Abstract
The molecular mechanisms responsible for opposing oncogenic and tumor-suppressor activities of NF-kB are obscure. Semi-quantitative immunohistochemistry of primary breast tumors using antibodies to RelA, the pleiotropic NF-kB factor, and Ki67 revealed a negative correlation between RelA levels and Ki67-index among ER +/HER2 − tumors [1]. Similarly, expression of AURKA, a marker for proliferation, negatively correlates with expression of NFKBIA, a surrogate for RelA expression and activity, in ER +/HER2 − tumors analyzed by The Cancer Genome Atlas [2], [3], [4]. Furthermore, conditional expression of RelA using a Tetracycline-inducible system in Human Mammary Epithelial Cells (HRA cells) caused proliferation arrest while withdrawal of Doxycycline (Dox) and suppression of RelA expression in arrested cells restored cell cycle progression [1]. To identify genes responsible for the negative relationship between RelA levels and proliferation, we performed genome-wide gene expression analysis of HRA cells under the following conditions: RelA un-induced, No Dox (ND); Dox induced for 24 h; Dox induced for 72 h; Dox induced for 24 h then Dox withdrawn for 48 h. The expression data was submitted to Gene Expression Ominibus (GEO) and the accession number is GSE65040. Analysis of the data identified cross-talk between basal RelA activity and the Interferon pathway mediated by IRF1, a target of RelA [5]. Activation of the Interferon pathway lead to down-regulation of CDK4 expression resulting in RB1 hypo-phosphorylation and suppression of cell cycle progression. The tumor-suppressor activity of NF-kB, specifically RelA, may stem from cross-talk with the Interferon pathway.
| Specifications | |
| Organism/cell line/tissue | Homo sapiens/mammary epithelial cells (HMEC)/mammary gland |
| Sex | Female |
| Sequencer or array type | PrimeView Human Gene Expression Array |
| Data format | Raw and normalized |
| Experimental factors | ND (No Dox), 24 + (Dox treatment for 24 h), 72 + (Dox treatment for 72 h) and DW (Dox Withdrawn; Dox treatment for 24 h and Dox withdrawn for 48 h) |
| Experimental features | HMEC conditionally expressing (Doxycycline inducible) RelA |
| Consent | Freely available |
| Sample source location | NA |
1. Direct link to gene expression data deposited in Gene Expression Omnibus (GEO)
2. Materials, methods and experimental design
2.1. Materials
Late passage P16neg hTERT immortalized Human Mammary Epithelial Cells (HMEC) were a gift from Jean Zhao [6]. The cells were cultured in DMEM-F12 (Life Technologies) supplemented with Insulin (10 μg/ml; Life Technologies), Epidermal Growth Factor (10 ng/ml, Peprotech), Cholera Toxin (1 ng/ml, Sigma Aldrich), Hydrocortisone (500 ng/ml, Sigma-Aldrich) and 0.6% FBS (Clontech Laboratories). Phoenix cells (Orbigen) were cultured in DMEM supplemented with 10% FBS (Clontech). Other chemicals used in the study were: Anti-Anti (Life Technologies) Doxycycline (Sigma-Aldrich), Neomycin (Sigma Aldrich), Puromycin (Invivogen), and miRNeasy mini kit (Qiagen). The PrimeView Human Gene Expression Array from Affymetrix was used to estimate genome-wide gene expression levels.
2.2. Methods
Retroviruses encoding the Tetracycline promoter transactivator (neomycin selection) and Flag-tagged RelA (Puromycin selection) were generated in Phoenix cells using standard protocols. HMEC were incubated with filtered culture supernatant containing virus particles and Polybrene (5 μg/ml, Millipore) for 12 h. Infected HMEC were selected using Neomycin (400 μg/ml) and Puromycin (1 μg/ml). Resulting cell line was designated HRA (HMEC harboring RelA) and pooled stable clones were used in the experiment. Dox inducible (1 μg/ml) expression of RelA and reduction of RelA expression after withdrawal of Dox were confirmed by Western blot [1].
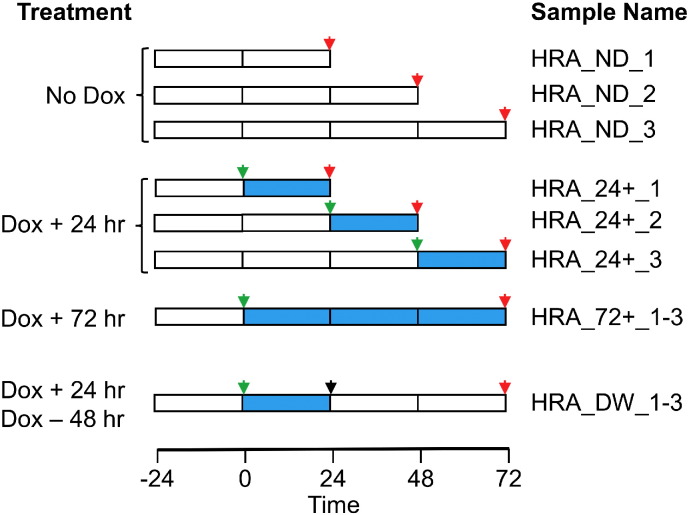
HRA cells were plated in 6 well plates and 24 h later, treated with Dox according to the scheme in Fig. 1 to generate triplicate samples for gene expression analysis. ND (No Dox; un-treated samples) were harvested 48, 72 and 96 h post-plating (indicated by red arrows in Fig. 1). To generate the 24 + samples, independent samples were treated with Dox 24, 48 and 72 h post-plating (indicated by green arrows) and harvested 24 h later (indicated by red arrows). The 72 + samples were treated with Dox 24 h post plating (green arrow) and harvested after 96 h of Dox treatment. The DW (Dox Withdrawn) sample was generated by treating cells with Dox 24 h post plating (green arrow), Dox withdrawn 24 h later (black arrow) and harvested after 48 h (red arrow). Culture medium in every plate was replaced with fresh medium containing Dox or devoid of Dox as required 24 h post-plating and every 24 h thereafter. For harvesting total RNA, the plates were transferred to ice, cells were washed with cold PBS and lysed using Trizol. Total RNA was purified using the miRNeasy mini kit from Qiagen using the manufacturer's protocol. The Affymetrix PrimeView array was used to estimate gene expression. RNA labeling, hybridization and scanning were performed at the Molecular Biology Core Facilities at Dana-Farber Cancer Institute according to manufacturer's protocol.
Fig. 1.
Schematic representation of Dox treatment of HRA cells to regulate RelA expression. Open boxes indicate the absence of Dox, blue boxes indicate the presence of Dox. Green arrows indicate times at which Dox was added, black arrow indicates withdrawal of Dox and red arrows indicate time at which each sample was harvested for RNA extraction. The names of triplicate samples are given for each treatment condition.
2.3. Gene expression analysis
Quality checks of the expression data cel files were performed using AffylmGUI in R and data from all arrays was confirmed suitable for downstream analysis [7]. The cel files were normalized using RMA in GenePattern [8]. The Brainarray chip definition file (version 18) based on ensemble gene (ENSG) was used for probe summarization [9]. Differentially expressed genes between pairs of samples were identified using LIMMA implemented in MEV [10]. Other downstream analysis performed using the dataset is described in a previous manuscript [1].
3. Conclusion
This dataset may be useful for identifying predictive biomarkers of response to CDK4/6 inhibitors e.g. Palbociclib, Abemaciclib, LEE011 and P1446A-05 in RB1 competent tumors where CDK4 is not amplified [11], [12], [13], [14].
Conflict of interest
The authors declare that no conflicting interests exist.
Acknowledgments
We thank the Molecular Biology Core Facilities at Dana-Farber Cancer Institute for generating gene expression data. The research was supported by the DF/HCC SPORE (CA89393) and the Women's Cancer Program at Dana-Farber Cancer Institute. BSK is a recipient of the Teri Brodeur and Hale Family Fellowships.
Contributor Information
Bose S. Kochupurakkal, Email: bose_kochupurakkal@dfci.harvard.edu.
J. Dirk Iglehart, Email: jiglehart@partners.org.
References
- 1.Kochupurakkal B.S., Wang Z.C., Hua T., Culhane A.C., Rodig S.J., Rajkovic-Molek K. RelA-induced interferon response negatively regulates proliferation. PLoS One. 2015;10(10):e0140243. doi: 10.1371/journal.pone.0140243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haibe-Kains B., Desmedt C., Loi S., Culhane A.C., Bontempi G., Quackenbush J. A three-gene model to robustly identify breast cancer molecular subtypes. J. Natl. Cancer Inst. 2012;104(4):311–325. doi: 10.1093/jnci/djr545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayden M.S., Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neph S., Stergachis A.B., Reynolds A., Sandstrom R., Borenstein E., Stamatoyannopoulos J.A. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150(6):1274–1286. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao J.J., Gjoerup O.V., Subramanian R.R., Cheng Y., Chen W., Roberts T.M. Human mammary epithelial cell transformation through the activation of phosphatidylinositol 3-kinase. Cancer Cell. 2003;3(5):483–495. doi: 10.1016/s1535-6108(03)00088-6. [DOI] [PubMed] [Google Scholar]
- 7.Wettenhall J.M., Simpson K.M., Satterley K., Smyth G.K. AffylmGUI: a graphical user interface for linear modeling of single channel microarray data. Bioinformatics. 2006;22(7):897–899. doi: 10.1093/bioinformatics/btl025. [DOI] [PubMed] [Google Scholar]
- 8.Reich M., Liefeld T., Gould J., Lerner J., Tamayo P., Mesirov J.P. GenePattern 2.0. Nat. Genet. 2006;38(5):500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 9.Dai M., Wang P., Boyd A.D., Kostov G., Athey B., Jones E.G. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 11.https://clinicaltrials.gov/ct2/results?term=Abemaciclib&Search=Search. (Access Date 2015 Nov 11).
- 12.https://clinicaltrials.gov/ct2/results?term=LEE011&Search=Search. (Access Date 2015 Nov 11).
- 13.https://clinicaltrials.gov/ct2/results?term=P1446A-05&Search=Search. (Access Date 2015 Nov 11).
- 14.https://clinicaltrials.gov/ct2/results?term=Palbociclib&Search=Search. (Access Date 2015 Nov 11).