Abstract
A database that integrates all the information required for biological processing is essential to be stored in one platform. We have attempted to create one such integrated database that can be a one stop shop for the essential features required to fetch valuable result. LmSmdB (L. major and S. mansoni database) is an integrated database that accounts for the biological networks and regulatory pathways computationally determined by integrating the knowledge of the genome sequences of the mentioned organisms. It is the first database of its kind that has together with the network designing showed the simulation pattern of the product. This database intends to create a comprehensive canopy for the regulation of lipid metabolism reaction in the parasite by integrating the transcription factors, regulatory genes and the protein products controlled by the transcription factors and hence operating the metabolism at genetic level.
Keywords: L.major, S.mansoni, Regulatory networks, Transcription factors, Database
1. Introduction
An integrated relational database of curated parasites namely L. major and S.mansoni, causative organism for leishmaniasis and schistosomiasis is hosted on the NCCS webportal (http://www.nccs.res.in/LmSmdb/). This database provides information from various ‘omics’ studies which integrates analyze and manage environments for systems biology research. This is possible with the advent of genome analysis tool that allows identification of genes related to pathogenesis and aid to determine genes as a diagnostic marker essential in metabolic pathways which may lead to drug target identification. Towards this end, we have compared the lipid metabolic pathways specifically for Leishmania major and Schistosoma mansoni by integrating the knowledge of genome sequences of the mentioned organisms. Molecular pathways that appear to be targeted during infection, and these results highlight pathways exhibiting responses specific for a given pathogen infection. The present database is an example of the usefulness of data integration techniques which enable the creation of a hypothesis generation platform for human diseases.
Identifying a drug target and modeling a disease network is straightforward if there is conservation between two species. This conservation between the species helps to search the homologs and orthologues in a model organism. On the basis of sequence conservation, function between two genes can be deciphered which estimates probability of conservation, but evolution does not always evolve parallel with respect to evolution of sequences. We therefore made an effort to integrate two orthologues which are evolutionarily related for extracting information useful for cataloging details of interaction network particularly a gene regulatory network (GRN) and a protein interaction network (PIN) giving an insight into the relationship of protein/genes within the network. A gene regulatory network (GRN) was built in order to find the genes that are essential and highly connected with their corresponding partners. The highly connected nodes in the GRN or the hubs indicate the importance of the node in the interaction and a probable target.
In the present database, we have laid our focus on the lipid metabolism of parasites that cause infectious diseases. It is noted from [3], [4] that lipid metabolism is important for maintenance of infectivity and viability of the parasite. Any change in the biochemical network results in a change in the intercellular lipid trafficking and change in the membrane composition. As lipids play an important role in the virulence and multiplication of the parasite, any approach to disrupt the lipid metabolism could result in a therapeutic strategy to stop parasite proliferation and growth [5]. In this regard, a database LmSmdB was created which studies the network of interactions, maps pathways across taxonomic branches and also incorporates data obtained from simulation studies. During the creation of LmSmdB we have simplified the process of integrating metabolic pathway interactions and protein sequence information for pathogen related studies. The main aim behind the construction of the metabolic network was to identify the important hubs which could act as drug targets in the network. We have also simplified the process of integrating metabolic pathway interactions and protein sequence information for pathogen related studies in order to address the problems related to drug target identification.
The parasite, Leishmania major, the causative organism for cutaneous leishmaniasis was fully sequenced in 2005 [1]. Similarly S. mansoni which causes schistosomiasis was sequenced in the year 2009. [2]. There are several databases like KEGG [8], BIOCYC [9], UNIPROT (The Uniprot Consortium) [10], NCBI [11] etc. that hold information about the proteins, genes and metabolites related to various biochemical pathways present in L.major and S.mansoni. In additions, several organism specific databases provide information on both metabolic and regulatory network whereas none of the said database provides simulation of the constructed network. In order to understand network architecture and simulation pattern of the biochemical network, we have simplified and streamlined the process of integration of molecules, genes, and protein structure in order to perform simulation of the built network. The contents on the server are exactly the same as the source code of the web page on the client side. It is the first known attempt for integrating the metabolic network and simulation of the network in a database. The different metabolic and regulatory networks were constructed using different tools [6]; the detailed information is discussed as below.
1.1. Metabolic network reconstruction
Metabolic networks of the lipid metabolism of L.major and S.mansoni genome were constructed using Cell designer 4.3 software [7], manually to curate network of the metabolism of an organism with all the genes, proteins and reactions assembled from a functionally annotated genome, biochemical data, and literature that are compiled into a stoichiometric matrix which serves as an opportunity for systematic identification of metabolic targets in the pathogen.
The regulatory network analysis of L.major and S.mansoni was dealt using the gene regulatory network interfaces with GeneNetWeaver. As the state of metabolic networks are sensed by association with the gene regulatory network, the initial data was drawn from KEGG database, Biocyc database and literature survey.
Different types of simulation like stochastic, deterministic and hybrid simulation was applied on the generated regulatory network. The advantage of using in-silico network is the ease to carry the perturbation experiments that can be easily simulated to produce expression data unlike in vivo experiments, which are usually expensive and time consuming. Moreover, both quantity and quality of the expression data generated can be controlled (e.g. by varying the amount of molecular and/or measurement noise).
2. Materials and methods
2.1. Database creation
WAMP (WAMPserver) a Windows web development environment allows the creation of web applications with Apache2, HTML and a MySQL database. WAMP 5 was used to create the LmSmdB, where HTML and PHP were used as an administrator to handle the administration of MySQL over the Web.
2.2. Querying database
The LmSmdB web page can be searched using keywords or queried using gene name or the NCBI/KEGG cluster identifier. A collection of gene aliases was assembled based on their availability on different databases. One of the key features of the database is its ability to extract data for several genes together in a batch, eliminating the need for cross referencing of the data from external databases. This batch search is useful for genomic studies where frequent updations associated with genes or proteins have to be examined frequently. The list of genes that serves as an input is in the form of a text file. This can be uploaded on the server or the list can be directly pasted into the search box. Grouping of the documents or web pages relevant to the query is done by the data sources and by the similarity searches among these documents. The entire architecture of LmSmdB database has been presented in Fig. 1. The similarity searches between two documents are based on the sequence of ‘words’ like ‘Leishmania’, or an abbreviation, like ‘Lmjr’, a sequence word ‘ATGCTGGG’ or ‘MEPQSDVL’, a number, or a combination of letters and numbers like ‘Smp_124730’. A binary vector is then built on the descriptive set of words which actually define the biological information that has been searched for. Vectors with three or lesser words above threshold are removed. Certain low complexity words like ‘ATGC’, ‘MEPQSQVL’, or ‘0123456789’, are considered as unique sets of symbols and are not considered in the vectors. The descriptive set of N words, is defined for the whole database as soon as it is updated. This set consists of ‘quality’ words that cover the maximum amount of documents such that N is nearly minimal with the quality of words relating to the relative stability of the corresponding component of the vector in similar documents. GIT record the changes to a file or set of files to recall a specific version.
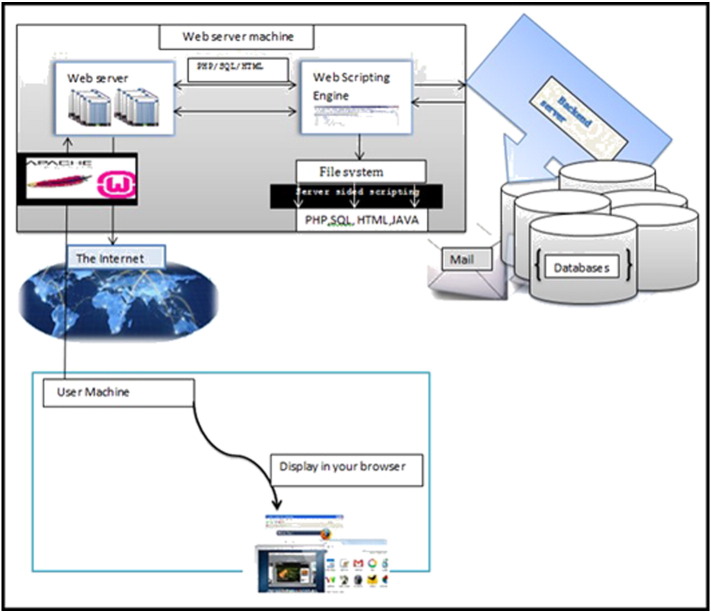
Fig. 1.
The architecture of LmSmdB database consists of a backend server, web-server machine and a user machine.
2.3. Description and structure of database
The database architecture is provided in Fig. 1 which consists of a backend server, a web server for displaying the result, the language in which the query is submitted is in SQL and JAVA. The server sided scripting is mostly about connecting websites to back end servers which helps to enable two way communication. The first way is the server to client communication where the web pages can be assembled from back end server output and the second is the client to server communication where the user enters the information to get an output. The database consists of tables, images and links to different databases.
3. Utility and discussions
LmSmdB is a convenient, graph based data integration system that captures, incorporates, and manages available data related to the functional importance of several genes and proteins. The database is structured in the form of a network that incorporates features of a given entity such as genes, proteins and pathways. The database is also capable of dynamically incorporating new sets of objects and their relation thus integrating data types like tables and sequences.
The protein information along with the compartmentalization is included in the database which links to the KEGG database. The GRN and the PIN are provided in the image format, the protein and genes included in the table format along with the link to other databases (KEGG). The outline of the database along with the work-flow is shown in Fig. 2.
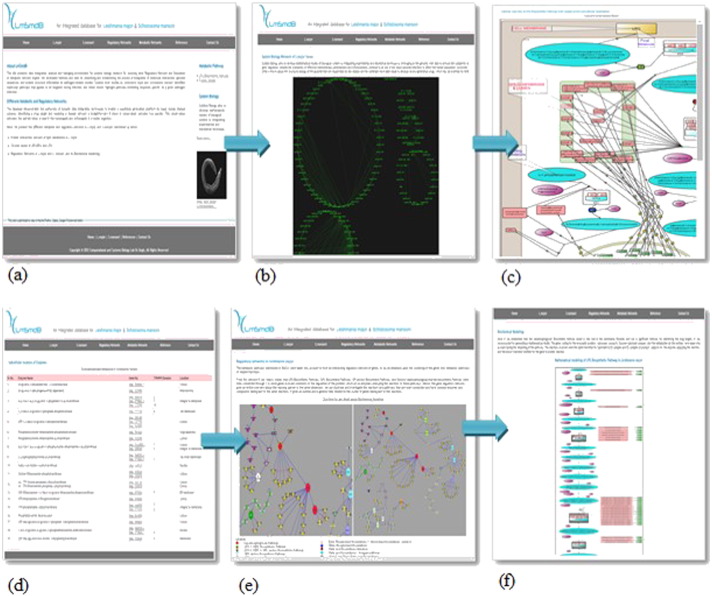
Fig. 2.
LmSmdB database report page (a) home page of the database, gives information on the disease leishmaniasis and schistosomiasis, metabolic networks, regulatory networks, references and contact details. (b) GRN of Schistosoma mansoni. (c) Metabolic network reconstruction for L. major. (d) List of proteins in the network. (e) The Regulatory network of L. major. (f) The Metabolic network reconstruction in Schistosoma mansoni.
The database architecture consists of a back end server and a web server for displaying the result. The language in which the query is submitted is in SQL, HTML and PHP. Server side script connects the websites to back end server to enable two way communication i.e. from client side where the web pages are assembled and the second is the client to server communication where the user enters the information to get an output. The internal database of the system is structured in the form of a network database which contains all the features assigned to bio entities like genes, proteins and pathways. New sets of objects along with their relations within a search are easily incorporated. This is done by integrating several data types like graphs, tables and sequences. Any problem arising in the data integration are dealt with via an ontology driven data mapping, multiple data annotation and heterogeneous data querying in order to enable integration of the user's data. The graphs obtained in Cell designer, the interactions are visualized in JAVA which depicts how a gene, metabolite and a protein are connected. We simplified and effectively integrated the molecular interaction in the form of protein as nodes and edges as interaction together with the structural information. Systems level studies on Leishmania major and Schistosoma mansoni identified molecular pathways that appear to be targeted during infection, and these results highlight pathways exhibiting responses specific for a given pathogen infection demonstrating the practical use of data integration techniques, to enable hypothesis generation platform for major human disease systems providing access to heterogeneous information that are of value to researchers targeting the tropical diseases.
4. Availability
LmSmdB is hosted on the webportal of National Centre for Cell Science (NCCS).
5. Data maintenance
LmSmdB will be continuously updated, through manual screening of new publications and literature review. In addition, we also openly welcome suggestions by researchers that include new or missing findings to be inserted in the database by contacting authors by email. Our contact information is reported in the ‘Contact Us’ page.
6. Conclusion
LmSmdB is the first online resource containing genomic information related to metabolic and regulatory network specifically for L. major and S. monsoni. It is designed as a common purpose framework for systems biology providing formalized graphic notation of biological systems structure and functioning, their visualization and simulations as well as access to databases with relevant data. A user can browse information regarding the disease caused by these two pathogens and the gene involved in the pathogenesis. By looking at LmSmdB, the user can also get the link to the KEGG pathway of particular gene, which is directly involved in the pathway or network.
Acknowledgments
The authors would like to thank Department of Biotechnology, Ministry of Science and Technology, Government of India for funding the work. (BT/PR3140/BID/7/379/2011). The authors would also like to thank the Director, National Centre for Cell Science (NCCS), Pune for supporting the Bioinformatics and High Performance Computing Facility at NCCS, Pune, India.
References
- 1.Ivens A.C. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;(5733):436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berriman M. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460(7253):352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niemela Bioinformatics and computational method for lipidomics. J. Chromatogr. B. 2009;877:2855–2862. doi: 10.1016/j.jchromb.2009.01.025. [DOI] [PubMed] [Google Scholar]
- 4.Chavali Systems analysis of metabolism in the pathogenic trypanosomatid Leishmania major. Mol. Syst. Biol. 2008;4:1–19. doi: 10.1038/msb.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh S., Shinde S. In: Stochastic simulation for biochemical reaction networks in infectious disease, medicinal chemistry and drug design. Ekinci D., editor. InTech; 2012. [Google Scholar]
- 6.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. (Nov) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Funahashi A., Tanimura N., Morohashi M., Kitano H. CellDesigner: a process diagram editor for gene-regulatory and biochemical network. Biosilico. 2003;1:159–162. [Google Scholar]
- 8.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular datasets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caspi R., Altman T., Dale J.M., Dreher K., Fulcher C.A., Gilham F., Kaipa P., Karthikeyan A.S., Kothari A., Krummenacker M., Latendresse M., Mueller L.A., Paley S., Popescu L., Pujar A., Shearer A.G., Zhang P., Karp P.D. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Uniprot consortium Reorganizing the protein space at the Universal Protein Resource Uniprot. Nucleic Acids Res. 2012;40:D71–D75. doi: 10.1093/nar/gkr981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geer L.Y., Marchler-Bauer A., Geer R.C., Han L., He J., He S., Liu C., Shi W., Bryant S.H. The NCBI BioSystems database. Nucleic Acids Res. 2010;D492-6 doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]