Abstract
The role of Homeobox transcription factors during fin and limb development have been the focus of recent work investigating the evolutionary origin of limb-specific morphologies. Here we characterize the expression of HoxD genes, as well as the cluster-associated genes Evx2 and LNP, in the paddlefish Polyodon spathula, a basal ray-finned fish. Our results demonstrate a collinear pattern of nesting in early fin buds that includes HoxD14, a gene previously thought to be isolated from global Hox regulation. We also show that in both Polyodon and the catshark Scyliorhinus canicula (a representative chondrichthyan) late phase HoxD transcripts are present in cells of the fin-fold and co-localize with And1, a component of the dermal skeleton. These new data support an ancestral role for HoxD genes in patterning the fin-folds of jawed vertebrates, and fuel new hypotheses about the evolution of cluster regulation and the potential downstream differentiation outcomes of distinct HoxD-regulated compartments.
One of the challenges of evolutionary morphology is to gain insight into how changes in developmental programs contribute to anatomical diversification in lineages descended from a common ancestor. Hox subfamily transcription factors are critical organizers of regional patterning during embryonic development, and have received much attention for their role in morphological evolution1,2,3,4,5,6,7. Invertebrates typically possess a single Hox cluster, whereas vertebrates possess at least four Hox clusters—designated as HoxA, B, C, and D—resulting from two rounds of whole genome duplication8. Additional duplication events further expanded the repertoire of Hox genes in several vertebrate lineages including lamprey (6 clusters: ref. 9), teleosts (8 ancestrally, with patterns of both loss and further duplication within derived clades; refs 10, 11, 12), and paddlefish, a non-teleost actinopterygian13. The genes within each Hox cluster fall into a series of paralogy groups based on sequence homology. Comparative genomic studies support the hypothesis that the ancestral vertebrate Hox cluster included 14 paralogues8,14,15. While there is significant experimental and clinical data describing paralogy groups 1–13, little is known about the role of Hox14 paralogues during development due to their loss from the genomes of teleosts and tetrapods thus far sampled8,13,15,16.
How paired fins gave rise to limbs during the invasion of land is one of the compelling questions in vertebrate evolution17,18. This morphological transition involved several key changes in appendage anatomy, including the loss of the dermal skeleton of the fin, and an expansion/remodeling of the distal endoskeleton to form an autopod with digits19,20 (Fig. 1). HoxA/D cluster genes are active during both fin and limb development, and over the last two decades, have been the focus of many studies aimed at gaining insight into the evolutionary origin of limb-specific morphologies21,22,23,24,25,26,27,28,29.
Figure 1. Pectoral appendage skeletons for representative vertebrates.
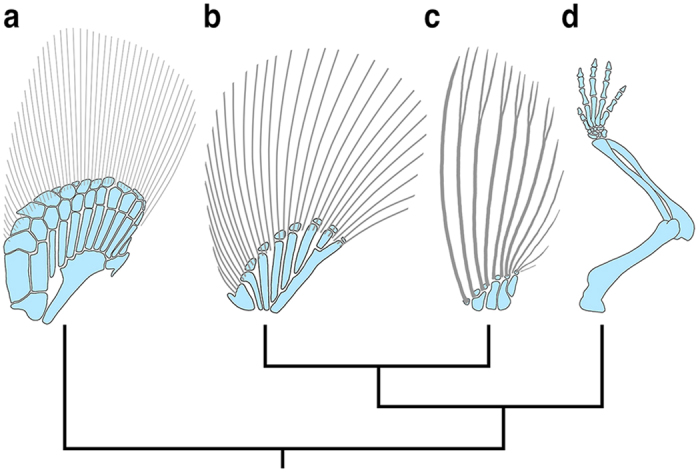
(a) Chondrichthyan (catshark - Scyliorhinus canicula). (b) Non-teleost actinopterygian (paddlefish - Polyodon spathula). (c) Teleost (zebrafish - Danio rerio). (d) Tetrapod (mouse – Mus musculus). Endochondral skeleton in blue, dermal skeleton in gray. Anterior is left, distal is up for all figures. Figures not to scale.
In tetrapods, the expression of HoxD genes are temporally and spatially dynamic during limb ontogeny, occurring in two phases, an early phase and a late phase, each driven by distinct regulatory elements positioned outside the cluster30,31,32. This has been particularly well-studied in mice, where during the early phase, transcripts of HoxD1–9 are present throughout the initial limb bud, followed by the sequential, posteriorly-nested activation of HoxD10–13 during bud outgrowth33. This first phase of collinear expression (i.e., HoxD13 is most posteriorly restricted) patterns the proximal limb (stylopod and zeugopod), and is predominantly regulated by enhancer elements positioned telomeric (3′) to the cluster33,34. In contrast, during the late phase of expression, HoxD9–13 are activated in the autopod, forming an inverse collinear arrangement (i.e., HoxD13 is most broadly expressed) that patterns the digits33. The enhancer elements that drive this late phase are interspersed across a region centromeric (5′) to the cluster and have been shown to regulate the expression of Evx2 and LNP, two non-Hox genes within the 5′ genomic neighborhood35,36,37. Together, these observations from mice underscore a topological switch between domains of chromatin interaction to pattern the zeugopod and autopod, respectively. Interestingly, one current model for limb development derived from these data posits that this regulatory transition creates a zone of low Hox dosage that manifests anatomically as the small, nodular bones of the wrist and ankle34,38.
Among non-tetrapod gnathostomes, the distribution of HoxD transcripts during paired fin development has been partially characterized in teleosts [zebrafish: refs 21, 24 and 39], a basal actinopterygian [paddlefish: ref. 22], a basal sarcopterygian [lungfish: ref. 40], and two chondrichthyans [catshark and skate: refs 23, 41 and 42]. These in situ studies reveal a conserved, collinear pattern of expression in the proximal region of early fin buds, similar to the first phase of HoxD expression in tetrapods18. In later stages of paired fin development, HoxD boundaries shift, suggesting a change in regulatory control. The dynamics of these shifts, however, vary with lineage. In zebrafish, a late phase of HoxD11a–13a expression appears to extend into the mesenchyme of the dermal fin-fold, creating a patterning compartment in which the anterior and posterior boundaries of each paralogue are in register24. In contrast, current data sets for paddlefish and catshark indicate that 5′ HoxD transcripts form an inverse collinear profile22,23,41. In paddlefish, this late phase overlaps early expression at the site of distal radial formation in the mid-fin, and has been interpreted as evidence of a deep developmental homology (sensu refs 43 and 44) between the distal radials and digits22 (but see ref. 38). In catshark, late phase HoxD transcripts localize to the distal margin of the fin23,41, and unlike actinopterygians are spatially separated from the early phase of expression45. Overall, these data from non-tetrapods suggest an evolutionary scenario in which distinct proximal and distal HoxD patterning compartments were already in place in the paired fins of ancestral gnathostomes18,23,45.
Recent studies have focused on testing whether the regulatory landscapes that control Hox expression in fins and limbs are functionally conserved. Significantly, this work has shown that several of the enhancer elements that drive digit expression in mice are represented in non-tetrapods17,25,29,35,37,46, and that a subset of these homology regions are not only active distally during paired fin development [zebrafish CsB: ref. 25; zebrafish Island I: ref. 29; Gar CsB and Island I: ref. 29], but when inserted into mice drive reporter expression in the proximal autopod [skate CsB: ref. 25; zebrafish CsB: ref. 25; pufferfish BAC clones: ref. 28] and digits [Gar CsB and Island I: ref. 29]. Additionally, there is evidence that trans-acting factors are at least partially conserved between fins and limbs, as a tetrapod specific element (mouse CsC) has been shown to drive distal fin expression when inserted into zebrafish26. Together, these results provide evidence that the origin of limb-specific morphologies involved modification of an ancient, conserved regulatory architecture for HoxD activation already in place in fins.
Current models for the fin to limb transition that integrate these data focus on the role of HoxD genes in patterning the endoskeleton, though with alternative perspectives on the relationship between cluster regulation and anatomical homology17,18,28,29,38,45. In these models, the fin-fold is considered a Hox-free zone, and its formation is thought to interrupt the ectodermal/mesenchymal signaling interactions that determine the relative size of a Hox-patterned endoskeletal mesenchyme17,23,24. Key to further informing models of appendage evolution is a better understanding of the phylogenetic distribution of HoxD expression relative to the proximo-distal fin compartments in basal gnathostomes. Herein, we characterize the expression of HoxD genes, as well as the cluster-related genes Evx2 and LNP, in a basal actinopterygian, the American paddlefish Polyodon spathula. Our results demonstrate a posteriorly restricted, collinear pattern of nesting in the early fin buds that includes HoxD14Beta, a gene previously thought to be insulated from canonical Hox regulation in the mesoderm14,47. Additionally, we show that in later stages of fin development, the collinear expression profile established in early fin buds is maintained along the site of distal radial formation, with no “autopodial-like” anterior expansion of HoxD13 (contra a previous report for Polyodon22), Evx2, or LNP. Most significantly, we observed a proximo-distal dynamic to HoxD expression in which late phase transcripts of both 3′ and 5′ paralogues extended into the fin-fold mesenchyme, co-localizing with And1, the elastoidin component of the actinotrichia48. In order to determine the phylogenetic distribution of this condition, we compared the expression domains of And1 and HoxD12 in a representative chondrichthyan, the lesser-spotted catshark Scyliorhinus canicula. These in situs revealed an overlap between distal HoxD and And1 expression, much like that of paddlefish, suggesting HoxD-positive cells contribute to fin-fold specific tissues in both taxa (the ceratotrichia and actinotrichia of chondrichthyans and actinopterygians, respectively). Our results support an ancestral role for HoxD genes in patterning the fin-fold, bringing new information to current models for fin/limb evolution and fueling novel hypotheses about the relationship between cluster regulation and appendage morphology.
Results
Expression of HoxD cluster genes in paddlefish paired fins
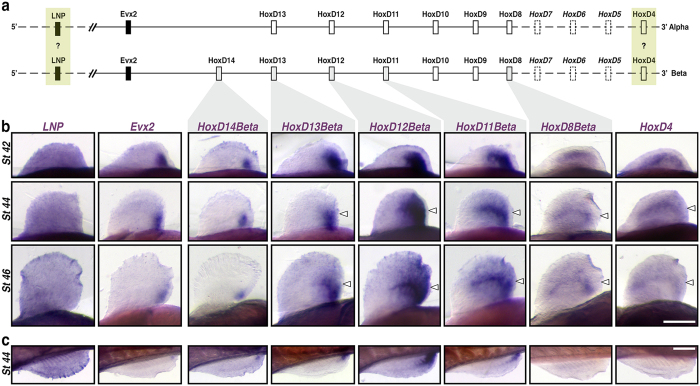
A whole genome duplication event occurred in the paddlefish lineage approximately 42 million years ago, resulting in the formation of unique Alpha and Beta HoxD clusters13 (Fig. 2a). In order to determine if both clusters are transcriptionally active during development, we surveyed our Polyodon transcriptome assembly using published genomic BAC sequences for specific HoxD Alpha and Beta paralogues13. These blast searches identified both Alpha and Beta sequences of HoxD8, HoxD12, and HoxD13, and Beta sequence of HoxD11. Unexpectedly, we also identified transcripts of HoxD14, a Beta cluster gene previously hypothesized to be inactive during appendage patterning in vertebrates14,47.
Figure 2. Expression of HoxD cluster genes in paddlefish paired fins.
(a) Schematic representation of Alpha and Beta HoxD clusters based on ref. 13. Gene key: Open boxes – Hox genes; closed boxes – non-Hox genes; solid lines – genes characterized and attributable to either Alpha and Beta clusters based on published BAC clones; yellow boxes – genes cloned but not attributable to a specific cluster; Dashed boxes – uncharacterized genes. (b) Pectoral fin whole-mount in situ hybridizations for LNP, Evx2, HoxD14Beta, HoxD13Beta, HoxD12Beta, HoxD11Beta, HoxD8Beta, and HoxD4 from stages 42 (early fin bud), 44 (onset of endoskeletal radial differentiation), and 46 (differentiated fin – onset of feeding larva)49. Pectoral fins in ventral view, anterior to the left, distal is up; Genes are shown in columns, and developmental stages in rows. Open arrowheads denote the position of distal radial formation along the A-P axis, where HoxD expression persists22 following outgrowth of the fin-fold. (c) Pelvic fin whole-mount in situ hybridizations comparable to (b) for stage 44. Pelvic fins in medial view, anterior to the left, distal is down. Scale bars = 200 nm.
In order to characterize the spatiotemporal expression dynamics of HoxD cluster genes during paired fin development, we performed a series of in situ hybridizations using probes targeted against either Alpha or Beta paralogues. Because the distribution of labeling appeared similar between genes from duplicated clusters (compare Fig. 2b and Supplemental Fig. 1), we limit the descriptions presented here to the HoxD Beta cluster only. It is worth noting, however, that we cannot rule out the possibility of probe cross reactivity due to high sequence similarity between a given set of paralogues (e.g., HoxD11Alpha and HoxD11Beta), and that positive staining may reflect their combined distribution of transcripts. Additionally, we could not assign an Alpha/Beta identity to the HoxD4 clone used in our analyses in the absence of available genomic sequence from the 3′ end of each duplicated cluster13.
In Stage 42 embryos, transcripts of HoxD4 and HoxD8Beta were detected along much of the length of the pectoral fin buds (Fig. 2b). In contrast, transcripts of HoxD11Beta–14Beta were more restricted in distribution, forming a collinear, posteriorly nested pattern of expression that mirrored the 3′–5′ arrangement of paralogues within the cluster (Fig. 2a,b). In more advanced embryos (Stages 44 and 46), the expression domains of HoxD4, HoxD8Beta, and HoxD11Beta–HoxD13Beta persisted in the middle of the fin following outgrowth of the distal fin and fin-fold (see arrowheads in Fig. 2b). Although an anterior expansion of HoxD13 expression in stage 46 fins has been reported in Polyodon (ref. 22 and see additional discussion in Supplemental Fig. 1), we observed that the collinear pattern of HoxD nesting established in early fin buds was maintained through later stages (Fig. 2b, Supplemental Fig. 1, and Supplemental Fig. 2). We also examined the distribution of LNP and Evx2 to infer if these non-Hox genes are coordinately regulated with 5′HoxD paralogues, as in mice where Evx2 and LNP co-localize with HoxD1335. In paddlefish, Evx2 expression was restricted to the posterior most region of the fin and appeared similar in distribution to HoxD14Beta (Fig. 2b). LNP, in contrast, was detected throughout the pectoral fins, with no distinct regionalization (Fig. 2b). Pelvic fins mirrored the 5′ HoxD and Evx2 expression patterns observed in pectoral fins (Fig. 2c). Transcripts of the more 3′-ward paralogues HoxD4 and HoxD8Beta, however, were difficult to detect in whole mount.
HoxD expression in the fin-fold compartment of paddlefish
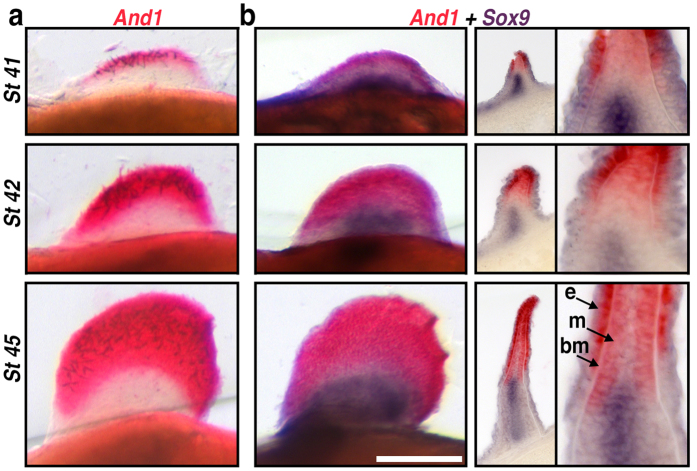
During limb development, HoxD genes are expressed in well-characterized proximal and distal zones that correspond to the stylopod/zeugopod and autopod, respectively. Given that fins are highly regionalized along the proximo-distal axis into endoskeletal radials (proximally) and a fin-fold/dermal skeleton (distally) (Fig. 1), we sought to characterize the distribution of HoxD transcripts relative to the early patterning zones that give rise to these morphologically distinct compartments. Actinodin genes encode non-collagenous (elastoidin) components of the actinotrichia and provide an early molecular marker for cells contributing to the fin-fold48. We identified a homologue of Actinodin1 (And1) in paddlefish, which we used for both single and double colorimetric in situs along with the pre-chondrogenic marker Sox9 to visualize the early endoskeletal radials (Fig. 3). In Stage 41 embryos, And1 transcripts were detected along the apical margin of the pectoral fin buds, forming a labeling boundary just distal to the Sox9 expression domain (Fig. 3a). In later stage fins (Stage 42–45), the relative position of the And1/Sox9 boundary was maintained despite considerable growth in both compartments. Cross sections through developing fins at these stages revealed And1 transcripts in both the fin-fold mesenchyme and the ectoderm adjacent to the basement membrane (Fig. 3b). Notably, at stage 45 the proximal margin of And1 labeling extended beyond the distal, lateral margins of Sox9 labeling, presaging the ultimate arrangement of the dermal fin supports and cartilaginous fin radials described for mature fins (see Fig. 3 in refs 49 and 50).
Figure 3. Actinodin1 expression in the fin-fold compartment of paddlefish.
(a,b) Pectoral fin in situ hybridizations in whole-mount, representative cross sections, and magnifications. Stages are shown in rows (a) Actinodin1 (And1), an early molecular marker for cells contributing to the fin-fold. And1 transcripts (red) appear in the presumptive distal fin and fin-fold of early fin buds (stage 41) and persist as the fin-fold elongates (stages 42 and 45). (b) Double in situs for the pre-chondrogenic marker Sox9 (purple) and And1 (red) reveal that endochondral and dermal compartments remain separate throughout fin development. Cross sections (and magnification) reveal And1 transcripts in both the fin-fold mesenchyme (m) and the ectoderm (e) adjacent to the basement membrane (bm). The slight proximo-distal overlap between And1 and Sox9 expression presages the arrangement of the dermal fin skeleton later in development. Scale bars = 200 nm.
Our And1/Sox9 data make visible the proximal boundary of the fin-fold patterning compartment at various stages of pectoral fin development, and show that prechondrogenic cells are excluded from the population of mesenchyme distal to this boundary. We next performed a series of double colorimetric in situs to determine if HoxD paralogues co-localize with And1 in the fin-fold mesenchyme. In whole mount pectoral fins at Stage 45, transcripts of HoxD11Beta–HoxD13Beta formed the same posteriorly-restricted pattern of nesting along the mid-fin (Fig. 4a) as in single in situs (Fig. 2b). Significantly, the expression of these 5′ HoxD paralogues extended distally to overlap with the And1 expression domain. Cross sections revealed differences in the profile of HoxD and And1 boundaries, depending on their position along the anterior-posterior axis (Fig. 4c). Whereas no 5′ HoxD transcripts were detected in the anterior portion of the fin, in more posterior sections HoxD11Beta–HoxD13Beta were expressed throughout the fin-fold mesenchyme, with a proximal boundary for strongest labeling that roughly aligned with that of And1 in the ectoderm. In sections near the caudal margin of the fin, no such alignment was observed, as HoxD11Beta–Hox13Beta labeling was continuous between the fin-fold and proximal fin mesenchyme.
Figure 4. HoxD expression in the fin-fold compartment of paddlefish.
(a,b) Pectoral fin double in situs for HoxD genes (purple) and And1 (red) in whole-mount (top row), representative cross sections (middle row) and magnifications of select sections (bottom row). Numbers mark planes of section. Genes are shown in columns. (a) Co-expression of 5′ Hox genes HoxD13Beta, HoxD12Beta, and HoxD11Beta (purple) with And1 (red) reveal the posteriorly-restricted pattern observed in 5′ HoxD single in situs (compare with Fig. 2b). Significantly, cross sections confirm that 5′ HoxD expression extends distally to overlap the And1 expression domain. 5′ HoxD transcripts were not detected in the anterior fin (sections labeled 1). More posterior sections reveal expression throughout the fin-fold mesenchyme, with a proximal boundary that roughly aligned with ectodermal And1 (sections labeled 2). In sections near the caudal margin of the fin, 5′ HoxD expression is continuous between the fin-fold and proximal fin mesenchyme (sections labeled 3). (b) Co-expression of the more 3′-ward HoxD genes HoxD8Beta, and HoxD4 (purple) with And1 (red) reveals transcripts of both Hox paralogues within the fin-fold mesenchyme and aligned proximally with the And1 labeling boundary in the ectoderm (sections labeled 1, 2). Anterior is left, distal is up for all whole mounts. Ventral is left, distal is up for all sections. Scale bars = 200 nm.
In mice, early phase HoxD patterning of the proximal limb is primarily driven by the 3′ telomeric landscape and involves the upregulation of paralogues along the length of the cluster (i.e., HoxD1–13)33,34. Regulatory control over HoxD expression then transitions to the 5′ centromeric landscape to pattern the autopod, where only HoxD9–13 (i.e., those paralogues neighboring the 5′ end of the cluster) are active33,37. In paddlefish, our in situ data demonstrate that 5′ HoxD paralogues are expressed in the fin-fold (Fig. 4a), which like the autopod in mice represents the distal-most compartment of the appendage. In order to determine if HoxD expression in the fin-fold is restricted to genes at the 5′ end of the cluster (similar to late phase autopod patterning), we compared the expression of And1 and the more 3′-ward HoxD paralogues HoxD4 and HoxD8Beta. In whole mount fins, strong And1 labeling in the ectoderm made it difficult to discern 3′ HoxD labeling within the fin-fold. In section, however, transcripts of both HoxD4 and HoxD8Beta were detected in the fin-fold mesenchyme (Fig. 4b), and like the 5′ paralogues examined (Fig. 4a), aligned proximally with the And1 labeling boundary in the ectoderm.
HoxD expression in the fin-fold compartment of chondrichthyans
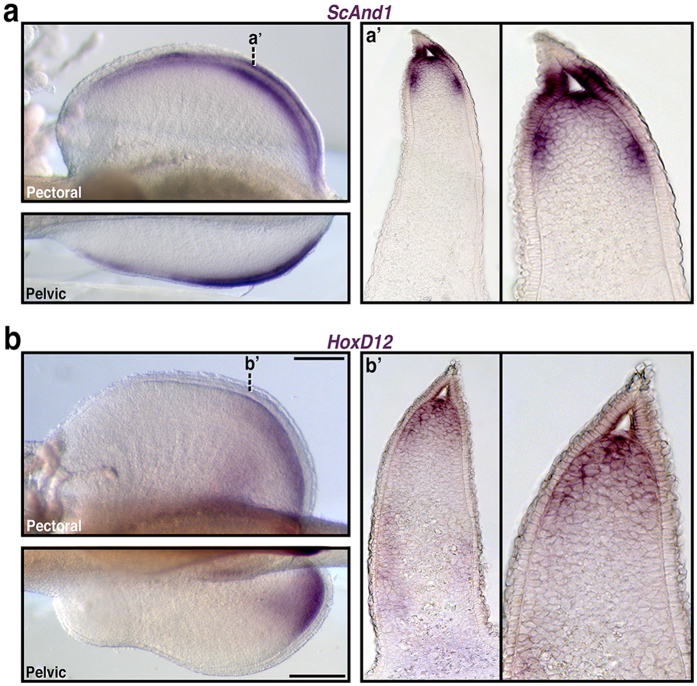
To test whether Hox patterning of the fin-fold compartment is a derived condition of paddlefish or a more general feature of gnathostomes, we extended our analysis to the lesser-spotted catshark Scyliorhinus canicula as a representative chondrichthyan. Chondrichthyan dermal fin skeletons are composed of ceratotrichia, which are histologically and chemically similar to actinotrichia51. Blast searches of the catshark transcriptome assembly returned a putative Actinodin homologue, which we identified as ScAnd1. Whole mount in situs revealed ScAnd1 expression in the distal pectoral, pelvic, and median fins (Fig. 5a and Supplemental Fig. 3), consistent with the distribution of labeling in actinopterygians (this study, and ref. 48). Cross sections through Stage 30 pectoral fins confirmed that ScAnd1 expression was both mesenchymal and ectodermal and marked the boundaries of the fin-fold (Fig. 5a).
Figure 5. HoxD expression in the fin-fold compartment of catshark.
(a,b) In situ hybridizations in whole-mount and representative cross sections. (a) In situ expression for the catshark Scyliorhinus canicula Actinodin1 homologue (ScAnd1), shown to be an early molecular marker for cells contributing to the fin-fold in teleosts. Whole-mount ScAnd1 expression in Stage 30 pectoral and pelvic fins is restricted to the distal fin. (a’) Cross section of ScAnd1 Stage 30 pectoral fins (and magnification) reveal ScAnd1 positive cells mark the boundaries of the fin-fold with both mesenchymal and ectodermal expression domains. (b) In situ gene expression for S. canicula HoxD12. Whole-mount HoxD12 expression in Stage 30 pectoral and pelvic fins form distinct proximal and distal domains. (b’) Cross section of HoxD12 Stage 30 pectoral fins (and magnification) reveal overlap with the distal domain of ScAnd1 (compare to a’), indicating HoxD positive cells in the fin-fold. For all whole mounts: anterior is left, distal is up in pectoral fins, distal is down in pelvic fins. Dashed lines correlate to plane of section in (a’,b’). Scale bars = 200 nm.
In catshark, 5′ HoxD genes are expressed in early and late waves, patterning the proximal and distal regions of the fin, respectively23. In order to determine if the distribution of late phase HoxD transcripts extends into the fin-fold compartment, we compared the expression of HoxD12 with our ScAnd1 results. In Stage 30 pectoral and pelvic fins, HoxD12 labeling formed distinct domains proximally and distally, consistent with previously published results23 (Fig. 5b). Significantly, cross sections showed considerable overlap between the distal domain of HoxD12 and ScAnd1, indicating HoxD expression in the fin-fold mesenchyme of catshark (Fig. 5b).
Discussion
In this study, we have characterized the expression of HoxD and HoxD cluster-related genes during paired fin development in the American Paddlefish Polyodon spathula. These results demonstrate an early collinear arrangement of 5′ HoxD nesting, similar to that of other vertebrates21,22,23,24,33 and consistent with a model in which the first phase of HoxD activation is part of a conserved pathway for establishing Shh in the ZPA and appendage asymmetry52,53,54,55. Our results also demonstrate that in more advanced fins, collinear HoxD nesting is maintained along the presumptive site of distal radial formation following outgrowth of the fin-fold. This persistent mid-fin expression resembles the later dynamics of early phase patterning in limbs, where HoxD expression in the nascent limb bud continues in the fore-limb forming territory following outgrowth of the autopod33,38. Notably, we did not observe an extended anterior sweep of expression of HoxD13 (contra a previous report22), HoxD14, or the cluster-related genes Evx2 or LNP, in any of the stages examined. These observations provide evidence that the inverse collinear profile considered a hallmark of digit patterning in tetrapods may not be present in paddlefish (or a more inclusive clade, see ref. 24).
In addition to early patterning of the proximal fin bud, our paddlefish in situ results reveal a late phase of HoxD expression that co-localizes with And1 in the mesenchyme of the fin-fold, a region absent of pre-chondrocytes. These new data, together with a report of late HoxD expression in the fin-fold of zebrafish24, suggest this character may be shared among actinopterygians. Chondrichthyans provide a critical outgroup for testing this hypothesis. Our data for catshark confirm the spatially distinct early and late phases of HoxD activation described by Freitas and colleagues23, and demonstrate overlap between the late distal phase of HoxD12 labeling and that of And1. In tetrapods, most studies of Hox function during limb development have focused on endoskeletal patterning and the effects of gene abrogation on the formation of the stylopod, zeugopod, and autopod56,57,58,59,60,61,62. Recent work, however, has shown that Hox genes have a broader role in limbs, and are required for normal muscle and tendon formation independent of skeletal phenotype62. While the anatomical outcome of HoxA/D loss in non-tetrapods remains elusive, the co-localization of HoxD and And1 labeling in a representative actinopterygian and chondrichthyan provide evidence of Hox-positive cells contributing to fin-fold specific tissues (i.e., the elastoidin component of the actinotrichia in paddlefish and ceratotrichia in catshark), and suggest an ancient role for Hox genes in integrating aspects of appendage formation beyond endoskeletal patterning.
Our unexpected find, the first report of appendicular expression of HoxD14Beta, is contra recent hypotheses that Hox14 paralogues are isolated from canonical regulation of the cluster14,47. Instead, our results demonstrate that transcripts of HoxD14Beta are posteriorly-restricted within the fins, and together with other cluster paralogues form a collinear arrangement of nesting, as would be expected with canonical early phase regulation. These results suggest an ancestral role for HoxD14 in appendage patterning in gnathostomes, one that has been lost in most lineages14,47 yet retained, at least in terms of expression, in paddlefish.
Current evidence from comparative gene expression, genomics, and transgenic assays support a model in which bimodal regulation of the HoxD cluster is partially conserved between fins and limbs25,26,28,29,46. The apparent antiquity of this regulatory strategy28, together with the data presented here, raise the intriguing and underexplored possibility that HoxD patterning of the proximal fin and fin-fold may reflect separate controls from the 3′ and 5′ landscapes, respectively. In tetrapods, late phase HoxD-related expression in the autopod is limited to HoxD9–13 (those paralogues nearest the 5′ end of the cluster) and the 5′ flanking genes Evx2 and LNP, both of which are co-regulated with HoxD1333,35. We predicted a similar restriction in cluster activity in the fin-folds of paddlefish, but found transcripts of the 3′-ward members HoxD4 and HoxD8Beta throughout the fin-fold mesenchyme. This difference, combined with a lack of anteriorized HoxD13, Evx2 and LNP, may reflect lineage specific variation in landscape/cluster interactions between tetrapods and paddlefish, or more generally actinopterygians24. Additional comparative data characterizing the expression of 3′ HoxD paralogues during fin development in a chondrichthyan will inform inferences about ancestral patterns of cluster activation, and fuel new hypotheses as to the regulatory dynamics of the 3′ and 5′ landscapes during appendage formation in non-tetrapods. Interestingly, current models for limb development posit that chromatin re-organization between early (proximal) and late (distal) HoxD expression creates an intermediate zone of low Hox dosage that manifests anatomically as the small bones of the wrist32,34,38. In fins, a series of small, endochondral bones (distal radials) often separate the proximal radials and fin-fold (e.g., Fig. 1). An ancestral role for HoxD genes in patterning the distal fin compartment makes it tempting to speculate that a similar mechanism may explain appendage morphologies in non-tetrapods. However, not all patterns meet the predictions of this model (e.g., strong HoxD labeling in the presumptive site of distal radial formation in paddlefish), suggesting that modulation of Hox dosage and its effects on cell behavior in the context of local signaling environments differ across lineages. Our results demonstrate that further comparative work, with an increasing focus on basal gnathostomes, will be necessary to elucidate the ancestral roles of Hox genes in appendage patterning, roles that may involve a great flexibility in skeletogenic differentiation outcomes.
Methods
Animal husbandry and staging
Embryos of the American Paddlefish Polyodon spathula were obtained from Osage Catfisheries Inc. (Osage Beach, MO, USA), and were raised at 18 °C in recirculating large-volume freshwater tanks (pH 7.5 ± 0.2, salinity of 1.0 ± 0.2ppt), fixed overnight in Carnoy’s solution, and dehydrated to 100% ethanol for storage at −20 °C. Embryos of the catshark Scyliorhinus canicula were produced at the Biological Model facility of the Roscoff Marine Station, and raised at 17 °C in oxygenated sea water, fixed for 16 hours in 4% PFA, and dehydrated to 100% Methanol prior to storage at −20 °C. Animals were staged according to refs 49, 63, 64, 65.
All experiments and animal care in this study were performed in accordance with the approved institutional guidelines and regulations of the Institutional Animal Care and Use Committee (IACUC) of Kennesaw State University (approved protocol #12-001).
Transcriptome
Paddlefish larvae from early pectoral fin development stages (stages 41–44)63 were preserved in RNAlater and then dissected into pectoral fin and reference tissue (operculum) samples. After RNA library construction, IlluminaTM sequencing of the transcriptome was performed by the Centre for Applied Genomics – Toronto (Toronto, Canada) producing a total of 65,682,405 pectoral fin raw reads (72 bp nonpaired). We used the Trinity program (default settings)66 to assemble 92,446 likely transcripts. Transcripts were annotated and validated against the National Centre for Biotechnology Information (NCBI) non-redundant (nr) database using BLAST (cut off E value of 10−5).
cDNA synthesis for cloning
RNA was isolated from Stage 26–46 paddlefish embryos with Trizol reagent (Invitrogen) plus the PureLinkTM Micro-to-Midi Total RNA Purification System (Invitrogen), and used as template to generate single-strand cDNA with the Superscript III First Strand Synthesis kit (Invitrogen), all per manufacturer’s instructions. Paddlefish primers were designed from transcriptome sequences as follows (F, forward; R, reverse): Evx2: CCGGAAGTCTTCCATACCCTTT (F), CCCTTCACACAACCTAACTGACG (R); Sox9: CTCGATCCCTTCATGAAGATGAC (F), GATGTGAGTTTGCTCAGATCCG (R); LNP: TCCTGAGGAATGGACTGCAA (F), GAGTCTTCCGAGCAGGATTTAGAA (R); HoxD4: TGGATCCTAAATTTCCACCTTGCG (F), GCACAGTTTGTAAATGCTGGCTG (R); HoxD8Beta: ACTACGACTGTCAGTTTCCACGG (F), CGTCCTTTGGTAGTGAAGATGGGAAT (R); HoxD12Alpha: TTTGAATTTCCCTAACCCGGACTC (F), GACGGCATTGTCATGTTTAAGTTG (R); HoxD12Beta: TTTGAATTACTCTTCCCCGGACTC (F), CATGGCTGGTTTGAGTCGACAG (R); HoxD14Beta: TATGGATGGATCCCAGTTCTCC (F), CGTCTCACCTGTCTTTCTGTCA (R); And1: CTCGTACCCTGCGTAGTTACTG (F); CTCACCAAGCCGCTGTAACC (R). Primers for catshark were as follows: HoxD12: TATTTCGCCAACCTGCGTCC (F), CTTGTTGGTCGCTCAGGTTCAG (R); ScAnd1: TGGCAGAGCAGAACCATGTG (F), GTCAGGATCTTGAACACCTTGGTC (R).
PCR products were cloned into pGEM®-T Easy Vector (Promega), which was purified and sequenced using Genscript DNA Sequencing Services. All sequences were analyzed using Unipro UGENE and BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Orthology for the Actinodin family members was confirmed using maximum likelihood in Mega6.06. GenBank accession numbers for this work are KU744647-KU744655. For in situ hybridization experiments, the entire PCR product obtained from each primer set was used as template for in vitro transcription (see below). Plasmids containing paddlefish fragments of HoxD11Alpha, HoxD11Beta, HoxD13Alpha, and HoxD13Beta were obtained through Life Technologies Gene Art Services and were based on published genomic BAC clones13. All new probes generated during this study ranged from approximately 480 to 900 base pairs in length. Paddlefish probes for HoxD4, HoxD8Beta, and HoxD14Beta were complementary to regions of coding exons one and two, including the homeodomain. Probes for HoxD11Alpha and HoxD11Beta were complementary to a region of coding exon one, coding exon two, including the homeodomain, and 3′UTR. Probes for HoxD12Alpha, HoxD12Beta, HoxD13Alpha, and HoxD13Beta were complementary to the first coding exon and excluded the homeodomain. Probes for Sox9, And1, and LNP were targeted against coding region. Probe for Evx2 was targeted against coding region and 3′ UTR. Probes HoxD13-EF527821 and ScHoxD12 were from Refs 22, and 23 respectively.
In situ hybridization
Plasmids were linearized for probe template using restriction enzymes (New England Biolabs, NEB) and any resultant 3′ overhangs were blunted using DNA Polymerase I, Large Klenow (NEB). Probe synthesis was carried out using SP6, T7, or T3 RNA polymerases (Promega) and either Digoxigenin or Fluorescein RNA labeling mixes (Roche) per manufacturer′s instructions. Whole-mount in situ hybridizations were as described in ref. 67 at a hybridization temperature of 69–70 °C, with the following modifications for double colorimetric in situs: Both Digoxigenin (Sox9, HoxD4, HoxD8Beta, HoxD11Beta–HoxD13Beta) and Fluorescein (And1) labeled probes were included during the hybridization step. Digoxigenin-labeled probes were detected with Anti-Digoxigenin-AP, Fab fragments (1:2000 dilution; Roche) and developed with BM-Purple (Roche). Following the first color reaction, embryos were incubated in 0.1M Glycine (pH 2.2) for 30 minutes to inactivate AP, and then washed thoroughly with MABT. Fluorescein-labeled probes were detected with Anti-Fluorescein-AP, Fab fragments (1:1000 dilution; Roche) and developed using Fast Red Tablets (Roche) per manufacturer’s instructions.
For paddlefish, the total number of antisense in situ specimens examined were as follows: HoxD4: St41–43 (n = 5), St44–45 (n = 8); St46–1dps (n = 4); HoxD8: St41–43 (n = 9), St44–45 (n = 4); St46–1dps (n = 3); HoxD11Alpha: St41–43 (n = 4), St44–45 (n = 3); St46–1dps (n = 4); HoxD11Beta: St41–43 (n = 11), St44–45 (n = 11); St46–1dps (n = 8); 2dps–3dps (n = 4); HoxD12Alpha: St41–43 (n = 7), St44–45 (n = 5); St46–1dps (n = 5); HoxD12Beta: St41–43 (n = 4), St44–45 (n = 3); St46–1dps (n = 5), 2dps–3dps (n = 3); HoxD13Alpha: St41–43 (n = 6), St44–45 (n = 12); St46–1dps (n = 13), 2dps–3dps (n = 5), 5–15dps (n = 5); HoxD13Beta: St41–43 (n = 7), St44–45 (n = 9); St46–1dps (n = 12), 2dps–3dps (n = 7), 5–15dps (n = 4); HoxD13EF527821: St41–43 (n = 4), St44–45 (n = 7); St46–1dps (n = 3); HoxD14Beta: St41–43 (n = 11), St44–45 (n = 13); St46–1dps (n = 10), 2dps–3dps (n = 3), 5–15dps (n = 3); Evx2: St41–43 (n = 10), St44–45 (n = 7); St46–1dps (n = 7), 2dps–3dps (n = 5), 5–15dps (n = 3); LNP: St41–43 (n = 6), St44–45 (n = 4); St46–1dps (n = 3); And1: St41–43 (n = 4), St44–45 (n = 5); And1 plus Sox9: St41–43 (n = 7), St44–45 (n = 10); And1 plus HoxD4: St44–45 (n = 7); And1 plus HoxD8Beta: St44–45 (n = 7); And1 plus HoxD11Beta: St44–45 (n = 6); And1 plus HoxD12Beta: St44–45 (n = 9); And1 plus HoxD13Beta: St44–45 (n = 8). For catshark the total number of in situ specimens examined were as follows: HoxD12: St28–31 (n = 3); And1: St28–31 (n = 3). Sense probe was used with stage-matched specimens as a negative control for antisense hybridization experiments (see Supplemental Fig. 4). Whole mount imaging was done using a Zeiss Discovery.V12 Stereo microscope equipped with an AxioCam MRc5 camera and Zen 2012 (blue edition) software.
Histology
Embryos used for sectioning were incubated in successive 10%, 20%, and 30% sucrose in PBS solutions for a minimum of 3 hours each at room temperature, and then transferred to a 1:1 solution of TBS Tissue Freezing Medium (Fisher): 30% Sucrose in PBS for 8 hours. Embryos were then embedded in TBS Tissue Freezing Medium in disposable plastic moulds and frozen for 10 minutes using isopentane chilled with liquid nitrogen. Embryos were cryosectioned at 16 μm on a Leica CM1850 cryostat, dried for two hours at 35 °C on a slide warmer, and cover-slipped with 25% glycerol in PBS. Sections were imaged using a Zeiss AxioImager.M2 compound microscope equipped with an Axiocam 503 color camera and Zen 2012 (blue edition) software.
Additional Information
How to cite this article: Tulenko, F. J. et al. HoxD expression in the fin-fold compartment of basal gnathostomes and implications for paired appendage evolution. Sci. Rep. 6, 22720; doi: 10.1038/srep22720 (2016).
Supplementary Material
Acknowledgments
We thank Osage Catfisheries, Inc. and the Kahrs family for their continued support of paddlefish research. We thank A. Burke, D. Menke, and S. Nowak for critical reading of the manuscript. This research was supported by a grant from the NSF (IOS 1144965 to M.C.D.) and by instrumentation supplied by the Georgia Research Alliance.
Footnotes
The authors declare no competing financial interests.
Author Contributions M.C.D. and F.J.T. conceived the project. M.C.D., F.J.T. and G.J.A. designed the experiments; S.E.S. designed and constructed the custom iBLAST server used to analyze transcriptome data. M.C.D. facilitated this project through aquaculture support and expertise for the paddlefish data. S.M. facilitated this project through aquaculture support and expertise for the chondrichthyan data. F.J.T., G.J.A. and J.L.M. performed all cloning, in situ hybridization assays, and imaging. F.J.T. and M.C.D. generated all figures and artwork and wrote the manuscript with significant contributions from G.J.A., J.L.M. and S.E.S.
References
- Duboule D. & Dollé P. The structural and functional organization of the murine HOX gene family resembles that of Drosophila homeotic genes. EMBO J. 8, 1497–1505 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A., Papalopulu N. & Krumlauf R. The murine and Drosophila homeobox gene complexes have common features of organization and expression. Cell 57, 367–378 (1989). [DOI] [PubMed] [Google Scholar]
- Burke A. C., Nelson C. E., Morgan B. A. & Tabin C. Hox genes and the evolution of vertebrate axial morphology. Development 121, 333–46 (1995). [DOI] [PubMed] [Google Scholar]
- Cohn M. J. & Tickle C. Developmental basis of limblessness and axial patterning in snakes. Nature 399, 474–479 (1999). [DOI] [PubMed] [Google Scholar]
- Duboule D. The rise and fall of Hox gene clusters. Development 134, 2549–2560 (2007). [DOI] [PubMed] [Google Scholar]
- Freitas R., GuangJun Z. & Cohn M. J. Evidence that mechanisms of fin development evolved in the midline of early verterbates. Nature 442, 1033–1037 (2006). [DOI] [PubMed] [Google Scholar]
- Head J. H. & Polly P. D. Evolution of the snake body form reveals homoplasy in amniote gene function. Nature. 520, 86–89 (2015). [DOI] [PubMed] [Google Scholar]
- Pascual-Anaya J., D’Aniello S., Kuratani S. & Garcia-Fernandez J. Evolution of Hox gene clusters in deuterostomes. BMC Dev. Biol. 13, 26 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta T. K. et al. Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). Proc. Natl. Acad. Sci. USA 110, 16044–16049 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers K. E. et al. Hox cluster duplication in the basal teleost Hiodon alosoides (Osteoglossomorpha). Theory Biosci . 128, 109–120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S. & Meyer A. The evolution and maintenance of Hox gene clusters in vertebrates and the teleost-specific genome duplication. Int. J. Dev. Biol. 53, 765–773 (2009). [DOI] [PubMed] [Google Scholar]
- Mungpakdee S. et al. Differential evolution of the 13 Atlantic salmon Hox clusters. Mol. Biol. Evol. 25, 1333–1343 (2008). [DOI] [PubMed] [Google Scholar]
- Crow K. D., Smith C. D., Cheng J. F., Wagner G. P. & Amemiya C. T. An independent genome duplication from Hox paralogs in the American paddlefish – a representative basal ray-finned fish and important comparative reference. Genome Biol. Evol . 4, 937–953 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S. et al. 2008. Noncanonical role of Hox14 revealed by its expression patterns in lamprey and shark. Proc. Natl. Acad. Sci. USA 105, 6679–6683 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers T. P. & Amemiya C. T. Evidence for a Hox14 paralog group in vertebrates. Curr. Biol. 14, R183–R184 (2004). [DOI] [PubMed] [Google Scholar]
- Lee A. P., Koh E. G., Tay A., Brenner S. & Venkatesh B. Highly conserved syntenic blocks at the vertebrate Hox loci and conserved regulatory elements within and outside Hox gene clusters. Proc. Natl. Acad. Sci. USA 103, 6994–6999 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider I. & Shubin N. The origin of the tetrapod limb: from expeditions to enhancers. Trends Genet. 29, 419–426 (2013). [DOI] [PubMed] [Google Scholar]
- Davis M. C. The Deep Homology of the Autopod: Insights from Hox Gene Regulation. Integr. Comp. Biol. 2, 224–232 (2013). [DOI] [PubMed] [Google Scholar]
- Shubin N. H. & Davis M. C. in Modularity in development and evolution (eds Schlosser G. & Wagner G.) Ch. 19, 429–440 (University of Chicago Press, 2004). [Google Scholar]
- Shubin N. H., Daeschler E. B. & Jenkins F. A. Jr. The pectoral fin of Tiktaalik roseae and the origin of the tetrapod limb. Nature 440, 764–771 (2006). [DOI] [PubMed] [Google Scholar]
- Sordino P., van der Hoeven F. & Duboule D. Hox gene expression in teleost fins and the origin of vertebrate digits. Nature 375, 678–681 (1995). [DOI] [PubMed] [Google Scholar]
- Davis M. C., Dahn R. D. & Shubin N. H. An autopodial-like pattern of Hox expression in the fins of a basal actinopterygian fish. Nature 447, 473–476. (2007). [DOI] [PubMed] [Google Scholar]
- Freitas R., Zhang G. & Cohn M. J. Biphasic Hoxd gene expression in shark paired fins reveals an ancient origin of the distal limb domain. PLoS ONE 15, e754 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn D. & Ho R. K. Tri-phasic expression of posterior Hox genes during development of pectoral fins in zebrafish: implications for the evolution of vertebrate paired appendages. Dev. Biol. 322, 220–233 (2008). [DOI] [PubMed] [Google Scholar]
- Schneider I. et al. Appendage expression driven by the Hoxd Global Control Region is an ancient gnathostome feature. Proc. Natl. Acad. Sci. USA 108, 12782–12786 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas R., Gómez-Marín C., Wilson J. M., Casares F. & Gómez-Skarmeta J. L. Hoxd13 contribution to the evolution of vertebrate appendages. Dev Cell. 23, 1219–1229 (2012). [DOI] [PubMed] [Google Scholar]
- Sheth R. et al. Hox genes regulate digit patterning by controlling the wavelength of a turing-type mechanism. Science 338, 1476–1480 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woltering J. M., Noordermeer D., Leleu M. & Duboule D. Conservation and divergence of regulatory strategies at Hox Loci and the origin of tetrapod digits. PLoS Biol 12, e1001773 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke A. R. et al. Deep conservation of wrist and digit enhancers in fish. Proc. Natl. Acad. Sci. USA 112, 803–808 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. E. et al. Analysis of Hox gene expression in the chick limb bud. Development 122, 1449–1466 (1996). [DOI] [PubMed] [Google Scholar]
- Spitz F. & Duboule D. Global control regions and regulatory landscapes in vertebrate development and evolution. Adv. Genet. 61, 175–205 (2008). [DOI] [PubMed] [Google Scholar]
- Andrey G. & Duboule D. SnapShot: Hox gene regulation. Cell 156, 856.e1 (2014). [DOI] [PubMed] [Google Scholar]
- Tarchini B. & Duboule D. Control of Hoxd genes’ collinearity during early limb development. Dev. Cell 10, 93–103 (2006). [DOI] [PubMed] [Google Scholar]
- Andrey G. et al. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science 340, 1234167 (2013). [DOI] [PubMed] [Google Scholar]
- Spitz F., Gonzalez F. & Duboule D. A global control region defines a chromosomal regulatory landscape containing the HoxD cluster. Cell 113, 405–417 (2003). [DOI] [PubMed] [Google Scholar]
- Gonzalez F., Duboule D. & Spitz F. Transgenic analysis of Hoxd gene regulation during digit development. Dev. Biol. 306, 847–859 (2007). [DOI] [PubMed] [Google Scholar]
- Montavon T. et al. A regulatory archipelago controls Hox genes transcription in digits. Cell 147, 1132–1145 (2011). [DOI] [PubMed] [Google Scholar]
- Woltering J. M. & Duboule D. The origin of digits: Expression patterns versus regulatory mechanism. Dev. Cell 18, 526–532 (2010). [DOI] [PubMed] [Google Scholar]
- Takamatsu N. et al. Duplicated Abd-B class genes in medaka hoxAa and hoxAb clusters exhibit differential expression patterns in pectoral fin buds. Dev. Genes Evol. 217, 263–273 (2007). [DOI] [PubMed] [Google Scholar]
- Johanson Z. et al. Fish fingers: digit homologues in sarcopterygian fish fins. J. Exp. Zool. B. Mol. Dev. Evol . 308, 757–68 (2007). [DOI] [PubMed] [Google Scholar]
- Sakamoto K. et al. Heterochronic shift in Hox-mediated activation of sonic hedgehog leads to morphological changes during fin development. PLoS One 4, e5121 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shaughnessy K. L., Dahn R. D. & Cohn M. J. Molecular development of chondrichthyan claspers and the evolution of copulatory organs. Nat Commun. Apr 14;6:6698 . doi: 10.1038/ncomms7698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubin N., Tabin C. & Carroll S. Fossils, genes and the evolution of animal limbs. Nature 388, 639–648 (1997). [DOI] [PubMed] [Google Scholar]
- Shubin N., Tabin C. & Carroll S. Deep homology and the origins of evolutionary novelty. Nature 457, 818–823 (2009). [DOI] [PubMed] [Google Scholar]
- Freitas R., Gómez-Skarmeta J. L. & Rodrigues P. N. New frontiers in the evolution of fin development. J Exp Zoolog B Mol Dev Evol. 322B, 540–552 (2014). [DOI] [PubMed] [Google Scholar]
- Amemiya C. T. et al. The African coelacanth genome provides insight into tetrapod evolution. Nature 496, 311–316 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiner N., Ericsson R., Meyer A. & Kuraku S. Revisiting the origin of the vertebrate Hox14 by including its relict sarcopterygian members. J. Exp. Zool. B. Mol. Dev. Evol . 316, 515–25 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang J. et al. Loss of fish actinotrichia proteins and the fin-to-limb transition. Nature 466, 234–237 (2010). [DOI] [PubMed] [Google Scholar]
- Davis M. C., Shubin N. H. & Force A. Pectoral fin and girdle development in the basal actinopterygians Polyodon spathula and Acipenser transmontanus. J. Morph . 262, 608–628 (2004). [DOI] [PubMed] [Google Scholar]
- Mabee P. M. & Noordsy M. Development of the paired fins in the paddlefish, Polyodon spathula. J. Morphol. 261, 334–344 (2004). [DOI] [PubMed] [Google Scholar]
- SantaMaria J. A., Santos R. L. & Becerra J. An antiserum against ceratotrichia (selachian) recognizes actinotrichia in teleost regenerating fins. Int. J. Dev. Biol . Suppl 1, 175S–176S (1996). [PubMed] [Google Scholar]
- Dahn R. D., Davis M. C., Pappano W. N. & Shubin N. H. Sonic hedgehog function in chondrichthyan fins and the evolution of appendage patterning. Nature 445, 311–314 (2007). [DOI] [PubMed] [Google Scholar]
- Zakany J., Kmita M. & Duboule D. A dual role for Hox genes in limb anterior-posterior asymmetry. Science 304, 1669–1672 (2004). [DOI] [PubMed] [Google Scholar]
- Kmita M. et al. Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature 435, 1113–1116 (2005). [DOI] [PubMed] [Google Scholar]
- Tarchini B., Dubuole D. & Kmita M. Regulatory constraints in the evolution of the tetrapod limb anterior-posterior polarity. Nature 443, 985–988 (2006). [DOI] [PubMed] [Google Scholar]
- Davis A. P., Witte D. P., Hsieh-Li H. M., Potter S. S. & Capecchi M. R. Absence of radius and ulna in mice lacking hoxa-11 and hoxd-11. Nature 375, 791–795 (1995). [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain C. et al. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development 122, 2997–3011 (1996). [DOI] [PubMed] [Google Scholar]
- Fromental-Ramain C. et al. Specific and redundant functions of the paralogous Hoxa-9 and Hoxd-9 genes in forelimb and axial skeleton patterning. Development. 122, 461–472 (1996). [DOI] [PubMed] [Google Scholar]
- Wellik D. M. & Capecchi M. R. Hox10 and Hox11 genes are required to globally pattern the mammalian skeleton. Science 301, 363–367 (2003). [DOI] [PubMed] [Google Scholar]
- Boulet A. M. & Capecchi M. R. Multiple roles of Hoxa11 and Hoxd11 in the formation of the mammalian forlimp zeugopod. Development 131, 299–309 (2004). [DOI] [PubMed] [Google Scholar]
- Zákány J. & Duboule D. The role of Hox genes during vertebrate limb development. Curr. Opin. Genet. Dev . 17, 359–366 (2007). [DOI] [PubMed] [Google Scholar]
- Swinehart I. T., Schlientz A. J., Quintanilla C. S., Mortlock D. P. & Wellik D. M. Hox11 genes are required for regional patterning and integration of muscle tendon and bone. Development 140, 4574–4582 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis W. E. & Grande L. Early development of the actinopterygian head. I. External development and staging of the paddlefish Polyodon spathula. J. Morphol. 213, 47–83 (1992). [DOI] [PubMed] [Google Scholar]
- Ballard W. W. & Needham R. G. Normal embryonic stages of Polyodon spathula (Walbaum). J. Morphol. 114, 465–477 (1964). [DOI] [PubMed] [Google Scholar]
- Ballard W. W., Mellinger J. & Lechenault H. A series of normal stages for development of scyliorhinus-canicula, the lesser spotted dogfish (Chondrichthyes, Scyliorhinidae). J. Exp. Zool. 267, 318–336 (1993). [Google Scholar]
- Grabherr M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrell M. S., Bemis W. E., Northcutt R. G., Davis M. C. & Baker C. V. H. Electrosensory ampullary organs are derived from lateral line placodes in bony fishes. Nat. Commun. 2, 496 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.