Abstract
AIM: To detect the expression of sal-like protein 4 (SALL4) and to explore its relationship with clinicopathological characteristics and prognosis of hepatocellular carcinoma (HCC).
METHODS: One hundred and twenty-six samples of HCC tissue, 44 of adjacent noncancerous cirrhotic tissue and 10 of liver hemangioma tissue, were obtained from patients who underwent hepatectomy for HCC at the Fourth Hospital of Hebei Medical University. None of the patients had received any form of treatment before the operation. After resection, all the tissues were fixed in 10% neutral formaldehyde and embedded in paraffin. Expression of SALL4 was detected by immunohistochemistry. Patients were followed up for postoperative survival until February 2014. The relationships between SALL4 expression level and clinicopathological data and prognosis of HCC were analyzed.
RESULTS: SALL4 expression was negative in the 10 samples of tissue from liver hemangioma, was weakly positive in the two samples from adjacent noncancerous cirrhotic tissue, and positive in 58 samples of HCC tissues. The differences were statistically significant (P < 0.05). Expression of SALL4 was higher in patients with higher α-fetoprotein (AFP) levels, portal vein tumor thrombus, and later clinical stage based on the Barcelona Clinic Liver Cancer classification (P < 0.05). Among patients with negative expression, weakly positive expression, positive expression, and strongly positive expression of SALL4, the median survival time was 39, 25, 23, and 9 mo, respectively (P < 0.001). When both AFP and SALL4 were detected, patients who were negative for both AFP and SALL4, SALL4-positive only, AFP-positive only, and positive for both AFP and SALL4, had a median survival time of 41, 38, 31, and 12 mo, respectively (P < 0.001).
CONCLUSION: Expression of SALL4 is relevant to the prognosis of HCC patients. Patients with higher expression levels of SALL4 and AFP have worse prognosis.
Keywords: Sal-like protein 4, Hepatocellular carcinoma, Survival analysis, α-Fetoprotein, Prognosis
Core tip: Sal-like protein 4 (SALL4) is expressed as an oncogene in a variety of tumors, including germ cell tumors, breast cancer, and α-fetoprotein (AFP)-producing gastric cancer. However, there are few reports about the association between SALL4 expression and hepatocellular carcinoma (HCC). In this study, we found that SALL4 was actively expressed in HCC cells with stem cell characteristics and played an important role in predicting the prognosis of HCC patients. In addition, joint detection of SALL4 and AFP may have a more profound significance for the prognosis of HCC patients.
INTRODUCTION
Hepatocellular carcinoma (HCC) is a primary liver cancer and accounts for > 90% of cases. As HCC patients have no specific symptoms in the early stage, most patients have lost the best opportunity for treatment (radical surgery) when diagnosed. Effects to improve the survival rate of HCC patients are limited by the drug intervention, radiotherapy, chemotherapy, and molecular targeted therapy. Thus far, the pathogenesis of HCC is still not clear. However, with the rapid development of genetics and molecular biology, researchers have begun to explore the pathogenesis of HCC at the molecular genetic level.
The occurrence of HCC is relevant to a variety of signaling pathways, including the Notch, Wnt, mitogen-activated protein kinase (MAPK), and hedgehog signaling pathways[1-4]. Sal-like protein 4 (SALL4) is a newly discovered proto-oncogene that is expressed in embryonic stem cells and liver stem cells but is not expressed in normal adult liver cells. Recent studies have found that expression of SALL4 can be detected in HCC specimens.
SALL4 is located on human chromosome 20q13 and is a zinc finger transcription factor that includes two subtypes: SALL4A and SALL4B. Mutation of SALL4 causes Okihiro syndrome and familial IVIC syndrome, which are autosomal dominant genetic diseases that manifest as multiple organ dysfunction[5,6]. SALL4 is highly expressed in embryonic stem cells and plays an important role in early embryogenesis, organogenesis, embryonic stem cell proliferation, and maintenance of pluripotency, interacting with octamer-binding transcription factor 4 (OCT4), SRY-box containing gene 2 (SOX2), and NANOG[7-9]. SALL4 is expressed in normal hematopoietic stem cells, and its expression decreases gradually with cell differentiation and maturation. High expression levels of SALL4 are related to certain hematopoietic malignancies and can lead to acute leukemia in transgenic mice[10]. In addition, acute and chronic myeloid leukemia, along with acute lymphocytic leukemia, is also associated with high levels of SALL4 expression[11], which suggests that SALL4 is a key regulator of leukemic cells[12]. SALL4 is expressed as an oncogene in a variety of tumors, including germ cell tumors, breast cancer, and α-fetoprotein (AFP)-producing gastric cancer[13-15], which indicates that SALL4 may be involved in the development of tumors[16]. Recent studies have found that SALL4 is actively expressed in HCC with stem cell characteristics and plays an important role in predicting the prognosis of HCC patients. Therefore, further studies of SALL4 may provide new therapeutic targets for HCC patients.
MATERIALS AND METHODS
Clinical HCC specimens
One hundred and twenty-six specimens of HCC tissues, 44 adjacent noncancerous cirrhotic hepatic tissues, and 10 liver hemangioma tissues were obtained from patients who underwent hepatectomy for HCC from May 2008 to December 2010 at the Fourth Hospital of Hebei Medical University, Shijiazhuang, China. The patients had not received any treatment before surgery. The samples were formalin-fixed, paraffin-embedded, and subjected to immunohistochemistry. The clinicopathology of all specimens had already been established by pathologists.
Follow-up
Patients were followed up by telephone or correspondence from the day of surgery to February 2014. The median follow up period was 23 mo (range from 0 to 70 mo). Among the 126 patients, 40 were alive and 11 were lost to follow-up. All patients gave their informed consent to this research.
Immunohistochemistry
After resection, all the tissues were fixed with 10% neutral formaldehyde and embedded in paraffin. Immunohistochemistry was performed using Maxvision. The primary antibody was purchased from Santa Cruz Biotechnology (Dallas, TX, United States) and used at a dilution of 1:80.
Statistical analysis
The data were analyzed using SPSS version 13.0 software (Chicago, IL, United States). Nonparametric tests were used to compare the expression levels of each group, and Spearman correlation was used for correlation analysis. Survival rate was calculated by the Kaplan-Meier and Cox Regression method, and the differences in survival rate were compared by the log-rank test. P < 0.05 was considered statistically significant.
RESULTS
Expression sites and form of SALL4
SALL4 was not expressed in liver hemangioma tissues (Figure 1A), and there was almost no expression in adjacent noncancerous cirrhotic tissues (Figure 1B). It was, however, partly expressed in HCC tissues with light yellow to brownish yellow granules in the nuclei (Figure 1C).
Figure 1.
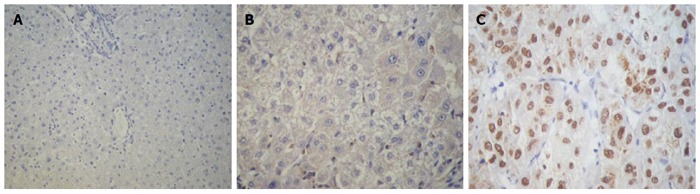
Expression of sal-like protein 4 in liver tissues. A: Expression of sal-like protein 4 (SALL4) in liver hemangioma tissue (Maxvision, magnification × 200); B: Expression of SALL4 in adjacent noncancerous cirrhotic tissue (Maxvision, magnification × 400); C: Expression of SALL4 in hepatocellular carcinoma tissue (Maxvision, magnification × 400).
Differential expression of SALL4 among experimental groups
Tests for the expression of SALL4 in 10 liver hemangioma tissues were all negative. Among the 44 specimens from adjacent noncancerous cirrhotic tissues; two were weakly positive, and 42 were negative. Among the 126 HCC tissue specimens; 68 were negative, and 58 were positive. There was no significant difference in the expression of SALL4 between liver hemangioma tissues and adjacent noncancerous cirrhotic tissues (P = 0.496). However, there was a significant difference between liver hemangioma and HCC tissue (P = 0.009) and between adjacent noncancerous hepatic tissue and HCC tissue (P < 0.001) (Table 1).
Table 1.
Expression of SALL4 in hepatocellular carcinoma tissue, non-cancerous cirrhotic tissue, and liver hemangioma tissue
| SALL4 | Hemangioma tissue (n = 10) | Adjacent non-cancerous cirrhotic tissue (n = 44) | HCC tissue (n = 126) |
| - | 10 | 42 | 68 |
| + | 0 | 2 | 27 |
| ++ | 0 | 0 | 21 |
| +++ | 0 | 0 | 10 |
| P value | 0.496 | 0 | 0.009 |
P value was obtained by non-parametric test. P = 0.496 represents hemangioma group vs adjacent non-cancerous cirrhotic group; P < 0.001 represents adjacent non-cancerous cirrhotic group vs HCC group; P = 0.009 represents HCC group vs hemangioma group. HCC: Hepatocellular carcinoma; SALL4: Sal-like protein 4.
Relationship between expression of SALL4 in HCC tissues and clinicopathological characteristics
Expression of SALL4 in HCC tissue was not related to histological differentiation but was positively correlated with clinical stage, as it was strongly expressed at a later clinical stage (P < 0.05). Expression of SALL4 was also positively correlated with Barcelona Clinic Liver Cancer (BCLC) classification and strongly expressed in patients in later BCLC B and C (P < 0.05) (Figure 2A and Table 2). Expression of SALL4 was not related to sex, age, hepatitis B surface antigen, hepatitis B e antibody, blood platelets, Child-Pugh score, or tumor diameter but was positively correlated with AFP, and SALL4 was strongly expressed in patients with higher AFP levels (P < 0.05). Expression of SALL4 was also higher in the presence of portal vein tumor thrombosis (P < 0.05) (Table 2).
Figure 2.
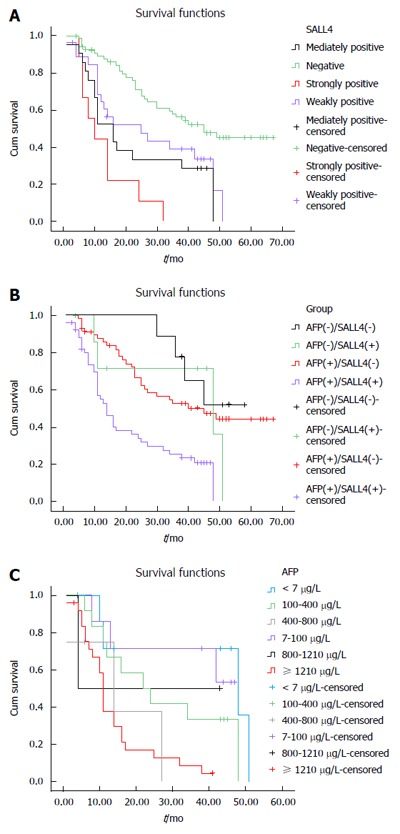
Survival analysis. A: Survival analysis correlated with expression of SALL4 in postoperative hepatocellular carcinoma patients; B: Survival analysis of postoperative hepatocellular carcinoma patients correlated with combinedα-fetoprotein level and SALL4 expression; C: Survival analysis of SALL4-positive hepatocellular carcinoma patients correlated with α-fetoprotein. AFP: α-fetoprotein; SALL4: Sal-like protein 4.
Table 2.
Relationship between the expression of SALL4 and clinical characteristics in hepatocellular carcinoma patients
| Parameters | SALL4 (-/+) | P value1 | P value2 |
| Clinical stages | |||
| Ia | 11/7 | ||
| Ib | 17/8 | ||
| IIa | 21/13 | ||
| IIb | 11/18 | ||
| IIIa | 8/12 | 0.034 | 0.006 |
| Histological differentiation | |||
| Well | 9/4 | ||
| Moderate | 30/23 | ||
| Poor | 29/31 | 0.344 | 0.069 |
| BCLC stages | |||
| A | 53/29 | ||
| B | 3/2 | ||
| C | 12/27 | 0.001 | 0.000 |
| Sex | |||
| Male | 57/50 | ||
| Female | 11/8 | 0.931 | 0.932 |
| Age (yr) | |||
| < 50 | 24/21 | ||
| ≥ 50 | 44/37 | 0.918 | 0.918 |
| Tumor diameter (cm) | |||
| < 3 | 6/6 | ||
| 3-5 | 22/12 | ||
| 5-10 | 24/22 | ||
| ≥ 10 | 16/18 | 0.176 | 0.042 |
| Platelet | |||
| (× 109/L) | |||
| < 100 | 9/11 | ||
| 100-200 | 47/32 | ||
| 200-300 | 9/12 | ||
| ≥ 300 | 3/3 | 0.231 | 0.863 |
| Vascular invasion | |||
| With | 11/21 | ||
| Without | 57/37 | 0.004 | 0.004 |
| HBsAg | |||
| Positive | 58/51 | ||
| Negative | 10/7 | 0.487 | 0.490 |
| HBeAb | |||
| Positive | 28/19 | ||
| Negative | 40/39 | 0.359 | 0.361 |
| Child-Pugh | |||
| Child A | 67/57 | ||
| Child B | 1/1 | 0.822 | 0.823 |
| AFP (μg/L) | |||
| < 7 | 9/7 | ||
| 7-100 | 26/8 | ||
| 100-400 | 10/12 | ||
| 400-800 | 5/4 | ||
| 800-1210 | 0/2 | ||
| ≥ 1210 | 18/25 | 0.013 | 0.003 |
P value was obtained by non-parametric test;
P value was obtained by Spearman correlation. AFP: α-fetoprotein; SALL4: Sal-like protein 4.
Prognostic significance of combined detection of AFP and SALL4
In HCC tissue, expression of SALL4 was correlated with prognosis of HCC patients. Patients with negative expression, weak positive expression, positive expression, and strong positive expression of SALL4 displayed a median survival time of 39, 25, 23, and 9 mo, respectively (P < 0.001) (Table 3). In the HCC group, patients with higher expression levels of SALL4 usually had shorter median survival time and worse prognosis (Figure 2B). In addition, on multivariable analysis, the result showed that clinical stages (P = 0.002), histological differentiation (P = 0.003), AFP (P = 0.046) , and SALL4 (P = 0.029), remained as significant independent predictors of prognosis of HCC patients (Table 4).
Table 3.
Survival time vs SALL4 expression in postoperative hepatocellular carcinoma patients
| SALL4 | Died/total | Median survival time (mo) | P value |
| - | 32/68 | 39 | |
| + | 18/27 | 25 | |
| ++ | 16/21 | 23 | |
| +++ | 9/10 | 9 | 0.000 |
SALL4: Sal-like protein 4.
Table 4.
Multivariable analysis results in postoperative hepatocellular carcinoma patients
| Parameters | P value | Relative risk | 95%CI |
| Clinical stages | 0.002 | 1.547 | 1.169-2.047 |
| histological differentiation | 0.003 | 1.823 | 1.218-2.728 |
| BCLC stage | 0.876 | 1.028 | 0.725-1.459 |
| AFP | 0.046 | 1.158 | 1.002-1.338 |
| Tumor size | 0.503 | 1.133 | 0.786-1.635 |
| Vascular invasive | 0.531 | 0.796 | 0.389-1.627 |
| SALL4 | 0.029 | 1.301 | 1.028-1.646 |
SALL4: Sal-like protein 4; AFP: α-fetoprotein.
There were nine cases in which both AFP and SALL4 were negative with a median survival time of 41 mos. There were seven cases with positive expression of SALL4 only, with a median survival time of 38 mo. There were 59 AFP-positive cases, with a median survival time of 31 mo. For the 51 cases that were both AFP and SALL4 positive, the median survival time was 12 mo. The differences among the groups were statistically significant (P < 0.001) (Table 5). The group that was both AFP and SALL4 positive had the worst prognosis (Figure 2B). When the expression of SALL4 was positive, the number of patients with AFP < 7 μg/L, 7-100 μg/L, 100-400 μg/L, 400-800 μg/L, 800-1210 μg/L, and ≥ 1210 μg/L was seven, eight, 12, four, two, and 25, respectively, and they had a median survival time of 30, 26, 14, 10, 24, and 7 mo, respectively. The differences among the groups were statistically significant (P = 0.003) (Figure 2C and Table 6). As AFP level increased, survival time decreased. When the expression of SALL4 was positive, AFP played a role in predicting the prognosis of HCC patients.
Table 5.
Survival time vs combined α-fetoprotein level and SALL4 expression in postoperative hepatocellular carcinoma patients
| Group | Cases | Median survival time(mo) | P value |
| AFP (-)/SALL4 (-) | 9 | 41 | |
| AFP (-)/SALL4 (+) | 7 | 38 | |
| AFP (+)/SALL4 (-) | 59 | 31 | |
| AFP (+)/SALL4 (+) | 51 | 12 | 0.000 |
AFP (-) < 7 μg/L; AFP (+) ≥ 7 μg/L. AFP: α-fetoprotein; SALL4: Sal-like protein 4.
Table 6.
Survival time vs α-fetoprotein level in SALL4-positive postoperative hepatocellular carcinoma patients
| AFP (μg/L) | Cases | Median survival time (mo) | P value |
| < 7 | 7 | 30 | |
| 7-100 | 8 | 26 | |
| 100-400 | 12 | 14 | |
| 400-800 | 4 | 10 | |
| 800-1210 | 2 | 24 | |
| ≥ 1210 | 25 | 7 | 0.003 |
AFP: α-fetoprotein.
DISCUSSION
Some researchers have pointed out that SALL4 is not expressed in HCC tissues, but is highly expressed in AFP-producing gastric carcinoma tissues, and this was used as a method for distinguishing the two types of tissues[15,16]. However, the present study showed that SALL4 was detectable in HCC tissues. The reason for the different results is that SALL4 is not expressed in all HCC tissues. The HCC tissues in which SALL4 was positively expressed had features in common with stem cells and were characterized by a highly invasive nature and were also resistant to chemotherapy and radiotherapy[17]. Some reports showed that SALL4 is expressed in the nucleus and cytoplasm. The immunohistochemical results of Oikawa et al[18] indicated that SALL4 is expressed in both the nucleus and cytoplasm. Zeng et al[17], who evaluated the effect of SALL4 overexpression, found little expression of SALL4 in HuH1 cells. They transfected plasmid constructs encoding SALL4 (pCMV6-SALL4) and identified the expression of two isoforms using these constructs. Evaluation of the subcellular localization of GFP-tagged SALL4 (pCMV6-SALL4-GFP) showed that it could be detected in both the cytoplasm and the nucleus. The overexpression of SALL4 in HuH1 cells resulted in the activation of spheroid formation and invasive capacities and upregulated the level of hepatic stem cell markers cytokeratin (CK)19, epithelial cell adhesion molecule, and CD44. However, knockdown of SALL4 with SALL4 short hairpin RNA resulted in a decrease in spheroid formation capacity and invasive capacity, which suggests that SALL4 has the potential to enhance tumor invasiveness.
A recent study suggested that SALL4 forms a nucleosome remodeling and deacetylase (NuRD) complex with histone deacetylases (HDACs) and potentially regulates the activity of HDAC[19]. Knockdown of SALL4 decreased HDAC activity, while overexpression of SALL4 increased the activity of HDAC and enhanced the chemosensitivity to HDAC inhibitor. That study also found that the SALL4-NuRD complex had the same start sites as phosphatase and tensin homolog protein (PTEN) and, through competitive binding with the PTEN binding site, the expression of PTEN was decreased. As a tumor suppressor gene, PTEN plays an important role in the development of a variety of tumors, and the PTEN/phosphoinositide 3 kinase (PI3K)/Akt signaling pathway is closely related to HCC.
A recent report[20] described a SALL4 peptide, containing 12 amino acids, that competitively inhibited the binding of SALL4-NuRD complex, thereby blocking the interaction between SALL4 and NuRD. In addition, SALL4 peptides decreased the viability of HCC cells and increased expression of PTEN, but had no interaction with SALL4-negative cells, which suggests that SALL4 peptides have no toxic effect on normal cells. In a mouse xenograft model, tumor sizes were significantly smaller in mice that received the SALL4 peptide compared with mice that did not receive it. These results suggest that the SALL4 peptide is expected to be a new breakthrough therapy for SALL4-positive HCC patients.
It has been reported that SALL4 is not detected in chronic hepatitis but is faintly detected in bile ductules and in hepatocytes at the interface of parenchymal and stromal cells in liver cirrhosis[18]. Overexpression of SALL4 results in increased expression of CK19, which is a specific gene of the bile duct, and knockdown of SALL4 inhibited expression of CK19. These results suggest that SALL4 plays an important role in promoting differentiation of hepatoblasts into bile duct cells[21]. The observed expression of SALL4 in bile duct cells suggests that the hepatoblasts are activated and differentiated into bile duct cells. In our study, SALL4 expression was detected at weak levels in adjacent noncancerous cirrhotic tissues, suggesting that cirrhosis is the early stage of liver cancer.
In this study, we report for the first time the combined detection of SALL4 and AFP, which enabled evaluation of the prognosis of HCC patients. Nowadays, AFP is used as a diagnostic marker. However, the diagnostic value of AFP is obscured by its low specificity and high sensitivity[22-24], and some researchers believe that AFP has no significant prognostic value for HCC patients. According to our results, joint detection of SALL4 and AFP allowed for evaluation of the prognostic value. The results showed that patients who were both SALL4 and AFP positive had the worst prognosis (Figure 2B and Table 5). Among the SALL4-positive patients, the mean survival time decreased with increasing AFP levels (Table 6). Gene expression analysis has demonstrated that genes that are upregulated during proliferation and metastasis are significantly enriched when SALL4 is highly expressed in HCC[19], which suggests that SALL4-positive HCC patients have greater malignant potential and worse prognosis. In our study, we confirmed that AFP is positively correlated with SALL4 (Table 2). SALL4 was positively expressed in patients with increased AFP levels. The higher the level of SALL4 expression, the higher the grade of malignancy of the tumor, which resulted in shorter survival time and worse prognosis. Therefore, AFP had a prognostic value for HCC patients who were SALL4 positive. However, due to the small sample size, we were unable to analyze the influence of different AFP levels when SALL4 expression was weakly positive, moderately positive, and strongly positive.
In conclusion, expression of SALL4 in HCC tissues was stronger than in liver hemangioma tissues and adjacent noncancerous cirrhotic tissues. The expression level of SALL4 was related to the AFP level, clinical stage, BCLC classification, and portal vein tumor thrombosis. SALL4 as an independent predictor affected the prognosis of HCC patients. Patients with higher expression levels of SALL4 and AFP have worse prognosis.
COMMENTS
Background
Hepatocellular carcinoma (HCC) is one of the most common digestive system tumors with a poor prognosis. Sal-like protein 4 (SALL4) is an oncogene that is not expressed in normal adult liver tissues but is partly detected in HCC. Moreover, its expression is relevant to the prognosis of HCC patients. In this study, immunohistochemical staining was used to explore the expression of SALL4. The results suggest that expression of SALL4 in HCC tissues is higher than in noncancerous hepatic tissues.
Research frontiers
This study independently evaluated the expression characteristics of SALL4 in HCC tissues, noncancerous cirrhotic tissues, and liver hemangioma tissues using immunohistochemistry. The results suggested that SALL4 is actively expressed in HCC tissues, more than in the other tissues, and is correlated with the prognosis of HCC patients.
Innovations and breakthroughs
SALL4 is expressed as oncogenes in a variety of tumors, including germ cell tumors, breast cancer, and α-fetoprotein (AFP)-producing gastric cancer. In this study, the authors found that SALL4 was actively expressed in HCC cells with stem cell characteristics and played an important role in predicting the prognosis of HCC patients. When SALL4 was expressed at higher levels, the prognosis of HCC patients was worse.
Applications
HCC is one of the most common malignant tumors, and when diagnosed, most people have lost the best opportunity for radical cure. AFP is now used as a diagnostic marker for HCC patients. However, its low specificity and high sensitivity are controversial. In this study, the authors found that SALL4 was partly detected in HCC patients, and high expression was related to prognosis. This is the first report on the combined detection of SALL4 and AFP, which may improve the diagnostic and prognostic value for HCC patients.
Terminology
SALL4 is a transcription factor located on human chromosome 20q13 and encodes a protein containing an eight zinc finger motif, including SALL4A and SALL4B subtypes. Epithelial cell adhesion molecule is a 40 kDa transmembrane glycoprotein encoded by GA-733-2 gene. It plays an important role in the process of epithelial cancer.
Peer-review
This article shows the expression of SALL4 in HCC tissue (n = 126), adjacent non-cancerous cirrhotic tissue (n = 44), and liver hemangioma tissue (n = 10) by immunohistochemical staining. The results indicate that the expression of SALL4 is actively expressed in HCC tissue and also associated with the prognosis of HCC patients. This may provide a more reliable way for the clinical diagnosis of HCC besides AFP alone. However, more clinical trials are needed to confirm the results.
Footnotes
Supported by the National Natural Science Foundation of China, No. 81072966/H2902; and the Natural Science Foundation of Hebei Province, China, No. C2011206134.
Institutional review board statement: This study was reviewed and approved by the Institutional Review Board of the Fourth Hospital of Hebei Medical University, Shijiazhuang, China.
Informed consent statement: All study participants, or their legal guardian, provided informed written consent prior to study enrollment.
Conflict-of-interest statement: The authors declare that there is no conflict of interest related to this study.
Data sharing statement: No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: July 6, 2015
First decision: July 19, 2015
Article in press: December 8, 2015
P- Reviewer: Facciorusso A S- Editor: Yu J L- Editor: Filipodia E- Editor: Wang CH
References
- 1.Lehwald N, Tao GZ, Jang KY, Sorkin M, Knoefel WT, Sylvester KG. Wnt-β-catenin signaling protects against hepatic ischemia and reperfusion injury in mice. Gastroenterology. 2011;141:707–718, 718.e1-5. doi: 10.1053/j.gastro.2011.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bejsovec A. Wnt pathway activation: new relations and locations. Cell. 2005;120:11–14. doi: 10.1016/j.cell.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 3.Nusse R. Wnts and Hedgehogs: lipid-modified proteins and similarities in signaling mechanisms at the cell surface. Development. 2003;130:5297–5305. doi: 10.1242/dev.00821. [DOI] [PubMed] [Google Scholar]
- 4.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Baradie R, Yamada K, St Hilaire C, Chan WM, Andrews C, McIntosh N, Nakano M, Martonyi EJ, Raymond WR, Okumura S, et al. Duane radial ray syndrome (Okihiro syndrome) maps to 20q13 and results from mutations in SALL4, a new member of the SAL family. Am J Hum Genet. 2002;71:1195–1199. doi: 10.1086/343821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paradisi I, Arias S. IVIC syndrome is caused by a c.2607delA mutation in the SALL4 locus. Am J Med Genet A. 2007;143:326–332. doi: 10.1002/ajmg.a.31603. [DOI] [PubMed] [Google Scholar]
- 7.Rao S, Zhen S, Roumiantsev S, McDonald LT, Yuan GC, Orkin SH. Differential roles of Sall4 isoforms in embryonic stem cell pluripotency. Mol Cell Biol. 2010;30:5364–5380. doi: 10.1128/MCB.00419-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanimura N, Saito M, Ebisuya M, Nishida E, Ishikawa F. Stemness-related factor Sall4 interacts with transcription factors Oct-3/4 and Sox2 and occupies Oct-Sox elements in mouse embryonic stem cells. J Biol Chem. 2013;288:5027–5038. doi: 10.1074/jbc.M112.411173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang J, Gao C, Chai L, Ma Y. A novel SALL4/OCT4 transcriptional feedback network for pluripotency of embryonic stem cells. PLoS One. 2010;5:e10766. doi: 10.1371/journal.pone.0010766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang P, Sun H, Liu YF, Wang GY, Yin YF. Expression of SALL4 and BMI-1 mRNA in acute leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2008;16:1271–1274. [PubMed] [Google Scholar]
- 12.Yang J, Chai L, Gao C, Fowles TC, Alipio Z, Dang H, Xu D, Fink LM, Ward DC, Ma Y. SALL4 is a key regulator of survival and apoptosis in human leukemic cells. Blood. 2008;112:805–813. doi: 10.1182/blood-2007-11-126326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao D, Humphrey PA, Allan RW. SALL4 is a novel sensitive and specific marker for metastatic germ cell tumors, with particular utility in detection of metastatic yolk sac tumors. Cancer. 2009;115:2640–2651. doi: 10.1002/cncr.24308. [DOI] [PubMed] [Google Scholar]
- 14.Bard JD, Gelebart P, Amin HM, Young LC, Ma Y, Lai R. Signal transducer and activator of transcription 3 is a transcriptional factor regulating the gene expression of SALL4. FASEB J. 2009;23:1405–1414. doi: 10.1096/fj.08-117721. [DOI] [PubMed] [Google Scholar]
- 15.Ushiku T, Shinozaki A, Shibahara J, Iwasaki Y, Tateishi Y, Funata N, Fukayama M. SALL4 represents fetal gut differentiation of gastric cancer, and is diagnostically useful in distinguishing hepatoid gastric carcinoma from hepatocellular carcinoma. Am J Surg Pathol. 2010;34:533–540. doi: 10.1097/PAS.0b013e3181d1dcdd. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda H, Sato Y, Yoneda N, Harada K, Sasaki M, Kitamura S, Sudo Y, Ooi A, Nakanuma Y. α-Fetoprotein-producing gastric carcinoma and combined hepatocellular and cholangiocarcinoma show similar morphology but different histogenesis with respect to SALL4 expression. Hum Pathol. 2012;43:1955–1963. doi: 10.1016/j.humpath.2011.11.022. [DOI] [PubMed] [Google Scholar]
- 17.Zeng SS, Yamashita T, Kondo M, Nio K, Hayashi T, Hara Y, Nomura Y, Yoshida M, Hayashi T, Oishi N, et al. The transcription factor SALL4 regulates stemness of EpCAM-positive hepatocellular carcinoma. J Hepatol. 2014;60:127–134. doi: 10.1016/j.jhep.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 18.Oikawa T, Kamiya A, Zeniya M, Chikada H, Hyuck AD, Yamazaki Y, Wauthier E, Tajiri H, Miller LD, Wang XW, et al. Sal-like protein 4 (SALL4), a stem cell biomarker in liver cancers. Hepatology. 2013;57:1469–1483. doi: 10.1002/hep.26159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Jeong HW, Kong N, Yang Y, Carroll J, Luo HR, Silberstein LE, Yupoma L. Stem cell factor SALL4 represses the transcriptions of PTEN and SALL1 through an epigenetic repressor complex. PLoS One. 2009;4:e5577. doi: 10.1371/journal.pone.0005577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yong KJ, Chai L, Tenen DG. Oncofetal gene SALL4 in aggressive hepatocellular carcinoma. N Engl J Med. 2013;369:1171–1172. doi: 10.1056/NEJMc1308785. [DOI] [PubMed] [Google Scholar]
- 21.Oikawa T, Kamiya A, Kakinuma S, Zeniya M, Nishinakamura R, Tajiri H, Nakauchi H. Sall4 regulates cell fate decision in fetal hepatic stem/progenitor cells. Gastroenterology. 2009;136:1000–1011. doi: 10.1053/j.gastro.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 22.Lee SS, Shin HS, Kim HJ, Lee SJ, Lee HS, Hyun KH, Kim YH, Kwon BW, Han JH, Choi H, et al. Analysis of prognostic factors and 5-year survival rate in patients with hepatocellular carcinoma: a single-center experience. Korean J Hepatol. 2012;18:48–55. doi: 10.3350/kjhep.2012.18.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shim JH, Yoon DL, Han S, Lee YJ, Lee SG, Kim KM, Lim YS, Lee HC, Chung YH, Lee YS. Is serum alpha-fetoprotein useful for predicting recurrence and mortality specific to hepatocellular carcinoma after hepatectomy? A test based on propensity scores and competing risks analysis. Ann Surg Oncol. 2012;19:3687–3696. doi: 10.1245/s10434-012-2416-1. [DOI] [PubMed] [Google Scholar]
- 24.Giannini EG, Marenco S, Borgonovo G, Savarino V, Farinati F, Del Poggio P, Rapaccini GL, Anna Di Nolfo M, Benvegnù L, Zoli M, et al. Alpha-fetoprotein has no prognostic role in small hepatocellular carcinoma identified during surveillance in compensated cirrhosis. Hepatology. 2012;56:1371–1379. doi: 10.1002/hep.25814. [DOI] [PubMed] [Google Scholar]


