Abstract
The Illumina's Infinium HumanMethylation450 (HM450) BeadChip array provides a simultaneous examination of DNA methylation status of more than 480,000 CpG sites in the human genome. Its relatively simple protocol is achieved by employing a hybridization methodology followed by single-base extension reactions. However, nucleotide variations among individuals in the hybridization probe sequences can affect the results, i.e. estimates of methylation levels. To investigate possible effects of maternal nutritional conditions on the extent of epigenetic alterations in utero, we examined genome-wide DNA methylation profiles of 33 chorionic villi samples collected in Japan (GEO accession number GSE62733), and revealed using Smirnov-Grubbs' outlier test that epigenetic alterations accumulate in placentas under adverse in utero environments. In that study, we compiled a list of HM450 probes overlapping with the reported nucleotide variants in the Phase 3 dataset (release 20130502) of the 1000 Genomes Project. We excluded the probes whose sequences overlapped with variants with minor allele frequency (MAF) higher than 1% in the Japanese population from identified methylation outliers, to diminish the number of outliers that could have been spuriously identified due to variants at/near the target CpG sites. We herein compiled lists of HM450 probes with MAF information of the African, European, American, South Asian and East Asian populations, in addition to the Japanese population. The provided lists are useful for methylome analyses for human populations using the HM450 BeadChip arrays.
Keywords: DNA methylation, Methylation BeadChip, Genetic variations, The 1000 Genomes Project, Minor allele frequency
| Specifications [standardized info for the reader] | |
| Organism/tissue | Homo sapiens/postpartum placentas (chorionic villi) |
| Sex | Females and males |
| Sequencer or array type | Illumina's Infinium HumanMethylation450 BeadChip array |
| Data format | Raw and analyzed |
| Experimental factors | Maternal gestational weight gain and growth of corresponding fetus |
| Experimental features | Outlier tests for genome-wide DNA methylation profiles of chorionic villi |
| Consent | Publicly available from NCBI GEO |
| Sample source location | Japan |
1. Direct link to deposited data
2. Experimental design, materials and methods
In our previous study, we investigated the possible effects of maternal nutritional conditions during pregnancy on the extent of methylation changes in the fetal genomes [1]. We collected 33 postpartum placentas from Japanese women and obtained chorionic villous tissues. Extracted genomic DNA samples were treated with the EpiTect Plus DNA Bisulfite Kit (Qiagen), and 300 ng of each sample was subjected to the Illumina's Infinium HumanMethylation450 (HM450) BeadChip arrays for methylome profiling [2]. The data obtained using the manufacturer's standard protocol have been submitted to NCBI GEO under accession number GSE62733.
We were cautious with genetic variations overlapping with the sequence intervals of the probes on the BeadChip array. Nucleotide variants at the target CpG site of a probe result in the loss of the target site, and those outside the target CpG site, are considered to impair hybridization and single primer extensions with various degrees depending on their distance from the target CpG site. As it was impractical cost-wise to determine the whole-genome sequences of all samples, we surveyed nucleotide variants overlapping with the probe intervals from the variant data (Phase 3 dataset) of the 1000 Genomes Project [3] available at the following FTP site, ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/. The whole set of probes on the HM450 BeadChip targets 482,421 CpG sites in the human genome. The genomic locations of the probes (50 bp in length) and their target CpG sites were determined as described previously [4].
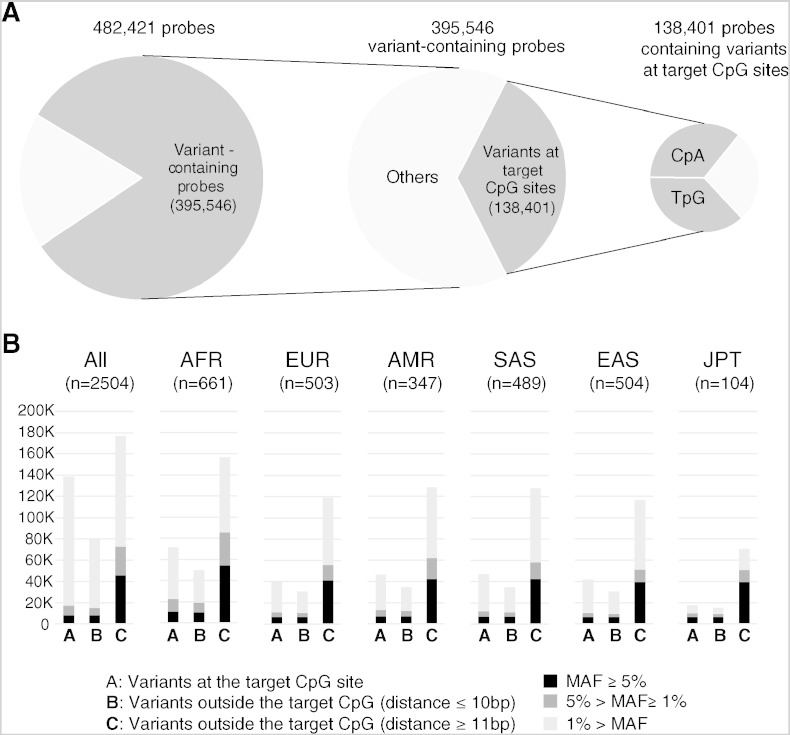
When all variants in the Phase 3 dataset were used without considering their allele frequencies in the populations, 395,546 (82.0%) out of 482,421 probes overlapped with at least one variant. Of the 395,546 probes, 138,401 probes (35.0%) overlapped with nucleotide variant(s) at their target CpG sites. Out of the 138,401 CpG sites, alternate dinucleotides of 100,523 sites (72.6%) were either TpG or CpA (Fig.1A), most of which were presumed to have been derived through spontaneous deamination of C in methylated CpG sites. The alternate TpG (or CpA if observed from the complementary strand) dinucleotide genomic sequence is indistinguishable from the bisulfite-converted form of unmethylated CpG, and can be spuriously identified to be hypomethylated. However, it should be noted that the majority of these variant-containing probes were found only in one to several individuals among all subjects (n = 2504). When only variants whose minor allele frequency (MAF) is higher than 1% among all subjects, the numbers of variant-containing probes and of variant-containing probes at their CpG target site dropped to 105,280 (21.8%) and 17,274 (3.6%), respectively (Fig.1B).
Fig. 1.
Numbers of HM450 probes whose genomic interval overlaps with nucleotide variants reported in the Phase 3 dataset of the 1000 Genomes Project. A. Out of all 482,421 probes that target a CpG site, 395,546 probes contained one or more reported variant(s) in their genomic intervals. Among them, 138,401 probes (35.0%) overlapped with nucleotide variant(s) at their target CpG sites. Out of the 138,401 sites, alternate dinucleotides of 100,523 sites (72.6%) were either TpG or CpA. B. Numbers of probes containing nucleotide variants detected among Japanese (JPT), African (AFR), European (EUR), American (AMR), South Asian (SAS), and East Asian (EAS) subjects, and among all subjects (n = 2500). In each panel, variant-containing probes were classified in three categories (A, B, and C) depending on the distance to the C (MAPINFO [4]) of the target CpG site. In each category, probes were further divided into three sub-categories depending on the minor allele frequency of the overlapping variant(s) and shown as stacked column charts (black, MAF ≥ 5%; dark gray, 5% > MAF ≥ 1%; light gray, 1% > MAF).
In our aforementioned study [1], we excluded CpG sites whose probes overlapped with variants with > 1% MAF in the Japanese population from a list of methylation outliers that were detected to be differentially methylated with statistical significance in one sample compared with the others by Smirnov–Grubbs' outlier tests. Since the exclusion criteria of variant-containing probes can vary depending on the aims of studies and the ethnic background of the populations enrolled, we extended our analysis and compiled lists of probes on the HM450 BeadChip array overlapping with nucleotide variants detected in the East Asian (EAS), American (AMR), African (AFR), European (EUR), and South Asian (SAS) populations in the 1000 Genomes Project (Supplementary Tables 1 and 2), and lists of nucleotide variants overlapping with HM450 probes (Supplementary Tables 3 and 4). Our lists are more updated than the Illumina-provided probe lists containing nucleotide variant information: HumanMethylation450_15017482_v.1.1.csv listing 89,678 probes as single nucleotide polymorphism (SNP)-containing in its “probe_SNPs” and “probe_SNPs_10” columns based on the information of NCBI dbSNP Build 131, and humanmethylation450_dbsnp137.snpupdate.table.v2.sorted.txt listing 273,660 probes as SNP-containing.
3. Discussion
When a spontaneous deamination occurs at an unmethylated cytosine site, it mutates the cytosine to uracil. Because uracil is not a canonical base of DNA, such mutation is immediately recognized and corrected by DNA repair mechanisms in vivo [5]. In contrast, deamination of methylated cytosine gives rise to thymine, which cannot be readily corrected. Although it might be repaired by a mismatch-specific thymine-DNA glycosylase, C-to-T (or G-to-A if observed from the complementary strand) is the most common single-nucleotide substitution in organisms with cytosine methylation [6]. By using 8.2 million SNPs available from dbSNP123 (released on November 3, 2004), Zhao and Zhang reported that the frequency of CpG dinucleotides at the polymorphic sites was 6.09 times higher than that in the human genome reference sequence [7]. Consistently, HM450 probes were found to contain nucleotide variants at their target CpG sites (2 bp) much more frequently than in the rest of the probe intervals (48 bp of Type I probes and 49 bp of Type II probes [2]) (Fig. 1).
We observed that hypomethylated outliers tended to coincide with variant-containing probes more often than hypermethylated ones [1], indicating that a significant fraction of the hypomethylated outliers was detected to be hypomethylated not due to methylation change but a nucleotide variant, which resulted in the loss of the target CpG site. In CpG islands (CGIs), CpG SNPs were reported to be 3.92-fold less frequent than in the human genome [7]. Nevertheless, we observed that hypermethylated outliers tended to be clustered in CGIs [1], indicating that such hypermethylated outliers were more likely to represent bona fide placental epigenetic alterations caused by adverse in utero environments.
As demonstrated in our previous study [1], the compiled lists of HM450 probes with nucleotide variant information presented in this study would be helpful for the methylome analyses, until we can readily determine the whole-genome sequences of all subjects.
Conflict of interest
The authors declare that there are no conflicts of interests.
Acknowledgments
This study was supported by a grant from the National Center for Child Health and Development (NCCHD) of Japan (24–3) to KN. Computation time was provided by the computer cluster Hitachi HA8000/RS210 at the Center for Regenerative Medicine, National Research Institute for Child Health and Development. The manuscript was proofread and edited by Dr. Julian Tang of the Department of Education for Clinical Research, NCCHD.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.gdata.2015.11.023.
Appendix A. Supplementary data
Supplementary material.
Supplementary tables.
References
- 1.Kawai T., Yamada T., Abe K., Okamura K., Kamura H., Akaishi R., Minakami H., Nakabayashi K., Hata K. Increased epigenetic alterations at the promoters of transcriptional regulators following inadequate maternal gestational weight gain. Sci Rep. 2015;5:14224. doi: 10.1038/srep14224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bibikova M., Barnes B., Tsan C., Ho V., Klotzle B., Le J.M., Delano D., Zhang L., Schroth G.P., Gunderson K.L., Fan J.B., Shen R. High density DNA methylation array with single CpG site resolution. Genomics. 2011;98(4):288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 3.1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526(7571):68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Price M.E., Cotton A.M., Lam L.L., Farré P., Emberly E., Brown C.I., Robinson W.P., Kobor M.S. Additional annotation enhances potential for biologically-relevant analysis of the Illumina Infinium HumanMethylation450 BeadChip array. Epigenetics Chromatin. 2013;6(1):4. doi: 10.1186/1756-8935-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindahl T. DNA repair enzymes. Annu. Rev. Biochem. 1982;51:61–87. doi: 10.1146/annurev.bi.51.070182.000425. [DOI] [PubMed] [Google Scholar]
- 6.Neddermann P., Gallinari P., Lettieri T., Schmid D., Truong O., Hsuan J.J., Wiebauer K., Jiricny J. Cloning and expression of human G/T mismatch-specific thymine-DNA glycosylase. J. Biol. Chem. 1996;271(22):12767–12774. doi: 10.1074/jbc.271.22.12767. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Z., Zhang F. Sequence context analysis of 8.2 million single nucleotide polymorphisms in the human genome. Gene. 2006;366(2):316–324. doi: 10.1016/j.gene.2005.08.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.
Supplementary tables.