Summary
Microarray platforms, enabling simultaneous measurement of many allergens with a small serum sample, are potentially powerful tools in allergy diagnostics. We report here the first study comparing a fully automated microarray system, the Microtest allergy system, with a manual microarray platform, Immuno‐Solid phase Allergen Chip (ISAC), and two well‐established singleplex allergy tests, skin prick test (SPT) and ImmunoCAP, all tested on the same patients. One hundred and three adult allergic patients attending the allergy clinic were included into the study. All patients were tested with four allergy test methods (SPT, ImmunoCAP, Microtest and ISAC 112) and a total of 3485 pairwise test results were analysed and compared. The four methods showed comparable results with a positive/negative agreement of 81–88% for any pair of test methods compared, which is in line with data in the literature. The most prevalent allergens (cat, dog, mite, timothy, birch and peanut) and their individual allergen components revealed an agreement between methods with correlation coefficients between 0·73 and 0·95. All four methods revealed deviating individual patient results for a minority of patients. These results indicate that microarray platforms are efficient and useful tools to characterize the specific immunoglobulin (Ig)E profile of allergic patients using a small volume of serum sample. The results produced by the Microtest system were in agreement with diagnostic tests in current use. Further data collection and evaluation are needed for other populations, geographical regions and allergens.
Keywords: allergen component, allergy test, ISAC, microarray, Microtest, sIgE
Introduction
Skin prick tests (SPT) and traditional extract‐based allergen‐specific immunoglobulin (Ig)E blood tests have been used for decades. Together with the clinical history, they help to confirm or refute the diagnosis of IgE‐mediated allergy in patients with symptoms such as rhino‐conjunctivitis, asthma, urticaria, anaphylaxis, eczema or suspected food allergies. There have been two major innovations in the field of specific IgE detection in the last 20 years: the introduction of microarray technology and the development of allergen components [i.e. detection of specific IgE against purified, native or recombinant proteins, also referred to as molecular allergy diagnostics or component resolved diagnostics (CRD)] 1, 2, 3, 4, 5.
Allergen components are increasingly being used routinely, and their main benefits in allergy diagnostics compared to traditional extract‐based allergens are their ability to: (a) increase the resolution of a test by distinguishing sensitization due to molecular cross‐reactivity from true co‐sensitization 2, 3, 6; and (b) enable more accurate prediction of the type of clinical reaction that may occur. For example, reactivity to stable storage proteins are associated more often with severe, systemic reactions, while reactivity to labile proteins such as profilin or PR‐10 proteins is associated more commonly with local, oral reactions, possibly as relevant epitopes may be degraded in the gastrointestinal tract 2, 3, 4.
CRD can be particularly helpful when investigating polysensitized patients 2, 3, 4.
Microarray technology has the potential to transform the future of allergy diagnostics, as it enables simultaneous assay of specific IgE against numerous target proteins from a small volume of patient sample 2, 7, 8, 9, 10, 11, 12. Microarrays containing common environmental and food allergens can help the clinician to obtain an overview of the specific IgE reactivity profile of the patient in order to ‘see the a broader picture’ and identify reactivity to unsuspected or hidden allergens 2, 3, 4, 5, 6, 13, 14, 15. Allergen components as well as whole allergen extracts can be immobilized on microarrays, combining two new powerful technologies: microarrays and CRD.
The aim of this study was to evaluate the performance of a novel automated microarray test platform, the Microtest Allergy System. The Microtest system consists of a self‐contained biochip, a reagent cartridge, software and an instrument. The instrument is fully automated and requires a small volume of serum/plasma sample (100 µl) for simultaneous measurement of specific IgE against an allergen panel of 26 common airborne and food allergen extracts and components.
The main difficulty in evaluating the performance of a new multiplex allergy test is the lack of a gold standard reference for comparison, the large number of allergens tested, together with differences in the allergen panel composition (allergen extracts and/or components) and source of raw material. Using food challenge as a gold standard is not feasible, considering that provocation tests with multiple allergens would be impossible in practice and formidably costly. We thus chose to pursue the practical alternative strategy of comparing results generated by the Microtest system with results from three other established allergy test methods: SPT and two in‐vitro blood tests, one singleplex test system (ImmunoCAP, manufactured by Phadia AB, Uppsala, Sweden) and one manual microarray platform [Immuno‐Solid phase Allergen Chip (ISAC) 112, manufactured by Phadia AB].
Regardless of which diagnostic method is used, it is important to use the clinical history to interpret the test results because a positive skin or blood test yields information on IgE sensitization, which is not always equivalent to clinical allergy.
Aim
The aim of this study was to evaluate the Microtest allergy system by comparing its test results with three other allergy test methods. A secondary aim was to develop a methodology to evaluate the performance of a microarray test, given the absence of the gold standard data from allergen challenge testing.
Methods
Patients
A total of 103 adult allergic patients referred to the specialist allergy clinic at the University Hospital of Wales were included in this retrospective study. The patients were selected adults whose symptoms were sufficiently complex to merit a microarray test, and comprised 78 females and 25 males with a mean age of 33 ± 13 years. The underlying clinical diagnoses were: hay fever/rhino‐conjunctivitis (23 patients), asthma (17 patients), food allergy (40 patients), urticaria (18 patients), eczema (15 patients), anaphylaxis (19 patients) and others (e.g. food intolerance (32 patients). Many of the patients had more than one of the above diagnoses. Data for each patient available from the clinical records and pathology systems included the clinical history, SPT, ImmunoCAP and ISAC results. Sera for all the measurements were drawn at the out‐patient clinics, separated within 24 h, and stored at −20°C until Microtest microarray assays were performed. Ethical permission was not required for assay evaluation.
SPT
A panel of 11 common allergens was tested on SPT, this being the standard practice of the clinical service. Additional allergens were also tested in some individuals, depending on their clinical history. Positive (histamine 10 mg/ml) and negative controls were tested according to guidelines 16. The largest diameter of the weal was measured. Absence of a weal was defined as a ‘negative’ test result. A weal diameter < 3 mm was defined as ‘low/uncertain’, a weal ranging from 3 to 6 mm was considered ‘moderate’ and a weal ≥ mm was defined as ‘high (see Table 2).
Table 2.
Definition of positive/negative and range classifications.
| Classification | Negative | Low | Moderate | High |
|---|---|---|---|---|
| SPT (mm) | < 1 | 1–2·9 | 3–5·9 | ≥ 6 |
| CAP (kUA/l) | < 0·35 | 0·35–1·0 | 1–15 | >15 |
| MT (kUA/l) | < 0·35 | 0·35–1·0 | 1–15 | >15 |
| ISAC (ISU) | < 0·35 | 0·35–1·0 | 1–15 | >15 |
| Negative | Positive | |||
SPT = skin prick test; MT = Microtest; CAP = ImmunoCAP; ISAC = Immuno‐Solid phase Allergen Chip.
The standard panel of SPT reagents used on all patients comprised histamine, negative control (saline), house dust mite, cat, dog, mixed grass pollens, mixed tree pollens, egg, milk, peanut, hazelnut, almond and walnut. Other reagents used were: Aspergillus, latex, cod, shrimp, tuna, birch pollen, kiwi, wasp venom, bee venom and horse. All reagents were purchased from Allergy Therapeutics (Worthing, UK) apart from almond, kiwi, Aspergillus and tuna, which were purchased from Stallergenes SA (Antony, France), and peanut, walnut, wasp, bee, latex, shrimp and horse from ALK‐Abelló (Reading, UK).
ImmunoCAP
ImmunoCAP tests (manufacturer: Phadia AB) were requested as an additional test for suspected allergen triggers on certain patients based on their clinical history and SPT results according to the routine practice in the out‐patient clinic. The Phadia 250 instrument was used according to the manufacturer's instructions for use in our CPA (Clinical Pathology Accreditation) Accredited Laboratory. Results are reported in kUA/l. Test results were classified as negative, low/uncertain, moderate or high, as defined in Table 2.
ISAC
Patients were tested on the ISAC 112 platform (manufacturer: Phadia AB) for a panel of 112 allergen components, according to the manufacturer's instructions for use.
Briefly, allergen components are immobilized in triplicate on a glass slide. When starting the assay the microarray is washed and the surface is activated. The assay includes a two‐step reaction: (1) a 30‐µl sample (serum or plasma) is applied onto the microarray reaction site; and (2) after incubation and washing, fluorescence‐labelled anti‐human IgE detection antibody is applied. After incubation, washing and drying the microarray is scanned and the image is processed using the microarray software analyser (MIA) software and test results are reported in semi‐quantitative ISU units 2, 17, 18.
Test results were classified as negative, low/uncertain, moderate or high, as defined in Table 2. To compare the component‐based ISAC method with the other methods that determine the IgE antibody concentration to the whole allergen extract, the ISU values of the corresponding components on ISAC were summed as described in Melioli et al. 17. All ISAC tests were run during 2011–14 using the 112 version of the biochip.
Microtest
All patients were tested on the platform according to the manufacturer's instructions. A 100‐μl serum sample was applied to the instrument for parallel measurement of a predefined panel of allergen extracts and components. Test results were classified as negative, low/uncertain, moderate or high as defined in Table 2 based on the calculated concentration in kUA/l. All sera were assayed between November 2013 and March 2014 using a research version of the Microtest Allergy System and included the following allergen sources: cat, dog, horse, house dust mite (Dermatophagoides pteronyssinus), German cockroach, birch, olive, timothy grass, rye grass, cultivated rye grass, Bermuda grass, Alternaria, egg, milk, peanut, hazelnut, wheat, soy, cod, shrimp, latex, bee venom and the following allergen components: Fel d 1, Ole e 1, Bet v 1, Phl p 5, Der p 1, Der p 2, Gal d 1, Ara h 2 and Cor a 1.
The basic principle of the assay is outlined below: allergen extracts and components are immobilized covalently in triplicate or more onto a precoated slide in the form of a matrix with > 150 spots. Each slide contains one microarray. The automated Microtest assay includes a three‐step reaction: (0) the microarray is washed and the surface is activated; 100 μl patient sample (serum or plasma) is added by the user through the reagent port. The fluid flows into the chamber where it is incubated for 1 h at ∼32°C. During this step each allergen spotted in the array reacts with the specific IgE in the patient sample. Following incubation, the Microtest instrument washes the sample away. (1) The Microtest instrument adds the primary detection antibody that binds to the allergen‐IgE complex. After 45 min incubation performed at room temperature the instrument washes away the secondary antibody in excess. (2) The instrument adds the secondary horseradish peroxide (HRP)‐conjugated antibody that detects the bound primary antibody complex. After 45 min incubation at room temperature, the instrument washes away the unbound secondary antibody. (3) The instrument adds the third and last reagent, a detection buffer to develop the immunocomplex‐specific fluorescence. After 20 min incubation, the Microtest flushes away the solution and then dries the incubation chamber of each biochip. The fluorescent signal is then read and analysed by the instrument, and a test result report is generated. The whole procedure takes approximately 4–4·5 h with approximately 10 min hands‐on time. Up to five patient samples can be assayed at the same time. An early version of the assay principle is outlined in 19.
Table 1 presents an overview of the four methods used in the study.
Table 1.
Overview of four allergy test methods.
| Microtest | ISAC | ImmunoCAP 250 | SPT | |
|---|---|---|---|---|
| Method | Multiplex | Multiplex | Singleplex | Singleplex |
| in‐vitro | in‐vitro | in‐vitro | in‐vivo | |
| Automated | Manual | Automated | Manual | |
| Where | In clinic or laboratory | Laboratory | Laboratory | Clinic |
| Time to result | ∼4 h | ∼4 h | ∼2 h | ∼25 min |
| Reported results/assay | 26 | 112 | 1 | 1 |
| Components measured/assay | 16 | 112 | 1 | Not available |
| Extracts measured/assay | 19 | 0 | 1 | 1 |
| Sample volume | 100 µl/26 allergens | 30 µl/112 allergens | 50 µl/allergen | 0 |
SPT = skin prick test; ISAC = Immuno‐Solid phase Allergen Chip.
Use of qualitative and quantitative data to express results
Test results were collected from the four test methods and the agreement between the methods was analysed quantitatively (in mm, kUA/l and ISU), semi‐quantitatively in class ranges (negative, low, moderate, high) and classified qualitatively as positive or negative, as shown in Table 2.
Using different agreement/concordance measures to analyse results
The agreement between assay systems was calculated in three ways: (a) test results were classified as either negative (negative or very low/uncertain) or positive (moderate or high) and the concordances in percentage between the four test methods were calculated. (b) Test results were classified into four classes (negative, low, moderate, high) and the sum of all determinations in agreement (the same class score) was divided by the total number of observations. (c) The correlation coefficient (Spearman's rank) between results given by different test methods was estimated. A correlation coefficient of 1·0 indicates a perfect correlation, while correlation coefficients below 0·5 indicate a poor correlation.
Microsoft Excel was used for the statistical analysis. The Spearman's rank correlation (which is used when data are not normally distributed as in our case) is defined as the Pearson's correlation coefficient on the ranks of the data. Because Spearman's rank correlation is not an available function in Excel, the rank order of the data was first calculated.
Results
Overall, 3845 pairs of test results were analysed and compared. The data were analysed and compared method‐to‐method by studying (1) the allergen components, (2) the allergen sources and (3) all test results together, as outlined below.
The patient group revealed a higher prevalence of aeroallergen sensitization (timothy 48%, mite (Dermatophagoides pteronyssinus) 36%, cat 25%, birch 24% and dog 16%) and foods from plant origin (peanut 22%) than foods from the animal kingdom (egg, milk, cod or shrimp all < 3%). All four test methods confirmed the difference in prevalence.
Analysis of allergen components
Allergen components on the Microtest chip were compared with the corresponding component on ISAC. Example plots are shown in Fig. 1. Spearman's correlation coefficients based on kUA/l and ISU units for the allergen components are presented in Table 3, and range between r = 0.83 and 0.95 depending on allergen studied. When both platforms measure the same form of the allergen, recombinant (r) or native (n), results are shown in Table 2. Similar response levels were observed on both platforms except for Phl p 5 and Der p 1, where Microtest demonstrated a slightly higher IgE response compared to ISAC.
Figure 1.
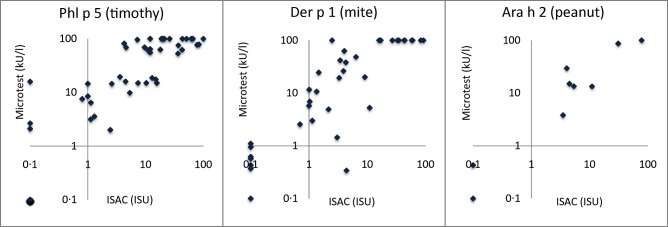
Allergen component results plotted for Immuno‐Solid phase Allergen Chip (ISAC) versus Microtest (MT) for Phl p 5 (timothy), Der p 1 (house dust mite) and Ara h 2 (peanut).
Table 3.
The correlation coefficient (Spearman) between Microtest and ISAC for allergen components on the Microtest chip together with number of samples > 0·35 kUA/l.
| Allergen | n > 0·35 | Corr. coeff. |
|---|---|---|
| rBet v 1 | 23 | 0·85 |
| rPhl p 5 | 41 | 0·95 |
| rOle e 1 | 23 | 0·83 |
| rFel d 1 | 32 | 0·89 |
| nDer p 1 | 31 | 0·93 |
| nDer p 2 | 34 | 0·95 |
* N total = 92 per component, including both negative and positive sample observations.
More data are needed for Ara h 2 (n >0·35 = 7, r = 0.94) and Gal d 1 (n >0·35 = 2) to provide a reliable estimate of the correlation coefficient. Cor a 1 showed a poor correlation coefficient, r = 0·55 (n >0·35 = 21), and a skewed distribution of results mainly below 2 kUA/l with few results above 5 kUA/l. More data are needed to investigate Cor a 1 further.
Analysis of allergen sources
A sufficient number of patients were tested with ImmunoCAP against timothy grass, house dust mite and peanut for a statistically meaningful comparison of all four test methods (> 15 positive IgE results/method). Figure 2 shows the ImmunoCAP (CAP) results versus ISAC/Microtest where class 0 results (< 0·35 kUA/l) were set to 0·1 kUA/l to allow logarithmic presentation of data. The corresponding correlation coefficients are presented in Table 4. A clear correlation, r = 0.73–0.92 depending on allergen studied, between the methods was observed with some deviating individual samples for all methods. The deviating samples where Microtest is negative while ImmunoCAP > 1 kUA/l were investigated in more detail (see below).
Figure 2.
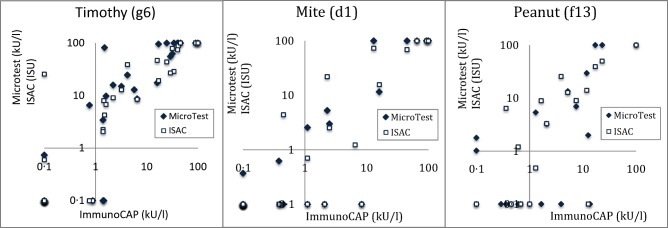
Microtest (MT) and Immuno‐Solid phase Allergen Chip (ISAC) results plotted versus ImmunoCAP for timothy, house dust mite and peanut.
Table 4.
Spearman's rank correlation coefficient (r) based on immunoglobulin (Ig)E concentration values between Microtest (MT), ImmunoCAP (CAP) and Immuno‐Solid phase Allergen Chip (ISAC).
| Allergen | r MT–CAP | r MT–ISAC | r CAP–ISAC |
|---|---|---|---|
| Mite | 0·80 | 0·89 | 0·84 |
| Timothy | 0·93 | 0·88 | 0·92 |
| Peanut | 0·73 | 0·86 | 0·80 |
| Birch | n.a. | 0.75 | n.a. |
| Cat | n.a. | 0.91 | n.a. |
| Dog | n.a. | 0.85 | n.a. |
n.a. = Not available.
In Fig. 2, timothy: one sample > 1 kUA/l on CAP went undetected by Microtest. This patient sample obtained the following results on the other methods: SPT negative, CAP = 1·45 kUA/l and monosensitized to Phl p 1 (8 ISU) on ISAC.
Another deviating sample showed the following IgE levels: ISAC = 25 kU/l, CAP negative, SPT not tested and Microtest = 0·74 kU/l. The ISAC profile for timothy showed no IgE binding to any of the timothy components except Phl p 1 (25 kU/l).
In Fig. 2, mite: three samples > 1 kUA/l on CAP went undetected by Microtest. Patient 1: CAP = 8 kUA/l, SPT = 2 mm and ISAC was negative; patient 2: CAP= 2 kUA/l, SPT = 4 mm and ISAC was negative; patient 3: CAP = 1 kUA/l, SPT = 1 mm and ISAC was negative.
In Fig. 2, peanut: four samples > 1 kUA/l went undetected by Microtest. Patient 1: CAP 13 kUA/l, SPT‐negative and ISAC result is not available; patient 2: CAP 3·9 kUA/l, SPT = 1 mm and ISAC was negative (Ara h 1,2,3,6,8,9‐negative but positive to profilin); patient 3: CAP = 1·65 kUA/l, SPT = 2 mm and ISAC single Ara h 9‐positive; patient 4: CAP = 1 kUA/l, SPT = 3 mm and ISAC‐negative on all peanut, profilin and CCD components. There was also one sample positive on Microtest (1·78 kU/l) but negative on ImmunoCAP, SPT and ISAC.
A sufficient number of test results were obtained to conduct a Microtest, ISAC and SPT (but not ImmunoCAP) comparison for birch, cat and dog. The numbers of available ImmunoCAP results were too few to provide a reliable analysis. Instead of plotting the quantitative results, which would be difficult to interpret for SPT results, the positive–negative concordances between the three test methods are shown in Table 5. The lowest number of pairwise patient samples were found for Microtest and SPT, with n = 84, 82 and 83 for birch, cat and dog, respectively. Table 5 includes timothy grass pollen as well; however, the number of pairwise samples is significantly lower as fewer patients were tested with ImmunoCAP compared to the other three test methods. ISAC versus Microtest results are shown in Fig. 3.
Table 5.
Positive/negative concordance in % between methods.
| Timothy | ||||
|---|---|---|---|---|
| (%) | SPT | CAP | Microtest | ISAC |
| SPT | 100 | 87 | 82 | 78 |
| CAP | 87 | 100 | 88 | 95 |
| MT | 82 | 88 | 100 | 83 |
| ISAC | 78 | 95 | 83 | 100 |
| n(min) = 34. | ||||
| Birch | ||||
|---|---|---|---|---|
| (%) | SPT | CAP | Microtest | ISAC |
| SPT | 100 | n.a. | 83 | 77 |
| CAP | n.a. | 100 | n.a. | n.a. |
| MT | 83 | n.a. | 100 | 88 |
| ISAC | 77 | n.a. | 88 | 100 |
| n(min) = 84; n.a. = not available. | ||||
| Cat | ||||
|---|---|---|---|---|
| (%) | SPT | CAP | Microtest | ISAC |
| SPT | 100 | n.a. | 83 | 88 |
| CAP | n.a. | 100 | n.a. | n.a. |
| MT | 83 | n.a. | 100 | 91 |
| ISAC | 88 | n.a. | 91 | 100 |
| n(min) = 82; n.a. = not available. | ||||
| Dog | ||||
|---|---|---|---|---|
| (%) | SPT | CAP | Microtest | ISAC |
| SPT | 100 | n.a. | 90 | 89 |
| CAP | n.a. | 100 | n.a. | n.a. |
| MT | 90 | n.a. | 100 | 95 |
| ISAC | 89 | n.a. | 95 | 100 |
n(min) = 83; n.a. = not available. SPT = skin prick test; MT = Microtest; CAP = ImmunoCAP; ISAC = Immuno‐Solid phase Allergen Chip.
Figure 3.
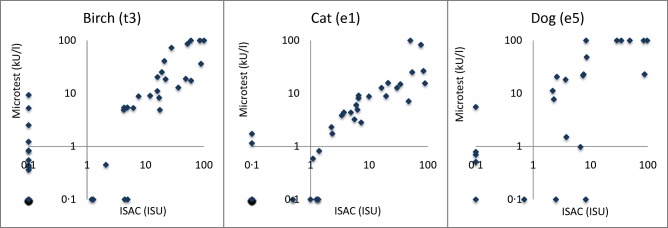
Microtest (MT) results plotted versus Immuno‐Solid phase Allergen Chip (ISAC) for birch, cat and dog.
Spearman's correlation coefficient (r) between Microtest, ImmunoCAP and ISAC based on IgE concentration in kUA/l and ISU is presented in Table 4.
The paucity of patients with a clinical diagnosis of egg, milk, cod, shrimp, wheat, soy and hazelnut allergy disabled any valid comparison of these allergens due to lack of statistical power.
All allergen results analysed together
The overall % concordance, ranging from 81 to 88%, based on positive/negative classification is shown in Table 6. The results are based on the following number of pairwise compared patient and allergen results: n (SPT–ImmunoCAP) = 197, n (SPT–Microtest) = 752, n (SPT–ISAC) = 824, n (ImmunoCAP–Microtest) = 204, n (ImmunoCAP–ISAC) = 233 and n (Microtest–ISAC) = 891.
Table 6.
Positive/negative % concordance between methods.
| (%) | SPT | CAP | Microtest | ISAC |
|---|---|---|---|---|
| SPT | 100 | 81 | 86 | 86 |
| CAP | 81 | 100 | 87 | 88 |
| MT | 86 | 87 | 100 | 86 |
| ISAC | 86 | 88 | 86 | 100 |
SPT = skin prick test; MT = Microtest; CAP = ImmunoCAP; ISAC = Immuno‐Solid phase Allergen Chip.
No significant differences in results or trends were obtained when analysing positive/negative classification, high/moderate/low/negative classification or specific IgE antibody concentration (kUA/l or ISU). Supporting information Table S1 shows a more detailed view of the results using semiquantitative range classification.
Discussion
The aim of this study was to evaluate the performance of the Microtest Allergy System by comparing it with other allergy test methods used in our laboratory. Serum‐specific IgE test and SPT are the two most commonly used methods of detecting or confirming allergen sensitization. SPTs are used widely and considered reliable and informative by allergy specialists 16, 20, 21. In contrast to SPT, blood tests are suitable for most types of patients, including those with severe eczema and those taking anti‐histamine medication 16, 22. Both methods have similar diagnostic value in terms of sensitivity and specificity, with both parameters varying with the clinical scenario and allergen tested 16, 20, 23. The concordance between STP and in‐vitro test results is reported commonly to range within 75–95%, depending on allergen, methods and population studied 16, 24. The IgE antibody concentration measured may vary between test methods and a few individual patient samples show deviating results 16, 17, 18, 23, 24, 25, 26, 27, 28, 29, 30.
The results obtained in this study indicate that the same situation applies to the microarray platforms studied here. Table 6 shows a concordance between 81 and 88% for any pair of methods, which compare favourably with other reported studies 16, 17, 18, 24, 25, 26, 29, 30. The individual allergens studied (peanut, mite, timothy, birch, cat and dog) showed a concordance between methods ranging from 77 to 95% (Table 5). Worth noting is that all four methods revealed deviating results for individual patient samples (see, for example, Fig. 2). The allergen components generally revealed a higher correlation coefficient than the corresponding whole allergen (e.g. r(Phl p 5) = 0.95 in Table 3, compared to r(timothy) = 0.88 in Table 4), which is logical and encouraging.
Different assay methods use different technologies to measure the concentration of the sIgE antibodies in the blood, and each method can potentially introduce different deviations or measure the IgE antibody population with different efficiency 20. The wide range of SPT methodologies and their qualitative nature complicate comparison of SPT results from different studies 16. Furthermore, it is difficult to compare studies based on SPT with those based on in‐vitro tests that passed a negative/positive concordance, due to the fundamentally different nature of the output from each test.
When comparing assay results using an allergen extract, e.g. milk, an error can be introduced, as the allergen components in milk will be represented in a relatively higher or lower (or even zero) concentration relative to a method which uses only allergen components. The allergen component composition of an extract may vary from producer to producer as well as from harvest to harvest 31, 32, 33. Furthermore the IgE sensitization profile differs between individuals on at least three different levels; from allergen source, to allergen component as well as on the epitope (binding site) level 34, 35, 36, 37. In addition to the differences in allergen composition the four allergy tests studied here use different methodology for assay, detection, calibration and estimation of results. Despite all these intrinsic assay differences and potential sources of error, we believe these results clearly show some key findings and an overall correlation between methods.
When comparing Microtest with the ISAC microarray from a user's point of view there are some significant similarities and differences. Microtest is a fully automated platform, while ISAC is a manual assay method. Both platforms have an allergen panel that covers (1) common inhalation allergens and allergic asthma triggers listed by the American Academy of Allergy, Asthma and Immunology (AAAAI), including indoor (mite and cockroach), pets (cat, dog and horse) and outdoor allergens (pollen and mould); and (2) common food allergens, which account for 90% of food allergic reactions, according to the US Food and Drug Administration (FDA) (milk, egg, peanuts, tree nuts, wheat, soy, fish and shellfish).
The Microtest allergen panel is simpler in comparison with the ISAC panel. It represents a trade‐off between allergen coverage and a manageable interpretation of the results to limit over‐reporting. The whole allergen extracts help to identify the allergen sources to which the patient is sensitized and improve the allergen coverage. The allergen components help to differentiate between specific and cross‐reactive sensitization in multi‐sensitized patients, and improve the specificity in plant‐derived food allergies.
Conversely, the ISAC panel holds 112 allergen components and no allergen extract, and is therefore a good tool for a detailed molecular IgE profile overview of patients. Some allergens are covered well by the components on the chip, e.g. timothy, cat, birch and peanut, while some clinically important allergen components from other allergen sources may not be included on the chip (e.g. for ragweed, mugwort, shrimp and cashew nut), which is something the user needs to bear in mind when interpreting the results. Studies comparing ISAC with ImmunoCAP generally show an 80–97% agreement, depending on allergen and population studied 17, 18, 25, 26, 27, 28.
It is relevant to ask what place microarray IgE tests have in the routine clinical setting. Current good clinical practice is to carry out allergy tests only when there is a clinical suspicion or a clinical history supportive of allergy. Therefore, the clinical utility and indication to test for food allergy may be questioned in patients presenting with asthma and allergic rhinitis but with no history of food allergy. The use of screening panels of allergen‐specific IgE without previous consideration of the history of the patient is generally not recommended 38.
One argument for panel testing (e.g. microarray) is that case history alone may overlook relevant inhalant or food allergens. Identifying the obvious precipitant allergen is not always sufficient, as the immune response to different allergens is cumulative and together may thus push the patient over the symptom‐onset threshold, especially during viral infections 39, 40. In addition, polysensitized patients with co‐morbidities are common, and the majority of patients (both children and adults) with asthma have underlying allergies 41, 42. Even a very comprehensive history may overlook many allergens’ relevance, especially in multi‐sensitized patients. Failure to identify the full complexity of a patient's allergenic response may prevent that patient being given appropriate avoidance advice and allergy management. For these patients, testing with a panel of common allergens may identify relevant allergies more reliably than traditional selection of suspected allergens according to the case history.
While panel testing may help to uncover unsuspected allergens it may, conversely, increase the complexity of result interpretation by generating sensitization data, which may have limited or no relevance to the final diagnosis. Therefore, platforms such as the 112‐allergen component ISAC microarray have been considered by some to generate unnecessary results that may be difficult to explain. It is known that sensitization and allergy are distinct, in that sensitization or atopy (in blood tests or SPT) do not necessarily imply that symptoms will occur upon exposure to that allergen. The dominant molecular and cellular mechanisms underlying this are uncertain. They are believed to involve blocking IgG4 antibodies, T regulatory cells, other influences on basophil activation and possibly other factors. Nevertheless, it is important to remember that a positive IgE response may also indicate the development of a symptomatic allergic immune response, as IgE antibodies can be detected long before the onset of clinical symptoms 16, 34, 35. The finding of sensitization when patients tolerate contact with those allergens requires explanation of the above to patients.
When managing a patient with clinical allergy in whom objective confirmation of the precipitating allergen(s) is indicated, the clinician is faced with a choice of which test(s) to perform. SPTs are straightforward to perform, give immediate results in the clinic and may suffice if they confirm the suspected diagnosis.
Should clinical doubt persist, or should the relative causal contribution of two or more allergens require further elucidation, blood tests constitute useful additional tests. Blood tests such as the ImmunoCAP are well established, and the level of antibodies measured by ImmunoCAP correlates with the probability of symptoms occurring following allergen exposure. When it is necessary to delineate the presence of allergic antibodies to only a few allergens, ImmunoCAP tests will be cheaper to perform than microarray assays. They may also yield results that are easier to interpret, as they are more familiar (given that cross‐reacting allergen components are not involved). They are well suited to many patients with a clear‐cut clinical history of allergy.
Conversely, Microarray tests are useful when there is uncertainty about the identity of the allergen(s) that may underlie the clinical presentation 2, 6, 13, 14, where it may be useful to obtain information about the reactivity to a broader spectrum of allergens, and where simultaneous testing of allergen components may help distinguish between, for example, pollen‐related oral allergy syndrome to nuts and true nut allergy.
The decision about which tests to request for any individual patient will be influenced by a number of factors, including the number of allergens that need to be investigated, the serum volume required (especially in young children), the cost of the test and the budget available. In general, if more than six to 10 allergens need testing, a microarray test may be preferable both for the extra amount of information obtained and for reasons of economy. Dedicated research studies modelling the cost‐effectiveness of using the traditional test strategy compared to microarray panel testing are needed to understand the overall costs involved and the precise circumstances (including the type of patient and the clinical scenarios) for which each test produces optimal clinical and economic results.
The first choice of investigation in National Health Service (NHS) clinical practice appears to be SPT, possibly for reasons of cost, ease of performance and immediate results. For clinicians in such practice, a single allergen‐specific IgE test constitutes the second‐line test for the great majority of patients, while microarray based tests may be reserved for those patients presenting a complex clinical profile, for whom testing against a broad panel is appropriate. However, many doctors do not use SPT in their clinical practice for various reasons.
For doctors and clinics that do not want to or cannot perform SPT, in‐vitro single allergen tests or microarray tests offer suitable and valuable alternatives. In particular, single allergen tests are most suitable in patients with clearly defined triggering allergens. For patients with several suspected allergic triggers or an unclear clinical history, microarray testing with a panel of allergens may offer measurable advantages through comprehensive IgE profiling of the patient. This approach is in line with the approach described in the WAO–ARIA–GA2LEN consensus document on molecular‐based allergy diagnostics, 2013 2.
In summary, the two microarray platforms studied, Microtest and ISAC, showed comparable results to the traditional singleplex methods ImmunoCAP and SPT. The most prevalent allergens (grass, birch, cat, dog, mite and peanut) were studied in more detail and all showed a good concordance between methods. Further data collection and evaluation of the platform is needed in patients with food allergies and less common allergies (e.g. latex), as well as patients drawn from other populations and geographical regions.
Disclosure
P. E. W. has received remuneration for attendance at a ThermoFisher Scientific advisory board. S. J. is supported by a NISCHR Fellowship and has received support for consulting, conferences and/or research from CSL Behring, Baxalta, BPL, Biotest, Octapharma, Shire, MEDA, ThermoFisher Scientific, UCB, Pharma, SOBI and NISCHR. T. E. has received support for consulting, conferences and/or research from ALK Abello, Allergy Therapeutics, Meda, Microtest, Novartis, Stallergenes and ThermoFisher Scientific. A. Ö. was employed at ThermoFisher Scientific/Phadia 2007‐2014. A. Ö., V. H. and F. B. are currently employed at Microtest Diagnostics.
Supporting information
Table S1. Classification concordance in % between methods.
Acknowledgements
P. E. W., T. E. S. and S. J. planned and designed the study, recruited the patients, provided laboratory facilities and performed all testing and collection of patient data and information. A. O., F. B. and V. H. performed the statistical data analysis and provided input to the discussion. P. E. W., A. O., S. J. and T. E. S. wrote the manuscript.
References
- 1. Valenta R, Lidholm J, Niederberger V et al The recombinant allergen‐based concept of component‐resolved diagnostics and immunotherapy (CRD and CRIT). Clin Exp Allergy 1999; 29:896–904. [DOI] [PubMed] [Google Scholar]
- 2. Canonica GW, Ansotegui IJ, Pawankar R et al WAO–ARIA–GA2LEN consensus document on molecular‐based allergy diagnostics. World Allergy Organ J 2013; 6:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sastre J. Molecular diagnosis in allergy. Clin Exp Allergy 2010; 40:1442–60. [DOI] [PubMed] [Google Scholar]
- 4. Treudler R, Simon JC. Overview of component resolved diagnostics. Curr. Allergy Asthma Rep 2013; 13:110–7. [DOI] [PubMed] [Google Scholar]
- 5. Borres M, Ebisawa M, Eigenmann P. Use of allergen components begins a new era in pediatric allergology. Pediatr Allergy Immunol 2011; 22:454–61. [DOI] [PubMed] [Google Scholar]
- 6. Passalacqua G, Melioli G, Bonifazi F et al The additional values of microarray allergen assay in the management of polysensitized patients with respiratory allergy. Allergy 2013; 68:1029–33. [DOI] [PubMed] [Google Scholar]
- 7. Harwanegg C, Hiller R. Protein microarrays for the diagnosis of allergic diseases: state‐of‐the‐art and future development. Clin Chem Lab Med 2005; 43:1321–6. [DOI] [PubMed] [Google Scholar]
- 8. Jahn‐Schmid B, Harwanegg C, Hiller R et al Allergen microarray: comparison of microarray using recombinant allergens with conventional diagnostic methods to detect allergen‐specific serum immunoglobulin E. Clin Exp Allergy 2003; 33:1443–9. [DOI] [PubMed] [Google Scholar]
- 9. Lupinek C, Wollmann E, Baar A et al Advances in allergen‐microarray technology for diagnosis and monitoring of allergy: The MeDALL allergen‐chip. Methods 2014; 66:106–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scala E, Alessandri C, Bernardi ML et al Cross‐sectional survey on immunoglobulin E reactivity in 23 077 subjects using an allergenic molecule‐based microarray detection system. Clin Exp Allergy 2010; 40:911–21. [DOI] [PubMed] [Google Scholar]
- 11. Ott H, Schröder CM, Stanzel S et al Microarray‐based IgE detection in capillary blood samples of patients with atopy. Allergy 2006; 61:1146–7. [DOI] [PubMed] [Google Scholar]
- 12. Shreffler WG. Microarrayed recombinant allergens for diagnostic testing. J Allergy Clin Immunol 2011; 127:843–9. [DOI] [PubMed] [Google Scholar]
- 13. Heaps A, Carter S, Selwood C et al The utility of the ISAC allergen array in the investigation of idiopathic anaphylaxis. Clin Exp Immunol 2014; 177:483–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gassner M, Gehrig R, Schmid‐Grendelmeier P. Hay fever as a Christmas gift. N Engl J Med 2013; 368:393–4. [DOI] [PubMed] [Google Scholar]
- 15. Carter S, Heaps A, Boswijk K et al Identification of clinically relevant allergens using the Phadia ISAC microarray in patients with idiopathic anaphylaxis. Clin Exp Allergy 2012; 42:1829–30. [Google Scholar]
- 16. Heinzerling L, Mari A, Bergmann K‐C et al The skin prick test – European standards. Clin Transl Allergy 2013; 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Melioli G, Bonifazi F, Bonini S et al The ImmunoCAP ISAC molecular allergology approach in adult multi‐sensitized Italian patients with respiratory symptoms. Clin Biochem 2011; 44:1005–11. [DOI] [PubMed] [Google Scholar]
- 18. Gadisseur R, Chapelle JP, Cavalier E. A new tool in the field of in‐vitro diagnosis of allergy: preliminary results in the comparison of ImmunoCAP250 with the ImmunoCAP ISAC. Clin Chem Lab Med 2011; 49:277–80. [DOI] [PubMed] [Google Scholar]
- 19. Dottorini T, Sole G, Nunziangeli L et al Serum reactivity profiling in an asthma affected cohort. PLoS One 2011; 6:e22319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cox L. Overview of serological specific IgE antibody testing in children. Curr Allergy Asthma Rep 2011; 11:447–53. [DOI] [PubMed] [Google Scholar]
- 21. Høst A, Andrae S, Charkin S et al Allergy testing in children: why, who, when and how? Allergy 2003; 58:559–69. [DOI] [PubMed] [Google Scholar]
- 22. Williams P, Sewell WA, Bunn C et al Clinical immunology review series: an approach to the use of the immunology laboratory in the diagnosis of clinical allergy. Clin Exp Immunol 2008; 153:10–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hamilton RG, Williams PB. Human IgE antibody serology: a primer for the practicing North American allergist/immunologist. J Allergy Clin Immunol 2010; 126:33–8. [DOI] [PubMed] [Google Scholar]
- 24. Calabria C, Dietrich J, Hagan L. Comparison of serum‐specific IgE (ImmunoCAP) and skin‐prick test results for 53 inhalant allergens in patients with chronic rhinitis. Allergy Asthma Proc 2009; 30:386–96. [DOI] [PubMed] [Google Scholar]
- 25. Lizaso M, García B, Tabar A et al Comparison of conventional and component resolved diagnostics by two different methods (Advia‐Centaur/Microarray‐ISAC) in pollen allergy. Ann Allergy Asthma Immunol 2011; 107:35–41. [DOI] [PubMed] [Google Scholar]
- 26. Ott H, Fölster‐Holst R, Merk HF et al Allergen microarrays: a novel tool for high‐resolution IgE profiling in adults with atopic dermatitis. Eur J Dermatol 2010; 20:54–61. [DOI] [PubMed] [Google Scholar]
- 27. Martínez‐Aranguren R, Lizaso MT, Goikoetxea MJ et al Is the Determination of Specific IgE against components using ISAC 112 a reproducible technique? PLOS ONE 2014; 9:e88394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cabrera‐Freitag P, Goikoetxea M. Gamboa PM et al A study of the variability of the in vitro component‐based microarray ISAC CDR 103 technique. J Investig Allergol Clin Immunol 2011; 21:414–5. [PubMed] [Google Scholar]
- 29. Szecsi PB, Stender S. Comparison of immunoglobulin E measurements on IMMULITE and ImmunoCAP in samples consisting of allergen‐specific mouse human chimeric monoclonal antibodies towards allergen extracts and four recombinant allergens. Int Arch Allergy Immunol 2013; 162:131–4. [DOI] [PubMed] [Google Scholar]
- 30. Want J, Godbold JH, Sampson HA. Correlation of serum allergy (IgE) tests performed by different assay systems. J Allergy Clin Immunol 2008; 121:1219–24. [DOI] [PubMed] [Google Scholar]
- 31. Focke M, Marth K, Valenta R et al Molecular composition and biological activity of commercial birch pollen allergen extract. Eur J Clin Invest 2009; 39:429–36. [DOI] [PubMed] [Google Scholar]
- 32. Focke M, Marth K, Valenta R et al Heterogeneity of commercial timothy grass pollen extracts. Clin Exp Allergy 2008; 38:1400–8. [DOI] [PubMed] [Google Scholar]
- 33. Barber D, Moreno C, Ledesma A et al Degree of olive pollen exposure and sensitization patterns. Clinical implications. J Investig Allergol Clin Immunol 2007; 17 (Suppl. 1):63–8. [PubMed] [Google Scholar]
- 34. Hatzler L, Panetta V, Lau S et al Molecular spreading and predictive value of preclinical IgE response to Phleum pratense in children with hay fever. J Allergy Clin Immunol 2012; 130:894–901. [DOI] [PubMed] [Google Scholar]
- 35. Onell A, Hjälle L, Borres MP. Exploring the temporal development of childhood IgE profiles to allergen components. Clin Transl Allergy 2012; 2:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tripodi S, Frediani T, Lucarelli S et al Molecular profiles of IgE to Phleum pratense in children with grass pollen allergy: implications for specific immunotherapy. J Allergy Clin Immunol 2012; 129:834–9. [DOI] [PubMed] [Google Scholar]
- 37. Schmid‐Grendelmeier P. Recombinant allergens. For routine use or still only science? Hautarzt 2010; 61:946–53. [DOI] [PubMed] [Google Scholar]
- 38. Eigenmann PA, Atanaskovic‐Markovic M, O'B Hourihane J et al Testing children for allergies: why, how, who and when. An updated statement of the European Academy of Allergy and Clinical Immunology (EAACI) Section on Pediatrics and the EAACI–Clemens von Pirquet Foundation. Pediatr Allergy Immunol 2013; 24:195–209. [DOI] [PubMed] [Google Scholar]
- 39. Wickman M. When allergies complicate allergies. Allergy 2005; 60:14–8. [DOI] [PubMed] [Google Scholar]
- 40. Murray CS, Poletti G, Kebadze T et al Study of modifiable risk factors for asthma exacerbations: virus infection and allergen exposure increase the risk of asthma hospital admissions in children. Thorax 2006; 61:376–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roberts G, Patel N, Levi‐Schaffer F et al Food allergy as a risk factor for life‐threatening asthma in childhood: a case–controlled study. J Allergy Clin Immunol 2003; 112:168–74. [DOI] [PubMed] [Google Scholar]
- 42. Arbes SJ, Gergen PJ, Vaughn B et al Asthma cases attributable to atopy: results from the Third National Health and Nutrition Examination Survey. J Allergy Clin Immunol 2007; 120:1139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Classification concordance in % between methods.


