Abstract
Increased expression of the human DEK proto-oncogene (DEK) gene has been associated with numerous human malignancies. The DEK protein is associated with chromatin reconstruction and gene transcription, and is important in cell apoptosis. The present study aimed to elucidate the role of DEK with regard to gastric adenocarcinoma tumor progression and patient prognosis. DEK protein expression was analyzed using immunohistochemistry in 192 tumors paired with adjacent non-cancerous gastric mucosa that had been surgically resected from patients with primary gastric adenocarcinoma. The association between DEK expression and the clinicopathological characteristics of the patients was evaluated using the χ2 test and Fisher's exact test. The survival rates of the patients were calculated using the Kaplan-Meier method. Cox analysis evaluated the association between the expression of DEK and the survival rate of the patients. The DEK protein was expressed in 84 patients with gastric adenocarcinoma (43.8%) and in 20 of the paired normal gastric mucosa tissues (11.5%). The DEK expression rate was found to be associated with tumor size (P=0.006), tumor grade (P=0.023), lymph node metastasis (P=0.018), serous invasion (P=0.026), tumor stage (P=0.001) and Ki-67 expression (P=0.003). Furthermore, patients with gastric adenocarcinoma that expressed DEK had decreased disease-free (log-rank, 16.785; P<0.0001) and overall (log-rank, 15.759; P<0.0001) survival rates compared with patients without DEK expression. Patients with late-stage gastric adenocarcinoma that expressed DEK exhibited a lower overall survival rate compared with patients without DEK expression (P=0.002). Additional analysis revealed that DEK expression was an independent prognostic factor for the prognosis of gastric adenocarcinoma (hazard ratio, 0.556; 95% confidence interval, 0.337–0.918; P=0.022). From the results of the present study, it can be concluded that the detection of DEK protein expression in gastric adenocarcinoma tissues may be important for the diagnosis and prognosis of patients, and may be a targeted therapy for the treatment of gastric adenocarcinoma.
Keywords: DEK proto-oncogene, immunohistochemistry, survival analysis, gastric adenocarcinoma
Introduction
Gastric adenocarcinoma is a common malignant tumor, and surgical resection is considered to be the most effective treatment (1,2). The incidence of gastric adenocarcinoma has declined over time, due to improving living standards, improvements in early diagnosis, advanced surgical techniques and combined therapy (surgery, chemotherapy and radiotherapy). However, distant metastasis and local recurrence are unavoidable in the majority of cases, and the survival rate and prognosis of gastric adenocarcinoma patients remains far from satisfactory (1). Despite advances in the elucidation of the molecular basis of this disease, the spectrum of genes that have altered expression in gastric adenocarcinoma and the roles these genes play remain unclear (2). Therefore, sensitive gastric adenocarcinoma biomarkers that predict the prognosis and guide effective targeted therapy of patients are required.
The DEK proto-oncogene (DEK) protein was identified in a subset of patients with acute myeloid leukemia as a fusion gene with nucleoporin 214 (3,4). In the majority of aggressive human tumors, including neuroblastoma, malignant glioma, melanoma, acute adult leukemia, and bladder and cervical cancer, the DEK gene frequently exhibits an increased expression level (5–9). In addition, autoantibodies to DEK have been identified in juvenile rheumatoid arthritis, systemic lupus erythematosus and sarcoidosis. Previous studies have suggested that the DEK protein is closely associated with the apoptosis of cells, since it inhibits p53-mediated apoptosis, it works in combination with the viral oncogenes, E6 and E7, to overcome senescence in cells, and it also promotes Harvey Rat Sarcoma Viral Oncogene Homolog-driven keratinocyte transformation (4,10–12).
A study by Han et al (13) demonstrated that the DEK protein was closely associated with the proliferation of serous ovarian tumor cells, and that the overexpression of DEK was significantly associated with the increased proliferating index of Ki-67. In the study, the demonstration that DEK expression is associated with the proliferative index of cells and cervical cancer is important for the diagnosis of precancerous lesions. Furthermore, according to the results from tumor tissue analysis, previous studies reported that DEK expression was associated with the development of human colorectal cancer, and was proposed as a novel molecular target for cancer treatment (14). However, to the best of our knowledge, there have been no studies to support the increase in DEK protein expression in patients with gastric adenocarcinoma. The present study used tissue microarrays (TMAs) to compare the expression of DEK in gastric adenocarcinoma samples and adjacent non-cancerous mucosa. In addition, the association between DEK protein expression, clinicopathological characteristics and patient survival rates was analyzed.
Materials and methods
Patient samples
A total of 192 cases of gastric adenocarcinoma were paired with adjacent noncancerous tissues from patients who underwent surgery between May 2004 and May 2007 at Dandong Central Hospital (Dandong, Liaoning, China). The patient cohort consisted of 148 men and 44 women, with a mean age of 49.7 years (range, 29–72 years). All the patients were diagnosed with gastric adenocarcinoma by pathological examination. Tumor stage was determined according to the 2010 American Joint Committee on Cancer Staging Manual (15), and as previously described (16), was demonstrated to be closely associated with the prognosis of the patients: Stage I–II, 122 patients and stage III–IV, 70 patients; well-differentiated tumor, 60 patients and poorly-differentiated tumor, 132 patients. The adjacent non-cancerous gastric mucosa tissues surrounding the tumor were also used in the present study. None of the patients received chemotherapy prior to surgery.
The study was approved by the Institutional Review Board and informed written consent was obtained from all the patients prior to sample collection.
TMAs
The TMAs were produced by Shanghai Xinchao Biological Technology Co., Ltd., (Shanghai, China) and were prepared as follows: Tissue cores (diameter of 1.5 mm) were extracted from paraffin-embedded tissues using a tissue array instrument (TMArrayer™, Organization Microarrayer; Pathology Devices Inc., Westminster, MD, USA) and the tissues were arranged regularly on a paraffin block. The tissue array blocks were heated at 52°C to fuse the tissue cores to the paraffin. The tissue array blocks were subsequently processed using a Leica RM2235 Manual Rotary Microtome (Leica Microsystems GmbH, Wetzlar, Germany; speed, 20 µm/rpm). Up to 80% of the tissue cores were fully exposed. The tissue array blocks were cut into 4-µm thick sections using the Leica RM2235 Microtome at a speed of 240 rpm. The cut tissue array blocks were attached to glass slides, which were heated to 60°C in an oven for 16 h and subsequently placed in a 5°C refrigerator until required.
Immunohistochemistry
For the immunohistochemical analysis, a streptavidin-alkaline phosphatase-labeling method was performed according to the following protocol: The paraffin biopsy slides were de-waxed and hydrated in a citrate antigen solution (Beijing Zhongshan Technology Co., Ltd., Beijing, China). The slides were subsequently rinsed in phosphate-buffered saline (PBS; Beijing Zhongshan Technology Co., Ltd.), placed in 30 ml/l hydrogen peroxide (Beijing Zhongshan Technology Co., Ltd.) for 10 min and normal serum was added to the slides at 37°C for 15 min. The following primary antibodies were added to the slides overnight at 4°C: Monoclonal mouse anti-DEK antibody (dilution, 1:50; catalog no. 610948; BD Biosciences, Franklin Lakes, NJ, USA) and monoclonal anti-Ki-67 antibody (dilution, 1:50; catalog no. MAB-0672; Fuzhou Maixin Biotech. Co., Ltd., Fuzhou, China). For the negative control, the antibody was replaced by PBS. The secondary antibody was goat anti-mouse immunoglobulin (Ig)G horseradish peroxidase-conjugated antibody (dilution, 1:500; catalog. no ZB-2305; Beijing Zhongshan Technology Co., Ltd.), which were incubated with the slides for 60 min at 37°C. Hematoxylin and eosin and 3,3′-diaminobenzidine were then added to the slides. DEK and Ki-67 exhibit nuclear staining. The expression index was determined by staining intensity and percentage of cells, according to the following criteria (17): Cells were scored as ‘−’ (negative, no or <5% positive cells), ‘+’ (0–50% positive cells), and ‘++’ (>50% positive cells). The positive descriptor (DEK overexpression) was assigned to ‘++’ scored cells. For survival analysis, DEK expression level was denoted as high expression or overexpression (‘++’) and low expression (‘−’ and ‘+’).
Follow-up observation
The 192 cancer patients were followed-up for survival. On the data of final follow-up (October 30, 2013), 70 patients had died while 122 patients remained alive. The median survival time was 63 months. The long interval between the final treatment date and conclusion of follow-up is responsible for the delay in manuscript submission.
Statistical analysis
Analysis was performed using SPSS version 17.0 (SPSS, Inc., Chicago, IL, USA). The association between DEK expression and the clinicopathological characteristics of the patients was evaluated using the χ2 test and Fisher's exact test. The survival rates of patients following tumor resection were calculated using the Kaplan-Meier method, and the difference between survival curves was analyzed using the log-rank test. Multivariant survival analysis was performed on all the characteristics using Cox proportional hazard regression model. P<0.05 was considered to indicate a statistically significant difference.
Results
Expression of DEK protein in gastric adenocarcinoma
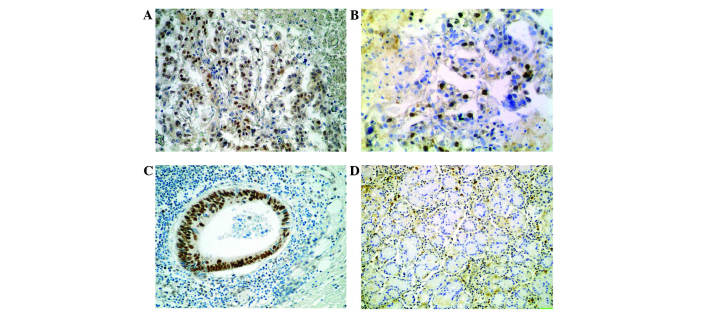
The DEK protein was identified as highly expressed in the cell nuclei of 84 gastric adenocarcinoma (43.8%) and 22 adjacent non-cancerous tissues (11.5%) using immunohistochemistry. There was consequently a significant difference between cells classified as overexpressing DEK in the gastric adenocarcinoma tissues and the adjacent non-cancerous gastric mucosa tissues (P<0.0001; Fig. 1).
Figure 1.
Immunohistochemical staining of DEK and Ki-67 protein. Positive (A) DEK and (B) Ki-67 expression in the adenocarcinoma tissues (magnification, ×200). (C) Postive DEK expression in the metastatic cancer cells in a lymph node (magnification, ×200). (D) No DEK expression in the normal gastric mucosa (magnification, ×100). DEK, DEK proto-oncogene.
Clinicopathological and prognostic significance of DEK expression
To elucidate the role of DEK in gastric adenocarcinoma progression, the present study investigated the association between DEK protein overexpression and the clinicopathological characteristics of the patients. Table I reveals that DEK overexpression was associated with tumor size, tumor grade, lymph node metastasis, serosal invasion, tumor stage and expression of Ki-67 (P=0.006, P=0.023, P=0.018, P=0.026, P=0.001 and P=0.003, respectively), but not with the age and gender of the patients. Furthermore, the patients with gastric adenocarcinoma with DEK overexpression had a lower overall survival rate compared with patients with no DEK overexpression (P<0.0001; Fig 2).
Table I.
Univariant analysis of the expression of DEK and various risk factors in 192 patients presenting with gastric adenocarcinoma.
| DEK expression | |||||
|---|---|---|---|---|---|
| Characteristics | Total, n | ++, n | −/+, n | HR (95% CI) | P-value |
| Total | 192 | 84 | 108 | ||
| Gender | 0.078 | ||||
| Male | 148 | 72 | 76 | ||
| Female | 44 | 12 | 32 | 2.526 (0.890–7.170) | |
| Age, years | 0.645 | ||||
| ≥50 | 113 | 51 | 62 | ||
| <50 | 79 | 33 | 46 | 1.147 (0.642–2.049) | |
| Tumor size, cm | 0.006a | ||||
| ≤5 | 108 | 34 | 74 | ||
| >5 | 84 | 50 | 34 | 0.312 (0.135–0.725) | |
| Tumor grade | 0.023b | ||||
| Good | 60 | 19 | 41 | ||
| Poor | 132 | 65 | 67 | 0.478 (0.251–0.908) | |
| LN metastasis | 0.018b | ||||
| Negative | 123 | 46 | 77 | ||
| Positive | 69 | 38 | 31 | 0.487 (0.268–0.887) | |
| Serosal invasion | 0.026b | ||||
| Negative | 127 | 48 | 77 | ||
| Positive | 65 | 36 | 29 | 0.502 (0.274–0.922) | |
| TNM stage | 0.001a | ||||
| I–II | 122 | 38 | 84 | ||
| III–IV | 70 | 46 | 24 | 0.236 (0.098–0.571) | |
| Ki-67 | 0.003a | ||||
| Negative | 74 | 18 | 56 | ||
| Positive | 118 | 66 | 52 | 0.253 (0.102–0.629) | |
P<0.01 and
P<0.05. DEK, DEK proto-oncogene; HR, hazards ratio; CI, confidence interval; LN, lymph node; TNM, tumor-node-metastasis.
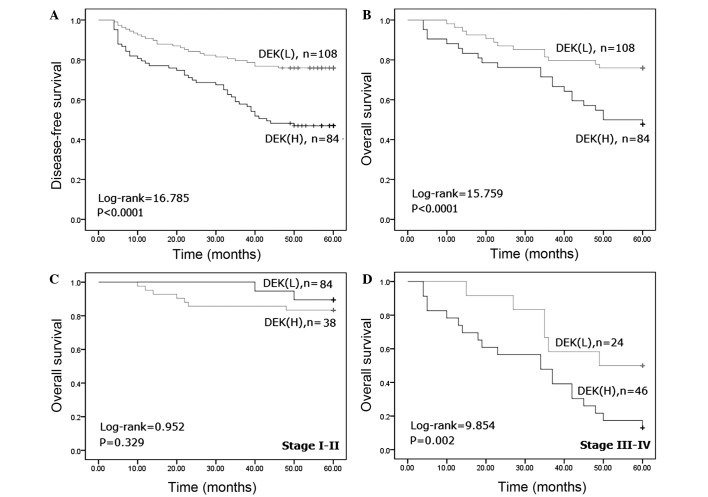
Figure 2.
Kaplan-Meier analysis of survival rates of patients with gastric adenocarcinoma in association with DEK protein expression. Patients with DEK overexpression had significantly lower (A) disease survival (log-rank, 16.785; P<0.0001) and (B) overall survival (log-rank, 15.759; P<0.0001) rates compared with patients with no DEK overexpression. (C) In patients with stage I–II gastric adenocarcinoma, the overall survival rate was not associated with DEK overexpression (log-rank, 0.952; P=0.329). (D) In patients with stage III–IV gastric adenocarcinoma, there was statistical significance between the overall survival rate and DEK overexpression (log-rank, 9.854; P=0.002). DEK, DEK proto-oncogene; L, low expression; H, high expression.
DEK expression is associated with the survival rate of patients with gastric adenocarcinoma
To additionally confirm the role of DEK overexpression in gastric adenocarcinoma progression, the present study analyzed the disease-free and overall survival rates for the 192 patients with gastric adenocarcinoma using Kaplan-Meier survival curves and demonstrated that patients with gastric adenocarcinoma that overexpressed DEK had lower disease-free (log-rank, 16.785; P<0.0001) and overall (log-rank, 15.759; P<0.0001) survival rates compared with patients without DEK overexpression (Fig. 2A and B). Furthermore, the present study analyzed the association between the DEK overexpression rate and the clinical stage of gastric adenocarcinoma to additionally substantiate the importance of DEK overexpression in gastric adenocarcinoma progression. The present analysis identified that patients with late-stage gastric adenocarcinoma that overexpressed DEK had a lower overall survival rate compared with patients with late-stage gastric adenocarcinoma that did not express DEK (P=0.002). However, the overall survival rate was not associated with the DEK expression status in patients with early-stage gastric adenocarcinoma (P=0.329) (Fig. 2C and D).
DEK overexpression is an independent prognostic factor in gastric adenocarcinoma
Univariate analysis for all significant variables demonstrated that patients that overexpressed DEK had a significantly lower overall survival rate compared with patients that did not overexpress DEK (hazards ratio (HR), 0.565; 95% confidence interval (CI), 0.369–0.866; P=0.009; Table II). Age, lymph node metastasis, serosal invasion and tumor stage were also associated with the overall survival rate of the patients. Additionally, multivariate analysis was performed using the Cox proportional hazards model. The results demonstrated that tumor stage was an independent prognostic factor for the survival rate of patients with gastric adenocarcinoma (HR, 0.663; 95% CI, 0.481–0.915; P=0.012; Table III). Notably, DEK overexpression was a significant independent prognostic factor in patients with gastric adenocarcinoma (HR, 0.556; 95% CI, 0.337–0.918; P=0.022; Table III).
Table II.
Univariate survival analyses of various characteristics in 192 patients with gastric adenocarcinoma using the Cox regression model.
| Characteristics | B | SE | Wald | HR (95% CI) | P-value |
|---|---|---|---|---|---|
| DEK | −0.571 | 0.218 | 6.862 | 0.565 (0.369–0.866) | 0.009a |
| Gender | −0.324 | 0.206 | 2.464 | 0.723 (0.483–1.084) | 0.116 |
| Age, years | −0.469 | 0.206 | 5.154 | 0.626 (0.418–0.938) | 0.023b |
| Tumor size, cm | −0.199 | 0.243 | 0.671 | 0.820 (0.509–1.319) | 0.413 |
| Tumor grade | −0.048 | 0.220 | 0.048 | 0.953 (0.619–1.467) | 0.826 |
| LN | −0.485 | 0.215 | 5.116 | 0.616 (0.404–0.937) | 0.024b |
| SI | −0.463 | 0.233 | 3.934 | 0.629 (0.398–0.995) | 0.047b |
| Tumor stage | −0.831 | 0.216 | 14.818 | 0.436 (0.285–0.665) | 0.000a |
| Ki-67 | −0.099 | 0.104 | 0.907 | 0.906 (0.739–1.111) | 0.341 |
P<0.01 and
P<0.05. DEK, DEK proto-oncogene; LN, lymph node metastasis; SI, serosal invasion; B, B coefficient; SE, standard error; HR, hazards ratio; CI, confidence interval.
Table III.
Multivariant survival analyses of various characteristics in 192 patients with gastric adenocarcinoma using Cox regression model.
| Characteristics | B | SE | Wald | HR (95% CI) | P-value |
|---|---|---|---|---|---|
| DEK | −0.587 | 0.256 | 5.275 | 0.556 (0.337–0.918) | 0.022a |
| Age, years | −0.388 | 0.209 | 3.445 | 0.679 (0.451–1.022) | 0.063 |
| LN | −0.331 | 0.223 | 2.193 | 0.718 (0.464–1.113) | 0.139 |
| SI | −0.257 | 0.253 | 1.028 | 0.774 (0.471–1.271) | 0.311 |
| Tumor stage | −0.411 | 0.164 | 6.244 | 0.663 (0.481–0.915) | 0.012a |
P<0.05. DEK, DEK proto-oncogene; LN, lymph node metastasis; SI, serosal invasion; B, B coefficient; SE, standard error; HR, hazards ratio; CI, confidence interval.
Discussion
The DEK gene is located on chromosome 6p22.3 (18); it is a highly conserved nuclear factor and the only member of its protein class. DEK is preferentially expressed in proliferating and malignant cells, which may result in 4–6 million copies per nucleus (19). DEK was originally identified as the target of a recurrent t(6;9) translocation in a subset of patients with acute myeloid leukemia (AML). The human DEK protein has been independently purified as a protein that modulates the topology of SV40 mini-chromosomes (20) and consists of 375 amino acids with 4 distinct acidic amino acids, and has a central SAP box DNA-binding domain and an additional carboxy-terminal DNA-binding region that partially overlaps with a multimerization domain. DEK is widely known as a proto-oncogene due to its involvement in the chromosomal translocation in patients with AML and its increased expression in multiple human malignancies (5). Kavanaugh et al (21) demonstrated that overexpression of DEK induces human keratinocyte transformation and that DEK-knockout mice are partially resistant to chemically induced papilloma formation.
Previous studies have revealed that DEK mRNA expression is increased in invasive ductal breast cancer, and has an increased gene expression in high-grade and late-stage breast cancer, therefore implicating it as a potential novel target in recurrent tumors (5,22–24). In addition, Shibata et al (25) identified that DEK overexpression affects the activity of global transcriptional regulators and is associated with tumor development and a poor prognosis of patients with high-grade neuroendocrine carcinoma. Our previous study indicated that DEK was additionally significantly correlated with the prognostic characteristics of patients with colorectal cancer, and DEK depletion by RNAi in SW-620 and HCT116 cells significantly decreased cell proliferation and increased cell apoptosis. Upregulation of DEK was involved in the p53/mouse double minute, B-cell lymphoma 2 family, and caspase signaling pathways (26). Notably, certain studies have revealed that DEK expression is associated with resistance to chemotherapeutic drugs, such as camptothecin, etoposide, neocarzinostatin and doxorubicin, which is often used to treat breast cancer (21,27,28).
The present results demonstrated that DEK is potentially important in the progression of gastric adenocarcinoma. In total, 43.8% (84/192) of patients with gastric adenocarcinoma clearly overexpressed the DEK protein. The present study revealed that DEK overexpression in patients with gastric adenocarcinoma was associated with the presence of large tumors, a poorer tumor grade, serosal invasion, lymph node metastasis, increased stage tumors and Ki-67 expression. Additional analysis demonstrated that DEK overexpression in gastric adenocarcinoma was associated with the survival rate of patients and was a significant independent predictor for the survival of patients with gastric adenocarcinoma and a high-stage tumor. Therefore, the present study identified DEK as a potential biomarker for gastric adenocarcinoma in tumor progression and prognosis. DEK may serve as a useful novel therapeutic biomarker, particularly for high-stage gastric adenocarcinoma. However, additional studies are required to confirm this conclusion.
Acknowledgements
The present study was supported by the Natural Science Foundation (grant no. 20140082) and the Doctoral Research Foundation of Eastern Liaoning University. (grant nos. 2014BZ0801 and 2014BZ0802).
References
- 1.Wang D, Wang L, Zhou J, Pan J, Qian W, Fu J, Zhang G, Zhu Y, Liu C, Wang C, Jin Z, He Z, Wu J, Shi B. Reduced expression of PTPRD correlates with poor prognosis in gastric adenocarcinoma. PLoS One. 2014;11:e113754. doi: 10.1371/journal.pone.0113754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang X, Takano Y, Zheng HC. The pathobiological features of gastrointestinal cancers (Review) Oncol Lett. 2012;3:961–969. doi: 10.3892/ol.2012.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.von Lindern M, Fornerod M, van Baal S, Jaegle M, de Wit T, Buijs A, Grosveld G. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol Cell Biol. 1992;12:1687–1697. doi: 10.1128/MCB.12.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wise-Draper TM, Allen HV, Jones EE, Habash KB, Matsuo H, Wells SI. Apoptosis inhibition by the human DEK oncoprotein involves interference with p53 functions. Mol Cell Biol. 2006;26:7506–7519. doi: 10.1128/MCB.00430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wise-Draper TM, Mintz-Cole RA, Morris TA, Simpson DS, Wikenheiser-Brokamp KA, Currier MA, Cripe TP, Grosveld GC, Wells SI. Overexpression of the cellular DEK protein promotes epithelial transformation in vitro and in vivo. Cancer Res. 2009;69:1792–1799. doi: 10.1158/0008-5472.CAN-08-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Q, Li Z, Lin H, Han L, Liu S, Lin Z. DEK overexpression in uterine cervical cancers. Pathol Int. 2008;58:378–382. doi: 10.1111/j.1440-1827.2008.02239.x. [DOI] [PubMed] [Google Scholar]
- 7.Orlic M, Spencer CE, Wang L, Gallie BL. Expression analysis of 6p22 genomic gain in retinoblastoma. Genes Chromosomes Cancer. 2006;45:72–82. doi: 10.1002/gcc.20263. [DOI] [PubMed] [Google Scholar]
- 8.Carro MS, Spiga FM, Quarto M, Di Ninni V, Volorio S, Alcalay M, Müller H. DEK expression is controlled by E2F and deregulated in diverse tumor types. Cell Cycle. 2006;5:1202–1207. doi: 10.4161/cc.5.11.2801. [DOI] [PubMed] [Google Scholar]
- 9.Datta A, Adelson ME, Mogilevkin Y, Mordechai E, Sidi AA, Trama JP. Oncoprotein DEK as a tissue and urinary biomarker for bladder cancer. BMC Cancer. 2011;11:234. doi: 10.1186/1471-2407-11-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohan S, Abdelwahab SI, Kamalidehghan B, Syam S, May KS, Harmal NS, Shafifiyaz N, Hadi AH, Hashim NM, Rahmani M, et al. Involvement of NF-κB and Bcl2/Bax signaling pathways in the apoptosis of MCF7 cells induced by a xanthone compound pyranocycloartobiloxanthone A. Phytomedicine. 2012;19:1007–1015. doi: 10.1016/j.phymed.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 11.Khaw AK, Yong JW, Kalthur G, Hande MP. Genistein induces growth arrest and suppresses telomerase activity in brain tumor cells. Genes Chromosomes Cancer. 2012;51:961–974. doi: 10.1002/gcc.21979. [DOI] [PubMed] [Google Scholar]
- 12.Lupescu A, Jilani K, Zbidah M, Lang F. Induction of apoptotic erythrocyte death by rotenone. Toxicology. 2012;300:132–137. doi: 10.1016/j.tox.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 13.Han S, Xuan Y, Liu S, Zhang M, Jin D, Jin R, Lin Z. Clinicopathological significance of DEK overexpression in serous ovarian tumors. Pathol Int. 2009;59:443–447. doi: 10.1111/j.1440-1827.2009.02392.x. [DOI] [PubMed] [Google Scholar]
- 14.Lin L, Piao J, Gao W, Piao Y, Jin G, Ma Y, Li J, Lin Z. DEK overexpression as an independent biomarker for poor prognosis in colorectal cancer. BMC Cancer. 2013;13:366–345. doi: 10.1186/1471-2407-13-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edge S, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC Cancer Staging Manual. 7th. New York, NY: Springer-Verlag New York; 2010. [Google Scholar]
- 16.Marchet A, Mocellin S, Ambrosi A, Morgagni P, Vittimberga G, Roviello F, Marrelli D, de Manzoni G, Minicozzi A, Coniglio A, et al. Validation of the new AJCC TNM staging system for gastric cancer in a large cohort of patients (n=2,155): Focus on the T category. Eur J Surg Oncol. 2011;37:779–785. doi: 10.1016/j.ejso.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 17.Köbel M, Weichert W, Crüwell K, Schmitt WD, Lautenschläger C, Hauptmann S. Epithelial hyaluronic acid and CD44v6 are mutually involved in invasion of colorectal adenocarcinomas and linked to patient prognosis. Virchows Arch. 2004;445:456–464. doi: 10.1007/s00428-004-1095-0. [DOI] [PubMed] [Google Scholar]
- 18.Lazăr D, Tăban S, Raica M, Sporea I, Cornianu M, Goldiş A, Vernic C. Immunohistochemical evaluation of the tumor neoangiogenesis as a prognostic factor for gastric cancers. Rom J Morphol Embryol. 2008;49:137–148. [PubMed] [Google Scholar]
- 19.Kappes F, Burger K, Baack M, Fackelmayer FO, Gruss C. Subcellular localization of the human proto-oncogene protein DEK. J Biol Chem. 2001;276:26317–26323. doi: 10.1074/jbc.M100162200. [DOI] [PubMed] [Google Scholar]
- 20.Killeen MR. Integrating a neurobiological systems approach into child neglect and abuse theory and practice. J Child Fam Nurs. 1999;2:406–407. [PubMed] [Google Scholar]
- 21.Kavanaugh GM, Wise-Draper TM, Morreale RJ, Morrison MA, Gole B, Schwemberger S, Tichy ED, Lu L, Babcock GF, Wells JM, et al. The human DEK oncogene regulates DNA damage response signaling and repair. Nucleic Acids Res. 2011;39:7465–7476. doi: 10.1093/nar/gkr454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinnedge Privette LM, McClaine R, Wagh PK, Wikenheiser-Brokamp KA, Waltz SE, Wells SI. The human DEK oncogene stimulates β-catenin signaling, invasion and mammosphere formation in breast cancer. Oncogene. 2011;30:2741–2752. doi: 10.1038/onc.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abba MC, Sun H, Hawkins KA, Drake JA, Hu Y, Nunez MI, Gaddis S, Shi T, Horvath S, Sahin A, Aldaz CM. Breast cancer molecular signatures as determined by SAGE: Correlation with lymph node status. Mol Cancer Res. 2007;5:881–890. doi: 10.1158/1541-7786.MCR-07-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/S1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shibata T, Kokubu A, Miyamoto M, Hosoda F, Gotoh M, Tsuta K, Asamura H, Matsuno Y, Kondo T, Imoto I, et al. DEK oncoprotein regulates transcriptional modifiers and sustains tumor initiation activity in high-grade neuroendocrine carcinoma of the lung. Oncogene. 2010;29:4671–4681. doi: 10.1038/onc.2010.217. [DOI] [PubMed] [Google Scholar]
- 26.Lin L, Piao J, Ma Y, Jin T, Quan C, Kong J, Li Y, Lin Z. Mechanisms underlying cancer growth and apoptosis by DEK overexpression in colorectal cancer. PLoS One. 2014;9:e111260. doi: 10.1371/journal.pone.0111260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kappes F, Fahrer J, Khodadoust MS, Tabbert A, Strasser C, Mor-Vaknin N, Moreno-Villanueva M, Bürkle A, Markovitz DM, Ferrando-May E. DEK is a poly(ADP-ribose) acceptor in apoptosis and mediates resistance to genotoxic stress. Mol Cell Biol. 2008;28:3245–3257. doi: 10.1128/MCB.01921-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khodadoust MS, Verhaegen M, Kappes F, et al. Melanoma proliferation and chemoresistance controlled by the DEK oncogene. Cancer Res. 2009;69:6405–6413. doi: 10.1158/0008-5472.CAN-09-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]