Abstract
Mesenchymal stromal cells (MSCs), sometimes called mesenchymal stem cells, are cultured cells able to give rise to mature mesenchymal cells such as adipocytes, osteoblasts, and chondrocytes, and to secrete a wide range of trophic and immunomodulatory molecules. Evidence indicates that pericytes, cells that surround and maintain physical connections with endothelial cells in blood vessels, can give rise to MSCs (da Silva Meirelles et al., 2008 [1]; Caplan and Correa, 2011 [2]). We have compared the transcriptomes of highly purified, human adipose tissue pericytes subjected to culture-expansion in pericyte medium or MSC medium, with that of human adipose tissue MSCs isolated with traditional methods to test the hypothesis that their transcriptomes are similar (da Silva Meirelles et al., 2015 [3]). Here, we provide further information and analyses of microarray data from three pericyte populations cultured in pericyte medium, three pericyte populations cultured in MSC medium, and three adipose tissue MSC populations deposited in the Gene Expression Omnibus under accession number GSE67747.
Keywords: Mesenchymal stromal cells, Mesenchymal stem cells, Pericytes, Microarrays
| Specifications | |
| Organism/cell line/tissue | Homo sapiens |
| Sex | Female |
| Sequencer or array type | Whole Human Genome Oligo Microarray chips (Agilent, G4112F and G4845A; design IDs 014,850 and 026,652, respectively) |
| Data format | Raw: TXT; normalized data: TXT, SOFT, MINiML |
| Experimental factors | Human adipose tissue mesenchymal stromal cells vs. human adipose tissue pericytes cultured under mesenchymal stromal cell conditions; human adipose tissue pericytes vs. human adipose tissue pericytes cultured under mesenchymal stromal cell conditions; human adipose tissue mesenchymal stromal cells vs. human adipose tissue pericytes cultured in pericyte medium. |
| Experimental features | The transcriptomes of highly purified, human adipose tissue-derived pericytes subjected to culture-expansion in pericyte medium (n = 3) were compared to those of human adipose tissue mesenchymal stromal cells (n = 3) and human adipose tissue pericytes cultured under mesenchymal stromal cell conditions (n = 3) to detect differentially expressed transcripts. |
| Consent | Informed consent was obtained from all tissue donors enrolled in the study. |
| Sample source location | Ribeirão Preto, São Paulo, Brazil |
1. Direct link to deposited data
2. Experimental design, materials and methods
2.1. Samples and sample donors
Adipose tissue was obtained from patients undergoing elective plastic surgery at the University Hospital of the School of Medicine of Ribeirão Preto, University of São Paulo at Ribeirão Preto, Brazil. All patients provided informed consent for the use of their biological material in this study. This study was approved by the Brazilian National Commission on Ethics in Research (CAAE 0054.0.004.000-08).
Adipose tissue pericytes were isolated from donors 1, 2 and 3 (and named cAT3G5Cs 1, 2, and 3, respectively), and adipose tissue mesenchymal stromal cells were isolated from donors 16, 17 and 18 (and named ATMSCs 16, 17, and 18, respectively). Adipose tissue pericytes from donors 1, 2, and 3 were also cultured under mesenchymal stromal cell conditions prior to transcriptomic analyses, and named cAT3G5Cs 1 DME10, cAT3G5Cs 2 DME10, and cAT3G5Cs 3 DME10, respectively. The samples and corresponding data used here were obtained in a previously published study [3]. All tissue donors were females. Tissue samples used to isolate the cells consisted of liposuction material with exception of the sample obtained from donor 17, which was a tissue fragment removed during dermolipectomy.
2.2. Microarray hybridization and scanning
RNA was extracted using TRIzol LS reagent (Life Technologies do Brasil Ltda, São Paulo, SP, Brazil), and cleaned up using the RNeasy mini kit (QIAGEN Biotecnologia Brasil Ltda, São Paulo, SP, Brazil) following the manufacturers' instructions. RNA was quantified using a NanoDrop 1000 spectrophotometer (ThermoScientific, Wilmington, DE).
Oligonucleotide microarrays from two 4 × 44K Whole Human Genome Microarray Kits, (G4112F, and G4845A; design IDs 014850, and 026652, respectively; Agilent Technologies, Santa Clara, CA), which contain probes for more than 41,000 gene transcripts, were used to analyze gene expression of the samples. A predetermined amount of control bacterial RNA from the One Color RNA Spike-In Kit (Agilent, 5188–5282) was added to total RNA prior to synthesis of complementary RNA (cRNA) and labeling with cyanine 3 (Cy3) using the One Color Quick Amp Labeling Kit (Agilent, 5190–0442). RNA was reverse-transcribed using oligo (dT) containing a promoter for RNA T7 polymerase. The resultant cDNA was purified, fragmented, and used as template for cRNA in vitro transcription using T7 RNA polymerase and nucleotides, which included Cy3-CTP for labeling. The cDNA obtained was purified using the Illustra RNAspin mini Kit (25‐0500‐71; GE Healthcare Life Sciences, Logan, UT). cDNA quantitation and labeling efficiency were determined using a NanoDrop 1000 spectrophotometer (ThermoScientific). Labeled cRNA was hybridized with microarray slides using the Gene Expression Hybridization Kit (Agilent, 5188–5242) in SureHyb hybridization chambers (Agilent, G2534A) for 17 h at 65 °C at 10 RPM in a hybridization oven (Agilent, G2545A). After hybridization, microarray slides were washed and dried. The slides were then scanned at 535 nm with a resolution of 5 μm/pixel using a DNA Microarray Scanner with Sure Scan High-Resolution Technology (Agilent). Expression data were extracted using Agilent's Feature Extraction software versions 8.5 or 11.5.
2.3. Microarray data consolidation
To compare data from the two microarray design IDs used in this study, one tab-delimited text file corresponding to each design was selected to define probes common to both using Microsoft Excel's VLOOKUP function after filtering out probes corresponding to controls. Since both designs contain a nonmatching number of repeated probes, the resulting probe list had duplicate probes removed by checking the option “unique records only” in Excel's advanced filter. The resulting unique probe list, which contained 18,561 probes, was used as a reference to remove probes (and their associated parameter values) which were not shared by both designs using Excel's VLOOKUP function after organizing probe names in ascending order. Non-unique probes were removed using Excel's advanced filter, and data files were used for downstream analyses.
2.4. Group comparisons
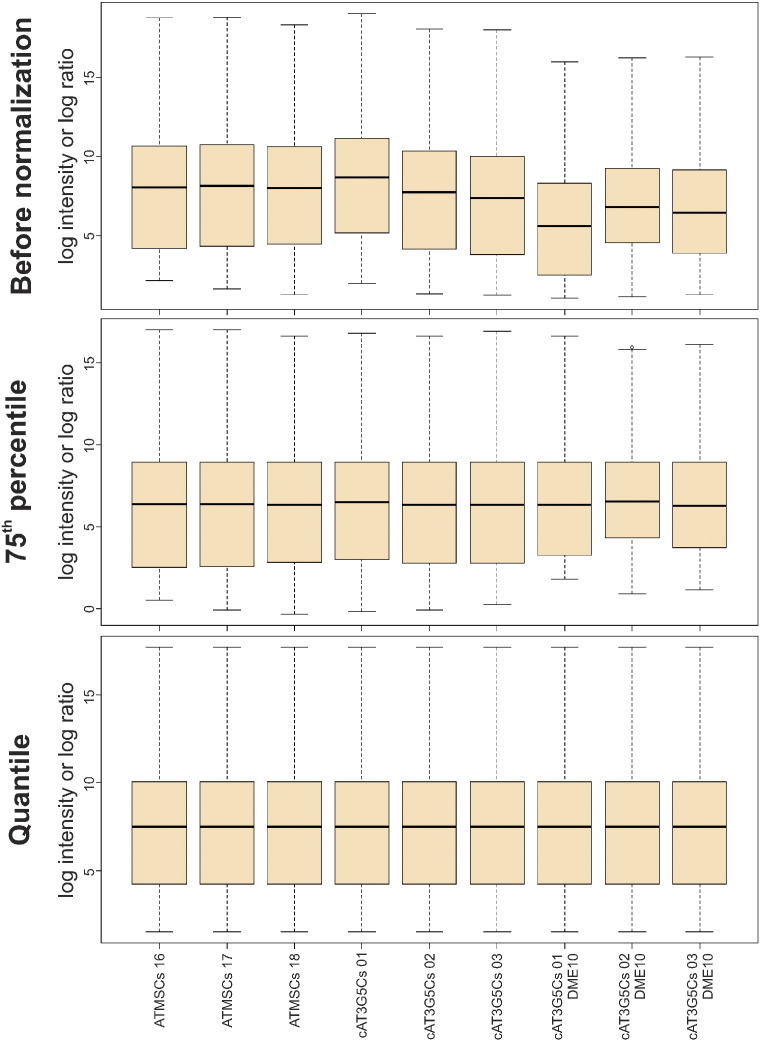
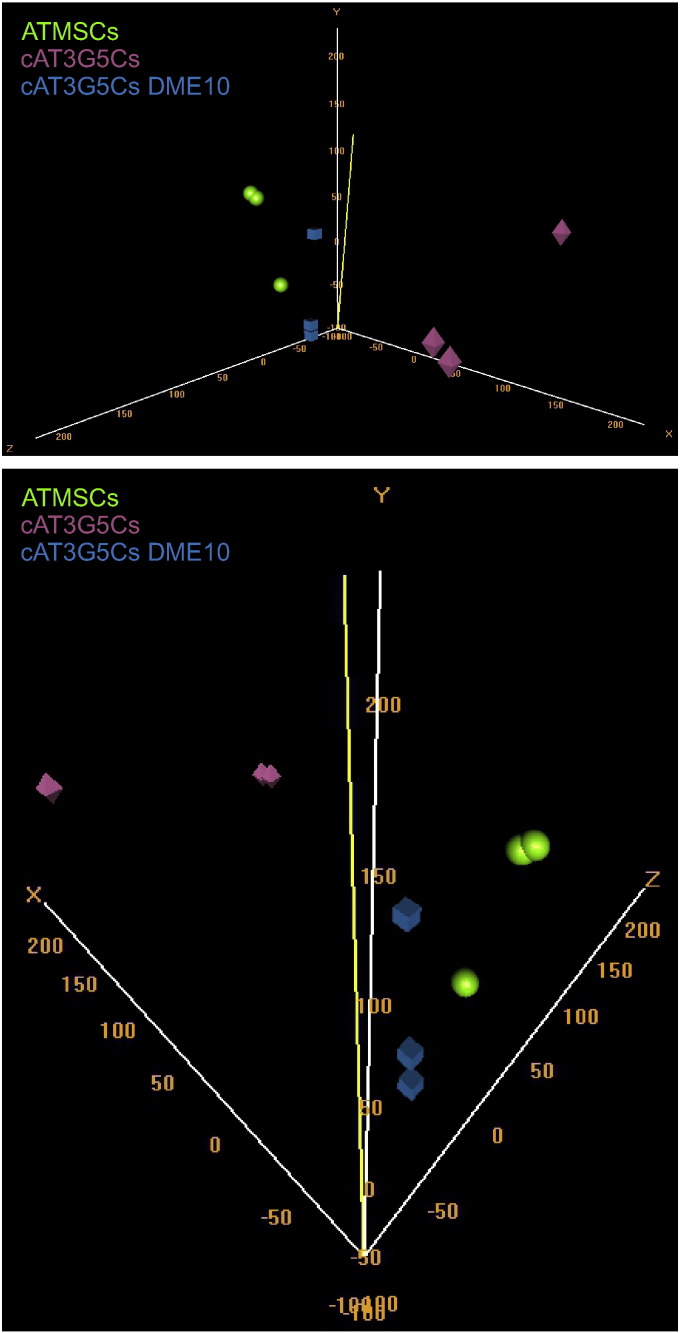
The following groups were compared to each other: cAT3G5Cs vs. cAT3G5Cs DME10, cAT3G5Cs vs. ATMSCs, and cAT3G5Cs vs. ATMSCs. Comparisons were made using the “class comparison between groups of arrays” tool in BRB-ArrayTools (version 4.3.1), available at http://linus.nci.nih.gov/BRB-ArrayTools.html. Data files were imported to BRB-ArrayTools using its general format importer tool. The unique ID was defined as the probe name, the intensity value was set as the processed signal of the green channel (gProcessedSignal column), the column gIsFeatNonUnifOL was set as a flag and the column gNumPix was set as the spot size descriptor. Expression data were converted to log2 values and subjected to quantile normalization [4], which proved to be superior to normalization using 75th percentile values (Fig. 1). Flagged spots and spots with a size less than 10 were removed, and genes that contained more than 50% of missing values were excluded. Normalized log2 expression values for each probe were averaged within each group. For three-dimensional visualization of the microarray data, the data were analyzed with Euclidean metric using the “Visualization of samples” tool in BRB-ArrayTools. Representative three-dimensional plots are shown in Fig. 2.
Fig. 1.
Boxplots showing the distribution of expression values of non-control probes in each consolidated microarray dataset before normalization (top panel), after normalization to the 75th percentile (middle panel), and after quantile normalization (bottom panel).
Fig. 2.
Three-dimensional scatterplot showing the distribution of consolidated microarray data of ATMSCs (green spheres), cAT3G5Cs (pink octahedrons), and cAT3G5Cs DME10 (blue cubes). A frontal view (top panel) and a top rear view (bottom panel) demonstrate the close proximity of ATMSCs and cAT3G5Cs DME10 as compared to cAT3G5Cs.
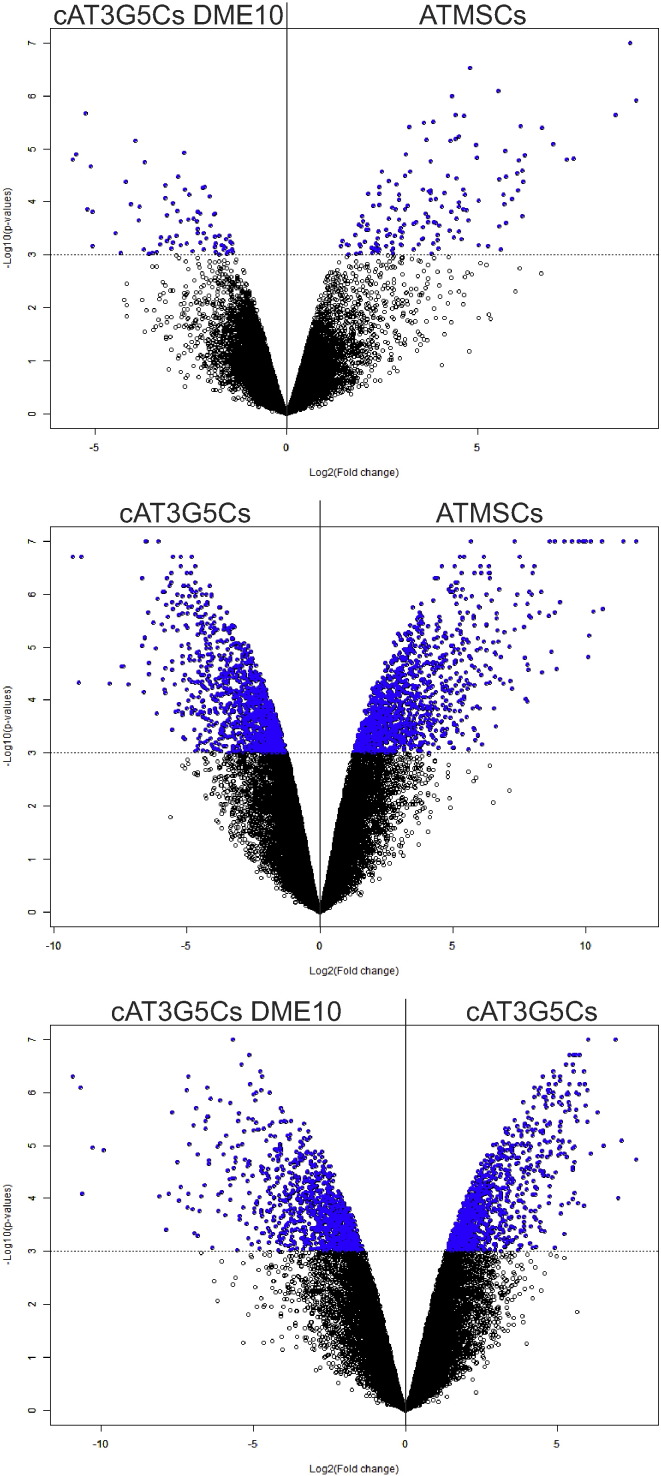
Groups were also compared by means of volcano plots after statistical analysis using the “Class comparison between groups of arrays” tool in BRB-ArrayTools, using default settings. For these analyses, data from the compared groups alone (cAT3G5Cs vs. cAT3G5Cs DME10, cAT3G5Cs vs. ATMSCs, or cAT3G5Cs vs. ATMSCs) were loaded into BRB-ArrayTools for each analysis, rather than loading the data from the three studied groups at once. The resulting volcano plots are shown in Fig. 3.
Fig. 3.
Volcano plots based on log2 fold-change against – log10 (p-value) showing the proportion of differentially expressed genes (blue dots) in cAT3G5Cs DME10 vs. ATMSCs (top panel), cAT3G5Cs vs. ATMSCs (middle panel), and cAT3G5Cs DME10 vs. cAT3G5Cs (bottom panel) comparison groups.
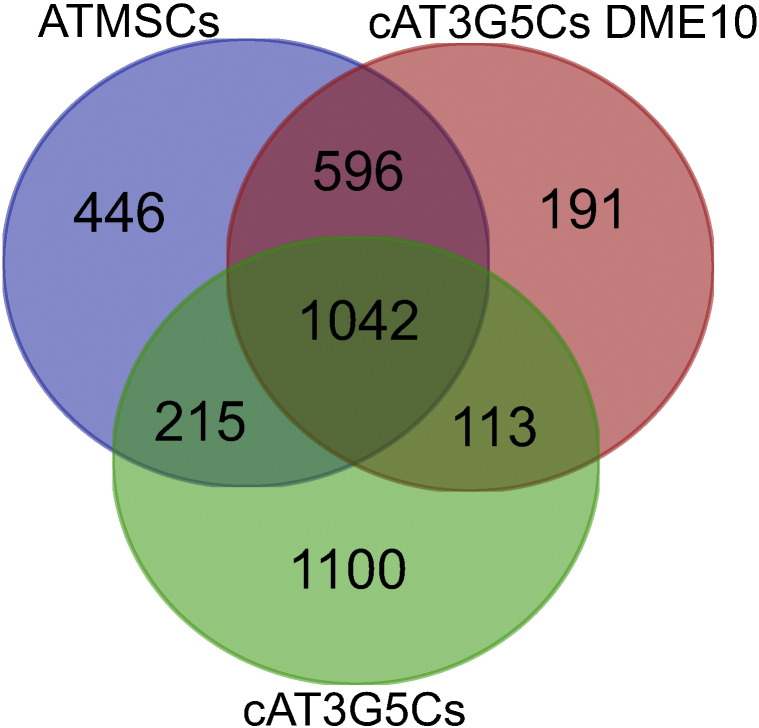
To determine genes whose expression was shared by the three cell populations under study, and to define genes uniquely expressed by them, cAT3G5Cs, cAT3G5Cs DME10, and ATMSCs (n = 3 each) were compared to a non-mesenchymal cell population, peripheral blood white blood cells (PBWBCs; GEO accessions GSM469524, GSM469528, and GSM469532), to determine genes differentially expressed by them. This was done by loading microarray data from these four cell populations into BRB-ArrayTools, and statistically comparing them pairwise using “Class comparison between groups of arrays” with default settings. Probes corresponding to transcripts whose levels were significantly higher in cAT3G5Cs (2470 probes), cAT3G5Cs DME10 (1942 probes), and ATMSCs (2299 probes) as compared to PBWBCs were filtered, and a Venn diagram was built using an online tool available at http://bioinformatics.psb.ugent.be/cgi-bin/liste/Venn/calculate_venn.htpl (Fig. 4).
Fig. 4.
Venn diagram of transcripts differentially expressed by ATMSCs, cAT3G5Cs DME10, and cAT3G5Cs when individually compared to peripheral blood white blood cells.
Given the high similarity between cAT3G5Cs DME10 and ATMSCs previously identified by us [3], we focused on the transcripts whose expression was shared exclusively by these two cell populations in the Venn diagram (596 probes), using transcripts shared exclusively by ATMSCs and cAT3G5Cs (215 probes), and by cAT3G5Cs DME10 and cAT3G5Cs (113 probes), for comparison purposes. It is noteworthy that the number of transcripts shared exclusively by cAT3G5Cs DME10 and ATMSCs is almost double the sum of the number of transcripts exclusively shared by the two other comparison groups, which attests she similarity between these two cell populations. The probe lists at these intersections of the Venn diagram were analyzed using the Functional Annotation Tool in Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 at http://david.abcc.ncifcrf.gov/home.jsp, using AGILENT_ID as identifier, and “Homo sapiens” as background. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and full Gene Ontology term lists (“ALL”) were selected for analysis. Pathways or Gene Ontology terms with a Benjamini–Hochberg-corrected P value less than 0.05 were considered significantly enriched in the gene lists analyzed. Results of Gene Ontology term analysis are shown in Table 1. Some of the terms enriched in the ATMSCs vs. cAT3G5Cs DME10 comparison group refer to biological processes related to development of skeleton and vasculature. The terms “contractile fiber part” and “contractile fiber” were among the cell component terms enriched in this comparison group, with encompassed genes directly involved in cell contraction such as MYH2, MYH2, ACTA2, MYL9, MYL2.
Table 1.
Gene ontology term enrichment analysis of transcripts whose differential expression, when compared to peripheral blood white blood cells, is shared by ATMSCs and cAT3G5Cs DME10, ATMSCs and cAT3G5Cs, and cAT3G5Cs DME10 and cAT3G5Cs, after exclusion of transcripts shared by these three cell populations.
| ATMSCs vs. cAT3G5Cs DME10 |
||||
|---|---|---|---|---|
| Biological process |
||||
| Term | Count | % | P value | Benjamini |
| GO:0007155—cell adhesion | 54 | 9.56 | 4.42E − 10 | 9.01E − 07 |
| GO:0022610—biological adhesion | 54 | 9.56 | 4.70E − 10 | 4.79E − 07 |
| GO:0048731—system development | 115 | 20.35 | 2.89E − 08 | 1.96E − 05 |
| GO:0048856—anatomical structure development | 122 | 21.59 | 3.08E − 08 | 1.57E − 05 |
| GO:0007275—multicellular organismal development | 130 | 23.01 | 3.46E − 07 | 1.41E-04 |
| GO:0032502—developmental process | 137 | 24.25 | 1.58E − 06 | 5.36E − 04 |
| GO:0001501—skeletal system development | 25 | 4.43 | 3.56E − 05 | 0.010316 |
| GO:0001568—blood vessel development | 21 | 3.72 | 5.14E − 05 | 0.013006 |
| GO:0048514—blood vessel morphogenesis | 19 | 3.36 | 6.88E − 05 | 0.015451 |
| GO:0001944—vasculature development | 21 | 3.72 | 7.21E − 05 | 0.01458 |
| GO:0032501—multicellular organismal process | 165 | 29.20 | 1.19E − 04 | 0.021743 |
| GO:0007156—homophilic cell adhesion | 14 | 2.48 | 1.57E − 04 | 0.02634 |
| GO:0048646—anatomical structure formation involved in morphogenesis | 25 | 4.43 | 1.65E − 04 | 0.025506 |
| GO:0009653—anatomical structure morphogenesis | 59 | 10.44 | 1.81E − 04 | 0.026015 |
| Cell component |
||||
|---|---|---|---|---|
| Term | Count | % | P value | Benjamini |
| GO:0031012—extracellular matrix | 30 | 5.31 | 6.05E − 07 | 2.04E-04 |
| GO:0005578—proteinaceous extracellular matrix | 28 | 4.96 | 1.38E − 06 | 2.33E − 04 |
| GO:0044421—extracellular region part | 55 | 9.74 | 5.91E − 06 | 6.66E − 04 |
| GO:0005576—extracellular region | 95 | 16.81 | 6.02E − 06 | 5.08E − 04 |
| GO:0016020—membrane | 258 | 45.66 | 1.61E − 04 | 0.010814 |
| GO:0015629—actin cytoskeleton | 20 | 3.54 | 5.07E − 04 | 0.028147 |
| GO:0044449—contractile fiber part | 12 | 2.12 | 5.82E − 04 | 0.027718 |
| GO:0043292—contractile fiber | 12 | 2.12 | 0.001032 | 0.042694 |
| Molecular function |
||||
|---|---|---|---|---|
| Term | Count | % | P value | Benjamini |
| GO:0005509—calcium ion binding | 56 | 9.91 | 3.69E − 07 | 2.36E − 04 |
| GO:0005515—protein binding | 282 | 49.91 | 3.54E − 05 | 0.01124 |
| GO:0008092—cytoskeletal protein binding | 33 | 5.84 | 3.73E − 05 | 0.007919 |
| GO:0019838—growth factor binding | 12 | 2.12 | 2.59E − 04 | 0.040574 |
| ATMSCs vs. cAT3G5Cs |
||||
|---|---|---|---|---|
| Biological process |
No significantly enriched GO terms |
|||
| Cell component |
||||
| Term | Count | % | P value | Benjamini |
| GO:0044424—intracellular part | 129 | 62.32 | 4.86E − 05 | 0.012024 |
| GO:0005737—cytoplasm | 97 | 46.86 | 1.28E − 04 | 0.015806 |
| GO:0005622—intracellular | 129 | 62.32 | 4.95E − 04 | 0.040248 |
| GO:0031966—mitochondrial membrane | 13 | 6.28 | 5.76E − 04 | 0.035234 |
| GO:0044429—mitochondrial part | 16 | 7.73 | 8.71E − 04 | 0.042462 |
| GO:0005743—mitochondrial inner membrane | 11 | 5.31 | 9.77E − 04 | 0.03974 |
| GO:0005740—mitochondrial envelope | 13 | 6.28 | 9.85E − 04 | 0.034442 |
| GO:0005739—mitochondrion | 23 | 11.11 | 0.001109 | 0.03394 |
| GO:0031967—organelle envelope | 16 | 7.73 | 0.001314 | 0.035725 |
| GO:0031975—envelope | 16 | 7.73 | 0.001357 | 0.033235 |
| GO:0044455—mitochondrial membrane part | 7 | 3.38 | 0.00155 | 0.0345 |
| GO:0019866—organelle inner membrane | 11 | 5.31 | 0.001679 | 0.034263 |
| Molecular function | No significantly enriched GO terms | |||
| cAT3G5Cs DME10 vs. cAT3G5Cs | ||||
|---|---|---|---|---|
| Biological process | No significantly enriched GO terms | |||
| Cell component | No significantly enriched GO terms | |||
| Molecular function | No significantly enriched GO terms | |||
Count: number of genes that match the indicated gene ontology term; %, percentage of genes relative to the total number of genes involved with the indicated gene ontology term; P value, parametric P value; Benjamini, P value as adjusted using the Benjamini–Hochberg method.
KEGG pathway analysis identified only one significantly enriched pathway among in the transcripts shared only between cAT3G5Cs DME10 and ATMSCs: hsa04610 – complement and coagulation cascades – with a Benjamini–Hochberg-corrected p-value of 6.42 × 10− 05 and a false discovery rate of 6.55 × 10− 04. Transcripts that matched this pathway were BDKRB2, F10, CFH, MASP1, A2M, C1S, BDKRB1, CFI, CFB, C1R, LOC653879, LOC100133511, C3, F8, and PLAU. No significantly enriched pathways were present in genes shared only by ATMSCs and cAT3G5Cs, and by cAT3G5Cs DME10 and cAT3G5Cs. These findings suggest that culture of ATMSCs and cAT3G5Cs in ATMSC medium renders these cells prone to contribute to blood coagulation and production of complement proteins.
3. Discussion
Here, we provide further details on methods that may help reproduce the analysis of microarray data from cultured pericytes from human adipose tissue cultured under pericyte-optimized conditions (cAT3G5Cs), pericytes cultured under mesenchymal stromal cell conditions (cAT3G5Cs DME10), and human adipose tissue-derived mesenchymal stromal cells (ATMSCs), along with further analyses of these data. These analyses highlight the importance of the normalization method used and reinforce our previously published findings, according to which pericytes cultured under ATMSC conditions exhibit a gene expression profile almost identical to that of ATMSCs [3]. The analyses shown here extend our previous findings regarding the high transcriptomic similarity between cultured pericytes and mesenchymal stromal cells, as observed in three-dimensional scatter plot analysis (Fig 2), volcano plots (Fig. 3), and Venn diagram analysis (Fig. 4). The Venn diagram shown in Fig. 4 was built with lists of transcripts differentially expressed by cAT3G5Cs, cAT3G5Cs DME10 or ATMSCs as compared to an unrelated cell population, peripheral blood white blood cells. Gene ontology term enrichment analysis of transcripts shared only between cAT3G5Cs DME10 and ATMSCs highlighted by this Venn diagram revealed an enrichment of transcripts directly involved with cell contraction. This finding drew our attention because stromal cells have been proposed to be vascular smooth muscle-like, and to follow a developmental program of vascular smooth muscle cell differentiation in culture [5]. Culture conditions seem to play a major role in this feature since cAT3G5Cs, which are cultured under pericyte conditions, do not express such a number of genes directly associated with cell contraction. Finally, when the transcripts shared only between cAT3G5Cs DME10 and ATMSCs (as assessed using the Venn diagram) were subjected to a pathway analysis using the KEGG database, they were found to be enriched for genes that code for proteins involved in complement and coagulation cascades. This finding not only emphasizes similarities between ATMSCs and pericytes cultured under ATMSC conditions, but also stresses the physiological association of these cell populations with blood vessels, as previously proposed [1], [2].
Acknowledgements
The authors are indebted to the team of Divisão de Cirurgia Plástica e Queimaduras of Hospital das Clínicas de Ribeirão Preto for providing adipose tissue samples, and Ms. Aline Magro Bueno, Amélia Goes de Araújo, and Patrícia Vianna Bonini Palma for the technical assistance. This work has been funded by the following grants: #2013/08135-2, São Paulo Research Foundation (FAPESP); USP #12.1.25441.01.2, Research Support of the University of São Paulo, CISBi-NAP; and grants #477806/2008-2, #573754/2008-0, and #131371/2011-8, Brazilian National Council for Scientific and Technological Development (CNPq).
References
- 1.da Silva Meirelles L., Caplan A.I., Nardi N.B. In search of the in vivo identity of mesenchymal stem cells. Stem Cells. 2008 Sep;26(9):2287–2299. doi: 10.1634/stemcells.2007-1122. [DOI] [PubMed] [Google Scholar]
- 2.Caplan A.I., Correa D. The MSC: an injury drugstore. Cell Stem Cell. 2011 Jul 8;9(1):11–15. doi: 10.1016/j.stem.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.da Silva Meirelles L., Maistro Malta T., de Deus Wagatsuma V.M., Palma P.V., Araújo A.G., Ribeiro Malmegrim K.C., Morato de Oliveira F., Panepucci R.A., Silva W.A., Jr., Kashima Haddad S., Covas D.T. Cultured human adipose tissue pericytes and mesenchymal stromal cells display a very similar gene expression profile. Stem Cells Dev. Aug 19 2015 doi: 10.1089/scd.2015.0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. Jan 22 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 5.Dennis J.E., Charbord P. Origin and differentiation of human and murine stroma. Stem Cells. 2002;20(3):205–214. doi: 10.1634/stemcells.20-3-205. [DOI] [PubMed] [Google Scholar]