Abstract
Background
The ARABIDOPSIS SKP1-LIKE1 (ASK1) protein functions as a subunit of SKP1-CUL1-F-box (SCF) E3 ubiquitin ligases. Previous genetic studies showed that ASK1 plays important roles in Arabidopsis flower development and male meiosis. However, the molecular impact of ASK1-containing SCF E3 ubiquitin ligases (ASK1-E3s) on the floral proteome and transcriptome is unknown.
Results
Here we identified proteins that are potentially regulated by ASK1-E3s by comparing floral bud proteomes of wild-type and the ask1 mutant plants. More than 200 proteins were detected in the ask1 mutant but not in wild-type and >300 were detected at higher levels in the ask1 mutant than in wild-type, but their RNA levels were not significantly different between wild-type and ask1 floral buds as shown by transcriptomics analysis, suggesting that they are likely regulated at the protein level by ASK1-E3s. Integrated analyses of floral proteomics and transcriptomics of ask1 and wild-type uncovered several potential aspects of ASK1-E3 functions, including regulation of transcription regulators, kinases, peptidases, and ribosomal proteins, with implications on possible mechanisms of ASK1-E3 functions in floral development.
Conclusions
Our results suggested that ASK1-E3s play important roles in Arabidopsis protein degradation during flower development. This study opens up new possibilities for further functional studies of these candidate E3 substrates.
Electronic supplementary material
The online version of this article (doi:10.1186/s12870-015-0571-9) contains supplementary material, which is available to authorized users.
Keywords: Arabidopsis, ASK1, E3 ubiquitin ligase, Mass spectrometry, Protein degradation, Proteomics, Transcriptomics
Background
The ubiquitin-proteasome system (UPS) plays important roles in targeted protein degradation, thereby regulating a variety of cellular processes [1–3]. Ubiquitination reactions are catalyzed by the sequential actions of E1 ubiquitin activating enzymes, E2 ubiquitin conjugating enzymes, and E3 ubiquitin ligases. Multiple ubiquitin molecules can be attached to the existing ubiquitin moieties on the protein substrates to form polyubiquitin chains and the polyubiquitinated proteins are usually then degraded by the 26S proteasome.
The UPS regulates many processes in plants, including development and biotic/abiotic stress responses [1, 3–5]. This broad spectrum of functions is made possible by the large number of genes encoding components in the UPS. Plants usually contain a few E1 enzymes, tens of E2 enzymes, and hundreds of E3 ligases, which determine substrate specificities. Therefore, the numerous E3 ligases can potentially ubiquitinate many proteins. Moreover, the modular design of multimeric E3 ubiquitin ligases including the SKP1-CUL1-F-box (SCF) complexes greatly expands the likely number of proteins that can be specifically ubiquitinated. The subunits of SCF complexes are encoded by multi-gene families, especially the F-box proteins, which are encoded by hundreds of genes in plants. Thus, the combination of these components can form various SCF complexes to ubiquitinate numerous substrate proteins.
Genetic studies indicate that plant F-box proteins are involved in hormone signaling pathways, self-incompatibility, developmental processes, and others. Among the F-box proteins important for hormone signaling, TRANSPORT INHIBITOR RESPONSE 1 (TIR1) is a receptor of auxin and the SCFTIR1 ubiquitin ligase facilitates the degradation of AUX/IAA proteins, which are repressors of auxin-induced gene expression [6–9]. The F-box protein CORONATINE INSENSITIVE 1 (COI1) has a similar mechanism in regulating jasmonic acid (JA) signaling; COI1 is a receptor of JA and SCFCOI1 destabilizes JAZ proteins, thereby releasing the transcription factor MYC2 for the activation of JA-responsive genes [10–12]. Other signaling pathways for hormones such as ethylene, gibberellic acid (GA), and abscisic acid (ABA) also require components of the UPS [5]. In addition, S-locus F-box proteins (SLFs) function as the pollen-specific determinants of self-incompatibility [13–16]. The F-box protein UNUSUAL FLORAL ORGANS (UFO) is important for normal meristem identity and floral organ development [17–19]. UFO can interact with LEAFY genetically to activate AP3 expression [20–22].
The Arabidopsis homolog of the yeast and human SKP1 genes, the ARABIDOPSIS SKP1-LIKE1 (ASK1), encodes an SCF subunit that bridges Cullin and F-box proteins [23]. It has been shown that ASK1 can interact with F-box proteins UFO [22, 24], COI1 [25], TIR1 [6], and others [24, 26, 27]. Since these F-box proteins have important roles in different pathways, ASK1, as a key component in SCFs, likely has crucial functions in many processes. This was suggested by previous genetic studies of the ask1 mutant, which has defects in male meiosis, floral organ development, and vegetative growth [23, 28–31]. Although a few substrates of SCFs have been identified in Arabidopsis, they are mainly specific to the well-studied F-box proteins described above. A large number of other ASK1-interacting F-box proteins and their substrates remain elusive, as do the biological pathways regulated by E3s containing ASK1.
Most of the known ubiquitin ligase substrates were identified by protein-protein interaction methods, usually when the F-box protein has a known function [10, 32–34]. Recently, mass spectrometry (MS)-based proteomics approaches have been increasingly applied in various areas including differential gene expression, post-translational modifications, disease marker discovery, as well as the identification of ubiquitin ligase substrates either by detection of ubiquitinated proteins [35, 36], or by comparing proteomes of wild-type (WT) and ubiquitin ligase mutants [37]. In this study, we used a proteomics approach, Multidimensional Protein Identification Technology (MudPIT), to identify floral proteins potentially regulated by ASK1 by comparing floral bud proteomes of WT and ask1 mutant plants. Furthermore, we performed comparative transcriptomics analysis of WT and ask1 floral buds to investigate the effect of ASK1 on gene expression. The integrated transcriptomics and proteomics analyses revealed that many proteins are potentially regulated by ASK1-E3s. We discuss several possible ways of how ASK1 might regulate protein stability and further downstream gene expression.
Results and discussion
Transcriptomic analysis of Ler and ask1 floral buds
To determine the effect of the ask1 mutation on the floral transcriptome, WT (Ler) and ask1 floral bud transcriptomes were analyzed using GeneChip Arabidopsis ATH1 Genome Array. The average values from Ler and ask1 microarrays were compared to find genes whose RNA levels differ by at least two fold and Student’s t-test p-value < 0.05. We found that 74 and 42 genes were up-regulated and down-regulated, respectively, in ask1 transcriptome compared with Ler (Additional files 1 and 2). We used agriGO [38] to determine if certain gene categories are over-represented in the up-/down-regulated genes in ask1. We found that genes are enriched in the GO categories of responsive to various stimuli or stresses (Fig. 1). Among the 42 down-regulated genes (including ASK1) in ask1, 19 genes are related to biotic/abiotic signaling pathways (Table 1), including hormone, light/circadian, temperature, salt, and other signaling pathways. Among the 74 up-regulated genes in ask1, 39 genes were annotated to be involved in response to various biotic/ abiotic signals (Table 2). The molecular functions of most of these genes are not well understood except for evidence from transcriptional responses to stimuli (e.g., COLD-REGULATED 15A/15B, DARK INDUCIBLE 10, SENESCENCE 1, etc.) and sequence homology with well-characterized proteins or protein domains (e.g., HAD superfamily acid phosphatase, JUMONJI DOMAIN CONTAINING 5, CONSTANS-LIKE 2, etc.).
Fig. 1.
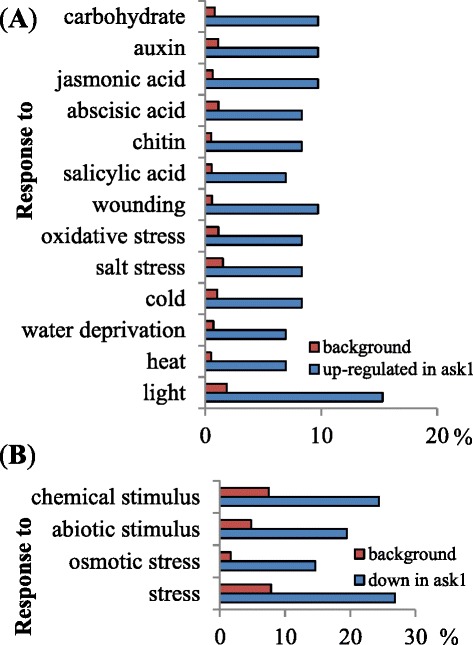
GO categories of stimulus/stress responsive genes enriched in the up-/down-regulated genes in the ask1 transcriptome. a GO categories of stimulus/stress responsive genes enriched in the up-regulated genes in the ask1 transcriptome. b GO categories of stimulus/stress responsive genes enriched in the down-regulated genes in the ask1 transcriptome. Background percentage (%) represents the proportion of all annotated genes of each GO category within the total genes in the ATH1 microarray. ask1 percentage (%) represents the proportion of up-/down-regulated genes in the ask1 transcriptome of each GO category within the total genes in the ATH1 microarray
Table 1.
Responsive genes down-regulated in the ask1 mutant transcriptome
| Gene ID | Gene name | Signaling pathways/responses |
|---|---|---|
| AT5G15960 | KIN1 | Cold and ABA |
| AT1G35720 | ANNEXIN 1 (ANNAT1) | Oxidative stress |
| AT2G42530 | COLD REGULATED 15B (COR15B) | Cold |
| AT5G42900 | COLD REGULATED GENE 27 (COR27) | Cold |
| AT2G42540 | COLD-REGULATED 15A (COR15A) | Cold |
| AT4G30650 | Low temperature and salt responsive protein | Low temperature and salt |
| AT5G20250 | DARK INDUCIBLE 10 (DIN10) | Light, sucrose |
| AT1G56220 | Dormancy/auxin associated | Dormancy/auxin |
| AT2G33830 | Dormancy/auxin associated | Dormancy/auxin |
| AT1G28330 | DORMANCY-ASSOCIATED PROTEIN-LIKE 1 | Dormancy |
| AT3G20810 | JUMONJI DOMAIN CONTAINING 5 (JMJD5) | Circadian |
| AT5G37260 | CIRCADIAN 1 (CIR1) | Circadian |
| AT4G35770 | SENESCENCE 1 (SEN1) | Phosphate starvation |
| AT3G17790 | PURPLE ACID PHOSPHATASE 17 (PAP17) | Phosphate starvation, and hydrogen peroxide |
| AT1G77120 | ALCOHOL DEHYDROGENASE 1 (ADH1) | Anaerobic response |
| AT2G39920 | HAD superfamily acid phosphatase | Cadmium ion |
| AT4G33020 | ZINC IRON PERMEASE (ZIP9) | Zinc ion |
| AT5G06870 | POLYGALACTURONASE INHIBITING PROTEIN 2 (PGIP2) | Fungal infection, Methyl jasmonate |
| AT2G05520 | GLYCINE-RICH PROTEIN 3 (GRP3) | ABA, salicylic acid, ethylene, desiccation |
Table 2.
Responsive genes up-regulated in the ask1 mutant transcriptome
| Gene ID | Gene name/description | Signaling pathways/responses |
| AT5G54490 | PINOID-BINDING PROTEIN 1 (PBP1) | Auxin |
| AT3G09870 | SAUR-like auxin-responsive protein | Auxin |
| AT5G61600 | ETHYLENE RESPONSE FACTOR 104 (ERF104) | Ethylene |
| AT4G34410 | REDOX RESPONSIVE TRANSCRIPTION FACTOR 1 | Ethylene |
| AT1G19180 | JASMONATE-ZIM-DOMAIN PROTEIN 1 (JAZ1) | Jasmonic acid |
| AT1G17380 | JASMONATE-ZIM-DOMAIN PROTEIN 5 (JAZ5) | Jasmonic acid |
| AT3G11480 | SABATH methyltransferase | Jasmonic acid, fungus, wounding |
| AT4G27280 | Calcium-binding EF-hand family protein | Karrikin |
| AT3G02380 | CONSTANS-LIKE 2 (COL2) | Light |
| AT3G22840 | EARLY LIGHT-INDUCABLE PROTEIN1 (ELIP1) | Light |
| AT4G14690 | EARLY LIGHT-INDUCIBLE PROTEIN 2 (ELIP2) | Light |
| AT3G17609 | HY5-HOMOLOG (HYH) | Light |
| AT3G59060 | PHYTOCHROME INTERACTING FACTOR 3-LIKE 6 | Light |
| AT5G59820 | RESPONSIVE TO HIGH LIGHT 41 (RHL41) | Light |
| AT2G30520 | ROOT PHOTOTROPISM 2 (RPT2) | Light |
| AT2G46830 | CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) | Circadian |
| AT1G01060 | LATE ELONGATED HYPOCOTYL (LHY) | Circadian |
| AT3G09600 | REVEILLE 8 (RVE8) | Circadian |
| AT3G12580 | HEAT SHOCK PROTEIN 70 (HSP70) | Heat |
| AT5G51440 | HSP20-like | Heat |
| AT2G31380 | SALT TOLERANCE HOMOLOGUE (STH) | Salt |
| AT1G27730 | SALT TOLERANCE ZINC FINGER (STZ) | Salt |
| AT3G55980 | SALT-INDUCIBLE ZINC FINGER 1 (SZF1) | Salt |
| AT2G33380 | RESPONSIVE TO DESICCATION 20 (RD20) | Desiccation |
| AT5G24660 | RESPONSE TO LOW SULFUR 2 (LSU2) | Sulfur deficiency |
| AT1G19640 | JASMONIC ACID CARBOXYL METHYLTRANSFERASE | Wounding, and methyljasmonate |
| AT5G64510 | TUNICAMYCIN INDUCED 1 (TIN1) | ER-stress, heat, light, hydrogen peroxide |
| AT5G57560 | TOUCH 4 (TCH4) | Mechanical stimulus |
| AT1G12110 | NITRATE TRANSPORTER 1.1 (NRT1.1) | Nitrate, water deprivation |
| AT1G61800 | GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2 (GPT2) | Glucose, sucrose, karrikin, nematode |
| AT2G46400 | WRKY DNA-BINDING PROTEIN 46 | Chitin |
| AT5G51190 | Ethylene response factor | Chitin, wounding |
| AT3G61190 | BON ASSOCIATION PROTEIN 1 (BAP1) | Chitin, cold, fungus, heat, jasmonic acid, salicylic acid, wounding |
| AT4G11280 | ACC SYNTHASE 6 (ACS6) | ABA, auxin, chitin, ethylene, jasmonic acid, oxidative stress, wounding |
| AT5G59310 | LIPID TRANSFER PROTEIN 4 (LTP4) | ABA, cold, salt, water deprivation |
| AT4G25100 | FE SUPEROXIDE DISMUTASE 1 (FSD1) | Cadmium ion, copper ion, oxidative stress |
| AT1G02930 | GLUTATHIONE S-TRANSFERASE 6 | Cadmium ion, oxidative stress, salt, water deprivation |
| AT3G21890 | B-box type zinc finger protein | UV-B, sucrose |
| AT2G37040 | PHE AMMONIA LYASE 1 (PAL1) | UV-B, karrikin, oxidative stress, wounding |
Nevertheless, several genes have been functionally characterized, including CIRCADIAN CLOCK ASSOCIATED 1 (CCA1), LATE ELONGATED HYPOCOTYL (LHY), JASMONATE-ZIM-DOMAIN PROTEIN 1 (JAZ1), and JAZ5. CCA1 and LHY encode Myb-like transcription factors that synergistically regulate circadian rhythm of Arabidopsis [39] and thus are important for coordinating internal physiological activities with external environmental cues. JAZ genes are induced by JA through a feedback loop involving JAZ proteins and the G-box-binding MYC2: JAZ proteins bind to and repress the activity of MYC2 in the absence of JA; upon perception of JA, JAZ proteins are degraded after ubiquitination by SCFCOI1 and the released MYC2 can activate transcription of downstream genes, including JAZ genes [40]. According to this feedback regulatory model, it is expected that the ask1 mutation would reduce SCF activities, allowing JAZ proteins to accumulate and repress MYC2 activity and thus reducing the JAZ transcript levels. However, we found that JAZ1 and JAZ5 transcript levels were unexpectedly higher in the ask1 mutant than in WT. This paradox suggests that an uncharacterized mechanism may be involved in modulating the JA signaling pathway. For example, ASK1-containing SCFs might facilitate the removal of a yet unidentified transcription activator that has the ability of inducing the expression of JAZ genes in the absence of JA; when ASK1 is mutated this transcriptional activator is stabilized, thereby inducing the expression of downstream genes including JAZ1 and JAZ5. Further studies are needed to uncover new aspects of these regulatory networks.
We then analyzed possible overrepresentation of cis-elements in the putative promoter regions of these up-/down-regulated genes in the ask1 transcriptome. The frequencies of 6-mer motifs within the 500 bp and 1000 bp putative promoter regions were determined using the Motif Analysis tool from The Arabidopsis Information Resource (TAIR) (Table 3). The G-box (CACGTG) is overrepresented in the putative promoter regions of up-/down-regulated genes, suggesting that corresponding genes might be regulated by G-box-binding transcription factors, which themselves or whose co-factors might be regulated by ASK1-E3 ligases, similar to the JAZ-MYC2 model. Some of these transcription factors or co-factors might be short-lived repressors; when they are stabilized in the absence of ASK1, their target genes are then down-regulated. Others may function as unstable activators, whose stabilization in the absence of ASK1 results in up-regulation of downstream genes. Alternatively, some transcription factors may have dual functions, both activation and repression, as is true for MYC2 [41, 42]. The fact that the genes whose promoters contain these cis-elements are altered in transcription in the ask1 mutant suggests that the protein levels of the corresponding transcription factors were changed in ask1. Another motif, GATAAG (I box), was enriched in the down-regulated genes in ask1. The I box was previously found to be enriched in promoters of light-regulated genes [43] and is required for Arabidopsis rbcS-1A expression [44]. Further experiments are required to test whether the putative cis-elements are functional and to identify cognate transcription factors that connect ASK1-E3 regulation with transcriptional changes.
Table 3.
Top five enriched cis-elements in the putative promoter regions of down-/up-regulated genes in the ask1 transcriptome
| Oligomer | # in query | # in genomic set | # of promoters in query with oligomer | # of promoters in genomic set with oligoMer | Binomial distribution p-value |
|---|---|---|---|---|---|
| 500 bp promoters of down-regulated genes in ask1 | |||||
| CACGTG | 30 | 7766 | 12/42 | 3253/33602 | 3.53E-04 |
| ACACGT | 23 | 7390 | 17/42 | 5609/33602 | 1.61E-04 |
| CGCAAA | 13 | 4569 | 13/42 | 3995/33602 | 6.16E-04 |
| GCCACG | 11 | 2914 | 10/42 | 2594/33602 | 8.46E-04 |
| GATAAG | 27 | 9179 | 19/42 | 7797/33602 | 9.09E-04 |
| 1000 bp promoters of down-regulated genes in ask1 | |||||
| CACGTG | 42 | 12404 | 16/42 | 5033/33602 | 1.57E-04 |
| AACTGT | 33 | 17175 | 28/42 | 13171/33602 | 2.04E-04 |
| GATAAG | 41 | 18464 | 28/42 | 13811/33602 | 4.93E-04 |
| ATTATG | 60 | 33774 | 35/42 | 20241/33602 | 8.37E-04 |
| CGTGTA | 22 | 7824 | 17/42 | 6717/33602 | 1.25E-03 |
| 500 bp promoters of up-regulated genes in ask1 | |||||
| CACGTG | 72 | 7766 | 24/74 | 3253/33602 | 4.95E-08 |
| ACACGT | 57 | 7390 | 30/74 | 5609/33602 | 7.19E-07 |
| ACGTGG | 30 | 5475 | 22/74 | 4404/33602 | 9.42E-05 |
| AAGTGG | 31 | 7504 | 27/74 | 6502/33602 | 2.63E-04 |
| ACACTC | 23 | 5788 | 23/74 | 5177/33602 | 3.37E-04 |
| 1000 bp promoters of up-regulated genes in ask1 | |||||
| CACGTG | 100 | 12404 | 30/74 | 5033/33602 | 6.83E-08 |
| ACACGT | 77 | 12599 | 38/74 | 9080/33602 | 5.14E-06 |
| ACGTGG | 46 | 9196 | 31/74 | 7047/33602 | 2.50E-05 |
| ATATTA | 177 | 65927 | 68/74 | 25116/33602 | 1.22E-04 |
| TGAGAC | 48 | 12304 | 36/74 | 10027/33602 | 2.98E-04 |
The enrichment of biotic/abiotic stress related genes in the up-/down-regulated genes in the transcriptome of ask1 mutant floral buds has several possible implications. First, the up-regulation of 39 biotic/abiotic stress related genes in ask1 floral buds (Table 2) suggests the expression of such genes might be tightly constrained to avoid unnecessary expression to ensure continuous and maximal allocation of resources to reproductive organs. In WT floral buds, the expression of these genes may be turned off due to degradation of positive transcriptional regulators by ASK1-E3-mediated ubiquitination, but stresses might block the degradation of such positive regulators. Second, the observation that 19 genes annotated as stress responsive were down-regulated in ask1 floral buds (Table 1) compared with WT floral buds suggests their involvement in normal flower development. Although these genes are annotated as responsive to biotic/abiotic signals, they could be triggered by endogenous signals such as programmed cell death (e.g., tapetum degeneration) and/or controlled dehydration during later stages of anther and pollen development [45]. However, the lack of cell-type-specific transcriptome information makes it difficult to determine the extent to which the transcriptome reprogramming for these developmentally-controlled processes resembles stress responses. In summary, ASK1-E3s might destabilize proteins that are involved in the complex regulations of signaling pathways in normal flower development or in response to external stimuli.
Proteomic analysis of Ler and ask1 floral buds
To probe the effect of ask1 on the floral proteome and to identify potential substrates of ASK1-E3s, we used a label-free proteomic method, MudPIT, to analyze floral bud proteomes of the ask1 mutant and Ler (Fig. 2). Total protein extracts of four Ler and five ask1 floral bud samples were digested in-solution with trypsin without pre-separation to maximize digestion of proteins with different properties (e.g., hydrophobicity and charges) and compartmentalization (cytosol, membrane, nucleus and organelles). MudPIT runs of the four Ler samples (Ler-1 ~ Ler_4) detected 2348, 2258, 1658, and 1400 proteins, respectively, with a false discovery rate (FDR) of <1 %, (Additional file 3). When the four datasets were merged, a total of 3220 non-redundant proteins were detected. MudPIT runs of the five ask1 samples (ask1_1 ~ ask1_5) detected 1780, 1441, 1959, 1007, and 363 proteins, respectively, with FDR < 1 %, (Additional file 3), for a total of 2916 non-redundant proteins. The ask1_5 run detected fewer proteins because the starting protein amount was about 20 % of the others to test whether a smaller amount of input protein extract could lead to different efficiency of protein detection. The test result did not show a huge difference in the detection efficiency when the amount of starting material was changed, i.e., the number of detected proteins is proportional to the starting protein amount. The 363 proteins detected in this test run were included in the total ask1 proteins, but excluded for comparison between individual runs with spectral counting normalization in the following sections.
Fig. 2.
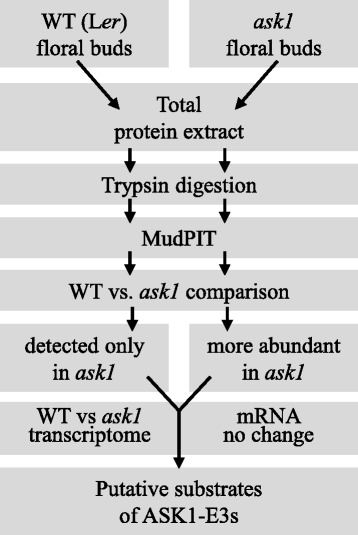
Workflow of the integrated proteomics and transcriptomics to identify putative ASK1-E3 substrates
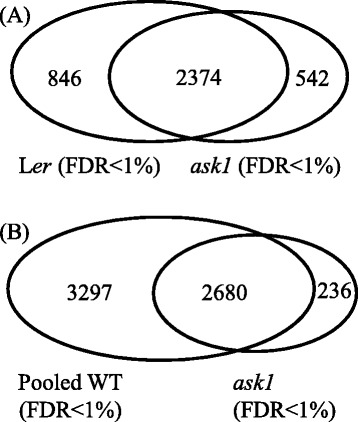
The stochastic sampling and detection sensitivity of MS-based proteomics results in the partial identification of the whole proteome in each experiment and partially overlapping datasets from different MS runs. Indeed, analysis of our Ler and ask1 samples produced partially overlapping sets of proteins (Fig. 3): 884 (27.5 %) of the 3220 total Ler proteins (FDR < 1 %) were detected by all four MS runs, 684 (17.2 %) proteins by three MS runs, 554 (21.2 %) proteins by two MS runs, and 1096 (34.1 %) proteins detected only once. Among the 2899 ask1 proteins (proteins unique to ask1_5 not included), 568 (19.6 %), 493 (17.0 %), 598 (20.6 %), and 1240 (42.8 %) proteins were detected by four, three, two and one of the four MS runs, respectively. Even the proteins detected once had high confidence (FDR < 1 %) resulting from very stringent MS detection and searching criteria and thus were regarded as detected.
Fig. 3.
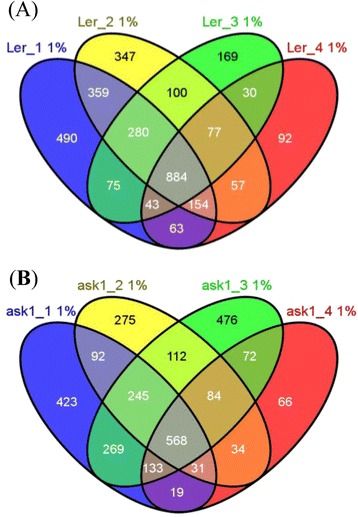
Independent proteomics samples detect partially overlapping sets of proteins. a Four Ler samples Ler_1 to Ler_4. b Four ask1 samples ask1_1 to ask1_4. The “1 %” after each sample name represents FDR < 1 %
Proteins of some cellular component categories (e.g., membrane proteins) that are usually considered to be difficult to be detected by MS without using detergents were well represented in our Ler and ask1 proteomes (Additional file 4), indicating that our method was able to detect proteins localized to the plasma membrane, organelle membrane, and nuclear envelope. One important improvement to minimize bias was that total protein extracts were digested by trypsin without separation into soluble or insoluble portions. Although insoluble proteins were often thought to be recalcitrant to enzyme digestion, extensive sonication, denaturing treatments, and prolonged tryptic digestion seem to have improved detection efficiency.
Detection of additional floral proteins compared with previous proteomics studies
Comparing our floral bud proteomes with previously published proteomics data, we detected many additional proteins (Fig. 4). WT floral bud proteins from two previous studies [46, 47] were combined into one dataset (named “previous WT”) containing 5461 non-redundant proteins (FDR < 1 %). Compared with the previous WT, additional 516 proteins were only detected in our Ler proteome dataset containing 3220 non-redundant proteins and additional 752 proteins were detected in our Ler + ask1 combined proteome dataset containing 3762 non-redundant proteins. The identification of these additional proteins indicates that proteomics detection has not been saturated. The development of more advanced MS technologies with the capacity of overcoming the huge dynamic range of proteins in biological samples is required to further push the proteomic identification closer to saturation. Until then, we have to keep in mind that there is still much room for improvement to our proteomics data and we need to be cautious when drawing conclusions from the current data.
Fig. 4.
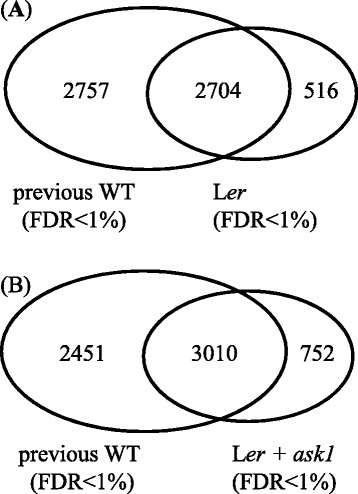
Detection of additional floral bud proteins than previous studies. a Comparison of WT floral bud proteins detected in previous studies (previous WT) and Ler from this study (FDR < 1 %). b Comparison of floral bud proteins detected in previous WT and total proteins detected in our Ler and ask1 samples (FDR < 1 %)
Proteins detected only in ask1 or higher in ask1
Comparison of our ask1 and Ler proteomes revealed that 542 proteins were found only in ask1 (Fig. 5a). These proteins have accumulated in ask1 but not WT, suggesting that they might be putative ASK1-E3 substrates. However, it remains possible that the accumulation of these proteins might be an indirect effect of the ask1 mutation. Since the limited number of MudPIT runs was not enough to saturate the proteome, we cannot rule out the possibility that some proteins detected only in ask1 could have been detected also in Ler if more MS runs had been done. In order to narrow down putative ASK1-E3 substrates, we combined our Ler floral bud proteome data with the previous WT data [46, 47] yielding a larger WT floral bud proteome dataset consisting of 5977 proteins (Pooled WT). This Pooled WT dataset represents the most comprehensive floral bud proteome currently available obtained by MS methods. The total contribution of our WT (Ler) floral bud proteome dataset to this Pooled WT proteome is about 53.9 % (3220/5977). By comparing ask1 and the Pooled WT proteomes, we found that 236 proteins were only detected in ask1 (ask1-only proteins); these are thus more likely to be ASK1-E3 substrates (Fig. 5b and Additional file 5).
Fig. 5.
Proteins only detected in ask1 but not in WT proteomes. a Comparison of Ler and ask1 proteomes from this study. b Comparison of the ask1 proteome with the pooled WT, a combined WT proteome from previous studies and this study
We examined RNA levels of these ask1-only proteins from microarray data to determine whether increased transcription contributed to the accumulation of these proteins. The RNA levels of most of these genes were not significantly different between ask1 and Ler (do not meet the criteria of two-fold cutoff and Student’s t-test p-value < 0.05), except that 11 genes lacked probes on the ATH microarray chip (Additional file 5). Therefore, at least 225 proteins seem to be controlled by ASK1-E3s at the protein level.
Because we used a label-free proteomics method, protein abundance cannot be directly compared between samples. One of the relative quantification approaches, spectral counting, has been demonstrated to show higher reproducibility, larger dynamic range, and stronger linear correlation with relative protein abundance than sequence coverage, peptide number, and ion chromatographic methods [46, 48–51]. Therefore, proteomics datasets of this study were normalized using the spectral counting method as reported [46] and the average values were compared between Ler and ask1 to find 322 proteins with higher abundance (1.5-fold cutoff) in ask1 (ask1-higher; Additional file 6). The previous WT data were not included because they were generated by different proteomics methods and difficult to compare quantitatively with our data. We extracted the RNA expression values from microarray data for these ask1-higher proteins to determine whether their elevated protein levels were due to increased transcript levels (Additional file 6). Only the RNA level of AT2G33380 was 2.2-fold higher in ask1 than Ler. The RNA levels of other genes were not significantly different between ask1 and Ler, except that 12 genes had no probes on the microarray chip, suggesting that at least 309 of ask1-higher proteins are probably regulated at the protein level.
ASK1 regulates abundance of regulatory proteins acting at multiple levels
GO categorization of the ask1-only and ask1-higher proteins shows that some molecular functional categories are overrepresented (Fisher test p-value < 0.05) (Fig. 6). Since regulatory proteins are often affected by the UPS, certain categories are of particular interest, including transcriptional regulators, kinases, and peptidases/proteases. Interestingly, many ribosomal proteins were also found to accumulate in ask1 indicating a possible role of the UPS in translational regulation or extraribosomal functions of ribosomal proteins.
Fig. 6.
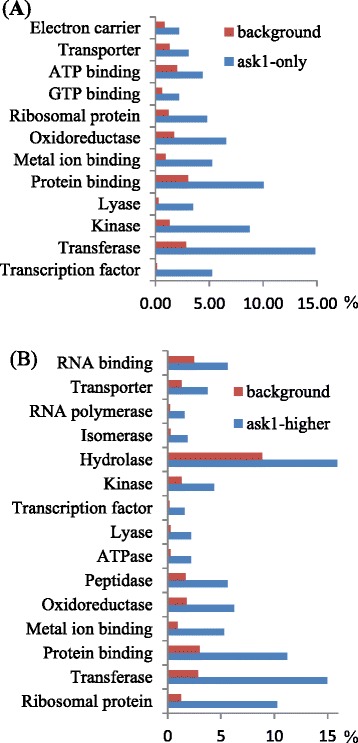
Overrepresented GO categories in proteins accumulated in the ask1 proteome. a Overrepresented GO categories in ask1-only proteins. b Overrepresented GO categories in ask1-higher proteins. Each “Background” percentage (%) represents the proportion of all annotated proteins of each GO category in the total proteins in the Arabidopsis thaliana genome (TAIR10 version). Each “ask1-only” or “ask1-higher” percentage (%) represents the proportion of proteins that were only detected in ask1 or with higher levels in ask1 of each GO category in the total proteins detected in the ask1 samples
ASK1-E3s affects the levels of transcriptional regulators
The ask1 mutation affected 19 transcription factors and chromatin remodelers, including some with functional information (Table 4). One of them was JAZ9, which might function as a repressor of MYC transcription factors in the JA signaling pathway [52], consistent with the SCFCOI1-dependent ubiquitination of JAZ proteins and subsequent degradation upon JA perception [10–12]. Another protein detected in ask1 was the basic helix-loop-helix (bHLH) transcription factor MYC3, which interacts with JAZ proteins and functions with MYC2 and MYC4 to activate JA responses [53, 54]. MYC3 binds to G-boxes [53], possibly regulating promoters with G-boxes, including some of the genes that exhibited altered expression in ask1, as supported by the enrichment of G-boxes in the promoters of the genes exhibiting altered expression in ask1 as described in the first section (Table 3). The accumulation of JAZ9 and MYC3 in the ask1 proteome raises the possibility that protein stability of both JAZs and MYCs is regulated by the UPS, adding another layer of regulation in the JA signaling pathway. In addition, eight zinc finger transcription factors and several transcription factors of other types (Table 4) also exhibited elevated protein levels in ask1, indicating that destabilization of these transcription factors by ASK1-E3s might be important for floral development.
Table 4.
Transcriptional regulators enriched in ask1-only or ask1-higher proteins
| Transcription factors | |
|---|---|
| Enriched in ask1-only proteins | |
| AT5G46760 | MYC3, basic helix-loop-helix (bHLH) transcription factor |
| AT1G70700 | JASMONATE-ZIM-DOMAIN PROTEIN 9 (JAZ9) |
| AT1G32360 | Zinc finger (CCCH-type) family protein |
| AT2G24500 | Zinc finger (C2H2-type) protein FZF |
| AT5G60850 | Zinc finger OBF BINDING PROTEIN 4 (OBP4) |
| AT3G61850 | Zinc finger DOF AFFECTING GERMINATION 1 (DAG1) |
| AT4G36620 | Zinc finger GATA TRANSCRIPTION FACTOR 19 (GATA19) |
| AT2G02540 | ZINC FINGER HOMEODOMAIN 3 (ZHD3) |
| AT5G15210 | ZINC FINGER HOMEODOMAIN 8 (ZHD8) |
| AT1G54830 | NUCLEAR FACTOR Y, SUBUNIT C3 (NF-YC3) |
| AT1G58100 | TCP DOMAIN PROTEIN 8 (TCP8) |
| AT3G10490 | NAC DOMAIN CONTAINING PROTEIN 52 (ANAC052) |
| AT4G02020 | Polycomb group protein SWINGER (SWN) |
| Enriched in ask1-higher proteins | |
| AT1G49480 | RELATED TO VERNALIZATION1 1 (RTV1) |
| AT1G76880 | Duplicated homeodomain-like superfamily protein |
| AT3G28920 | ZINC FINGER HOMEODOMAIN 9 (ZHD9) |
| AT3G48430 | RELATIVE OF EARLY FLOWERING 6 (REF6); JUMONJI DOMAIN-CONTAINING PROTEIN 12 (JMJ12) |
| AT4G35570 | HIGH MOBILITY GROUP B5 (HMGB5) |
| AT4G38130 | HISTONE DEACETYLASE 1 (HD1);HISTONE DEACETYLASE19 |
The ask1 mutation also caused an increased level of a Polycomb group protein, SWINGER (SWN) (Table 4), which interacts with other Polycomb group proteins to repress expression of FLOWERING LOCUS C (FLC) and controls the initiation of endosperm development [55–57]. The elevated SWN protein level is expected to repress FLC expression. Although our ask1 and Ler floral bud transcriptome data did not show significant difference of FLC expression, our unpublished anther transcriptome data showed that averaged microarray values of FLC are 68 and 251 (raw microarray data normalized by R package RMA) in ask1 and Ler anther transcriptomes, respectively, i.e., the FLC level is lower in ask1 anthers than that in Ler anthers (regular Student’s t-test p-value = 0.04). The down-regulation of FLC in the ask1 anther transcriptome suggests that degradation of the SWN protein in developing anthers normally derepress its target genes including FLC. Another affected chromatin remodeling protein is RELATIVE OF EARLY FLOWERING 6 (REF6), which is a histone H3 lysine 27 demethylase [58] and positively regulates flowering and brassinosteroid signaling [59]. Our results suggest that ASK1-E3s may modulate gene transcription by facilitating the degradation of both sequence-specific DNA binding transcription factors and chromatin remodelers.
ASK1-E3 affects ribosomal proteins
Interestingly, many ribosomal proteins were identified only or with higher levels in ask1 (Table 5), indicating that ASK1-E3s may also have a role in translational regulation. Several ribosomal proteins have been genetically studied: NUCLEAR FUSION DEFECTIVE 3 is required for polar nuclei fusion during female gametophyte development [60]; PIGGYBACK1 influences leaf vascular patterning [61]; OLIGOCELLULA7 is involved in ribosome biogenesis and organ size control [62]; and POINTED FIRST LEAF2 plays a role in early leaf development [63]. Ribosomal proteins might be regulated by ASK1-E3s either for ribosome turnover, or for extraribosomal regulatory purposes. It was suggested that ribosomal proteins can be ubiquitinated for selective degradation of ribosomes by autophagy [64, 65]. So, the accumulation of ribosomal proteins in ask1 might result from the failure of ubiquitination by ASK1-E3s and subsequent autophagy. A previous study showed that F-BOX PROTEIN 7 (FBP7), which interacts with ASK1, is required for efficient translation under temperature stress conditions, but the substrate of this F-box protein is not identified [66]. It is possible that F-box proteins, such as FBP7, may regulate translation by ubiquitination of specific ribosomal proteins. Our results suggest that ASK1-E3s might be extensively involved in the regulation of translation.
Table 5.
Ribosomal proteins enriched in ask1-only and ask1-higher proteins
| Ribosomal proteins | |
|---|---|
| Enriched in ask1-only proteins | |
| AT5G02610 | Ribosomal L29 |
| AT1G26880 | Ribosomal protein L34e |
| AT4G25890 | 60S acidic ribosomal protein family |
| AT5G67510 | Translation protein SH3-like family protein, large ribosomal subunit |
| AT5G39850 | Ribosomal protein S4 |
| AT5G43640 | Ribosomal protein S19 |
| AT4G34555 | Ribosomal protein S25 |
| AT3G61110 | Ribosomal protein S27 |
| AT1G31817 | Mitochondrial 50S ribosomal L21, NUCLEAR FUSION DEFECTIVE 3 |
| AT2G38140 | Plastid-specific ribosomal protein 4 (PSRP4) |
| Enriched in ask1-only proteins | |
| AT1G07830 | Ribosomal protein L29 family protein |
| AT1G15930 | Ribosomal protein L7Ae/L30e/S12e/Gadd45 family protein |
| AT1G26910 | Ribosomal protein L16p/L10e family protein |
| AT1G27400 | Ribosomal protein L22p/L17e family protein |
| AT1G41880 | Ribosomal protein L35Ae family protein |
| AT1G67430 | Ribosomal protein L22p/L17e family protein |
| AT1G69620 | RIBOSOMAL PROTEIN L34 (RPL34) |
| AT1G78630 | Ribosomal protein L13 family protein, EMBRYO DEFECTIVE 1473 (EMB1473) |
| AT2G27530 | Ribosomal protein L10aP, PIGGYBACK1 (PGY1) |
| AT3G07110 | Ribosomal protein L13 family protein |
| AT3G54210 | Ribosomal protein L17 family protein |
| AT3G59540 | Ribosomal L38e protein family |
| AT5G23900 | Ribosomal protein L13e family protein |
| AT5G27850 | Ribosomal protein L18e/L15 superfamily protein |
| AT5G39740 | RIBOSOMAL PROTEIN L5B (RPL5B); OLIGOCELLULA 7 (OLI7) |
| AT2G28830 | PLANT U-BOX 12 (PUB12) with ribosomal protein L10e/L16 domain |
| AT1G74970 | RIBOSOMAL PROTEIN S9 (RPS9) |
| AT2G40510 | Ribosomal protein S26e family protein |
| AT2G40590 | Ribosomal protein S26e family protein |
| AT3G04920 | Ribosomal protein S24e family protein |
| AT3G13120 | Ribosomal protein S10p/S20e family protein |
| AT3G56340 | Ribosomal protein S26e family protein |
| AT4G00100 | RIBOSOMAL PROTEIN S13A (RPS13A); POINTED FIRST LEAF 2 (PFL2) |
| AT4G33865 | Ribosomal protein S14p/S29e family protein |
| AT4G39200 | Ribosomal protein S14p/S29e family protein |
| AT5G04800 | Ribosomal S17 family protein |
| AT5G15200 | Ribosomal protein S4 |
| AT5G28060 | Ribosomal protein S24e family protein |
| AT5G52650 | RNA binding Plectin/S10 domain-containing protein |
| AT3G16080 | Zinc-binding ribosomal protein family protein |
| ATCG00800 | Chloroplast ribosomal protein S3, RESISTANCE TO PSEUDOMONAS SYRINGAE 3 |
| ATCG01240 | 30S chloroplast ribosomal protein S7, RIBOSOMAL PROTEIN S7 (RPS7.2) |
| AT1G07320 | Plastid RIBOSOMAL PROTEIN L4 (RPL4); EMBRYO DEFECTIVE 2784 (EMB2784) |
Alternatively, ribosomal proteins may have extraribosomal functions, which are exemplified by the previous findings that several ribosomal proteins can block the ubiquitination of the tumor suppressor p53 upon ribosomal stress [67]. Therefore, the ribosomal proteins accumulated in ask1 might function as regulatory proteins which themselves may be regulated by ubiquitin-mediated proteolysis. Further studies of genetic and molecular interactions between ribosomal proteins and E3 ubiquitin ligases are required for elucidating the role of UPS in translational regulation.
It is difficult to rule out the possibility that the accumulation of these ribosomal proteins might be a side effect of the ask1 mutation. Since the ask1 is a stable mutant, it is possible that long-term and large-scale disturbance of protein degradation might impose an intracellular stress which in turn affects the ribosome biosynthesis, activity or turnover. Therefore, more research is needed to elucidate the link between protein degradation and ribosomal protein functions.
ASK1-E3s affects regulators of protein activities
A number of kinases accumulated in the ask1 proteome (Table 6), including some that are important for plant responses to various stimuli. Among them, CALCIUM-DEPENDENT PROTEIN KINASE 6 (CPK6) is a positive regulator of salt/drought stress tolerance [68], methyl jasmonate signaling in guard cells [69], and ABA regulation of guard cell ion channels [70]. In addition, LYSM DOMAIN RECEPTOR-LIKE KINASE 1 (LYSM RLK1) is involved in chitin-mediated plant innate immunity [71, 72], and MAP KINASE KINASE 2 (MKK2) regulates cold and salt stress signaling and innate immunity [73–75]. Our results indicate that ASK1-E3s normally destabilize these kinases during normal flower development, possibly to suppress biotic/abiotic stress responses in the absence of stimuli.
Table 6.
Kinases enriched in ask1-only and ask1-higher proteins
| Kinases | |
|---|---|
| Enriched in ask1-only proteins | |
| AT2G17290 | CALCIUM-DEPENDENT PROTEIN KINASE 6 (CPK6) |
| AT4G21940 | CALCIUM-DEPENDENT PROTEIN KINASE 15 (CPK15) |
| AT5G45190 | Cyclin T partner CYCT1;5 |
| AT3G48750 | Cyclin-dependent kinase CELL DIVISION CONTROL 2 (CDC2) |
| AT4G29810 | MAP KINASE KINASE 2 (MKK2) |
| AT3G29160 | SNF1-RELATED PROTEIN KINASE 1.2 (SnRK1.2) |
| AT5G63650 | SNF1-RELATED PROTEIN KINASE 2.5 (SNRK2.5) |
| AT4G26100 | CASEIN KINASE 1 (CK1) |
| AT4G35780 | ACT-like protein tyrosine kinase |
| AT5G49470 | PAS domain-containing protein tyrosine kinase |
| AT5G11020 | Protein kinase superfamily protein |
| AT5G24010 | Protein kinase superfamily protein |
| AT5G57610 | Protein kinase superfamily protein |
| AT5G43020 | Leucine-rich repeat protein kinase family protein |
| AT3G21630 | LYSM DOMAIN RECEPTOR-LIKE KINASE 1 (LYSM RLK1) |
| AT3G14350 | STRUBBELIG-RECEPTOR FAMILY 7 (SRF7) |
| AT4G33240 | 1-phosphatidylinositol-3-phosphate (PtdIns3P) 5-kinase |
| Enriched in ask1-higher proteins | |
| AT1G31910 | GHMP kinase family protein |
| AT2G18170 | MAP KINASE 7 (ATMPK7) |
| AT2G27970 | CDK-SUBUNIT 2 (CKS2) |
| AT3G02880 | Leucine-rich repeat protein kinase family protein |
| AT4G21210 | PPDK REGULATORY PROTEIN (RP1) |
| AT4G35230 | BR-SIGNALING KINASE 1 (BSK1) |
Also affected are two cell cycle regulators, cyclin-dependent kinase CELL DIVISION CONTROL 2 (CDC2) and a Cyclin T protein, CYCT1;5. CDC2 is required for male gametogenesis [76]. CYCT1;5 is a subunit of cyclin-dependent kinase C complexes involved in cauliflower mosaic virus infection, plant growth and development [77]. The accumulation of cell cycle regulators in ask1 may affect mitosis and/or meiosis, as suggested by the findings that ask1 mutant plants have reduced cell numbers and defective male meiosis [23, 28–31]. Timely removal of cell cycle regulators is likely to be an important part of ASK1 function in regulating plant development.
We also found that SNF1-RELATED PROTEIN KINASE 1.2 (SnRK1.2)/SNF1 KINASE HOMOLOG 11 (KIN11), which was reported to be degraded during phosphate starvation [78], accumulated in the ask1 proteome. SnRK1.2/KIN11 was also shown to interact with ASK1 [79]. These findings imply that ASK1 might directly recruit SnRK1.2/KIN11 to Cul1 without an F-box protein, resulting in ubiquitination and degradation of SnRK1.2/KIN11. Alternatively, an F-box protein might also interact with ASK1 and SnRK1.2/KIN11, forming an SCF complex for ubiquitinating SnRK1.2/KIN11.
Other kinases are largely unknown, but might have important functions in signal perception and transduction. For example, AT5G43020 and AT3G14350 contain transmembrane domains and could be membrane receptor kinases. In summary, the increased levels of these kinases suggest that ASK1-E3s negatively regulate levels of these protein kinases to control cell cycle, plant immunity, hormone signaling, and other processes.
ASK1-E3 affects regulators of protein stability
The peptidase category is enriched in the ask1-higher proteins (Table 7), indicating that ASK1-E3s may regulate degradation of peptidases, which in turn affect protein processing or turnover. Four peptidases (AT1G53850, AT5G66140, AT1G77440, and AT3G60820) are isoforms of 20S proteasome alpha/beta subunits, indicating that the proteasome core complex may also be regulated by UPS. Two ubiquitin-specific proteases UBIQUITIN-SPECIFIC PROTEASE5 (UBP5) and UBP6 were also detected in ask1-higher proteins, suggesting that deubiquitinases, which antagonize protein ubiquitination, might also be regulated by the UPS. The BRI1 SUPPRESSOR 1 (BRS1), a secreted serine carboxypeptidase, is involved in brassinosteroid signaling possibly by processing some proteins [80]. Other peptidases are largely unknown except information from expression and homology. Peptidases/proteases may normally be subject to negative regulation by ASK1-E3s, thus coupling peptidase-mediated protein processing or degradation with the UPS.
Table 7.
Peptidases enriched in ask1-higher proteins
| Peptidases | |
|---|---|
| AT1G01300 | Eukaryotic aspartyl protease family protein |
| AT1G79720 | Eukaryotic aspartyl protease family protein |
| AT1G02305 | Cysteine proteinases superfamily protein |
| AT3G62940 | Cysteine proteinases superfamily protein |
| AT5G43060 | Granulin repeat cysteine protease family protein, ESPONSIVE TO DEHYDRATION 21B (RD21B) |
| AT4G30610 | SERINE CARBOXYPEPTIDASE 24 PRECURSOR (SCPL24); BRI1 SUPPRESSOR 1 (BRS1) |
| AT4G30810 | SERINE CARBOXYPEPTIDASE-LIKE 29 (SCPL29) |
| AT1G13270 | METHIONINE AMINOPEPTIDASE 1B (MAP1C) |
| AT3G14067 | Subtilase family protein |
| AT5G04710 | Zn-dependent exopeptidases superfamily protein |
| AT5G05740 | S2P-like putative metalloprotease, ETHYLENE-DEPENDENT GRAVITROPISM-DEFICIENT AND YELLOW-GREEN-LIKE 2 (EGY2) |
| AT2G40930 | UBIQUITIN-SPECIFIC PROTEASE 5 (UBP5) |
| AT1G51710 | UBIQUITIN-SPECIFIC PROTEASE 6 (UBP6) |
| AT1G53850 | 20S PROTEASOME ALPHA SUBUNIT E1 (PAE1) |
| AT5G66140 | 20S PROTEASOME ALPHA SUBUNIT D2 (PAD2) |
| AT1G77440 | 20S PROTEASOME BETA SUBUNIT C2 (PBC2) |
| AT3G60820 | 20S PROTEASOME BETA SUBUNIT F1 (PBF1) |
Possible ways that ASK1 regulates gene expression
By integrative analysis of transcriptome and proteome data, we found that ASK1-E3s might regulate gene expression at multiple steps, ranging from transcriptional, translational, to post-translational regulations. ASK1-E3s may destabilize transcription repressors or activators to derepress or inactivate gene transcription, respectively (Fig. 7a). In the absence of ASK1, the accumulation of these transcriptional repressors or activators results in down-regulation or up-regulation of gene transcription, respectively. However, we cannot rule out the possibility that the altered transcriptome and proteome might be indirect consequences of the ask1 mutation.
Fig. 7.
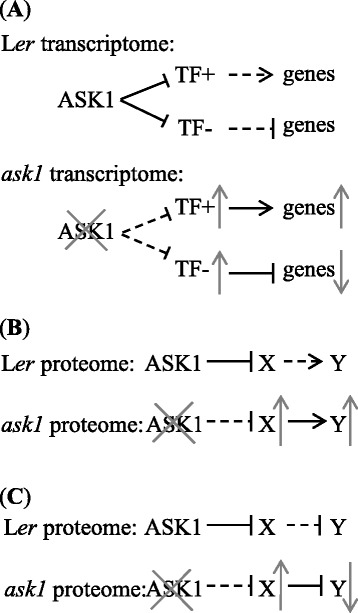
Possible mechanisms of transcriptome and proteome regulations by ASK1-E3s. a ASK1-E3s may regulate gene transcription by destabilizing transcription factors. The transcription factors are stabilized in ask1 mutant and activate or repress downstream gene transcription. TF+, transcriptional activators; TF-, transcriptional repressors. b ASK1-E3s might destabilize substrate X, which positively regulates the abundance of target proteins Y. In the ask1 mutant proteome, ASK1-E3 substrate X and their target protein Y accumulate. c ASK1-E3s might destabilize substrate X, which negatively regulates the abundance of target protein Y. In the ask1 mutant proteome, ASK1-E3 substrate X accumulates but target protein Y decreases. Bars, negative regulation; horizontal arrows, positive regulation; dashed gray bars and horizontal arrows, missing regulations; upward arrows, increase in abundance; downward arrows, decrease in abundance
The proteins accumulated in ask1 might be direct substrates of ASK1-E3s, or stabilized by ASK1-E3 substrates (Fig. 7b). For example, ubiquitin-specific proteases UBP5 and UBP6, which accumulate in the ask1 proteome (Table 7), might be substrates of ASK1-E3s; UBP5 and UBP6 could deubiquitinate and prevent degradation of ubiquitinated proteins, whose protein levels are then increased in ask1. An example in human is the herpesvirus-associated ubiquitin-specific protease (HAUSP), which stabilizes a tumor suppressor p53 by deubiquitination [81]. Ribosomal proteins may share a similar mechanism: accumulation of ribosomal proteins in ask1 may increase protein synthesis; alternatively, if ribosomal proteins have extraribosomal regulatory functions, they may stabilize some proteins in a similar way as those stabilizing p53 in human [67].
In another possible scenario, ASK1-E3s may destabilize some proteolytic enzymes (e.g., E3 ubiquitin ligases or peptidases), which can degrade other proteins (Fig. 7c), forming a double negative regulation cascade. The accumulation of such proteolytic enzymes in ask1 may cause reduced levels of their proteolytic substrates. Proteasome subunits and peptidases that accumulate in ask1 may be involved in degradation of their substrate proteins, which could be detected with lower levels in ask1 proteome (Additional file 7). However, it remains difficult to identify these proteolytic substrates due to lack of functional information of the proteolytic enzymes.
There are probably more proteins regulated by ASK1-E3s than those identified in this study. For example, the substrates of the well-studied F-box proteins, TIR1 and COI1, were not detected except JAZ9 (Table 4). One possible reason is that, due to technical limitations, MS might not have uncovered proteins with low-level and/or spatiotemporally restricted expression (e.g, the putative UFO substrate, LEAFY, which is mainly expressed in the inflorescence meristem and floral meristem [20–22]), and when the substrates of F-box proteins are subject to degradation. Another important reason is the functional redundancies among the 21 ASK family members in Arabidopsis. Since the ASK1 gene is expressed throughout the plant with higher levels in growing organs, its mutation is expected to cause more defects in many plant organs. However, the actual defects are milder than the expected, probably due to the redundancy among the ASK family members. The ASK2 gene is the most closely related gene to ASK1. The single mutant of ask2 is similar to WT plants, but the ask1 ask2 double mutant has severe defects in embryo development and is lethal soon after seed germination [82]. This suggests that the redundancy of ASK1 with ASK2, and possibly other ASK proteins, probably has masked some aspects of the ASK1 function. In other words, some ASK1-E3 substrates might also be ubiquitinated by SCFs containing other ASK proteins (e.g., ASK2-E3s), and thus would be unable to accumulate in the ask1 proteome. This might also explain why most of the well-known substrates of F-box proteins (TIR1 and COI1) were not identified in our MS data.
For example, one new aspect of ASK1 functions was revealed by our reanalysis of female fertility of the ask1 mutant, which was reported to be female fertile in previous studies [23, 28–31]. We loaded excess WT (Ler) pollen onto stigmas of the ask1 mutant, the dysfunctional tapetum 1 (dyt1) mutant (as a control with male sterility and female fertility) [83], and Ler (as a self-pollination control), and finally we counted mature seeds from every silique (Additional file 8). The result clearly showed that the pollinated ask1 pistils yielded significantly fewer seeds (16.0 seeds/silique on average) than Ler (52.5 seeds/silique on average) and dyt1 (52.0 seeds/silique on average) (Student’s t-test p-value < 0.01); while the pollinated dyt1 pistils yielded similar numbers of seeds as Ler (Student’s t-test p-value > 0.05). This finding suggests a previously unrecognized role of ASK1 in female reproductive development in Arabidopsis. Studying the masked aspects of ASK1 functions will need tissue-specific silencing of multiple ASK family members, or tissue-specific ASK1 complementation within the ask1 ask2 double mutant or higher order mutants. In addition, characterization of the ubiquitinated proteome may identify potential substrates of E3 ubiquitin ligases and ubiquitination sites within each protein, providing additional clues about ASK1 function in related processes.
Conclusions
Protein degradation is an integral part of various biological processes. The UPS is of particular interest since it selectively degrades proteins, including many key regulators of many cellular pathways [1–3]. However, searching for specific substrates of E3 ubiquitin ligases has been difficult probably due to rapid degradation of substrate proteins once they have been polyubiquitinated by E3 ubiquitin ligases, relatively weak interaction between E3s and substrates, narrow spatiotemporal window where the E3-substate interaction occurs, and others.
In this study, we have searched for potential E3 substrates by using an Arabidopsis mutant that lacks the functional ASK1 gene encoding a key component of SCF-type E3 ubiquitin ligases and that has developmental defects, particularly in floral organs including petals and anthers [23, 28–31]. We employed a MS-based method, MudPIT, to explore floral bud proteomes and detected 2916 and 3220 proteins in ask1 and WT proteomes, respectively. By comparing the ask1 proteome with a pooled WT floral bud proteome (our WT floral bud proteome combined with two published WT floral bud proteomes), we found 236 proteins that are unique to the ask1 proteome and 322 proteins with higher levels in the ask1 proteome. The accumulation of these proteins in the absence of ASK1-E3s suggests that they may be targeted by ASK1-E3s for degradation in WT. Our transcriptomics analysis of ask1 and WT floral buds showed that the transcripts of genes encoding the proteins accumulated in the ask1 proteome are not significantly affected by the ask1 mutation, suggesting that these proteins are regulated at the protein level and thus are more likely to be candidate substrates of ASK1-E3s. Functional categorization revealed that many of the potential substrates of ASK1-E3s are involved in regulation of transcription, translation, protein phosphorylation, and protein degradation. This indicates a multifaceted role of ASK1 in regulating plant development. Much more work is required to validate these candidate E3 substrates and to investigate their specific molecular functions.
Methods
Plant materials and growth conditions
The Arabidopsis thaliana ecotype Landsberg erecta (Ler) and ask1 mutant within the Ler background [23] were used. Plants were grown on soil (Metro-Mix 360, Sun Gro Horticulture, Bellevue, WA) in a growth room with a temperature of 23 °C and long day conditions (16 h light and 8 h dark). The ask1 mutant plants were selected from the progeny of ASK1/ask1 heterozygous plants by their abnormal phenotypes including reduced plant size compared with WT plants of the same age, reduced number and/or reduced size of petals, sterile anthers, short filaments, and short siliques. Clusters of unopened floral buds from the primary inflorescences (from inflorescence meristem to the biggest unopened bud) of the ask1 mutant and Ler were collected from plants with about 5 open flowers.
Microarray analysis
Ler and ask1 floral bud total RNA was extracted using the NucleoSpin® RNA Plant kit (MACHEREY-NAGEL, Bethlehem, PA). RNA quality analysis was performed on the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA), controlled by the Agilent 2100 Expert software, using the Plant RNA Nano assay following the RNA 6000 Nano kit protocol. Microarray was performed using the GeneChip Arabidopsis ATH1 Genome Array (Affymetrix, Santa Clara, CA) in the Penn State Genomics Core Facility – University Park, PA. Three biological replicates of ask1 and four biological replicates of Ler were performed (Additional file 9).
Data analysis was conducted as previously described with some modifications [84]. Microarray datasets (.CEL files) were normalized by R package RMA and exported as Excel files. Microarray signal values were averaged from biological replicates of each genotype and compared between ask1 and Ler to find differentially expressed genes which show at least 2-fold differences in RNA levels and p-value < 0.05 (regular Student’s t-test). GO categorization was conducted using the Singular Enrichment Analysis (SEA) from agriGO [38]. The Affymetrix ATH1 Genome Array (GPL198) was selected as the background reference which contains 22479 annotated genes. The statistical test was set to Fisher and significance level set to 0.05.
Protein extraction with trichloroacetic acid/acetone method
The protein extraction method was modified from a previous study [85]. Floral buds were ground thoroughly in liquid nitrogen with mortars and pestles and the powder was suspended in -20 °C Acetone with 10 % w/v Trichloroacetic Acid (TCA) and 0.07 % (v/v) β-Mercaptoethanol (1 ml for 0.3 g of tissue powder). After being incubated for 2 h (or overnight) at -20 °C, the protein suspension was centrifuged for 15-20 min at 14,000 rpm. The supernatant was removed and the protein pellet was resuspended and washed with 1 ml of -20 °C Acetone containing 0.07 % (v/v) β-Mercaptoethanol followed by centrifugation for 15-20 min at 14,000 rpm. This washing step was repeated until the pellet was almost white. The protein pellet was vacuum dried for 5-10 min and stored at -20 °C or immediately used for trypsin digestion.
In-solution trypsin digestion of protein extract
About 20-30 mg of crude protein extract from the TCA/Acetone method was resuspended in 1 ml of rehydration buffer [100 mM NH4HCO3, 10 mM Dithiothreitol (DTT), 10 % (v/v) Acetonitrile] and sonicated for 5 times, 20 s each time, duty cycle 40 %, power 3 using a Branson Sonifier S-450A (Branson Ultrasonics, Danbury, CT). Proteins were denatured at 60 °C for 45–60 min and alkylated by 50 mM Iodoacetamide at 37 °C for 30 min in dark. 40 μl of 1 M DTT was added to quench the alkylation reaction. Alkylated proteins were digested by 20 μg of Trypsin Gold, Mass Spectrometry Grade (Promega, Madison, WI) for 16-18 h at 37 °C with moderate shaking. The remaining indigestible debris was removed by centrifugation at 12,000 rpm for 10 min. The supernatant was transferred to a new 1.5 ml tube and centrifuged again to remove residual debris. The supernatant was transferred to a new 1.5 ml tube and was adjust to pH 3.0 with glacial acetic acid. The peptide solution was vacuum dried completely to evaporate off NH4HCO3 and acetonitrile. The pellet was resuspended in 200 μl of H2O and vacuum dried. Three repeats of resuspension and drying were performed in total. Finally the peptides were analyzed in the Proteomics and Mass Spec Core Facility, College of Medicine, Pennsylvania State University, Hershey, PA.
Mass spectrometry analysis/MudPIT
Trypsin-digested peptide samples were analyzed by MudPIT according to the 2D LC-MALDI separation and analysis procedures published previously using a 4800 proteomic analyzer MALDI TOF/TOF tandem system (Applied Biosysems) [86] except several modifications. The ProteinPilot software version 4.2 was used to perform protein identification by searching MS spectra against the protein database which included the Arabidopsis thaliana protein list TAIR10_pep_20101214, 156 common human and lab contaminants (ABSciex_ContaminantDB_20070711), and a reverse “decoy” version of the protein database itself (concatenated Reverse Decoy Database). Proteins with local FDR < 1 % were accepted as detected (Additional files 10, 11, 12, 13, 14, 15, 16, 17, and 18).
Proteomics data analysis
We combined proteins detected in ask1 samples into the ask1 proteome, and combined proteins detected in Ler into the Ler proteome. We first compared our ask1 and Ler proteomes to find proteins that are only detected in ask1 samples. We also obtained previously published proteomics data of wild-type Arabidopsis thaliana floral buds [46, 47] and combined them into a “previous WT” proteome containing 5461 non-redundant proteins (FDR < 1 %). Comparison of our Ler and ask1 proteomes with the previous WT proteome resulted in the finding of additional floral proteins in our data. We combined our Ler floral bud proteome with the previous WT proteome [46, 47] to a “Pooled WT” proteome consisting of 5977 non-redundant proteins. Comparison of our ask1 proteome with the Pooled WT proteome led to the identification of proteins that are considered as “ask1-only” proteins.
Since each sample was analyzed by MudPIT individually without labeling and multiplexing, the abundance of each protein cannot be directly compared across different samples. Instead, the relative abundance of each protein in a sample was normalized using the spectral counting method as previously described [46, 48–51, 87]. The following formula is used to calculate the spectral counting values which represent the normalized relative abundance of proteins:
The “Measured spectra of protein K” is the number of actually detected MS spectra that specifically match to the protein K.
The “Measured spectra of all proteins in dataset” is the sum of the measured spectra of proteins in one sample.
The “Theoretical peptides of all proteins in dataset” is the total number of the in silico tryptic peptides of all proteins detected in one sample. The in silico tryptic digestion was carried out using the tool “digest” from the Galaxy platform (https://usegalaxy.org/). Since trypsin normally does not cut after lysine (K) or arginine (R) residues if it is followed by a Proline (P), we specified these sites as non-cut sites. Partial digestion and fragments containing more than one potential cut site were not included. Peptides containing at least 6 amino acid residues were considered as theoretical peptides.
The “Theoretical peptides of protein K” is the number of tryptic peptides of a protein K that was determined in the above “Theoretical peptides of all proteins in dataset”.
For a protein detected in both ask1 and Ler samples, its spectral counting values were averaged across ask1 and Ler samples, respectively. Then the average spectral counting values of a protein in ask1 and Ler samples were compared. Proteins whose average spectral counting value in ask1 samples is at least 1.5-fold of that in Ler samples were considered as “ask1-higher” proteins.
Availability of supporting data
The data sets supporting the results of this article are included within the article and its additional file. The raw microarray datasets were deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) with the accession number GSE42841.
Acknowledgements
We thank Anne Stanley (Proteins and Mass Spectrometry Core Research Facility, Penn State College of Medicine, Hershey, PA) for mass spectrometry technical services; and Craig Praul (Director of Expression Analysis, Penn State Genomics Core Facility–University Park, PA) for microarray technical services. This work was supported by a US Department of Energy grant (DE-FG02-02ER15332) to H.M. and by the Department of Biology Huck Institutes of the Life Sciences, Pennsylvania State University (University Park, PA) and Fudan University (Shanghai, China).
Abbreviations
- ASK1
ARABIDOPSIS SKP1-LIKE1
- SCF
SKP1-CUL1-F-box
- ASK1-E3s
ASK1-containing SCF E3 ubiquitin ligases
- UPS
Ubiquitin-proteasome system
- TIR1
TRANSPORT INHIBITOR RESPONSE 1
- COI1
CORONATINE INSENSITIVE 1
- JA
Jasmonic acid
- GA
Gibberellic acid
- ABA
Abscisic acid
- SLFs
S-locus F-box proteins
- UFO
UNUSUAL FLORAL ORGANS
- MS
Mass spectrometry
- MudPIT
Multidimensional Protein Identification Technology
- CCA1
CIRCADIAN CLOCK ASSOCIATED 1
- LHY
LATE ELONGATED HYPOCOTYL
- JAZ1
JASMONATE-ZIM-DOMAIN PROTEIN 1
- TAIR
The Arabidopsis Information Resource
- FDR
False discovery rate
- WT
Wild-type
- bHLH
Basic helix-loop-helix
- SWN
SWINGER
- FLC
FLOWERING LOCUS C
- REF6
RELATIVE OF EARLY FLOWERING 6
- FBP7
F-BOX PROTEIN 7
- CPK6
CALCIUM-DEPENDENT PROTEIN KINASE 6
- LYSM RLK1
LYSM DOMAIN RECEPTOR-LIKE KINASE 1
- MKK2
MAP KINASE KINASE 2
- CDC2
Cell Division Cycle 2
- SnRK1.2
SNF1-RELATED PROTEIN KINASE 1.2
- KIN11
SNF1 KINASE HOMOLOG 11
- UBP
UBIQUITIN-SPECIFIC PROTEASE
- BRS1
BRI1 SUPPRESSOR 1
- HAUSP
Herpesvirus-associated ubiquitin-specific protease
- DYT1
DYSFUNCTIONAL TAPETUM 1
- Ler
Landsberg erecta
- SEA
Singular Enrichment Analysis
- NCBI
National Center for Biotechnology Information
- GEO
Gene Expression Omnibus
- TCA
Trichloroacetic Acid
- DTT
Dithiothreitol
Additional files
Genes up-regulated in the ask1 mutant microarray compared with L er.
Genes down-regulated in the ask1 mutant microarray compared with L er.
Proteins detected in individual proteomics samples with FDR < 1 %.
Three membrane protein categories are well represented in the detected L er and ask1 proteins (FDR < 1 %).
ask1 -only proteins with microarray values.
ask1 -higher proteins with spectral counting and microarray values.
ask1 -lower proteins with spectral counting and microarray values.
The ask1 mutant has reduced female fertility.
Normalized microarray data of three biological replicates of ask1 and four biological replicates of L er floral buds.
Mass spectrometry data of ask1 _1 sample exported as .zip file.
Mass spectrometry data of ask1 _2 sample exported as .zip file.
Mass spectrometry data of ask1 _3 sample exported as .zip file.
Mass spectrometry data of ask1 _4 sample exported as .zip file.
Mass spectrometry data of ask1 _5 sample exported as .zip file.
Mass spectrometry data of L er_ 1 sample exported as .zip file.
Mass spectrometry data of L er_ 2 sample exported as .zip file.
Mass spectrometry data of L er_ 3 sample exported as .zip file.
Mass spectrometry data of L er_ 4 sample exported as .zip file.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
DL and HM conceived and designed the experiments. DL and WN performed the experiments. DL and BAS analyzed the data. DL wrote and revised the manuscript and HM edited the manuscript. All authors read and approved the final manuscript.
Contributor Information
Dihong Lu, Email: ludihong123@gmail.com.
Weimin Ni, Email: wzb1@berkeley.edu.
Bruce A. Stanley, Email: bstanley@psu.edu
Hong Ma, Email: hongma@fudan.edu.cn.
References
- 1.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55(1):555–90. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 2.Sorokin AV, Kim ER, Ovchinnikov LP. Proteasome system of protein degradation and processing. Biochemistry (Moscow) 2009;74(13):1411–42. doi: 10.1134/S000629790913001X. [DOI] [PubMed] [Google Scholar]
- 3.Schwechheimer C, Willige BC, Zourelidou M, Dohmann EM. Examining protein stability and its relevance for plant growth and development. Methods Mol Biol. 2009;479:147–71. doi: 10.1007/978-1-59745-289-2_10. [DOI] [PubMed] [Google Scholar]
- 4.Dreher K, Callis J. Ubiquitin, hormones and biotic stress in plants. Ann Bot. 2007;99(5):787–822. doi: 10.1093/aob/mcl255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stone SL, Callis J. Ubiquitin ligases mediate growth and development by promoting protein death. Curr Opin Plant Biol. 2007;10(6):624–32. doi: 10.1016/j.pbi.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 6.Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 1999;13(13):1678–91. doi: 10.1101/gad.13.13.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature. 2001;414(6861):271–6. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 8.Kepinski S, Leyser O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):446–51. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 9.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435(7041):441–5. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 10.Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCFCOI1 complex during jasmonate signalling. Nature. 2007;448(7154):661–5. doi: 10.1038/nature05960. [DOI] [PubMed] [Google Scholar]
- 11.Xu LH, Liu FQ, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang DF, Xie DX. The SCFCOl1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell. 2002;14(8):1919–35. doi: 10.1105/tpc.003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devoto A, Nieto-Rostro M, Xie D, Ellis C, Harmston R, Patrick E, Davis J, Sherratt L, Coleman M, Turner JG. COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 2002;32(4):457–66. doi: 10.1046/j.1365-313X.2002.01432.x. [DOI] [PubMed] [Google Scholar]
- 13.Hua Z, Meng X, Kao TH. Comparison of Petunia inflata S-Locus F-box protein (Pi SLF) with Pi SLF like proteins reveals its unique function in S-RNase based self-incompatibility. Plant Cell. 2007;19(11):3593–609. doi: 10.1105/tpc.107.055426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hua Z, Kao TH. Identification and characterization of components of a putative petunia S-locus F-box-containing E3 ligase complex involved in S-RNase-based self-incompatibility. Plant Cell. 2006;18(10):2531–53. doi: 10.1105/tpc.106.041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fields AM, Wang N, Hua Z, Meng X, Kao TH. Functional characterization of two chimeric proteins between a Petunia inflata S-locus F-box protein, PiSLF2, and a PiSLF-like protein, PiSLFLb-S2. Plant Mol Biol. 2010;74(3):279–92. doi: 10.1007/s11103-010-9672-x. [DOI] [PubMed] [Google Scholar]
- 16.Lai Z, Ma W, Han B, Liang L, Zhang Y, Hong G, Xue Y. An F-box gene linked to the self-incompatibility (S) locus of Antirrhinum is expressed specifically in pollen and tapetum. Plant Mol Biol. 2002;50(1):29–42. doi: 10.1023/A:1016050018779. [DOI] [PubMed] [Google Scholar]
- 17.Ingram GC, Goodrich J, Wilkinson MD, Simon R, Haughn GW, Coen ES. Parallels between UNUSUAL FLORAL ORGANS and FIMBRIATA, genes controlling flower development in Arabidopsis and Antirrhinum. Plant Cell. 1995;7(9):1501–10. doi: 10.1105/tpc.7.9.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilkinson MD, Haughn GW. UNUSUAL FLORAL ORGANS controls meristem identity and organ primordia fate in Arabidopsis. Plant Cell. 1995;7(9):1485–99. doi: 10.1105/tpc.7.9.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levin JZ, Meyerowitz EM. UFO: an Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell. 1995;7(5):529–48. doi: 10.1105/tpc.7.5.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chae E, Tan QK, Hill TA, Irish VF. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development. 2008;135(7):1235–45. doi: 10.1242/dev.015842. [DOI] [PubMed] [Google Scholar]
- 21.Lee I, Wolfe DS, Nilsson O, Weigel D. A LEAFY co-regulator encoded by UNUSUAL FLORAL ORGANS. Curr Biol. 1997;7(2):95–104. doi: 10.1016/S0960-9822(06)00053-4. [DOI] [PubMed] [Google Scholar]
- 22.Samach A, Klenz JE, Kohalmi SE, Risseeuw E, Haughn GW, Crosby WL. The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 1999;20(4):433–45. doi: 10.1046/j.1365-313x.1999.00617.x. [DOI] [PubMed] [Google Scholar]
- 23.Yang M, Hu Y, Lodhi M, McCombie WR, Ma H. The Arabidopsis SKP1-LIKE1 gene is essential for male meiosis and may control homologue separation. Proc Natl Acad Sci U S A. 1999;96(20):11416–21. doi: 10.1073/pnas.96.20.11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takahashi N, Kuroda H, Kuromori T, Hirayama T, Seki M, Shinozaki K, Shimada H, Matsui M. Expression and interaction analysis of Arabidopsis Skp1-related genes. Plant Cell Physiol. 2004;45(1):83–91. doi: 10.1093/pcp/pch009. [DOI] [PubMed] [Google Scholar]
- 25.Dai L, Xu L, Huang D, Li X, Luo K, Guan C. ASK1 physically interacts with COI1 and is required for male fertility in Arabidopsis. Sci China Ser C: Life Sci. 2002;45(6):631–6. doi: 10.1007/BF02879751. [DOI] [PubMed] [Google Scholar]
- 26.Woo HR, Chung KM, Park JH, Oh SA, Ahn T, Hong SH, Jang SK, Nam HG. ORE9, an F-box protein that regulates leaf senescence in Arabidopsis. Plant Cell. 2001;13(8):1779–90. doi: 10.1105/tpc.13.8.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han L, Mason M, Risseeuw EP, Crosby WL, Somers DE. Formation of an SCFZTL complex is required for proper regulation of circadian timing. Plant J. 2004;40(2):291–301. doi: 10.1111/j.1365-313X.2004.02207.x. [DOI] [PubMed] [Google Scholar]
- 28.Zhao D, Yu Q, Chen M, Ma H. The ASK1 gene regulates B function gene expression in cooperation with UFO and LEAFY in Arabidopsis. Development. 2001;128(14):2735–46. doi: 10.1242/dev.128.14.2735. [DOI] [PubMed] [Google Scholar]
- 29.Zhao D, Yang X, Quan L, Timofejeva L, Rigel NW, Ma H, Makaroff CA. ASK1, a SKP1 homolog, is required for nuclear reorganization, presynaptic homolog juxtaposition and the proper distribution of cohesin during meiosis in Arabidopsis. Plant Mol Biol. 2006;62(1-2):99–110. doi: 10.1007/s11103-006-9006-1. [DOI] [PubMed] [Google Scholar]
- 30.Zhao D, Yang M, Solava J, Ma H. The ASK1 gene regulates development and interacts with the UFO gene to control floral organ identity in Arabidopsis. Dev Genet. 1999;25(3):209–23. doi: 10.1002/(SICI)1520-6408(1999)25:3<209::AID-DVG4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 31.Zhao D, Han T, Risseeuw E, Crosby WL, Ma H. Conservation and divergence of ASK1 and ASK2 gene functions during male meiosis in Arabidopsis thaliana. Plant Mol Biol. 2003;53(1-2):163–73. doi: 10.1023/B:PLAN.0000009273.81702.b5. [DOI] [PubMed] [Google Scholar]
- 32.Jin J, Ang XL, Shirogane T, Wade Harper J. Identification of substrates for F-box proteins. Methods Enzymol. 2005;399:287–309. doi: 10.1016/S0076-6879(05)99020-4. [DOI] [PubMed] [Google Scholar]
- 33.Tan MK, Lim HJ, Harper JW. SCF(FBXO22) regulates histone H3 lysine 9 and 36 methylation levels by targeting histone demethylase KDM4A for ubiquitin-mediated proteasomal degradation. Mol Cell Biol. 2011;31(18):3687–99. doi: 10.1128/MCB.05746-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An F, Zhao Q, Ji Y, Li W, Jiang Z, Yu X, Zhang C, Han Y, He W, Liu Y, et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell. 2010;22(7):2384–401. doi: 10.1105/tpc.110.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Igawa T, Fujiwara M, Takahashi H, Sawasaki T, Endo Y, Seki M, Shinozaki K, Fukao Y, Yanagawa Y. Isolation and identification of ubiquitin-related proteins from Arabidopsis seedlings. J Exp Bot. 2009;60(11):3067–73. doi: 10.1093/jxb/erp134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maor R, Jones A, Nuhse TS, Studholme DJ, Peck SC, Shirasu K. Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol Cell Proteomics. 2007;6(4):601–10. doi: 10.1074/mcp.M600408-MCP200. [DOI] [PubMed] [Google Scholar]
- 37.Burande CF, Heuze ML, Lamsoul I, Monsarrat B, Uttenweiler-Joseph S, Lutz PG. A label-free quantitative proteomics strategy to identify E3 ubiquitin ligase substrates targeted to proteasome degradation. Mol Cell Proteomics. 2009;8(7):1719–27. doi: 10.1074/mcp.M800410-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38(Web Server issue):W64–70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu SX, Knowles SM, Andronis C, Ong MS, Tobin EM. CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009;150(2):834–43. doi: 10.1104/pp.108.133272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448(7154):666–71. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- 41.Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S. A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell. 2005;17(7):1953–66. doi: 10.1105/tpc.105.032060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell. 2007;19(7):2225–45. doi: 10.1105/tpc.106.048017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR. An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci U S A. 1988;85(19):7089–93. doi: 10.1073/pnas.85.19.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Donald RG, Cashmore AR. Mutation of either G box or I box sequences profoundly affects expression from the Arabidopsis rbcS-1A promoter. EMBO J. 1990;9(6):1717–26. doi: 10.1002/j.1460-2075.1990.tb08295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rogers HJ. Programmed cell death in floral organs: how and why do flowers die? Ann Bot. 2006;97(3):309–15. doi: 10.1093/aob/mcj051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science. 2008;320(5878):938–41. doi: 10.1126/science.1157956. [DOI] [PubMed] [Google Scholar]
- 47.Feng B, Li L, Zhou X, Stanley B, Ma H. Analysis of the Arabidopsis floral proteome: detection of over 2 000 proteins and evidence for posttranslational modifications. J Integr Plant Biol. 2009;51(2):207–23. doi: 10.1111/j.1744-7909.2008.00787.x. [DOI] [PubMed] [Google Scholar]
- 48.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 49.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76(14):4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 50.Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, Washburn MP. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40(4):303–11. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A. 2006;103(50):18928–33. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mosblech A, Thurow C, Gatz C, Feussner I, Heilmann I. Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 2011;65(6):949–57. doi: 10.1111/j.1365-313X.2011.04480.x. [DOI] [PubMed] [Google Scholar]
- 53.Fernandez-Calvo P, Chini A, Fernandez-Barbero G, Chico JM, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011;23(2):701–15. doi: 10.1105/tpc.110.080788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng Z, Sun L, Qi T, Zhang B, Peng W, Liu Y, Xie D. The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol Plant. 2011;4(2):279–88. doi: 10.1093/mp/ssq073. [DOI] [PubMed] [Google Scholar]
- 55.Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA. The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci U S A. 2006;103(39):14631–6. doi: 10.1073/pnas.0606385103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang D, Tyson MD, Jackson SS, Yadegari R. Partially redundant functions of two SET-domain polycomb-group proteins in controlling initiation of seed development in Arabidopsis. Proc Natl Acad Sci U S A. 2006;103(35):13244–9. doi: 10.1073/pnas.0605551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development. 2004;131(21):5263–76. doi: 10.1242/dev.01400. [DOI] [PubMed] [Google Scholar]
- 58.Lu FL, Cui X, Zhang SB, Jenuwein T, Cao XF. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet. 2011;43(7):715–U144. doi: 10.1038/ng.854. [DOI] [PubMed] [Google Scholar]
- 59.Yu X, Li L, Guo M, Chory J, Yin Y. Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci U S A. 2008;105(21):7618–23. doi: 10.1073/pnas.0802254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Portereiko MF, Sandaklie-Nikolova L, Lloyd A, Dever CA, Otsuga D, Drews GN. NUCLEAR FUSION DEFECTIVE1 encodes the Arabidopsis RPL21M protein and is required for karyogamy during female gametophyte development and fertilization. Plant Physiol. 2006;141(3):957–65. doi: 10.1104/pp.106.079319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pinon V, Etchells JP, Rossignol P, Collier SA, Arroyo JM, Martienssen RA, Byrne ME. Three PIGGYBACK genes that specifically influence leaf patterning encode ribosomal proteins. Development. 2008;135(7):1315–24. doi: 10.1242/dev.016469. [DOI] [PubMed] [Google Scholar]
- 62.Fujikura U, Horiguchi G, Ponce MR, Micol JL, Tsukaya H. Coordination of cell proliferation and cell expansion mediated by ribosome-related processes in the leaves of Arabidopsis thaliana. Plant J. 2009;59(3):499–508. doi: 10.1111/j.1365-313X.2009.03886.x. [DOI] [PubMed] [Google Scholar]
- 63.Ito T, Kim GT, Shinozaki K. Disruption of an Arabidopsis cytoplasmic ribosomal protein S13-homologous gene by transposon-mediated mutagenesis causes aberrant growth and development. Plant J. 2000;22(3):257–64. doi: 10.1046/j.1365-313x.2000.00728.x. [DOI] [PubMed] [Google Scholar]
- 64.Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10(5):602–10. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- 65.Tagwerker C, Flick K, Cui M, Guerrero C, Dou Y, Auer B, Baldi P, Huang L, Kaiser P. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol Cell Proteomics. 2006;5(4):737–48. doi: 10.1074/mcp.M500368-MCP200. [DOI] [PubMed] [Google Scholar]
- 66.Calderon-Villalobos LI, Nill C, Marrocco K, Kretsch T, Schwechheimer C. The evolutionarily conserved Arabidopsis thaliana F-box protein AtFBP7 is required for efficient translation during temperature stress. Gene. 2007;392(1-2):106–16. doi: 10.1016/j.gene.2006.11.016. [DOI] [PubMed] [Google Scholar]
- 67.Zhou X, Hao Q, Liao J, Zhang Q, Lu H. Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene. 2013;32(3):388–96. doi: 10.1038/onc.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu J, Tian YS, Peng RH, Xiong AS, Zhu B, Jin XF, Gao F, Fu XY, Hou XL, Yao QH. AtCPK6, a functionally redundant and positive regulator involved in salt/drought stress tolerance in Arabidopsis. Planta. 2010;231(6):1251–60. doi: 10.1007/s00425-010-1122-0. [DOI] [PubMed] [Google Scholar]
- 69.Munemasa S, Hossain MA, Nakamura Y, Mori IC, Murata Y. The Arabidopsis calcium-dependent protein kinase, CPK6, functions as a positive regulator of methyl jasmonate signaling in guard cells. Plant Physiol. 2011;155(1):553–61. doi: 10.1104/pp.110.162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mori IC, Murata Y, Yang Y, Munemasa S, Wang YF, Andreoli S, Tiriac H, Alonso JM, Harper JF, Ecker JR, et al. CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca(2+)-permeable channels and stomatal closure. PLoS Biol. 2006;4(10) doi: 10.1371/journal.pbio.0040327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gimenez-Ibanez S, Ntoukakis V, Rathjen JP. The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant Signal Behav. 2009;4(6):539–41. doi: 10.4161/psb.4.6.8697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iizasa E, Mitsutomi M, Nagano Y. Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J Biol Chem. 2010;285(5):2996–3004. doi: 10.1074/jbc.M109.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teige M, Scheikl E, Eulgem T, Doczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell. 2004;15(1):141–52. doi: 10.1016/j.molcel.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 74.Brader G, Djamei A, Teige M, Palva ET, Hirt H. The MAP kinase kinase MKK2 affects disease resistance in Arabidopsis. Mol Plant Microbe Interact. 2007;20(5):589–96. doi: 10.1094/MPMI-20-5-0589. [DOI] [PubMed] [Google Scholar]
- 75.Gao M, Liu J, Bi D, Zhang Z, Cheng F, Chen S, Zhang Y. MEKK1, MKK1/MKK2 and MPK4 function together in a mitogen-activated protein kinase cascade to regulate innate immunity in plants. Cell Res. 2008;18(12):1190–8. doi: 10.1038/cr.2008.300. [DOI] [PubMed] [Google Scholar]
- 76.Iwakawa H, Shinmyo A, Sekine M. Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 2006;45(5):819–31. doi: 10.1111/j.1365-313X.2005.02643.x. [DOI] [PubMed] [Google Scholar]
- 77.Cui X, Fan B, Scholz J, Chen Z. Roles of Arabidopsis cyclin-dependent kinase C complexes in cauliflower mosaic virus infection, plant growth, and development. Plant Cell. 2007;19(4):1388–402. doi: 10.1105/tpc.107.051375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fragoso S, Espindola L, Paez-Valencia J, Gamboa A, Camacho Y, Martinez-Barajas E, Coello P. SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiol. 2009;149(4):1906–16. doi: 10.1104/pp.108.133298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Farras R, Ferrando A, Jasik J, Kleinow T, Okresz L, Tiburcio A, Salchert K, del Pozo C, Schell J, Koncz C. SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J. 2001;20(11):2742–56. doi: 10.1093/emboj/20.11.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li J, Lease KA, Tax FE, Walker JC. BRS1, a serine carboxypeptidase, regulates BRI1 signaling in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2001;98(10):5916–21. doi: 10.1073/pnas.091065998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416(6881):648–53. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 82.Liu F, Ni W, Griffith ME, Huang Z, Chang C, Peng W, Ma H, Xie D. The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell. 2004;16(1):5–20. doi: 10.1105/tpc.017772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development. 2006;133(16):3085–95. doi: 10.1242/dev.02463. [DOI] [PubMed] [Google Scholar]
- 84.Feng B, Lu D, Ma X, Peng Y, Sun Y, Ning G, Ma H. Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J. 2012;72(4):612–24. doi: 10.1111/j.1365-313X.2012.05104.x. [DOI] [PubMed] [Google Scholar]
- 85.Damerval C, De Vienne D, Zivy M, Thiellement H. Technical improvements in two-dimensional electrophoresis increase the level of genetic variation detected in wheat-seedling proteins. Electrophoresis. 1986;7(1):52–4. doi: 10.1002/elps.1150070108. [DOI] [Google Scholar]
- 86.Zhao Z, Zhang W, Stanley BA, Assmann SM. Functional proteomics of Arabidopsis thaliana guard cells uncovers new stomatal signaling pathways. Plant Cell. 2008;20(12):3210–26. doi: 10.1105/tpc.108.063263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhu W, Smith JW, Huang CM. Mass spectrometry-based label-free quantitative proteomics. J Biomed Biotechnol. 2010;2010:840518. doi: 10.1155/2010/840518. [DOI] [PMC free article] [PubMed] [Google Scholar]


