Abstract
Mangrove sediments were collected from major mangrove stands on the Red Sea Coast of Saudi Arabia. Forty five isolates belonging to 12 genera were purified and five isolates as well as their consortium were found to be able to grow in association with petroleum oil as sole carbon source under in vitro conditions. The isolated strains were identified based on internal transcribed spacer (ITS) rDNA sequence analysis. The fungal strains with the greatest potentiality to degrade diesel oil, without developing antagonistic activity, were identified as Alternaria alternata, Aspergillus terreus, Cladosporium sphaerospermum, Eupenicillium hirayamae and Paecilomyces variotii. As compared to the controls, these fungi accumulated significantly higher biomass, produced extracellular enzymes and liberated larger volumes of CO2. These observations with GC–MS data confirm that these isolates displayed rapid diesel oil bioremoval and when used together as a consortium, there was no antagonistic activity.
Keywords: Diesel fuel, ITS, Biodegradation, Mangrove sediments, Saudi Arabia
1. Introduction
Diesel fuel obtained by the distillation of crude oil has a carbon range between C8 and C26 (Adam and Duncan, 1999) with high content of polyaromatic hydrocarbons (Wang et al., 1990). The toxicity of diesel to ecosystems is more as compared to crude oil, because of diesel’s higher content of light-hydrocarbons. Although diesel is a commonly used fuel for vehicles and machines, it is recognized as a serious threat to ecosystems (Jagtap et al., 2014).
Diesel hydrocarbons can accumulate in food chains at various levels where they disrupt biochemical or physiological processes of many organisms; thus causing carcinogenesis of some organs, mutagenesis in the genetic material, and impairment in reproductive capacity, and hemorrhages in exposed population (Janani Prathiba et al., 2014).
Bioremediation has proved to be the most promising, practical and economical method for the complete mineralization of hydrocarbons to carbon dioxide and water (Wang et al., 2015).
The need for remediating polluted areas has induced development of new technologies to detoxify contaminants not only through chemical or physical methods, but through biological techniques as well. Bioremediation comprises a set of technologies that make the removal of contaminants possible or render them less harmful by means of biological activity (Silva et al., 2015).
Many isolated bacterial and fungal species have been reported to be capable of biodegrading petroleum hydrocarbons and even polynuclear aromatic hydrocarbons effectively (Márquez-Rocha et al., 2005). Laboratory studies provide greater control and manipulation in providing a basis to distinguish between biotic and abiotic processes, and to determine the optimal conditions for biodegradation (Li et al., 2008).
It is known that petroleum hydrocarbons can be removed by microorganisms such as fungi belonging to the genera Aspergillus, Penicillium, Fusarium, Amorphotheca, Neosartorya, Paecilomyces, Talaromyces, Graphium, yeasts which includes Candida, Yarrowia and Pichia and microalgae (Chaillan et al., 2004). This study attempted to assess the ability of fungi isolated from mangrove stands on the Red Sea Coast of Saudi Arabia to degrade diesel oil under in vitro conditions.
2. Materials and methods
2.1. Reagents
Clean diesel oil was procured from Saudi Aramco. Diethyl either (BDH) used as solvent for preparation of samples for GC–Mass analysis was obtained from the local market.
2.2. Collection of samples
Samples for isolation of fungi were collected form mangrove stands located near Jeddah and Jazan cities on the Red Sea Coast of Saudi Arabia and Farasan Island 40 km off the shore. Floating debris, tidal water, and sediment under the mangrove plants were collected as samples for isolation of fungi. The debris consisted of dead fallen leaves, pieces of pneumatophores, bark and wood of mangrove plants. The samples were collected in sterilized specimen tubes and were brought to lab in ice.
2.3. Isolation and purification of fungi
Fungi were isolated from wood debris by splitting the specimen into smaller pieces and directly laying on a synthetic mineral salt medium (MSM) formulated to simulate seawater conditions based on the compositions of Ameen et al. (2014) with some modification. Composition of the medium per liter was: MgSO4 246.5 mg/l, FeSO4·7H2O 5.56 mg/l, ZnSO4·7H2O 0.29 mg/l, MnSO4·H2O 0.34 mg/l, CuSO4·5H2O 0.025 mg/l, NH4Cl 5.35 mg/l, KCl 7.46 mg/l, CaCl2·2H2O 1.47 mg/l, NaCl 5.84 mg/l, COCl2·6H2O 0.027 mg/l, KH2PO4 136 mg/l, Na2MoO4 24 mg/l, and dextrose 20 g/l. The medium was gelled with 15 g/l agar and the pH adjusted to 5.5 before autoclaving. Tidal water and sediment suspension were layered on the same medium by the standard dilution method. The plates were incubated at 30 ± 2 °C for 5–7 days. Pure cultures were obtained by sub culturing inoculum from young colonies developing on the initial medium.
2.4. Identification of fungal isolates
The selected pure fungal isolates were subcultured from the MSM agar plates and inoculated into the potato dextrose agar (PDA) to harvest more mycelia. Total genomic DNA of the isolates was extracted according to the method of Wiese et al. (2011). The isolated strains were identified on the basis of internal transcribed spacer (ITS) rDNA sequence analysis. For sequence analysis, the ITS1-5.8S-ITS4 rDNA gene of the fungus was amplified using PCR with primer set pITS1 (5′ TCCGTAGGTGAACCTGCCG-3′) and pITS4 (5′ TCCTCCGCTTATTGATATGC-3′) (Al-Nasrawi, 2012). The 550-bp amplicon obtained was cloned and sequenced. DNA sequence of the positive clones with 18S rDNA gene fragment was compared with those available on the database using the BLAST program at the National Center for Biotechnology Information (NCBI) and analyzed.
2.5. Biomass determination on diesel fuel hydrocarbons
Fungal biomass was determined by filtering the culture broth through Whatman No. 1 filter paper. Recovered biomass samples were weighed and dried in pre-weighed aluminum foil at 60° C to a constant weight and the l dry mass was obtained by subtraction. Three replicate flasks were maintained for each fungal isolate. For each treatment, three flasks with corresponding inoculum but without diesel fuel substrate were retained as controls. Gain in biomass under each treatment and the corresponding control was recorded; and difference between gain in treatment and control was considered to be due to biodegradation activity of the fungus.
2.6. Enzyme assays
Five fungal isolates, which showed an increase in total dry weight under treatment were co-cultivated with diesel fuel in replicates along with the controls as mentioned above. As a separate treatment, consortium of the five above isolates was also tested for enzyme activity. After 4 weeks of incubation, enzyme level in the medium was assayed for controls and treatments. Cultures were centrifuged at 10,000 rpm, 4 °C; the pellet consisting of fungal cells was discarded and enzyme level was determined in the extracellular fluids. Catalase activity was determined according to Aebi (1983) by measuring decomposition of H2O2 and decline in absorbance at 240 nm in a 3 min period. The reaction mixture contained 50 mM phosphate buffer (pH 7.0), 15 mM H2O2, and 0.1 ml of enzyme extract in final volume of 3 ml. Laccase was estimated by oxidation of 2,2-Azino-bis-3-ethyl-benzthiazoline-6-sulfonic acid (ABTS) according to Novotny et al. (1999) using 0.1 mM ABTS in the reaction buffer of 100 mM sodium tartrate (pH 4.5) with 50 μl culture filtrate. One unit (U) of laccase activity was defined as the production of 1 μmol product per min at 30 °C and pH 4.5. Manganese-dependent peroxidase (MnP) was estimated by using 0.01% phenol in the presence of 0.1 mM H2O2 and 1 mM MnSO4 in 100 mM sodium tartrate (pH 4.5); while lignin peroxidase (LiP) was determined by the oxidation of 2 mM veratryl alcohol in 100 mM sodium tartrate (pH 4.5) with 0.4 mM H2O2 (Paszcymski et al., 1988). All enzymes assayed in this study were expressed as U/ml.
2.7. Evolution of CO2
Fungal isolates and diesel fuel were co-cultivated as mentioned above and quantity of CO2 evolved was estimated for an incubation period of 4 weeks. Three flaks were maintained for each isolate together with corresponding control flasks inoculated with the fungus but devoid of diesel. Volumetric and gravimetric estimation of CO2 evolved during 4-week incubation was performed using Sturm test (Sturm, 1973). For gravimetric analysis sterile air was sequentially passed through 1 M KOH solution to remove atmospheric CO2 and then through the treatment flasks. The bubbling air provided aeration for the fungal activity and at the same time allowed any CO2 evolving from the fungal activity to dissolve readily in the broth. The test was performed at room temperature (26 ± 2 °C). Amount of CO2 dissolved in the broth was estimated by adding 100 ml of 0.1 M BaCl2 that formed precipitate of barium carbonate; and CO2 released was gravimetrically calculated by measuring the weight of the precipitate. Difference in the values obtained between control and test bottles was recorded.
For volumetric analysis, the dissolved carbon dioxide present in the medium was estimated by titration (Sturm, 1973). The broth was filtered to remove fungal mass and the diesel fuel, then 25 ml filtrate was taken in a conical flask to which 0.05 ml of 0.1 N thiosulphate solution was added. After the addition of 2 drops of methyl orange indicator, solution was titrated against 0.02 M sodium hydroxide solution. End point appeared as a change in color from orange red to yellow. After this, two drops of phenolphthalein indicator were added and titration was continued until pink color was observed. Volumes of the titrant used were noted and the amount of CO2 evolved was calculated using the formula:
where A = volume of NaOH titrant in ml, B = normality of NaOH, and V = volume of sample in ml.
2.8. Preparation of diesel samples for GC–MS analysis
For sample preparation, 5 ml of sample was transferred into a 50 ml separatory funnel, and 5 ml of diethyl ether was added to it (Sanyaolu et al., 2012). The sample was shaken vigorously for about 2 mins with periodic venting to release vapor pressure. The organic layer was allowed to separate for 10 mins and was recovered into the 50 ml beaker. The aqueous layer was re-extracted twice with 2 ml of the extractant. The combined extract was dried by passing through the funnel containing the anhydrous sodium sulfate. The dried extract was concentrated with a stream of nitrogen gas.
PerkinElmer Mass Spectrometer with a HP-5MS column was used for analysis using the method of Wu et al. (2010) with minor modification. For analysis, a 30 mm – 0.25 mm (internal dia.) – 0.25 μm (particle size) fuse-silica capillary column (5MS – HP Inc.) was employed. The column temperature program was set as follows: 100 °C hold for 1 min, 15 °C/min to 160 °C at 5 °C/min to 300 °C hold for 7 min. The GC injector was held isothermally at 280 °C with a split less period of 3 min. The solvent delay time was set at 5 min. Helium was used as the carrier gas, at a flow rate of 1 ml/min by using electronic pressure control. The GC/MS interface temperature was maintained at 280 °C. The MS was operated in electron impact (EI) ionization mode with electron energy of 70 eV, and scan ranged from 50 to 500 amu (atom to mass unit) to determine appropriate masses for selected ion monitoring. The MS ion source and mass filter (quad) temperatures were held at 230 °C and 150 °C respectively. To minimize the baseline shifting after a derivatizing reagent peak, the signal was turned off as soon as the derivatizing reagent appeared and turned on again after the derivatizing reagent was eluted. To increase sensitivity, selected ion monitoring (SIM) mode was used to quantitatively analyze the peak, and the molecular ion was detected and quantified with ion loss of the methyl group (mass 190 for lactone), the dwell time was 0.1 s, and the scan cycle was 4.26/s. All samples in the present study were analyzed in triplicates.
3. Results and discussion
3.1. Isolation and identification of isolates
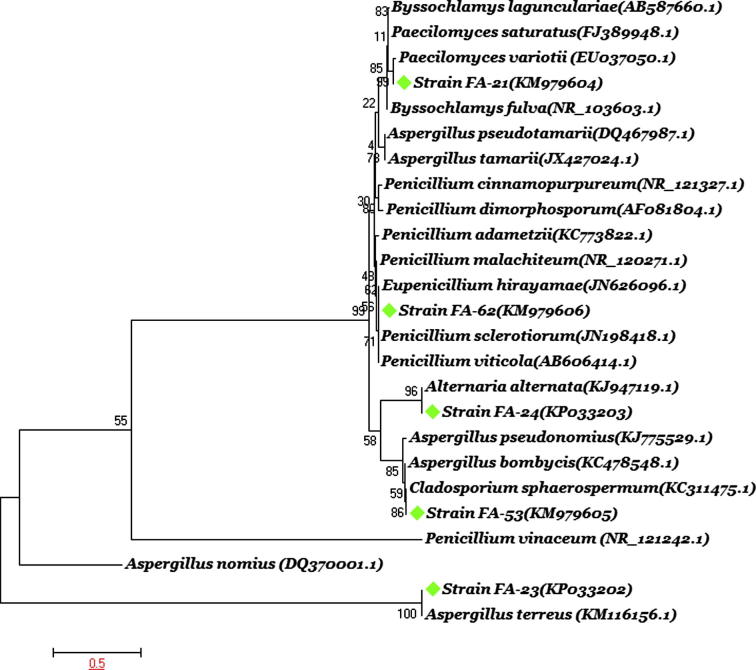
Forty five isolates were recovered from the samples. These included eight species each of Aspergillus and Penicillium; three species of Candida; and nine individual species of Acremonium, Alternaria, Emericella, Eurotium, Exophiala, Geosmithia, Paecilomyces, Pichia, and Cladosporium. These isolates were identified on the basis of morphology and phylogenetic analyses based on internal transcribed spacer (ITS) regions. Five isolates FA-21 (Paecilomyces variotii), FA-23 (Aspergillus terreus), FA-24 (Alternaria alternata), FA-53(Cladosporium sphaerospermum) and FA-62 (Eupenicillium hirayamae) as well as their consortium were selected for further assessment of biodegradation potential on the basis of superior growth and biomass accumulation during co-cultivation with diesel fuel. The sequences of the partial 18S rDNA gene fragments amplified from these strains were compared with the sequence data in the GenBank by an online alignment search (Table 1). The results indicated that the partial 18S rDNA sequence of isolates FA-24 and FA-53 was 100% identical to that of A. alternata and C. sphaerospermum (Accession No. KJ947119.1 and KC311475.1 respectively); while the partial 18S rDNA sequence of isolates FA-21, FA-23 and FA-62 was 99% identical to that of Paecilomyces variotii, Aspergillus terreus and Eupenicillium hirayamae respectively (Accession No. EU037050.1, KM116156.1 and JN626096.1). The sequences were submitted to NCBI GenBank and accession numbers were obtained. A phylogenetic tree based on the comparison of sequences shows overall relatedness of the identified isolates with other fungal genotypes (Fig. 1). Molecular analysis of fungal rDNA at the sequence level provides a powerful technique for assessing fungal diversity at the genus level; and PCR primers play a crucial role in the molecular assessment of environmental microbes. The specificity of the primer pairs is vital in this context to allow selective or enriching amplifications of fungal rDNA genes from environmental DNA (Pang and Mitchell, 2005).
Table 1.
Sequence-based identification of active fungal isolates.
| No. | Isolation code | Accession numbers | Closely related fungal sequence | Identity (%) | Coverage |
|---|---|---|---|---|---|
| 1 | FA-21 | KM979604 | Paecilomyces variotiiEU037050.1 | 99 | 507/513 |
| 2 | FA-23 | KP033202 | Aspergillus terreus KM116156.1 | 99 | 365/370 |
| 3 | FA-24 | KP033203 | Alternaria alternata KJ947119.1 | 100 | 488/488 |
| 4 | FA-53 | KM979605 | Cladosporium sphaerospermum KC311475.1 | 100 | 504/504 |
| 5 | FA-62 | KM979606 | Eupenicillium hirayamaeJN626096.1 | 99 | 481/482 |
Figure 1.
Phylogenetic dendrogram of fungal strains based on the ITS rDNA sequence. Numbers following the names of the strains are accession numbers of published sequences. The tree was constructed by neighbor-joining algorithm using maximum composite likelihood model. Bootstrap percentages from 1000 replicates are shown.
3.2. Dry weight accumulation by fungi under treatment with diesel fuel
During the fungal degradation of diesel, the investigated inocula grew within 15–20 days to form colonies of variable size and appearance. Profuse growth of Eupenicillium hirayamae, C. sphaerospermum, A. terreus, P. variotii and their consortium respectively was noticed around the diesel fuel (Fig. 2). These isolates showed greater accumulation of biomass as compared to their corresponding controls (Table 2). E. hirayamae gained the maximum weight of 43.4% followed by C. sphaerospermum (40%); whereas minimum weight gain (28%) was recorded in A. alternata. Several other studies, which have demonstrated biodegradation potential of different fungi, have also reported biomass accumulation under similar conditions. Hasan (2014) has reported a significant gain in fresh weight of Aspergillus niger and Rhizopus stolonifer in 10% kerosene broth, accumulating 0.530 g and 0.522 g dry weight respectively. A study by Lotfinasabasl et al. (2012) recorded that A. niger showed the largest colony diameter on medium with 20% kerosene among A. terreus, Rhizopus sp. and Penicillium sp.
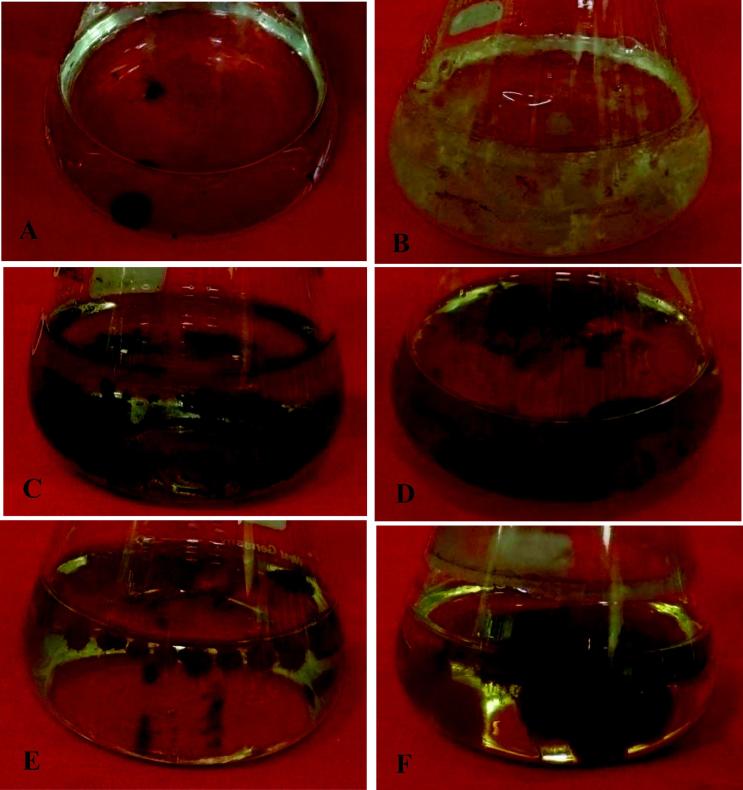
Figure 2.
Fungal growth in association with diesel fuel as carbon source: (A) Cladosporium sphaerospermum showing weak growth without carbon source (Control); (B) Eupenicillium hirayamae; (C) Cladosporium sphaerospermum; (D) Aspergillus terreus; (E) Paecilomyces variotii; and (F) Consortium of fungal strains – showing profuse growth with diesel.
Table 2.
Dry weight accumulation by fungal isolates during cultivation with diesel.
| Fungal isolate | Biomass (dry weight) comparison after 4-week co-cultivation⁎ |
|||
|---|---|---|---|---|
| Controls (g/l) | Treatments (g/l) | Gain via biodegradation |
||
| Weight (g/l) | (%) | |||
| Alternaria alternata | 0.642 ± 0.010 | 0.823 ± 0.058 | 0.181 | 28 |
| Aspergillus terreus | 0.625 ± 0.011 | 0.831 ± 0.046 | 0.206 | 32.9 |
| Cladosporium sphaerospermum | 0.599 ± 0.048 | 0.839 ± 0.092 | 0.240 | 40 |
| Eupenicillium hirayamae | 0.591 ± 0.049 | 0.848 ± 0.048 | 0.257 | 43.4 |
| Paecilomyces variotii | 0.594 ± 0.038 | 0.778 ± 0.045 | 0.184 | 30.9 |
| Consortium | 0.662 ± 0.027 | 1.034 ± 0.059 | 0.372 | 56 |
Data represent mean of three replicates ± Standard Deviation.
3.3. Enzyme activity of fungal isolates
Production of laccase, MnP, LiP and catalase enzymes was recorded in most of the control and treatment cultures of the five tested fungi as well as their consortium (Table 3). E. hirayamae produced the highest level of catalase (14.07 ± 0.99 U/ml) followed by C. sphaerospermum (11.4 ± 0.74 U/ml) which corroborates well with biomass accumulation in the same order (Table 2). Results showed that the activity of the enzymes had increased considerably as compared to the controls. Consortium of the five tested fungi showed remarkably higher level of enzymes as compared to the individual fungi. This may be due to synergistic effect of the pooled genotypes.
Table 3.
Enzyme activity of fungal isolates during co-cultivation with diesel as a sole carbon source.
| Fungal isolate | Laccase (U/ml) |
MnP (U/ml) |
LiP (U/ml) |
CAT (U/ml) |
||||
|---|---|---|---|---|---|---|---|---|
| Controls⁎ | Treatments | Controls | Treatments | Controls | Treatments | Controls | Treatments | |
| Alternaria alternata | 2.6 ± 0.84 | 6.4 ± 1.52 | 6.15 ± 1.58 | 11.25 ± 1.18 | ND | ND | ND | 6.17 ± 0.76 |
| ND | ND | ND | ||||||
| ND | ND | ND | ||||||
| Aspergillus terreus | 1.6 ± 0.61 | 4.49 ± 0.59 | 1.51 ± 0.73 | 4.08 ± 0.42 | 1.56 ± 0.65 | 3.53 ± 0.630 | 1.95 ± 0.92 | 7.73 ± 0.56 |
| Cladosporium sphaerospermum | 2.34 ± 1.20 | 3.13 ± 1.17 | ND | 4.08 ± 0.42 | 1.87 ± 1.08 | 8.32 ± 0.93 | 1.80 ± 0.66 | 11.4 ± 0.74 |
| ND | ||||||||
| ND | ||||||||
| Eupenicillium hirayamae | ND | 2.40 ± 0.54 | 2.56 ± 0.53 | 8.96 ± 1.12 | 2.38 ± 0.75 | 11.34 ± 1.16 | 1.88 ± 0.87 | 14.07 ± 0.99 |
| ND | ||||||||
| ND | ||||||||
| Paecilomyces variotii | ND | 2.88 ± 1.58 | 1.78 ± 0.63 | 3.97 ± 0.15 | ND | ND | 1.37 ± 0.65 | 7.83 ± 0.65 |
| ND | ND | ND | ||||||
| ND | ND | ND | ||||||
| Consortium | 9.8 ± 2.48 | 15.11 ± 0.84 | 20.27 ± 2.9 | 22.95 ± 2.49 | 6.04 ± 195. | 14.48 ± 2.06 | 6.33 ± 2.18 | 37.23 ± 8.53 |
Data represent mean of three replicates ± Standard Deviation; ND, not detected.
A microbial consortium provides a greater spectrum of enzyme activity in bioremoval since it involves metabolic expression of microorganisms belonging to each distinct taxon. Silva et al. (2015) reported that the microbial consortium consisting of bacteria and yeasts from a polluted environment showed high ability to degrade diesel oil constituents and maintenance of appropriate conditions led to transformation of the oily substances into less toxic compounds. Bioremediation of a high mass polymer and hydrocarbons has to be facilitated by extracellular enzymes released by the acting microorganism (Mohan and Srivastava, 2010, Zheng et al., 2005). Hence, elevated enzyme levels in our treatment broths were direct indication of the degenerative activity. Wu et al. (2010) investigated biodegradation potential of Fusarium solani strains against anthracene (ANT) and benz[a]anthracene (BAA) and recorded laccase to be the only active enzyme and found no traces of MnP and LiP. In general, level of catalase was highest among the four enzymes followed by laccase and LiP. Level of the four enzymes was conspicuously higher in cultures growing in association with diesel oil as compared to the controls; probably because, here the enzymes were in greater demand for hydrocarbon breakdown reactions. Enhanced activity of three enzymes, including catalase, was recorded by Mohsenzadeh et al. (2012) by the fungal strains, Acromonium sp., Alternaria sp., A. terreus and Penicillium sp., in the broth with different concentrations of petroleum pollutants. High activity of catalase was also reported in the soil microorganisms in petroleum-polluted soils (Ugochukwu et al., 2008). To the wood-decaying fungi, the average rates of polycyclic aromatic hydrocarbon bioremediation have been correlated with average activity of ligninolytic enzymes (Eibes et al., 2006). Ali et al. (2012) have reported that A. terreus isolated from Orman Garden soil and Penicillium chrysogenum isolated from Wadi Degla protectorate soil, exhibited lignin peroxidase and manganese peroxidase activity during bioremediation study of some PAHs, which is concurrent with our observations.
3.4. CO2 evolution due to fungal activity on diesel fuel
No significant difference was observed between CO2 evolution estimates taken by volumetric and gravimetric methods; therefore, data were merged as means of the two procedures for each fungal isolate (Table 4). E. hirayamae showed maximum enhancement of CO2 emission (72%) followed by C. sphaerospermum (65%) and A. terreus (61.8%). This trend closely matched with the trends of biomass accumulation and enzyme activity suggesting that a variable degree of bioremediation of diesel hydrocarbons has taken place by fungal activity reflected in corroborative levels of enzyme production, biomass accumulation, and CO2 liberation.
Table 4.
CO2 evolution during treatment of fungal isolates with diesel.
| Fungal isolate | Comparison of CO2 emission after 4-week co-cultivation⁎ |
|||
|---|---|---|---|---|
| Controls (g/l) | Treatments (g/l) | Enhancement due to biodegradation |
||
| Weight (g/l) | (%) | |||
| Alternaria alternata | 0.460 ± 0.003 | 0.684 ± 0.041 | 0.224 | 48.6 |
| Aspergillus terreus | 0.435 ± 0.005 | 0.704 ± 0.025 | 0.269 | 61.8 |
| Cladosporium sphaerospermum | 0.432 ± 0.018 | 0.715 ± 0.017 | 0.283 | 65 |
| Eupenicillium hirayamae | 0.448 ± 0.045 | 0.771 ± 0.050 | 0.323 | 72 |
| Paecilomyces variotii | 0.445 ± 0.004 | 0.669 ± 0.050 | 0.224 | 50.3 |
| Consortium | 0.493 ± 0.043 | 0.891 ± 0.070 | 0.398 | 80.7 |
Data represent mean of three replicates ± Standard Deviation.
Several CO2 evolution testd are used to study the assimilation of polymeric carbon (Muller et al., 1992, Zee et al., 1994, Chandra and Rustgi, 1998, Calmon et al., 2000); and liberation of carbon dioxide during the degradation of petroleum hydrocarbons can be used as a reliable indication of the fungal activity in the medium. Vanishree et al. (2014) evaluated the amount of CO2 released during biodegradation of petroleum hydrocarbons as an indicator of the activity of Aspergillus sp. Balba et al. (1998) have experimentally shown that mineralization studies involving measurements of total CO2 production could provide excellent information on the biodegradation of hydrocarbons in contaminated soils.
3.5. GC–MS analysis
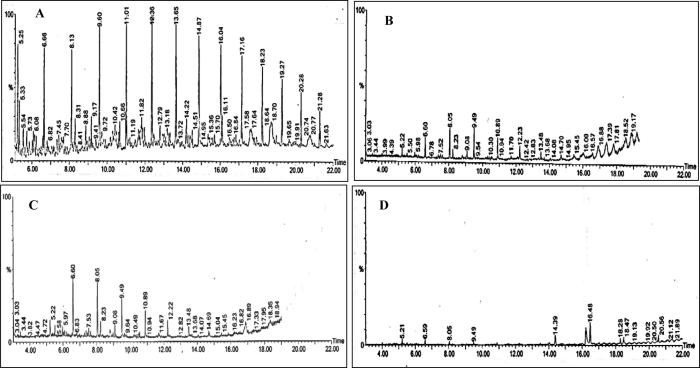
The gas chromatogram of diesel substrate retrieved from the inoculated medium at 0 and 30 days is shown in Fig. 3. It was found that the fungal strains as well as their consortium were efficient oil degraders. They completely degraded the main hydrocarbons (Tritetracontane, 1,2-benzenedicarboxylicacid,mono (2-ethylhexyl) ester and Tetrapentacontane 1,54-dibromo) present in diesel fuel leading to production of short chain compounds. Treated diesel substrate showed a decrease in the area of major peaks indicating breakdown of the main compounds; while new peaks appearing in these samples represented breakdown products or presumed metabolites. This phenomenon demonstrated the ability of the tested fungi to biodegrade multiple hydrocarbon compounds; which was not recorded in the control samples. This can be attributed to utilization of high mass fractions in diesel by the fungi for metabolic processes. Li et al. (2008) also used GC-data to analyze the biodegradation of diesel oil by filamentous fungi and found that Cladosporium strongly degraded diesel with a degradation ratio of up to 34% after 5 day treatment. Our results provided enough evidence that the tested fungal isolates could effectively degrade the diesel substrate.
Figure 3.
GC–MS analysis of diesel substrate: (A) Chromatogram of diesel before treatment (control); (B) Diesel compounds degraded by Eupenicillium hirayamae; (C) Chromatograph of diesel treated with Cladosporium sphaerospermum and (D) Chromatograph showing bio-removal of diesel components by consortium of fungi.
4. Conclusion
Five fungal isolates mentioned above displayed rapid diesel degradation ability, and when used together as a consortium, there was a synergistic effect that enhanced the degradation process. We also conclude that the laccase, MnP, LiP and catalase enzymes have a role in diesel fuel and alkane bio-removal by mangrove fungi.
Conflict of interest
None declared.
Acknowledgements
This work was supported by King Saud University, Deanship of Scientific Research, College of Science Research Center.
Footnotes
Peer review under responsibility of King Saud University.
References
- Adam G., Duncan H.J. Effect of diesel fuel on growth of selected plant species. Environ. Geochem. Health. 1999;21:353–357. [Google Scholar]
- Aebi H. Catalase. In: Bergmeyer H.U., editor. Methods of Enzymatic Analysis. third ed. Verlag Chemie; Weinheim: 1983. pp. 273–286. [Google Scholar]
- Ali M., Khalil N.M., Abd El-Ghany M. Biodegradation of some polycyclic aromatic hydrocarbons by Aspergillus terreus. Afr. J. Microbiol. Res. 2012;6:3783–3790. [Google Scholar]
- Al-Nasrawi H. Biodegradation of crude oil by fungi isolated from Gulf of Mexico. J. Biorem. Biodegrad. 2012;3:147. [Google Scholar]
- Ameen F., Moslem M.A., Hadi S., Al-Sabri A. Biodegradation of cellulosic materials by marine fungi isolated from South Corniche of Jeddah, Saudi Arabia. J. Pure Appl. Microbiol. 2014;8:3617–3626. [Google Scholar]
- Balba M.T., Al-Avadhi N., Daher R.A.l. Bioremediation of oil-contaminated soil microbiological methods for feasibility assessment and field evaluation. J. Microbiol. Methods. 1998;32:15–16. [Google Scholar]
- Calmon A.L., Bresson D., Maurel V.B., Feuilloley P., Silvestre F. An automated test for measuring polymer biodegradation. Chemosphere. 2000;41:645–651. doi: 10.1016/s0045-6535(99)00491-9. [DOI] [PubMed] [Google Scholar]
- Chaillan F., Fleche A Le., Bury E. Identification and biodegradation potential of tropical aerobic hydrocarbon degrading microorganisms. Res. Microbiol. 2004;155:587–595. doi: 10.1016/j.resmic.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Chandra R., Rustgi R. Biodegradable polymers. Prog. Polym. Sci. 1998;23:1273–1335. [Google Scholar]
- Eibes G., Cajthaml T., Moreira M., Feijoo G., Lema J. Enzymatic degradation of anthracene, dibenzothiophene and pyrene by manganese peroxidase in media containing acetone. Chemosphere. 2006;64:408–414. doi: 10.1016/j.chemosphere.2005.11.075. [DOI] [PubMed] [Google Scholar]
- Hasan I. Biodegradation of kerosene by Aspergillus niger and Rhizopus stolonifer. Appl. Environ. Microbiol. 2014;2:31–36. [Google Scholar]
- Jagtap S.S., Woo S.M., Kim T.S., Dhiman S.S., Kim D., Lee J. Phytoremediation of diesel-contaminated soil and saccharification of the resulting biomass. Fuel. 2014;116:292–298. [Google Scholar]
- Janani Prathiba G., Keerthi K., Aparna D., Sourav B., Priyadarsini R.I. Molecular identification of the isolated diesel degrading bacteria and optimization studies. J. Biochem. Technol. 2014;5:727–730. [Google Scholar]
- Li Y., Liu H., Tian Z., Zhu L., Wu Y., Tang H. Diesel pollution biodegradation: synergetic effect of Mycobacterium and filamentous fungi. Biomed. Environ. Sci. 2008;21:181–187. doi: 10.1016/S0895-3988(08)60026-4. [DOI] [PubMed] [Google Scholar]
- Lotfinasabasl S., Gunale V.R., Rajurkar N.S. Assessment of petroleum hydrocarbon degradation from soil and tarball by fungi. Biosci. Disc. 2012;3:186–192. [Google Scholar]
- Márquez-Rocha F.J., Olmos-Soto J., Rosano-Hernández M.C. Determination of the hydrocarbon-degrading metabolic capabilities of tropical bacterial isolates. Int. Biodeterior. Biodegrad. 2005;55:17–23. [Google Scholar]
- Mohan S.K., Srivastava T. Microbial deterioration and degradation of polymeric materials. J. Biochem. Technol. 2010;2:210–215. [Google Scholar]
- Mohsenzadeh F., Chehregani Rad A., Akbari M. Evaluation of oil removal efficiency and enzymatic activity in some fungal strains for bioremediation of petroleum-polluted soils. Iran. J. Environ. Health Sci. Eng. 2012;9:26. doi: 10.1186/1735-2746-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller R.J., Augusta J., Pantke M. An inter laboratory investigation into biodegradation of plastics; Part I: A modified Sturm-test. Mater. Org. 1992;27:179–189. [Google Scholar]
- Novotny C., Erbanova P., Sasek V., Kubatova A., Cajthaml T., Lang E., Krahl J., Zadrazil F. Extracellular oxidative enzyme production and PAH removal in soil by exploratory mycelium of white rot fungi. Biodegradation. 1999;10:159–168. doi: 10.1023/a:1008324111558. [DOI] [PubMed] [Google Scholar]
- Pang K.L., Mitchell J.I. Molecular approaches for assessing fungal diversity in marine substrata. Bot. Mar. 2005;48:332–347. [Google Scholar]
- Paszcymski A., Crawford R., Huynh V.B. Manganese peroxidase of Phanerochaete chrysosporium: purification. Methods Enzymol. 1988;161:264–271. [Google Scholar]
- Sanyaolu A.A., Sanyaolu V.T., Kolawole-Joseph O.S., Jawando S.S. Biodeterioration of premium motor spirit (PMS) by fungal species. Int. J. Sci. Nat. 2012;3(2):276–285. [Google Scholar]
- Silva D.P., Cavalcanti D.D., de Melo E.V. Bio-removal of diesel oil through a microbial consortium isolated from a polluted environment. Int. Biodeterior. Biodegr. 2015;97:85–89. [Google Scholar]
- Sturm R.N.J. Biodegradability of nonionic surfactants: screening test for predicting rate and ultimate biodegradation. J. Oil Chem. Soc. 1973;50:159. doi: 10.1007/BF02640470. [DOI] [PubMed] [Google Scholar]
- Ugochukwu K.C., Agha N.C., Ogbulie J.N. Lipase activities of microbial isolates from soil contaminated with crude oil after bioremediation. Afr. J. Biotechnol. 2008;7:2881–2884. [Google Scholar]
- Vanishree M., Thatheyus A.J., Ramya D. Biodegradation of petrol using Aspergillus sp. Annu. Res. Rev. Biol. 2014;6:914–923. [Google Scholar]
- Wang X., Yu X., Bartha R. Effect of bioremediation on polycyclic aromatic hydrocarbon residues in soil. Environ. Sci. Technol. 1990;24:1086–1089. [Google Scholar]
- Wang X., Wang X., Liu M. Adsorption–synergic biodegradation of diesel oil in synthetic seawater by acclimated strains immobilized on multifunctional materials. Mar. Pollut. Bull. 2015;92:195–200. doi: 10.1016/j.marpolbul.2014.12.033. [DOI] [PubMed] [Google Scholar]
- Wiese J., Ohlendorf B., Blümel M., Schmaljohann R., Imhoff J.F. Phylogenetic identification of fungi isolated from the marine sponge Tethya aurantium and identification of their secondary metabolites. Mar. Drugs. 2011;9:561–585. doi: 10.3390/md9040561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Luo Z., Vrijmoed L. Biodegradation of anthracene and benz[a]anthracene by two Fusarium solani strains isolated from mangrove sediments. Bioresour. Technol. 2010;101:9666–9672. doi: 10.1016/j.biortech.2010.07.049. [DOI] [PubMed] [Google Scholar]
- Zee M.V.D., Sutsma L., Tan G.B., Tournois H., De Wit D. Assessment of biodegradation of water insoluble polymeric materials in aerobic and anaerobic aquatic environments. Chemosphere. 1994;28:1757–1771. [Google Scholar]
- Zheng Y., Yanful K.E., Bassi A.S. A review of plastic waste biodegradation. Crit. Rev. Biotechnol. 2005;25:243–250. doi: 10.1080/07388550500346359. [DOI] [PubMed] [Google Scholar]