Abstract
Background
Recently, conformational activation of ADAMTS13 was identified. This mechanism showed the evolution from a condensed and inhibited conformation, in which the proximal MDTCS and distal T2-CUB2 domains are in close contact with each other, to an activated structure due to ding with the von Willebrand factor (VWF).
Objectives
Identification of cryptic epitope/exosite exposure after conformational activation and of sites of flexibility in ADAMTS13.
Methods
The activating effect of 25 anti-T2-CUB2 antibodies was studied in the FRETS-VWF73 and the vortex assay. Cryptic epitope/exosite exposure was determined in ELISA and VWF binding assay. The molecular basis for flexibility was hypothesized through RADAR analysis, tested in ELISA using deletion variants and visualized using electron microscopy.
Results
Eleven activating anti-ADAMTS13 antibodies, directed against the T5-CUB2 domains, were identified in the FRETS-VWF73 assay. RADAR analysis identified three linker regions in the distal domains. Interestingly, identification of an antibody recognizing a cryptic epitope in the metalloprotease domain confirmed the contribution of these linker regions to conformational activation of the enzyme. The proof of flexibility around both the T2 and metalloprotease domains by electron microscopy furthermore supported this contribution. In addition, cryptic epitope exposure was identified in the distal domains, as activating anti-T2-CUB2 antibodies increased the binding to folded VWF up to ~3-fold.
Conclusion
Conformational activation of ADAMTS13 leads to cryptic epitope/exosite exposure in both proximal and distal domains, subsequently inducing increased activity. Furthermore, three linker regions in the distal domains are responsible for flexibility and enable the interaction between the proximal and the T8-CUB2 domains.
Keywords: ADAMTS13 protein, allosteric regulation, autoantibodies, protein conformation, von Willebrand factor
INTRODUCTION
Enzymes regulating blood coagulation and thrombosis are activated through proteolytic digestion of their zymogen forms. However, ADAMTS13 (A Disintegrin And Metalloprotease with ThromboSpondin type-1 repeats, member 13), an essential enzyme in the preservation of the balance between thrombus formation at the site of a vascular injury and spontaneous clot formation, lacks such a zymogen form. Consequently, its activity needs to be regulated in a different way.
The substrate of ADAMTS13, von Willebrand factor (VWF), initiates thrombus formation under high shear conditions and is synthesized as ultra-large (> 10,000 kDa) multimers (UL-VWF) by endothelial cells and megakaryocytes [1–3]. ADAMTS13 reduces the size of these UL-VWF multimers, as these are highly reactive in binding platelets and induce spontaneous microthrombus formation if not correctly processed [1]. A deficiency in ADAMTS13 leads to the microangiopathic disease thrombotic thrombocytopenic purpura (TTP).
ADAMTS13 exerts its proteolytic activity only when VWF undergoes a conformational change. Under normal physiological conditions, ADAMTS13 is not able to cleave the VWF scissile bond (Tyr1605-Met1606), which is hidden in the central β-sheet of the folded VWF A2 domain [4–6]. Exposure of this scissile bond is facilitated by force-induced unfolding of the A2 domain, thereby allowing cleavage. Unfolding of VWF occurs in flowing blood under high shear stress [7,8], in platelet-decorated VWF strings anchored to the endothelial surface [9,10] and in a growing thrombus [11]. Binding of ADAMTS13 to VWF, prior to cleavage, occurs through a multistep process. ADAMTS13 is composed of a metalloprotease (M), disintegrin-like (D), thrombospondin type-1 repeat (T), cysteine-rich (C), spacer (S), seven additional T and two CUB domains. The distal (carboxyterminal (C-terminal)) T2-CUB2 domains are important in binding to folded VWF [12–14], while multiple exosites in the catalytic proximal (aminoterminal (N-terminal)) MDTCS domains are crucial in binding to the unfolded A2 domain and perfect positioning of its catalytic site towards the cleavage site (reviewed by Crawley et al. [15]).
Interestingly, conformational activation of ADAMTS13 by VWF [16] and by activating anti-T2-CUB2 domain monoclonal antibodies (mAbs) [16,17] has been recently demonstrated. ADAMTS13 was shown to adopt both inactive and activated conformations. The inactive structure was characterized by interactions between the distal and proximal domains and appeared condensed in EM, while the reduced interaction in the activated structure appeared more elongated in EM. Conformational activation was furthermore in line with the observed hyperactivity of the enzyme devoid of the distal domains [18–20].
In this study, the mechanism of conformational activation was further investigated. First, this was performed through development of novel activating anti-T2-CUB2 mAbs and through identification of a novel anti-metalloprotease domain mAb recognizing an epitope hidden in the inactive condensed form of ADAMTS13. Second, regions responsible for flexibility were discovered that could account for the observed conformational activation of ADAMTS13.
METHODS
Generation of anti-ADAMTS13 mAbs and Fab fragments
A subset of the mAbs and their development have been described previously (Fig. 1A) [16,17,21–26]. Abs were biotinylated using the EZ-Link™ Sulfo-NHS-SS-Biotin kit (Pierce Biotechnology, Rockford, IL, A) and Fab fragments were prepared as described [27].
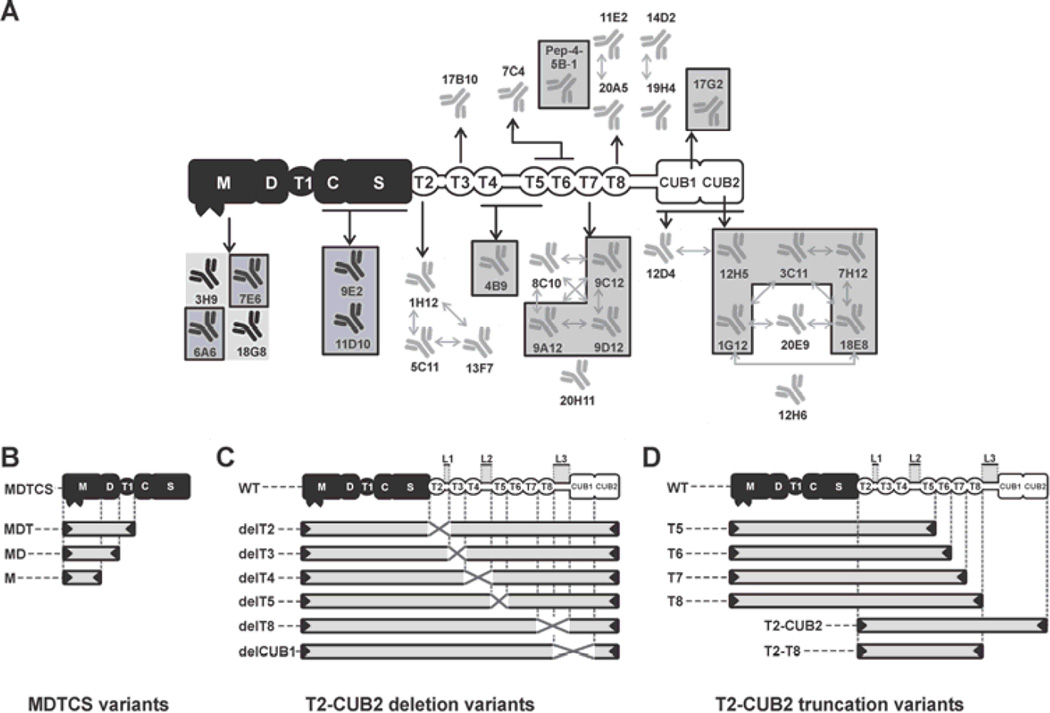
Fig. 1. Graphical representation of ADAMTS13 variants and epitope overview of the developed anti-ADAMTS13 mAbs.
The proximal MDTCS domains are represented in black and include a metalloprotease (M), disintegrin-like (D), thrombospondin type-1 repeat (T1), cysteine-rich (C) and spacer (S) domain. Distal (T2-CUB2) domains are represented in white and consist of seven thrombospondin type-1 repeats (T2 up to T8) and two CUB (Complement component C1r/C1s, Urinary epidermal growth factor (Uegf) and Bone morphogenic protein-1) domains. (A) Epitope overview of the reported and newly developed (indicated by a box) anti-ADAMTS13 mAbs. Abs with overlapping epitopes are indicated with arrows. (B, C and D) Graphical representation of the ADAMTS13 variants used in this study. (B) MDTCS variants MDTCS (W688X), MDT (Q499X), MD (W387X) and M (P285X). (C) WT and T2-CUB2 deletion (‘del’) variants. The linker regions between the T2–T3 domains (Linker 1), the T4–T5 domains (Linker 2) and the T8-CUB1 domains (Linker 3) are indicated by L1, L2 and L3 respectively. Deleted domains and linker regions are indicated by a cross. (D) T2-CUB2 truncation variants.
Construction, expression and purification of ADAMTS13 variants
Recombinant wild type human ADAMTS13 (WT rADAMTS13) was produced and purified as described [12]. MDTCS variants (Fig. 1B) were generated and expressed as described [26]. The T2-CUB2 deletion (Fig. 1C) and truncation (Fig. 1D) variants were constructed starting from the tetracycline-inducible pcDNA™4T/O vector (Invitrogen, Carlsbad, CA, USA) containing cDNA encoding WT ADAMTS13 with a C-terminal His- and V5-tag [28]. Deletion variants were prepared using inverse polymerase chain reaction (PCR). Sequences were verified (GATC Biotech AG, Konstanz, Germany) and T2-CUB2 deletion and truncation variants were expressed using the inducible T-REx system (Invitrogen), with exception of T2–T8 and T2-CUB2 that were expressed in CHO Flp-In cells (Invitrogen). Conditioned medium containing ADAMTS13 variants (except T2-CUB2 and T2–T8) was purified as described [12]. T2-CUB2 and T2–T8 were purified using a Ni2+-Sepharose Fast Flow column (GE Healthcare, Waukesha, WI, USA).
Epitope mapping of anti-ADAMTS13 mAbs
The mAbs were initially mapped against MDTCS and T2-CUB2. Binding to ADAMTS13 was used as a reference. Purified mAbs were coated individually on a 96-well microtiter plate. After blocking (3% milk in PBS), plates were incubated with a serial dilution of MDTCS, T2-CUB2 or ADAMTS13, starting at 15 nM (1 h at 37 °C). Binding was detected with the HRP-labeled anti-V5 mAb (Invitrogen, 1/3000 in PBS, 0.3% milk). Colorimetric development was done using OPD and H2O2 and was stopped using 4 M sulfuric acid, after which absorbance (490 nm) was determined. Epitope mapping of anti-MDTCS mAbs was refined using MDTCS variants (Fig. 1B) as described above, except that expression medium was used and that binding was detected with the biotinylated mAb 3H9 (3H9bio). Secondary detection was performed with HRP-conjugated streptavidin (1/10,000 in PBS, 0.3% milk). Epitopes of the anti-T2-CUB2 mAbs were mapped using the T2-CUB2 variants (Fig. 1C and D). Detection was done with anti-V5-HRP. Competition of the anti-T2-CUB2 mAbs directed against the same domain was tested by adding biotinylated mAbs (EC50), together with a dilution of the respective non-biotinylated mAb, to ADAMTS13 (8.6 nM), captured with 3H9 (5 µg/mL). Binding was detected with HRP-conjugated streptavidin.
Static ADAMTS13 proteolytic activity assay
The influence of anti-T2-CUB2 mAbs and their Fab fragments on ADAMTS13 activity was analysed using the FRETS-VWF73 assay (pH 7.4) [29].
Shear-dependent ADAMTS13 proteolytic activity assay
The effect of activating mAbs on cleavage of VWF under high shear conditions was analysed in the vortex assay [14,30]. ADAMTS13 (50 nM) in absence or presence of EDTA (10 mM) or mAb (40 µg/mL) was added to pVWF (8 µg/mL) under vortexing at 2500 rpm for 50 s. The multimeric pattern was determined as described [31].
The effect of activating mAbs on cleavage of platelet-decorated endothelial cell-anchored VWF strings under low shear conditions was analysed in the VWF string assay (shear rate of 250 s−1). The assay was performed as described [10] with minor modifications. After perfusion with platelets, coverslips were perfused with ADAMTS13 (1.25 nM) with or without the respective mAb (10 µg/mL) for 135 s.
ADAMTS13-VWF binding assay
The effect of anti-T2-CUB2 mAbs and their Fab fragments on binding of ADAMTS13 to folded VWF was studied in a binding assay [32]. Briefly, purified VWF (pVWF, 10 µg/mL, Haemate®P, CSL Behring GmbH, Marburg, Germany) was captured by the in-house developed anti-VWF A1 mAb 6D1. Subsequently, a dilution of anti-T2-CUB2 mAb (initially 10 µg/mL) and ADAMTS13 (18 nM), preincubated at 37 °C for 30 min (20 mM Tris-HCl (pH 7.8), 5 mM EDTA, 0.2% BSA and 1% mouse serum), was added. Binding was detected with mAbs 11D10bio and 18G8bio. Secondary detection was done as mentioned above.
Identification of T domains
The ADAMTS13 amino acid sequence (NCBI, Bethesda, MD, USA) was compared with the consensus sequence of thrombospondin type-1 repeat of thrombospondin-1 (PDB, La Jolla, CA, USA) using RADAR (Rapid Automatic Detection and Alignment of Repeats) analysis (EMBL-EBI, Cambridge, UK).
ADAMTS13-6A6 binding assay
Binding of ADAMTS13 (with or without the addition of an anti-T2-CUB2 Fab) or a T2-CUB2 variant to mAb 6A6 was studied using ELISA. 6A6 was coated at 5 µg/mL. After blocking, ADAMTS13 (15 nM) (pre-incubated for 30 min at 37 °C with or without a serial dilution of the respective Fab (10 µg/mL)) or a T2-CUB2 variant (15 nM) was added (1.5 h at 37 °C). Binding was detected with anti-V5-HRP.
Protein purification, negative staining, electron microscopy image collection and data processing
Fab fragments of mAbs 3H9 (3H9 Fab) and 5C11 (5C11 Fab) were prepared through papain digestion [27]. Fabs were purified on a MonoQ column (GE Healthcare) and eluted with a 0–100 mM NaCl gradient (30 min) in 20 mM Tris-HCl (pH 8.5). Fractions were analysed using SDS-PAGE and identified Fabs were pooled, concentrated, loaded on Superose6 (GE Healthcare) and eluted. Fabs were mixed with ADAMTS13 in a 4× molar excess for ≥ 1 h before loading on the electron microscopy (EM) grid. Negative staining, EM image collection and data processing were performed as described [33–35].
Statistical analysis
Slopes of the FRETS-VWF73 assay were fitted through linear regression. Data of the ADAMTS13-VWF binding assay were fitted with a one-site specific binding equation. Means were compared with the reference, using the Student’s t-test (parametric, unpaired, 1-tailed (VWF string assay) or 2-tailed). statistical applications were performed using the GraphPad Prism v5.03 software (GraphPad Software, Inc., San Diego, CA, USA).
RESULTS
Development and epitope mapping of anti-ADAMTS13 mAbs
We developed 14 new anti-ADAMTS13 mAbs by immunizing Balb/C mice with rADAMTS13. Binding of the MDTCS (Fig. 1B), T2-CUB2 deletion (Fig. 1C) and T2-CUB2 truncation (Fig. 1D) variants of ADAMTS13 to the respective mAb was performed using ELISA. Epitopes of the mAbs were subsequently determined by comparison of the binding and non-binding ADAMTS13 variants. Supplementary Fig. 1 (Fig. S1) shows the detailed epitope mapping of representative anti-ADAMTS13 mAbs and also includes the detailed epitope mapping of our previously generated mAbs. Fig. 1A shows a schematic overview of the epitopes of all developed mAbs.
All mAbs with an epitope in the same domain were checked for identical or overlapping epitopes using a competition assay. Cross-competing mAbs are represented by arrows in Fig. 1A. All anti-T2 mAbs had overlapping epitopes, hence representing one group of mAbs recognizing a similar region. Both anti-T7 and anti-T8 mAbs were divided into two groups, each recognizing different epitopes in the T7 (8C10, 9A12, 9C12 & 9D12 vs 20H11) and the T8 (11E2 & 20A5 vs 14D2 & 19H4) domain respectively. The anti-CUB mAbs could also be divided into several groups, each recognizing different epitopes (e.g. anti-CUB mAbs 12H6 and 17G2 did not cross compete with any other anti-CUB mAb). However, some mAbs have complex patterns of cross-reactivity (e.g. 18E8 did compete with both 3C11 and 20E9, but 3C11 did not compete with 20E9), which indicates that competing mAbs can have nonidentical, partially overlapping epitopes.
Conformational activation of ADAMTS13 by mAbs against the T5-CUB2 domains
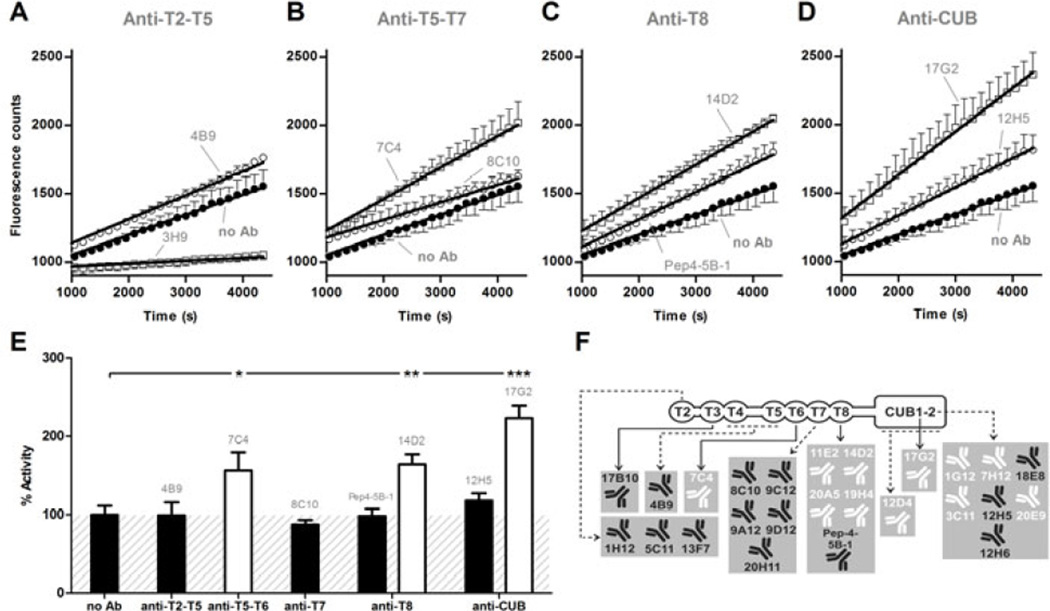
We previously reported that 5 mAbs directed against the T2-CUB2 domains stimulated the proteolytic activity of ADAMTS13 more than 1.5 times in the static FRETS-VWF73 assay [16,17]. These observations supported the hypothesis of allosteric activation of ADAMTS13. Here, we update these data and show that among our 10 newly developed anti-T2-CUB2 mAbs, 4 mAbs (anti-CUB1 mAb 17G2 and anti-CUB2 mAbs 1G12, 3C11 and 7H12) also stimulated the proteolytic activity. In our large panel of anti-T2-CUB2 mAbs (in total 25 mAbs), we have now identified 11 specific anti-T5-CUB2 mAbs that increased the proteolytic activity of plasma ADAMTS13 more than 1.3 times and up to two-fold (Fig. 2A–E). This activation was specific as the remaining anti-T2-CUB2 mAbs proved unable to stimulate the cleavage of the FRETS-VWF73 substrate (Fig. 2E). An overview of the functional effect of each mAb is given in Fig. 2F and Table S1. Fab fragments of activating mAbs (11E2 Fab and 12D4 Fab) increased the proteolytic activity equally well as their respective divalent mAbs (data not shown), hereby excluding the possibility that the increased activity could be due to crosslinking of ADAMTS13.
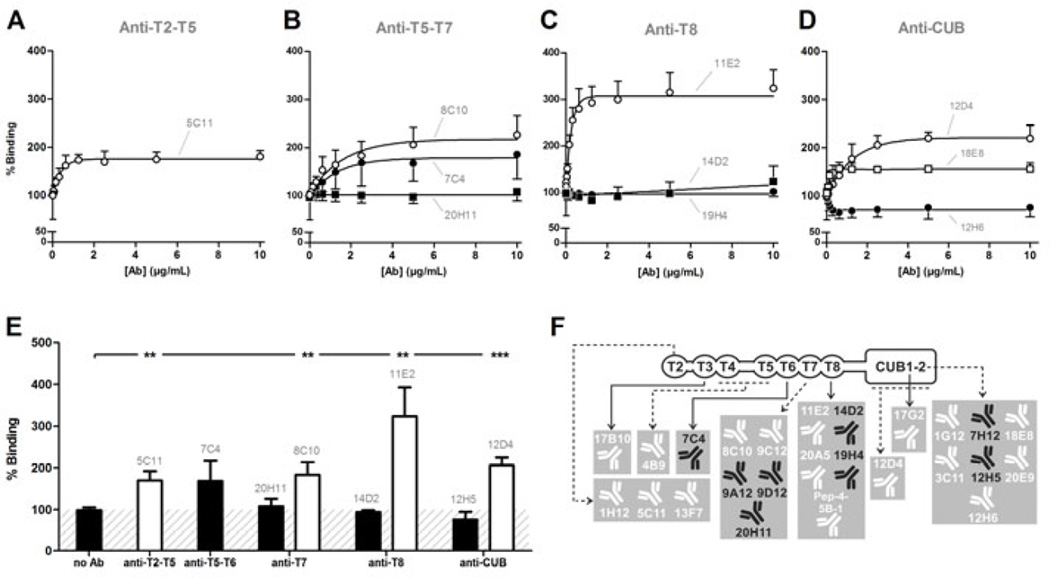
Fig. 2. Activation of ADAMTS13 by anti-T2-CUB2 mAbs under static conditions.
(A–E) The influence of anti-T2-CUB2 mAbs on the activity of plasma ADAMTS13 was measured using the static FRETS-VWF73 assay. Inhibiting anti-metalloprotease domain mAb 3H9 [23] was used as a negative control and the absence of a mAb (‘no Ab’) as a positive control. (A–D) Linear regression curves were determined for each anti-T2-CUB2 mAb. Non-functional mAbs 4B9 (A), 8C10 (B), Pep4-5B-1 (C) and 12H5 (D) are representative for all non-functional anti-T2-T5, anti-T5-T7, anti-T8 and anti-CUB mAbs, respectively. Likewise, activating mAbs 7C4 (B), 14D2 (C) and 17G2 (D) are representative for all activating anti-T5-T7, anti-T8 and anti-CUB mAbs, respectively. Error bars represent the SD of three independently performed experiments. (E) The activity rate of each anti-T2-CUB2 mAb was calculated. In line with A-D, representative values for the non-functional anti-T2-T5, anti-T7, anti-T8 and anti-CUB mAbs are given by mAbs 4B9, 8C10, Pep4-5B-1 and 12H5 respectively. For the functional anti-T5-T6, anti-T8 and anti-CUB mAbs, representative values are given by mAbs 7C4, 14D2 and 17G2 respectively. Differences between experiments in presence (e.g. 4B9) and absence of the mAb were statistically analysed using the Student t-test (P < 0.05 (*), P < 0.01 (**) and P < 0.001 (***)). The condition where no mAb was added was set as 100% activity. Error bars represent the SD of three independently performed experiments. (F) Overview of the functionality of all anti-T2-CUB2 mAbs (for detailed results, see Table S1). Non-functional and activating mAbs are represented in black and white, respectively.
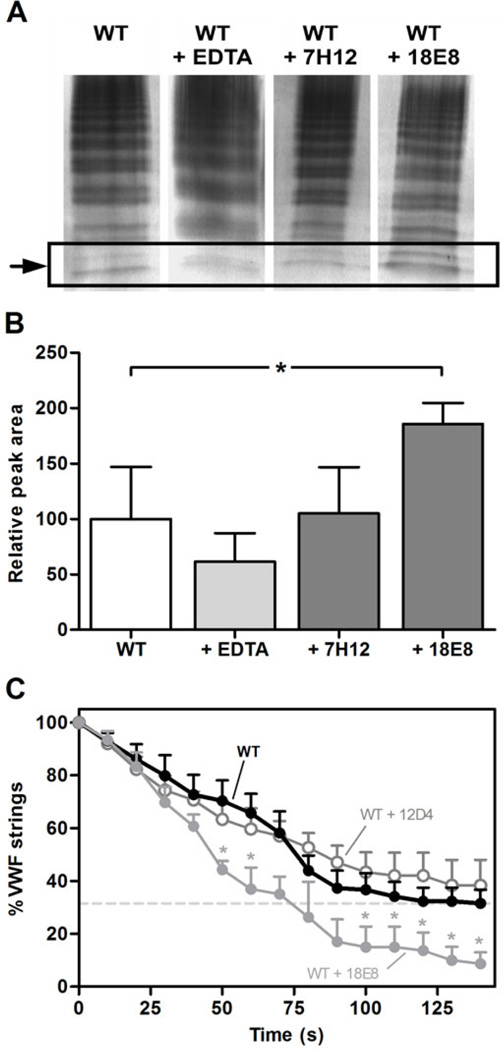
The activating effect of the mAbs was additionally tested under flow conditions. Under the high shear conditions (estimated shear rate of more than 12,000 s−1 [14]) of the vortex assay, we observed a 2-fold increase in ADAMTS13 proteolytic activity, induced by the anti-CUB2 mAb 18E8 (Fig. 3A–B), which is in line with the findings of Muia et al. [16]. Under the low shear conditions (shear rate of 250 s−1 [10]) of the VWF string assay, mAb 18E8 also proved its activating effect (Fig. 3C). Furthermore, these effects were specific as the anti-CUB2 mAb 7H12 and 12D4 exerted no effect in respectively the vortex (Fig. 3A–B) and VWF string assay (Fig. 3C).
Fig. 3. Activation of ADAMTS13 by anti-T2-CUB2 mAbs under shear conditions.
(A and B) Anti-CUB2 mAb 18E8 increases the proteolytic activity of ADAMTS13 under high shear conditions in the vortex assay. Recombinant ADAMTS13 (50 nM), in the absence or the presence of EDTA (10 mM) or anti- CUB2 mAbs 7H12 or 18E8 (both at 40 µg/mL), was added to pVWF (8 mg/mL) under constant vortexing at 2500 rpm for 50 s. Samples were loaded on a 2.5% SDS IEF agarose gel and the VWF multimers were visualized using the AP-labeled polyclonal anti-VWF Ab. (A) Cleavage product was identified as an additional band below the low molecular weight multimers (indicated by the arrow). (B) Quantification of the cleavage product was done using the ImageJ 1.48v software. A relative peak area of ‘100’ was attributed to the average intensity of the cleavage product in the presence of WT ADAMTS13. Differences were statistically analysed using the Student t-test. Error bars represent the SD of four independently performed experiments. (C) Anti-CUB2 mAb 18E8 increases the proteolytic activity of ADAMTS13 under low shear conditions in the VWF string assay. Platelet-decorated VWF strings were visualized using fluorescence microscopy on the surface of BOECs (Blood Outgrowth Endothelial Cells) perfused with washed DIOC6-labeled platelets at a shear rate of 250 s−1. Disappearance of the VWF strings was followed in fuction of time in the presence of 12D4, 18E8 (both at 10 µg/mL) or WT ADAMTS13 (1.25 nM) alone. Differences were statistically analysed using the Student t-test. Error bars represent the SEM of three independently performed experiments.
Conformational activation of ADAMTS13 exposes a cryptic site in its metalloprotease domain
As conformational activation of ADAMTS13 implies that distal T2-CUB2 domains are in close contact with the proximal MDTCS domains in the inactive conformation, we checked whether our panel of anti-ADAMTS13 mAbs contained mAbs that were directed against cryptic epitopes in the proximal domains.
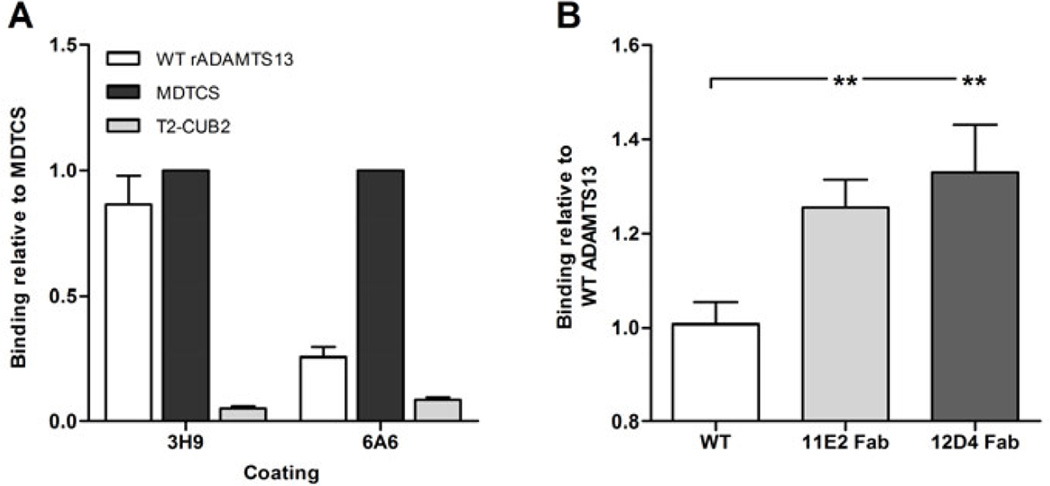
Therefore, we developed an immuno assay that allowed measurement of binding of either WT ADAMTS13 or MDTCS to the respective coated mAb. Interestingly, one mAb (anti-metalloprotease domain mAb 6A6) efficiently captured MDTCS but not WT ADAMTS13 (Fig. 4A). In contrast, anti-metalloprotease domain mAb 3H9 captured both ADAMTS13 and MDTCS equally well (Fig. 4A). Hence, these data suggest that the epitope of mAb 6A6 is shielded in WT ADAMTS13 by the distal domains.
Fig. 4. The cryptic epitope of anti-metalloprotease domain mAb 6A6 is exposed by conformational activation of ADAMTS13.
Binding of rADAMTS13, MDTCS and T2-CUB2 to 3H9 and/or 6A6 (both anti-metalloprotease domain mAbs) was tested in ELISA. Binding was detected with the anti-V5-HRP mAb. (A) Binding of rADAMTS13, MDTCS and T2-CUB2 to 3H9 and 6A6. Binding was calculated relative to the binding of MDTCS. (B) Binding of ADAMTS13, pre-incubated with the respective anti- T8 and anti-CUB mAb or Fab fragment, to 6A6. Binding was calculated relative to the binding of rADAMTS13. Differences were statistically analysed using the Student t-test. Error bars represent the SD of at least three independent experiments.
Conformational activation of ADAMTS13, induced by Fab fragments of activating mAbs 11E2 and 12D4, also resulted in the exposure of the 6A6 epitope in WT ADAMTS13 (Fig. 4B), hereby further supporting the hypothesis that 6A6 has a cryptic epitope in the metalloprotease domain that is exposed upon conformational activation of ADAMTS13.
Conformational activation of ADAMTS13 leads to exposure of cryptic sites in its distal domains
Exposure of cryptic exosites and epitopes in the proximal domains by conformational activation of ADAMTS13 subsequently implies exposure of cryptic exosites in the distal domains. However, a mAb recognizing a cryptic epitope in the distal domains was not yet identified from our current panel of mAbs. We hypothesized that when distal domains bind folded VWF [12,13], conformational activation of ADAMTS13 might expose additional VWF binding exosites and increase the affinity of binding to folded VWF. Therefore, binding of ADAMTS13 to folded VWF was studied in the presence or absence of anti-T2-CUB2 mAbs.
A subset of anti-T2-CUB2 mAbs indeed significantly increased the binding to folded VWF. Addition of anti-T2 (1H12, 5C11 and 13F7), anti-T3 (17B10), anti-T4-T5 (4B9), anti-T7 (8C10 and 9C12), anti-T8 (11E2, 20A5 and Pep4-5B-1) and anti-CUB (1G12, 3C11, 12D4, 12H6, 17G2, 18E8 and 20E9) mAbs led to a significant increase in binding (up to more than three-fold, Table S2) (Fig. 5A–E). The increase in ADAMTS13 binding was unlikely to be due to crosslinking of ADAMTS13 by the IgG mAbs because non-functional anti-T2-CUB2 mAbs were also identified (Fig. 5A–E). An overview of the functional effect of each mAb is given in Fig. 5F and Table S2. In addition, although 12D4 Fab exerted no stimulatory effect, 11E2 Fab did stimulate the binding to folded VWF. However, the magnitude of this effect was significantly reduced compared to mAb 11E2, due to the reduced affinity of 11E2 Fab for ADAMTS13 (data not shown).
Fig. 5. Anti-T2-CUB2 mAbs stimulate the binding of ADAMTS13 to folded VWF.
Folded pVWF was captured on the anti-VWF A1 domain mAb 6D1. Recombinant ADAMTS13, pre-incubated with the respective mAb (½ dilution series), was added to captured pVWF and detected with biotinylated anti-MDTCS mAbs and HRP-labeled streptavidin. The absence of mAb (‘no Ab’) was set as 100% binding. Error bars represent the SD of at least three independent experiments. Non-functional mAbs 7C4 and 20H11 (B), 14D2 and 19H4 (C) and 12H5 (D) are representative for all non-functional anti-T5-T6, anti-T7, anti-T8 and anti-CUB mAbs, respectively. Likewise, activating mAbs 5C11 (A), 8C10 (B), 11E2 (C), 12D4 and 18E8 (D) are representative for all activating anti-T2-T5, anti-T7, anti-T8 and anti-CUB mAbs, respectively. (E) The percentage binding induced by each anti-T2-CUB2 mAb at 10 µg/mL was calculated and compared with the percentage binding in the absence of the mAb, using the Student t-test. In line with A–D, mAbs 7C4, 20H11, 14D2 and 12H5 represent respectively the non-functional anti-T5-T6, anti-T7, anti-T8 and anti-CUB mAbs. In addition, all functional anti-T2-T5, anti-T7, anti-T8 and anti-CUB mAbs are represented by mAbs 5C11, 8C10, 11E2 and 12D4 respectively. Error bars represent the SD of three independently performed experiments. (F) Overview of the functional effect of all anti-T2-CUB2 mAbs (for details, see Table S2). Non-functional and activating mAbs are represented in black and white, respectively.
In summary, anti-T2-CUB2 mAbs stimulate the binding of ADAMTS13 to folded VWF. As T5-CUB2 domains have been shown to be involved in binding to folded VWF [12,13], conformational activation of ADAMTS13 by activating mAbs exposes additional exosites in the distal domains.
Flexibility in ADAMTS13 accounts for conformational activation
The identification of activating mAbs revealed exposure of cryptic exosites in both the proximal and distal domains. Furthermore, conformational activation implies the existence of specific regions that provide the observed flexibility of the enzyme.
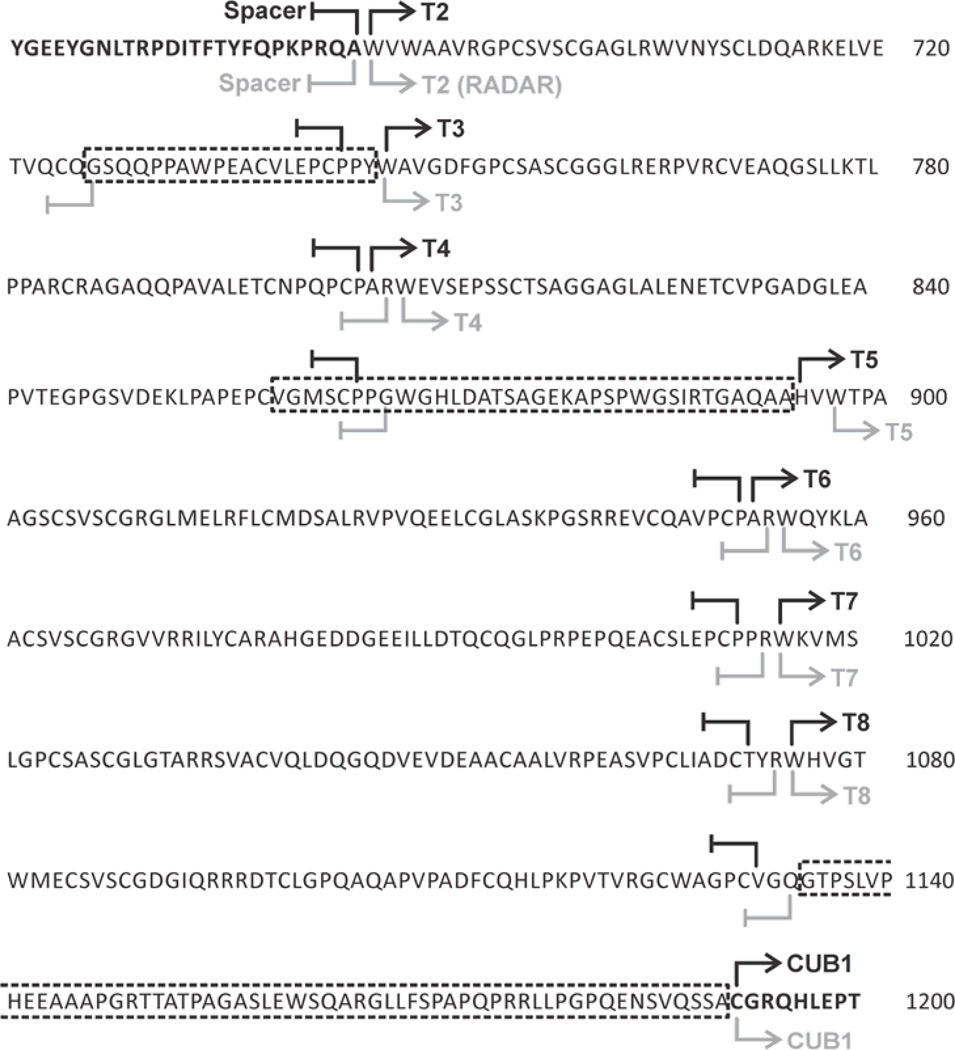
We hypothesized that linker regions between the T domains might account for the flexibility of ADAMTS13. Indeed, Carlson et al. [36] and Calzada et al. [37] showed that 4 additional residues in the linker region between the T1–T2 domains of thrombospondin-1 are likely to affect the flexibility of the protein. Hence, we used RADAR analysis to identify linker regions between the T domains of ADAMTS13. Through comparison of the amino acid sequence of ADAMTS13 with the consensus amino acid sequence of the thrombospondin type-1 repeats, we confirmed the boundaries of the seven T repeats in T2-CUB2 (Fig. 6), one linker region (L2) between the T4–T5 domains and another linker region (L3) between the T8-CUB1 domains (Fig. 1C) [38]. Interestingly, RADAR also identified a smaller third linker region (L1) between the T2–T3 domains (Fig. 6).
Fig. 6. The distal part of ADAMTS13 contains three linker regions.
The amino acid sequence of the distal domains is represented. Linker regions (framed with dotted lines) are located between the T2 and T3 domains (L1), the T4 and T5 domains (L2), and the T8 and CUB1 domains (L3). The current boundaries, defined by Zheng et al. [38], are represented in black and the boundaries defined by RADAR analysis are represented in gray.
Linker regions in the distal domains are responsible for flexibility
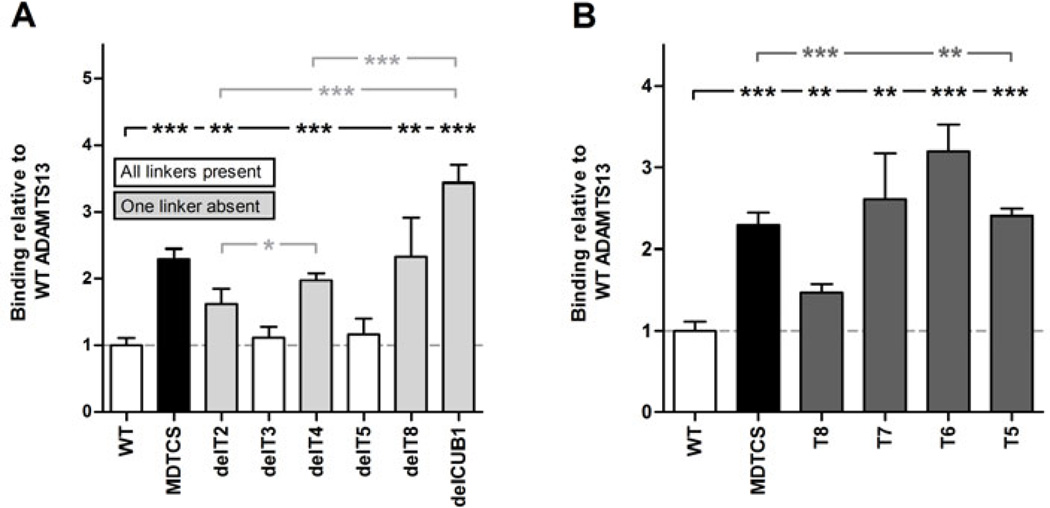
If linker regions indeed contribute to the flexibility of the distal domains, we hypothesized that some ADAMTS13 variants with deleted linker regions might have a less flexible (i.e. more elongated/active) conformation compared to WT ADAMTS13. Hence, binding of all available T2-CUB2 truncation and deletion variants (Fig. 1C and D) was investigated in the ADAMTS13-6A6 binding assay. Indeed, deletion variants devoid of one linker region (delT2, delT4, delT8 and delCUB1) showed increased binding to 6A6 compared to WT, while variants containing all linker regions (delT3 and delT5) did not (Fig. 7A). In addition, the amino acid length of the linker regions was positively correlated with the magnitude of binding to 6A6 (e.g. deletion of L3 (delT8 and delCUB1) led to a higher increase in binding than deletion of L2 (delT4) and L1 (delT2) (Fig. 7A). Hence removal of linker regions between distal domains leads to a more unshielded conformation of ADAMTS13.
Fig. 7. Exposure of the 6A6 cryptic epitope by removal of linker regions between the distal domains or deletion of T8-CUB2 domains.
Binding of rADAMTS13 and its variants to anti-metalloprotease mAb 6A6 was tested in ELISA. Binding was detected with the anti-V5-HRP mAb and calculated relative to the binding of rADAMTS13. (A) Binding of rADAMTS13, MDTCS and the T2-CUB2 deletion variants to 6A6. White bars represent rADAMTS13 and its variants which contain all three linker regions (L1, L2 and L3), while gray bars represent rADAMTS13 variants devoid of one of the three linker regions (see Fig. 1). (B) Binding of rADAMTS13, MDTCS and the T2-CUB2 truncation variants to 6A6. Differences were statistically analysed using the Student t-test. Error bars represent the SD of at least three independent experiments.
The identification of mAb 6A6, recognizing a cryptic epitope in the proximal metalloprotease domain, allowed us to provide biochemical evidence that the distal T7-CUB2 domains shield the proximal MDTCS domains. Our previous small angle X-ray scattering (SAXS) data showed that the T7-CUB2 domains could shield the active site and substrate binding sites in the proximal MDTCS domains [16]. Hence, gradual deletion of the distal domains from the C-terminus to the N-terminus was expected to result in increased binding to 6A6. Indeed, deletion of the CUB domains (T8 variant) significantly increased binding to 6A6 compared to WT (Fig. 7B). However, this increase was lower than the increase induced by deletion of all distal domains (MDTCS) (Fig. 7B), suggesting only partial exposure of the cryptic epitope in the T8 variant. Interestingly, deletion of the T8 domain resulted in complete exposure of the 6A6 cryptic epitope, as the variants devoid of the T8 domain bound 6A6 equally well (T5 and T7) or better (T6) than MDTCS (Fig. 7B). Consequently, both T8 and CUB domains are crucial in shielding the cryptic 6A6 epitope present in the proximal domains.
Electron microscopy reveals flexibility around the T2 and metalloprotease domains
Currently, little experimental information is available on the structure of full-length ADAMTS13. Quick-freeze deep-etch EM and negative stain EM showed many different conformations of ADAMTS13 with both condensed and elongated forms [16,17], implying a highly flexible structure. However, neither structural details nor the position of the proximal and distal domains were revealed in these studies.
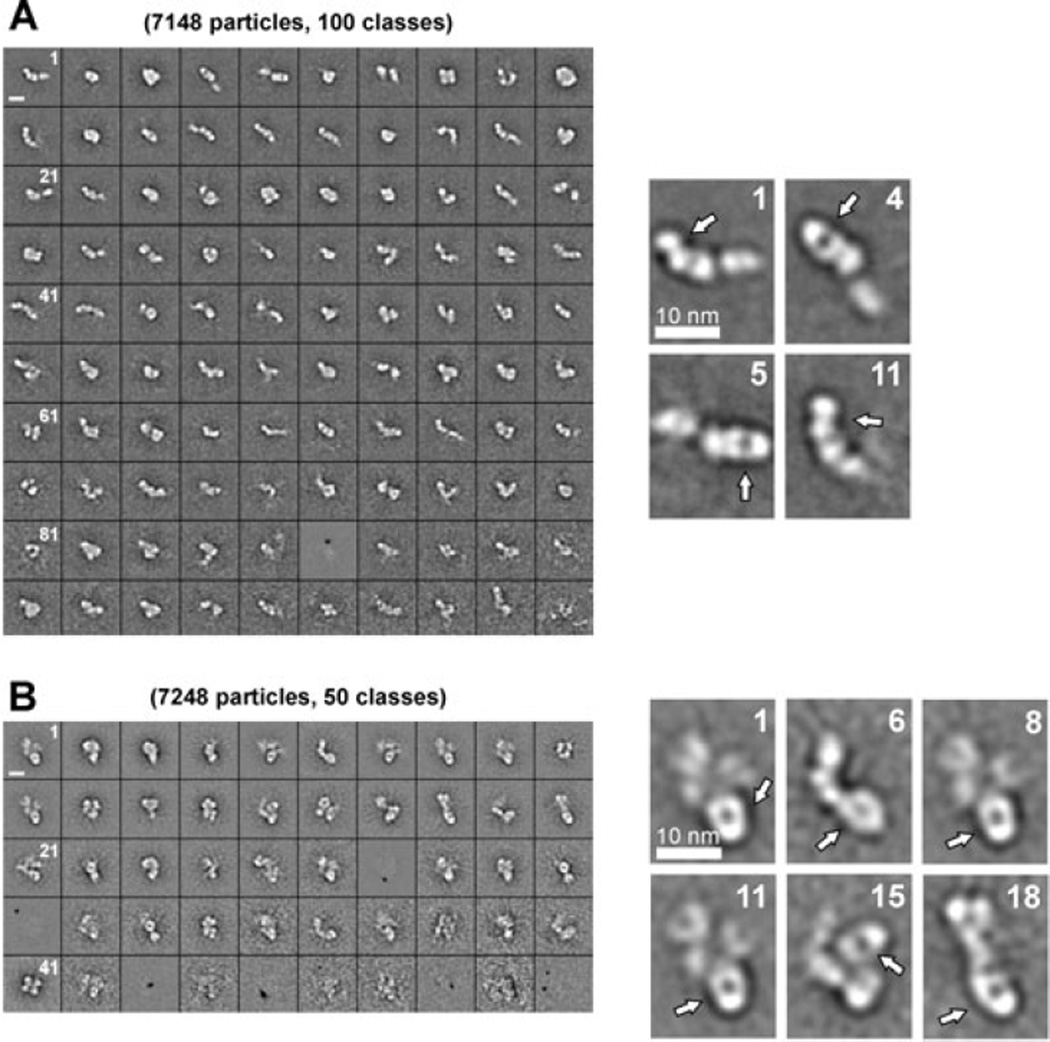
EM of purified rADAMTS13 indeed revealed the presence of a large number of different structures (Fig. S2). Therefore, in order to localize proximal and distal parts of ADAMTS13, EM images were made of ADAMTS13 in complex with the anti-metalloprotease 3H9 Fab fragment or the anti-T2 5C11 Fab fragment (Fig. 8). Class averages of ADAMTS13-3H9 Fab complexes allowed clear identification of the Fab fragment bound to the metalloprotease domain. For example, in class averages 1 and 11, the 3H9 Fab fragment is bound to the grid on its side, while in class averages 4 and 5, it is bound flat to the grid (Fig. 8A). Despite the clear observation of the metalloprotease domain, the domains C-terminally located from this domain could not be clearly identified, indicating a degree of flexibility around this domain. In line with 3H9 Fab, the anti-T2 5C11 Fab was clearly visible in class averages 1, 6, 8, 11, 15 and 18 (Fig. 8B). The structure surrounding the Fab fragments could not be well-defined and impaired identification of the domains N-terminally and C-terminally located from the T2 domain.
Fig. 8. EM analysis reveals flexibility around the T2 and metalloprotease domain.
EM images of purified rADAMTS13, pre-incubated with anti-metalloprotease domain 3H9 Fab (A) or anti-T2 5C11 Fab (B), were made and respectively 100 and 50 class averages of the different structures of ADAMTS13 were selected. For both conditions, magnified class averages are shown. The respective Fab fragment was easily identified due to its characteristic round shape (indicated with the white arrow). (A) Domains C-terminal from the metalloprotease domain were difficult to identify, icating flexibility around this domain. (B) Domains N- and C-terminal from T2 are visible but could not be identified, also indicating flexibility around the T2 domain. Scale bars, 10 nm.
The observed flexibility around the T2 domain supports the biochemical data showing that linker regions in the distal domains contribute to the flexibility of ADAMTS13. Additionally, EM reveals flexibility around the metalloprotease domain.
DISCUSSION
ADAMTS13 is expressed as a constitutively active enzyme whose activity is tightly regulated through conformational changes in its substrate VWF. Additional regulatory mechanisms of ADAMTS13 activity have already been described. On the one hand, both Gao et al. [20] and Di Stasio et al. [39] showed, using the short VWF73 substrate, that the proteolytic activity of ADAMTS13 is negatively regulated once sufficient concentrations of C-terminal VWF product (Met1606-Arg1668) are formed after cleavage of the Tyr1605-Met1606 scissile bond. On the other hand, the mechanism of substrate-induced disruption of the interaction between the proximal and distal domains of ADAMTS13, leading to allosteric activation of the enzyme, was recently described [16,17]. In this study, we further characterized the mechanism of conformational activation and identified flexible regions that allow the enzyme to adopt condensed inactive and elongated active conformations.
Among 31 anti-ADAMTS13 mAbs, including 14 unreported, we found 11 that activate the cleavage of FRETS-VWF73. Interestingly, the epitopes of these mAbs were all located in the C-terminal T5-CUB2 domains. The mAb-induced conformational activation of ADAMTS13 was additionally studied under high and low shear conditions, using respectively the vortex and the VWF string assay. Here, the increased activity induced by mAb 18E8 in both assays suggests a contribution of this mechanism under shear conditions.
Conformational activation of ADAMTS13 was previously linked to shielding of the proximal MDTCS domains by the distal T2-CUB2 domains [16,17] and consequently implies the existence of cryptic epitopes in both proximal and distal domains. Indeed, screening of all anti-MDTCS mAbs identified the anti-metalloprotease domain mAb 6A6 that had the ability to distinguish the condensed from the elongated conformations. This unique mAb bound WT ADAMTS13 ~4 times less efficiently than MDTCS, indicating that its epitope is hidden by the distal domains when ADAMTS13 adopts a condensed conformation. Addition of Fab fragments of activating mAbs significantly increased the binding to 6A6, hereby confirming the mechanism of mAb-induced conformational activation. Second, binding of ADAMTS13 to folded VWF is known to depend on its distal domains [12,13]. The existence of cryptic epitopes in the distal domains was also studied and confirmed as 17 anti-T2-CUB2 mAbs increased the binding of ADAMTS13 to folded VWF. Interestingly, mAbs 12D4 [16] and 18E8, which increased the activity under high shear conditions, also significantly increased binding to folded VWF (respectively ~2 and ~1.5-fold), while mAbs 7H12 and 19H4 [16], which exerted no effect under shear, also did not increase binding to folded VWF. These data show that mAb-induced exposure of additional exosites in the distal domains leads to increased proteolysis under shear conditions.
Conformational activation of ADAMTS13 suggests the presence of flexible regions in the enzyme. In line with the finding that linker regions between T domains of thrombospondin-1 likely affect flexibility [36,37], we here hypothesized that three linker regions between the distal T domains (Fig. 1C) contribute to the flexibility of ADAMTS13. This was confirmed as ADAMTS13 variants devoid of one of its linkers adopted a more elongated conformation than variants where all three linkers were present (Fig. 7A). In addition, our EM images of full-length ADAMTS13 in complex with the anti-T2 Fab fragment revealed flexibility around the T2 domain, hereby confirming the presence of the ible linker region L1. EM analysis also revealed flexibility around the metalloprotease domain. A linker region of 11 amino acids, connecting the metalloprotease and disintegrin-like domains, was identified in ADAMTS13, based on the crystal structures of ADAMTS1, 4 and 5 [40–43]. Hence, this linker is likely responsible for the observed flexibility in our EM data. This flexibility might also explain why the crystal structure of the DTCS domains but not of the MDTCS domains of ADAMTS13 has been resolved [43].
Co-immunoprecipitation experiments revealed an interaction between the CUB and spacer domains [17]. SAXS data supported the CUB-spacer interaction and additionally revealed interactions between the T8 and the remaining MDTC domains [16,44]. Our study provides novel biochemical evidence supporting the T8 interaction with the MDTC domains, as the cryptic epitope of the antimetalloprotease domain mAb 6A6 became accessible when the T8 to CUB2 domains were deleted (Fig. 7B).
In conclusion, we have identified new features in the mechanism of ADAMTS13 activity regulation through conformational activation. Three linker regions between the distal domains were shown to contribute to the flexibility of these domains, allowing shielding of the proximal domains. Furthermore, of all distal domains, both T8 and CUB domains are the main contributors in this shielding mechanism. In addition, we showed that VWF- or mAb-induced conformational activation results in cryptic exosite exposure in both proximal and distal domains, leading to increased proteolytic activity of ADAMTS13 under both static and shear conditions.
Supplementary Material
Acknowledgments
This work was funded by a Ph.D. grant of the Agency for Innovation by Science and Technology (IWT, www.iwt.be) Flanders, Belgium (IWT-SB/111507, awarded to L. Deforche) and a grant from the ‘Fonds voor Wetenschappelijk Onderzoek’ (FWO, www.fwo.be) Flanders Belgium (G.0548.11, awarded to K. Vanhoorelbeke). This work was also supported by National Institutes of Health Grants (grants.nih.gov/) R01 HL72917, R01 HL89746, U54 HL112303 and T32 HL007088 (awarded to J.E. Sadler). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We thank I. Pareyn for the development of the mAbs.
K. Vanhoorelbeke reports grants from ‘Fonds voor Wetenschappelijk Onderzoek’: Flanders Belgium, during the conduct of the study.
J. E. Sadler reports grants from National Institutes of Health, during the conduct of the study and personal fees from Baxter HealthCare, BioMarin and XO1 Limited outside the submitted work. In addition, J. E. Sadler has a patent for Fluorogenic substrate for ADAMTS13. U.S. Patent Number 8,663,912 issued.
L. Deforche reports grants from The Agency for Innovation by Science and Technology: Flanders, Belgium, during the conduct of the study.
Footnotes
ADDENDUM
L. Deforche and K. Vanhoorelbeke conceived and designed the experiments; L. Deforche, E. Roose, A. Vandenbulcke, N. Vandeputte, L. Z. Mi and K. Vanhoorelbeke performed the experiments; L. Deforche, E. Roose, T. A. Springer, L. Z. Mi and K. Vanhoorelbeke analysed the data; J. Muia, J. E. Sadler, K. Soejima and H. Rottensteiner contributed to reagens, materials and analysis tools; H. B. Feys, J. E. Sadler, H. Deckmyn and S. F. De Meyer critically reviewed the manuscript; L. Deforche and K. Vanhoorelbeke wrote the manuscript. All authors read and approved the final version of the manuscript
DISCLOSURE
H. Rottensteiner is an employee of Baxter Innovations GmbH..
REFERENCES
- 1.Sadler JE. von Willebrand factor. J Biol Chem. 1991;266:22777–22780. [PubMed] [Google Scholar]
- 2.Wagner DD, Olmsted JB, Marder VJ. Immunolocalization of von Willebrand protein in Weibel-Palade bodies of human endothelial cells. J Cell Biol. 1982;95:355–360. doi: 10.1083/jcb.95.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sporn LA, Chavin SI, Marder VJ, Wagner DD. Biosynthesis of von Willebrand protein by human megakaryocytes. J Clin Invest. 1985;76:1102–1106. doi: 10.1172/JCI112064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X, Halvorsen K, Zhang C-Z, Wong WP, Springer TA. Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science. 2009;324:1330–1334. doi: 10.1126/science.1170905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu T, Lin J, Cruz MA, Dong J, Zhu C. Force-induced cleavage of single VWFA1A2A3 tridomains by ADAMTS-13. Blood. 2010;115:370–378. doi: 10.1182/blood-2009-03-210369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ying J, Ling Y, Westfield LA, Sadler JE, Shao J-Y. Unfolding the A2 domain of von Willebrand factor with the optical trap. Biophys J. 2010;98:1685–1693. doi: 10.1016/j.bpj.2009.12.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai HM, Sussman II, Nagel RL. Shear stress enhances the proteolysis of von Willebrand factor in normal plasma. Blood. 1994;83:2171–2179. [PubMed] [Google Scholar]
- 8.Shim K, Anderson PJ, Tuley EA, Wiswall E, Sadler JE. Platelet-VWF complexes are preferred substrates of ADAMTS13 under fluid shear stress. Blood. 2008;111:651–657. doi: 10.1182/blood-2007-05-093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turner N, Nolasco L, Dong J-F, Moake J. ADAMTS-13 cleaves long von Willebrand factor multimeric strings anchored to endothelial cells in the absence of flow, platelets or conformation-altering chemicals. J Thromb Haemost. 2009;7:229–232. doi: 10.1111/j.1538-7836.2008.03209.x. [DOI] [PubMed] [Google Scholar]
- 10.De Ceunynck K, Rocha S, Feys HB, De Meyer SF, Uji-i H, Deckmyn H, Hofkens J, Vanhoorelbeke K. Local elongation of endothelial cell-anchored von Willebrand factor strings precedes ADAMTS13 protein-mediated proteolysis. J Biol Chem. 2011;286:36361–36367. doi: 10.1074/jbc.M111.271890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sadler JE. Von Willebrand factor, ADAMTS13, and thrombotic thrombocytopenic purpura. Blood. 2008;112:11–18. doi: 10.1182/blood-2008-02-078170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feys HB, Anderson PJ, Vanhoorelbeke K, Majerus EM, Sadler JE. Multi-step binding of ADAMTS-13 to von Willebrand factor. J Thromb Haemost. 2009;7:2088–2095. doi: 10.1111/j.1538-7836.2009.03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanardelli S, Chion ACK, Groot E, Lenting PJ, McKinnon TAJ, Laffan MA, Tseng M, Lane DA. A novel binding site for ADAMTS13 constitutively exposed on the surface of globular VWF. Blood. 2009;114:2819–2828. doi: 10.1182/blood-2009-05-224915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang P, Pan W, Rux AH, Sachais BS, Zheng XL. The cooperative activity between the carboxyl-terminal TSP1 repeats and the CUB domains of ADAMTS13 is crucial for recognition of von Willebrand factor under flow. Blood. 2007;110:1887–1894. doi: 10.1182/blood-2007-04-083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crawley JTB, de Groot R, Xiang Y, Luken BM, Lane DA. Unraveling the scissile bond: how ADAMTS13 recognizes and cleaves von Willebrand factor. Blood. 2011;118:3212–3221. doi: 10.1182/blood-2011-02-306597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muia J, Zhu J, Gupta G, Haberichter SL, Friedman KD, Feys HB, Deforche L, Vanhoorelbeke K, Westfield LA, Roth R, Tolia NH, Heuser JE, Sadler JE. Allosteric activation of ADAMTS13 by von Willebrand factor. Proc Natl Acad Sci U S A. 2014;111:18584–18589. doi: 10.1073/pnas.1413282112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.South K, Luken BM, Crawley JTB, Phillips R, Thomas M, Collins RF, Deforche L, Vanhoorelbeke K, Lane Da. Conformational activation of ADAMTS13. Proc Natl Acad Sci. 2014;111:18578–18583. doi: 10.1073/pnas.1411979112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tao Z, Wang Y, Choi H, Bernardo A, Nishio K, Sadler JE, López JA, Dong J-F. Cleavage of ultralarge multimers of von Willebrand factor by C-terminal-truncated mutants of ADAMTS-13 under flow. Blood. 2005;106:141–143. doi: 10.1182/blood-2004-11-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ai J, Smith P, Wang S, Zhang P, Zheng XL. The proximal carboxyl-terminal domains of ADAMTS13 determine substrate specificity and are all required for cleavage of von Willebrand factor. J Biol Chem. 2005;280:29428–29434. doi: 10.1074/jbc.M505513200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao W, Anderson PJ, Majerus EM, Tuley EA, Sadler JE. Exosite interactions contribute to tension-induced cleavage of von Willebrand factor by the antithrombotic ADAMTS13 metalloprotease. Proc Natl Acad Sci U S A. 2006;103:19099–19104. doi: 10.1073/pnas.0607264104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feys HB, Vandeputte N, Palla R, Peyvandi F, Peerlinck K, Deckmyn H, Lijnen HR, Vanhoorelbeke K. Inactivation of ADAMTS13 by plasmin as a potential cause of thrombotic thrombocytopenic purpura. J Thromb Haemost. 2010;8:2053–2062. doi: 10.1111/j.1538-7836.2010.03942.x. [DOI] [PubMed] [Google Scholar]
- 22.Feys HB, Liu F, Dong N, Pareyn I, Vauterin S, Vandeputte N, Noppe W, Ruan C, Deckmyn H, Vanhoorelbeke K. ADAMTS-13 plasma level determination uncovers antigen absence in acquired thrombotic thrombocytopenic purpura and ethnic differences. J Thromb Haemost. 2006;4:955–962. doi: 10.1111/j.1538-7836.2006.01833.x. [DOI] [PubMed] [Google Scholar]
- 23.Feys HB, Roodt J, Vandeputte N, Pareyn I, Lamprecht S, van Rensburg WJ, Anderson PJ, Budde U, Louw VJ, Badenhorst PN, Deckmyn H, Vanhoorelbeke K. Blood. Vol. 116. American Society of Hematology; 2010. Thrombotic thrombocytopenic purpura directly linked with ADAMTS13 inhibition in the baboon (Papio ursinus) pp. 2005–2010. [DOI] [PubMed] [Google Scholar]
- 24.Igari A, Nakagawa T, Moriki T, Yamaguchi Y, Matsumoto M, Fujimura Y, Soejima K, Murata M. Identification of epitopes on ADAMTS13 recognized by a panel of monoclonal antibodies with functional or non-functional effects on catalytic activity. Thromb Res. 2012;130:e79–e83. doi: 10.1016/j.thromres.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 25.Uemura M, Tatsumi K, Matsumoto M, Fujimoto M, Matsuyama T, Ishikawa M, Iwamoto T-A, Mori T, Wanaka A, Fukui H, Fujimura Y. Localization of ADAMTS13 to the stellate cells of human liver. Blood. 2005;106:922–924. doi: 10.1182/blood-2005-01-0152. [DOI] [PubMed] [Google Scholar]
- 26.Soejima K, Nakamura H, Hirashima M, Morikawa W, Nozaki C, Nakagaki T. Analysis on the molecular species and concentration of circulating ADAMTS13 in Blood. J Biochem. 2006;139:147–154. doi: 10.1093/jb/mvj013. [DOI] [PubMed] [Google Scholar]
- 27.Cauwenberghs N, Meiring M, Vauterin S, van Wyk V, Lamprecht S, Roodt JP, Novák L, Harsfalvi J, Deckmyn H, Kotzé HF. Antithrombotic effect of platelet glycoprotein Ib-blocking monoclonal antibody Fab fragments in nonhuman primates. Arterioscler Thromb Vasc Biol. 2000;20:1347–1353. doi: 10.1161/01.atv.20.5.1347. [DOI] [PubMed] [Google Scholar]
- 28.Anderson PJ, Kokame K, Sadler JE. Zinc and calcium ions cooperatively modulate ADAMTS13 activity. J Biol Chem. 2006;281:850–857. doi: 10.1074/jbc.M504540200. [DOI] [PubMed] [Google Scholar]
- 29.Kokame K, Nobe Y, Kokubo Y, Okayama A, Miyata T. FRETS-VWF73, a first fluorogenic substrate for ADAMTS13 assay. Br J Haematol. 2005;129:93–100. doi: 10.1111/j.1365-2141.2005.05420.x. [DOI] [PubMed] [Google Scholar]
- 30.Han Y, Xiao J, Falls E, Zheng XL. A shear-based assay for assessing plasma ADAMTS13 activity and inhibitors in patients with thrombotic thrombocytopenic purpura. Transfusion. 2011;51:1580–1591. doi: 10.1111/j.1537-2995.2010.03020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Meyer SF, Vandeputte N, Pareyn I, Petrus I, Lenting PJ, Chuah MKL, VandenDriessche T, Deckmyn H, Vanhoorelbeke K. Restoration of plasma von Willebrand factor deficiency is sufficient to correct thrombus formation after gene therapy for severe von Willebrand disease. Arterioscler Thromb Vasc Biol. 2008;28:1621–1626. doi: 10.1161/ATVBAHA.108.168369. [DOI] [PubMed] [Google Scholar]
- 32.McKinnon TAJ, Chion ACK, Millington AJ, Lane DA, Laffan MA. N-linked glycosylation of VWF modulates its interaction with ADAMTS13. Blood. 2008;111:3042–3049. doi: 10.1182/blood-2007-06-095042. [DOI] [PubMed] [Google Scholar]
- 33.Mi L-Z, Lu C, Li Z, Nishida N, Walz T, Springer TA. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat Struct Mol Biol. 2011;18:984–989. doi: 10.1038/nsmb.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y-F, Eng ET, Nishida N, Lu C, Walz T, Springer TA. EMBO J. Vol. 30. EMBO Press; 2011. A pH-regulated dimeric bouquet in the structure of von Willebrand factor; pp. 4098–4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X, Xie C, Nishida N, Li Z, Walz T, Springer TA. Requirement of open headpiece conformation for activation of leukocyte integrin alphaXbeta2. Proc Natl Acad Sci U S A. 2010;107:14727–14732. doi: 10.1073/pnas.1008663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlson CB, Lawler J, Mosher DF. Structures of thrombospondins. Cell Mol Life Sci. 2008;65:672–686. doi: 10.1007/s00018-007-7484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calzada MJ, Kuznetsova SA, Sipes JM, Rodrigues RG, Cashel JA, Annis DS, Mosher DF, Roberts DD. Calcium indirectly regulates immunochemical reactivity and functional activities of the N-domain of thrombospondin-1. Matrix Biol. 2008;27:339–351. doi: 10.1016/j.matbio.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng X, Chung D, Takayama TK, Majerus EM, Sadler JE, Fujikawa K. Structure of von Willebrand factor-cleaving protease (ADAMTS13), a metalloprotease involved in thrombotic thrombocytopenic purpura. J Biol Chem. 2001;276:41059–41063. doi: 10.1074/jbc.C100515200. [DOI] [PubMed] [Google Scholar]
- 39.Di Stasio E, Lancellotti S, Peyvandi F, Palla R, Mannucci PM, De Cristofaro R. Mechanistic studies on ADAMTS13 catalysis. Biophys J. 2008;95:2450–2461. doi: 10.1529/biophysj.108.131532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerhardt S, Hassall G, Hawtin P, McCall E, Flavell L, Minshull C, Hargreaves D, Ting A, Pauptit RA, Parker AE, Abbott WM. Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains. J Mol Biol. 2007;373:891–902. doi: 10.1016/j.jmb.2007.07.047. [DOI] [PubMed] [Google Scholar]
- 41.Shieh H-S, Mathis KJ, Williams JM, Hills RL, Wiese JF, Benson TE, Kiefer JR, Marino MH, Carroll JN, Leone JW, Malfait A-M, Arner EC, Tortorella MD, Tomasselli A. High resolution crystal structure of the catalytic domain of ADAMTS-5 (aggrecanase-2) J Biol Chem. 2008;283:1501–1507. doi: 10.1074/jbc.M705879200. [DOI] [PubMed] [Google Scholar]
- 42.Mosyak L, Georgiadis K, Shane T, Svenson K, Hebert T, McDonagh T, Mackie S, Olland S, Lin L, Zhong X, Kriz R, Reifenberg EL, Collins-Racie LA, Corcoran C, Freeman B, Zollner R, Marvell T, Vera M, Sum P-E, Lavallie ER, et al. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 2008;17:16–21. doi: 10.1110/ps.073287008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akiyama M, Takeda S, Kokame K, Takagi J, Miyata T. Crystal structures of the noncatalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor. Proc Natl Acad Sci U S A. 2009;106:19274–19279. doi: 10.1073/pnas.0909755106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu J, Muia J, Tolia NH, Westfield LA, Sadler JE. Blood. Vol. 124. American Society of Hematology; 2014. A Folded ADAMTS13 Conformation Identified By Small-Angle X-Ray Scattering Can Account for Allosteric Regulation By Distal Thrombospondin-1 and CUB Domains; p. 107. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.