Abstract
Chronic lymphocytic leukemia (CLL) is the most common adult leukemia. While therapeutic antibodies show clinical activity in CLL patients, resistance inevitably develops resulting in treatment failure. Identifying mechanisms of antibody resistance and methods to reduce resistance would be valuable in managing CLL. Monocyte derived cells (MDCs), also known as nurse like cells (NLCs) in CLL [1], [2], are known to be crucial components of the CLL microenvironment network and following “maturation” in in vitro culture systems are able to provide support for the survival of the malignant B cells from CLL patients. In addition to their protective role, MDCs are key effector cells in mediating responses to therapeutic antibody therapies [3]. We have determined that macrophages from patients with early stable CLL are able to elicit superior cytotoxic response to therapeutic antibodies than macrophages derived from patients with progressive CLL. We have exploited this unique finding to gain insight into antibody resistance. Thus, we have profiled monocytes on day 0 and MDCs on day 7 from antibody sensitive and antibody resistant CLL patients (GEO accession number GEO: GSE71409). We show that there are no significant differences in transcriptomes from the monocytes or MDCs derived from sensitive or resistant patient samples. However, we show that MDCs acquire an M2-like macrophage transcriptomic signature following 7 days culture regardless of whether they were derived from sensitive or resistant patient samples.
Keywords: Chronic lymphocytic leukemia, Monocyte derived cells, Antibody resistance, Microarray
| Specifications | |
|---|---|
| Organism | Homo sapiens |
| Sex | Not applicable |
| Sequencer or array type | Illumina HumanHT-12 V4.0 expression beadchip |
| Data format | Raw and normalized data in plain text format. |
| Experimental factors | Monocytes on day 0 vs. monocyte derived cells (MDCs) on day 7 |
| Experimental features | Expression profiling of CD14 positive monocytes isolated from CLL patient PBMCs on day 0 and from MDCs from CLL PBMCs cultured for seven days to identify differences between the MDCs derived from sensitive or resistant CLL patients and differences in MDCs pre and post in vitro culture |
| Consent | All samples were obtained after informed consent |
| Sample source location | Brisbane, Australia |
1. Direct link to deposited data
Deposited data can be found here: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE71409
2. Data
Whole genome transcriptomics data from CD14 positive monocytes isolated from CLL patient peripheral blood mononuclear cells (PBMCs) for four patients and monocyte derived cells from CLL PBMCs cultured for seven days for seven patients.
3. Experimental design, materials and methods
3.1. Patient samples and cell purification
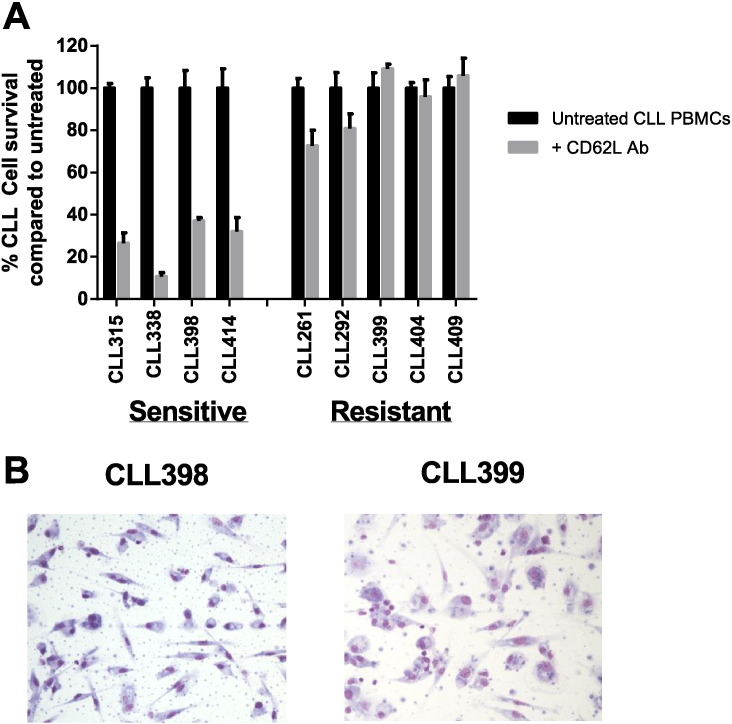
Peripheral blood from CLL patients was collected after informed consent according to protocols approved by the Princess Alexandra Hospital (PAH) Human Research Ethics Committee. Patient characteristics are outlined in Table 1. PBMCs were isolated by density-gradient centrifugation over Histopaque-1077 and red blood cells lysed using ammonium chloride. To determine antibody sensitivity, CLL PBMCs were cultured in RPMI + 10% fetal calf serum in the presence of LEAF Purified anti-human CD62L antibody (DREG56; 0.1 μg/ml; Biolegend) and CLL cell survival examined after seven days using trypan blue exclusion [4]. CLL patients' responses were classified as sensitive if survival ≤ 40% or resistant if survival ≥ 70% of untreated cultures (Fig. 1A).
Table 1.
Patient clinical features.
| Patient code | Age | Sex | CD38 | Zap 70 | IgVH status | FISH |
|---|---|---|---|---|---|---|
| CLL261 | 40 | M | Negative | Positive | Unmutated | 13q deletion & 17p deletion |
| CLL292 | 77 | M | Positive | Positive | ND | 11q deletion & 13q deletion |
| CLL315 | 75 | F | Negative | Positive | Mutated | Trisomy 12 |
| CLL338 | 69 | F | Negative | Positive | Mutated | ND |
| CLL395 | 70 | F | Negative | Positive | ND | Normal karyotype |
| CLL398 | 59 | F | Negative | Negative | Mutated | 13q deletion |
| CLL399 | 41 | M | Positive | Positive | Mutated | 17p deletion + monosomy 12 |
| CLL404 | 88 | M | Positive | Positive | Unmutated | 13q deletion |
| CLL409 | 72 | M | Negative | Positive | Mutated | 13q deletion |
| CLL414 | 67 | M | Positive | Positive | Unmutated | 17p deletion & 13q deletion |
Fig. 1.
Differential sensitivity to CD62L antibody in vitro. (A) CLL PBMCs were cultured in the presence of anti-human CD62L antibody (DREG56; 0.1 μg/ml) and cell survival examined after 7 days using trypan blue exclusion. CLL cell survival was normalized to untreated CLL PBMCs survival. (B) CLL PBMCs were cultured for 7 days before non-adherent cells were removed by gentle agitation and MDCs were visualized by Giemsa staining (magnification = 20 ×). A representative image from one sensitive and one resistant patient is shown.
For day 0 samples, monocytes were isolated from CLL PBMCs using MACS cell separation and CD14 MicroBeads according to manufacturer's instructions (Miltenyi Biotec). Purity of samples was examined by flow cytometry and all samples ≥ 85% based on CD14 positivity. For day 7 monocyte-derived cells (MDCs), CLL PBMCs were maintained in RPMI 10% FCS for seven days to allow maturation of MDCs and MDCs isolated after removal of non-adherent cells. Representative images showing MDC morphology from an antibody sensitive and antibody resistant CLL patient is shown in Fig. 1B. CD14 positive monocytes from 2 sensitive and 2 resistant patients and MDCs from 4 sensitive and 3 resistant CLL patients are used in this study.
3.2. Gene expression analysis
Total RNA from monocytes and MDCs was extracted with Bioline Isolate II RNA Micro kit in accordance with the prescribed protocol provided with the kit (Bioline). The integrity of the isolated RNA was determined using Agilent RNA 6000 Pico kit (Agilent Technologies) on an Agilent Bioanalyzer. Microarray profiling was performed using Illumina HumanHT-12 v4 Expression BeadChips (Illumina) as per the manufacturer's protocol. Probe intensities have been background corrected using negative control probes and quantile normalized using the neqc [5] function of the limma [6] package.
Microarray data have been uploaded to Gene Expression Omnibus under the reference: GSE71409. The original CEL files from GEO accession GSE5099 [7], [8] were downloaded. These contain gene expression data for freshly isolated monocytes at day 0, macrophages at day 3 and day 7, Interferon Gamma and LPS treated macrophages and IL-4 treated macrophages. These experiments were performed on the Affymetrix Human Genome U133A Array. Arrays from GSE5099 and the current study were preprocessed and normalized with platform specific tools: the Affymetrix arrays were normalized using the RMA algorithm [9], [10] and the Illumina arrays were quantile normalized after applying the VST algorithm [11], [12]. Large platform specific batch effects were observed between the two datasets, so an unsupervised empirical Bayes method was used to remove these batch effects after as implemented in the ‘virtualArray’ package [13]. The two datasets were merged on common genes after multiple probes that mapped to the same gene were collapsed by taking the median expression value per sample. Genes that had no expression detected in any sample across both studies (as indicated with a detection p-value < 0.01) were removed from the data.
4. Conclusion
MDCs isolated from PBMCs of CLL patients are frequently referred to as nurse like cells and are known to promote the survival of malignant B cells in culture and to mediate responses to therapeutic antibodies [1], [2], [3]. We have compared the transcriptomic profiles of MDCs that are sensitive to, or resistant to, therapeutic antibodies.
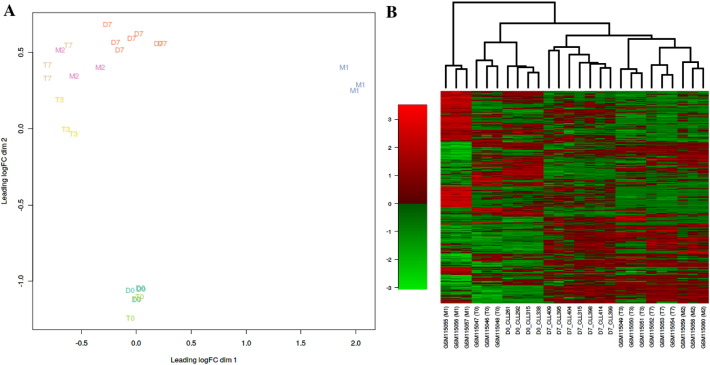
Transcriptomic profiling of freshly isolated monocytes and 7 day adherent MDCs from 4 freshly isolated and 7 adherent (7 day) patient cultures failed to find statistically significant differentially expressed transcripts that discriminate between MDCs derived from sensitive vs resistant patients. However, after normalization and correction for platform-specific batch effects, we were able to compare our dataset to that of a previously published dataset of MDCs cultured for 0, 3 or 7 days [7]. Clustering of our transcript data and that of Martinez et al. [7] reveals marked similarities between the two datasets (Fig. 2A & B). The day 0 monocytes of both studies cluster together and are more similar to one another than any other cell type. Moreover, while the day 7 adherent MDCs from the current study, form their own distinct cluster they share similarities with the day 3, day 7 and M2 macrophages of Martinez, et al. [7]. These data confirm that following 7 days culture our MDCs are enriched for macrophages/NLCs which display an M2-like transcript profile.
Fig. 2.
CLL MDCs display characteristics similar to M2 macrophages. Microarray data from monocytes on day 0 and MDCs on day 7 from sensitive and resistant patients were compared to those from GEO accession GSE5099. Datasets were merged on common genes and the program ‘virtualArray’ was applied to remove platform specific batch effects using an unsupervised empirical Bayes algorithm. (A) multidimensional scaling plot and (B) heatmap of the top 500 genes with the most variable expression. Both figures show a similar pattern: the day 0 monocytes of the current study are most similar to the day 0 monocytes obtained by Martinez et al. (ref. 7) while the day 7 macrophages of the current study cluster close to the day 7 macrophages and M2 macrophages of Martinez et al. (ref. 7). M1 cells have a gene expression profile that is dissimilar to any other cell type either in the previously published study or the current study.
References
- 1.Burger J.A., Tsukada N., Burger M., Zvaifler N.J., Dell'Aquila M., Kipps T.J. Blood-derived nurse-like cells protect chronic lymphocytic leukemia B cells from spontaneous apoptosis through stromal cell-derived factor-1. Blood. 2000;96(8):2655–2663. [PubMed] [Google Scholar]
- 2.Tsukada N., Burger J.A., Zvaifler N.J., Kipps T.J. Distinctive features of “nurselike” cells that differentiate in the context of chronic lymphocytic leukemia. Blood. 2002;99(3):1030–1037. doi: 10.1182/blood.v99.3.1030. [DOI] [PubMed] [Google Scholar]
- 3.Ysebaert L., Fournie J.J. Genomic and phenotypic characterization of nurse-like cells that promote drug resistance in chronic lymphocytic leukemia. Leuk. Lymphoma. 2011;52(7):1404–1406. doi: 10.3109/10428194.2011.568078. [DOI] [PubMed] [Google Scholar]
- 4.Burgess M., Gill D., Singhania R., Cheung C., Chambers L., Renyolds B.A. CD62L as a therapeutic target in chronic lymphocytic leukemia. Clin. Cancer Res. 2013;19(20):5675–5685. doi: 10.1158/1078-0432.CCR-13-1037. [DOI] [PubMed] [Google Scholar]
- 5.Shi W., Oshlack A., Smyth G.K. Optimizing the noise versus bias trade-off for Illumina whole genome expression BeadChips. Nucleic Acids Res. 2010;38(22) doi: 10.1093/nar/gkq871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7) doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinez F.O., Gordon S., Locati M., Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 2006;177(10):7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 8.Solinas G., Schiarea S., Liguori M., Fabbri M., Pesce S., Zammataro L. Tumor-conditioned macrophages secrete migration-stimulating factor: a new marker for M2-polarization, influencing tumor cell motility. J. Immunol. 2010;185(1):642–652. doi: 10.4049/jimmunol.1000413. [DOI] [PubMed] [Google Scholar]
- 9.Gautier L., Cope L., Bolstad B.M., Irizarry R.A. Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20(3):307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 10.Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 11.Du P., Kibbe W.A., Lin S.M. Lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 12.Lin S.M., Du P., Huber W., Kibbe W.A. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36(2) doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heider A., Alt R. virtualArray: a R/bioconductor package to merge raw data from different microarray platforms. BMC Bioinformatics. 2013;14:75. doi: 10.1186/1471-2105-14-75. [DOI] [PMC free article] [PubMed] [Google Scholar]