara L, Bai D. Somatic mutations in the connexin 40 gene (GJA5) in atrial fibrillation. N Engl J Med 2006;354:2677–2688. [DOI] [PubMed] [Google Scholar]
- 8.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K, Palsson A, Blondal T, Sulem P, Backman VM, Hardarson GA, Palsdottir E, Helgason A, Sigurjonsdottir R, Sverrisson JT, Kostulas K, Ng MC, Baum L, So WY, Wong KS, Chan JC, Furie KL, Greenberg SM, Sale M, Kelly P, MacRae CA, Smith EE, Rosand J, Hillert J, Ma RC, Ellinor PT, Thorgeirsson G, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 2007;448:353–357. [DOI] [PubMed] [Google Scholar]
- 9.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, Gulcher J, Mathiesen EB, Njølstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Kucera G, Stubblefield T, Carter S, Roden D, Ng MC, Baum L, So WY, Wong KS, Chan JC, Gieger C, Wichmann HE, Gschwendtner A, Dichgans M, Kuhlenbäumer G, Berger K, Ringelstein EB, Bevan S, Markus HS, Kostulas K, Hillert J, Sveinbjörnsdóttir S, Valdimarsson EM, Løchen ML, Ma RC, Darbar D, Kong A, Arnar DO, Thorsteinsdottir U, Stefansson K. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet 2009;41:876–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, Schnabel RB, Bis JC, Boerwinkle E, Sinner MF, Dehghan A, Lubitz SA, D'Agostino RB Sr, Lumley T, Ehret GB, Heeringa J, Aspelund T, Newton-Cheh C, Larson MG, Marciante KD, Soliman EZ, Rivadeneira F, Wang TJ, Eiríksdottir G, Levy D, Psaty BM, Li M, Chamberlain AM, Hofman A, Vasan RS, Harris TB, Rotter JI, Kao WH, Agarwal SK, Stricker BH, Wang K, Launer LJ, Smith NL, Chakravarti A, Uitterlinden AG, Wolf PA, Sotoodehnia N, Köttgen A, van Duijn CM, Meitinger T, Mueller M, Perz S, Steinbeck G, Wichmann HE, Lunetta KL, Heckbert SR, Gudnason V, Alonso A, Kääb S, Ellinor PT, Witteman JC. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet 2009;41:879–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, Sinner MF, de Bakker PI, Mueller M, Lubitz SA, Fox E, Darbar D, Smith NL, Smith JD, Schnabel RB, Soliman EZ, Rice KM, Van Wagoner DR, Beckmann BM, van Noord C, Wang K, Ehret GB, Rotter JI, Hazen SL, Steinbeck G, Smith AV, Launer LJ, Harris TB, Makino S, Nelis M, Milan DJ, Perz S, Esko T, Köttgen A, Moebus S, Newton-Cheh C, Li M, Möhlenkamp S, Wang TJ, Kao WH, Vasan RS, Nöthen MM, MacRae CA, Stricker BH, Hofman A, Uitterlinden AG, Levy D, Boerwinkle E, Metspalu A, Topol EJ, Chakravarti A, Gudnason V, Psaty BM, Roden DM, Meitinger T, Wichmann HE, Witteman JC, Barnard J, Arking DE, Benjamin EJ, Heckbert SR, Kääb S. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet 2010;42:240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Müller-Nurasyid M, Krijthe BP, Lubitz SA, Bis JC, Chung MK, Dörr M, Ozaki K, Roberts JD, Smith JG, Pfeufer A, Sinner MF, Lohman K, Ding J, Smith NL, Smith JD, Rienstra M, Rice KM, Van Wagoner DR, Magnani JW, Wakili R, Clauss S, Rotter JI, Steinbeck G, Launer LJ, Davies RW, Borkovich M, Harris TB, Lin H, Völker U, Völzke H, Milan DJ, Hofman A, Boerwinkle E, Chen LY, Soliman EZ, Voight BF, Li G, Chakravarti A, Kubo M, Tedrow UB, Rose LM, Ridker PM, Conen D, Tsunoda T, Furukawa T, Sotoodehnia N, Xu S, Kamatani N, Levy D, Nakamura Y, Parvez B, Mahida S, Furie KL, Rosand J, Muhammad R, Psaty BM, Meitinger T, Perz S, Wichmann HE, Witteman JC, Kao WH, Kathiresan S, Roden DM, Uitterlinden AG, Rivadeneira F, McKnight B, Sjögren M, Newman AB, Liu Y, Gollob MH, Melander O, Tanaka T, Stricker BH, Felix SB, Alonso A, Darbar D, Barnard J, Chasman DI, Heckbert SR, Benjamin EJ, Gudnason V, Kääb S. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet 2012;44:670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lubitz SA, Lunetta KL, Lin H, Arking DE, Trompet S, Li G, Krijthe BP, Chasman DI, Barnard J, Kleber ME, Dörr M, Ozaki K, Smith AV, Müller-Nurasyid M, Walter S, Agarwal SK, Bis JC, Brody JA, Chen LY, Everett BM, Ford I, Franco OH, Harris TB, Hofman A, Kääb S, Mahida S, Kathiresan S, Kubo M, Launer LJ, Macfarlane PW, Magnani JW, McKnight B, McManus DD, Peters A, Psaty BM, Rose LM, Rotter JI, Silbernagel G, Smith JD, Sotoodehnia N, Stott DJ, Taylor KD, Tomaschitz A, Tsunoda T, Uitterlinden AG, Van Wagoner DR, Völker U, Völzke H, Murabito JM, Sinner MF, Gudnason V, Felix SB, März W, Chung M, Albert CM, Stricker BH, Tanaka T, Heckbert SR, Jukema JW, Alonso A, Benjamin EJ, Ellinor PT. Novel genetic markers associate with atrial fibrillation risk in Europeans and Japanese. J Am Coll Cardiol 2014;63:1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sinner MF, Tucker NR, Lunetta KL, Ozaki K, Smith JG, Trompet S, Bis JC, Lin H, Chung MK, Nielsen JB, Lubitz SA, Krijthe BP, Magnani JW, Ye J, Gollob MH, Tsunoda T, Müller-Nurasyid M, Lichtner P, Peters A, Dolmatova E, Kubo M, Smith JD, Psaty BM, Smith NL, Jukema JW, Chasman DI, Albert CM, Ebana Y, Furukawa T, Macfarlane PW, Harris TB, Darbar D, Dörr M, Holst AG, Svendsen JH, Hofman A, Uitterlinden AG, Gudnason V, Isobe M, Malik R, Dichgans M, Rosand J, Van Wagoner DR; METASTROKE Consortium; AFGen Consortium, Benjamin EJ, Milan DJ, Melander O, Heckbert SR, Ford I, Liu Y, Barnard J, Olesen MS, Stricker BH, Tanaka T, Kääb S, Ellinor PT. Integrating genetic, transcriptional, and functional analyses to identify 5 novel genes for atrial fibrillation. Circulation 2014;130:1225–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet 2013;45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett BM, Cook NR, Conen D, Chasman DI, Ridker PM, Albert CM. Novel genetic markers improve measures of atrial fibrillation risk prediction. Eur Heart J 2013;34:2243–2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parvez B, Shoemaker MB, Muhammad R, Richardson R, Jiang L, Blair MA, Roden DM, Darbar D. Common genetic polymorphism at 4q25 locus predicts atrial fibrillation recurrence after successful cardioversion. Heart Rhythm 2013;10:849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benjamin Shoemaker M, Muhammad R, Parvez B, White BW, Streur M, Song Y, Stubblefield T, Kucera G, Blair M, Rytlewski J, Parvathaneni S, Nagarakanti R, Saavedra P, Ellis CR, Patrick Whalen S, Roden DM, Darbar RD. Common atrial fibrillation risk alleles at 4q25 predict recurrence after catheter-based atrial fibrillation ablation. Heart Rhythm 2013;10:394–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shoemaker MB, Bollmann A, Lubitz SA, Ueberham L, Saini H, Montgomery J, Edwards T, Yoneda Z, Sinner MF, Arya A, Sommer P, Delaney J, Goyal SK, Saavedra P, Kanagasundram A, Whalen SP, Roden DM, Hindricks G, Ellis CR, Ellinor PT, Darbar D, Husser D. Common genetic variants and response to atrial fibrillation ablation. Circ Arrhythm Electrophysiol 2015;8:296–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Husser D, Adams V, Piorkowski C, Hindricks G, Bollmann A. Chromosome 4q25 variants and atrial fibrillation recurrence after catheter ablation. J Am Coll Cardiol 2010;55:747–753. [DOI] [PubMed] [Google Scholar]
- 21.Parvez B, Vaglio J, Rowan S, Muhammad R, Kucera G, Stubblefield T, Carter S, Roden D, Darbar D. Symptomatic response to antiarrhythmic drug therapy is modulated by a common single nucleotide polymorphism in atrial fibrillation. J Am Coll Cardiol 2012;60:539–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi E-K, Park JH, Lee J-Y, Nam CM, Hwang MK, Uhm J-S, Joung B, Ko Y-G, Lee M-H, Lubitz SA, Ellinor PT, Pak H-N. Korean Atrial Fibrillation (AF) Network: genetic variants for AF do not predict ablation success. J Am Heart Assoc 2015;4:e002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikpay M, Goel A, Won HH, Hall LM, Willenborg C, Kanoni S, Saleheen D, Kyriakou T, Nelson CP, Hopewell JC, Webb TR, Zeng L, Dehghan A, Alver M, Armasu SM, Auro K, Bjonnes A, Chasman DI, Chen S, Ford I, Franceschini N, Gieger C, Grace C, Gustafsson S, Huang J, Hwang SJ, Kim YK, Kleber ME, Lau KW, Lu X, Lu Y, Lyytikäinen LP, Mihailov E, Morrison AC, Pervjakova N, Qu L, Rose LM, Salfati E, Saxena R, Scholz M, Smith AV, Tikkanen E, Uitterlinden A, Yang X, Zhang W, Zhao W, de Andrade M, de Vries PS, van Zuydam NR, Anand SS, Bertram L, Beutner F, Dedoussis G, Frossard P, Gauguier D, Goodall AH, Gottesman O, Haber M, Han BG, Huang J, Jalilzadeh S, Kessler T, König IR, Lannfelt L, Lieb W, Lind L, Lindgren CM, Lokki ML, Magnusson PK, Mallick NH, Mehra N, Meitinger T, Memon FU, Morris AP, Nieminen MS, Pedersen NL, Peters A, Rallidis LS, Rasheed A, Samuel M, Shah SH, Sinisalo J, Stirrups KE, Trompet S, Wang L, Zaman KS, Ardissino D, Boerwinkle E, Borecki IB, Bottinger EP, Buring JE, Chambers JC, Collins R, Cupples LA, Danesh J, Demuth I, Elosua R, Epstein SE, Esko T, Feitosa MF, Franco OH, Franzosi MG, Granger CB, Gu D, Gudnason V, Hall AS, Hamsten A, Harris TB, Hazen SL, Hengstenberg C, Hofman A, Ingelsson E, Iribarren C, Jukema JW, Karhunen PJ, Kim BJ, Kooner JS, Kullo IJ, Lehtimäki T, Loos RJ, Melander O, Metspalu A, März W, Palmer CN, Perola M, Quertermous T, Rader DJ, Ridker PM, Ripatti S, Roberts R, Salomaa V, Sanghera DK, Schwartz SM, Seedorf U, Stewart AF, Stott DJ, Thiery J, Zalloua PA, O'Donnell CJ, Reilly MP, Assimes TL, Thompson JR, Erdmann J, Clarke R, Watkins H, Kathiresan S, McPherson R, Deloukas P, Schunkert H, Samani NJ, Farrall M; CARDIoGRAMplusC4D Consortium. A comprehensive 1000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ripke S, Neale BM, Corvin A, Walters JT, Farh KH, Holmans PA, Lee P, Bulik-Sullivan B, Collier DA, Huang H, Pers TH, Agartz I, Agerbo E, Albus M, Alexander M, Amin F, Bacanu SA, Begemann M, Belliveau RA Jr, Bene J, Bergen SE, Bevilacqua E, Bigdeli TB, Black DW, Bruggeman R, Buccola NG, Buckner RL, Byerley W, Cahn W, Cai G, Campion D, Cantor RM, Carr VJ, Carrera N, Catts SV, Chambert KD, Chan RC, Chen RY, Chen EY, Cheng W, Cheung EF, Chong SA, Cloninger CR, Cohen D, Cohen N, Cormican P, Craddock N, Crowley JJ, Curtis D, Davidson M, Davis KL, Degenhardt F, Del Favero J, Demontis D, Dikeos D, Dinan T, Djurovic S, Donohoe G, Drapeau E, Duan J, Dudbridge F, Durmishi N, Eichhammer P, Eriksson J, Escott-Price V, Essioux L, Fanous AH, Farrell MS, Frank J, Franke L, Freedman R, Freimer NB, Friedl M, Friedman JI, Fromer M, Genovese G, Georgieva L, Giegling I, Giusti-Rodríguez P, Godard S, Goldstein JI, Golimbet V, Gopal S, Gratten J, de Haan L, Hammer C, Hamshere ML, Hansen M, Hansen T, Haroutunian V, Hartmann AM, Henskens FA, Herms S, Hirschhorn JN, Hoffmann P, Hofman A, Hollegaard MV, Hougaard DM, Ikeda M, Joa I, Julià A, Kahn RS, Kalaydjieva L, Karachanak-Yankova S, Karjalainen J, Kavanagh D, Keller MC, Kennedy JL, Khrunin A, Kim Y, Klovins J, Knowles JA, Konte B, Kucinskas V, Ausrele Kucinskiene Z, Kuzelova-Ptackova H, Kähler AK, Laurent C, Keong JL, Lee SH, Legge SE, Lerer B, Li M, Li T, Liang KY, Lieberman J, Limborska S, Loughland CM, Lubinski J, Lönnqvist J, Macek M Jr, Magnusson PK, Maher BS, Maier W, Mallet J, Marsal S, Mattheisen M, Mattingsdal M, McCarley RW, McDonald C, McIntosh AM, Meier S, Meijer CJ, Melegh B, Melle I, Mesholam-Gately RI, Metspalu A, Michie PT, Milani L, Milanova V, Mokrab Y, Morris DW, Mors O, Murphy KC, Murray RM, Myin-Germeys I, Müller-Myhsok B, Nelis M, Nenadic I, Nertney DA, Nestadt G, Nicodemus KK, Nikitina-Zake L, Nisenbaum L, Nordin A, O'Callaghan E, O'Dushlaine C, O'Neill FA, Oh SY, Olincy A, Olsen L, Van Os J, Pantelis C, Papadimitriou GN, Papiol S, Parkhomenko E, Pato MT, Paunio T, Pejovic-Milovancevic M, Perkins DO, Pietiläinen O, Pimm J, Pocklington AJ, Powell J, Price A, Pulver AE, Purcell SM, Quested D, Rasmussen HB, Reichenberg A, Reimers MA, Richards AL, Roffman JL, Roussos P, Ruderfer DM, Salomaa V, Sanders AR, Schall U, Schubert CR, Schulze TG, Schwab SG, Scolnick EM, Scott RJ, Seidman LJ, Shi J, Sigurdsson E, Silagadze T, Silverman JM, Sim K, Slominsky P, Smoller JW, So HC, Spencer CA, Stahl EA, Stefansson H, Steinberg S, Stogmann E, Straub RE, Strengman E, Strohmaier J, Stroup TS, Subramaniam M, Suvisaari J, Svrakic DM, Szatkiewicz JP, Söderman E, Thirumalai S, Toncheva D, Tosato S, Veijola J, Waddington J, Walsh D, Wang D, Wang Q, Webb BT, Weiser M, Wildenauer DB, Williams NM, Williams S, Witt SH, Wolen AR, Wong EH, Wormley BK, Xi HS, Zai CC, Zheng X, Zimprich F, Wray NR, Stefansson K, Visscher PM, Adolfsson R, Andreassen OA, Blackwood DH, Bramon E, Buxbaum JD, Børglum AD, Cichon S, Darvasi A, Domenici E, Ehrenreich H, Esko T, Gejman PV, Gill M, Gurling H, Hultman CM, Iwata N, Jablensky AV, Jönsson EG, Kendler KS, Kirov G, Knight J, Lencz T, Levinson DF, Li QS, Liu J, Malhotra AK, McCarroll SA, McQuillin A, Moran JL, Mortensen PB, Mowry BJ, Nöthen MM, Ophoff RA, Owen MJ, Palotie A, Pato CN, Petryshen TL, Posthuma D, Rietschel M, Riley BP, Rujescu D, Sham PC, Sklar P, St Clair D, Weinberger DR, Wendland JR, Werge T, Daly MJ, Sullivan PF, O'Donovan MC. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014;511:421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen P-S, Chen LS, Fishbein MC, Lin S-F, Nattel S. Role of the autonomic nervous system in atrial fibrillation: pathophysiology and therapy. Circ Res 2014;114:1500–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heijman J, Voigt N, Nattel S, Dobrev D. Cellular and molecular electrophysiology of atrial fibrillation initiation, maintenance, and progression. Circ Res 2014;114:1483–1499. [DOI] [PubMed] [Google Scholar]
- 27.Andrade J, Khairy P, Dobrev D, Nattel S. The clinical profile and pathophysiology of atrial fibrillation: relationships among clinical features, epidemiology, and mechanisms. Circ Res 2014;114:1453–1468. [DOI] [PubMed] [Google Scholar]
- 28.International HapMap Consortium. The International HapMap Project. Nature 2003;426:789–796. [DOI] [PubMed] [Google Scholar]
- 29.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004;306:636–640. [DOI] [PubMed] [Google Scholar]
- 31.Roadmap Epigenomics Consortium, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJ, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin H, Dolmatova EV, Morley MP, Lunetta KL, McManus DD, Magnani JW, Margulies KB, Hakonarson H, Del Monte F, Benjamin EJ, Cappola TP, Ellinor PT. Gene expression and genetic variation in human atria. Heart Rhythm 2014:11:266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gore-Panter SR, Hsu J, Hanna P, Gillinov AM, Pettersson G, Newton DW, Moravec CS, Van Wagoner DR, Chung MK, Barnard J, Smith JD. Atrial fibrillation associated chromosome 4q25 variants are not associated with PITX2c expression in human adult left atrial appendages. PLoS ONE 2014;9:e86245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Wit E, de Laat W. A decade of 3C technologies: insights into nuclear organization. Genes Dev 2012;26:11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Ferrante TC, Terry R, Turczyk BM, Yang JL, Lee HS, Aach J, Zhang K, Church GM. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc 2015;10:442–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JH, Daugharthy ER, Scheiman J, Kalhor R, Yang JL, Ferrante TC, Terry R, Jeanty SSF, Li C, Amamoto R, Peters DT, Turczyk BM, Marblestone AH, Inverso SA, Bernard A, Mali P, Rios X, Aach J, Church GM. Highly multiplexed subcellular RNA sequencing in situ. Science 2014;343:1360–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riley G, Syeda F, Kirchhof P, Fabritz L. An introduction to murine models of atrial fibrillation. Front Physiol 2012;3:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nishida K, Michael G, Dobrev D, Nattel S. Animal models for atrial fibrillation: clinical insights and scientific opportunities. Europace 2009;12:160–172. [DOI] [PubMed] [Google Scholar]
- 39.Prather RS, Lorson M, Ross JW, Whyte JJ, Walters E. Genetically engineered pig models for human diseases. Annu Rev Anim Biosci 2013;1:203–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, Gonzales APW, Li Z, Peterson RT, Yeh J-RJ, Aryee MJ, Joung JK. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015;523:481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doetschman T, Azhar M. Cardiac-specific inducible and conditional gene targeting in mice. Circ Res 2012;110:1498–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milan DJ, Macrae CA. Zebrafish genetic models for arrhythmia. Prog Biophys Mol Biol 2008;98:301–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verkerk AO, Remme CA. Zebrafish: a novel research tool for cardiac (patho)electrophysiology and ion channel disorders. Front Physiol 2012;3:255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahida S, Mills RW, Tucker NR, Simonson B, Macri V, Lemoine MD, Das S, Milan DJ, Ellinor PT. Overexpression of KCNN3 results in sudden cardiac death. Cardiovasc Res 2014;101:326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaese S, Verheule S. Cardiac electrophysiology in mice: a matter of size. Front Physiol 2012;3:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leyton-Mange JS, Mills RW, Macri VS, Jang MY, Butte FN, Ellinor PT, Milan DJ. Rapid cellular phenotyping of human pluripotent stem cell-derived cardiomyocytes using a genetically encoded fluorescent voltage sensor. Stem Cell Rep 2014;2:163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moretti A, Bellin M, Welling A, Jung CB, Lam JT, Bott-Flügel L, Dorn T, Goedel A, Höhnke C, Hofmann F, Seyfarth M, Sinnecker D, Schömig A, Laugwitz K-L. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med 2010;363:1397–1409. [DOI] [PubMed] [Google Scholar]
- 48.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, Pirruccello JP, Muchmore B, Prokunina-Olsson L, Hall JL, Schadt EE, Morales CR, Lund-Katz S, Phillips MC, Wong J, Cantley W, Racie T, Ejebe KG, Orho-Melander M, Melander O, Koteliansky V, Fitzgerald K, Krauss RM, Cowan CA, Kathiresan S, Rader DJ. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature 2010;466:714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kapoor A, Sekar RB, Hansen NF, Fox-Talbot K, Morley M, Pihur V, Chatterjee S, Brandimarto J, Moravec CS, Pulit SL, Pfeufer A, Mullikin J, Ross M, Green ED, Bentley D, Newton-Cheh C, Boerwinkle E, Tomaselli GF, Cappola TP, Arking DE, Halushka MK, Chakravarti A. An enhancer polymorphism at the cardiomyocyte intercalated disc protein NOS1AP locus is a major regulator of the QT interval. Am J Hum Genet 2014;94:854–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van den Boogaard M, Smemo S, Burnicka-Turek O, Arnolds DE, van de Werken HJG, Klous P, McKean D, Muehlschlegel JD, Moosmann J, Toka O, Yang XH, Koopmann TT, Adriaens ME, Bezzina CR, de Laat W, Seidman C, Seidman JG, Christoffels VM, Nobrega MA, Barnett P, Moskowitz IP. A common genetic variant within SCN10A modulates cardiac SCN5A expression. J Clin Invest 2014;124:1844–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bauer DE, Kamran SC, Lessard S, Xu J, Fujiwara Y, Lin C, Shao Z, Canver MC, Smith EC, Pinello L, Sabo PJ, Vierstra J, Voit RA, Yuan G-C, Porteus MH, Stamatoyannopoulos JA, Lettre G, Orkin SH. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 2013;342:253–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spieler D, Kaffe M, Knauf F, Bessa J, Tena JJ, Giesert F, Schormair B, Tilch E, Lee H, Horsch M, Czamara D, Karbalai N, von Toerne C, Waldenberger M, Gieger C, Lichtner P, Claussnitzer M, Naumann R, Müller-Myhsok B, Torres M, Garrett L, Rozman J, Klingenspor M, Gailus-Durner V, Fuchs H, Hrabě de Angelis M, Beckers J, Hölter SM, Meitinger T, Hauck SM, Laumen H, Wurst W, Casares F, Gómez-Skarmeta JL, Winkelmann J. Restless legs syndrome-associated intronic common variant in Meis1 alters enhancer function in the developing telencephalon. Genome Res 2014;24:592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patwardhan RP, Hiatt JB, Witten DM, Kim MJ, Smith RP, May D, Lee C, Andrie JM, Lee S-I, Cooper GM, Ahituv N, Pennacchio LA, Shendure J. Massively parallel functional dissection of mammalian enhancers in vivo. Nat Biotechnol. 2012;30:265–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melnikov A, Murugan A, Zhang X, Tesileanu T, Wang L, Rogov P, Feizi S, Gnirke A, Callan CG, Kinney JB, Kellis M, Lander ES, Mikkelsen TS. Systematic dissection and optimization of inducible enhancers in human cells using a massively parallel reporter assay. Nat Biotechnol 2012;30:271–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Melnikov A, Zhang X, Rogov P, Wang L, Mikkelsen TS. Massively parallel reporter assays in cultured mammalian cells. J Vis Exp 2014;90:e51719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kheradpour P, Ernst J, Melnikov A, Rogov P, Wang L, Zhang X, Alston J, Mikkelsen TS, Kellis M. Systematic dissection of regulatory motifs in 2000 predicted human enhancers using a massively parallel reporter assay. Genome Res 2013;23:800–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sandelin A, Alkema W, Engström P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 2004;32:D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012;40:D930–D934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002;18:333–334. [DOI] [PubMed] [Google Scholar]
- 60.Matys V, Fricke E, Geffers R, Gössling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos D-U, Land S, Lewicki-Potapov B, Michael H, Münch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res 2003;31:374–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein–DNA interactions. Nucleic Acids Res 2009;37:D77–D82. [DOI] [PMC free article] [PubMed] [Google Scholar]