Abstract
Mechanical stresses on the myocyte nucleus have been associated with several diseases and potentially transduce mechanical stimuli into cellular responses. Although a number of physical links between the nuclear envelope and cytoplasmic filaments have been identified, previous studies have focused on the mechanical properties of individual components of the nucleus, such as the nuclear envelope and lamin network. The mechanical interaction between the cytoskeleton and chromatin on nuclear deformability remains elusive. Here, we investigated how cytoskeletal and chromatin structures influence nuclear mechanics in cardiac myocytes. Rapid decondensation of chromatin and rupture of the nuclear membrane caused a sudden expansion of DNA, a consequence of prestress exerted on the nucleus. To characterize the prestress exerted on the nucleus, we measured the shape and the stiffness of isolated nuclei and nuclei in living myocytes during disruption of cytoskeletal, myofibrillar, and chromatin structure. We found that the nucleus in myocytes is subject to both tensional and compressional prestress and its deformability is determined by a balance of those opposing forces. By developing a computational model of the prestressed nucleus, we showed that cytoskeletal and chromatin prestresses create vulnerability in the nuclear envelope. Our studies suggest the cytoskeletal–nuclear–chromatin interconnectivity may play an important role in mechanics of myocyte contraction and in the development of laminopathies by lamin mutations.
Keywords: Prestress, nuclear mechanics, atomic force microscopy, muscular thin film, myocyte
Introduction
Mechanical behavior of the nucleus has gained recent attention as a potential link between mechanical stimuli and gene expression.1–3 Mutations in lamins, the structural proteins of the nuclear envelope, yield a diversity of pathologies.4 Ruptured nuclei are observed particularly in muscle and cardiac tissues of patients with laminopathy.5,6 This tissue specificity suggests that the nucleus may have a structural role within the cell in addition to simple compartmentalization. Elucidation of this role is difficult due to the complex,7 and still controversial,8 micromechanical environment of the nucleus.
The potential for the nucleus to serve as a mechanosensor and its role in the mechanism of disease in laminopathies have prompted several studies of nuclear mechanics. The material properties of the nuclear envelope of isolated nuclei have been measured by micropipette aspiration9,10 and atomic force microscopy (AFM) indentation.11 Others have dissected the relative contributions of nuclear envelope proteins to nuclear stiffness within in vitro fibroblast laminopathy models.12,13 Physical interactions of the nucleus and the cytoskeleton were recently revealed suggesting that the nuclear mechanics can be influenced by the cytoskeleton.14,15 Tremblay et al. showed actin and microtubule filaments play critical roles in regulating the nuclear deformation in response to substrate strain.16 However, it still remains unclear how subcellular structures including the cytoskeleton and chromatin regulate the mechanical behaviors of the nucleus.
The nucleus in cardiac myocytes deforms during normal cardiac contraction making nuclear deformability relevant to cardiac muscle function. We observed the rupture of the nucleus in living cells when the nuclear membrane was disrupted, which was similar to the rupture of isolated nuclei shown in Mazumder and Shivashankar.17 We hypothesized that prestress generated by cytoskeletal and chromatin structures plays an important role in determining the stress distribution on the nuclear membrane in live cardiac myocytes. We measured the influence of the myofibrils on nuclear deformation both during cardiac contraction and diastole. We have pharmacologically disrupted actin filaments and microtubules to elucidate their contribution to nuclear shape and deformability. The experimental results for nuclei in cells were compared to those for isolated nuclei, which are free of prestress generated from cytoskeletal architecture. The role of chromatin on nuclear mechanics was also characterized by performing experiments after modifying chromatin structure by histone hyper-acetylation. By developing a computational model of the nucleus, we found that cytoskeletal prestress contributes to a non-uniform distribution of stress along the nuclear envelope. Our results suggest that interplay between the cytoskeleton, nuclear envelope, and chromatin plays an important role in determining the structure and mechanical properties of the nucleus.
Materials and methods
Cardiac myocyte harvest and culture
All procedures performed were conducted according to the guidelines of the Harvard University Animal Care and Use Committee. Ventricular myocytes were isolated from two-day-old Sprague Dawley rats as previously described.18 Briefly, excised ventricular tissue was agitated in a 0.1% trypsin solution cooled to 4℃ for approximately 14 h. Trypsinized ventricles were dissociated into their cellular constituents via serial exposure to a 0.1% solution of collagenase type II at 37℃ for 2 min. The dissociated cell solution was passed through a nylon mesh with 40 µm pores to remove any non-digested tissue. The cell solution was then serially pre-plated in tissue culture flasks twice for 45 min each time to enrich the myocyte portion of the cell population. Isolated myocytes were seeded onto coverslips with patterned fibronectin substrates in culture medium consisting of Medium 199 base supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA), 10 mM HEPES (Invitrogen), 0.1 mM MEM non-essential amino acids (Invitrogen), 20 mM glucose (Sigma Aldrich, St. Louis, MO), 2 mM l-glutamine (Invitrogen), 1.5 µM vitamin B-12 (Sigma), and 50 U/mL penicillin (Sigma). After 24 h of plating the cells were rinsed with PBS to remove any dead or non-adhered cells. On the second day of culture, the serum concentration of the medium was reduced to 2%. All experiments were performed after three days of cell culture.
Micropatterning extracellular matrix proteins on culture substrates
Cardiac myocytes were cultured onto 15 µm wide lines of extracellular matrix proteins as previously described18–20 to mimic cellular morphology found in vivo. Briefly, polydimethylsiloxane (PDMS; Sylgard 184, Dow Corning, Midland, MI) was spun onto glass coverslips and cured overnight at 60℃. A photolithographic mask was designed using AutoCAD software (Autodesk, San Rafeal, CA) and used to make negative templates of the patterns on silicon wafers coated with SU 8-2 photoresist (MicroChem Corp., Newton, MA). Stamps were prepared by polymerizing PDMS on the patterned silicon wafer and inked for 1 h with extracellular matrix protein fibronectin (FN, Invitrogen). Immediately after drying by air, the stamps were microcontacted with the PDMS-coated glass coverslips for FN transfer. The stamped coverslips were rinsed with a 1% solution of Pluronic F-127 (BASF, Florham Park, NJ) to prevent cells from attaching to non-FN-coated regions.
Isolation of nuclei
Nuclei from cultured cardiac myocytes were extracted and purified similar to previous techniques10,17 with the following modifications. Myocytes were cultured on PDMS with 15 µm wide lines of micropatterned FN proteins. After trypsinization, they were washed two times with PBS and resuspended in a buffer of 10 mM HEPES (pH 7.4), 10 mM KCl, and 1.5 mM MgCl2. The cells were incubated at room temperature and then on ice for 5 min each. Afterwards, 0.5% Triton-X was added to the cell solution and mixed well using a vortex before incubation on ice for another 5 min. Nuclei were separated from cellular debris by centrifugation at 1200 r/min for 5 min. The resulting supernatant was discarded and the pellet of nuclei was resuspended in a physiological buffer consisting of 130 mM KCl, 1.5 mM MgCl2, 10 mM Na2HPO4, 1 mM Na2ATP, and 1 mM DTT. To potentiate the replenishment of ATP, 5 mM creatine phosphate and 0.05 mg/mL creatine phosphokinase were also added. Isolated nuclei were seeded onto PDMS-coated substrates treated with 0.01% poly-l-lysine for an hour to allow the nuclei to adhere to the surface.
AFM
Indentation, height, and force measurements were made with an atomic force microscope. The optical lever sensitivity of tipless silicon nitride AFM cantilevers (k∼ 60 pN/nm, Veeco, Plainview, NY) was calibrated by deflection of the cantilever against glass in ambient air. Tip stiffness was calibrated thermally according to the Sader method21 in air in MFP-3D software (Asylum Research, Santa Barbara, CA) on an MFP-3D-BIO AFM (Asylum Research) integrated with an Axiovert 200 inverted microscope (Zeiss). Nuclei in intact cells or isolated nuclei were located by fluorescent signal of 4'-6-diamindino-2-pheylinodole, dihydrochloride (DAPI, Invitrogen) and then probed from above with the AFM. Indentations were performed at a constant speed of 5 µm/s because mechanical response of the nucleus can vary depending on the speed due to its viscoelasticity. Experiments with intact, live cells and isolated nuclei were performed in specialized buffers: for live cells, normal Tyrode’s solution (137 mM NaCl, 5.4 mM KCl, 1.2 mM MgCl2, 1 mM CaCl2, 20 mM HEPES, pH = 7.4) was used and for isolated nuclei, experiments were performed in an intracellular buffer bath (130 mM KCl, 1.5 mM MgCl2, 10 mM Na2HPO4, 1 mM Na2ATP, 1 mM DTT, 5 mM creatine phosphate, and 0.05 mg/mL creatine phosphokinase, pH = 7.4).
Fluorescence staining and microscopy
For live cell staining during experiments, chromatin and nuclei were labeled by addition of 35 µM DAPI in culture medium for 5 min. Live cells were imaged with a CCD camera (Cascade 512 b Coolsnap, Roper Scientific, Tucson, AZ) integrated with IPLab (BD Biosciences, Rockville, MD) on a Zeiss Axiovert 200 inverted fluorescence microscope and illuminated with an X-Cite 120 lamp (Exfo Life Sciences, Mississauga, Ontario). For fixed cells, fixation and staining occurred as follows. Cultured cells were washed with warmed PBS and then incubated in 4% paraformaldehyde in PBS for 10 min at 37℃. After three washes with PBS, the primary staining solution was added to the coverslips for 1 h. Depending on the molecule of interest, this included DAPI, fluorescein-conjugated phalloidin (Invitrogen), anti-α-actinin antibody raised in mouse (Sigma), antidesmin antibody raised in rabbit (Abcam, Cambridge MA), or anti-β-tubulin antibody raised in mouse (Developmental Studies Hybridoma Bank, Iowa City, IA). After three washes with PBS, the secondary staining solution was added, which included tetramethylrhodamine-conjugated goat-anti-mouse antibody (Invitrogen) and/or fluorescein-conjugated goat-anti-rabbit antibody (Invitrogen). In staining isolated nuclei, both primary and secondary antibodies were diluted in an intracellular buffer. Anti-Lamin B1 antibody raised in rabbit (Abcam) and antinucleoporin p62 antibody raised in mouse (BD Biosciences, Rockville, MD) were incubated with isolated nuclei for 1 h. Samples were visualized with a CCD camera (Coolsnap, Roper Scientific) integrated with IPLab on a Leica 6000DMIb (Leica, Wetzlar, Germany) inverted fluorescence microscope.
Pharmacological interventions
Non-sarcomeric actin filaments were disrupted by incorporation of 10 µM cytochalasin D (Sigma) to culture medium for 24 h. Microtubules were disrupted by addition of 33 µM nocodazole (Sigma) to culture medium for 2 h. Chromatin was decondensed by addition of 0.66 mM trichostatin A (TSA, Sigma) to culture medium for 4 h. Contractility was inhibited with addition of 10 µM blebbistatin (Sigma) to culture medium for 4 h. Rapid expansion of DNA by digestion of histone and DNA structural proteins was induced by treatment with 0.25% trypsin–EDTA (Sigma). Rho proteins were inhibited by incubating cells with 42 nM purified C3 transferase (Cytoskeleton, Denver, CO) for 4 h.
Muscular thin film (MTF) assay
Prestress applied to the nucleus was estimated by using the MTF assay developed in our lab.22 Briefly, poly(N-isopropylacrylamide) (PIPAAm; Polysciences, Inc.) was spin coated onto 25 mm glass cover slips and PDMS was spin coated on top of the PIPAAm. After curing the PDMS coverslips at 65℃ for 4 h, FN was patterned as 15 µm wide lines using microcontact printing. Isolated myocytes were seeded on the substrates and cultured in an incubator at 37℃ and 5% CO2.
After culturing for four days after seeding, coverslips were removed from the incubator and transferred into a Petri dish filled with 37℃ normal Tyrode’s solution. The MTFs were cut using a straight-blade razor. By cooling the Tyrode’s solution to room temperature, the PIPAAm dissolved and the MTF was released from the coverslip. The MTF was adhered to a stainless steel post-coated with polytetrafluoroethylene via hydrophobic interactions. Spontaneous contraction of the MTF was recorded on a stereomicroscope (Model MZ6, Leica microsystems, Inc.). Assuming the MTF as a plane strain beam consisting of a passive PDMS layer and a contractile cell monolayer, stresses generated by the cells were calculated by measuring the MTF’s radius of curvature (See Alford et al.22 for details).
Finite element modeling
Deformation and stress of the nuclear lamina were simulated using COMSOL Multiphysics 3.5 (COMSOL, Inc., Burlington, MA). The nucleus was modeled as a thin 3D axisymmetric ellipsoid whose stress-free configuration corresponds with the average dimensions measured in extracted nuclei (z = 13.3 µm, r = 10.15 µm) with a thickness of 25 nm. The large-strain constitutive behavior of the lamina is unknown, but it is assumed here to be nearly incompressible with the form
| (1) |
where W is the strain-energy density; I is the first strain invariant; A, B, and C are material parameters; κ is the bulk modulus; J is the elastic volume ratio; and p is a penalty variable.23 A, B, C, and κ are assumed to be 83.3, 10, 83.3 kPa, and 1 MPa, respectively, in order to have the small strain (<10%) modulus approximate the measured 1 MPa Young’s modulus of the lamina,9 while also including the strain stiffening, common in structural proteins.24–26 The nuclear interior is modeled as a pressure. The internal surface of the lamina is assumed to be under a positive pressure of 1 kPa, exerted by the compressed DNA. The external surface is under a complex traction exerted by axially aligned actin fibers and randomly oriented microtubules. The ith component of the traction (ti) is defined as
| (2) |
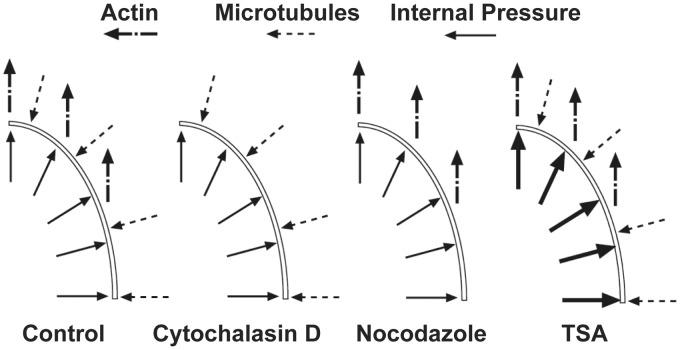
where i = r,z are the axes along the short and long axes of the nucleus, α and β are the values of the tension and compression due to the surrounding F-actin and microtubules, respectively. F is the deformation tensor, J is the determinant of F, and n is the unit vector normal to the undeformed surface. α and β are assumed to be 3 and 1.5 kPa, respectively, based on the experimental results of the MTF experiments, detailed in “Results” section. To simulate treatment with cytochalasin D, α was set to zero. To simulate nocodazole treatment, α and β were set as 4.5 kPa and zero, respectively, to account for microtubule depolymerization and increased activity of Rho proteins.27,28 TSA treatment was simulated by raising the internal pressure to 3 kPa. Finally, to simulate the nuclear rupture, the internal pressure was set to 3 kPa.29 For the in situ nucleus, we assume that actin and microtubules are intact so α and β remain unchanged. For the extracted nucleus, we let α = β = 0.
Nuclear envelope stress is reported as Von Mises stress, defined as
| (3) |
where J2 is the second deviatoric stress invariant.
Analysis
All data and images were analyzed, statistics calculated, and plots rendered in MatLab (Mathworks, Natick, MA). Schematic images were produced in Adobe Illustrator (Adobe, San Jose, CA). Statistical comparisons were performed using one-way analysis of variance, followed by Tukey’s least significant difference procedure to determine statistically different groups.
Results
Intracellular nuclear rupture
Previously, Shivashankar and colleagues29 reported rapid decondensation of chromatin and rupture of the nuclear membrane and lamin network in isolated cell nuclei and hypothesized that compaction is an important factor in controlling nuclear stability. We serendipitously replicated this result in intact cardiac myocytes within an anisotropic, 2D engineered cardiac tissue. To cause the nuclear rupture, we treated live DAPI-stained myocytes with trypsin that is known as a general protease. Trypsin application caused cells to detach from the substrate, relaxing the nuclei. Nuclear rupture was observed in myocytes which still adhered to the substrate with their nuclei subject to prestress (Figure 1(a)). In the experiments using TSA which is more specific in dissociating DNA, we could not observe nuclear rupture similar to what was seen with trypsin treatment.
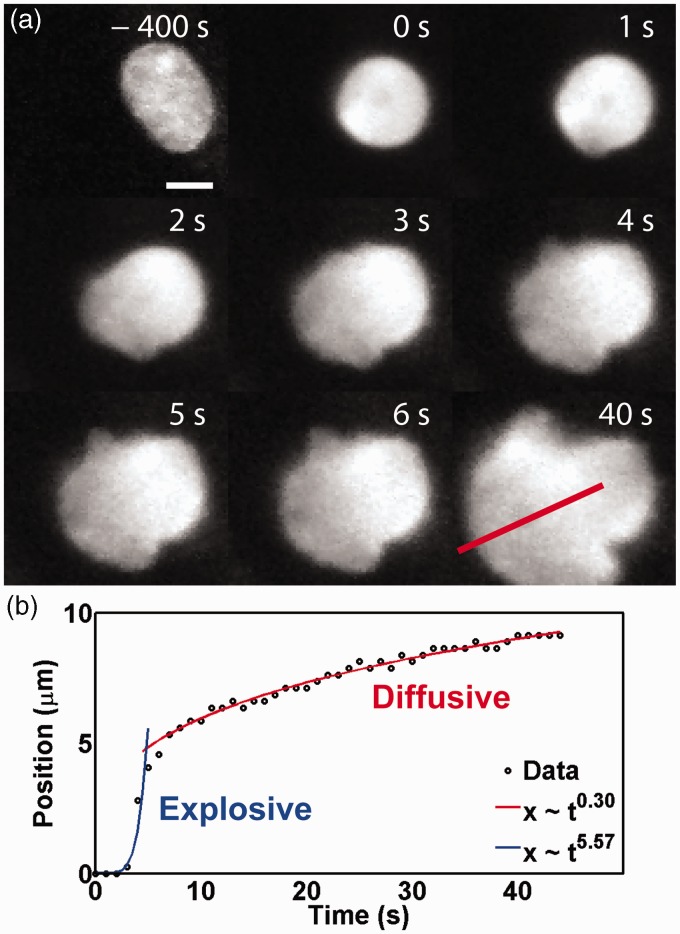
Figure 1.
(a) Prolonged treatment with 0.25% trypsin causes rapid expansion of DAPI-labeled chromatin and subsequent nuclear rupture. The location of the front of the escaping chromatin was tracked along a path normal to the surface of the undisturbed nucleus (red trace in last panel of a). Scale bar represents 5 µm. (b) The front expands in two distinct phases, the first an active outward entropic expansion (blue curve) and the second from passive diffusion (red curve). Time is zero-referenced to the beginning of nuclear failure
The expansion of the front of DNA was measured along an axis perpendicular to the surface of the undisturbed nuclear envelope and fit to a power law of the form x∼ tu, in which x represents the front position and t denotes time. The expulsion of chromatin from these localized failures is biphasic; at first, explosive, expanding with time raised to the u = 10.27 ± 7.81 (std, n = 20) power, then diffusive, with the front moving with u = 1.06 ± 0.57 (std, n = 20) (Figure 1(b)).
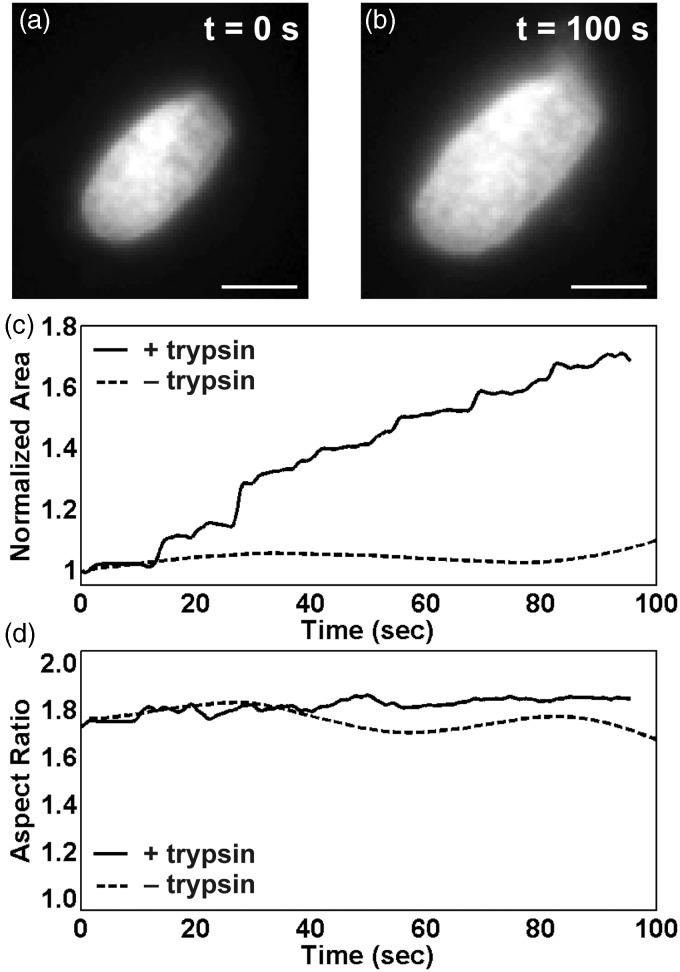
Application of trypsin to isolated nuclei (Figure 2(a) and (b)) caused a nuclear swelling defined by an increase in area (Figure 2(c)) but with no significant change in nuclear aspect ratio (Figure 2(d)). Compared to the nuclear rupture observed in live cells, the nuclear swelling of isolated nuclei occurred for a longer time without a significant change of nuclear shape.
Figure 2.
An isolated nucleus (a) treated with trypsin causing decondensation of DNA expands in a longer timescale maintaining the nuclear shape (b) unlike nuclear rupture in live myocytes. Scale bar represents 5 µm. The area of the nucleus expands (c) with no change in aspect ratio (d)
The pressure generated by rapid expansion of chromatin caused distinctly different behaviors in intact and isolated nuclei. Inside the myocyte, nuclear failure initiated at a point of stress concentration resulted in rupture as opposed to a uniform and slow nuclear swelling as we and others29 have observed in isolated nuclei. Given our observations, we hypothesized that prestress imposed by the cytoskeleton and chromatin renders the nucleus vulnerable to the observed failure and is responsible for the nuclear rupture observed during DNA decondensation.
Probing nuclear deformability with AFM indentation
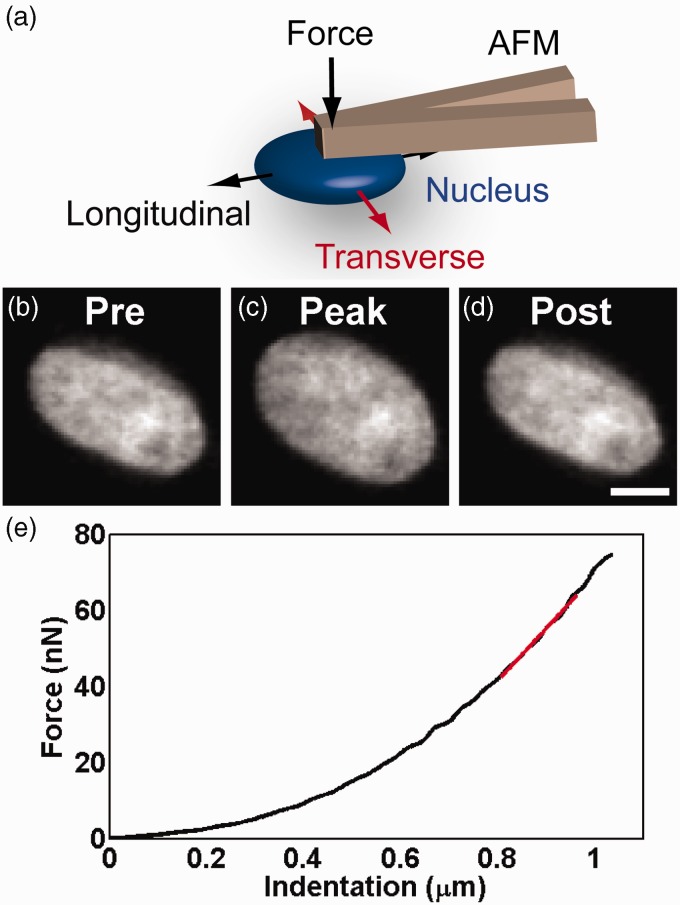
To understand how the cytoskeleton and chromatin can influence nuclear mechanics, we cultured cardiac myocytes on micropatterned substrates and non-destructively indented their nuclei with tipless AFM cantilevers to measure stiffness. Forces with magnitude 40–80 nN were applied normal to the substrate over the nucleus (Figure 3(a)), causing reversible deformation of the nucleus in the plane of substrate (Figure 3(b) to (d)). AFM indentation consistently produced a force–indentation curve with initial non-linearity followed by a linear region at higher forces, from which we calculated nuclear stiffness as the slope (Figure 3(e)). Although the measured stiffness is not just a material property of the nucleus itself, it represents the ability of the nucleus to deform within its cellular microenvironment as demonstrated in Krause et al.30
Figure 3.
(a) Schematic diagram of indentation on a nucleus by an atomic force microscope cantilever. Force is applied downward toward the substrate, deforming the nucleus and causing expansion in the plane of the substrate visualized by fluorescence of DAPI-labeled chromatin (pre-indentation b, peak indentation c, post-indentation d). Scale bar represents 5 µm. (e) The force and distance of indentation normal to the plane of the substrate are recorded and used to compute stiffness
Myocyte myofibrils can stabilize nuclear deformation
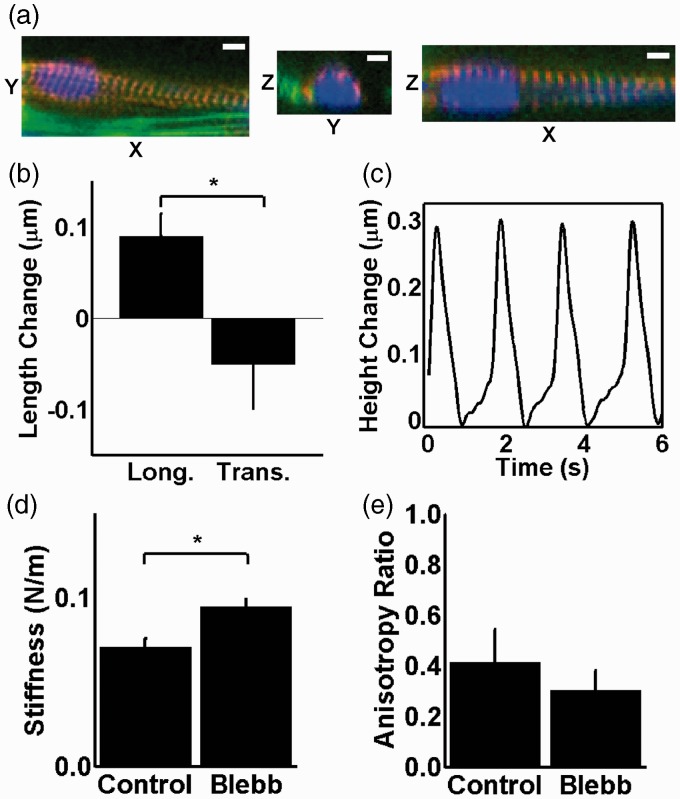
Myofibrils are serially aligned sarcomeres that shorten to induce myocyte contraction. Because the nucleus in cardiac myocytes is surrounded by myofibrils (Figure 4(a)), we asked if spontaneous myofibril shortening affects nuclear mechanics. To investigate this, we monitored morphological changes of the nucleus by DAPI fluorescence during spontaneous contraction. We observed a mean extension of the longitudinal axis of 0.09 ± 0.02 µm and shortening of the transverse axis of 0.05 ± 0.05 µm (Figure 4(b)). Despite large cell-to-cell variability, we find that the nucleus is consistently stretched along its long axis during contraction. This suggests that, as myofibrils contract, they apply tension not only to adjacent cells and their substrates but also to the nucleus. Coincident with this change in morphology, contraction also induces displacement of the nucleus away from the plane of the substrate, as measured by AFM deflection. A temporal profile of this motion reveals periodic displacements with an average amplitude of 154 ± 107 nm (std, n = 20) at an approximate velocity of 1 µm/s (Figure 4(c)). The mean height of nuclei from substrates was 2.18 ± 0.98 µm at diastole (std, n = 20). Together, these data indicate that the physical connection of the nucleus to the cytoskeleton in cardiac myocytes has particular consequences such that the contraction stretches the nucleus in the direction parallel to the myofibrils, causing shortening in the transverse axis and modest protrusion normal to the plane of the substrate.
Figure 4.
(a) Confocal microscopy 3D slices of a cardiac myocyte with labeled DNA (blue), actin (green), and α-actinin (red) illustrating the myocyte nucleus enveloped by the myofibrils. Scale bar represents 5 µm. (b) The change in dimensions of the nucleus during spontaneous contraction was measured from fluorescent images of contracting cells (n = 96). (c) The out of plane displacement of the nucleus induced by spontaneous contraction of a cardiac myocyte was measured by deflection of an AFM cantilever. Crossbridge inhibitor blebbistatin causes an increase in nuclear stiffness (d) and in the asymmetry of deformation during indentation (e). * denotes statistically different means with p < 0.05
This cytoskeletal–nuclear coupling suggests that myofibrils may influence nuclear deformability. AFM indentation revealed that nuclei of myocytes treated with blebbistatin are stiffer than those in untreated myocytes (Figure 4(d)). Blebbistatin inhibits myosin heavy chain ATPases. The increased stiffness of nucleus may be attributed to inactivation of myosins which can relieve stress of actin networks by allowing actin filaments sliding.31 This effect is also evident in the geometry of deformation during indentation captured within the anisotropy ratio, defined as the ratio of the percent change in length of the nucleus in the longitudinal direction to the percent change in length in the transverse. The blebbistatin treatment caused the length changes in the longitudinal and the transverse direction to decrease from 2.1 to 0.9% and 7.5 to 3.8%, respectively. The decreased anisotropy ratio (Figure 4(e)) indicates that deformation in the longitudinal direction, in the direction of the myofibrils, becomes more difficult relative to deformation orthogonal to the myofibrils. In the absence of blebbistatin, actin–myosin interactions stabilize nuclear deformation by allowing deformation along the myofibril direction. However, when nuclear integrity is disrupted by lamin mutations, the myofibrils may also contribute to nuclear failure by providing stabilization especially in the longitudinal direction and not in the transverse and hence create non-uniform stress in the nuclear envelope.
The cytoskeleton prestresses the nucleus, affecting its ability to deform
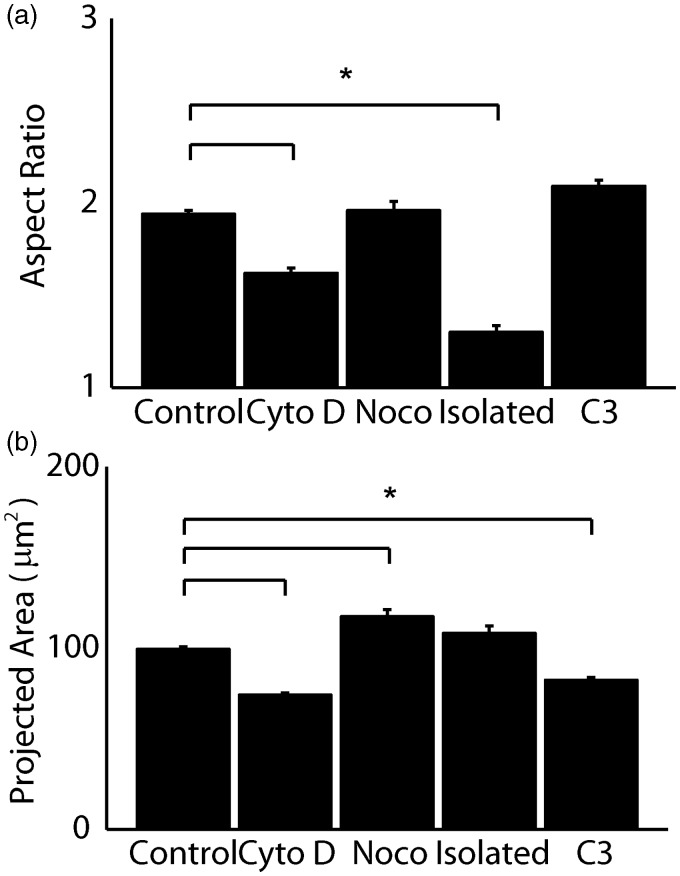
The cytoskeleton is a prestressed network and is physically integrated with the nuclear envelope. Thus, we sought to determine whether the nucleus is subject to prestresses similar to the cytoskeleton. The nuclear aspect ratio within intact cardiac myocytes was compared to that of nuclei extracted and seeded onto substrates (Figure 5(a)). Removal of the nucleus from its cellular microenvironment causes a decrease in aspect ratio, implying that the cytoskeleton actively exerts force to stretch the nucleus along the longitudinal axis of the cell. To resolve the contributions of individual cytoskeletal elements in the intact myocyte to this prestress, we applied cytochalasin D and nocodazole to disrupt actin filaments and microtubules, respectively, by shifting the polymers’ dynamic equilibria toward depolymerization.32 Cytochalasin D treatment causes the aspect ratio of the nucleus to significantly decrease (Figure 5(a)), while also decreasing the area of the nucleus (Figure5(b)), implying that actin filaments are responsible for stretching the nucleus in the longitudinal direction of the myocyte. Nocodazole has no effect on the nuclear aspect ratio; however, it does cause the area of the nucleus to increase (Figure 5(b)), an effect which may be due to the dissolution of the microtubule organizing center (MTOC), which in striated muscle is organized in a cage-like structure around the nucleus and may restrain its physical dimensions. A previous report suggests enhanced activity of Rho when microtubules are chemically disrupted.27 Thus, in a separate experiment we measured the changes in nuclear area when Rho was inhibited with C3 transferase and saw no significant change in the aspect ratio (Figure 5(a)), but a slight decrease in the projected area relative to controls (Figure 5(b)). To summarize, the nuclear area was reduced when the tensile load was reduced by chemical dissolution of the actin cytoskeleton or Rho inhibition and the area increased when the microtubule network, bearing compressive loads,33 was chemically disrupted. These results suggest sensitivity of the nuclear mechanical state to the architecture and prestress of the cytoskeletal network.
Figure 5.
The aspect ratio (a) and area (b) of intact (n = 756), cytochalasin D-treated (n = 209), nocodazole-treated (n = 102), isolated (n = 46), and C3 transferase-treated (n = 167) nuclei were measured by fluorescent labeling with DAPI. * denotes statistically different means with p < 0.001
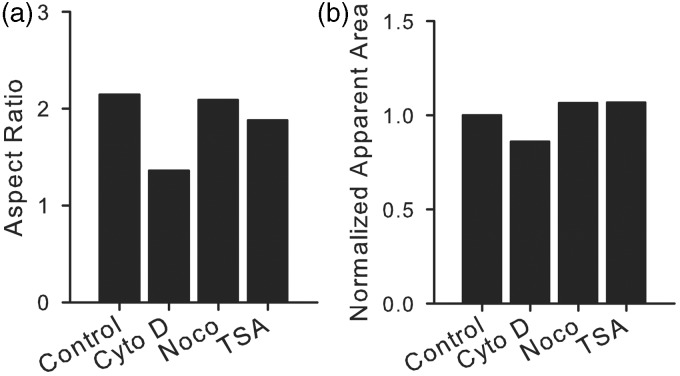
Figure 10.
Computational results. Effect of the load in Figure 8 on the aspect ratio (a) and area (b) of the nucleus
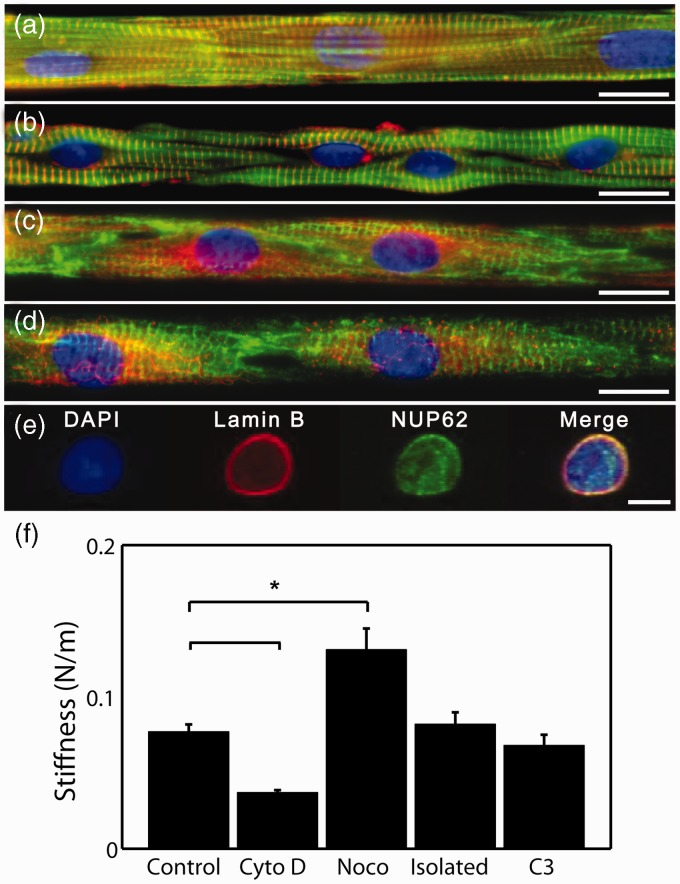
While the ability of the nucleus to deform is commonly believed to be due to the mechanical properties of the nuclear envelope, our data suggest that connectivity with the cell cytoskeleton may also influence nuclear deformability. Therefore, we assessed the contribution of individual constituents of the cytoskeleton to the measured values of nuclear stiffness. At the concentration and incubation duration employed here, cytochalasin D disrupts only non-sarcomeric actin filaments, as previously reported.34 This allows the contribution of polymerized actin to be assessed in the presence of structurally intact myofibrils. While intact, the myofibrils visualized by actin and α-actinin staining appear to have collapsed from their normal linear parallel conformation (Figure 6(a)) into a wavy pattern (Figure 6(b)). The dense microtubule networks normally observed (Figure 6(c)) are absent after nocodazole treatment, with only remnants of the MTOC visible (Figure 6(d)). After isolating nuclei from cardiac myocytes, immunostaining of the nuclear pore complex and nuclear envelope demonstrated that isolated nuclei remain physically intact (Figure 6(e)).
Figure 6.
Micropatterned cardiac myocytes with cytoskeletal elements labeled: actin (green) and α-actinin (red) (a, b) and desmin (green) and β-tubulin (red) (c, d). (b) Non-sarcomeric actin filaments were disrupted with application of 10 µM cytochalasin D for 24 h causing alteration of sarcomeric actin (green) and α-actinin (red). (d) The microtubule (red) network is disrupted by application of 1 µM nocodazole for 24 h while the desmin (green) intermediate filament network remains intact. Scale bar represents 15 µm. (e) An isolated nucleus stained with DAPI (blue), Lamin B1 (red), and nucleoporin p62 (green) antibodies. Scale bar represents 5 µm. (f) The stiffness of nuclei was measured by AFM indentation under normal conditions, “Control” (n = 29), cytoskeletal disruption, “Cyto D” (n = 25) and “Noco” (n = 39), free of cytoskeletal elements, “Isolated” (n = 33), and inhibition of Rho proteins, “C3” (n = 18). * denotes statistically different means with p < 0.001
We measured the stiffness of the nucleus after these chemical disruptions of the cytoskeleton in the intact myocyte and in the isolated nuclei (Figure 6(f)). Depolymerization of actin filaments resulted in a 47% loss of apparent stiffness of the nucleus during indentation. Thus, actin fibers and the asymmetric tension they apply resist outward transverse deformation of the nucleus. We also measured the effect of the microtubule network on nuclear deformability by applying nocodazole. As a result, depolymerization of microtubules increased the apparent stiffness of the nucleus by 86%. Microtubules apply compression to the nucleus, which competes with the tension applied by actin, as seen earlier. Thus, when microtubules are disrupted, the nucleus enlarges, relieving some of its prestress. Further expansion driven by indentation is therefore difficult and requires stretching of the nuclear envelope polymer network. We measured a 12% decrease in nuclear stiffness when inhibiting Rho with C3 transferase. The cumulative effect of the cytoskeleton on nuclear stiffness can be investigated by comparing the mechanical response of the nucleus to AFM indentation within the myocyte to its response when isolated from the cell by dissolution of the membrane and cytoskeleton and extracting the nucleus. Although structure or physical properties of the nucleus might be altered by the extracting protocol, the stiffness of isolated nuclei was measured to be higher than that of intact nuclei with cytochalasin D treatment, lower than that of intact nuclei with nocodazole treatment, and similar to that of intact nuclei without any treatment. These results suggest that both compression and tension applied to the nucleus by the cytoskeleton have a considerable influence over the ability of the nucleus to deform and the nucleus is under a balance of those opposing forces.
Chromatin structure affects nuclear mechanics
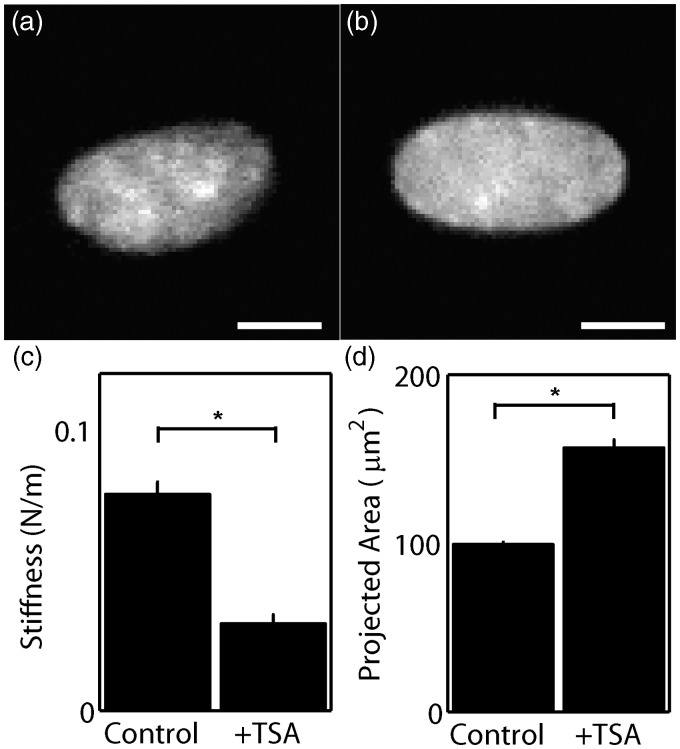
We asked how the internal structure of the nucleus affects nuclear mechanics. Chromatin tightly interfaced with histones provides structural rigidity to the nucleoplasm.35 Acetylation of histones mediates the association of DNA with histones and is maintained by an equilibrium of acetylation and deacetylation enzymatic activity.36 While histone acetyltransferases acetylates lysine on histones, histone deacetylases (HDACs) remove acetyl groups from histones, allowing interaction with DNA. The influence of the association of chromatin with histones on nuclear deformability can be tested by application of the HDAC-inhibitor trichostatin A (TSA).37 Prior to indentation by AFM, TSA was added to the culture medium of cardiac myocytes for 4 h. The relaxation of the association of DNA with histones produced a smoother texture of DAPI-stained nuclei in cultured myocytes (Figure 7(a) and (b)), reflecting a shift toward less condensed DNA. Indentation revealed that the dissociation of DNA from histones by TSA results in a 56% decrease in nuclear stiffness (Figure 7(c)), which is similar to the results observed in Krause et al.30 This may be attributed to decondensation of the nucleus by DNA unwinding or dissociation from histones, as indicated by a significant increase in nuclear area after treatment with TSA (Figure 7(d)). These results suggest that the nuclear shape and stiffness are a function of both the polymer networks inside and outside of the nuclear envelope.
Figure 7.
Chromatin was decondensed by application of 0.66 mM trichostatin A (TSA) for 4 h. DAPI-labeled nuclei without (a) and with TSA treatment (b) were imaged. Scale bar represents 5 µm. (c) The stiffness of nuclei treated with TSA (n = 37) was compared to untreated (n = 36). (d) Nuclear area in TSA treated myocytes (n = 64) was compared to the area in untreated myocytes (n = 756). * denotes statistically different means with p < 0.001
Prestress creates vulnerability in the nuclear envelope
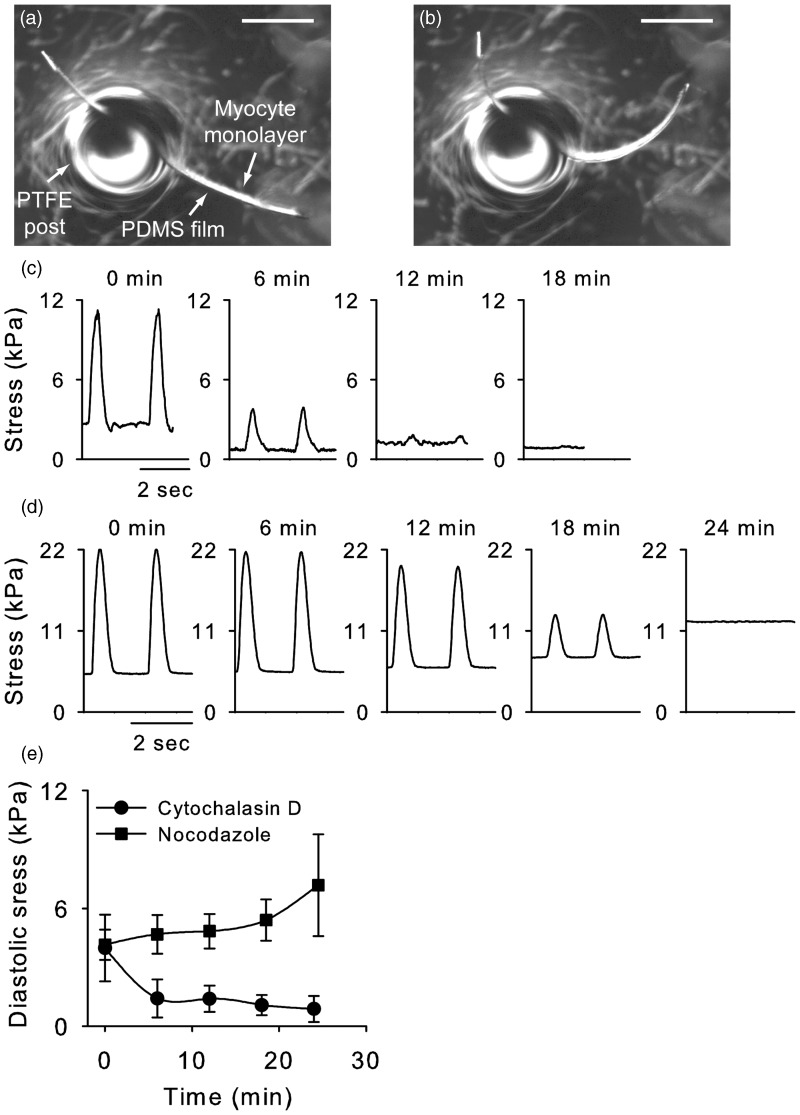
To further understand the effects of the cytoskeleton and chromatin prestress on nuclear mechanics, we developed a numerical model of the nucleus. The nucleus is modeled as a thin axisymmetric ellipsoid whose main load bearing structure is the nuclear lamina. Load is applied to the nucleus as a uniform internal pressure by DNA, external pressure by microtubules, and uniaxial pressure by actin microfilaments, as detailed in “ Materials and methods” section (Figure 8). To determine the values of the stresses applied to the nucleus, we employed an MTF assay in combination with pharmacological interventions. During diastole, the MTF shows a baseline curvature due to the resting tension (Figure 9(a)). The radius of curvature decreases during systole, as a result of contractile forces by cardiac myocytes (Figure 9(b)). MTF contractions were monitored until spontaneous contractions stopped after adding 10 µM cytochalasin D and 33 µM nocodazole. Cytochalasin D treatment significantly decreased both peak systolic and diastolic stress in a short time (Figure 9(c)). In contrast, nocodazole treatment did not change peak stress in the short time and increased diastolic stress (Figure 9(d)). At the end of experiments, diastolic stress increased by 3.0 kPa in nocodazole treatment while it decreased by 3.1 kPa in cytochalasin D treatment (Figure 9(e)). We assume the changes in diastolic stress correspond to cytoskeletal prestress generated by actin filaments and microtubules in cytochalasin D and nocodazole experiments, respectively.
Figure 8.
Computational model for the nucleus under the load applied by polymerized actin, microtubules, and DNA on the nuclear envelope
Figure 9.
A muscular thin film of cardiac myocytes during diastole (a) and systole (b). Scale bar represents 1 mm. Contractile stress of MTFs after cytochalasin D (c) and nocodazole (d) treatment. (e) Average changes in diastolic stress by cytochalasin D (n = 3) and nocodazole treatment (n = 3)
The nucleus model was developed by using the results obtained in MTF experiments with the assumption that the nucleus bears the same stress as the cytoskeleton. Within the model, pharmacological interventions are represented as removal or increase of cytoskeletal stress or internal pressure, consistent with the mechanism of the respective intervention (Figure 8, see “Materials and methods” section for details). When the MTF-determined actin and microtubule stress values were implemented in the model, stress modulation due to pharmacological intervention resulted in changes in nuclear aspect ratio and area consistent with those observed experimentally (Figure 10). Thus, the model suggests that cytoskeletal and chromatin prestress may explain the experimentally observed changes in nuclear morphology. Further, we performed a parameter study in the model and found that reasonable values of the material parameters and applied stress do not significantly change the qualitative results of the model (Supporting Information), demonstrating the robustness of this hypothesis.
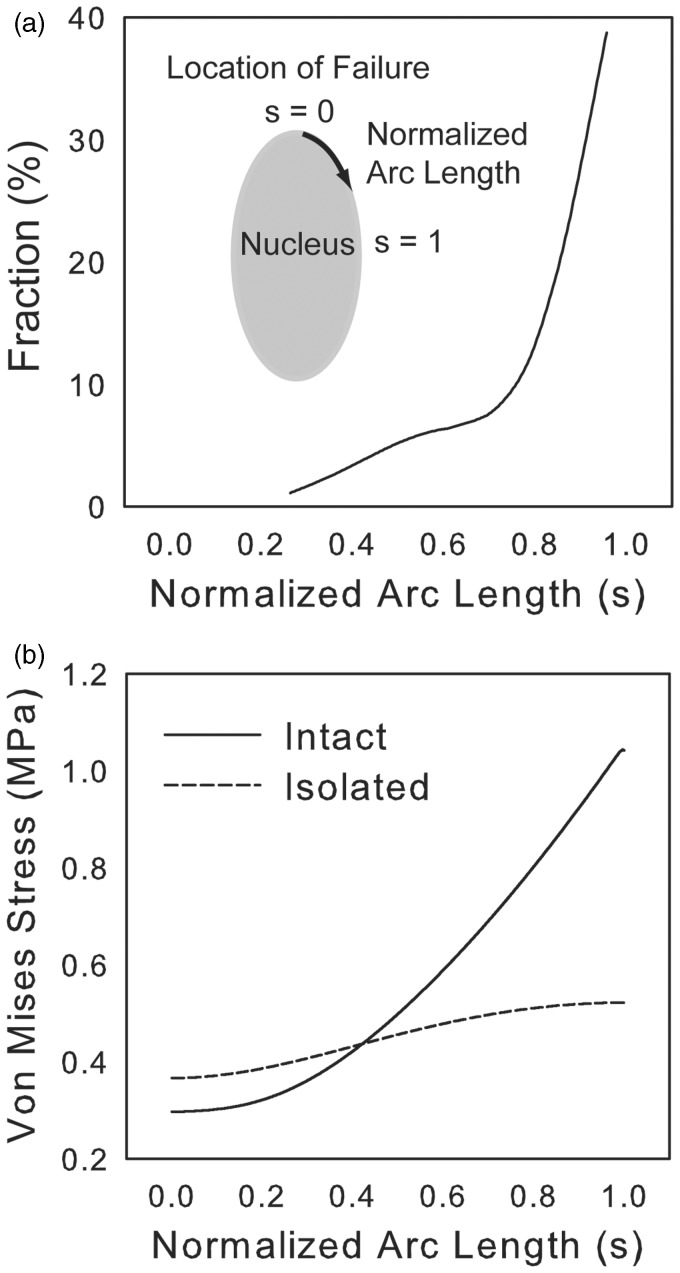
We experimentally observed localization of nuclear rupture preferentially occurring along the transverse face of the nucleus, as opposed to the longitudinal (Figure 11(a)). Because material failure begins as a localization of stress, the model was implemented to predict accumulation of stress and biased sites of failure within the nuclear envelope. Using the model, we calculated the Von Mises stress of the lamina, which represents the total distortion energy of the nuclear envelope. The nucleus will begin to yield as Von Mises stress approaches the yield strength of the lamina. Thus, increased Von Mises stress is indicative of an increased likelihood of nuclear rupture.
Figure 11.
(a) Histogram of the initial location of failure in trypsin-induced nuclear rupture. Normalized arc length (s) equals zero at the most longitudinal point on the nucleus and one at the most transverse (inset). The incidence of failure is plotted for 77 nuclei which had 81 points of failure. (b) Von Mises stress distribution as a function of normalized arc length for an isolated (no cytoskeletal forces) and intact nucleus
Our model predicts that nuclear geometry and external loads applied by the cytoskeleton lead to stress concentrations on the transverse face of the nucleus. In the isolated nucleus, due to geometry, the internal pressure pushing outward creates a concentration of stress along the transverse side of the nuclear envelope (Figure 11(b)). Therefore, the consistency of failure observed experimentally along the transverse face may be due to high stress state on that face. Greater asymmetry in nuclear shape causes a larger localization of stress upon loading from expanding DNA. Intact nuclei, where prestress maintains a larger nuclear aspect ratio than that of isolated nuclei, will have a more pronounced stress concentration. Additionally, the orthotropic traction field applied in the direction of the long axis of the cell by actin fibers induces increased tension at the transverse wall of the nucleus, leading to a sharply increased Von Mises stress (Figure 11(b)) and susceptibility to failure at that point. Thus, the prestress imposed on the nucleus by the cytoskeleton and by chromatin can have detrimental consequences, such as this vulnerability exploited through extreme loading by expansion of chromatin and weakening of the nuclear membrane.
Discussion
The cell nucleus may have a functional role in maintaining the mechanical integrity of the cell. Recent studies have focused on the molecular constituents of nuclear structure, with the implication that their dysfunction can result in cellular and tissue dysfunction.1 While the material properties and structural configuration of the nuclear envelope are critical determinant of nuclear deformation, there are additional structures and processes, equally critical, which affect nuclear mechanics. By using the pharmaceutical agents of cytochalasin D and nocodazole to disrupt actin and microtubule cytoskeletal networks, respectively, we found that the prestress they impose on the nuclear envelope alters nuclear morphology and deformability. We expect a stabilizer of cytoskeletal networks will cause the opposite effect on the nuclear mechanics compared to a destabilizer. For example, a treatment with Taxol that prevents depolymerization of microtubules will cause both the nuclear size and stiffness to decrease due to the maintained compression stress by microtubules. We have also observed that chromatin structure, specifically the association of DNA with histones as mediated by HDACs, also impacts nuclear mechanics and the state of prestress in which the nucleus is held. The implications of chromatin structure on nuclear mechanics may be particularly dramatic if the decondensation of chromatin occurs when the nuclear envelope becomes fragile due to lamin mutations. The cumulative effect of the cytoskeleton and chromatin prestress is an asymmetric nuclear shape and non-uniform stress distribution on the nuclear envelope. These might cause the nucleus to be vulnerable to changes in the mechanical environment of the heart, which are caused by both physiological and pathological processes.
There is evidence for advantages to maintaining prestress within the cell. In general, a prestressed cytoskeletal network provides stiffness and stability to the cell, vital to cellular survival, structure reorganization, motility, and contraction.38–40 Tensile cytoskeletal fibers are efficient transducers of mechanical signals over long distances, possibly used for signaling between substrate, cell periphery, and the nucleus.41 On the other hand, we demonstrated that cytoskeletal prestress can cause the intercellular nuclear rupture when the nuclear structure is disrupted by trypsin. As a non-specific protease, trypsin disrupted the nuclear membrane and diffused into the nucleus to cleave histones and other structural proteins including the lamin network. However, decondensation of chromatin itself by TSA treatment was not sufficient to cause the membrane rupture with the following rapid escape of chromatin. It suggests that the nuclear rupture is a consequence of the combination of defects in the nuclear membrane, expansion of the chromatin, and cytoskeletal tensional prestress. The nuclear membrane ruptures at the transverse face of the nucleus, where stress is concentrated, and cytoskeletal tension lead to a rapid outburst of genetic material from the nucleus in a short timescale. Despite the fact that the cell nucleus is large compared to other subcellular structures, the mechanical role of the nucleus within the cell is still poorly understood. The physical integration of the cytoskeleton, nucleus, and intranuclear structures,15,41 as well as some of the molecular constituents7,14 of these connections, has been previously studied. However, the biomechanical implications of the integration of the nucleus, which is a prestressed continuous network spanning ∼ 10 µm, to the ∼10 nm sized fibers of the cytoskeleton in cellular mechanics remain unknown. There are interesting implications for the inclusion of the nucleus in the cytoskeletal network as a mechanical conduit for mechanotransduction. The nuclear lamina is a non-linear elastic network, as are other intermediate filaments and many biopolymers.26 Rheological experiments on gels of reconstituted lamin B1 protein also indicate that lamins are non-linear polymers that strain harden.42 As the cytoskeleton actively deforms the nucleus to adopt a different morphology, it may be simultaneously changing the apparent stiffness of the strain-hardening nucleus. The nucleus can then be viewed as a malleable component of the cytoskeletal network with a tunable stiffness. As the cytoskeleton can adapt its contractility to external stimuli,43 the adjustment of nuclear stiffness is another potential cellular response.
The influence of the cytoskeleton and chromatin on nuclear mechanics provides insight into how mutations in nuclear envelope structural proteins result in disease. There is some evidence that suggests mutations in the lamin proteins disrupt the stability of nuclear structure. Recent experiments found that cells lacking lamins A/C gene (LMNA) showed large nuclear deformation under mechanical strain.12,13 These defective nuclear mechanics can cause a direct rupture of the nucleus or interrupt cellular functions indirectly through nuclear mechanotransduction. Due to the mechanical interconnectivity seen here between the nucleus and the cytoskeleton, localized defects in the nuclear membrane can propagate to dysfunction elsewhere in the cell, both to the internal chromatin or externally to the cytoskeleton. Our conception of nuclear mechanics should not be limited to quantification of nuclear stiffness itself but should also include understanding interactions of the nuclear envelope with the cytoskeleton and chromatin.
In summary, we investigated how cytoskeletal prestress regulates nuclear shape and stiffness in cardiac myocytes. We observed the nuclear rupture as a result of imbalanced prestress exerted on the nucleus when the structural integrity of the nuclear membrane was damaged by a protease. Through AFM measurements with pharmacological intervention, we showed nuclear deformability is altered by tensional and compressional prestress generated by the cytoskeleton and chromatin architecture. Our experimental results suggest that the mechanical behavior of the nucleus is determined by the interplay between the cytoskeleton, nuclear envelope, and chromatin. We also developed a computational model of the nucleus under prestress and found that prestress generates stresses on the nuclear envelope, which might be vulnerable to internal/external forces during laminopathies.
Acknowledgements
This work has been supported by the Nanoscale Science and Engineering Center of the National Science Foundation under NSF award number PHY-0117795, the Harvard Materials Research Science and Engineering Center under NSF award number DMR-0213805, and NIH grant 1 R01 HL079126-01A2 and Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science and Technology (NRF-2012R1A1A1042311).
Authors’ contributions
All authors participated in the design, interpretation of the studies and analysis of the data, and review of the manuscript; HL and WJA performed the experiments, PWA developed the computational model. MLM, AWF, SPS, and JAG analyzed data and isolated cells for experiments, KKP, HL, WJA, and PWA prepared the manuscript.
References
- 1.Dahl KN, Ribeiro AJS, Lammerding J. Nuclear shape, mechanics, and mechanotransduction. Circ Res 2008; 102: 1307–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingber DE. Cellular mechanotransduction: putting all the pieces together again. FASEB J 2006; 20: 811–27. [DOI] [PubMed] [Google Scholar]
- 3.Maxwell CA, Hendzel MJ. The integration of tissue structure and nuclear function. Biochem Cell Biol 2001; 79: 267–74. [PubMed] [Google Scholar]
- 4.Worman HJ, Bonne G. “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res 2007; 313: 2121–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Vos WH, Houben F, Kamps M, Malhas A, Verheyen F, Cox J, Manders EM, Verstraeten VL, van Steensel MA, Marcelis CL, van den Wijngaard A, Vaux DJ, Ramaekers FC, Broers JL. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum Mol Genet 2011; 20: 4175–86. [DOI] [PubMed] [Google Scholar]
- 6.Gupta P, Bilinska ZT, Sylvius N, Boudreau E, Veinot JP, Labib S, Bolongo PM, Hamza A, Jackson T, Ploski R, Walski M, Grzybowski J, Walczak E, Religa G, Fidzianska A, Tesson F. Genetic and ultrastructural studies in dilated cardiomyopathy patients: a large deletion in the lamin A/C gene is associated with cardiomyocyte nuclear envelope disruption. Basic Res Cardiol 2010; 105: 365–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crisp M, Burke B. The nuclear envelope as an integrator of nuclear and cytoplasmic architecture. FEBS Lett 2008; 582: 2023–32. [DOI] [PubMed] [Google Scholar]
- 8.Pederson T. Half a century of “the nuclear matrix”. Mol Biol Cell 2000; 11: 799–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahl KN, Kahn SM, Wilson KL, Discher DE. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J Cell Sci 2004; 117: 4779–86. [DOI] [PubMed] [Google Scholar]
- 10.Rowat AC, Lammerding J, Ipsen JH. Mechanical properties of the cell nucleus and the effect of emerin deficiency. Biophys J 2006; 91: 4649–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaziri A, Lee H, Mofrad MRK. Deformation of the cell nucleus under indentation: mechanics and mechanisms. J Mater Res 2006; 21: 2126–35. [Google Scholar]
- 12.Lammerding J, Fong LG, Ji JY, Reue K, Stewart CL, Young SG, Lee RT. Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem 2006; 281: 25768–80. [DOI] [PubMed] [Google Scholar]
- 13.Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest 2004; 113: 370–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol 2006; 172: 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci USA 1997; 94: 849–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tremblay D, Andrzejewski L, Leclerc A, Pelling AE. Actin and microtubules play distinct roles in governing the anisotropic deformation of cell nuclei in response to substrate strain. Cytoskeleton 2013; 70: 837–48. [DOI] [PubMed] [Google Scholar]
- 17.Mazumder A, Shivashankar GV. Gold-nanoparticle-assisted laser perturbation of chromatin assembly reveals unusual aspects of nuclear architecture within living cells. Biophys J 2007; 93: 2209–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feinberg AW, Feigel A, Shevkoplyas SS, Sheehy S, Whitesides GM, Parker KK. Muscular thin films for building actuators and powering devices. Science 2007; 317: 1366–70. [DOI] [PubMed] [Google Scholar]
- 19.Tan JL, Liu W, Nelson CM, Raghavan S, Chen CS. Simple approach to micropattern cells on common culture substrates by tuning substrate wettability. Tissue Eng 2004; 10: 865–72. [DOI] [PubMed] [Google Scholar]
- 20.Adams WJ, Pong T, Geisse NA, Sheehy SP, Diop-Frimpong B, Parker KK. Engineering design of a cardiac myocyte. J Comput Aided Mater Des 2007; 14: 19–29. [Google Scholar]
- 21.Sader JE, Larson I, Mulvaney P, White LR. Method for the calibration of atomic-force microscope cantilevers. Rev Sci Instrum 1995; 66: 3789–98. [Google Scholar]
- 22.Alford PW, Feinberg AW, Sheehy SP, Parker KK. Biohybrid thin films for measuring contractility in engineered cardiovascular muscle. Biomaterials 2010; 31: 3613–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alford PW, Taber LA. Computational study of growth and remodelling in the aortic arch. Comput Methods Biomech Biomed Eng 2008; 11: 525–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardel ML, Nakamura F, Hartwig JH, Crocker JC, Stossel TP, Weitz DA. Prestressed F-actin networks cross-linked by hinged filamins replicate mechanical properties of cells. Proc Natl Acad Sci USA 2006; 103: 1762–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shah JV, Janmey PA. Strain hardening of fibrin gels and plasma clots. Rheol Acta 1997; 36: 262–8. [Google Scholar]
- 26.Storm C, Pastore JJ, MacKintosh FC, Lubensky TC, Janmey PA. Nonlinear elasticity in biological gels. Nature 2005; 435: 191–4. [DOI] [PubMed] [Google Scholar]
- 27.Chitaley K, Webb RC. Microtubule depolymerization facilitates contraction of vascular smooth muscle via increased activation of RhoA/Rho-kinase. Med Hypotheses 2001; 56: 381–5. [DOI] [PubMed] [Google Scholar]
- 28.Zhou J, Kim HY, Wang JH, Davidson LA. Macroscopic stiffening of embryonic tissues via microtubules, RhoGEF and the assembly of contractile bundles of actomyosin. Development 2010; 137: 2785–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazumder A, Roopa T, Basu A, Mahadevan L, Shivashankar GV. Dynamics of chromatin decondensation reveals the structural integrity of a mechanically prestressed nucleus. Biophys J 2008; 95: 3028–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krause M, Te Riet J, Wolf K. Probing the compressibility of tumor cell nuclei by combined atomic force-confocal microscopy. Phys Biol 2013; 10: 065002–065002. [DOI] [PubMed] [Google Scholar]
- 31.Humphrey D, Duggan C, Saha D, Smith D, Kas J. Active fluidization of polymer networks through molecular motors. Nature 2002; 416: 413–6. [DOI] [PubMed] [Google Scholar]
- 32.Goddette DW, Frieden C. Actin polymerization. The mechanism of action of cytochalasin D. J Biol Chem 1986; 261: 15974–80. [PubMed] [Google Scholar]
- 33.Brangwynne CP, MacKintosh FC, Kumar S, Geisse NA, Talbot J, Mahadevan L, Parker KK, Ingber DE, Weitz DA. Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement. J Cell Biol 2006; 173: 733–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothen-Rutishauser BM, Ehler E, Perriard E, Messerli JM, Perriard JC. Different behaviour of the non-sarcomeric cytoskeleton in neonatal and adult rat cardiomyocytes. J Mol Cell Cardiol 1998; 30: 19–31. [DOI] [PubMed] [Google Scholar]
- 35.Hameed FM, Soni GV, Krishnamurthy H, Shivashankar GV. Probing structural stability of chromatin assembly sorted from living cells. Biochem Biophys Res Commun 2009; 385: 518–22. [DOI] [PubMed] [Google Scholar]
- 36.Sengupta N, Seto E. Regulation of histone deacetylase activities. J Cell Biochem 2004; 93: 57–67. [DOI] [PubMed] [Google Scholar]
- 37.Vanhaecke T, Papeleu P, Elaut G, Rogiers V. Trichostatin A-like hydroxamate histone deacetylase inhibitors as therapeutic agents: toxicological point of view. Curr Med Chem 2004; 11: 1629–43. [DOI] [PubMed] [Google Scholar]
- 38.Ingber DE. Tensegrity II. How structural networks influence cellular information processing networks. J Cell Sci 2003; 116: 1397–1408. [DOI] [PubMed] [Google Scholar]
- 39.Stamenovic D. Effects of cytoskeletal prestress on cell rheological behavior. Acta Biomater 2005; 1: 255–62. [DOI] [PubMed] [Google Scholar]
- 40.Steward RL, Jr, Cheng CM, Wang DL, LeDuc PR. Probing cell structure responses through a shear and stretching mechanical stimulation technique. Cell Biochem Biophys 2010; 56: 115–24. [DOI] [PubMed] [Google Scholar]
- 41.Hu SH, Chen JX, Butler JP, Wang N. Prestress mediates force propagation into the nucleus. Biochem Biophys Res Commun 2005; 329: 423–8. [DOI] [PubMed] [Google Scholar]
- 42.Panorchan P, Schafer BW, Wirtz D, Tseng Y. Nuclear envelope breakdown requires overcoming the mechanical integrity of the nuclear lamina. J Biol Chem 2004; 279: 43462–7. [DOI] [PubMed] [Google Scholar]
- 43.Discher DE, Janmey P, Wang Y-L. Tissue cells feel and respond to the stiffness of their substrate. Science 2005; 310: 1139–43. [DOI] [PubMed] [Google Scholar]