Abstract
Malaria is a major public health burden throughout the world. Resistance to the antimalarial drugs has increased the mortality and morbidity rate that is achieved so far through the malaria control program. Monitoring the drug resistance to the available antimalarial drugs helps to implement effective drug policy, through the in vivo efficacy studies, in vitro drug susceptibility tests and detection of molecular markers. It is important to understand the mechanism of the antimalarial drugs, as it is one of the key factors in the emergence and spread of drug resistance. This review summarizes the commonly used antimalarial drugs, their mechanism of action and the genetic markers validated so far for the detection of drug-resistant parasites.
Keywords: Antimalarial drugs, drug resistance markers, malaria, Plasmodium falciparum, Plasmodium vivax
INTRODUCTION
Malaria is one of the deadliest diseases which claim millions of life throughout the world. Five known species of Plasmodium genus that causes malaria in human are Plasmodium falciparum, Plasmodium vivax, Plasmodium malariae, Plasmodium ovale, and Plasmodium knowlesi. Of these, P. falciparum is the most virulent parasite, with high mortality and morbidity rate. The discovery of synthetic drug chloroquine (CQ) in the 1940s helped to treat, prevent and eradicate malaria through National Malaria Control and Eradication Programme throughout the world in 1950s.[1,2] However, the therapeutic efficacy of CQ and efforts to eradicate malaria worldwide were diminished due to the occurrence of CQ resistance. The failure of these eradication programs led to re-emergence of malaria and spread of CQ-resistant parasite in Southeast Asia and South America.[1,2,3,4,5] Due to the lack of potent and affordable drug for malaria treatment, the spread of CQ-resistant parasite to Africa around 1980s claimed 2–3-fold increase in malaria-related death.[5,6] Hence, CQ was replaced with sulfadoxine/pyrimethamine (SP) as the first-line of treatment for malaria; however, parasite became resistant to SP and spread widely.[7,8,9] Use of antimalarial drugs as combination therapy instead of monotherapy has been in practice to increase the efficacy of drug and to delay the emergence of drug resistance parasite. Since then, artemisinin-based combination therapies (ACTs) has been most widely and effectively used for malaria treatment. Recently, resistance to artemisinin has been reported in Southeast Asia increasing the global alarm for malaria treatment and control.[10,11]
The emergence of antimalarial drug-resistant parasite will not only demise the malaria control efforts achieved but to look for new treatment regimes to treat and control the disease. Thus, an in-depth knowledge to understand the mechanism of action of antimalarial drug used and the molecular marker involved for the surveillance of drug-resistant parasite will aid in designing an effective drug policy in all malaria endemic regions and affected countries.
EPIDEMIOLOGY
WHO estimated malaria burden worldwide with approximately 1.2 billion people at risk of infection and its distribution as shown in Figure 1.[12] Globally, 198 million malaria cases have been reported in 2013 with 584,000 deaths indicating a decrease in malaria infection and mortality rate since 2000.[12] WHO African Region contributed 90% of all malaria deaths worldwide, and 78% of deaths reported belong to children <5 years age.[12]
Figure 1.
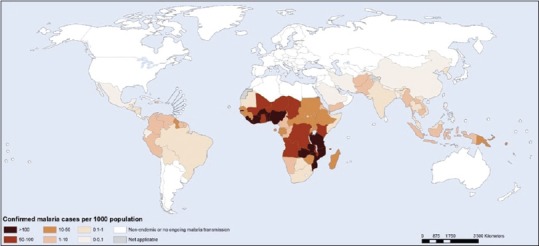
Malaria transmission and its distribution worldwide. World map adapted from “World Malaria Report 2014” illustrating the malaria transmission in various countries in 2013
In the 10 malaria-endemic countries from South East Asia region, about 352 million people are at high risk for malaria, and there was a decrease in malaria reported cases from 2.9 to 1.5 million during 2000–2013.[12] India, Indonesia, and Myanmar were found to carry a heavy load with 55%, 21%, and 21% of malaria cases, respectively, contributing to 97% cases totally in 2013.[12]
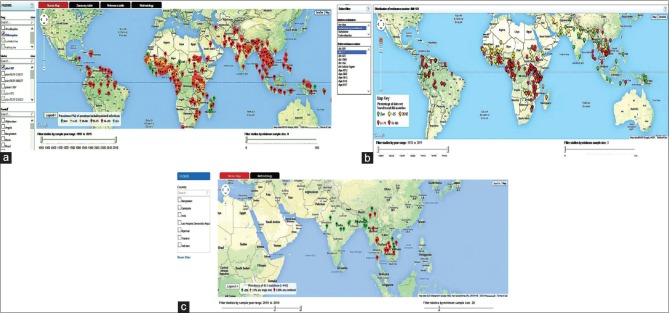
Antimalarial drug resistance has been observed for P. falciparum, P. vivax, and P. malariae globally.[13] Resistance to various antimalarial drugs (quinolones and antifolate family) has been reported in P. falciparum and artemisinin derivate resistance has also been documented recently in western Cambodia.[13,14] Figure 2a–c illustrate the worldwide surveillance of antimalarial drug resistance obtained from WWARN Molecular Surveyor.[15] The WWARN Molecular Surveyor website (http://www.wwarn.org/tracking-resistance) exhibits a range of tools and filters to access the spread of molecular markers associated with various antimalarials. In P. vivax, resistance to CQ, primaquine, mefloquine (MQ), and SP has been reported from various regions of the world and is emerging rapidly. CQ-resistant P. malariae has been reported recently in South Sumatra, Indonesia.[16]
Figure 2.
Prevalence of drug resistance distributed in various regions and countries. Antimalarial drug resistance surveillance obtained from WWARN Molecular Surveyor: (a) exhibiting the chloroquine drug resistance with incidence of Plasmodium falciparum chloroquine resistance transporter K76T mutation, (b) presenting the frequency of Pfdhfr gene with 51I mutation in association with resistance to sulfadoxine-pyrimethamine drug, (c) the prevalence of Kelch 13 propeller mutation for artemisinin resistance in the Southeast Asia region
Based on the National Vector Borne Disease Control Programme surveillance in India, the malaria cases has reduced from 2 million annually in the 1990s to 1 million in 2012, with a declining trend since 2002.[17] However, the P. falciparum cases reported has increased to 50.01% in 2012 from 39% in 1995.[17] In India, CQ resistance was first reported from Karbi-Anglong district, Assam, in 1973.[18] Since then, resistance to commonly used antimalarial drugs (CQ, MQ, and SP) has been reported for P. falciparum across the country and National Drug Policy recommends the use of ACT as first-line of treatment for P. falciparum since 2010.[19,20] Resistance to CQ has also been reported for P. vivax in few cases.[21,22]
ANTIMALARIALS USED AGAINST MALARIA
Based on the mode of action, the antimalarial drugs were mainly classified into three groups: quinolone, antifolate, and artemisinin derivatives. Table 1 summarizes the widely used antimalarials adapted from “Drug resistance in malaria”[23] and modified indicating their mechanism of action and validated molecular markers for drug resistance. Most of the antimalarial drugs target the asexual erythrocytic stages of the parasite, hence called as blood schizonticidal drugs.[24] Tissue schizonticidal drugs target the hypnozoites (dormant stage of the parasite) in the liver whereas gametocytocidal drugs kill the sexual stages of the parasite in the bloodstream.[24] These antimalarial drugs have a different mode of action and mechanism within the parasite that was explained in Table 1 and illustrated in Figure 3. Due to the emergence and spread of CQ and SP resistant parasite, antimalarials are being administrated as combination therapies. Table 2 depicts the currently available ACT drugs. In ACT, each antimalarial drug targets different mechanism of action within the parasite, thereby decreasing the emergence of the multidrug-resistant parasite. Understanding the mechanism of action of drugs will depict in the identification of the specific molecular marker for drug resistance and thereby monitoring the level and spread of resistance can be carried out through the drug-resistant markers. Thus, continuous assessment of markers and therapeutic efficacy studies in malaria endemic regions will help to detect the resistant parasite, and also to understand the degree and extent of resistance associated with a particular population. These studies will aid in timely changes for an effective drug policy when the treatment failure rate exceeds 10% at the end of follow-up based on the WHO guidelines.[25]
Table 1.
Antimalarial drugs used as monotherapy. Summary of the most commonly used antimalarial drugs along with their mechanism of action and validated molecular markers to determine their drug susceptibility/resistance. Most of the antimalarial drugs targets the asexual blood stages of the parasite, whereas primaquine targets the hypnozoites
Figure 3.
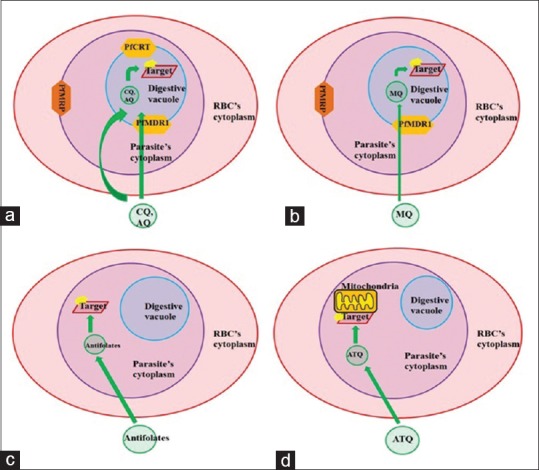
Proposed model for the mechanism and target localization of the antimalarial drugs (a and b) the 4-aminoquinolones such as chloroquine and amodiaquine and the amino alcohol derivatives such as mefloquine, quinine binds to the β-hematin molecule and inhibits the heme detoxification pathway in the parasiteæs digestive vacuole. (c) The antifolate derivatives target the Phdhps and Plasmodium falciparum bifunctional dihydrofolate reductase-thymidylate synthase gene involved in the folate biosynthesis in the cytoplasm of the parasite. (d) Atovaquone binds to the cytochrome b and interferes the electron transport mechanism in the mitochondria of the parasite
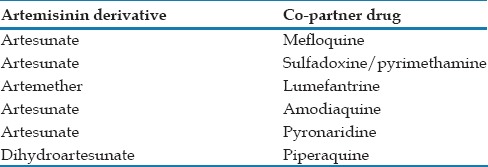
Table 2.
Commonly used artemisinin-based combination therapies drugs. The currently available combination of artemisinin derivative with the copartner drug used for the drug-resistant parasites
DRUG RESISTANCE GENETIC MARKERS
Identification of potent drug-resistant molecular marker is an important tool to determine the emergence and spread of antimalarial drug resistance worldwide. Genetic crosslink and mapping studies of parasites assisted in identifying the genetic markers for drug resistance. Crosslink studies between the CQ-sensitive and resistant strains localized a 36 kb segment on chromosome 7 of P. falciparum.[26] Further molecular analyses revealed mutation in the P. falciparum chloroquine resistance transporter (Pfcrt) gene associated strongly with CQ-susceptible and resistant strains in both laboratory and field studies.[27,28,29,30] Antifolate drugs, SP, inhibits P. falciparum dihydropteroate synthetase (PfDHPS) and P. falciparum bifunctional dihydrofolate reductase-thymidylate synthase (PfDHFR) enzyme involved in folate biosynthesis pathway. In P. falciparum, biochemical and genetic cross-linkage studies allowed the identification of mutation in Pfdhps and Pfdhfr gene involved in the reduced drug susceptibility of SP respectively.[31,32,33,34,35] Whole genome sequencing of the artemisinin-resistant parasite revealed mutation in the Kelch 13 (K13) propeller protein associated with the artemisinin resistance in both clinical and field isolates.[11,36,37] Molecular marker detection by polymerase chain reaction for drug resistance: Overcome the in vivo and in vitro tests used, many isolates and markers screened in a short time, samples collected and stored for prolonged duration can also be studied.[38] The various drug-resistant markers identified for P. falciparum and P. vivax antimalarial drugs are discussed below; thus emergence and spread of resistance can determine easily, and necessary drug policy decisions for malaria treatment be made at the right time.
PLASMODIUM FALCIPARUM CHLOROQUINE RESISTANCE TRANSPORTER
Pfcrt gene has 13 exons, localized to chromosome 7 encoding 424 amino acid transmembrane protein with a molecular mass of 48.6 kDa.[27] PfCRT protein belongs to the drug/metabolite transporter superfamily and chloroquine resistance transporter-like transporter family with 10 putative transmembrane domain spanning the digestive vacuole membrane of the parasite.[39] Several studies comparing the wild and mutant PfCRT allele expression showed less CQ accumulation in mutant PfCRT when compared to wild PfCRT[40,41] and this difference in CQ accumulation is due to the active transport mechanism of mutant PfCRT in the resistant parasite.[42,43] Recently, recombinant CQ-sensitive protein (PfCRT3D7) and CQ-resistant protein (PfCRTDd2, PfCRT7G8, PfCRTK76T [with K76T mutation in the 3D7 gene background]) were purified and found that both the CQ-sensitive and resistant protein transport CQ molecule. In addition, higher CQ transport activity was observed in CQ-resistant protein whereas higher accumulation of CQ measured in CQ-sensitive protein by proteoliposome study. Thus, the CQ-resistant variants showed decreased affinity toward CQ with increased transport activity, which lead to less accumulation of CQ in the digestive vacuole, conferring CQ resistance.[44]
Mutation in Pfcrt gene plays a significant role in determining the CQ resistance and its phenotype. The K76T mutation is the primary determinant of CQ resistance and susceptibility.[27] K76T mutation located in the first transmembrane domain of PfCRT protein, where the positively charged lysine residue is replaced by neutrally charged threonine residue at 76th position, could allow the efflux of diprotonated CQ out of the digestive vacuole by active transport.[39] Most common mutation in other regions (C72S, M74I, N75E, A220S, Q271E, N326S, I356T, and R371I) also confer resistance, but only in association with K76T mutation.
Variation in the PfCRT protein influences antimalarial drug susceptibility and resistance to quinine, amodiaquine (AQ), piperaquine, and lumefantrine.[45,46,47,48] CQ shows cross-resistance with AQ and quinine mainly mediated by 76T, whereas lumefantrine exhibits an inverse cross-resistance having reduced susceptibility in association with wild-type K76.[5] The PfCRT mutations at 72–76 codons confer higher resistance to CQ and medium level AQ resistance in Southeast Asia and Africa, whereas linked with greater AQ resistance in South America.[38] Thus, K76T mutation in PfCRT protein is a potent molecular marker for the antimalarial drug, depending on their previous use in the region.
PLASMODIUM FALCIPARUM MULTIDRUG RESISTANCE PROTEIN 1
The P. falciparum multidrug resistance protein 1 (Pfmdr1) gene located on chromosome 5 has one exon, encoding for P-glycoprotein homolog 1 protein with 1419 amino acid and 162.25 kDa molecular mass.[49,50] PfMDR1 protein is a transmembrane protein, with two domains each consisting of 6 helical transmembrane domains and a nucleotide binding fold region that act as a site for ATP binding, present in the digestive vacuole of the parasite similar to PfCRT protein and belongs to the ATP-binding cassette (ABC) superfamily.[49,50] Polymorphism, amplification, and variation in mRNA expression level of the Pfmdr1 gene have involved in resistance to various antimalarials and emergence of multi-drug resistance parasites.[50]
Mutation in Pfmdr1 gene at the following position (N86Y, Y184F, S1034C, N1042D, and D1246Y) have been reported to involve in determining the drug susceptibility to CQ, quinine, MQ, halofantrine, lumefantrine, and artemisinin.[38,51,52,53] PfMDR1 mutations at N86Y and N1042D position have associated with AQ resistance.[54] K76T and A220S mutation in the Pfcrt gene and N86Y mutation in the Pfmdr1 gene associated with high resistance to CQ in field isolates. In addition, copy number variation of Pfmdr1 gene has been linked to higher level of resistance to quinine, MQ, halofantrine, lumefantrine, and artemisinin.[55]
PLASMODIUM FALCIPARUM MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN
P. falciparum multidrug resistance-associated protein (Pfmrp) gene has one exon located on chromosome 1 belonging to ABC transporter family, similar to PfMDR1.[56] PfMRP is a transmembrane protein localized to the plasma membrane of the parasite encoding 1822 amino acid with a molecular mass of 214.44 kDa and is predicted to have two nucleotide binding domains and two membrane-spanning domains with each consisting of 6 helical transmembrane domains.[56,57] MRP helps in the transport of organic anionic substrates such as oxidized glutathione, glucuronate, sulfate conjugates, and also in drug transport.[56] Two mutations at position Y191H and A437S in PfMRP were found associated with CQ and quinine resistance.[58] Genetic knockout of Pfmrp gene in the resistant parasite, showed high sensitivity to various antimalarial drugs such as CQ, quinine, primaquine, piperaquine, and artemisinin, whereas more accumulation of CQ and quinine observed in the sensitive parasite.[57] Thus, PfMRP is involved in varying the antimalarial response to resistance but not in determining the drug resistance mainly, and also hypothesized that PfMRP protein effluxes various metabolites and drugs out of the parasite in association with other transporters.[57]
PLASMODIUM FALCIPARUM SODIUM HYDROGEN EXCHANGER
P. falciparum contains a candidate gene Pfnhe1 in chromosome 13 with two exons, coding for sodium hydrogen exchanger (Na+/H+ exchanger or PfNHE) protein associated with quinine resistance.[59] PfNHE1 is a transmembrane protein localized in the plasma membrane of the parasite with 1920 amino acid of 226 kDa molecular mass and predicted to have 12 transmembrane domains.[60] The role of PfNHE was not understood fully but hypothesized that it is involved in active efflux of protons to maintain pH 7.4 within the parasite, in response to acidification by anaerobic glycolysis, the primary energy source for the parasite.[61]
In field and in vitro culture studies, polymorphism in the microsatellite ms470 region exhibited a decrease in quinine susceptibility with an increase in DNNND repeat motif whereas increase in quinine susceptibility observed with a rise in NHNDNHNNDDD motif.[62] Three mutations at 790, 894, and 950 codons and polymorphism in the microsatellite region (msR1 and ms3580) showed no association with quinine resistance.[62] Thus, repeat polymorphism in Pfnhe1 gene may be used as a valid genetic marker to determine the quinine resistance in some regions[5] and resistance to quinine also mediated by other genetic markers such as Pfcrt, Pfmrd1, and Pfmrp.[63]
PLASMODIUM FALCIPARUM BIFUNCTIONAL DIHYDROFOLATE REDUCTASE-THYMIDYLATE SYNTHASE
The Pfdhfr-ts gene has one exon located on chromosome 4 encoding for PfDHFR protein with an appropriate molecular mass of 71.73 kDa and 608 amino acids in length. It is a bifunctional enzyme involved in two main folate metabolic activity: the biosynthesis of dTMP by thymidylate synthase activity and the reduction of dihydrofolate into tetrahydrofolate by dihydrofolate reductase activity.[64] The folate mechanism of PfDHFR enzyme inhibited by the action of antifolate drugs such as pyrimethamine and cycloguanil, thus reducing the production of pyrimidine for DNA replication.[65]
Pyrimethamine resistance has mainly associated with point mutation in the PfDHFR protein at S108D codon, further mutation at N51I, C59N, and I164L positions strengthen their resistance besides amplification of gene.[38,66,67,68] Double mutation at A16V and S108T positions in PfDHFR linked with the resistance of P. falciparum to cycloguanil.[69]
PLASMODIUM FALCIPARUM DIHYDROPTEROATE SYNTHETASE
The Pfdhps gene located on chromosome 8 with three exons that encode for PfDHPS protein consisting of 706 amino acid and 83.37 kDa molecular weight. PfDHPS enzyme catalyzes the reaction of the p-aminobenzoic acid (PABA) with a pterin derivative in synthesizing dihydrofolate, a folate precursor that is essential for the synthesis of pyrimidine in the parasite.[64] This catalytic enzyme action to synthesize dihydrofolate inhibited by sulfa drugs (sulfadoxine and dapsone), which act as an analog to PABA.[5,64]
Five mutations in the PfDHPS protein (S436A/F, A437G, L540E, A581G, and A613T/S) have linked with resistance to sulfadoxine in P. falciparum.[38,68] Mutation at 436, 581, and 613 codons are associated with higher level of resistance, whereas mutation at 437 and 540 contribute a low level of resistance with modulation effects in association with other mutation in PfDHPS.[69] Since the antimalarial drug resistance as monotherapy has emerged, sulfadoxine is always provided in combination with pyrimethamine, known as SP or Fansidar, and resistance to SP have associated with point mutation in both Pfdhfr and Pfdhps gene.[5,31,38,67]
CYTOCHROME B
Cytochrome b (cytb) gene is a subunit of cytochrome bc1 complex, which catalyses the transfer of electrons across the inner mitochondrial membrane to maintain the electrochemical potential of the membrane.[70] It is a mitochondrial membrane protein belonging to cytb family with 376 amino acid and 43.37 KDa molecular mass. It is predicted to have 10 putative helical transmembrane domains spanning the mitochondrial inner membrane of the parasite.
The antimalarial drug atovaquone binds to the ubiquinol binding site of cytb, thus disrupting the electrochemical potential of the mitochondrial membrane, which is fatal for the parasite survival.[70] Ubiquinol binding site is a highly conserved region of the protein and mutation in this region confers atovaquone resistance.[71,72] Single mutation at Y268N/S/C codon in the cytb gene associated with resistance to atovaquone in P. falciparum field isolates.[38]
KELCH 13
K13 protein has one exon located on the chromosome 13 with 726 amino acid and 83.66 kDa molecular mass. The C-terminal region of K13 protein has six kelch motifs consisting of beta sheets that folded into a propeller domain and mutation in this region is predicted to disrupt the domain scaffold and alter its function.[36,73] The kelch family proteins have diverse cellular functions, such as in organizing and interacting with other proteins.[36,73] Furthermore, several studies are required to understand the function of K13 protein, various mutation effects on the protein and the mechanism of drug action concerning protein fully.
Recently, the point mutation in the propeller region of K13 protein has been identified as a key determinant for artemisinin resistance in P. falciparum. Nonsynonymous polymorphism at Y493H, R539T, I543T, and C580Y position observed in the kelch repeat region of K13 propeller domain have associated with higher resistance to artemisinin.[36] M476I mutation in the kelch repeat no. 2 and D56V mutation in PF3D7_0110400 gene increased the artemisinin resistance in Tanzania.[36] In cultured and field isolates, mutation at these codons F446I, Y493H, P574L, R539T, and C580Y have contributed to a higher degree of resistance to artemisinin, and the frequency of C580Y allele mutation is higher and takes longer time for parasite clearance when compared to variation in other sites.[11,36] Thus, polymorphism in K13 propeller protein is a potent molecular marker in determining the emergence and spread of artemisinin-resistant P. falciparum.[36]
DRUG RESISTANCE IN VIVAX MALARIA
Malaria infection due to P. vivax is prevalent in many areas of the world except Africa, where P. falciparum infection remains higher.[12] The primary treatment for P. vivax consists of two drugs mainly: CQ (blood schizonticide) to eliminate the asexual blood stages of the parasite and primaquine (tissue schizonticide) to remove the dormant live stage (hypnozoites) of the parasite, which causes relapse in malaria infection.[12] Resistance to CQ in P. vivax emerged first in Papua New Guinea and Indonesia in the late 1980s, since then it has been reported in many parts of the world.[74] WHO recommends the use of ACT for P. vivax infection in areas where CQ resistance has been documented.[12]
The quest to find the molecular drug-resistant markers in P. vivax was based on the known molecular determinant markers in P. falciparum.[5] Increased susceptibility to CQ in P. vivax has strongly associated with the Y976F mutation in Pvmdr1 gene, which is a homolog of Pfmdr1.[75,76,77] Pvcrt gene in P. vivax, a homolog of Pfcrt, have not been associated with CQ resistance unlike in P. falciparum where the mutation in this gene exhibit a higher degree of resistance.[78] MQ resistance in P. vivax has associated with amplification of Pvmdr1 gene.[77,79] In addition, mutation at Y976F position of PvMRD1 in vitro has been observed with resistance to MQ and artesunate, which requires further clinical studies.[77,79]
The point mutation at F57L/I, S58R, T61M, and S117T/N codons of Pvdhfr gene have linked with pyrimethamine resistance and treatment failure in P. vivax.[80,81,82] The wild type residue V585 in Pvdhps gene showed innate resistance to sulfadoxine and is enhanced by the mutation in A383G and A553G codons in PvDHPS, which is similar to PfDHPS mutation at codons A347G and A581G.[83] In summary, drug resistance genetic markers contribute in determining the emergence and spread of resistance in P. vivax.
CONCLUSION
Emergence and spread of antimalarial drug resistance constitute a major threat toward the treatment of malaria and if not handled properly, could reverse the malaria control program and containment achieved so far worldwide. Till date, drug resistance has been reported mainly for P. falciparum and P. vivax. Antimalarial combination therapy targeting different mechanism of action could prolong the emergence and spread of drug-resistant parasites. Understanding the site of action and mechanism of the antimalarials is an important tool to identify drug-resistant marker, to prevent the development of drug resistance further and in the development of new antimalarial drugs/vaccines. The molecular markers of drug resistance play a vital role in the detection of resistance in clinical and field isolates when compared to the in vivo efficacy studies and in vitro tests. Thus, earlier detection of drug-resistant parasites in clinical isolates will aid in employing immediate and appropriate treatment that in turn reduces treatment failure and thereby mortality, and also prevents the spread of resistance. Hence, continuous monitoring and surveillance of drug-resistant molecular markers in malaria endemic regions is important in determining and assisting an effective national drug policy for malaria treatment. Therefore, more research is necessary to find new antimalarial drugs/vaccines for multidrug resistance parasites and in identification and validation of genetic markers for multidrug resistance, thereby containment and treatment of malaria can be achieved hand in hand.
Financial support and sponsorship
This work was supported by the Institute Research Grant from Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER), Pondicherry, India. The first author acknowledges the Indian Council of Medical Research (ICMR), New Delhi, for providing senior research fellowship. We thank Manochitra Kumar and Shashiraja Padukone for revising the manuscript critically.
Conflicts of interest
There are no conflicts of interest.
REFERENCES
- 1.Klein EY. Antimalarial drug resistance: A review of the biology and strategies to delay emergence and spread. Int J Antimicrob Agents. 2013;41:311–7. doi: 10.1016/j.ijantimicag.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farooq U, Mahajan RC. Drug resistance in malaria. J Vector Borne Dis. 2004;41:45–53. [PubMed] [Google Scholar]
- 3.Ballou WR, Hoffman SL, Sherwood JA, Hollingdale MR, Neva FA, Hockmeyer WT, et al. Safety and efficacy of a recombinant DNA Plasmodium falciparum sporozoite vaccine. Lancet. 1987;1:1277–81. doi: 10.1016/s0140-6736(87)90540-x. [DOI] [PubMed] [Google Scholar]
- 4.Payne D. Spread of chloroquine resistance in Plasmodium falciparum. Parasitol Today. 1987;3:241–6. doi: 10.1016/0169-4758(87)90147-5. [DOI] [PubMed] [Google Scholar]
- 5.Petersen I, Eastman R, Lanzer M. Drug-resistant malaria: Molecular mechanisms and implications for public health. FEBS Lett. 2011;585:1551–62. doi: 10.1016/j.febslet.2011.04.042. [DOI] [PubMed] [Google Scholar]
- 6.Trape JF. The public health impact of chloroquine resistance in Africa. Am J Trop Med Hyg. 2001;64(1-2 Suppl):12–7. doi: 10.4269/ajtmh.2001.64.12. [DOI] [PubMed] [Google Scholar]
- 7.Clyde DF, Shute GT. Resistance of Plasmodium falciparum in Tanganyika to pyrimethamine administered at weekly intervals. Trans R Soc Trop Med Hyg. 1957;51:505–13. doi: 10.1016/0035-9203(57)90039-1. [DOI] [PubMed] [Google Scholar]
- 8.Roper C, Pearce R, Bredenkamp B, Gumede J, Drakeley C, Mosha F, et al. Antifolate antimalarial resistance in Southeast Africa: A population-based analysis. Lancet. 2003;361:1174–81. doi: 10.1016/S0140-6736(03)12951-0. [DOI] [PubMed] [Google Scholar]
- 9.Nair S, Williams JT, Brockman A, Paiphun L, Mayxay M, Newton PN, et al. A selective sweep driven by pyrimethamine treatment in Southeast Asian malaria parasites. Mol Biol Evol. 2003;20:1526–36. doi: 10.1093/molbev/msg162. [DOI] [PubMed] [Google Scholar]
- 10.Geneva, Switzerland: WHO Press; 2014. WHO. Status Report on Artemisinin Resistance: September, 2014. [Google Scholar]
- 11.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, et al. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2014;371:411–23. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geneva, Switzerland: WHO Press; 2014. WHO. World Malaria Report. [Google Scholar]
- 13.2nd ed. Geneva, Switzerland: WHO Press; 2010. WHO. Guidelines for the Treatment of Malaria. [Google Scholar]
- 14.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM. Artemisinin Resistance in Cambodia (ARC) Study Consortium. Evidence of artemisinin-resistant malaria in Western Cambodia. N Engl J Med. 2008;359:2619–20. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 15.WWARN. Molecular Surveyor pfmdr1 & pfcrt; 2015. [Last accessed on 2015 May 12]. Available from: http://www.wwarn.org/molecular/surveyor .
- 16.Maguire JD, Sumawinata IW, Masbar S, Laksana B, Prodjodipuro P, Susanti I, et al. Chloroquine-resistant Plasmodium malariae in South Sumatra, Indonesia. Lancet. 2002;360:58–60. doi: 10.1016/S0140-6736(02)09336-4. [DOI] [PubMed] [Google Scholar]
- 17.National Vector Borne Disease Control Programme, Delhi. [Last accessed on 2015 May 12]. Available from: http://www.nvbdcp.gov.in/
- 18.Sehgal PN, Sharma MI, Sharma SL. Resistance to chloroquine in falciparum malaria in Assam State, India. J Commun Disord. 1973;5:175–80. [Google Scholar]
- 19.New Delhi: Directorate of National Vector Borne Disease Control Programme; 2013. Government of India. National Drug Policy on Malaria. [Google Scholar]
- 20.Parija SC, Praharaj I. Drug resistance in malaria. Indian J Med Microbiol. 2011;29:243–8. doi: 10.4103/0255-0857.83906. [DOI] [PubMed] [Google Scholar]
- 21.Garg M, Gopinathan N, Bodhe P, Kshirsagar NA. Vivax malaria resistant to chloroquine: Case reports from Bombay. Trans R Soc Trop Med Hyg. 1995;89:656–7. doi: 10.1016/0035-9203(95)90432-8. [DOI] [PubMed] [Google Scholar]
- 22.Baird JK. Chloroquine resistance in Plasmodium vivax. Antimicrob Agents Chemother. 2004;48:4075–83. doi: 10.1128/AAC.48.11.4075-4083.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bloland PB. Geneva, Switzerland: WHO; 2001. Drug Resistance in Malaria. [Google Scholar]
- 24.Bruce-Chwatt LJ. Classification of antimalarial drugs in AQ2 relation to different stages in the life-cycle of the parasite: Commentary on a diagram. Bull World Health Organ. 1962;27:287–90. [PMC free article] [PubMed] [Google Scholar]
- 25.Geneva, Switzerland: WHO Press; 2010. WHO. Global Report on Antimalarial Drug Efficacy and Drug Resistance: 2000-2010. [Google Scholar]
- 26.Wellems TE, Walker-Jonah A, Panton LJ. Genetic mapping of the chloroquine-resistance locus on Plasmodium falciparum chromosome 7. Proc Natl Acad Sci U S A. 1991;88:3382–6. doi: 10.1073/pnas.88.8.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–71. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baro NK, Callaghan PS, Roepe PD. Function of resistance conferring Plasmodium falciparum chloroquine resistance transporter isoforms. Biochemistry. 2013;52:4242–9. doi: 10.1021/bi400557x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ecker A, Lehane AM, Clain J, Fidock DA. PfCRT and its role in antimalarial drug resistance. Trends Parasitol. 2012;28:504–14. doi: 10.1016/j.pt.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durand R, Jafari S, Vauzelle J, Delabre JF, Jesic Z, Le Bras J. Analysis of pfcrt point mutations and chloroquine susceptibility in isolates of Plasmodium falciparum. Mol Biochem Parasitol. 2001;114:95–102. doi: 10.1016/s0166-6851(01)00247-x. [DOI] [PubMed] [Google Scholar]
- 31.Cowman AF, Morry MJ, Biggs BA, Cross GA, Foote SJ. Amino acid changes linked to pyrimethamine resistance in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum. Proc Natl Acad Sci U S A. 1988;85:9109–13. doi: 10.1073/pnas.85.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson DS, Walliker D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci U S A. 1988;85:9114–8. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang P, Lee CS, Bayoumi R, Djimde A, Doumbo O, Swedberg G, et al. Resistance to antifolates in Plasmodium falciparum monitored by sequence analysis of dihydropteroate synthetase and dihydrofolate reductase alleles in a large number of field samples of diverse origins. Mol Biochem Parasitol. 1997;89:161–77. doi: 10.1016/s0166-6851(97)00114-x. [DOI] [PubMed] [Google Scholar]
- 34.Triglia T, Cowman AF. The mechanism of resistance to sulfa drugs in Plasmodium falciparum. Drug Resist Updat. 1999;2:15–9. doi: 10.1054/drup.1998.0060. [DOI] [PubMed] [Google Scholar]
- 35.Gregson A, Plowe CV. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol Rev. 2005;57:117–45. doi: 10.1124/pr.57.1.4. [DOI] [PubMed] [Google Scholar]
- 36.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois AC, Khim N, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–5. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tun KM, Imwong M, Lwin KM, Win AA, Hlaing TM, Hlaing T, et al. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: A cross-sectional survey of the K13 molecular marker. Lancet Infect Dis. 2015;15:415–21. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed MB, Saliba KJ, Caruana SR, Kirk K, Cowman AF. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–9. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- 39.Martin RE, Kirk K. The malaria parasite's chloroquine resistance transporter is a member of the drug/metabolite transporter superfamily. Mol Biol Evol. 2004;21:1938–49. doi: 10.1093/molbev/msh205. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez CP, McLean JE, Rohrbach P, Fidock DA, Stein WD, Lanzer M. Evidence for a pfcrt-associated chloroquine efflux system in the human malarial parasite Plasmodium falciparum. Biochemistry. 2005;44:9862–70. doi: 10.1021/bi050061f. [DOI] [PubMed] [Google Scholar]
- 41.Yayon A, Cabantchik ZI, Ginsburg H. Identification of the acidic compartment of Plasmodium falciparum-infected human erythrocytes as the target of the antimalarial drug chloroquine. EMBO J. 1984;3:2695–700. doi: 10.1002/j.1460-2075.1984.tb02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sanchez CP, Rohrbach P, McLean JE, Fidock DA, Stein WD, Lanzer M. Differences in trans-stimulated chloroquine efflux kinetics are linked to PfCRT in Plasmodium falciparum. Mol Microbiol. 2007;64:407–20. doi: 10.1111/j.1365-2958.2007.05664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez CP, Stein WD, Lanzer M. Is PfCRT a channel or a carrier? Two competing models explaining chloroquine resistance in Plasmodium falciparum. Trends Parasitol. 2007;23:332–9. doi: 10.1016/j.pt.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 44.Juge N, Moriyama S, Miyaji T, Kawakami M, Iwai H, Fukui T, et al. Plasmodium falciparum chloroquine resistance transporter is a H -coupled polyspecific nutrient and drug exporter. Proc Natl Acad Sci U S A. 2015;112:3356–61. doi: 10.1073/pnas.1417102112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Menard D, Yapou F, Manirakiza A, Djalle D, Matsika-Claquin MD, Talarmin A. Polymorphisms in pfcrt, pfmdr1, dhfr genes and in vitro responses to antimalarials in Plasmodium falciparum isolates from Bangui, Central African Republic. Am J Trop Med Hyg. 2006;75:381–7. [PubMed] [Google Scholar]
- 46.Echeverry DF, Holmgren G, Murillo C, Higuita JC, Björkman A, Gil JP, et al. Short report: Polymorphisms in the pfcrt and pfmdr1 genes of Plasmodium falciparum and in vitro susceptibility to amodiaquine and desethylamodiaquine. Am J Trop Med Hyg. 2007;77:1034–8. [PubMed] [Google Scholar]
- 47.Muangnoicharoen S, Johnson DJ, Looareesuwan S, Krudsood S, Ward SA. Role of known molecular markers of resistance in the antimalarial potency of piperaquine and dihydroartemisinin in vitro. Antimicrob Agents Chemother. 2009;53:1362–6. doi: 10.1128/AAC.01656-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sisowath C, Petersen I, Veiga MI, Mårtensson A, Premji Z, Björkman A, et al. In vivo selection of Plasmodium falciparum parasites carrying the chloroquine-susceptible pfcrt K76 allele after treatment with artemether-lumefantrine in Africa. J Infect Dis. 2009;199:750–7. doi: 10.1086/596738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foote SJ, Thompson JK, Cowman AF, Kemp DJ. Amplification of the multidrug resistance gene in some chloroquine-resistant isolates of P. falciparum. Cell. 1989;57:921–30. doi: 10.1016/0092-8674(89)90330-9. [DOI] [PubMed] [Google Scholar]
- 50.Duraisingh MT, Cowman AF. Contribution of the pfmdr1 gene to antimalarial drug-resistance. Acta Trop. 2005;94:181–90. doi: 10.1016/j.actatropica.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 51.Sidhu AB, Valderramos SG, Fidock DA. pfmdr1 mutations contribute to quinine resistance and enhance mefloquine and artemisinin sensitivity in Plasmodium falciparum. Mol Microbiol. 2005;57:913–26. doi: 10.1111/j.1365-2958.2005.04729.x. [DOI] [PubMed] [Google Scholar]
- 52.Sisowath C, Strömberg J, Mårtensson A, Msellem M, Obondo C, Björkman A, et al. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem) J Infect Dis. 2005;191:1014–7. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 53.Pickard AL, Wongsrichanalai C, Purfield A, Kamwendo D, Emery K, Zalewski C, et al. Resistance to antimalarials in Southeast Asia and genetic polymorphisms in pfmdr1. Antimicrob Agents Chemother. 2003;47:2418–23. doi: 10.1128/AAC.47.8.2418-2423.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sá JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, et al. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci U S A. 2009;106:18883–9. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sidhu AB, Uhlemann AC, Valderramos SG, Valderramos JC, Krishna S, Fidock DA. Decreasing pfmdr1 copy number in Plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J Infect Dis. 2006;194:528–35. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klokouzas A, Tiffert T, van Schalkwyk D, Wu CP, van Veen HW, Barrand MA, et al. Plasmodium falciparum expresses a multidrug resistance-associated protein. Biochem Biophys Res Commun. 2004;321:197–201. doi: 10.1016/j.bbrc.2004.06.135. [DOI] [PubMed] [Google Scholar]
- 57.Raj DK, Mu J, Jiang H, Kabat J, Singh S, Sullivan M, et al. Disruption of a Plasmodium falciparum multidrug resistance-associated protein (PfMRP) alters its fitness and transport of antimalarial drugs and glutathione. J Biol Chem. 2009;284:7687–96. doi: 10.1074/jbc.M806944200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mu J, Ferdig MT, Feng X, Joy DA, Duan J, Furuya T, et al. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol Microbiol. 2003;49:977–89. doi: 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- 59.Ferdig MT, Cooper RA, Mu J, Deng B, Joy DA, Su XZ, et al. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol Microbiol. 2004;52:985–97. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- 60.Bennett TN, Patel J, Ferdig MT, Roepe PD. Plasmodium falciparum Na+/H+exchanger activity and quinine resistance. Mol Biochem Parasitol. 2007;153:48–58. doi: 10.1016/j.molbiopara.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bosia A, Ghigo D, Turrini F, Nissani E, Pescarmona GP, Ginsburg H. Kinetic characterization of Na+/H+antiport of Plasmodium falciparum membrane. J Cell Physiol. 1993;154:527–34. doi: 10.1002/jcp.1041540311. [DOI] [PubMed] [Google Scholar]
- 62.Briolant S, Pelleau S, Bogreau H, Hovette P, Zettor A, Castello J, et al. In vitro susceptibility to quinine and microsatellite variations of the Plasmodium falciparum Na+/H+exchanger (Pfnhe-1) gene: The absence of association in clinical isolates from the Republic of Congo. Malar J. 2011;10:37. doi: 10.1186/1475-2875-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ekland EH, Fidock DA. Advances in understanding the genetic basis of antimalarial drug resistance. Curr Opin Microbiol. 2007;10:363–70. doi: 10.1016/j.mib.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Foote SJ, Cowman AF. The mode of action and the mechanism of resistance to antimalarial drugs. Acta Trop. 1994;56:157–71. doi: 10.1016/0001-706x(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 65.Hyde JE. Exploring the folate pathway in Plasmodium falciparum. Acta Trop. 2005;94:191–206. doi: 10.1016/j.actatropica.2005.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thaithong S, Ranford-Cartwright LC, Siripoon N, Harnyuttanakorn P, Kanchanakhan NS, Seugorn A, et al. Plasmodium falciparum: Gene mutations and amplification of dihydrofolate reductase genes in parasites grown in vitro in presence of pyrimethamine. Exp Parasitol. 2001;98:59–70. doi: 10.1006/expr.2001.4618. [DOI] [PubMed] [Google Scholar]
- 67.Urdaneta L, Plowe C, Goldman I, Lal AA. Point mutations in dihydrofolate reductase and dihydropteroate synthase genes of Plasmodium falciparum isolates from Venezuela. Am J Trop Med Hyg. 1999;61:457–62. doi: 10.4269/ajtmh.1999.61.457. [DOI] [PubMed] [Google Scholar]
- 68.Wernsdorfer WH, Noedl H. Molecular markers for drug resistance in malaria: Use in treatment, diagnosis and epidemiology. Curr Opin Infect Dis. 2003;16:553–8. doi: 10.1097/00001432-200312000-00007. [DOI] [PubMed] [Google Scholar]
- 69.Hyde JE. Drug-resistant malaria – An insight. FEBS J. 2007;274:4688–98. doi: 10.1111/j.1742-4658.2007.05999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barton V, Fisher N, Biagini GA, Ward SA, O’Neill PM. Inhibiting Plasmodium cytochrome bc1: A complex issue. Curr Opin Chem Biol. 2010;14:440–6. doi: 10.1016/j.cbpa.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 71.Gil JP, Nogueira F, Strömberg-Nörklit J, Lindberg J, Carrolo M, Casimiro C, et al. Detection of atovaquone and Malarone resistance conferring mutations in Plasmodium falciparum cytochrome b gene (cytb) Mol Cell Probes. 2003;17:85–9. doi: 10.1016/s0890-8508(03)00006-9. [DOI] [PubMed] [Google Scholar]
- 72.Korsinczky M, Chen N, Kotecka B, Saul A, Rieckmann K, Cheng Q. Mutations in Plasmodium falciparum cytochrome b that are associated with atovaquone resistance are located at a putative drug-binding site. Antimicrob Agents Chemother. 2000;44:2100–8. doi: 10.1128/aac.44.8.2100-2108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sibley CH. Understanding drug resistance in malaria parasites: Basic science for public health. Mol Biochem Parasitol. 2014;195:107–14. doi: 10.1016/j.molbiopara.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 74.Whitby M, Wood G, Veenendaal JR, Rieckmann K. Chloroquine-resistant Plasmodium vivax. Lancet. 1989;2:1395. doi: 10.1016/s0140-6736(89)92002-3. [DOI] [PubMed] [Google Scholar]
- 75.Suwanarusk R, Russell B, Chavchich M, Chalfein F, Kenangalem E, Kosaisavee V, et al. Chloroquine resistant Plasmodium vivax: In vitro characterisation and association with molecular polymorphisms. PLoS One. 2007;2:e1089. doi: 10.1371/journal.pone.0001089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Russell B, Chalfein F, Prasetyorini B, Kenangalem E, Piera K, Suwanarusk R, et al. Determinants of in vitro drug susceptibility testing of Plasmodium vivax. Antimicrob Agents Chemother. 2008;52:1040–5. doi: 10.1128/AAC.01334-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suwanarusk R, Chavchich M, Russell B, Jaidee A, Chalfein F, Barends M, et al. Amplification of pvmdr1 associated with multidrug-resistant Plasmodium vivax. J Infect Dis. 2008;198:1558–64. doi: 10.1086/592451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nomura T, Carlton JM, Baird JK, del Portillo HA, Fryauff DJ, Rathore D, et al. Evidence for different mechanisms of chloroquine resistance in 2 Plasmodium species that cause human malaria. J Infect Dis. 2001;183:1653–61. doi: 10.1086/320707. [DOI] [PubMed] [Google Scholar]
- 79.Imwong M, Pukrittayakamee S, Pongtavornpinyo W, Nakeesathit S, Nair S, Newton P, et al. Gene amplification of the multidrug resistance 1 gene of Plasmodium vivax isolates from Thailand, Laos, and Myanmar. Antimicrob Agents Chemother. 2008;52:2657–9. doi: 10.1128/AAC.01459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Pécoulas PE, Tahar R, Ouatas T, Mazabraud A, Basco LK. Sequence variations in the Plasmodium vivax dihydrofolate reductase-thymidylate synthase gene and their relationship with pyrimethamine resistance. Mol Biochem Parasitol. 1998;92:265–73. doi: 10.1016/s0166-6851(97)00247-8. [DOI] [PubMed] [Google Scholar]
- 81.Marfurt J, de Monbrison F, Brega S, Barbollat L, Müller I, Sie A, et al. Molecular markers of in vivo Plasmodium vivax resistance to amodiaquine plus sulfadoxine-pyrimethamine: Mutations in pvdhfr and pvmdr1. J Infect Dis. 2008;198:409–17. doi: 10.1086/589882. [DOI] [PubMed] [Google Scholar]
- 82.Leartsakulpanich U, Imwong M, Pukrittayakamee S, White NJ, Snounou G, Sirawaraporn W, et al. Molecular characterization of dihydrofolate reductase in relation to antifolate resistance in Plasmodium vivax. Mol Biochem Parasitol. 2002;119:63–73. doi: 10.1016/s0166-6851(01)00402-9. [DOI] [PubMed] [Google Scholar]
- 83.Korsinczky M, Fischer K, Chen N, Baker J, Rieckmann K, Cheng Q. Sulfadoxine resistance in Plasmodium vivax is associated with a specific amino acid in dihydropteroate synthase at the putative sulfadoxine-binding site. Antimicrob Agents Chemother. 2004;48:2214–22. doi: 10.1128/AAC.48.6.2214-2222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]